Significance
Cas12a is a type V CRISPR-Cas endonuclease that relies on a programmable guide RNA to bind specific DNA sequences, and like most CRISPR-Cas systems, it suffers from spurious off-target binding. Improving the specificity of Cas12a is an important step toward its broader use as a tool for biomedical research, but a fundamental understanding of the biophysical determinants of nonspecific Cas12a binding is still lacking. Here, we developed a massively parallel CRISPR interference assay to reveal the thermodynamic factors involved in Cas12a off-target binding. Since our approach uses a parameter-free thermodynamic model that is broadly applicable to any RNA-guided endonucleases, our work should enhance our understanding of on- and off-target binding when applied to other CRISPR-Cas systems.
Keywords: molecular biophysics, CRISPR, Escherichia coli, statistical mechanics, transcriptional regulation
Abstract
The versatility of CRISPR-Cas endonucleases as a tool for biomedical research has led to diverse applications in gene editing, programmable transcriptional control, and nucleic acid detection. Most CRISPR-Cas systems, however, suffer from off-target effects and unpredictable nonspecific binding that negatively impact their reliability and broader applicability. To better evaluate the impact of mismatches on DNA target recognition and binding, we develop a massively parallel CRISPR interference (CRISPRi) assay to measure the binding energy between tens of thousands of CRISPR RNA (crRNA) and target DNA sequences. By developing a general thermodynamic model of CRISPR-Cas binding dynamics, our results unravel a comprehensive map of the energetic landscape of nuclease-dead Cas12a (dCas12a) from Francisella novicida as it inspects and binds to its DNA target. Our results reveal concealed thermodynamic factors affecting dCas12a DNA binding, which should guide the design and optimization of crRNA that limits off-target effects, including the crucial role of an extended protospacer adjacent motif (PAM) sequence and the impact of the specific base composition of crRNA–DNA mismatches. Our generalizable approach should also provide a mechanistic understanding of target recognition and DNA binding when applied to other CRISPR-Cas systems.
CRISPR and its associated genes are part of an adaptive immunity system used to combat phage infections in bacteria and archaea (1). The system consists of two main components: a CRISPR array, which contains repetitive sequences called repeats and variable sequences called spacers, and Cas genes, which facilitate spacer acquisition and the destruction of foreign DNA and RNA. Mature CRISPR RNAs (crRNAs) derived from the CRISPR array can in turn program Cas nucleases to recognize and cleave DNA targets in which nucleic acid sequence is complementary with the guide portion of the crRNA and proximal to a PAM (protospacer adjacent motif) site. Due to their simple and programmable nature, the nucleases of class 2 CRISPR systems, particularly Cas9 (type II) and Cas12 (type V), have been the subject of intense research interest for the purposes of genome editing (2–4), programmable gene regulation utilizing a catalytically dead CRISPR nuclease (dCas) (5–7), and nucleic acid detection (8, 9).
While CRISPR has already revolutionized many areas of research from fundamental biomedical sciences to synthetic biology to disease diagnostics, a fundamental understanding of the underlying factors affecting CRISPR-Cas off-target binding is still lacking. This is especially important for the further development of nondestructive base editors based on nuclease-dead CRISPR-Cas proteins (10, 11) because off-target binding, which may not entirely correlate with DNA cleavage (12–14), needs to be reduced to a minimum level to prevent unintended base changes. While several in silico models (15–20) have been developed to predict the binding affinity of RNA guided CRISPR-Cas proteins using data from in vitro biochemical assays (21–24) or in vivo indel frequencies (12–14, 25–27), these approaches only provide empirical interpretations of CRISPR-Cas DNA binding and often fail to yield a conceptual understanding of the underlying factors involved in CRISPR-Cas binding. Furthermore, it can be difficult to extract quantitative binding affinity measurements from in vivo indel frequencies due to the inherent CRISPR-Cas binding inefficiencies associated with cellular physiological factors such as cell type, chromatin state, and delivery method (28–30). Thus, there is a critical need for fundamental models that can help unravel the sequence-dependent determinants of CRISPR-Cas target recognition and DNA binding affinity.
To elucidate determinants of CRISPR-Cas12a off-target binding, we combine a thermodynamic model of Cas12a binding with rationally designed CRISPR interference (CRISPRi) assays to map the binding energy landscape of a type V CRISPR-Cas system from Francisella novicida (F. novicida Cas12a [FnCas12a]) as it searches for its DNA target. Our approach, inspired by biophysical models of CRISPR-Cas cleavage activity (31–33) and recently developed massively parallel multiplexed assays (34–37), aims to directly measure the energetic and thermodynamic determinants of CRISPR-Cas binding. In other words, our assays exclude sources of variation in DNA cleavage activity caused by unknown physiological factors (28–30) by only focusing on the steps leading to final DNA cleavage step. Furthermore, our predictive framework is not limited to FnCas12a and can be applied to any other CRISPR-Cas systems, which should in turn facilitate the development of predictive models of target recognition and binding efficiency for type II (Cas9) and type V (Cas12) RNA-guided CRISPR-Cas proteins.
Results
Thermodynamic Model of Nuclease-Dead Cas12a (dCas12a) Binding.
DNA cleavage by CRISPR-Cas endonucleases may be hindered by factors (28–30) other than the specific crRNA–DNA sequence, and it is important to disentangle these effects to gain a deeper understanding of off-target binding mechanisms. We thus hypothesize that the variability in indel formation observed in live cells may not entirely originate from differences in Cas12a’s cleavage activity, which is caused by the specific crRNA–DNA sequence targeted, but also from sequence-dependent PAM attachment efficiencies and the existence of crRNA–target DNA mismatches. We directly investigate this hypothesis by asking whether the steps leading to a ternary complex formation play a role in CRISPR-Cas off-target binding kinetics.
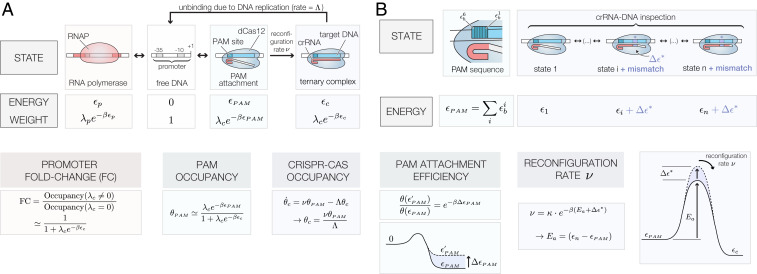
To formalize this approach and to obtain a deeper understanding of the energetic landscape of Cas12a as it inspects and associates with its DNA target, we developed a general model of CRISPR-Cas binding dynamics to determine how crRNA–DNA mismatches affect target recognition and binding. This model (Fig. 1 and SI Appendix) is based on recent structural biology and single-molecule studies (38, 39) which revealed that DNA hydrolysis by Cas12a occurs in three discrete stages: “PAM attachment,” where Cas12a latches onto a PAM site; “crRNA–DNA inspection,” where Cas12a forms a partial crRNA–DNA hybrid; and “reconfiguration,” where the protein forms a ternary complex and undergoes a conformal change that exposes its catalytic residues. While the final DNA cleaving step occurs after approximately 1 min under the conditions tested in ref. 38, Cas12a molecules with inactivated nuclease sites remain stably bound to their DNA target for more than 500 s. Hence, the reconfiguration step effectively has no detectable off rate, suggesting that DNA cleavage may be inevitable (given enough time) after Cas12a has reached this stably bound ternary state. The same stability has also been observed in single-molecule Cas9 experiments (37).
