Abstract
The capacities for neural stem cells (NSCs) self-renewal with differentiation are need to be precisely regulated for ensuring brain development and homeostasis. Recently, increasing number of studies have highlighted that long non-coding RNAs (lncRNAs) are associated with NSC fate determination during brain development stages. LncRNAs are a class of non-coding RNAs more than 200 nucleotides without protein-coding potential and function as novel critical regulators in multiple biological processes. However, the correlation between lncRNAs and NSC fate decision still need to be explored in-depth. In this review, we will summarize the roles and molecular mechanisms of lncRNAs focusing on NSCs self-renewal, neurogenesis and gliogenesis over the course of neural development, still more, dysregulation of lncRNAs in all stage of neural development have closely relationship with development disorders or glioma. In brief, lncRNAs may be explored as effective modulators in NSCs related neural development and novel biomarkers for diagnosis and prognosis of neurological disorders in the future.
Keywords: Neural stem cells, LncRNA, Self-renewal, Neural differentiation, Neurogenesis, Gliogenesis, Neurological disorders
Introduction
In the central nervous system (CNS), stringently regulatory mechanism is essential for proper neural stem cells (NSCs) related development and functions. Recently, epigenetic modulators, especially long non-coding RNAs (lncRNAs) are found to be crucial for the maintenance of NSCs related biological activity. This review aims to introduce the functions and regulatory mechanism of lncRNAs in NSCs self-renewal and differentiated into neurons or/and glial cells.
Neural stem cells
As a dynamic organ, vertebrate brain possesses the capacity of structural plasticity upon a variety of physiological, pathological and pharmacological stimuli owing to proliferation and differentiation ability of NSCs [1, 2]. Amazing huge number of neurons and glial cells constituting the cortex are generated from the differentiation of NSCs, which are able to self-renewal and major yield three forms of neural cells including neurons, astrocytes and oligodendrocytes in the brain [3], that the process of producing neurons and glial cells are termed neurogenesis and gliogenesis, respectively [4].
NSCs use symmetric divisions for adult neural precursor/progenitor cells (NPCs) amplification and asymmetric divisions for sequentially producing the right quantity of neurons and/or ultimately transition to gliogenesis sustaining postnatally [1, 4]. The largest NSCs niches throughout life are predominantly located in the subventricular zone (SVZ) nearby the lateral wall of the lateral ventricles and the sub-granular zone (SGZ) of hippocampus dentate gyrus (DG) [5]. The neuroblasts from NSCs in SVZ migrate along the rostral migratory stream (RMS) to the olfactory bulb (OB), where they terminally differentiate into local interneurons [6]; meanwhile, neuroblasts from NSCs in SGZ migrate short distances into the granule cell layer and mature into neurons, then integrate into functional circuit [7].
NSCs persist in the embryonic stage and in the specific area of the brain during adulthood [8]. However, it is still controversy whether adult neurogenesis occurs and even persist throughout lifetime. Some studies implied that proliferating progenitors and young neurons in the dentate gyrus (DG) sharply declined in the first year of life, only a few isolated young neurons were detected in the young and no young neurons were observed in DG [9, 10], whereas some others considered as hundreds of new neurons generated in each hippocampus/day in adult humans [11], and similar numbers of intermediate neural progenitors and thousands of immature neurons in the DG from young to older [12]. Undoubtedly, the generation of a certain number of neuronal progenitors from NSCs and then differentiation into neurons and/or glial cells are associated with brain development and changed in neurological disorders [13].
LNCRNAs
As the rapid progress of next-generation sequencing technologies, a large amount of lncRNAs were discovered and identified as essential modulators in fundamental biological processes, although they were initially considered as “noise” of genome. LncRNAs are a sub-class of non-coding RNAs transcripts longer than 200 nucleotides with 5ʹm7G caps and 3ʹ poly (A) in tails, which are generated by RNA polymerase II but lack canonical protein-coding capacity [14, 15]. Clark et al found that majority of lncRNAs exhibit widely stabilities similar to that of mRNA, while the mean value of lncRNA half-life was 4.8 h that slightly less than the mean value of protein-coding transcripts (7.7 h) [16].
LncRNAs originate from various gene coding or non-coding locations including intergenic regions, introns, enhancers, promoters, exons, either with a partial overlap with protein-coding exons in both directions [17, 18]. They organize gene expression in the context by recruitment of regulatory proteins, modulation and modification of chromosomes at transcriptional level, controlling RNA splicing, acting as a “sponge” of miRNAs to regulate RNA degradation at post-transcriptional level and also participate in cytoplasm and nuclear trafficking or cell differentiation [18, 19].
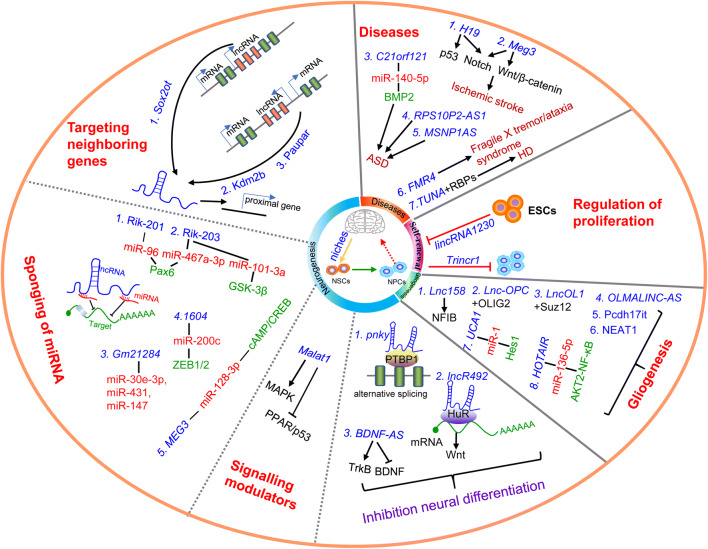
Accounting for 40% differentially expressed lncRNAs in human genome are specific to the brain, which involve in 4000–20,000 lncRNA genes [20]. LncRNAs have been reported be located in different brain cell types, such as neuron, glial cells and vascular cells, and playing crucial biofunction in the different brain cells [21]. In addition, lncRNAs are abundantly expressed in the particular NSCs generated regions of SVZ, DG or Striatum, which implies the crucial functional roles of lncRNAs in NSCs self-renewal, pluripotency, proliferation and differentiation [22–24]. The major goal of this article is to demonstrate the cell type-specific expression and functions of lncRNAs focusing on NSCs self-renewal, neurogenesis and gliogenesis over the course of neural development (Fig. 1).
Fig. 1.
