Significance
DNA recombinases Dmc1 and Rad51 are both required during meiosis in most eukaryotes. Although they share high identity in amino acid sequence and biochemical properties, the mechanism of the requirement is unclear. The presynaptic filament formation is the first committed step in the process of homologous recombination. In this study, we combined the ensemble-based and the single-molecule experiments to dissect the assembly mechanism of Dmc1. Our results show that Dmc1 recombinases possess evolutionarily conserved nucleation preference that allows 5′-to-3′ filament formation. Moreover, Rad51 patches on single-stranded DNA stimulate the assembly of Dmc1, demonstrating the ability of Rad51 to facilitate the filament formation of Dmc1 in meiosis. The results provide a molecular rationale of why both recombinases are required in meiosis.
Keywords: Dmc1, recombinase filament assembly, Rad51, nucleation
Abstract
Dmc1 recombinases are essential to homologous recombination in meiosis. Here, we studied the kinetics of the nucleoprotein filament assembly of Saccharomyces cerevisiae Dmc1 using single-molecule tethered particle motion experiments and in vitro biochemical assay. ScDmc1 nucleoprotein filaments are less stable than the ScRad51 ones because of the kinetically much reduced nucleation step. The lower nucleation rate of ScDmc1 results from its lower single-stranded DNA (ssDNA) affinity, compared to that of ScRad51. Surprisingly, ScDmc1 nucleates mostly on the DNA structure containing the single-stranded and duplex DNA junction with the allowed extension in the 5′-to-3′ polarity, while ScRad51 nucleation depends strongly on ssDNA lengths. This nucleation preference is also conserved for mammalian RAD51 and DMC1. In addition, ScDmc1 nucleation can be stimulated by short ScRad51 patches, but not by EcRecA ones. Pull-down experiments also confirm the physical interactions of ScDmc1 with ScRad51 in solution, but not with EcRecA. Our results are consistent with a model that Dmc1 nucleation can be facilitated by a structural component (such as DNA junction and protein–protein interaction) and DNA polarity. They provide direct evidence of how Rad51 is required for meiotic recombination and highlight a regulation strategy in Dmc1 nucleoprotein filament assembly.
Homologous recombination (HR) is indispensable for maintaining genome integrity and producing genetic diversity. To initiate HR, recombinase assembles on single-stranded DNA (ssDNA) often generated by the DNA end resection process from double-strand break (DSB) sites to form nucleoprotein filament. The nucleoprotein filament then engages the duplex DNA template for homology search. Once homology is located, recombinase-driven DNA pairing forms a displacement loop (D-loop) for the subsequent strand-exchange process. Thus, the assembly of nucleoprotein filament is the first committed step in the recombination pathway and has been subject to tight regulation (1, 2).
In most eukaryotic cells, two recombinases, Rad51 and Dmc1, are responsible for the HR process. These two recombinases share ∼45% amino acid identity (3–5) and have some similar biochemical properties. For example, both Rad51 and Dmc1 bind three nucleotides (nt) of ssDNA per promoter to form a right-handed helical filament in an adenosine triphosphate (ATP)-dependent manner, and both stabilize strand-exchange intermediates in three-nucleotide steps (6–10).
In addition to the similarities between Dmc1 and Rad51, differences exist. Dmc1 is present only in meiosis while Rad51 is expressed in both meiotic and mitotic cells (2, 8). Dmc1 is shown to have a better tolerance in mismatch during meiotic recombination (6, 11–14). Also, during meiosis, the enzymatic activity of Rad51 is inhibited by Hed1 (15–17), but Rad51 is suggested to function as an accessory factor for Dmc1-mediated D-loop formation (18). Early cytological studies showed that Rad51 and Dmc1 foci are adjacent but partially offset, leading to a model that Rad51 and Dmc1 form separated filaments on two ends of the meiotic breaks during the nucleoprotein filament assembly (19, 20). Interestingly, recent superresolution imaging shows that yeast Rad51 and Dmc1 filaments bind to the same DNA end in vivo (21). In vitro biochemical studies also confirm that Rad51 and Dmc1 spontaneously form segregated homotypic filaments (22). Rad51 is known to be required for Dmc1 assembly (23, 24); however, how Dmc1 forms a homotypic filament with Rad51 and how this process takes place remains mostly uncharacterized.
In this work, we studied the nucleoprotein filament assembly of purified Saccharomyces cerevisiae Dmc1, using both ensemble-based assays and real-time single-molecule experiments. Direct side-by-side characterization of ScDmc1 and ScRad51 recombinases showed clear kinetic differences in their nucleoprotein filament assembly. ScDmc1 has much reduced ssDNA affinity, and its nucleation is much slower compared to ScRad51. Surprisingly, we found that the slow Dmc1 nucleation can be stimulated by Rad51. This provides a model of how Rad51 can participate and stimulate Dmc1 nucleoprotein filament assembly and likely reflects the role of Rad51 in meiosis.
Results
Nucleoprotein Filaments of ScDmc1 Are Less Stable Than Those of ScRad51.