Fig. 1.
Thermodynamic model of dCas12a binding. (A) Thermodynamic states, energies, and Boltzmann weights of a dCas12a (, where = Boltzmann constant, T = temperature) for the PAM attachment, crRNA–DNA inspection, and reconfiguration steps. The fold change (FC), PAM occupancy , and CRISPR-Cas occupancy depend on the effective PAM energy , the CRISPR-Cas binding energy , and the DNA replication rate . All expressions assume that binding occurs in the weak promoter () and weak PAM binding () limits, where is the fugacity of a dCas12a molecule with chemical potential . (B) Internal base-dependent energies define a PAM-specific binding energy . In this model, the PAM-specific binding energy between two targets with energy that differs by scales their relative PAM attachment efficiency by . The presence of crRNA–target DNA mismatches increases the effective activation energy by a mismatch energy and scales the effective reconfiguration rate by a multiplicative factor equal to .
We first describe the probability that a Cas12a molecule loaded with a crRNA sequence will bind to a free, unobstructed target DNA sequence. Specifically, we use an approach based on thermodynamic models of transcriptional control (40–42) and transition-state theory (SI Appendix has details) to derive an expression for the CRISPR-Cas occupancy , which is defined as the fraction of time that a DNA target will be occupied by dCas12a endonuclease. In addition, since DNA replication forks seem to be the only process that can kick nuclease-dead Streptococcus pyogenes Cas9 (SpCas9) off of its DNA binding site (43), we assume that dCas12a unbinding occurs through a similar process (i.e., DNA duplication machinery kicks off dCas12a at a rate equal to , the cell’s duplication rate). Using these assumptions, is given by
| [1] |
where is the PAM occupancy and is the rate at which dCas12a will form a stable ternary complex after it encounters a PAM site (the reconfiguration rate).
We next compare occupancies of targets that vary by a few base determinants (Fig. 1B). In this framework, the propensity of a given crRNA to target to bind to an off-target DNA region compared with its intended target is simply given by the different energetic contributions of that specific off-target location. For instance, two identical DNA targets that possess different PAM sequences have effective binding energies that differ by , which in turn, translates into a reduction of the attachment probability by a factor equal to (the Boltzmann factor). Similarly, the presence of mismatches may alter the crRNA–DNA duplex energy by , which in turn, also yields a change in relative binding probabilities. Hence, the relative binding affinity between two targets that have different PAM sites or between an intended target and an off-target candidate is simply given by the binding sites’ Boltzmann weight
| [2] |
Our framework shares similarities with the uCRISPR model recently developed by Zhang et al. (33) that employs a unified energetic analysis to predict SpCas9 cleavage activity. However, instead of testing our model using in vivo indel measurements performed in human cells [which can be imprecise due to cellular physiological factors (28–30)], we use a massively parallel CRISPRi assay to directly measure the sequence-specific PAM binding energies and the energetic costs associated with crRNA–DNA mismatches in Escherichia coli bacteria.
Context Dependence of FnCas12a CRISPR Interference.
In order to test our thermodynamic model and further explore how dCas12a binds to its DNA target in E. coli, we developed a highly compact 175-bp-long genetic inverter inserted into a low-copy number plasmid (pSC101) containing a catalytically dead nuclease FnCas12a (Fig. 2A). The inverter element consists of a constitutive promoter driving the transcription of a crRNA followed by two rho-independent terminators. Located immediately downstream of two terminators is the output promoter, which contains a built-in PAM site followed by a DNA target within the promoter or after the promoter’s +1 location.
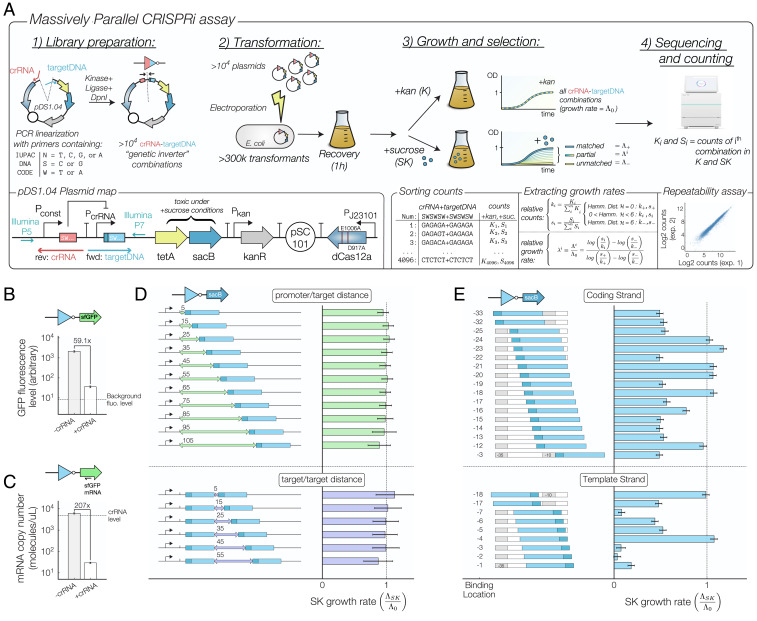
Fig. 2.
(A) Experimental workflow. More than different crRNA–target DNA combinations are assembled in parallel using PCR primers containing degenerate International Union of Pure and Applied Chemistry (IUPAC) DNA codes and cloned into electrocompetent cells (>300,000 transformants). Plasmid map: Each construct consists of an inverter element based on crRNA–target DNA binding and each inverter represses an output promoter driving a tetA-sacB cassette (tetA = tetracycline resistance protein, sacB = Bacillus subtilis levansucrase). dCas12a binding efficiency is assessed by comparing growth rates under control (K) and sucrose (SK) conditions. The relative growth under SK conditions is rescaled with respect to fully matched combinations (i.e., crRNA–target DNA sequences with Hamming distance = 0, which grow at a rate = growth rate under K conditions) and mismatched combinations ( = 6, which grow at a rate = 0). While the library construction is prone to some biases during the amplification and sequencing steps, a high level of repeatability is observed between experiments that started with the same assembled library (Lower Right). (B) Fluorescence measurement of a genetic inverter with matched crRNA–target DNA sequences shows a 59.1-fold decrease in superfolder GFP (sfGFP) protein level. (C) Digital PCR measurements of sfGFP mRNA level reveal a 207-fold decrease in mRNA expression in the presence of a matching crRNA sequence. A large concentration of crRNA (4,890 molecules per microliter of cell extract) also ensures that crRNAs are present at saturating conditions. (D) Systematic analysis of the growth rate under sucrose conditions when one (Upper) or two (Lower) DNA targets are located after the +1 promoter location. (E) Impact of dCas12a binding on the SK growth rate when the DNA target overlaps with the −35 and −10 regions of promoter. Error bars are calculated using an LOESS fit (44, 45) of the mean/variance relationship between experimental replicates of the fold change.
