Significance
In the extracellular milieu, soluble ligands must be delivered at the right time and place to their target cell surface receptors; however, the extracellular control mechanisms underlying these events remain unclear. We propose that remodeling of type I collagen spatiotemporally regulates pigment epithelium-derived factor (PEDF), a secreted glycoprotein, which promotes either cell survival or death, depending on the context. The crystal structure of PEDF in complex with the heterotrimeric collagen peptide revealed that it recognizes a cryptic site on type I collagen containing a lysyl residue susceptible for intermolecular cross-linking. Therefore, the receptor-binding activity of PEDF is regulated by collagen maturation and cross-linking. This study provides insights that will contribute to understanding the multifaceted nature of PEDF functions.
Abstract
Dynamic remodeling of the extracellular matrix affects many cellular processes, either directly or indirectly, through the regulation of soluble ligands; however, the mechanistic details of this process remain largely unknown. Here we propose that type I collagen remodeling regulates the receptor-binding activity of pigment epithelium-derived factor (PEDF), a widely expressed secreted glycoprotein that has multiple important biological functions in tissue and organ homeostasis. We determined the crystal structure of PEDF in complex with a disulfide cross-linked heterotrimeric collagen peptide, in which the α(I) chain segments—each containing the respective PEDF-binding region (residues 930 to 938)—are assembled with an α2α1α1 staggered configuration. The complex structure revealed that PEDF specifically interacts with a unique amphiphilic sequence, KGHRGFSGL, of the type I collagen α1 chain, with its proposed receptor-binding sites buried extensively. Molecular docking demonstrated that the PEDF-binding surface of type I collagen contains the cross-link–susceptible Lys930 residue of the α1 chain and provides a good foothold for stable docking with the α1(I) N-telopeptide of an adjacent triple helix in the fibril. Therefore, the binding surface is completely inaccessible if intermolecular crosslinking between two crosslink-susceptible lysyl residues, Lys9 in the N-telopeptide and Lys930, is present. These structural analyses demonstrate that PEDF molecules, once sequestered around newly synthesized pericellular collagen fibrils, are gradually liberated as collagen crosslinking increases, making them accessible for interaction with their target cell surface receptors in a spatiotemporally regulated manner.
Pigment epithelium-derived factor (PEDF), which was first discovered in cultured human fetal pigment epithelium and subsequently identified in a wide variety of tissues and organs throughout the body, is classified as a serine protease inhibitor (serpin) superfamily protein (1). Although to date, PEDF has exhibited no protease inhibitory activity (2), this secreted protein alternatively exerts multiple important biological functions, including neurotrophic, cell differentiating, antiangiogenic, and antitumor activities (1, 3). It is now recognized as a promising therapeutic candidate, particularly for cancer (4). Multiple cell surface receptors, including the 80-kDa calcium-independent phospholipase A2 (iPLA2)ζ, which belongs to the PLA2/nutrin/patatin-like phospholipase domain-containing 2 (PNPLA2) family, and the nonintegrin 37/67-kDa laminin receptor, have been proposed to interact with this multifunctional protein (3, 4). In addition, several binding epitopes of PEDF, including two distinctive receptor-binding regions, a 34-aa segment (Asp44–Asn77) and a 44-aa segment (Val78–Thr121), which exhibit antiangiogenic and proangiogenic/neurotrophic activities, respectively, have been identified and extensively studied (5). Several reports have suggested that the complexities of PEDF receptor binding are cell context-dependent, and that the existence of several target receptors, even in a single cell phenotype, may lead to contradictory cellular responses (i.e., cell survival or death) depending on the concentration of PEDF as well as the activation state of the target cell in vitro (5–7). These findings indicate that PEDF actions are extracellularly regulated in vivo to maintain physiological homeostasis; however, the mechanistic details of this regulation are largely unknown.
The crystal structure of human PEDF shows that it adopts a typical serpin fold but has unique asymmetric charge distributions composed of a positively charged surface in its C-terminus region and a negatively charged surface on the opposite side (8). This surface feature is reflected in the ability of PEDF to bind a variety of extracellular matrix (ECM) components. In particular, the negatively charged region is important for collagen binding (9, 10), while the positively charged region is responsible for interactions with glycosaminoglycans, including heparin/heparan sulfate and hyaluronan (11–13). As in the case of other serpins and growth factors (14, 15), ECM binding is essential for the biological functions of PEDF. Indeed, a cancer cell xenograft experiment using nude mice demonstrated that the collagen-binding null mutant of PEDF completely lost its antiangiogenic and antitumor properties, although the mutant retained binding ability to its target receptors (16). Using site-specific photocrosslinking coupled with matrix metalloproteinase-1 (MMP-1) digestion, we found that PEDF selectively binds to the C-terminal 1/4 fragment of type I collagen (17). The binding site was further narrowed down using a series of synthetic homotrimeric peptides containing portions of the type I collagen sequence rich in lysyl and arginyl residues, with which a crucial PEDF-binding site was unexpectedly identified in the C-terminal–specific amphipathic region (KGHRGFSGL; residues 930 to 938), in the type I collagen α1 chain (18). This remarkable sequence selectivity in collagen recognition suggests that the PEDF–collagen interaction is not simply dictated by a nonspecific ionic interaction hypothesized to be required for its deposition in the ECM but could have additional functional implications in expressing and regulating the biological activities of PEDF.
To further investigate this PEDF–collagen interaction, we designed and synthesized heterotrimeric collagen peptides, consisting of two α1 and one α2 chain segments of type I collagen, and elucidated the atomic details of PEDF interaction with the heterotrimeric type I collagen tripe helix. Using this structural information, we propose the mechanisms by which PEDF selectively recognizes a cryptic site on type I collagen α1 chain containing the crosslink-susceptible Lys930 residue. Molecular docking revealed that the PEDF-binding surface is completely masked by the interaction with a nonhelical N-telopeptide segment of the α1 chain via formation of intermolecular crosslinks in mature type I collagen fibrils. Therefore, this crosslink-coupled regulation of PEDF binding permits spatiotemporal action on specific cells that actively remodel their pericellular type I collagen fibrils.
Results
Design and Synthesis of Heterotrimeric Collagen Peptides.