The recombinases Rad51 and Dmc1 assemble on ssDNA to form nucleoprotein filaments during the first step of homologous recombination (25–27). As nucleoprotein filament assembly is the first committed step and orchestrates the following repair process, the rate and stability of filaments dictate the recombination efficiency. Here, we directly compared, side-by-side, the stability of nucleoprotein filaments of budding yeast S. cerevisiae Rad51 and Dmc1 by measuring how nucleoprotein filaments are protected against nuclease degradation. When recombinases bind to the ssDNA, the assembled recombinase-ssDNA complex is protected from nucleolytic degradation (Fig. 1A). Rad51 and Dmc1 were incubated separately with 80-nt ssDNA substrates (3-μM nucleotides) for 5 min to form nucleoprotein filaments in the presence of ATP. Benzonase, an endonuclease, was used to challenge the recombinase nucleoprotein filaments. Approximately 2-μM Rad51 is sufficient to form stable nucleoprotein filaments without degradation product upon benzonase digestion (Fig. 1B). In contrast, 6-μM Dmc1 is required for full protection (Fig. 1C). With 3-μM nucleotides used, the stoichiometric 1-μM of Rad51 leads to 51 ± 5.4% of filament assembly, but the same concentration of Dmc1 results only in 10 ± 1.6% of assembly (Fig. 1D). This shows the lower ssDNA affinity of Dmc1, compared to Rad51.
Fig. 1.
ScDmc1 nucleoprotein filament is less stable than ScRad51. (A) Schematic of the endonuclease protection assay. The 5′-32P–labeled 80-mer ssDNA (3-μM nucleotides) was incubated with yeast ScRad51 or ScDmc1 for 5 min at 37 °C in the presence of 1 mM ATP before being challenged by benzonase endonuclease. (B and C) ScRad51 (B) or ScDmc1 (C) nucleoprotein filament formation at different concentrations is monitored by 10% polyacrylamide gel. The 32P-label is denoted by the asterisk. (D) The quantification of B and C was graphed for a direct comparison. The error bars represent the SD calculated based on at least three independent experiments.
Nucleation Step of ScDmc1 Occurs More Slowly Than That of ScRad51.
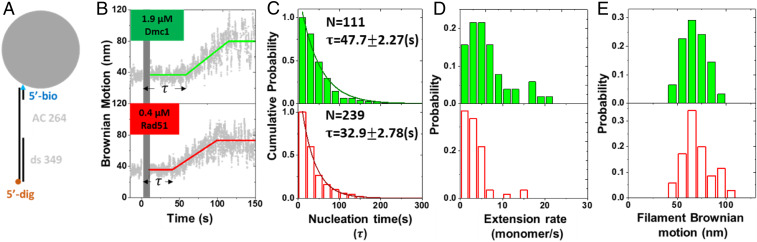
During the nucleoprotein assembly process, nucleation has been shown to be the rate-limiting step (28–30). Here, we used the previously developed single-molecule tethered particle motion (smTPM) experiments to determine the nucleation rate, extension rate, and assembled filament length on gap ssDNA substrates in real time (Fig. 2) (28, 29, 31, 32). When recombinases assemble on DNA, the increase in DNA contour length leads to the apparent DNA tether length increase (32, 33), as reflected by the increase in bead Brownian motion (BM) amplitude. Therefore, the real-time measurement of BM directly monitors the kinetics of nucleoprotein filament assembly process and has been used to study RecA and Rad51 recombinases (28, 32, 33). We used 349/264 DNA substrates, containing a 264-nt-long, secondary-structure–free, AC-only ssDNA region, a 349-bp-long double-stranded DNA handle, and a bead-labeled oligo to directly monitor Rad51 or Dmc1 nucleoprotein filament assembly (SI Appendix, Fig. S1). Under the reaction condition and timescale used here, recombinases nucleate and assemble on the ssDNA region of this DNA substrate (SI Appendix, Fig. S2). The typical assembly time courses of Rad51 and Dmc1 are shown in Fig. 2B, and they are similar to those of the filament assembly of Escherichia coli RecA (29) and mouse RAD51 (28). The similar nucleation times (“τ” in Fig. 2C)—the dwell time between the protein addition and apparent continuous BM increase of ScRad51 and ScDmc1—result from a striking difference in recombinase concentration used. The amount of 1.9 μM of ScDmc1 leads to the nucleation time of 47.7 ± 2.27 s, but only 0.4 μM of ScRad51 can nucleate in 32.9 ± 2.78 s (Fig. 2C). As nucleation time depends on recombinase concentration, this reflects the much slower nucleation rate of ScDmc1. In fact, attempts to use the same concentration of both recombinases make it experimentally not feasible. We also determined the extension rate and assembled filament length based on the BM time courses (29, 32, 33). The extension rate (Fig. 2D) and assembled filament length (Fig. 2E) between Rad51 and Dmc1 are not statistically significant in difference. Control experiments using 349-bp fully double-strand DNA (dsDNA) substrates returned with less than 5% of tethers with BM increase (SI Appendix, Fig. S2), confirming that BM increase events result from the interaction of recombinases and ssDNA, and the differences in nucleation times are due to the lower ssDNA affinity of ScDmc1.
Fig. 2.