We first sought to investigate effectiveness of dCas12a-mediated CRISPRi by measuring protein and messenger RNA (mRNA) levels of a simple inverter driving superfolder GFP (sfGFP) expression. The inverter constitutively expresses a crRNA targeting a DNA binding region located at the promoter’s −19 position. Fig. 2B shows that fluorescence levels for constructs containing fully matched crRNA–target DNA combinations (Hamming distance = 0) were 59.1 times lower than those with a mismatched crRNA–target DNA combination ( = 6). Additionally, mRNA transcript levels measured using digital droplet PCR resulted in a 207-fold reduction in mRNA transcript levels when a matching crRNA is expressed (Fig. 2C). Both of these results confirm that FnCas12a can repress RNA transcription (7) with a surprisingly high CRISPR-Cas occupancy equal to 99.5%.
Next, we tested how dCas12a interferes with RNA transcription under various configurations (Fig. 2 D and E) by inserting a library of several thousand simple inverter constructs in front of a tetA-sacB cassette. Since sacB is counterselectable in the presence of sucrose (46) (SI Appendix, Fig. S2), the genetic inverters that efficiently repress RNA transcription will be enriched in the population when grown under sucrose conditions (SK). Thus, we can evaluate the ability of an RNA-guided FnCas12a to prevent transcription by comparing the number of times that each construct is present in the whole population for control (K) and SK conditions using the MiSeq or iSeq 100 platform from Illumina. The relative change in the population fraction is then used to find the effective growth rate of every construct in each condition. While selection experiments are also performed under tetracycline-selective (TK) media, the counterselection experiment (SK) yields more useful information because binding affinity and FnCas12a occupancy are directly related to each construct’s growth rate (SI Appendix has a complete description of this method).
Fig. 2 D, Upper and SI Appendix, Fig. S3 show that CRISPRi occurs efficiently when the dCas12a target is located after the output promoter’s +1 transcription initiation site because the growth rate under SK conditions is close to its maximum value () regardless of the location of the DNA binding site. Interestingly, while interference measurements performed using SpCas9 revealed that a second binding site results in suppressive combinatorial effects that multiplicatively increase CRISPRi efficiency (5), the existence of a second PAM + target DNA sequence does not improve CRISPRi efficiency beyond what is achieved by a single target (Fig. 2 D, Lower and SI Appendix, Fig. S3).
Next, we tested dCas12a’ s ability to interfere with RNA transcription initiation by introducing a PAM + target DNA sequence within the promoter sequence. In particular, we tested several inverter constructs with PAM + target DNA sequence that was located at different positions within the promoter’s −35 and −1 locations, testing both the coding and template strands without altering conserved promoter regions (Fig. 2E). Our results show that CRISPRi through promoter occlusion is efficient for most targets on both the coding and template strands, although the effective repression rate is more variable than what has been reported for CRISPR-Cas9 interference (6). Growth under SK conditions is also lowest when the target DNA is located on the promoter’s template strand at locations −1, −2, −3, and −7 with respect to the transcription initiation site, which suggests that RNA:DNA hybrids on the nontemplate strand display a decreased effectiveness in preventing RNA transcription initiation.
dCas12a Binding Energies Depend on an Extended PAM Sequence.
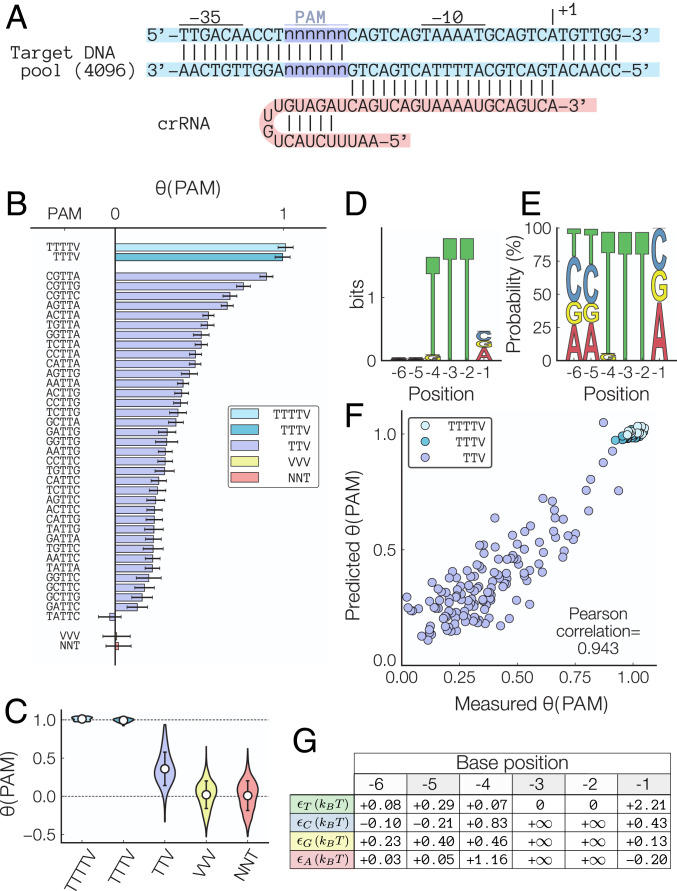
Having demonstrated the validity of our massively parallel CRISPRi assay to test multiple genetic inverter combinations, we next investigated the impact of a PAM sequence on the dCas12a binding. We first tested the sequence determinant of the PAM attachment step using an oligo pool containing a degenerate 5’-NNNNNN-3’ motif for a target DNA sequence located at the promoter’s −19 position (Fig. 3A) targeted by a single crRNA (target DNA sequence = CAGTCAGTAAAATGCAGTCA). Since previous work has shown that the PAM motif required for FnCas12a DNA cleavage is TTV (3), we nevertheless tested all sequences containing up to six bases of upstream context using 4,096 PAM site variants in a single experiment. These extra bases turn out to be very important: Fig. 3B shows that, while TTV is a suitable PAM site, its attachment efficiency is lower than an extended TTTV PAM site (Fig. 3B). In both individual and aggregate measurements, we observe that DNA binding to a DNA target proximal to a TTTV PAM site is 2.8 times more efficient than a TTV PAM site (Fig. 3C). This result is also confirmed by the bias toward TTTV PAM sites in the information content (Fig 3D) and the base-specific probability density in SK conditions (Fig. 3E).
Fig. 3.