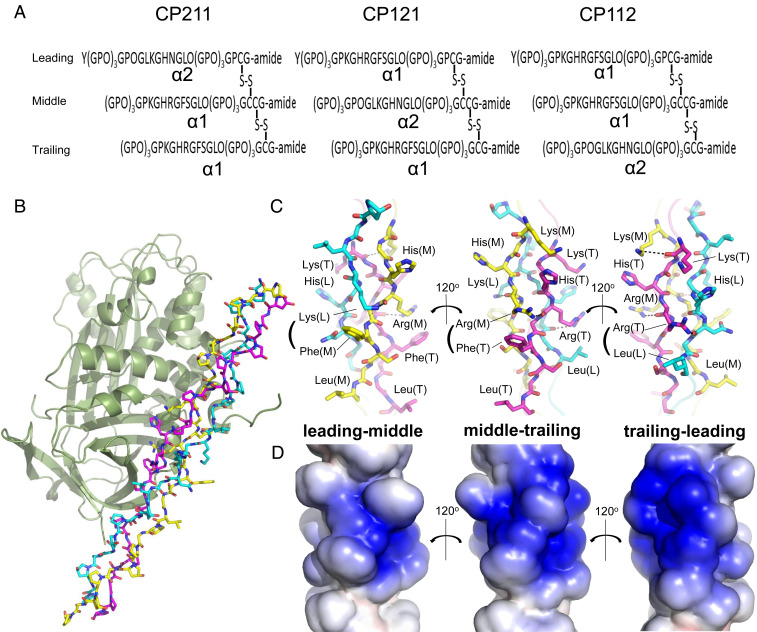
The exact chain staggering of native type I collagen is still unknown; thus, we designed and synthesized heterotrimeric type I collagen peptides in which all three structural isomers—α2α1α1, α1α2α1, and α1α1α2—were represented (Fig. 1A and SI Appendix, Fig. S1). Each peptide consisted of sequences corresponding to the conserved PEDF-binding region (residues 930 to 938) (18) of type I collagen (guest regions) flanked by glycyl-prolyl-hydroxyproline (Gly-Pro-Hyp) triplet repeats (host regions) for stabilization of the triple helical structure (19). We achieved staggering by one residue through selective Cys pairing at the C-terminal region. The resulting host-guest heterotrimeric collagen peptides with α2 chain segments in the leading, middle, and trailing positions were designated CP211, CP121, and CP112, respectively. These peptides share similar triple-helical stabilities, with melting temperatures (Tm) of ∼40 °C (SI Appendix, Fig. S2). Sedimentation velocity analytical ultracentrifugation (SV-AUC) and isothermal titration calorimetry (ITC) demonstrated that PEDF interacts with all of these heterotrimeric peptides with a stoichiometry of 1:1; however, the three peptides have different binding affinities depending on the chain registry (Table 1 and SI Appendix, Figs. S3 and S4). CP211 showed higher binding affinity for PEDF compared with the other peptides, comparable to the affinity (Kd = ∼0.13 μM) observed for the interaction between PEDF and native type I collagen (10).
Fig. 1.
Structural determination of PEDF in complex with synthetic heterotrimeric type I collagen peptide. (A) Designed disulfide-crosslinked heterotrimeric collagen peptides with three combinations of chain arrangements, in which the α2 chain segment derived from a sequence alignment with the previously identified PEDF-binding sequence of type I collagen α1 chain is positioned at the leading, middle, or trailing position (i.e., α2α1α1, α1α2α1, or α1α1α2), designated CP211, CP121, and CP112, respectively. (B) The PEDF-CP211 complex shown as a cartoon for PEDF and a stick model for CP211. The leading (L), middle (M), and trailing (T) chains in the triple helix are colored light blue, yellow, and magenta, respectively. (C) Stick model of the guest region of CP211 shown in three different orientations, each rotated by 120° about the vertical axis; highlighted leading-middle chains, middle-trailing chains, and trailing-leading chains are shown (left to right). Basic (Lys, His, and Arg) and hydrophobic (Phe and Leu) residues are labeled. Hydrogen-bonding interactions are shown as dotted lines. (D) Electrostatic potential surface representation of the guest region of CP211. The three different orientations, each rotated by 120° about the vertical axis, are shown as in Fig. 1C.
Table 1.
Binding parameters of PEDF to collagen peptides from SV-AUC and ITC analyses
| Peptide | SV-AUC | ITC | ||||
| Kd/μM | Kd/μM | n | ΔG/kcal mol−1 | ΔH/kcal mol−1 | TΔS/kcal mol−1 | |
| CP211 | 0.3 | 0.24 | 0.94 | −8.9 | 2.00 | 10.88 |
| CP121 | 4.1 | 3.85 | 0.83 | −7.3 | 3.78 | 11.04 |
| CP112 | 0.7 | 1.12 | 0.89 | −8.0 | 2.91 | 10.89 |
Crystal Structure of Mouse PEDF in Complex with Collagen Peptide CP211.
Our attempts to crystallize PEDF-collagen peptide complexes produced single crystals of the mouse PEDF-CP211 complex that allowed us to determine its structure at 2.47-Å resolution (Fig. 1B and SI Appendix, Table S1). From the electron density map, we were able to locate and discriminate each CP211 chain position except for the terminal regions, which had low electron densities, possibly due to conformational flexibility (SI Appendix, Fig. S5). The PEDF structure in the complex was easily superimposed onto the structure of apo human PEDF (8) and had an overall Cα root mean square deviation value of 0.40 Å, indicating no significant conformational change on binding to the collagen triple helix (SI Appendix, Fig. S6).
Of note, the PEDF-binding region of type I collagen is an imino-poor region known as the micro-unfolding domain, due to the low predicted thermal stability of its triple helix (20). However, CP211 in the complex has a canonical triple-helical structure with a helix parameter close to that of the 7/2 helical conformation (19), except at the two boundaries between the host and guest sequences, where the helix slightly bends to maximize contact with PEDF (SI Appendix, Fig. S7). The guest region includes an amphiphilic surface comprising two parts: one composed of the basic residues Lys, His, and Arg and the other composed of hydrophobic residues Phe and Leu (Fig. 1 C and D). The Lys and Arg of one chain form hydrogen bonds with the backbone carbonyls of adjacent chains to stabilize interchain interactions of the triple helix, alleviating electric repulsion among these basic residues.
Because of the characteristic one-residue stagger of the collagen triple helix, the side chain of the residue at the Yaa position in the sequence -Gly-Xaa-Yaa-Gly-Xaa′-Yaa′- is in close proximity to those at the Xaa and Xaa′ positions of adjacent chains. In CP211, interchain Yaa-Xaa′ stabilizing interactions were additionally observed between Lys (leading chain, L) and Phe (middle chain, M) residues, as well as between Arg (M) and Phe (trailing chain, T), each of which uniquely form an interchain cation–π interaction that connects the basic and hydrophobic portions of the collagen peptide (Fig. 1C, Left and Middle).
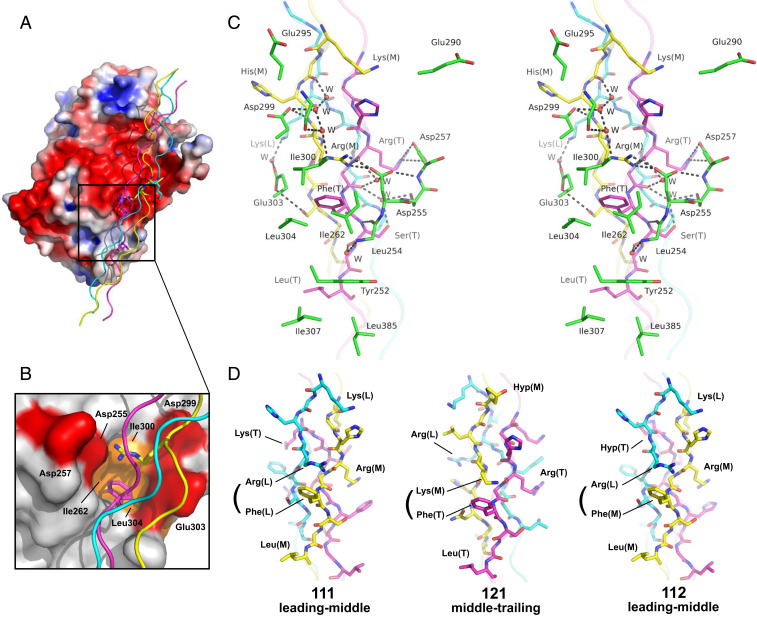
Using the heterotrimeric collagen peptide, we confirmed that PEDF specifically recognizes one particular surface of the type I collagen triple helix composed of an amphiphilic α1 chain region, KGHRGFSGL, at the middle and trailing positions (Fig. 1C, Middle). As proposed previously (10), PEDF recognizes type I collagen through a highly acidic surface (Fig. 2A). This surface faces a cluster of basic residues on consecutive α1 chain segments in the middle and trailing chains (Fig. 1 C and D, Middle). It does not interact with the less-basic α2 chain segment, OGLKGHNGL, in the leading chain. Although this electrostatic interaction contributes to the PEDF–collagen interaction, we found that direct contacts are made primarily in the binding cleft located at the boundary between the acidic and hydrophobic surfaces, which is consistent with the amphiphilic nature of the PEDF-binding sequence of the α1 chain (Fig. 2 A–C). The amphiphilic binding cleft of PEDF grasps the interchain Arg-Phe pair from two consecutive α1 chains (Fig. 2 B and C). The bottom floor of this cleft is surrounded by hydrophobic residues (Ile262, Ile300, and Leu304) that can accommodate the bulky aromatic ring of Phe (T), while the side chain of Arg (M) forms a salt bridge with Asp255 in the upper region of the cleft. These interactions are further stabilized by a salt bridge between Asp257 and Arg (T), and a network of hydrogen bonds among Asp299, His (M), and Arg (M) (Fig. 2C). This characteristic binding mode of PEDF recognizing an interchain Arg-Phe pair is unique among the protein-collagen peptide complexes reported to date (19, 20) and is consistent with previous mutational studies demonstrating that Asp255, Asp257, and Asp299 of mouse PEDF are crucial for the native type I collagen interaction (9, 16).