Dmc1 shows slower assembly kinetics than Rad51. (A) The 349/264 DNA substrate used in single-molecule nucleoprotein filament assembly experiments includes a duplex handle, a secondary-structure–free ssDNA (AC264) and a bead-labeled oligo. (B) Exemplary assembly time courses of ScDmc1 (1.9 μM, Top, green) and ScRad51 (0.4 μM, Bottom, red) nucleoprotein filament assembly. Recombinases were introduced at time 0 with the gray shaded bar representing the experimental dead time (<10 s) due to buffer exchange. It takes time before a BM change occurs (nucleation time, double-headed arrow), followed by a continuous BM increase (extension) to reach an assembled nucleoprotein filament. Note that nearly fivefold higher ScDmc1 concentration is required for the similar nucleation time observed for ScRad51. (C) Cumulative histograms of nucleation time (τ) for ScDmc1 and ScRad51 are fitted to single exponential decays. The nucleation times are 47.7 ± 2.27 s for 1.9-μM Dmc1 (n = 111) and 32.9 ± 2.78 s for 0.4-μM Rad51 (n = 239). The assembly process takes place in the presence of 1 mM ATP. The error bars of nucleation time are the SD by bootstrapping 5,000 times. (D) Histograms of the filament extension rate shown in monomer/s. Similar extension rates were observed. (E) Histograms of extended BM value, indicative of the nucleoprotein filament length at the end of the first assembly event (collected when the first stable BM was observed, after 200 s in B).
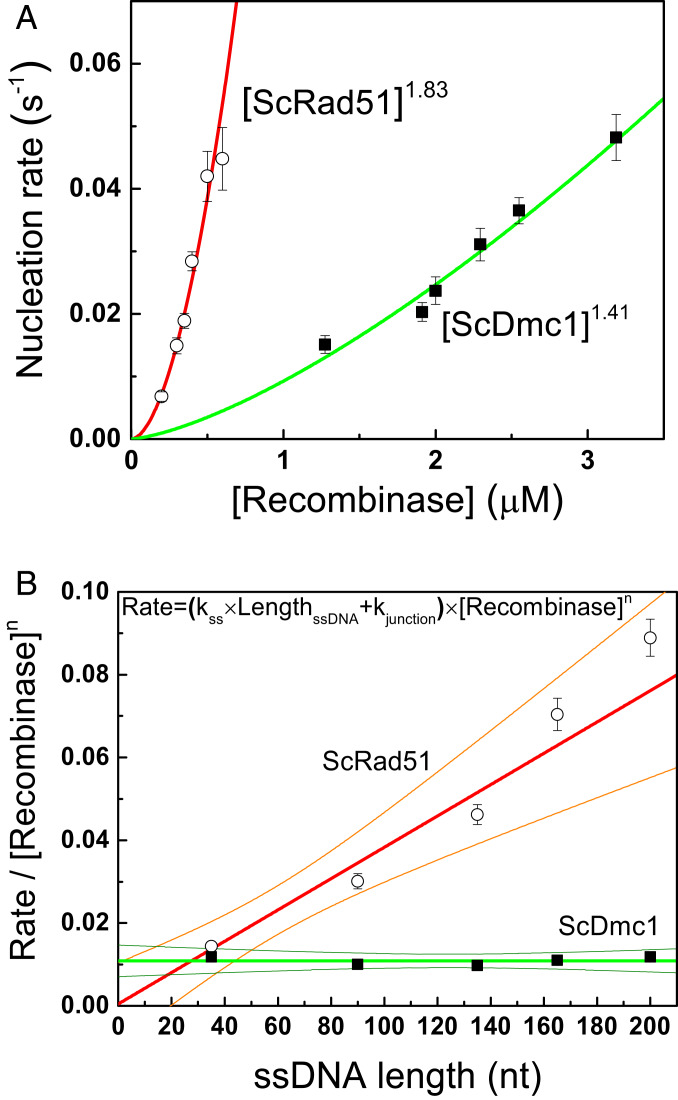
During the nucleation process, recombinases first assemble on ssDNA to form a stable nucleus before assembling into continuous and long nucleoprotein filaments. The difference in nucleating cluster sizes may account for the apparent nucleation rate differences seen in Fig. 2. Concentration dependence of recombinases on the nucleation rates determines the fundamental nucleating cluster size. We have previously determined the nucleation unit of mouse RAD51 to be 3 (28). Nucleation rates at different ScRad51 and ScDmc1 recombinase concentrations are determined using the same 349/264 substrate (SI Appendix, Fig. S3). Power-law dependence of concentrations on the nucleation rate leads to 1.83 ± 0.17 for ScRad51 and 1.41 ± 0.17 for ScDmc1 (Fig. 3A). Therefore, both ScRad51 and ScDmc1 require two recombinases to form a stable nucleating cluster.
Fig. 3.
Dmc1 and Rad51 have the same nucleation unit during filament assembly, but different ssDNA affinities. (A) Nucleation rates of ScDmc1 and ScRad51 nucleoprotein filaments measured at different recombinase concentrations using 349/264 DNA substrates and 1 mM ATP. Similar power-law fittings of recombinase concentration return with n = 1.41 ± 0.17 for Dmc1 and n = 1.83 ± 0.17 for ScRad51. Nucleation times were determined from more than 50 assembled events of at least three independent experiments (SI Appendix, Fig. S3). The error bars of nucleation rate are the SD by bootstrapping 5,000 times. (B) Nucleation rates of ScDmc1 and ScRad51 have different dependences on ssDNA lengths of DNA substrates (351/dTn, n = 35, 90, 135, 165, and 200). Nucleation rates at each DNA length were determined at the 1.9-μM ScDmc1 or 0.4-μM ScRad51 (SI Appendix, Fig. S4). Apparent nucleation rates vary linearly with the ssDNA length, but with different slopes for ScDmc1 and ScRad51. The error bars of nucleation rate are the SD by bootstrapping 5,000 times.