PAM attachment efficiencies. (A) Sequence of the PAM site occupancy library. (B) Measured PAM occupancies for all six-base PAM sites normalized by the PAM occupancy of the (T)TTTV consensus sequence. V = C, G, or A. Error bars = aggregated LOESS fit of the mean/variance relationship between experimental replicates of the fold change. (C) Aggregated PAM site occupancies display a preference for the (T)TTTV consensus sequence while still allowing binding to occur for TTV sequences. Error bars = SD. (D) Bit content and (E) probability density of the SK-selected PAM site libraries show a strong preference for NNNTTV PAM sites. N = T, C, G, or A. (F) Predicted using the base-dependent binding energy expression shows a strong agreement with the measured occupancies (Pearson correlation = 0.943). (G) Fitted values for position- and base-dependent binding energies in units of for PAM sites of the form TTV ( = Boltzmann constant, T = temperature). Details about the model and an expression for the binding energies for (T)TTTV PAM sites are included in SI Appendix. Note that this model only considers PAM sequences of the form NNNTTN by constraining .
Our results agree with recent work (47), which demonstrated that FnCas12a does exhibit activity in mammalian cells but only when used with a TTTV PAM site. It is important to note that, while Zetsche et al. (3) showed that a TTV PAM site seems to be sufficient to induce FnCas12a cleavage, it seems to be the least efficient motif that permits DNA binding (which could explain why FnCas12a was found to be ineffectual for mammalian cell editing using a TTV PAM site). Hence, our results suggest that PAM sites with an extended TTTV sequence should be prioritized when seeking potential FnCas12a DNA targets for CRISPRi, gene editing, nucleic acid detection, or other applications.
Expanding on this result, we next used the measured attachment efficiencies to develop a predictive model that takes into account the full six-base PAM site context to predict the attachment efficiency. Specifically, our thermodynamics model predicts that the effective PAM site attachment energy is additive, meaning that the PAM binding energy of an arbitrary sequence is given by , where is the specific binding energy of a base of type = (T,C,G,A) at location = (1…6). In this case, the relative PAM binding energy between two targets () is related to the relative growth rate under SK condition according to .
We developed a predictive model of PAM attachment efficiency by first using an initial set of values for each extracted from the PAM-specific growth rates and optimizing the model for 1,000 additional steps to minimize the measured–predicted mean square error (SI Appendix has details). Our model is able to accurately describe the variability in PAM attachment efficiencies observed in Fig. 3B, and its predictions for the relative PAM site occupancies agree with the measured attachment efficiencies (Fig. 3F) (Pearson correlation = 0.943). These results suggest that PAM attachment is well described by our thermodynamic model, and the optimized energetic contribution of each base located at position is shown in Fig. 3G. Hence, to ensure that the DNA target with the most efficient PAM site is selected when designing and optimizing a crRNA sequence for DNA binding or other gene editing application, the relative performance of each PAM sequence should be evaluated on a sequence-specific manner using the base-dependent binding energies provided in Fig. 3G.
Off-Target dCas12a Binding Depends Additively on Mismatch Energy.
To better understand the impact of crRNA–DNA mismatches on dCas12a binding, we next examined how a mismatch affects the effective activation energy (Fig. 1B) that is required for dCas12a to form a stable ternary complex. Indeed, even though a PAM site is present and dCas12a attaches itself to DNA, the additional energy associated with a crRNA–DNA mismatch can prevent DNA unzipping if insufficient homology is found. According to our model, the reconfiguration step occurs at a rate , where is the base-dependent energy cost associated with a single mismatch at location . Thus, the location-specific energy costs associated with individual mismatches could be directly obtained by measuring the reconfiguration rate of crRNA–DNA sequences that possess the same PAM sequence but with a crRNA that differs from the target DNA by one or more bases.
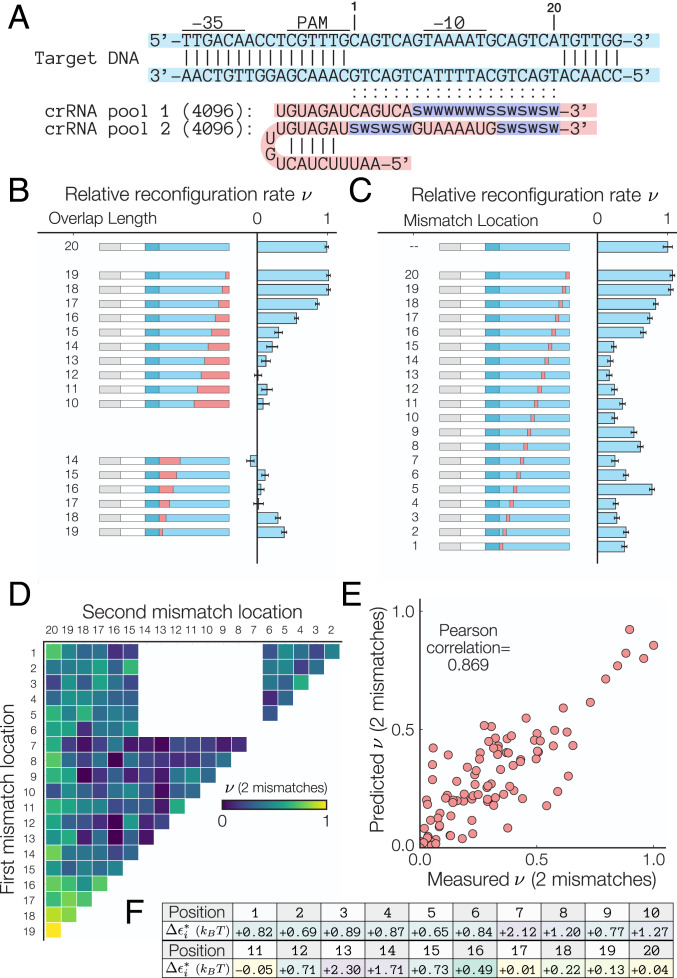
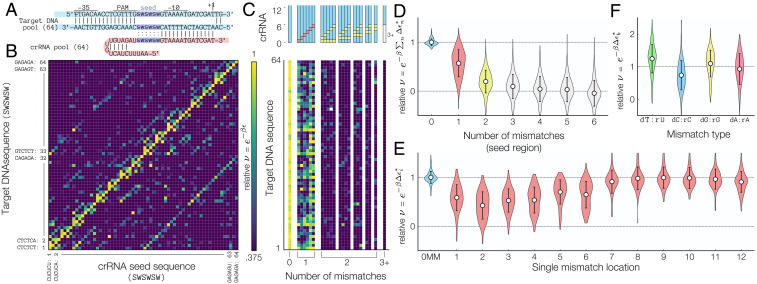
To test this, we used two different crRNA pools (Fig. 4A) to measure the mismatch-dependent reconfiguration rate . Each oligo pool consists of 4,096 different primer sequences generated by specifying degenerate DNA codes in the primer sequence (e.g., W = A or T, S = G or C), allowing us to test multiple mismatch combinations in a single experiment. Using the degenerate DNA codes S and W ensures that all crRNA sequences maintained the same GC content. In Fig. 4B, we tested the impact of “truncated” crRNAs (i.e., a crRNA with distal sequence that is noncomplementary to its target DNA) and “gapped” crRNAs (i.e., a crRNA with sequence that is noncomplementary to its target DNA for bases 1 to 6). Consistent with other work performed in Cas12a (26, 27), our results show that optimal reconfiguration rates occur for truncated crRNAs that possess more than 15 bases of homology. Furthermore, no significant binding was detected for gapped crRNAs with sequences that contain more than two mismatches.