Fig. 2.
Interfacial interactions of the PEDF-CP211 complex. (A) Electrostatic potential surface representation of PEDF and cartoon representation of CP211. The residues important for PEDF recognition are shown as stick models. The principal binding interface is indicated as a rectangular box. (B) Closeup view of the collagen-binding cleft of PEDF. Hydrophobic and acidic residues are colored orange and red, respectively. The interchain Arg-Phe pair is shown as a stick model. (C) Stereoview of the interaction surface of the complex. Hydrogen bonds are shown as black dotted lines. The water molecules involved in the hydrogen-bonded networks are shown as red spheres. (D) Modeling of the PEDF-binding region of homotrimeric or heterotrimeric collagen peptides. Schematics of the possible PEDF-binding face of the α1 chain homotrimer at leading-middle chains (Left), the peptide CP121 at middle-trailing chains (Middle), and the peptide CP112 at leading-middle chains (Right), that would contact PEDF if it binds to the interchain Yaa-Xaa′ pair, that is, Arg(L)-Phe(M) for α1 chain homotrimer, Lys(M)-Phe(T) for CP121, and Arg(L)-Phe(M) for CP112.
The amphiphilic nature of the collagen-binding cleft suitably accommodates an interchain Yaa-Xaa′ residue pair, in which the Yaa position and the Xaa′ position of the adjacent chain are basic and hydrophobic residues, respectively. In addition to the interchain Arg(M)-Phe(T) pair, two such pairs, Lys(L)-Phe(M) and Arg(T)-Leu(L), are found at the other two faces of the triple helix (Fig. 1C, Left and Right). Neither of these faces would cause steric clashes on binding to PEDF; however, replacement of Arg or Phe in the Arg-Phe pair by Lys or Leu should substantially weaken the interactions due to the loss of hydrogen-bonding networks and the reduction of hydrophobic contacts, respectively, makes these faces less dominant for the interaction. Therefore, PEDF has evolved to interact with the interchain Arg-Phe pair, formed solely by two α1 chains at consecutive positions.
Molecular modeling showed that the constellation of residues in the PEDF-binding face of CP211 can be fully reconstituted in one face comprising leading-middle chains of the α1 chain homotrimer (Fig. 2D, Left, and SI Appendix, Fig. S8). Consistently, the binding mode observed in the PEDF-CP211 complex is in excellent agreement with previous alanine-scanning experiments on the PEDF-binding motif, IKGHRGFSGL, of the α1 chain (18). These experiments showed that both Arg and Phe are essential for PEDF binding and that Lys and Leu are also important, with modest affinity reductions on their substitution. The remaining residues, Ile, His, and Ser, had no effect on PEDF binding. Concerning the roles for Lys and Leu, while our structure showed that Leu(T) in the trailing chain forms hydrophobic contacts with Tyr252, Ile307, and Leu385 of PEDF that stabilize the interfacial interaction (Fig. 2C), unexpectedly, lysyl residues do not make specific salt bridges or engage in direct hydrogen bonding with PEDF. Nevertheless, two lysyl residues might be involved in long-range electrostatic interactions: Lys(L) in the leading chain forms a water-mediated hydrogen bond with Glu303 of PEDF, and Lys(M) in the middle chain points toward Glu290 and Glu295 in an acidic patch of PEDF (Fig. 2C). These observations are also consistent with previous mutational experiments on PEDF showing that substitutions of these glutamic acid residues reduce PEDF–collagen interaction, suggesting a contribution of these lysyl residues to PEDF binding (9).
Since the binding affinity of CP211 for PEDF is comparable to that of natural type I collagen, and the interactions observed in the PEDF-CP211 complex are in agreement with previous mutational experiments on PEDF that mutated all the Asp and Glu residues in the acidic patch of PEDF (9), we conclude that the binding mode observed in the PEDF-CP211 complex is biologically relevant and provides important information for understanding the role of type I collagen in the regulation of PEDF activity.
Chain Registry-Dependent PEDF Binding of Heterotrimeric Collagen Peptides.
The SV-AUC and ITC experiments clearly showed that the PEDF-binding affinities of heterotrimeric collagen peptides depend on the chain registry and are in the following order: CP211 > CP112 >> CP121 (Table 1 and SI Appendix, Figs. S3 and S4). To gain structural insight into the mode of PEDF binding to these peptides, we constructed molecular models for the peptides CP121 and CP112, based on the CP211 triple helical structure (Fig. 2D and SI Appendix, Fig. S8). Since the sequence of the α2 chain segment contains neither Arg nor Phe, interchain Arg-Phe pair is not formed for all possible binding faces of CP121; thus, the interfacial interactions in the PEDF-CP211 complex cannot be reconstituted. One likely binding face found in the middle-trailing chains provides an interchain Lys(M)-Phe(T) pair that can be recognized by PEDF without any obvious steric clash (Fig. 2D, Middle). Although some of key interacting residues in the PEDF-CP211 complex are conserved at the binding face, the replacement of Arg to Lys in the Arg-Phe pair apparently reduces the number of electrostatic and hydrogen bond interactions with PEDF, which may decrease the affinity of CP121 more than that of CP211. Furthermore, the Lys(M) in the middle chain involved in long-range electrostatic interactions in the PEDF-CP211 complex is replaced by Hyp, which also decreases the PEDF-binding affinity.
In contrast, the constellation of key interacting residues, including the interchain Arg-Phe pair in the PEDF-CP211 complex, was almost completely reconstituted at one face comprising leading-middle chains of CP112 (Fig. 2D, Right). Nevertheless, a slight reduction in affinity was observed (Table 1). We reasoned that this reduction in affinity for CP112 relative to CP211 is explained by the presence of Hyp(T) in the trailing chain, corresponding to Lys(L) in CP211, resulting in the loss of water-mediated hydrogen bonding with Glu303, which contributes to the PEDF-binding of CP211. These decreased affinities are consistent with the ITC data showing unfavorable enthalpy changes for CP121 and CP112 relative to that of CP211 on binding to PEDF (Table 1).