We then tested how nucleation rates vary with the ssDNA lengths. Longer ssDNA provides more potential nucleating sites and reduces nucleation times. It is known that both ScRad51 and ScDmc1 share similar amino sequences and have the same DNA footprint of 3 nt for each monomer (6, 34–37). We thus compared nucleation times of 351/dTn gap DNA substrates containing different ssDNA lengths of n = 35, 90, 135, 165, and 200 at the fixed 0.4-μM ScRad51 and 1.9-μM ScDmc1 concentration, respectively (SI Appendix, Fig. S4). As expected, the nucleation times of ScRad51 reduce as ssDNA length increases (372.2, 178.5, 116.1, 76.1, and 60.4 s for dT35, 90, 135, 165, and 200, respectively, at the constant 0.4-μM ScRad51 concentration; SI Appendix, Fig. S4). Surprisingly, there is no apparent change in nucleation times for ScDmc1 at these ssDNA lengths (36.1, 40.6, 41.6, 37.1 and 34.5 s, at the constant 1.9-μM ScDmc1 concentration; SI Appendix, Fig. S4). The nucleation rate is proportional to the product of ssDNA concentration (in nucleotides) and recombinase concentration to the n-th power. We thus plot the normalized rate (the observed rate divided by the n-th power of recombinase concentration) as a function of ssDNA lengths, given that we used the fixed amount of ssDNA molecules in the experiments, as shown in Fig. 3B. In this case, n is used as 1.83 for ScRad51 and 1.41 for ScDmc1, based on the fitted parameters of Fig. 3A. The plot expects a linear dependence, and it is apparent for ScRad51. However, the normalized rate is nearly constant for ScDmc1 at different ssDNA lengths. Plotting the same sets of data but using n = 2 for both ScRad51 and ScDmc1 (SI Appendix, Fig. S5) results in a similar trend (fitting parameters listed in SI Appendix, Table S2). Considering that 351/dTn substrates include duplex DNA handle (351 bp), ssDNA (dTn), and ss/dsDNA junctions and that recombinases do not initiate nucleation and assembly on the duplex segment under experimental conditions used here, nucleation can take place at either ssDNA or the ss/dsDNA junction. The same nucleation times at different ssDNA lengths observed for ScDmc1 suggests that ScDmc1 could preferentially nucleate on the ss/dsDNA junctions. Linear fitting to Fig. 3B returns the slope as the nucleation rate constant in ssDNA and the y-intercept as the nucleation rate constant at the ss/dsDNA junction. Thus, the nucleation rate constant of ssDNA for ScRad51 is 33-fold higher than that of ScDmc1 (SI Appendix, Table S2), confirming a much higher ssDNA affinity of ScRad51 than ScDmc1 (Fig. 2). However, the junction nucleation rate constant of ScRad51 is almost zero and that of ScDmc1 is ∼20-fold higher than ScRad51.
Dmc1 Preferentially Nucleates on the ss/dsDNA Junction with Polarity.
The dramatic differences in nucleation site preference of ScRad51 and ScDmc1 shown in Fig. 3 predict that increasing amounts of junctions in DNA substrates while keeping the total length of ssDNA will not alter the Rad51 nucleation time, but will reduce the nucleation time for Dmc1. To test this hypothesis, we engineered 23- to 24-nt random sequences into the dTn substrates and annealed with oligos of complementary sequence to generate an additional one or two junctions in the 351/dT(45)2 and 351/dT(45)3 substrates (SI Appendix, Fig. S1).
Both 351/dT90 and 351/dT(45)2 substrates contain 90-nt ssDNA. ScRad51 nucleates at almost the same time (232.9 s and 229.9 s, Fig. 4A and SI Appendix, Fig. S6 A and B) for these two substrates. Surprisingly, nucleation time of ScDmc1 reduces twofold from 83.2 s (351/dT90) to 41.8 s [351/dT(45)2], when the junction numbers double as shown in Fig. 4B. When the junction number triples in 351/dT135 and 351/dT(45)3, the nucleation time of ScRad51 remains constant (152.0 and 154.5 s, Fig. 4A and SI Appendix, Fig. S6 E and F), but that of ScDmc1 reduces nearly threefold (83.2 and 25.1 s, Fig. 4B and SI Appendix, Fig. S6 G and H). Note that experiments using different ScDmc1 concentration (1.1 μM) again showed no ssDNA length dependence (90 vs. 135 nt, Fig. 4B and SI Appendix, Fig. S6 C and G). The same set of experiments was also carried out in a high-salt buffer containing 150 mM K+ (SI Appendix, Fig. S7) and the same observation that ScDmc1 preferentially nucleates at ss/dsDNA junctions was made.
Fig. 4.
Dmc1 and Rad51 show different nucleation preference for ssDNA. DNA substrates containing different numbers of ssDNA gaps show a faster nucleation rate for ScDmc1 but have no effect on ScRad51. Nucleation times of ScDmc1 (B) reduce in proportion to the amounts of ssDNA gaps but remain the same for ScRad51 (A). Nucleation time was determined at the 1.1-μM ScDmc1 or 0.4-μM ScRad51 in the presence of 1 mM ATP. The error bars of nucleation time are the SD by bootstrapping 5,000 times.