Fig. 4.
Mismatch binding efficiencies. (A) Sequence of the single-target mismatch libraries. Reconfiguration rates (B) for truncated and gapped crRNAs and (C) for crRNA–target DNA sequences with a single mismatch normalized by the fully matched ( = 0) crRNA–target DNA combination suggest that binding in the presence of mismatches is highly dependent on the location and total number of crRNA–target DNA mismatches. Error bars = LOESS fit of the mean/variance relationship between experimental replicates of the fold change. (D) Experimental measured normalized reconfiguration rates for crRNA with two mismatches. (E) Predicted two-mismatch using the mismatch-specific reconfiguration rate is in strong agreement with the measured (Pearson correlation = 0.869). Details about the model are included in SI Appendix. (F) Fitted model values for the location-dependent binding energies for single mismatches in units of ( = Boltzmann constant, T = temperature). Note that the binding energy depends nonmonotonically on the distance from the PAM site.
Next, we measured the reconfiguration rate for crRNA containing a single mismatch (Fig. 4C). The presence of a single mismatch can decrease the configuration rate by up to 82% when the mismatch occurs in the first 17 bases of the crRNA. Consistent with prior observations by Kim et al. (19), the energy cost of a single mismatch does not increase monotonically with distance from the PAM site, suggesting that other contextual determinants other than position affect the reconfiguration rate . Furthermore, the presence of mismatches located in the last three bases of the crRNA does not impede DNA binding, confirming other work performed using in vivo indel measurements (26, 27), which demonstrated that crRNA–DNA mismatches negatively impact dCas12a binding but only in the seed (bases 1 to 6) and the beginning of the distal region (bases 7 to 12).
Next, we analyzed how the presence of two mismatches impacts the reconfiguration rate. Since the energetic contributions of single mismatches are additive in our model, we anticipated that the two-mismatch reconfiguration rate is related to the single mismatch energies according to . To test this, we developed a predictive model that uses the single-base mismatch energies to predict . Fig. 4D shows the experimentally measured, location-dependent reconfiguration rate . Using an approach similar to the one used to predict PAM attachment efficiencies, we derived baseline values for the location-dependent binding energy. While the initial Pearson correlation between the predicted and baseline energy values was initially fairly low (P = 0.769), the predicted values for the two-mismatch reconfiguration rate are in agreement with the measured rates after the 1,000 optimization steps (P = 0.869) (Fig. 4E). Our results confirm that the energetic impacts of individual mismatches are additive, and location-dependent binding energy costs reported in Fig. 4F should be incorporated into models that aim to predict off-target binding.
High-Throughput Cross-Talk Assays Reveal Position- and Nucleotide-Specific Energy Costs.
We next asked how both crRNA and DNA variations in the first six bases of the PAM-proximal seed region affected the reconfiguration rate . We performed multiplexed CRISPRi assays using two oligo pools, each containing 128 different sequences, to test the pairing between all possible crRNA–DNA sequences of the form SWSWSW or WSWSWS in a single step (S = C or G, W = T or A). Once again, those pairings were chosen to maintain all crRNA–DNA sequences at a fixed GC content. This approach covers a large combinatorial space between the spacer–target sequences and produces a comprehensive cross-talk map between 16,384 possible crRNA–DNA combinations (Fig. 5A). While we also performed the same analysis on the crRNA distal region (bases 7 to 12) (SI Appendix, Fig. S6), only the SW quadrant of the seed region is shown in Fig. 5B (SI Appendix, Figs. S4–S6 show the full cross-talk maps).
Fig. 5.
Massively parallel determination of crRNA–target DNA binding. (A) Sequence of the multiplexed mismatch assays for Cas12’s seed region (bases 1 to 6). (B) Cross-talk map of the reconfiguration rate for (SWSWSW) (SWSWSW) data subset normalized by the average of all crRNA–target DNA combinations (full dataset is shown in SI Appendix, Fig. S4). Note that the off-diagonal elements represent a crRNA–DNA target that differs by a single mismatch ( = 1). A complete Hamming distance map is shown in SI Appendix, Fig. S5. (C) Target DNA-dependent normalized reconfiguration rate for SWSWSW sequences containing zero, one, two, or three or more mismatches. (D) Aggregated for crRNA–target DNA combinations containing between one and six mismatches shows that a stable ternary complex may still occur in the presence of up to three mismatches in the seed region. (E) Aggregated position-dependent for crRNA–target DNA combinations containing a single mismatch shows that dCas12a can tolerate single mismatches. (F) The reconfiguration rate for base-specific mismatches shows that dT:rU and dG:rG mismatches are tolerated at a higher level than dC:rC and dA:rA mismatches, revealing nucleotide-specific mismatch energy costs for dCas12a binding. Error bars = SD.
The cross-talk maps show that fully matching crRNA–DNA sequences (i.e., those along the main diagonal of Fig. 5B and in the first column in Fig. 5C) have the highest . Interestingly, the relative reconfiguration rate for all fully matched crRNA–DNA targets falls within a very narrow range of 1.00 0.06 (mean SD), suggesting that the specific base composition of the seed region does not have a large impact on DNA binding. This contrasts with in vivo multiplexed DNA cleavage assays for Cas12a variants that do show significant sequence dependence on cleavage activity (15, 19, 20). In addition, while SpCas9 binding and cleavage activity have different sequence specificities (12–14), we do not observe significant discrepancies between the binding and cleavage assays performed using catalytically active FnCas12a nuclease (SI Appendix, Fig. S7). Hence, our approach may provide a more accurate representation of dCas12a’s binding energy landscape because our approach excludes any source of variation caused by unknown cellular physiological factors by only investigating a small but comprehensive portion of all possible crRNA–target DNA sequences that possess the same GC content.
To further understand how a single mismatch affects the reconfiguration rate, we considered how varies as a function of the number and location of mismatches present. We first show in Fig. 5D that no significant binding was observed for sequences containing more than four mismatches in the seed region. Our analysis, however, reveals that formation of a stable ternary complex does occur in the presence of one, two, or three mismatches (P = 1 × , 8 × , and 1 × , respectively; null hypothesis = no binding will occur for one, two, or three mismatches). It is important to note that, by performing aggregate measurement across thousands of crRNA and DNA sequences, our results confer a much stronger statistical predictive power than other assays that only test a limited number of crRNA–DNA partners. In addition, we also show in Fig. 5E that mismatches have the greatest impact when located within the first six bases of the seed region. Sensitivity to a mismatch decreases with distance from the PAM site, and mismatches located in the distal region (bases 7 to 12) only minimally impact DNA binding.