Schematic representation the regulatory networks of lncRNAs in major biology activities during brain developmental process. It was depicted in details that lncRNAs contribute to NSCs self-renewal, neurogenesis/neuronal differentiation, neurodevelopmental disorders and gliogenesis major emerging as sponging of miRNAs, crucial regulators in signaling pathway and targeting proximal gene expression
The effect of lncRNA on NSCs/NPCs self-renewal and proliferation capacity
As one type of multipotent cells, NSCs possess a significant capacity for proliferation and self-renewal, which are essential for maintenance of CNS homeostasis [25], moreover, they also can be derived from totipotent stem cells and various pluripotent cells in vitro [26, 27]. Although the underling regulatory mechanism still remains unknown, recently, some evidence implied that lncRNA may emerge as a modulator in NSCs self-renewal and proliferation (Table 1). For instance, overexpression of lncRNA Trincr1 (TRIM71 interacting long noncoding RNA 1) suppressed the self-renewal of NPCs via restraining fibroblast growth factors (FGF)/extracellular signal regulated kinase (ERK) signaling pathway, which is essential for cell self-renewal [28, 29]. Furthermore, NPCs were increasingly transplanted from pluripotent stem cells for treatment neurological development disorders [26, 30]. LincRNA1230 was able to markedly block mouse ESCs transformation into NPCs, mechanistically, it restrained the combination of WD repeat domain 5 (Wdr5) to the promoter regions of neural lineage-associated genes via reducing enrichment of the H3K4me3 (tri-methylation of histone3 lysine4) modification at these loci [31].
Table 1.
The functional characterization of lncRNAs on neural related stem cells self-renewal
| LncRNA name | Mechanism | Biological function | References |
|---|---|---|---|
| Trincr1 | Restrain FGF/ERK signalling | Suppress NPCs self-renewal | [28, 29] |
| LincRNA1230 | Interact with Wdr5 | Block ESCs transformation into NPCS | [31] |
| Linc00115 | Activated by TGF-β, miR-200 s/ZEB1, miR200s/ZNF596/EZH2/STAT3 | Promote GSC self-renewal | [33] |
| TUG1 | Activated by Notch, sponging of miR-145 and recruiting polycomb via YY1 | Promote GSC self-renewal | [34] |
| Linc00152 | miR-103a-3p/FEZF1 axis | Promote GSC proliferation | [35] |
| ZNF281 | NF-κB1 pathway | Inhibit GSC self-renewal | [36] |
| TALNEC2 | Regulated by E2F1, miR-21/miR-191 | Promote GSC self-renewal | [37] |
| Linc01198 | Enhancing the NEDD4-1-dependent inhibition of PTEN | Promote glioma proliferation | [38] |
Glioblastoma stem-like cells (GSCs) exhibit the stemness properties of stem cell including self-renewing capability and multipotency [32], which progression and self-renewal are able to be modulated by lncRNAs. LncRNA linc00115 activated by Transforming Growth Factor-β (TGF-β), lncRNA TUG1 activated by notch receptor (Notch), linc00152 and lncRNA ZNF281 were newly identified lncRNAs that participated in controlling self-renewal and proliferation in GSCs via sponging of miR-200 s, sponging of miR-145, sponging of miR-103a-3p and targeting NF-κB1 signalling pathway, respectively [33–36]. Moreover, lncRNA tumor associated lncRNA expressed in chromosome 2 (TALNEC2) and linc01198 were also found to promote self-renewal and progression of GSCs [37, 38]. Thus, lncRNA maybe a novel potential therapeutic strategy for glioblastoma therapy.
The function of lncRNAs in neurogenesis/neural differentiation
Neurogenesis is a dynamic process associated with NSCs and NPCs differentiation into newborn neurons, which integrate into the local neural network in the mammalian CNS [39]. This process is a well-orchestrated sequence of complex biological and molecular event major including NPCs or NSCs proliferation and differentiation during pre- and post-natal brain development, which major occurs in SVZ and SGZ in DG of hippocampus [6, 40, 41].
Recently, as a partial of the mammalian genome, lncRNAs have emerged as crucial epigenetic regulatory elements implicated in NPCs or NSCs differentiation and neural development [40, 41]. Intricate expression regulation of lncRNAs in time and space is a crucial event in the developing CNS [42]. A quantity of lncRNAs are present abundantly in certain neurogenic cell-types or the specific functions in development and cellular identity in the nervous system [43, 44]. Furthermore, 13 lncRNAs temporally exhibiting neurogenic cell types specificity and hallmarks of RNA processing, have been noted in the purified neural cell types across consecutive time course overlap critical events in neurogenesis in Drosphila, proving that expressions of lncRNAs are highly dynamic and demarcates particular subpopulations within neurogenic cell types in CNS [45]. Interestingly, lncRNAs showed an adaptive function in the evolution of neurogenesis due to selective loss in the evolutionary time [46]. Actually, lncRNA was a key determinant in NSCs or NPCs during cell-fate determination, moreover, distinct lncRNA types are involved in the different situations of neurogenic precursor or stem cell differentiation. For instance, divergent lncRNAs have partiality for neuronal differentiation while sense downstream lncRNAs are more associated with astrocytic differentiation in NPCs or NSCs [47]. Moreover, lncRNAs can also participate in regulation the fate of NSC differentiation into glia and neurons in the physiology and pathological condition [48, 49]. In hence, it is still need to be discussed that lncRNAs how to play a role in neurogenesis/neuronal differentiation or gliogenesis (Table 2). Elaborating the underlying regulatory mechanism of neural differentiation is benefits for studying neural development or acquiring NSCs for diseases treatment.
Table 2.
The roles of lncRNAs on NSCs differentiation/neurogenesis and oligodendrogenesis
| LncRNA name | Mechanism | Biological function | References |
|---|---|---|---|
| Sox2ot | Link with Sox2, interact with YY1 | Prohibit NSCs proliferation and advance neuronal differentiation | [54] |
| RMST | Target Sox2 | Promote neurogenesis | [56] |
| Kdm2b | Bind with hnRNPAB and activate Kdm2b gene expression | Promote neurogenesis | [57] |
| Paupar | Bind with local genes-Pax6 and KAP1 | Promote neurogenesis | [58, 59] |
| Gm21284 | Interact with miR-30e-3p, miR-431 and miR-147 | Inhibit NSCs proliferation while promote NSCs differentiation | [63] |
| 1604 | miR-200c/ZEB1/2 axis | Promote neural differentiation | [61] |
| Rik-201 | Activated by C/EBPβ, miR-96/Sox6 | Enhance neural differentiation | [64] |
| Rik-203 | miR-467a-3p/Sox6, miR-101-3a/GSK-3β | Enhance neural differentiation | [64, 65] |
| Malat1 | Activate ERK/MAPK, inhibit PPAR/p53 | Promote neural differentiation | [67] |
| Pnky | Interact with PTBP1 | Inhibit neural differentiation and neurogenesis | [3, 68] |
| lncR492 | Interact with HuR and activate Wnt signalling | Inhibit neural differentiation | [69] |
| BDNF-AS | Targeting TrkB signaling pathway | Inhibit eNSCs-derived neurite outgrowth | [70] |
| UCA1 | miR-1/Hes1 | Promote NSCs differentiation to astrocyte but neuron | [48] |
| OPC | Regulated by OLIG2 | Promote oligodendrogenesis | [87] |
| lncOL1 | Form a complex with Suz12 | Promote oligodendrogenesis | [90] |
| lnc158 | Promote NFIB expression | Promote oligodendrogenesis | [91] |
| Pcdh17it | Oligodendrogenesis marker | [92] | |
| OLMALIN/-AS | Regulate oligodendrocyte maturation | [93] |
The function of lncRNAs on promoting neuronal differentiation
LncRNAs meet a requirement for neuronal differentiation and are considered to be indispensable for neurogenesis. They are specific to the brain region, especially SVZ, DG or OB, highly expressed during neuronal differentiation, and always exert their functions via interacting with transcription factor or neighboring genes on the same chromosome, acting as competing endogenous RNA (ceRNA) of miRNA and target protein or emerging as key signaling pathway modulators.