We then asked whether this junction nucleation preference of ScDmc1 has specific polarity, as there are junctions that allow ScDmc1 to extend on ssDNA in the 5′-to-3′ or 3′-to-5′ polarity. Polarity is referred by the ssDNA strand that recombinases bind. To test this, we annealed 351/dT(45)2 substrates with oligo 8 or 9 (SI Appendix, Fig. S1) that has an additional 3-nt dT overhang at either the 3′- or 5′-end. For example, in the case of 351/dT(45)2-5′-flap, there is only one ss/dsDNA junction in the bottom to allow a 5′-to-3′ extension, but there are two junctions (middle and top) to allow a 3′-to-5′ extension. In the 351/dT(45)2-3′-flap, the case is opposite. Surprisingly, ScDmc1 nucleation time reduces only in the 351/dT(45)2-3′-flap substrate, but remains the same for the 351/dT(45)2-5′-flap substrate (Fig. 5A and SI Appendix, Fig. S8 A–D). Therefore, only the 5′ ss/ds junction stimulates Dmc1 to nucleate and assemble in the 5′-to-3′ polarity.
Fig. 5.
Both yeast and mouse Dmc1 prefer assembling on 5′ ssDNA/dsDNA junctions. DNA substrates containing the ssDNA gap with an additional dT3 flap at the 3′-end enhance Dmc1 nucleation, but the dT3 flap at the 5′-end shows no effect. The total ssDNA length is 90 nt for all four substrates used here. (A) Nucleation time determined for 1.7-μM ScDmc1 in the presence of 1 mM ATP. (B) Nucleation time determined for 5-μM mDMC1 in the presence of 1 mM ATP. The error bars of nucleation time are the SD by bootstrapping 5,000 times.
Nucleation measurements were also determined for mouse DMC1 using the same set of DNA substrates (Fig. 5B and SI Appendix, Fig. S8 E–H). Surprisingly, mDMC1 also showed the same nucleation stimulation (∼1.7-fold) on the additional 5′ junction. We also confirmed that mRAD51 prefers to nucleate on ssDNA (SI Appendix, Fig. S9), as seen in ScRad51. The similar nucleation preference observed for both ScDmc1 and mDMC1 suggests that this nucleation preference on the junction with a defined polarity is an evolutionarily conserved character and likely reflects as an intrinsic property of Dmc1 recombinases during nucleoprotein filament assembly.
Dmc1 Assembly Is Stimulated by Rad51 Patches.
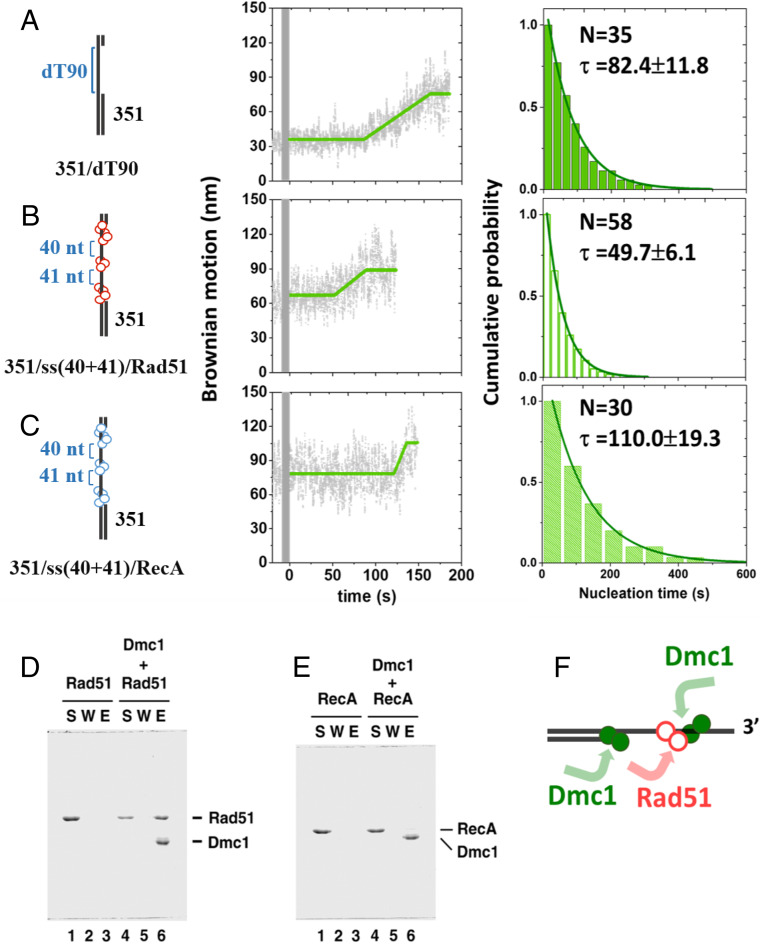
The observation of the 5′ ss/dsDNA junction as a preferred nucleation site for Dmc1 recombinases leads us to speculate that efficient Dmc1 nucleation requires 1) a structural component (as seen in the ss/dsDNA junction) and 2) defined ssDNA polarity. Rad51 has been previously suggested to interact with Dmc1 during meiosis for stimulation of strand-exchange activity (18). We tested the possibility of whether Rad51 can function as a structural component during Dmc1 filament assembly. Since Rad51 has much higher ssDNA affinity than Dmc1 (Figs. 2 and 3), Rad51 can readily bind to ssDNA during filament formation, and thus, the Rad51–ssDNA complex could act as a docking site to direct Dmc1 nucleation, potentially through the Rad51–Dmc1 interactions. To test this hypothesis, we prepared ssDNA containing short ScRad51 patches to see if ScDmc1 assembly can be stimulated by ScRad51. To prepare ScRad51 patches, we used DNA substrates [351/dT(37+29+36), SI Appendix, Fig. S1] containing long ssDNA of dT37-random40-dT29-random41-dT36 sequence and annealed four complementary oligos containing one to two mismatches (oligos 10 to 13 of ∼20 nt long each) to expose only the dT37/dT29/dT36 ssDNA region for ScRad51 binding on the slide (SI Appendix, Fig. S10A, I). After ScRad51 binding in the presence of AMP-PNP (II), excess oligos 14 to 17 (complementary to oligos 10 to 13) were introduced in the reaction chamber to compete out of oligos 10 to 13, leaving two ssDNA segments (40 and 41 nt, III). Each of the steps was