The correct chain registration of type I collagen has been a long-standing issue in the field of collagen research and is still being debated. Several studies have proposed contradictory models, in which the α2 chain is occupied at all possible positions, leading (21), middle (22), or trailing (23). From their recent binding experiments with heterotrimeric collagen peptides and collagen-binding proteins, including von Willebrand factor and discoidin domain receptor 2, Jalan et al. (24) concluded that the α2 chain in the trailing position is the most plausible form of type I collagen. For correct chain registry, our current structural analyses and previous mutational data indicate that positioning the α2 chain in the middle is not feasible, but could not conclusively identify the location of the α2 chain, at either the leading or trailing position, due to the nearly identical interfacial interactions between PEDF and these two isomers.
Specific PEDF-Binding Site on Type I Collagen.
An analysis of the type I collagen sequence showed that the RGF sequence, which potentially contributes to an interchain Arg-Phe pair in the triple helical structure, is rare in a sequence of >300 repeats of Gly-Xaa-Yaa and is found at only five sites outside of the C-terminal PEDF-binding region of the α1 chain (SI Appendix, Figs. S9 and S10). However, four of these sites have Pro or Hyp at the position next to Phe, rendering them unfavorable for interaction with PEDF, as the backbone carbonyl of Leu254 of PEDF forms a hydrogen bond with the backbone amide of Ser(T) of the collagen peptide in the PEDF-CP211 complex (Fig. 2C). Therefore, the imino acid is sterically unfavorable at this position. The remaining site at the α1 chain N terminus, KGHRGFSGL (residues 87 to 95), is identical to the C-terminal PEDF-binding site (residues 930 to 938) of the α1 chain. However, previous photocrosslinking experiments have demonstrated that PEDF primarily binds the collagenase-derived C-terminal 1/4 fragment of type I collagen (17). This discrepancy may be due to a difference between the posttranslational modifications at Lys87 and Lys930 of the α1 chain; Lys87 is almost fully glycosylated, while Lys930 is not (25, 26). The PEDF-CP211 structure clearly indicates that a bulky glycosyl moiety hampers complex formation due to steric clash and electrostatic repulsion if it is attached to hydroxylated Lys(M) of CP211 (SI Appendix, Fig. S11). These observations explain why PEDF selectively recognizes the C-terminal–specific portion of the type I collagen triple helix.
PEDF Binds to a Surface of Type I Collagen Masked by Collagen Crosslinking.
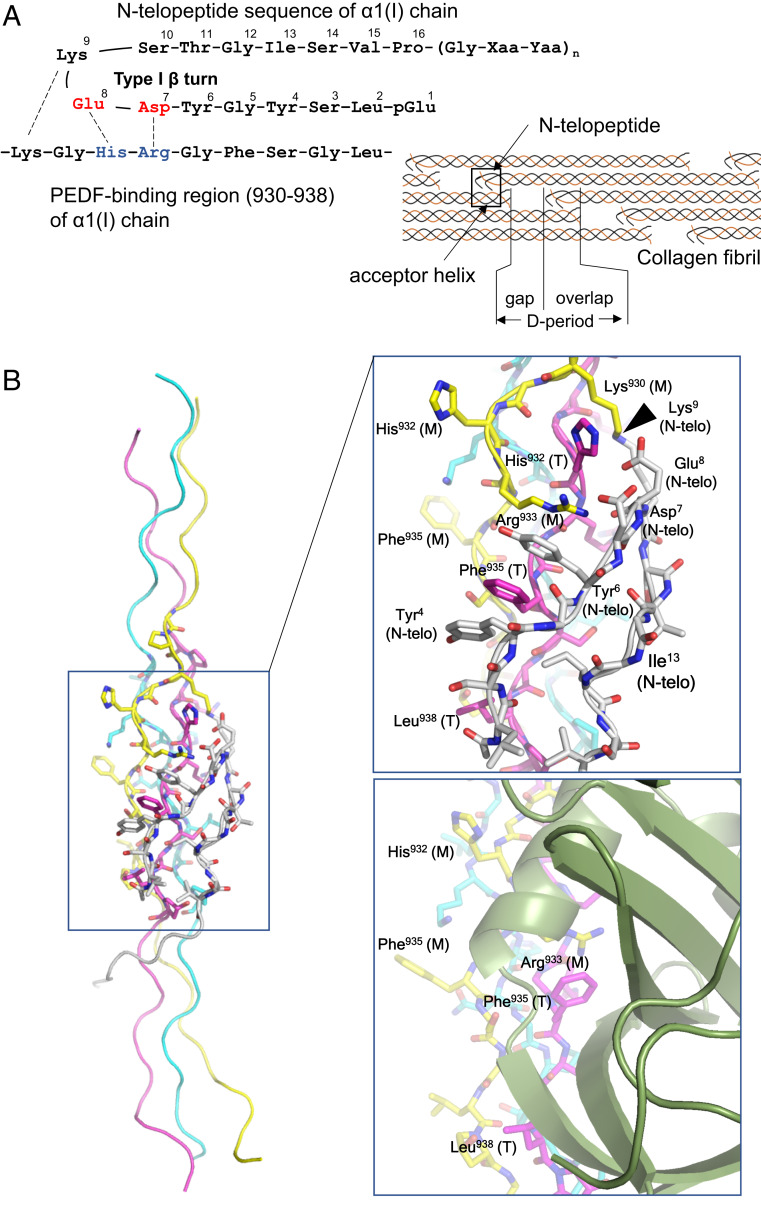
The PEDF-binding region of type I collagen that contains the crosslink-susceptible Lys930 is known to play an important role in reciprocal interactions with the nonhelical N-telopeptide segment of the α1 chain in D-periodic type I collagen fibrils (Fig. 3A) (25, 27). Previous NMR and modeling studies have indicated that the N-telopeptide segment in solution tends to form a type I β-turn structure, in which the hydrophobic pocket contains two tyrosyl and one leucyl residues (Leu2, Tyr4, and Tyr6) at one side and one isoleucyl and one valyl residues (Ile13 and Val15) at the opposite side that stabilize the turn structure, while polar residues Asp7, Glu8, and Lys9 are located in the middle of the turn and are exposed to the solvent (27–29). This amphipathic β-turn structure is suitable for interaction with the PEDF-binding region (hereinafter termed the acceptor helix), which has a similar amphipathic and complementary electrostatic surface feature (Fig. 3A) (27, 29).
Fig. 3.
Docking model of CP211 and the N-telopeptide segment of type I collagen. (A, Left) Schematic of interactions between the N-telopeptide and the PEDF-binding region of the α1 chain of type I collagen. (A, Right) Schematic of the axial arrangement of type I collagen molecules in the D-periodic fibril. The N-telopeptide segment and the PEDF-binding region (also termed the “acceptor helix”) are indicated by arrows. (B, Left) Overall structure of the CP211–N-telopeptide docking model. (B, Right, Top) Closeup view of the CP211–N-telopeptide interaction surface. The leading (L), middle (M), and trailing (T) chains in the triple helix are colored light blue, yellow, and magenta, respectively. The N-telopeptide segment is in white. Some amino acid residues of the guest region of CP211, located at the interface between CP211 and N-telopeptide, are labeled and numbered according to their positions in the type I collagen sequence. In addition to electrostatic interactions, the intercalation of Phe935 with two tyrosyl residues of the N-telopeptide, which is stabilized by aromatic ring stacking, is a hallmark of the interaction. The site of intermolecular crosslinking is indicated by an arrowhead. (Right, Bottom) Closeup view of the PEDF-CP211 interaction surface shown for comparison.