Effect on neighboring genes expression
LncRNAs are able to control neural development via influencing proximal protein-coding genes expressions. LncRNA Sox1 overlapping transcript (Sox1ot) and Sox2 overlapping transcript (Sox2ot) locating in nucleus are evolutionarily conserved lncRNAs that transcriptionally overlaps the Sox1 and Sox2, respectively, and considered as crucial modulators in the developing brain [50, 51]. Sox1 and Sox2 are transcription factors which associated with maintaining the stemness of pluripotent stem cells and NSCs [52]. Sox1ot and Sox2ot are highly expressed during neural development and link with Sox1 and Sox2 expression levels, respectively [53, 54]. Androgen receptor (AR),a transcription regulator in early embryonic stage, modulates lncRNA Sox2ot expression by interacting with Sox2 upstream DNA binding region at transcription level [55]. LncRNA Sox2ot prohibited NSCs proliferation and advanced neuronal differentiation by interacting with the transcriptional regulator YY1, which bind to CpG island in the Sox2 locus to suppress the expression of Sox2 [54]. LncRNA rhabdomyosarcoma 2-associated transcript (RMST) was considered to be indispensable for neurogenesis when its absence lead to more than 1000 genes differentially expression major via facilitating Sox2 binding the promoter regions and regulating the target genes [56]. In addition, lncRNA Kdm2b divergently transcribed from the same promoter bidirectional with Kdm2b and dispersed chromatin environment via binding hnRNPAB, then activated its nearby coding gene-Kdm2b expression for facilitating neuronal differentiation in early neurogenesis cortical projection neurons [57]. LncRNA Paupar divergently transcription from upstream of Pax6 participates in regulation of neural differentiation and OB neurogenesis via binding with local genes-Pax6 and KAP1 in cis-manner, as well as modulation the activity of various transcriptional regulatory elements on different chromosomes distinguishing from its synthesis locus and alteration of chromatin occupancy and H3K9me3 deposition [58, 59].
Acting as ceRNA of miRNA
MiRNAs as a group of short non-coding RNAs (approximately ~ 22 nucleotides long) suppressing coding gene translation are abundant in the nervous system and participate in all stage of neural differentiation during brain development [60]. LncRNAs could participate in neural development via emerging as ceRNA of miRNA and indirectly regulate gene expression in the cytoplasm [61, 62]. Several differentially expressed lncRNAs interacting with miR-30e-3p, miR-431 and miR-147 were determined by microarray analysis in hippocampal pool. Among these lncRNAs, Gm21284 was identified by function as a ceRNA to enhance the proportion of CHAT-positive cells during NSCs differentiation [63]. LncRNA 1604 silencing suppressed neural differentiation through acting as sponge of miR-200c to regulate the key transcription factors zinc finger E-box binding homeobox 1/2 (ZEB1/2) [61]. Moreover, lncRNA transcript could generate several variants to execute functions in neurogenesis. LncRNA C130071C03 Riken variants-Rik-201 and Rik-203 were also considered as modulators in the developing brain via being activated by neurogenesis related transcript protein CCAAT/enhancer-binding protein β (C/EBPβ). Suppression of Rik-201 and Rik-203 restrained neural differentiation via acting as ceRNAs of miR-96 and miR-467a-3p, respectively, which deinhibition restricted the expression of neural differentiation-related gene Sox6 [64]. Sevoflurane was also reported to attenuating the expression of lncRNA Rik-203 that resulted in the release of miR-101-3a but lessening Glycogen Synthase Kinase-3β (GSK-3β) level, ultimately inhibited neural differentiation [65]. Furthermore, miR-128-3p is abundantly expressed in brain and emerges as a key modulator in neural differentiation, which overexpression prohibited neuron but enhanced gliocytes differentiation. LncRNA MEG3 participated in promotion of neuron differentiation via emerging as a negative modulator of miR-128-3p while elevated by the cAMP/response element-binding protein (CREB) pathway [66].
Emerging as key signalling pathway modulators
LncRNAs could also contribute to neural differentiation emerging as a pivotal member of signaling pathway. Neurite outgrowth is a core event in early neuronal differentiation and regeneration stage. The lncRNA Metastasis-associated lung adenocarcinoma transcript1 (Malat1) was indispensable for neurite growth. Knockdown of Malat1 blocked neurite outgrowth but advanced cell death via suppression Mitogen-Activated Protein Kinase (MAPK) signaling pathway comparable with stimulation of Peroxisome proliferator-activated receptor (PPAR) and p53 signalling pathway [67].
The role of lncRNAs on repressing neuronal differentiation
Compare to the above lncRNAs which be highly expressed and promoted neuronal differentiation, some other neuronal lncRNAs were revealed to be down-regulated in CNS and blocked neuronal differentiation. LncRNA Pnky, being considered as the first known neuronal development inhibitor as its expression was decreased when V-SVZ NSCs differentiation into neuronal cells, forming a complex with splicing factor and RNA-binding protein (RBP)-polypyrimidine tract-binding protein (PTBP1) participated in regulation of NSCs differentiation to neurons via controlling alternative splicing. Knockdown of either pnky or PTBP1 expression could strengthen neurogenesis, which both elicited a splicing program in cultured post-natal V-SVZ NSCs to mature neurons [3, 68]. Maria et al discovered the lncRNA lncR492 as a lineage-specific inhibitor of neuroectodermal differentiation through interaction with mRNA binding protein HuR and activation of Wnt signaling pathway [69]. Furthermore, the enhanced expression of lncRNA brain derived neurotrophic factor antisense (BDNF-AS), which is an antisense RNA that inhibition of BDNF expression in neural growth, was able to inhibit neurite growth in ketamine treated mouse embryonic NSC-derived neurons via activating potassium uptake system protein (TrkB) signaling pathway [70].