confirmed by the BM distribution (SI Appendix, Fig. S10B). Finally, ScDmc1 recombinases were introduced in the presence of ATP to monitor the filament assembly in real time. When 1.1-μM ScDmc1 was used, the nucleation time on dT90 DNA substrate (351/dT90 substrate) was 82.4 s (Fig. 6A). Surprisingly, in the presence of ScRad51 patches (351/ss[40+41]/Rad51 substrate), the nucleation time reduced almost twofold to 49.7 s (Fig. 6B). In the ScRad51 patch substrates that we prepared, only the bottom two Rad51 patches provided the required DNA polarity for ScDmc1 to extend on the 5′-to-3′ polarity. An almost twofold reduction in nucleation time is consistent with two ScRad51 patches created. Control experiments using the same substrate and preparation procedure, but using E. coli RecA recombinases, returned with the slower ScDmc1 nucleation time of ∼110.0 s (Fig. 6C). The no-change or even slower nucleation time in the presence of EcRecA patches suggests that Dmc1 nucleation likely requires species-specific interaction. In fact, pull-down experiments showed that ScDmc1 physically interacts with ScRad51 (Fig. 6D), but not with EcRecA (Fig. 6E). Analysis of ScDmc1 extension rate on these three substrates (Fig. 6 A–C) revealed no significant statistical difference (SI Appendix, Fig. S11), reflective of the same ScDmc1 extension step in these DNA substrates.
Fig. 6.
Nucleation of ScDmc1 nucleoprotein filament assembly is stimulated by ScRad51, but not by EcRecA. In the presence of short discontinuous patches of ScRad51 on ssDNA, assembly of ScDmc1 is stimulated. (A) In 351/dT90 DNA substrate, exemplary BM time course of ScDmc1 assembly and the nucleation time histogram show the nucleation time of 82.4 s. (B) Experiments of preformed ScRad51 patches on 351/ss(40+41)/Rad51 DNA substrates show the reduced nucleation time of 49.7 s. (C) Experiments of preformed EcRecA patches on 351/ss(40+41)/RecA DNA substrates show the longer nucleation time of 110.0 s. (D–E) His6-tagged ScDmc1 incubated with ScRad51 (D) or EcRecA (E) was pulled down by TALON resins followed by wash and SDS elution. The supernatant (S), wash (W), and elution (E) were resolved by 12% SDS/PAGE and stained with Coomassie blue. (F) Proposed model for assembly of Dmc1 during nucleoprotein filament formation. The error bars of nucleation time are the SD by bootstrapping 5,000 times.
All experiments shown here are done in the absence of Ca2+ ions. Experiments carried out in the presence of Ca2+ ions also confirmed that Rad51 patches stimulate Dmc1 nucleation (SI Appendix, Fig. S12). Our results are consistent with the model that the ScRad51 patch on ssDNA serves as a structural component to interact with ScDmc1 and function as a ScDmc1 docking site during its rate-limited nucleation step.
Discussion
As shown from the ensemble-based experiments, ScDmc1 nucleoprotein filament is less protected from nuclease degradation and is thus less stable than the ScRad51. Using single-molecule experiments, we showed that both ScRad51 and ScDmc1 have the same nucleation size of two protomers (n = 2), but ScDmc1 has much reduced ssDNA affinity. Interestingly, ScDmc1 prefers to nucleate on the specific structural component, as in the ss/dsDNA junction with the required DNA polarity, so it can extend in the 5′-to-3′ direction. Dmc1 requirements on the structural component and ssDNA polarity are conserved evolutionarily, as seen in ScDmc1 and mDMC1, implicative of their important functional roles. Surprisingly, we showed that the ScRad51-ssDNA patch can function as a structural component to stimulate ScDmc1 nucleation (Fig. 6F). Therefore, our work provides evidence of how the nucleoprotein filament assembly of ScDmc1 can be stimulated by ScRad51.