We next considered whether the type of mismatch affects in Fig. 5F. Surprisingly, we find that single crRNA–DNA mismatches of the form dC:rC decrease by an additional 26% on average. In contrast, dT:rU and dG:rG mismatches are tolerated and increase the reconfiguration rate by 24 and 9.5%, respectively, compared with all types of single-base mismatch. This effect can be visualized in Fig. 5B, where off-diagonal elements that correspond to a single mismatch in the sixth location are more prominent in the lower right quadrant than those in the upper left quadrant (the upper left quadrant corresponds to a dC:rC mismatch, while the lower right quadrant corresponds to dG:rG mismatches). Insensitivity to wobble-transition mismatches has been previously reported in SpCas9 (21, 48) and Cas12a from Acidaminococcus sp. BV3L6 (AsCas12a) (19), but other work in AsCas12a found no significant effect due to a transversion mismatch (19), suggesting that tolerance to transversion mismatches may be unique to FnCas12a.
Discussion
We have established that massively parallel CRISPRi assays with their ability to rapidly measure thousands of different crRNA–target DNA variants in parallel are a viable method to assess dCas12a binding efficiencies. Our results reveal the fundamental relationship between crRNA–DNA interactions and the underlying energy landscape that dictates binding behavior of dCas12a. One major outcome of this study is that binding of DNA by CRISPR-Cas12a endonuclease does not strongly depend on the specific crRNA sequence used (at least within the set of tested sequences, which were kept at 50% GC content). Rather, variance in DNA binding affinities depends on the PAM sequence, the presence of mismatches, and the type of mismatch present. Indeed, the propensity of identical DNA targets to be recognized by a CRISPR-Cas nuclease matching crRNA may be significantly different depending on their respective PAM sequence. Similarly, the absolute number of mismatches in the seed region of a crRNA–DNA hybrid is more important than their specific location, and mismatches that occur after base 17 do not significantly affect binding affinity. Our results also show that dT:rU and dG:rG mismatches are tolerated to a greater degree than dA:rA and dC:rC mismatches.
Beyond that, the power of our approach also resides in our ability to use a parameter-free statistical mechanics framework to extract thermodynamic determinants of dCas12a binding. Importantly, our results are not specific to nuclease-dead CRISPR-Cas endonucleases—we confirm in SI Appendix, Fig. S7 that the same behavior is observed for catalytically active Cas12a nuclease—and our approach should foster the development of predictive, parameter-free biophysical models of on- and off-target binding affinities and DNA cleavage activities. In addition, because CRISPR-Cas systems are very common among prokaryotes (1), there is a need for the rapid and efficient characterization of newly sequenced CRISPR-Cas systems that may display enhanced target differentiation capabilities or alternative PAM site compositions. We anticipate that this method will also provide a mechanistic understanding of the thermodynamic determinants of DNA target recognition and binding affinities in uncharacterized CRISPR-Cas endonucleases and other nucleic acid binding enzymes.
Because our method is applicable to both the catalytically active and dead versions of the nuclease, it should also lead to improvements in a vast range of CRISPR applications, including in vivo gene editing, programmable repression, and nucleic acid detection. Our multiplexed approach is particularly applicable to the advancement of dCas-based gene circuit elements, which can be used to create complex circuits that behave orthogonally, operating independently without cross-talk (49–53). Furthermore, our approach can expedite the rational design of enhanced CRISPR nucleases and facilitate the development of CRISPR-Cas variants with greater specificity, improved proofreading capabilities, or increased activities (54–59).
Materials and Methods
Assembly of the CRISPR-Cas12a Plasmid Backbone.
Unless indicated otherwise, all experiments were conducted using a plasmid backbone, which constitutively expresses dCas12a (F. novicida) and tetA-sacB. This plasmid was assembled using standard Gibson assembly techniques from components sourced from several other plasmids (pY003 [pFnCpf1_delta Cas] was a gift from Feng Zhang, Broad Institute of Harvard and MIT, Cambridge, MA [Addgene plasmid 69974], pTKLP-tetA was a gift from Thomas Kuhlman, University of Illinois at Urbana–Champaign, Urbana, IL [Addgene plasmid 71325], and pKM154 was a gift from Kenan Murphy, University of Massachusetts Medical School, Worcester, MA [Addgene plasmid 13036]) using a backbone derived from pUA66 (60). FnCas12a was made to be catalytically inactive via two mutations, D917A and E1006A, performed using New England Biolabs (NEB)’s Q5 site-directed mutagenesis kit. The landing pad sequence needed for Illumina sequencing was inserted using an IDT gBlock gene fragment (Dataset S1). The entire plasmid sequence (pDS1.04) can be found in Dataset S1.
Design of PAM and Guide RNA (gRNA) Mismatch Assays.
In order to test the effects of PAM and gRNA mismatches at a large scale, we created a highly compact dCas12a repressing element such that target and gRNA properties could be changed with a single site-directed mutagenesis. The sequence of this compact element can be found in Dataset S1.
Assembly of Plasmid Libraries.
Our method of exploring CRISPRi is predicated on the use of large, randomized oligos in order to produce many mismatch combinations via site-directed mutagenesis. Oligos for PCR-based assembly of different guide:target variants were purchased from Thermo Fisher; oligos containing randomized bases were polyacrylamide gel electrophoresis (PAGE) purified, and all others were ordered as desalted oligo plates. Oligonucleotide sequences are listed in Dataset S1. PAGE-purified oligos were ordered phosphorylated by the manufacturer. Unphosphorylated oligos from plates were pooled together (according to their forward–reverse directions), and phosphate groups were added using Thermo Fisher’s T4 Polynucleotide Kinase. Pooled or randomized phosphorylated oligos were used to insert multiple crRNA and target DNA combinations in a single PCR step. Likely due to the large size of the insertion, we had a significant amount of difficulty finding parameters that resulted in complete PCR products. Parameters that worked were found serendipitously and include a high molar ratio of template to primers and extremely long (15-min+) extension times. PCR was done exclusively using Q5 hot start DNA polymerase from NEB. For cloning of single constructs, ligation and phosphorylation were accomplished using the Kinase + Ligase + DpnI mix from NEB’s site-directed mutagenesis kit. In the multiplexed experiments (except when noted below), ligation was accomplished using NEB’s ElectroLigase using 100 ng of DNA from the PCR purified using Zymo’s ZymoPURE Miniprep kit. Ligation was done according to the manufacturer’s instructions, with a 60-min incubation time at 25○C and a 15-min inactivation step at 65○C. Ligated product was either used immediately for transformation or frozen for future use. The catalytically active Cas12a experiment was cloned using a library derived from the kanamycin-selected control in the catalytically dead experiment since this was of known good coverage for all mismatch combinations. D917A and E1006A mutations in dCas12a in pDS1.04 were reverted using site-directed mutagenesis, and the catalytically restored Cas12a was inserted into the linearized backbone with all 4,096 variants in lieu of the catalytically dead CRISPR via assembly with NEB Hifi DNA assembly Master Mix. Insertions for the promoter–target and target–target spacing experiments were done using two rounds of PCR: the first one to add a functioning inverter element and the second one to add one or two PAM + target DNA sequences. Primer sequences are listed in Dataset S1.