The effect of lncRNA on neurodevelopmental disorder via targeting NSCs/NPCs proliferation and differentiation
Addition to be as critical determinant for neuronal differentiation or neurogenesis in NSCs or NPCs, lncRNAs are also acting as pivotal regulatory molecules in several neurodevelopmental related diseases including Huntington’s disease (HD) [71], Autism spectrum disorder (ASD) [72], Angelman syndrome (AS) [73], vascular disorders induced ischemic stroke [74, 75]. LncRNA Tcl1 Upstream Neuron-Associated lincRNA (TUNA) was found to be associated with HD, which function for maintenance of pluripotency and neural differentiation by interaction with three RBPs [71]. Furthermore, lncRNA FMR4 originating from Fragile X mental retardation 1 (FMR1) locus, which aberrant expansion causes autism [76], was able to improve hNPCs development, furthermore, dysregulation of FMR4 contributed to the pathophysiology Fragile X syndrome and/or Fragile X tremor/ataxia syndrome [77]. In addition, lncRNA ribosomal protein S10 pseudogene 2 anti-sense 1 (RPS10P2-AS1), moesin pseudogene 1antisense (MSNP1AS) and FMR4 were identified that contributed to another neurodevelopmental disorder-Autism spectrum disorder (ASD) risk [72, 77–79]. The expression of RPS10P2-AS1 was elevated in postmortem temporal cortex of patients with ASD as well as in NPCs upon to ASD-associated diesel particular matter, which implied the close relationship between RPS10P2 with ASD risk [78]. LncRNA MSNP1AS expression was elevated in the postmortem cerebral cortex of individuals with ASD, which was mimicked by overexpression of MSNP1AS in human NPCs reduced neurite number and neurite length by disrupting moesin protein level, when knockdown of MSNP1AS blocked 318 genes expression, most of which participating in chromatin organization and immune response [72, 79]. Moreover, lncRNA C21orf121 overexpression promoted conversion of stem cells from human exfoliated deciduous teeth into neuronal cells via acting as ceRNA of miR-140-5p to regulate BMP2 expression, which may provide a new therapeutic tool for ASD [80] (Table 3).
Table 3.
The major roles of lncRNAs in neurodevelopmental disorders
| LncRNA name | Mechanism | Biological function | Disease | References |
|---|---|---|---|---|
| TUNA | Interact with RBPs | Promote neural differentiation | HD | [71] |
| FMR4 | Derived from FMR1 locus | Promote hNPCs proliferation | Fragile X syndrome | [77] |
| RPS10P2-AS1 | Interact with RPS10P2 | ASD | [78] | |
| MSNP1AS | Regulate chromatin organization and immune response related gene | Inhibit neural differentiation | ASD | [72, 79] |
| C21orf121 | miR-140-5p/BMP2 | Promote neurogenesis | ASD | [80] |
| H19 | p53/Notch1 pathway | Block neurogenesis | Ischemic stroke | [74] |
| Meg3 | Notch or Wnt/β-catenin signaling pathway, miR-128-3p/ATRA/cAMP/CREB axis | Promote neurogenesis/neural differentiation | Ischemia-reperfusion injury | [75, 82] |
| NEAT1 | Associated with Wnt signaling | Promote oligodendrogenesis | Schizophrenia | [94] |
| HOTAIR | miR-136-5p/AKT2-NF-κB | Demyelination | [95] |
Furthermore, lncRNAs are involved in angiogenesis that NPCs in SVZ and SGZ migration to ischemic zone for restoration of blood supply after ischemic stroke [81]. One of the earliest identified lncRNA H19 executed a negative function in chronic regeneration to inhibit neurogenesis process after ischemia stroke via inhibition p53/Notch1 signalling pathway [74]. Similarly, lncRNA Meg3 also played a reversely effect on brain injury recovery and its absence improved nervous tissue impairment and promoted angiogenesis by triggering Notch pathway and Wnt/β-catenin signaling pathway [75, 82].
The role of lncRNAs in modulation of gliogenesis
As well known, except for neurons, NSCs can gradually alter their characteristics to generate astrocytes and oligodendrocytes in the CNS, which termed as “gliogenesis” [83, 84]. At initial phase of cortical development, NSCs or NPCs sequentially produce deep layer neurons followed by surficial layer neurons; at later phase, NSCs suspend neurogenesis and shift to gliogenesis to gain gliogenic competence [84, 85]. The timing of NSCs transition from neurogenesis to gliogenesis must be stringently controlled to ensure proper cortical development [84, 86]. Several lncRNAs are considered as crucial modulators during neuronal-glial fate specification and oligodendrocyte lineage maturation. LncRNA human urothelial carcinoma associated 1 (UCA1) was able to decide the differentiation direction of NSCs, when UCA1 silencing prohibited NSCs proliferation and differentiation to astrocyte but strengthen to neuron due to the enhancement of miR-1 expression but decrease expression of its target gene-Hes1 [48]. Moreover, some lncRNAs showed key functions for the fate of NSCs differentiate into glia and neurons exposure to hyperthermia [49]. Dong et al. screened 5000 lncRNAs to predict their roles in brain development and identified a highly conserved lncRNA-lnc-OPC. The expression of lnc-OPC, which upstream regulatory elements interaction with OLIG2, was dramatically enhanced in oligodendrocyte precursor cells (OPC) and contributed to OPC differentiation and oligodendrogenesis [87].
Myelination by oligodendrocytes is a vital event in the development and function of CNS and can be regulated by genetic and epigenetic factors including lncRNAs [88, 89]. The dynamic expression profiles of lncRNAs at different phases of oligodendrocyte growth were determined and then picked out a conserved chromatin-associated lncRNA-lncOL1. The gain of function of lncOL1 enhanced precocious oligodendrocyte differentiation in neural development via forming a complex with a component of polycomb repressive complex 2 (Suz12), which is an oligodendrocyte maturation promoter [90]. Overexpression of lnc158 in NSCs promoted several oligodendrocyte-related genes expressions and strengthened induction of oligodendrocyte lineage differentiation via positively modulation of an organ development regulatory factor-nuclear factor-IB (NFIB) [91]. In addition, an immature OL-specific lncRNA Pcdh17it was proved to be a novel biomarker for newborn immature OLs in the brain development [92]. Interestingly, lncRNA oligodendrocyte maturation-associated long intervening non-coding RNA (OLMALINC) has an identical expression type with its antisense counterpart, OLMALINC-AS, both abundantly expressed in the white matter of human frontal cortex and played vital roles in regulation of human oligodendrocyte maturation related genes [93]. In addition, oligodendrocyte-related abnormalities associated with developmental disorder including schizophrenia and demyelination are also regulated by lncRNAs [94, 95]. The expression levels of lncRNA NEAT1 was reduced in the brain of patients with schizophrenia and loss of NEAT1 influenced multiple oligodendrocytes cell differentiation related genes that caused population of oligodendrocytes-lineage cells diminishment during brain development [94]. LncRNA HOTAIR acting as ceRNA of miR-136-5p and AKT2-NF-κB was repressed and unbeneficial for repair impairment of cuprizone-induced demyelination [95]. Thus, lncRNAs indeed plays important functions in modulation of OL mature and oligodendrogenesis during brain development stage.