Our computational docking of the α1(I) N-telopeptide segment on the CP211 structure provides a plausible model for binding between the N-telopeptide and the acceptor helix, in which all the previously predicted hydrophobic and polar interactions are reasonably formed only at one particular surface, including the interchain Arg-Phe pair (Fig. 3B, Left and Right Top). Such docking would also bring Lys930 of the acceptor helix and Lys9 of the N-telopeptide in close proximity, thereby enabling crosslinking between them, along with the conversion of the Lys9 residue to an aldehyde by lysyl oxidase (LOX) (25). Intriguingly, this N-telopeptide–recognizing surface comprising middle-trailing chains of type I collagen corresponds to the surface recognized by PEDF (Fig. 3B, Right Bottom). Therefore, once covalent crosslinking is accomplished, PEDF is no longer able to interact with collagen fibrils. These results suggest that secreted PEDF favors newly synthesized, premature type I collagen fibrils rather than mature ones. This preference in collagen binding is consistent with recent atomic force microscopy experiments on the binding of PEDF to mature type I collagen fibrils by Cauble et al. (30), who reported that binding of exogeneous PEDF was not observed on or within the mature type I collagen fibrils, while it could bind to the reconstituted type I collagen fibrils with axial D-spacings. Given that the difference between these two fibrils is ascribed to the presence or absence of intermolecular crosslinks catalyzed by LOX, these observations are now consistent with our proposed mechanisms of PEDF binding to and liberation from type I collagen fibrils, in which the masking of the PEDF-binding site by intermolecular crosslinking is a hallmark of the regulation.
Comparison with Other Collagen-Binding Serpins.
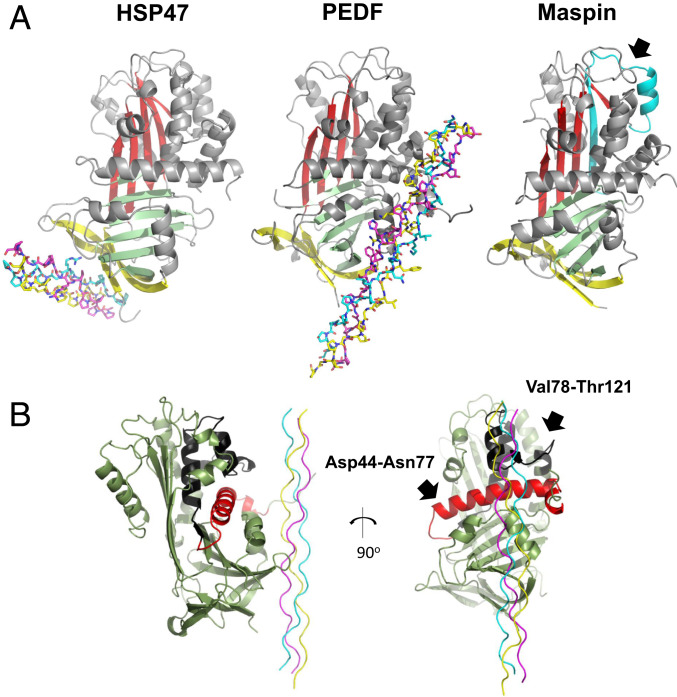
In addition to PEDF, other serpin family proteins, including heat shock protein 47 (HSP47) and maspin, are known to interact with type I collagen (31, 32). These proteins share the typical serpin fold composed of three β-sheets (A, B, and C) and nine α-helices. However, the collagen-binding regions of these serpins and their means of recognizing their target collagen sequences are significantly different; PEDF binds to the KGHRGFSGL sequence of collagen, with its amphipathic binding cleft located between hH and hG, while HSP47 recognizes an arginine at the Yaa position of an Gly-Xaa-Yaa triplet in the collagen triple helix via its β-sheet C (Fig. 4A and SI Appendix, Fig. S12).
Fig. 4.
Comparison of collagen-binding modes among collagen-binding serpins. (A) Crystal structure of the HSP47 complex with the homotrimeric collagen peptide Ac-(PPG)2-PRG-(PPG)2-NH2 (15-R8 CMP) (PDB ID code 4AU2) (Left), PEDF complex with collagen peptide CP211 (Middle) and maspin in an apo state (PDB ID code 1XU8) are shown. The leading (L), middle (M), and trailing (T) chains in the triple helix are colored in light blue, yellow, and magenta, respectively. Three β-sheets, A, B, and C, are shown in red, green, and yellow, respectively. α-helices and loop regions are shown in gray. The previously reported collagen-binding region (residues 84 to 112) of maspin is indicated by an arrow and shown in cyan. (B) Relative orientation of PEDF against CP211 in the complex. The previously proposed cell surface receptor-binding regions, a 34-aa segment and a 44-aa segment (5) that are completely masked by CP211 are colored red and black, respectively.
Although the complex structure of the maspin-collagen peptide has not been reported and its binding sequence on type I collagen remains unidentified, the collagen-binding region on maspin is reported to localize to residues 84 to 112, which are located opposite the collagen-binding region of HSP47 and PEDF (32, 33) (Fig. 4A). In addition, key residues critical for collagen recognition, such as Arg222 and Asp385 of HSP47 and Asp255, Asp257, and Asp299 of PEDF, are not highly conserved among these serpins (9, 31, 33) (SI Appendix, Fig. S12), suggesting that collagen-binding properties were acquired independently during evolution and are critical for biological function (34). Of note, the complex structure determined in this study demonstrates that two previously proposed receptor-binding regions of PEDF, 34-residue (Asp44–Asn77) and 44-residue (Val78–Thr121) segments (5), are completely masked on collagen binding (Fig. 4B), implying the role of type I collagen in modulating receptor binding of PEDF.
Discussion
We have elucidated the structural basis of type I collagen recognition by PEDF and demonstrated that the association of complementary amphiphilic molecular surfaces is highly sequence-specific. Using heterotrimeric peptides, we found that PEDF recognizes a characteristic epitope containing crosslink-susceptible Lys930 of a type I collagen α1 chain, which is masked during the process of collagen maturation and crosslinking. Since PEDF plays important regulatory roles in physiological homeostasis, its unique collagen-binding feature is highly relevant to its well-known spatiotemporal actions on specific cells, such as angiogenic tip cells embedded in or intruded into a type I collagen-rich interstitial matrix (35, 36). Such cells exist primarily under hypoxic conditions, have a characteristic shape with filopodia, and require active remodeling of the microenvironment through degradation, synthetization, and posttranslational modification of type I collagen by multiple enzymes, including lysyl hydroxylase and LOX (37, 38). These processes result in highly dense and linearized pericellular collagen fibrils at the leading edge of filopodia, where LOX is reportedly concentrated (39). The fact that cellular PEDF is down-regulated and degraded extracellularly by proteases under hypoxic conditions raises the question of how PEDF targets such cells (3). However, newly synthesized pericellular type I collagen fibrils strongly attract perfusing and/or exogenous PEDF molecules from distant cells under normoxic conditions and provide stabilization that prevents hypoxia-driven protease digestion of PEDF. This preference in collagen binding explains the observed localization of PEDF in remodeling tissues and organs (40, 41).