As functional and structural homologs, Rad51 and Dmc1 recombinases are thought to behave similarly. Despite the high amino acid identity of Rad51 and Dmc1 (∼45%) (3–5), their biochemical properties are surprisingly different. ScDmc1 is slow in nucleation and prefers to nucleate on the ssDNA site at the duplex junctions, while ScRad51 has much higher ssDNA affinity and nucleates on ssDNA. These differences do not result from the nucleation sizes, as both ScRad51 and ScDmc1 were determined to nucleate in dimers with numbers similar to 2.4 protomers measured for human RAD51 (38) and 3 protomers for mouse RAD51 (28). Even though amino acid sequences in DNA-binding domains are mostly similar for both Rad51 and Dmc1 recombinases, there exist some Rad51- and Dmc1-specific locations, such as those recently identified in DNA-binding loops (L1 and L2) (11). The amino acid differences found in L1 and L2 loops are thought to implicate in the difference of mismatch tolerance of Rad51 and Dmc1 recombinases (11). ScRad51 also has an extended N-terminal segment. It would be interesting to identify the specific amino acid of ScRad51 and ScDmc1 responsible for the observed differences in ssDNA affinity and nucleation preference.
Dmc1 nucleation requires ssDNA with defined polarity, likely reflecting the Dmc1’s extension in a 5′-to-3′ direction during filament assembly. This is consistent with the 3′-terminated ssDNA overhang generated from the end-recessing enzymes involved in the recombination pathway (1, 34, 39–44). Our data do not support the 3′-to-5′ filament growth of Dmc1, or at least that growth during the nucleating cluster stage in the 3′-to-5′ polarity is significantly slower than in the 5′-to-3′ one. During the recombinase nucleoprotein filament assembly, the rate-limited nucleation event is followed by fast extension. As gel-based biochemical experiments assay for stable filament formation or strand-exchange product, they lack sensitivity and time resolution to detect the nucleation events observed in single-molecule experiments. Single-molecule real-time measurements used here are thus powerful and essential in elucidating the mechanistic details during filament assembly.
Compared to ScRad51, ScDmc1’s nucleating rate constant in ssDNA is ∼33-fold lower (SI Appendix, Table S2), suggesting that ScDmc1 is very inefficient in nucleating an open ssDNA region by itself. We showed that ScDmc1 nucleation can be stimulated by a structural component, the ss/dsDNA junction in DNA substrates used here. This structural component can serve as a docking site to interact with Dmc1 to increase its ssDNA affinity. Considering that there is only one such site in the recessed DNA, this ss/dsDNA junction preference could be devastating for Dmc1 function, especially because recombination progression requires functional Dmc1 nucleoprotein filaments. If the ssDNA overhang is long, the slow kinetics could further attenuate Dmc1 function. Instead, slow nucleation kinetics of Dmc1 reflects a key and major regulatory step for Dmc1 function during meiosis, as other proteins can effectively stimulate Dmc1 filament assembly. Using preassembled ScRad51 patches, we clearly demonstrated that ScRad51 stimulates ScDmc1 nucleation. However, preformed EcRecA patches did not stimulate ScDmc1 nucleation. Pull-down experiments directly showed the physical interaction of ScDmc1 with ScRad51, but not with EcRecA. Therefore, the ScRad51 stimulation of the ScDmc1 filament assembly must happen through specific protein–protein interaction with ScDmc1. Earlier work suggests that catalytically inactive Rad51 serves as an accessory protein in Dmc1 strand-exchange activity (18). It is likely that Rad51 stimulates Dmc1’s recombination activity by functioning as the structural component during the Dmc1 filament assembly step. In this regulatory role, Rad51 must assemble effectively in the ssDNA region, consistent with the fast kinetics and high ssDNA affinity observed here. Effective regulation also requires Rad51 to form patches on ssDNA, thereby creating sufficient Dmc1-docking sites. Our results directly provide the molecular basis of the previously identified Rad51-Dmc1 homotypic filaments seen in vitro and in vivo (21, 22). It would be interesting to see whether other accessory proteins, such as Mei5-Sae3, Rdh54, and others, also participate in this complex regulatory process.
Here we provide biochemical evidence of how Dmc1 assembly can be stimulated by Rad51. Our finding shows that Dmc1 nucleoprotein filament assembly is a key regulatory step and provides hints on how Rad51 and other accessory proteins could act to stimulate and regulate Dmc1 function in meiosis. Understanding the molecular details of how other accessory proteins act on the Dmc1 assembly, individually or synergistically, will help to elucidate the regulation network of Dmc1 recombinases during meiosis.
Materials and Methods
Protein Expression and Purification.
The expression and purification procedures of ScDmc1 were performed as described previously (45). Briefly, His6-tagged Dmc1 expression plasmid was induced in E. coli Rosetta cells while OD600 reached 0.6 by 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and then harvested after a 3-h incubation at 37 °C. For protein purification, all of the purification steps were carried out at 4 °C, and all of the following buffers contained 0.1 mM Na3VO4, 2 mM ATP, and 2 mM MgCl2. As previously, Dmc1-containing lysate was purified in order with TALON affinity resin (Clontech), Heparin Sepharose column (GE Healthcare), and Mono Q column (GE Healthcare). Finally, the Dmc1-containing fractions were pooled and concentrated in a Centricon-30 (Millipore). The concentrated preparation was divided into small aliquots and stored at −80 °C.
For obtaining mouse DMC1, the expression and purification procedures for human DMC1 were followed as previously published (46). Briefly, His6-tagged DMC1 expression plasmid was induced in RecA-deficient E. coli cells (strain BLR), and then the obtaining cell paste was clarified and purified in order with TALON affinity resin, Source Q column (GE Healthcare), macrohydroxyapatite column (GE Healthcare), and Mono Q column. Finally, the DMC1-containing fractions were pooled and concentrated in a Centricon-30. The concentrated preparation was divided into small aliquots and stored at −80 °C.