Conclusion
Neural development related to NSCs/NPCs is considered as a huge complicate biological event. As advanced large-scale genome-wide sequencing analysis, more tissue-specific expression of lncRNAs were identified as essential modulators in fundamental neural developing biological processes. Most of their function remains to be explored, more novel lncRNAs and their molecular mechanisms remain to be found and probed in-depth yet. This review has described in detail the dramatically functional roles of lncRNAs in regulation of NSCs/NPCs self-renewal, proliferation and differentiation into neuron or glial cells, moreover, dysregulation of lncRNAs in all stage of neural development have closely relationship with development disorders or glioma. This suggest that lncRNAs have great potential to be applied in diagnosis, prognosis and treatment of neurodevelopmental disorders, still more, based on the features of their structural motifs, stability, easy-detectable and gene regulatory network, lncRNAs might be also employed as potential selection bio-markers for identifying or screening suitable NPCs/NPCs. With the deep-going study in the future, it will open a new era of lncRNA based NSCs proliferation and differentiation regulatory mode and neural development disorders therapy targets.
Acknowledgements
Not applicable.
Abbreviations
- ASD
Autism spectrum disorder
- AR
Androgen receptor
- AS
Angelman syndrome
- BDNF
Brain-derived neurotrophic factor
- CREB
cAMP/response element-binding protein
- C/EBPβ
CCAAT/enhancer-binding protein β
- DG
Dentate gyrus
- ERK
Extracellular signal regulated kinase
- ESCs
Embryonic stem cells
- FMR1
FMR4 originating from Fragile X mental retardation 1
- GSK-3β
Glycogen synthase kinase-3β
- GSCs
Glioblastoma stem-like cells
- HD
Huntington’s disease
- LncRNAs
Long non-coding RNAs
- Malat1
Metastasis-associated lung adenocarcinoma transcript1
- MAPK
Mitogen-activated protein kinase
- MEG3
Maternally expressed gene 3
- MSNP1AS
Moesin pseudogene 1 antisense
- NFIB
Nuclear factor-IB
- NSCs
Neural stem cells
- Notch
Notch receptor
- NPCs
Neural precursor/progenitor cells
- OB
Olfactory bulb
- OPC
Oligodendrocyte precursor cells
- OLMALINC
Oligodendrocyte maturation-associated long intervening non-coding RNA
- PPAR
Peroxisome proliferator-activated receptor
- PTBP1
RNA-binding protein (RBP)-polypyrimidine tract-binding protein
- RMS
Rostral migratory stream
- RMST
Rhabdomyosarcoma 2-associated transcript
- RPS10P2-AS1
Ribosomal protein S10 pseudogene 2 anti-sense 1
- SVZ
Subventricular zone
- SGZ
Sub-granular zone
- Sox1ot
Sox1 overlapping transcript
- Sox2ot
Sox2 overlapping transcript
- Suz12
Polycomb repressive complex 2
- TALNEC2
Tumor associated lncRNA expressed in chromosome 2
- Trincr1
TRIM71 interacting long noncoding RNA 1
- TGF-β
Transforming growth factor-β
- TUNA
Tcl1 upstream neuron-associated lincRNA
- UCA1
Urothelial carcinoma associated 1
- Wdr5
WD repeat domain 5
- ZEB1/2
Zinc finger E-box binding homeobox 1/2
Authors’ contribution
YZ major designed, carried out and wrote the manuscript. HL, QZ and YZ revised and corrected the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Number 31900694).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bao H, Song J. Treating brain disorders by targeting adult neural stem cells. Trends Mol Med. 2018;24:991–1006. doi: 10.1016/j.molmed.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kempermann G, Gage FH, Aigner L, Song H, Curtis MA, Thuret S, Kuhn HG, Jessberger S, Frankland PW, Cameron HA, Gould E, Hen R, Abrous DN, Toni N, Schinder AF, Zhao X, Lucassen PJ, Frisen J. Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell. 2018;23:25–30. doi: 10.1016/j.stem.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz C, Salinas RD, Zarabi H, Kriegstein AR, Lim DA. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fawal MA, Davy A. Impact of metabolic pathways and epigenetics on neural stem cells. Epigenet Insights. 2018;11:2516865718820946. doi: 10.1177/2516865718820946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morizur L, Chicheportiche A, Gauthier LR, Daynac M, Boussin FD, Mouthon MA. Distinct molecular signatures of quiescent and activated adult neural stem cells reveal specific interactions with their microenvironment. Stem Cell Rep. 2018;11:565–577. doi: 10.1016/j.stemcr.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonaguidi MA, Song J, Ming GL, Song H. A unifying hypothesis on mammalian neural stem cell properties in the adult hippocampus. Curr Opin Neurobiol. 2012;22:754–761. doi: 10.1016/j.conb.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ninkovic J, Gotz M. Fate specification in the adult brain–lessons for eliciting neurogenesis from glial cells. BioEssays. 2013;35:242–252. doi: 10.1002/bies.201200108. [DOI] [PubMed] [Google Scholar]
- 9.Sorrells SF, Paredes MF, Cebrian-Silla A, Sandoval K, Qi D, Kelley KW, James D, Mayer S, Chang J, Auguste KI, Chang EF, Gutierrez AJ, Kriegstein AR, Mathern GW, Oldham MC, Huang EJ, Garcia-Verdugo JM, Yang Z, Alvarez-Buylla A. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature. 2018;555:377–381. doi: 10.1038/nature25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis CV, Suh LS, Rodriguez ML, Kril JJ, Sutherland GT. Human adult neurogenesis across the ages: an immunohistochemical study. Neuropathol Appl Neurobiol. 2016;42:621–638. doi: 10.1111/nan.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, Bostrom E, Westerlund I, Vial C, Buchholz BA, Possnert G, Mash DC, Druid H, Frisen J. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tobin MK, Musaraca K, Disouky A, Shetti A, Bheri A, Honer WG, Kim N, Dawe RJ, Bennett DA, Arfanakis K, Lazarov O. Human hippocampal neurogenesis persists in aged adults and Alzheimer’s disease patients. Cell Stem Cell. 2019;24(974–982):e3. doi: 10.1016/j.stem.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 15.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur H, Sarmah D, Saraf J, Vats K, Kalia K, Borah A, Yavagal DR, Dave KR, Ghosh Z, Bhattacharya P. Noncoding RNAs in ischemic stroke: time to translate. Ann N Y Acad Sci. 2018;1421:19–36. doi: 10.1111/nyas.13612. [DOI] [PubMed] [Google Scholar]