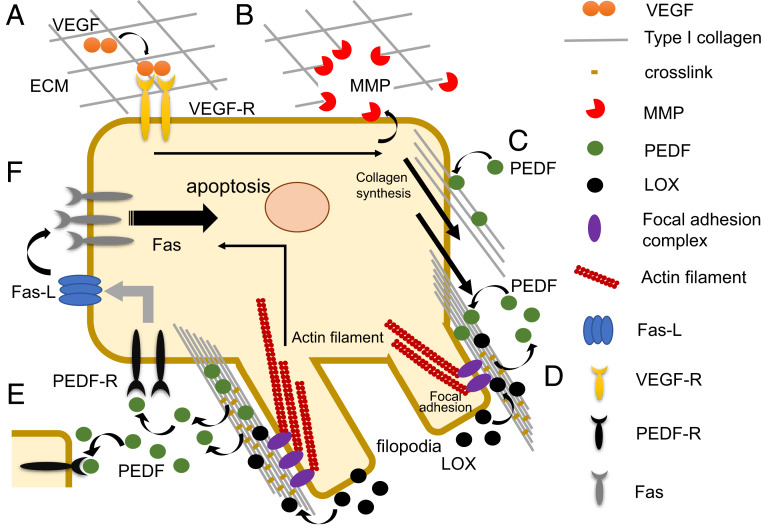
While collagen remodeling may help concentrate PEDF in the pericellular region, the complex structure determined in this study also indicates that PEDF activity on target receptors is initially restricted by the collagen triple helix, which is positioned on the two receptor-binding regions to hinder their interactions (Fig. 4B), implying a potential role for type I collagen as a rheostat regulating PEDF signaling. As crosslinking increases, the number of PEDF molecules, including those once sequestered and gradually liberated from collagen fibrils, also increases. These molecules interact with nearby target receptors until the fibrils are fully matured. This crosslink-coupled activation allows PEDF to operate as a unique functional switch during cell differentiation in a spatially and temporally regulated manner. One such example is found in its selective antiangiogenic activity during vascular remodeling, which may be important for maintaining a critical vessel density during physiological events, such as wound healing (36, 40). As illustrated in Fig. 5, hypoxia-driven vessel sprouting initiated by vascular endothelial growth factor (VEGF) signaling accompanies not only differentiation of endothelial cells (ECs) into a tip-cell phenotype, but also transiently enhances Fas death receptor surface expression, thereby sensitizing the cell to apoptosis (36).
Fig. 5.
Proposed mechanism of PEDF activation by type I collagen remodeling and maturation. On triggering by excessive growth factors such as VEGF induced under hypoxic conditions (A), cells (e.g., endothelial cells) up-regulate the expression of MMP as well as collagen syntheses to actively remodel their pericellular ECM (B and C). The newly synthesized and highly dense pericellular collagens initially attract perfusing and/or exogenous PEDF molecules produced under normoxic conditions (C), while elevated expression of LOX in response to hypoxia at that site enhances collagen crosslinking, thereby promoting filopodium formation via integrin clustering and focal adhesion formation. These processes liberate PEDF molecules once sequestered at the crosslinking sites of premature collagen fibrils and allow them to act on target receptors (PEDF-R) present on the cell or cells in close proximity for which Fas-L is up-regulated (D and E). Because VEGF signaling reportedly up-regulates surface expression of the Fas receptor at specific times, this crosslink-coupled action of PEDF spatiotemporally activates the Fas/FasL signaling cascade, resulting in selective apoptosis of target cells (F).
Given that PEDF is known to up-regulate Fas ligand (FasL) in multiple cell phenotypes (35, 36), the selective accumulation and crosslink-coupled action of PEDF likely promotes the coordinated up-regulation of FasL at cell tips or nearby sprouting vessels (Fig. 5). This spatial and temporal action is critical for determining EC cell fate, since a previous study has shown that when signals from VEGF achieve levels capable of counteracting signals generated by Fas–FasL interaction, the activity of PEDF is no longer apoptotic and instead promotes cell proliferation (6).
Ongoing bidirectional cell-ECM crosstalk, termed dynamic reciprocity, is important for directly or indirectly determining cell fate (42). Although the molecular mechanisms of how this orchestrates extracellular ligand-mediated cell signaling are just beginning to be elucidated (43), we propose a unique regulatory role for type I collagen remodeling in PEDF signaling, based on structural evidence showing that PEDF selectively recognizes the cryptic site of type I collagen, the availability of which is dynamically regulated by collagen maturation and crosslinking. This knowledge regarding the extracellular dynamics between the spatiotemporal system and the control signal network provides clues that will aid the design of therapeutic agents for diseases involving complicated mechanisms related to the pericellular matrix.
Materials and Methods
Reagents and Peptide Synthesis.
Amino acids, amino acid derivatives, resins, and S-3-nitro-2-pyridinesulfenyl chloride (NpySCl) were obtained from Novabiochem or Wako Pure Chemical Industries. 2,2ʹ-Dithiodipyridine (PySSPy) was obtained from Sigma-Aldrich. The three steric isomers CP211, CP121, and CP112, mimicking the PEDF-binding region of type I collagen, were synthesized according to the literature (44, 45). In brief, all peptide chains (i.e., the leading, middle, and trailing chains) were synthesized by 9-fluorenylmethoxycarbonyl–based solid-phase peptide synthesis. The leading and trailing chains were synthesized with an S-trityl (Trt) group for the thiol of cysteine. The S-Trt group was converted to thiol when the resin-linked peptide chains were deprotected and cleaved from the resin. The middle chain was synthesized with two different protecting groups for the thiols of cysteines, the S-Trt and S-acetamidomethyl (Acm) groups. The middle chain S-Trt group was converted to S-pyridinesulfenyl (Spy) by treatment with PySSPy during deprotection and cleavage from the resin. This S-Spy–protecting cysteine was reacted with the leading-chain thiol. The remaining S-Acm group was activated by S-3-nitro-2-pyridinesulfenyl chloride (NpyS) and reacted with the trailing-chain thiol. The resulting heterotrimer was purified by reverse-phase high-performance liquid chromatography (SI Appendix, Fig. S1). To determine the molecular masses of target compounds, the obtained peptides were subjected to electrospray ionization-time of flight-mass spectrometry (Bruker micrOTOF-2focus; Bruker Daltonics) (SI Appendix, Fig. S1).
The conformational study of these steric isomers was performed by circular dichroism (CD) spectroscopy (SI Appendix, Fig. S2). In brief, all CD spectra were recorded on a J-820 CD spectropolarimeter (JASCO). Peptides were dissolved in 50 mM Tris⋅HCl pH 7.5 and 100 mM NaCl at a 0.5 mg/mL concentration, and their ellipticity units per mole of amino acid residues were estimated at 4 °C. To assess the thermal stability of the triple helix, the thermal melting curve was monitored by observation of the [Θ]225 value with increasing temperature from 4 °C to 85 °C at 18 °C/h. The Tm value was estimated from the curve data as described previously (45).
Protein Expression and Purification.
Expression and purification of recombinant mouse PEDF were performed as described previously (18). In brief, Escherichia coli JM109 expressing the glutathione S-transferase (GST)-mature form of the mouse PEDF (Asn21–Thr417) fusion protein were lysed in PBS containing 1 M NaCl, 2 mM EDTA, 0.1 mg/mL lysozyme, and 0.2% (vol/vol) Triton X-100. After sonication and clearing by centrifugation, the lysate was loaded onto a glutathione Sepharose 4B column (GE Healthcare). To purify GST-PEDF, the fusion protein was eluted using 20 mM Tris⋅HCl pH 8.0, 1 M NaCl, 2 mM EDTA, and 10 mM glutathione. The GST tag of the fusion protein was cleaved by Factor Xa (GE Healthcare). The resulting protein was further purified with a Sephacryl S-200 HR size-exclusion column (GE Healthcare).
Analytical Ultracentrifugation.