Electroporation of Plasmid Libraries.
In order to achieve the transformation efficiencies required for good statistical coverage of all mismatch combinations in our multiplexed experiments, we used electroporation of our CRISPR mismatch libraries; 1 L of electroligated product was added to 25 L Lucigen Endura ElectroCompetent cells and then, electroporated at 1,400 V (BTX ECM399 Device). Cells were recovered in 2 mL of Lucigen recovery media as in ref. 61. Following the 1-h recovery, the full 2 mL were transferred to 23 mL of Terrific Broth (TB) with kanamycin in a 50-mL tube. TB was made by autoclaving 23.8 g of VWR’s TB powder with 2 mL of glycerol and 500 mL of purified water. Since the Endura cells are so densely packed, the resulting recovery product has a nonzero optical density (OD) of roughly 0.3. After the tubes reached an OD of 1.0 (approximately 8 h at 37 ○C, 225 rpm), each pair of tubes was combined in a flask, and 1 mL of that product was used to inoculate each of the selection conditions.
Sucrose and Tetracycline Selection.
Inoculated selection media (100 mL) were grown in 250-mL flasks (37 ○C, 225 rpm) until they reached an OD of 1.0 and then, cooled to 4○C prior to plasmid extraction. Unselective media (the control condition) are TB with kanamycin (50 g/mL). Tetracycline-selective media (TK; indicating both kanamycin and tetracycline) were produced in the same way, adding tetracycline at a concentration of 10 g/mL. Sucrose-selective media (SK) was produced by combining 10 mL of an autoclaved sucrose premix solution (22.5 g sucrose in 37.5 mL water) with a TB premix solution such that the resulting solution contains 4.5% (wt/vol) sucrose. Plasmid extraction was done using Zymo’s ZymoPURE II Midiprep kit according to the manufacturer’s instructions. Plasmids were then eluted in elution buffer and stored at −20○C prior to indexing for next generation sequencing. While Li et al. (46) utilize dual sensitivity to both sucrose and fusaric acid, we found no selective advantage due to the use of fusaric acid and did not utilize it beyond preliminary experiments.
Next Generation Sequencing and Analysis.
Our method is made possible by the inclusion of sequences flanking the inverter site of interest (the pDS1.04 sequence) to which Illumina indexing primers can bind. This allows us to lift out purely the sequences of interest using PCR, skipping most traditional library preparation steps. Indexes were added to our samples using primers from NEB’s NEBNext Multiplex Oligos for Illumina (Index Primers Set 1) using NEBNext Q5 Hot Start HiFi PCR Master Mix or NEBNext Ultra II Q5 Master Mix. Sequencing was performed either using Illumina’s MiSeq System from the Cornell Genomics Facility (150-bp kit, paired ends 2 75 bp) or an Illumina iSeq instrument in our own laboratory (2 150-bp run). Due to the extremely low complexity of these libraries, a 10% PhiX spike in was used in both cases. Results were analyzed using scripts written in Python, which can be made available on request. Only reads that perfectly matched the correct design in the sequencing window were counted in the final result to calculate the relative fraction of each construct in the sequenced populations.
Fluorescence Measurements of Protein Fold Change.
In initial fluorescence measurement experiments, plasmids containing dCas12a, guide RNA sequence, and a green fluorescent protein (GFP) target were transformed into NEB’s 5-alpha Competent E. coli (high efficiency) and recovered in SOC according to the manufacturer’s instructions. Initial assessment of repression efficacy was made by visual inspection of cells grown on Luria–Bertani (LB) plates. The sequences of these plasmids can be found in Dataset S1.
Quantitative measurements of fluorescence (used to produce Fig. 2B) were made using a Synergy H1 Hybrid Multi-Mode Microplate Reader produced by BioTek. Reported fold change corresponds to asymptotic fold change observed after roughly 5 h of growth in 200 L TB at 37 °C. GFP fluorescence measurements are corrected by subtracting out the measured green emittance from cells at the same OD, which entirely lack GFP.
Droplet Digital PCR Measurement of mRNA Fold Change.
mRNA fold change was measured using droplet digital PCR measurements. Transformed cells were grown in 20 mL TB for 12 h at 37 °C, and 300 L were then used for RNA extraction using Zymo’s Direct-zol RNA MiniPrep. Genomic DNA was removed using Thermo’s TURBO DNA-free Kit, and 10 ng of cleaned RNA was then used as a template for complementary DNA (cDNA) production utilizing the ProtoScript II Reverse Transcriptase kit from NEB and primer RT_GFP_Rev from Dataset S1. Droplet generation was done using a QX200 Droplet Generator produced by Bio-Rad. PCR amplification was done using a C1000 Touch Thermal Cycler (Bio-Rad) utilizing EvaGreen Supermix and following the manufacturer’s instructions. Primers corresponding to the GFP target are listed in Dataset S1. Results are read out on a Qx200 Droplet Reader. Data analysis from droplet digital PCR was completed using QuantaSoft software made available by the Cornell Genomics Center.
Measurement of Cell Growth as a Function of Sucrose and Tetracycline Concentration.
The Synergy H1 microplate reader was used to produce growth curves for cell growth in the presence of sucrose and tetracycline. Cells with sacB (pDS1.04) were tested with varying concentrations of sucrose, and cells lacking tetA were tested against varying concentrations of tetracycline. Cells were grown in 200 L TB at 37 °C. Growth rates reported in SI Appendix, Fig. S2 are the result of a logistic curve fit to the optical density measurement fixed such that each curve has a constant starting OD.
PAM Site Sequence Logo.
Since sequencing coverage was in excess of 100× for most sequences, all sequences were still detected under SK conditions, including those that had a growth rate. Hence, to generate the sequence logo and final base density in Fig. 3 D and E that were not tainted with those sequences, simulated counts were used instead of the measured counts . These simulated counts were computed from K condition counts according to , where T = 17.5/, an arbitrary growth time, and is the growth rate of each PAM sequence (SI Appendix). Then, sequence logos were computed from , where is the relative frequency of base at position and .
Statistical Analysis and CI Evaluation.
While it is prohibitive to replicate next generation sequencing experiments, there are independent replicates within a single experiment with different selection conditions from which we can extract a variance as a function of the number of counts from next generation sequencing. Specifically, independent replicates are sourced from the K and TK selection conditions with six mismatches in the seed region, for which we expect there to be no effective repression by dCas12a. We utilize the transformation previously used by other authors for RNA sequencing counts (45). This transformation is then fit using locally estimated scatterplot smoothing (LOESS) (44) via its python implementation (62). Variance falls with the log of the number of counts (as would be expected from Poisson statistics) but then asymptotes for large counts.