- 19.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen R, Xu X, Huang L, Zhong W, Cui L. The regulatory role of long noncoding RNAs in different brain cell types involved in ischemic stroke. Front Mol Neurosci. 2019;12:61. doi: 10.3389/fnmol.2019.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zywitza V, Misios A, Bunatyan L, Willnow TE, Rajewsky N. Single-cell transcriptomics characterizes cell types in the subventricular zone and uncovers molecular defects impairing adult neurogenesis. Cell Rep. 2018;25(2457–2469):e8. doi: 10.1016/j.celrep.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Deng B, Cheng X, Li H, Qin J, Tian M, Jin G. Microarray expression profiling in the denervated hippocampus identifies long noncoding RNAs functionally involved in neurogenesis. BMC Mol Biol. 2017;18:15. doi: 10.1186/s12867-017-0091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu L, Zhu J, Liu Y, Chen Y, Li Y, Huang L, Chen S, Li T, Dang Y, Chen T. Methamphetamine induces alterations in the long non-coding RNAs expression profile in the nucleus accumbens of the mouse. BMC Neurosci. 2015;16:18. doi: 10.1186/s12868-015-0157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosseinkhani M, Shirazi R, Rajaei F, Mahmoudi M, Mohammadi N, Abbasi M. Engineering of the embryonic and adult stem cell niches. Iran Red Crescent Med J. 2013;15:83–92. [Google Scholar]
- 26.Erceg S, Ronaghi M, Stojkovic M. Human embryonic stem cell differentiation toward regional specific neural precursors. Stem Cells. 2009;27:78–87. doi: 10.1634/stemcells.2008-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariano ED, Teixeira MJ, Marie SK, Lepski G. Adult stem cells in neural repair: current options, limitations and perspectives. World J Stem Cells. 2015;7:477–482. doi: 10.4252/wjsc.v7.i2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li YP, Duan FF, Zhao YT, Gu KL, Liao LQ, Su HB, Hao J, Zhang K, Yang N, Wang Y. A TRIM71 binding long noncoding RNA Trincr1 represses FGF/ERK signaling in embryonic stem cells. Nat Commun. 2019;10:1368. doi: 10.1038/s41467-019-08911-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- 30.Chicha L, Smith T, Guzman R. Stem cells for brain repair in neonatal hypoxia-ischemia. Childs Nerv Syst. 2014;30:37–46. doi: 10.1007/s00381-013-2304-4. [DOI] [PubMed] [Google Scholar]
- 31.Wang C, Li G, Wu Y, Xi J, Kang J. LincRNA1230 inhibits the differentiation of mouse ES cells towards neural progenitors. Sci China Life Sci. 2016;59:443–454. doi: 10.1007/s11427-016-5008-7. [DOI] [PubMed] [Google Scholar]
- 32.Cheng L, Huang Z, Zhou W, Wu Q, Donnola S, Liu JK, Fang X, Sloan AE, Mao Y, Lathia JD, Min W, McLendon RE, Rich JN, Bao S. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang J, Yu B, Li Y, Zhang W, Alvarez AA, Hu B, Cheng SY, Feng H. TGF-beta-activated lncRNA LINC00115 is a critical regulator of glioma stem-like cell tumorigenicity. EMBO Rep. 2019;20:e48170. doi: 10.15252/embr.201948170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, Shibata T, Naito M, Kim HJ, Miyata K, Kataoka K, Kondo Y. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun. 2016;7:13616. doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu M, Xue Y, Zheng J, Liu X, Yu H, Liu L, Li Z, Liu Y. Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol Cancer. 2017;16:110. doi: 10.1186/s12943-017-0677-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Li XT, Li JC, Feng M, Zhou YX, Du ZW. Novel lncRNA-ZNF281 regulates cell growth, stemness and invasion of glioma stem-like U251s cells. Neoplasma. 2019;66:118–127. doi: 10.4149/neo_2018_180613N391. [DOI] [PubMed] [Google Scholar]
- 37.Brodie S, Lee HK, Jiang W, Cazacu S, Xiang C, Poisson LM, Datta I, Kalkanis S, Ginsberg D, Brodie C. The novel long non-coding RNA TALNEC2, regulates tumor cell growth and the stemness and radiation response of glioma stem cells. Oncotarget. 2017;8:31785–31801. doi: 10.18632/oncotarget.15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen WL, Chen HJ, Hou GQ, Zhang XH, Ge JW. LINC01198 promotes proliferation and temozolomide resistance in a NEDD4-1-dependent manner, repressing PTEN expression in glioma. Aging. 2019;11:6053–6068. doi: 10.18632/aging.102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vieira MS, Goulart VAM, Parreira RC, Oliveira-Lima OC, Glaser T, Naaldijk YM, Ferrer A, Savanur VH, Reyes PA, Sandiford O, Rameshwar P, Ulrich H, Pinto MCX, Resende RR. Decoding epigenetic cell signaling in neuronal differentiation. Semin Cell Dev Biol. 2019;95:12–24. doi: 10.1016/j.semcdb.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Ayana R, Singh S, Pati S. Decoding crucial LncRNAs implicated in neurogenesis and neurological disorders. Stem Cells Dev. 2017;26:541–553. doi: 10.1089/scd.2016.0290. [DOI] [PubMed] [Google Scholar]
- 41.Yao B, Jin P. Unlocking epigenetic codes in neurogenesis. Genes Dev. 2014;28:1253–1271. doi: 10.1101/gad.241547.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goff LA, Groff AF, Sauvageau M, Trayes-Gibson Z, Sanchez-Gomez DB, Morse M, Martin RD, Elcavage LE, Liapis SC, Gonzalez-Celeiro M, Plana O, Li E, Gerhardinger C, Tomassy GS, Arlotta P, Rinn JL. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2015;112:6855–6862. doi: 10.1073/pnas.1411263112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 44.Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, He D, Weissman JS, Kriegstein AR, Diaz AA, Lim DA. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCorkindale AL, Wahle P, Werner S, Jungreis I, Menzel P, Shukla CJ, Abreu RLP, Irizarry RA, Meyer IM, Kellis M, Zinzen RP. A gene expression atlas of embryonic neurogenesis in Drosophila reveals complex spatiotemporal regulation of lncRNAs. Development. 