SV-AUC measurements were performed with an Optima XL-I analytical ultracentrifuge (Beckman Coulter) equipped with a four-hole An60 Ti rotor at 20 °C using 12-mm double-sector charcoal-filled epon centerpieces with quartz windows. The mouse PEDF and each collagen peptide were dialyzed extensively against 50 mM Tris⋅HCl pH 7.5 or 100 mM NaCl. Aside from the apo PEDF, the mixtures of PEDF and each collagen peptide were prepared in multiple molar ratios (0.2, 0.5, 1, 3, and 5), all with a final concentration of 5 μM PEDF. Data were collected at 42,000 rpm using absorbance optics with a radial increment of 0.003 cm. The distribution of the sedimentation coefficient was analyzed using the c(s) method of the program SEDFIT (46). The positions of the meniscus and the cell bottom were floated during the fitting. The range of sedimentation coefficients for fitting was 0 to 15 S with a resolution of 300. The frictional ratio was initialized at 1.2 and floated during the fitting. The partial molar volume, buffer density, and viscosity were calculated using SEDNTERP (http://www.rasmb.org/sednterp). The molar ratio dependency of the weight-averaged sedimentation coefficient of PEDF-CP211 mixture was fitted with the A + B ↔ AB model using program SEDPHAT (47).
Isothermal Titration Calorimetry.
ITC experiments were performed with an iTC200 isothermal titration calorimeter (GE Healthcare). All samples were dissolved in a buffer containing 50 mM Tris⋅HCl pH 7.5 and 100 mM NaCl. For each experiment, PEDF solution (30 μM) was loaded into the cell, and sample solution containing one of the three heterotrimeric collagen peptides (CP211, CP121, or CP112) was loaded into the syringe (0.35 mM). All titrations were performed by sequentially titrating 1 µL of syringe solution for the first titration point and 2 µL for all subsequent titration points, at 120-s intervals at 20 °C. Thermograms were analyzed using Origin 7.0 software provided by the manufacturer.
Crystallization.
Crystals of the mouse PEDF-CP211 complex were grown by the cocrystallization method. The PEDF stock solution was diluted to 15 mg/mL, mixed with 3.3 mg/mL CP211, and dialyzed against 25 mM Tris⋅HCl pH 8.0 and 100 mM NaCl. Initial crystallization conditions were screened by the sitting-drop vapor diffusion method at 4 °C using Crystal Screen 1 and 2 (Hampton Research), Wizard Screen I and II (Emerald Biosystems), and JBScreen Classic 1 to 4 (Jena Bioscience). Crystallization drops were prepared by mixing 0.8 µL of protein solution with 0.8 µL of reservoir solution, then were equilibrated against 40 µL of reservoir solution. Preliminary needle-shaped crystals of PEDF-CP211 were grown using a reservoir solution of 100 mM CAPS pH 10.5, 1,200 mM sodium phosphate, 800 mM potassium phosphate, and 200 mM lithium sulfate at 4 °C. By seeding with these initial crystals, large crystals were obtained from a mixture of 1.0 µL of protein solution and 1.0 µL of reservoir solution consisting of 90 mM CAPS pH 10.5, 1,080 mM sodium phosphate, 720 mM potassium phosphate, and 180 mM lithium sulfate at 4 °C.
Data Collection, Structure Determination, and Refinement.
Diffraction data from the mouse PEDF-CP211 complex crystals were collected at SPring-8 on beamline BL26B2 (Hyogo) using a CCD MX225 detector (Rayonix) at −173 °C. The crystals were cryoprotected using a reservoir solution of 50% (vol/vol) glycerol and flash-cooled in a liquid nitrogen gas stream at −173 °C. The data from the PEDF-CP211 complex crystal were processed with the HKL2000 software package (48). The structure of the PEDF-CP211 complex was determined by the molecular replacement method using the PHASER program of the PHENIX suite (49). The human PEDF structure (Protein Data Bank [PDB] ID code 1IMV) was used as a search model. After several rounds of refinement using the phenix.refine program of the PHENIX suite (49), interpretable electron densities for CP211 were readily identified around the negatively charged surface previously identified as a collagen-binding region of PEDF. The quality of these densities allowed us to identify each chain position in the heterotrimeric collagen peptide structure without ambiguity (SI Appendix, Fig. S5). All the residues for the guest region were appropriately modeled, while the electron densities corresponding to both terminal regions of CP211 were not well defined, so that several of the terminal residues, including bridged Cys residues, could not be modeled. For PEDF, nearly all residues were successfully modeled, except the N terminus (residues 21 to 36) and a portion of the solvent-exposed reactive center loop (residues 372 to 382), due to the disorder of these regions of the crystal. The final model was built using Coot (50) and refined with phenix.refine (49). The geometries of the final models were analyzed with MolProbity (51). All data collection and refinement statistics are provided in SI Appendix, Table S1.
Computational Docking of the α1(I) N-Telopeptide Segment on CP211 Triple Helix.
To construct the initial model, the structure of bovine α1-α2-α1 collagen in the N-telopeptide docked model (27) was replaced with the structure of CP211 determined as described above. After manual adjustment of the side chain conformations to optimize surface complementarity using the molecular sculpting function of PyMOL (https://pymol.org/2/), the steric clashes in the initial structure were removed by energy minimization using GROMACS version 5.1.2 with the AMBER ff14SB force field (52, 53). The molecular topology was modified to involve the lysinonorleucine structure formed by crosslinking between lysine residues in the middle chain and the telopeptide. A simulation box, which has triclinic periodic boundary conditions around the CP211-N-telopeptide complex model with an edge distance of 1.0 nm, was filled with water molecules using the simple point charge model.
Computational Modeling of the Glycosylated CP211 Triple Helix.
The structure was modeled with the topology of the α-D-glucopyranosyl-(1→2)-β-D-galactopyranosyl (Glc-Gal) moiety, composed by the Glycam Carbohydrate Builder (http://glycam.org/). The Glc-Gal moiety was introduced on the δ-carbon atom of the lysyl residue at the middle chain of CP211. To remove close van der Waals contacts in the initial structure, energy minimization using GROMACS version 5.1.2 with the AMBER ff14SB and GLYCAM06 force field was performed (52, 53).
Supplementary Material
Acknowledgments
We thank K. Takagi and M. Uranagase for structural studies and helpful discussions. The synchrotron X-ray diffraction experiments were performed with the approval of the SPring-8 Proposal Review Committee (2014A1344, 2014B1234, 2015B2101, 2017A2589, and 2017B2589).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors of the PEDF-CP211 complex have been deposited in the worldwide Protein Data Bank, www.rcsb.org (PDB ID code 6LOS).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004034117/-/DCSupplemental.