Data Availability.
The raw fastq files from sequencing are available at SRA accession no. PRJNA549693, and data analysis scripts are available at https://github.com/lambert-lab/Massively-parallel-dCas12a-assays.
Supplementary Material
Acknowledgments
We thank members of the laboratory of G.L. for providing feedback on the manuscript. We thank Feng Zhang, Thomas Kuhlman, and Kenan Murphy for providing us with plasmids containing the Cpf1/FnCas12a, tetA, and sacB coding sequences as well as James Sethna for helpful discussions. This work was supported by NIH Grant 1R35 GM133759 Maximizing Investigators’ Research Award and the Alfred P. Sloan Foundation. Sequencing was performed by the Biotechnology Resource Center (BRC)’s Genomics Facility at Cornell University’s Institute of Biotechnology.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.B.K. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in Sequence Read Archive database (SRA accession no. PRJNA549693) and data analysis scripts are available in GitHub at https://github.com/lambert-lab/Massively-parallel-dCas12a-assays.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918685117/-/DCSupplemental.
References
- 1.Mohanraju P., et al. , Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353, aad5147 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R., Doudna J. A., Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Zetsche B., et al. , Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zetsche B., et al. , Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi L. S., et al. , Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bikard D., et al. , Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S. K., et al. , Efficient transcriptional gene repression by type V-A CRISPR-cpf1 from Eubacterium eligens. ACS Synth. Biol. 6, 1273–1282 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Chen J., et al. , CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gootenberg J. S., et al. , Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komor A. C., Kim Y. B., Packer M. S., Zuris J. A., Liu D. R., Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaudelli N. M., et al. , Programmable base editing of AT to GC in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuscu C., Arslan S., Singh R., Thorpe J., Adli M., Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 32, 677–683 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Wu X., et al. , Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32, 670–676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Geen H., Henry I. M., Bhakta M. S., Meckler J. F., Segal D. J., A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 43, 3389–3404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doench J. G., et al. , Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haeussler M., et al. , Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tycko J., Myer V., Hsu P., Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol. Cell 63, 355–370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H. K., et al. , In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods 14, 153–159 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Kim H. K., et al. , Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat. Biotechnol. 36, 239–241 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A., DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szczelkun M. D., et al. , Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. U.S.A. 111, 9798–9803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu B. X., Hansen L. L., Artiles K. L., Nonet M. L., Fire A. Z., Landscape of target:guide homology effects on Cas9-mediated cleavage. Nucleic Acids Res. 42, 13778–13787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh D., et al. , Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc. Natl. Acad. Sci. U.S.A. 115, 5444–5449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan J., et al. , Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res. 24, 1009–1012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleinstiver B. P., et al. , Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34, 869–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D., et al. , Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Singh R., Kuscu C., Quinlan A., Qi Y., Adli M., Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 43, e118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H., et al. , Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25, 1147–1157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong N., Liu W., Wang X., WU-CRISPR: Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16, 218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farasat I., Salis H. M., A biophysical model of CRISPR/Cas9 activity for rational design of genome editing and gene regulation. PLoS Comput. Biol. 12, e1004724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein M., Eslami-Mossallam B., Arroyo D. G., Depken M., Hybridization kinetics explains CRISPR-Cas off-targeting rules. Cell Rep. 22, 1413–1423 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Zhang D., Hurst T., Duan D., Chen S. J., Unified energetics analysis unravels SpCas9 cleavage activity for optimal gRNA design. Proc. Natl. Acad. Sci. U.S.A. 116, 8693–8698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T., et al. , Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat. Commun. 9, 2475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo J., et al. , Improved sgRNA design in bacteria via genome-wide activity profiling. Nucleic Acids Res. 46, 7052–7069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall R., et al. , Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol. Cell 69, 146–157.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyle E. A., et al. , High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc. Natl. Acad. Sci. U.S.A. 114, 5461–5466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeon Y., et al. , Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat. Commun. 9, 2777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stella S., et al. , Conformational activation promotes CRISPR-Cas12a catalysis and resetting of the endonuclease activity. Cell 175, 1856–1871.e21 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Brewster R., et al. , The transcription factor titration effect dictates level of gene expression. Cell 156, 1312–1323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinert F. M., Brewster R. C., Rydenfelt M., Phillips R., Kegel W. K., Scaling of gene expression with transcription-factor fugacity. Phys. Rev. Lett. 113, 258101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landman J., Brewster R. C., Weinert F. M., Phillips R., Kegel W. K., Self-consistent theory of transcriptional control in complex regulatory architectures. PLoS One 12, e0179235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones D. L., et al. , Kinetics of dCas9 target search in Escherichia coli. Science 357, 1420–1424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cleveland W. S., Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 74, 829–836 (1979). [Google Scholar]
- 45.Law C. W., Chen Y., Shi W., Smyth G. K., voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X., Thomason L. C., Sawitzke J. A., Costantino N., Court D. L., Positive and negative selection using the tetA-sacB cassette: Recombineering and P1 transduction in Escherichia coli. Nucleic Acids Res. 41, e204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tóth E., et al. , Mb- and FnCpf1 nucleases are active in mammalian cells: Activities and PAM preferences of four wild-type Cpf1 nucleases and of their altered PAM specificity variants. Nucleic Acids Res. 46, 10272–10285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai S. Q., et al. , GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jusiak B., Cleto S., Perez-Piñera P., Lu T. K., Engineering synthetic gene circuits in living cells with CRISPR technology. Trends Biotechnol. 34, 535–547 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Didovyk A., Borek B., Tsimring L., Hasty J., Transcriptional regulation with CRISPR-Cas9: Principles, advances, and applications. Curr. Opin. Biotechnol. 40, 177–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Didovyk A., Borek B., Hasty J., Tsimring L., Orthogonal modular gene repression in Escherichia coli using engineered CRISPR/Cas9. ACS Synth. Biol. 5, 81–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen A. A. K., Voigt C. A., Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 10, 763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cress B. F., et al. , Rapid generation of CRISPR/dCas9-regulated, orthogonally repressible hybrid T7-lac promoters for modular, tuneable control of metabolic pathway fluxes in Escherichia coli. Nucleic Acids Res. 44, 4472–4485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J. S., et al. , Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slaymaker I. M., et al. , Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleinstiver B. P., et al. , High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Casini A., et al. , A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 36, 265–271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu J. H., et al. , Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kleinstiver B. P., et al. , Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaslaver A., et al. , A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3, 623–628 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Shalem O., et al. , Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cappellari M., et al. , The ATLAS3d project XX. Mass-size and mass- distributions of early-type galaxies: Bulge fraction drives kinematics, mass-to-light ratio, molecular gas fraction and stellar initial mass function. Mon. Not. R. Astron. Soc. 432, 1862–1893 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw fastq files from sequencing are available at SRA accession no. PRJNA549693, and data analysis scripts are available at https://github.com/lambert-lab/Massively-parallel-dCas12a-assays.