2019;146:dev175265. doi: 10.1242/dev.175265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewitus E, Huttner WB. Neurodevelopmental LincRNA microsyteny conservation and mammalian brain size evolution. PLoS ONE. 2015;10:e0131818. doi: 10.1371/journal.pone.0131818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prajapati B, Fatma M, Maddhesiya P, Sodhi MK, Fatima M, Dargar T, Bhagat R, Seth P, Sinha S. Identification and epigenetic analysis of divergent long non-coding RNAs in multilineage differentiation of human neural progenitor cells. RNA Biol. 2019;16:13–24. doi: 10.1080/15476286.2018.1553482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng J, Yi D, Liu Y, Wang M, Zhu Y, Shi H. Long nonding RNA UCA1 regulates neural stem cell differentiation by controlling miR-1/Hes1 expression. Am J Transl Res. 2017;9:3696–3704. [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, Deng Y, Duan D, Sun S, Ge L, Zhuo Y, Yuan T, Wu P, Wang H, Lu M, Xia Y. Hyperthermia influences fate determination of neural stem cells with lncRNAs alterations in the early differentiation. PLoS ONE. 2017;12:e0171359. doi: 10.1371/journal.pone.0171359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Askarian-Amiri ME, Seyfoddin V, Smart CE, Wang J, Kim JE, Hansji H, Baguley BC, Finlay GJ, Leung EY. Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PLoS ONE. 2014;9:e102140. doi: 10.1371/journal.pone.0102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kan L, Israsena N, Zhang Z, Hu M, Zhao LR, Jalali A, Sahni V, Kessler JA. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev Biol. 2004;269:580–594. doi: 10.1016/j.ydbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 52.Zhang S, Cui W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J Stem Cells. 2014;6:305–311. doi: 10.4252/wjsc.v6.i3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahmad A, Strohbuecker S, Tufarelli C, Sottile V. Expression of a SOX1 overlapping transcript in neural differentiation and cancer models. Cell Mol Life Sci. 2017;74:4245–4258. doi: 10.1007/s00018-017-2580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knauss JL, Miao N, Kim SN, Nie Y, Shi Y, Wu T, Pinto HB, Donohoe ME, Sun T. Long noncoding RNA Sox2ot and transcription factor YY1 co-regulate the differentiation of cortical neural progenitors by repressing Sox2. Cell Death Dis. 2018;9:799. doi: 10.1038/s41419-018-0840-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tosetti V, Sassone J, Ferri ALM, Taiana M, Bedini G, Nava S, Brenna G, Di Resta C, Pareyson D, Di Giulio AM, Carelli S, Parati EA, Gorio A. Transcriptional role of androgen receptor in the expression of long non-coding RNA Sox2OT in neurogenesis. PLoS ONE. 2017;12:e0180579. doi: 10.1371/journal.pone.0180579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 57.Li W, Shen W, Zhang B, Tian K, Li Y, Mu L, Luo Z, Zhong X, Wu X, Liu Y, Zhou Y. Long noncoding RNA LncKdm2b regulates cortical neuronal differentiation by cis-activating Kdm2b. Protein Cell. 2020;11(3):161–186. doi: 10.1007/s13238-019-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33:296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pavlaki I, Alammari F, Sun B, Clark N, Sirey T, Lee S, Woodcock DJ, Ponting CP, Szele FG, Vance KW. The long non-coding RNA Paupar promotes KAP1-dependent chromatin changes and regulates olfactory bulb neurogenesis. EMBO J. 2018;37(10):e98219. doi: 10.15252/embj.201798219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi Y, Zhao X, Hsieh J, Wichterle H, Impey S, Banerjee S, Neveu P, Kosik KS. MicroRNA regulation of neural stem cells and neurogenesis. J Neurosci. 2010;30:14931–14936. doi: 10.1523/JNEUROSCI.4280-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weng R, Lu C, Liu X, Li G, Lan Y, Qiao J, Bai M, Wang Z, Guo X, Ye D, Jiapaer Z, Yang Y, Xia C, Wang G, Kang J. Long noncoding RNA-1604 orchestrates neural differentiation through the miR-200c/ZEB axis. Stem Cells. 2018;36:325–336. doi: 10.1002/stem.2749. [DOI] [PubMed] [Google Scholar]
- 62.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng X, Li H, Zhao H, Li W, Qin J, Jin G. Function and mechanism of long non-coding RNA Gm21284 in the development of hippocampal cholinergic neurons. Cell Biosci. 2019;9:72. doi: 10.1186/s13578-019-0336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang L, Xue Z, Yan J, Wang J, Liu Q, Jiang H. LncRNA Riken-201 and Riken-203 modulates neural development by regulating the Sox6 through sequestering miRNAs. Cell Prolif. 2019;52:e12573. doi: 10.1111/cpr.12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang L, Yan J, Liu Q, Xie Z, Jiang H. LncRNA Rik-203 contributes to anesthesia neurotoxicity via microRNA-101a-3p and GSK-3beta-mediated neural differentiation. Sci Rep. 2019;9:6822. doi: 10.1038/s41598-019-42991-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao Y, Zhang R, Wei G, Dai S, Zhang X, Yang W, Li X, Bai C. Long non-coding RNA maternally expressed 3 increases the expression of neuron-specific genes by targeting miR-128-3p in all-trans retinoic acid-induced neurogenic differentiation from amniotic epithelial cells. Front Cell Dev Biol. 2019;7:342. doi: 10.3389/fcell.2019.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen L, Feng P, Zhu X, He S, Duan J, Zhou D. Long non-coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. J Cell Mol Med. 2016;20:2102–2110. doi: 10.1111/jcmm.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grammatikakis I, Gorospe M. Identification of neural stem cell differentiation repressor complex Pnky-PTBP1. Stem Cell Investig. 2016;3:10. doi: 10.21037/sci.2016.03.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winzi M, Casas Vila N, Paszkowski-Rogacz M, Ding L, Noack S, Theis M, Butter F, Buchholz F. The long noncoding RNA lncR492 inhibits neural differentiation of murine embryonic stem cells. PLoS ONE. 2018;13:e0191682. doi: 10.1371/journal.pone.0191682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng X, Lin C, Li Y, Ye J, Zhou J, Guo P. Long noncoding RNA BDNF-AS regulates ketamine-induced neurotoxicity in neural stem cell derived neurons. Biomed Pharmacother. 2016;82:722–728. doi: 10.1016/j.biopha.2016.05.050. [DOI] [PubMed] [Google Scholar]
- 71.Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, Head SR, Duester G, Dong PD, Rana TM. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeWitt JJ, Hecht PM, Grepo N, Wilkinson B, Evgrafov OV, Morris KV, Knowles JA, Campbell DB. Transcriptional gene silencing of the autism-associated long noncoding RNA MSNP1AS in human neural progenitor cells. Dev Neurosci. 2016;38:375–383. doi: 10.1159/000453258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen PF, Hsiao JS, Sirois CL, Chamberlain SJ. RBFOX1 and RBFOX2 are dispensable in iPSCs and iPSC-derived neurons and do not contribute to neural-specific paternal UBE3A silencing. Sci Rep. 2016;6:25368. doi: 10.1038/srep25368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J, Cao B, Zhao H, Gao Y, Luo Y, Chen Y, Feng J. Long noncoding RNA H19 prevents neurogenesis in ischemic stroke through p53/Notch1 pathway. Brain Res Bull. 2019;150:111–117. doi: 10.1016/j.brainresbull.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 75.You D, You H. Repression of long non-coding RNA MEG3 restores nerve growth and alleviates neurological impairment after cerebral ischemia-reperfusion injury in a rat model. Biomed Pharmacother. 2019;111:1447–1457. doi: 10.1016/j.biopha.2018.12.067. [DOI] [PubMed] [Google Scholar]