References
- 1.Tombran-Tink J., Barnstable C. J., PEDF: A multifaceted neurotrophic factor. Nat. Rev. Neurosci. 4, 628–636 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Becerra S. P., Sagasti A., Spinella P., Notario V., Pigment epithelium-derived factor behaves like a noninhibitory serpin. Neurotrophic activity does not require the serpin reactive loop. J. Biol. Chem. 270, 25992–25999 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Dawson D. W., et al. , Pigment epithelium-derived factor: A potent inhibitor of angiogenesis. Science 285, 245–248 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Becerra S. P., Notario V., The effects of PEDF on cancer biology: Mechanisms of action and therapeutic potential. Nat. Rev. Cancer 13, 258–271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filleur S., et al. , Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Res. 65, 5144–5152 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Hutchings H., Maitre-Boube M., Tombran-Tink J., Plouët J., Pigment epithelium-derived factor exerts opposite effects on endothelial cells of different phenotypes. Biochem. Biophys. Res. Commun. 294, 764–769 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Apte R. S., Barreiro R. A., Duh E., Volpert O., Ferguson T. A., Stimulation of neovascularization by the anti-angiogenic factor PEDF. Invest. Ophthalmol. Vis. Sci. 45, 4491–4497 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Simonovic M., Gettins P. G., Volz K., Crystal structure of human PEDF, a potent anti-angiogenic and neurite growth-promoting factor. Proc. Natl. Acad. Sci. U.S.A. 98, 11131–11135 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasui N., et al. , Dual-site recognition of different extracellular matrix components by anti-angiogenic/neurotrophic serpin, PEDF. Biochemistry 42, 3160–3167 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Meyer C., Notari L., Becerra S. P., Mapping the type I collagen-binding site on pigment epithelium-derived factor. Implications for its antiangiogenic activity. J. Biol. Chem. 277, 45400–45407 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Alberdi E., Hyde C. C., Becerra S. P., Pigment epithelium-derived factor (PEDF) binds to glycosaminoglycans: Analysis of the binding site. Biochemistry 37, 10643–10652 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Alberdi E. M., Weldon J. E., Becerra S. P., Glycosaminoglycans in human retinoblastoma cells: Heparan sulfate, a modulator of the pigment epithelium-derived factor-receptor interactions. BMC Biochem. 4, 1–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becerra S. P., et al. , Pigment epithelium-derived factor binds to hyaluronan. Mapping of a hyaluronan binding site. J. Biol. Chem. 283, 33310–33320 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gettins P. G., Serpin structure, mechanism, and function. Chem. Rev. 102, 4751–4804 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Macri L., Silverstein D., Clark R. A., Growth factor binding to the pericellular matrix and its importance in tissue engineering. Adv. Drug Deliv. Rev. 59, 1366–1381 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Hosomichi J., Yasui N., Koide T., Soma K., Morita I., Involvement of the collagen I-binding motif in the anti-angiogenic activity of pigment epithelium-derived factor. Biochem. Biophys. Res. Commun. 335, 756–761 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Yasui N., Koide T., Collagen-protein interactions mapped by phototriggered thiol introduction. J. Am. Chem. Soc. 125, 15728–15729 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Sekiya A., Okano-Kosugi H., Yamazaki C. M., Koide T., Pigment epithelium-derived factor (PEDF) shares binding sites in collagen with heparin/heparan sulfate proteoglycans. J. Biol. Chem. 286, 26364–26374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bella J., Collagen structure: New tricks from a very old dog. Biochem. J. 473, 1001–1025 (2016). [DOI] [PubMed] [Google Scholar]
- 20.An B., Lin Y. S., Brodsky B., Collagen interactions: Drug design and delivery. Adv. Drug Deliv. Rev. 97, 69–84 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Piez K. A., Trus B. L., Sequence regularities and packing of collagen molecules. J. Mol. Biol. 122, 419–432 (1978). [DOI] [PubMed] [Google Scholar]
- 22.Orgel J. P., Irving T. C., Miller A., Wess T. J., Microfibrillar structure of type I collagen in situ. Proc. Natl. Acad. Sci. U.S.A. 103, 9001–9005 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brondijk T. H. C., Bihan D., Farndale R. W., Huizinga E. G., Implications for collagen I chain registry from the structure of the collagen von Willebrand factor A3 domain complex. Proc. Natl. Acad. Sci. U.S.A. 109, 5253–5258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jalan A. A., et al. , Chain alignment of collagen I deciphered using computationally designed heterotrimers. Nat. Chem. Biol. 16, 423–429 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamauchi M., Sricholpech M., Lysine post-translational modifications of collagen. Essays Biochem. 52, 113–133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terajima M., et al. , Glycosylation and cross-linking in bone type I collagen. J. Biol. Chem. 289, 22636–22647 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malone J. P., George A., Veis A., Type I collagen N-telopeptides adopt an ordered structure when docked to their helix receptor during fibrillogenesis. Proteins 54, 206–215 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Otter A., Kotovych G., Scott P. G., Solution conformation of the type I collagen alpha-1 chain N-telopeptide studied by 1H NMR spectroscopy. Biochemistry 28, 8003–8010 (1989). [DOI] [PubMed] [Google Scholar]
- 29.Helseth D. L. Jr, Lechner J. H., Veis A., Role of the amino-terminal extrahelical region of type I collagen in directing the 4D overlap in fibrillogenesis. Biopolymers 18, 3005–3014 (1979). [Google Scholar]
- 30.Cauble M., et al. , Microstructure-dependent binding of pigment epithelium- derived factor (PEDF) to type I collagen fibrils. J. Struct. Biol. 199, 132–139 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Widmer C., et al. , Molecular basis for the action of the collagen-specific chaperone Hsp47/SERPINH1 and its structure-specific client recognition. Proc. Natl. Acad. Sci. U.S.A. 109, 13243–13247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blacque O. E., Worrall D. M., Evidence for a direct interaction between the tumor suppressor serpin, maspin, and types I and III collagen. J. Biol. Chem. 277, 10783–10788 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Law R. H., et al. , The high-resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix. J. Biol. Chem. 280, 22356–22364 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Silverman G. A., et al. , The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 276, 33293–33296 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Zhang H., et al. , PEDF and 34-mer inhibit angiogenesis in the heart by inducing tip cells apoptosis via up-regulating PPAR-γ to increase surface FasL. Apoptosis 21, 60–68 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Volpert O. V., et al. , Inducer-stimulated Fas targets activated endothelium for destruction by anti-angiogenic thrombospondin-1 and pigment epithelium-derived factor. Nat. Med. 8, 349–357 (2002). [DOI] [PubMed] [Google Scholar]
- 37.De Smet F., Segura I., De Bock K., Hohensinner P. J., Carmeliet P., Mechanisms of vessel branching: Filopodia on endothelial tip cells lead the way. Arterioscler. Thromb. Vasc. Biol. 29, 639–649 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Kirkpatrick N. D., Andreou S., Hoying J. B., Utzinger U., Live imaging of collagen remodeling during angiogenesis. Am. J. Physiol. Heart Circ. Physiol. 292, H3198–H3206 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Erler J. T., et al. , Lysyl oxidase is essential for hypoxia-induced metastasis. Nature 440, 1222–1226 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Wietecha M. S., et al. , Pigment epithelium-derived factor as a multifunctional regulator of wound healing. Am. J. Physiol. Heart Circ. Physiol. 309, H812–H826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quan G. M., et al. , Localization of pigment epithelium-derived factor in growing mouse bone. Calcif. Tissue Int. 76, 146–153 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Bissell M. J., Hall H. G., Parry G., How does the extracellular matrix direct gene expression? J. Theor. Biol. 99, 31–68 (1982). [DOI] [PubMed] [Google Scholar]
- 43.Dong X., et al. , Force interacts with macromolecular structure in activation of TGF-β. Nature 542, 55–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ottl J., Moroder L., Disulfide-bridged heterotrimeric collagen peptides containing the collagenase cleavage site of type I collagen. Synthesis and conformational properties. J. Am. Chem. Soc. 121, 653–661 (1999). [Google Scholar]
- 45.Shah N. K., Ramshaw J. A., Kirkpatrick A., Shah C., Brodsky B., A host-guest set of triple-helical peptides: Stability of Gly-X-Y triplets containing common nonpolar residues. Biochemistry 35, 10262–10268 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Schuck P., Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dam J., Velikovsky C. A., Mariuzza R. A., Urbanke C., Schuck P., Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s). Biophys. J. 89, 619–634 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams C. J., et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abraham M. J., et al. , GROMACS: High-performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015). [Google Scholar]
- 53.Maier J. A., et al. , ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.