- 76.Oostra BA, Willemsen R. A fragile balance: FMR1 expression levels. Hum Mol Genet. 2003;12(Spec No 2):R249–R257. doi: 10.1093/hmg/ddg298. [DOI] [PubMed] [Google Scholar]
- 77.Peschansky VJ, Pastori C, Zeier Z, Wentzel K, Velmeshev D, Magistri M, Silva JP, Wahlestedt C. The long non-coding RNA FMR4 promotes proliferation of human neural precursor cells and epigenetic regulation of gene expression in trans. Mol Cell Neurosci. 2016;74:49–57. doi: 10.1016/j.mcn.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 78.Bilinovich SM, Lewis K, Grepo N, Campbell DB. The long noncoding RNA RPS10P2-AS1 is implicated in autism spectrum disorder risk and modulates gene expression in human neuronal progenitor cells. Front Genet. 2019;10:970. doi: 10.3389/fgene.2019.00970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DeWitt JJ, Grepo N, Wilkinson B, Evgrafov OV, Knowles JA, Campbell DB. Impact of the autism-associated long noncoding RNA MSNP1AS on neuronal architecture and gene expression in human neural progenitor cells. Genes. 2016;7:76. doi: 10.3390/genes7100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J, Zhang ZY, Yu H, Yang AP, Hu PF, Liu Z, Wang M. Long noncoding RNA C21orf121/bone morphogenetic protein 2/microRNA-140-5p gene network promotes directed differentiation of stem cells from human exfoliated deciduous teeth to neuronal cells. J Cell Biochem. 2018;120(2):1464–1476. doi: 10.1002/jcb.27313. [DOI] [PubMed] [Google Scholar]
- 81.Osman AM, Porritt MJ, Nilsson M, Kuhn HG. Long-term stimulation of neural progenitor cell migration after cortical ischemia in mice. Stroke. 2011;42:3559–3565. doi: 10.1161/STROKEAHA.111.627802. [DOI] [PubMed] [Google Scholar]
- 82.Liu J, Li Q, Zhang KS, Hu B, Niu X, Zhou SM, Li SG, Luo YP, Wang Y, Deng ZF. Downregulation of the long non-coding RNA Meg3 promotes angiogenesis after ischemic brain injury by activating notch signaling. Mol Neurobiol. 2017;54:8179–8190. doi: 10.1007/s12035-016-0270-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Naruse M, Ishizaki Y, Ikenaka K, Tanaka A, Hitoshi S. Origin of oligodendrocytes in mammalian forebrains: a revised perspective. J Physiol Sci. 2017;67:63–70. doi: 10.1007/s12576-016-0479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ohtsuka T, Shimojo H, Matsunaga M, Watanabe N, Kometani K, Minato N, Kageyama R. Gene expression profiling of neural stem cells and identification of regulators of neural differentiation during cortical development. Stem Cells. 2011;29:1817–1828. doi: 10.1002/stem.731. [DOI] [PubMed] [Google Scholar]
- 85.Benito-Munoz M, Matute C, Cavaliere F. Adenosine A1 receptor inhibits postnatal neurogenesis and sustains astrogliogenesis from the subventricular zone. Glia. 2016;64:1465–1478. doi: 10.1002/glia.23010. [DOI] [PubMed] [Google Scholar]
- 86.Lein ES, Belgard TG, Hawrylycz M, Molnar Z. Transcriptomic perspectives on neocortical structure, development, evolution, and disease. Annu Rev Neurosci. 2017;40:629–652. doi: 10.1146/annurev-neuro-070815-013858. [DOI] [PubMed] [Google Scholar]
- 87.Dong X, Chen K, Cuevas-Diaz Duran R, You Y, Sloan SA, Zhang Y, Zong S, Cao Q, Barres BA, Wu JQ. Comprehensive identification of long non-coding RNAs in purified cell types from the brain reveals functional LncRNA in OPC fate determination. PLoS Genet. 2015;11:e1005669. doi: 10.1371/journal.pgen.1005669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emery B, Lu QR. Transcriptional and epigenetic regulation of oligodendrocyte development and myelination in the central nervous system. Cold Spring Harb Perspect Biol. 2015;7:a020461. doi: 10.1101/cshperspect.a020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zuchero JB, Barres BA. Intrinsic and extrinsic control of oligodendrocyte development. Curr Opin Neurobiol. 2013;23:914–920. doi: 10.1016/j.conb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He D, Wang J, Lu Y, Deng Y, Zhao C, Xu L, Chen Y, Hu YC, Zhou W, Lu QR. lncRNA functional networks in oligodendrocytes reveal stage-specific myelination control by an lncOL1/Suz12 complex in the CNS. Neuron. 2017;93:362–378. doi: 10.1016/j.neuron.2016.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Y, Guo B, Yang R, Xiao Z, Gao X, Yu J, Li S, Luo Y. A novel long noncoding RNA lnc158 promotes the differentiation of mouse neural precursor cells into oligodendrocytes by targeting nuclear factor-IB. NeuroReport. 2018;29:1121–1128. doi: 10.1097/WNR.0000000000001083. [DOI] [PubMed] [Google Scholar]
- 92.Kasuga Y, Fudge AD, Zhang Y, Li H. Characterization of a long noncoding RNA Pcdh17it as a novel marker for immature premyelinating oligodendrocytes. Glia. 2019;67:2166–2177. doi: 10.1002/glia.23684. [DOI] [PubMed] [Google Scholar]
- 93.Mills JD, Kavanagh T, Kim WS, Chen BJ, Waters PD, Halliday GM, Janitz M. High expression of long intervening non-coding RNA OLMALINC in the human cortical white matter is associated with regulation of oligodendrocyte maturation. Mol Brain. 2015;8:2. doi: 10.1186/s13041-014-0091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Katsel P, Roussos P, Fam P, Khan S, Tan W, Hirose T, Nakagawa S, Pletnikov MV, Haroutunian V. The expression of long noncoding RNA NEAT1 is reduced in schizophrenia and modulates oligodendrocytes transcription. NPJ Schizophr. 2019;5:3. doi: 10.1038/s41537-019-0071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duan C, Liu Y, Li Y, Chen H, Liu X, Chen X, Yue J, Zhou X, Yang J. Sulfasalazine alters microglia phenotype by competing endogenous RNA effect of miR-136-5p and long non-coding RNA HOTAIR in cuprizone-induced demyelination. Biochem Pharmacol. 2018;155:110–123. doi: 10.1016/j.bcp.2018.06.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.