To express ScRad51, the RecA-minus E. coli cells (BLR strain) harboring the plasmid (pLant2B-Rad51) that expressed Rad51 were grown in Luria broth at 37 °C until OD600 reached 0.6, and, at this point, were induced by addition of 0.1 mM IPTG and harvested after further incubation at 16 °C for 20 h. The purification procedure of Rad51 was followed with serial chromatographic columns as previously (9). Briefly, clarified lysate was precipitated by ammonium sulfate. The dissolved Rad51 pellet was fractionated serially in Sepharose Q column, macrohydroxyapatite column, and Source Q column. The Rad51-containing fractions were pooled, concentrated, and stored at −80 °C. The expression and purification procedures of mouse RAD51 were as described previously (47). E. coli RecA protein was purchased from New England Biolabs.
DNA Substrates for Protection Assays.
To prepare 5′-end, 32P-labeled 80-mer ssDNA for endonuclease protection assay, [γ-32P]ATP (PerkinElmer) was coupled to the 5′-end of oligo 1 (see SI Appendix, Table S1 for sequence) using polynucleotide kinase (New England Biolabs). The unincorporated nucleotide was removed by a Spin 6 column (Bio-Rad).
Endonuclease Protection Assay.
Indicated amount (0–6 μM) of His6-tagged Dmc1 or Rad51 was incubated with 5′ γ-32P–labeled 80-mer oligo 1 ssDNA (3 μM nucleotides) in buffer A (35 mM Tris⋅HCl pH 7.5, 1 mM dithiothreitol [DTT], 2.5 mM MgCl2, 50 mM KCl, and 100 ng/μL bovine serum albumin [BSA]) containing 1 mM ATP at 37 °C for 5 min. Following this, 5 units of benzonase (Sigma-Aldrich) was added to reaction mixtures (final volume 10 μL) and further incubated at 37 °C for 10 min. The reaction mixtures were terminated with a 2.5-μL stop mixture containing 240 mM ethylenediaminetetraacetic acid (EDTA), 0.2% sodium dodecyl sulfate (SDS), and 3.2 μg proteinase K at 37 °C for 15 min and then resolved in 10% polyacrylamide gel with TBE buffer (89 mM Tris, 89 mM borate, 2 mM EDTA, pH 8.0). Then the gel was dried, and the DNA species were quantified by a phosphor imaging system (Bio-Rad).
Affinity Pulldown.
Rad51 (3 μg) or RecA (3 μg) were incubated with His6-tagged Dmc1 (4.5 μg) in a 30-μL reaction buffer B (25 mM Tris⋅HCl, pH 7.5, 10% glycerol, 0.01% Igepal, 2 mM β-mercaptoethanol, 5 mM imidazole, and 150 mM KCl) for 20 min at 37 °C. After being mixed with 30 μL of TALON affinity resin for 20 min at 37 °C to capture His6-tagged Dmc1 and associated recombinases, the supernatants were separated from resin spin-down. The resins were washed three times in a 30-μL reaction buffer and then treated with 30 μL 2% SDS at 55 °C to elute proteins. The supernatant, the final wash, and the SDS eluate were analyzed by 12% SDS–polyacrylamide gel electrophoresis (PAGE) with Coomassie Blue staining.
DNA Substrates for Single-Molecule Experiments.
All DNA substrates are illustrated in SI Appendix, Fig. S1. The gap 349/264 DNA substrate, containing a duplex DNA handle of 349 bp, a single-stranded DNA gap of 264 nt containing AC repeating sequence without secondary structure, and a terminal 24-bp duplex for specific-labeling to beads were prepared as described previously (48). Briefly, it was prepared by annealing three ssDNA oligos (637, 349, and 24 nt) together. The 637- and 349-nt ssDNA were prepared using PCR reactions containing a phosphate-modified primer and an OH- or digoxigenin-modified primer and then followed by Lambda exonuclease (NEB) digestion to remove the phosphate-labeled strands to generate the desired ssDNA.
The dT gap 351/(35, 90, 135, 165, 200) DNA substrates were prepared by the ligation of three components: 1) the 351/331-bp DNA containing a 20-nt 5′-overhang; 2) oligo 2 containing n-nt polydT sequence (n = 35, 90, 135, 165, and 200 nt) sandwiched by a 20-nt at the 5′-end and 19 nt at the 3′-end; and 3) a 19-nt 5′-biotin–labeled oligo 3. A 351/331-bp dsDNA (component 1) with a 20-nt 5′ overhang was prepared using PCR, using one primer containing an abasic site 20 nt from the 5′-end and one digoxigenin-modified primer (28). Oligo 2 is treated with T4 polynucleotide kinase (NEB) to generate a 5′-phosphate–modified end before subsequent ligation. All three components were mixed, annealed, and ligated using T4 ligase (NEB) to generate the desired 351/(35, 90, 135, 165, 200 nt) gap substrates.
The dT(45)n (n = 2 or 3) DNA substrates were prepared similarly as for 351/dTn, except that 351-/331-bp DNA was ligated with oligo 4 or 5 containing two or three dT45 that were sandwiched by 24 nt [for dT(45)2] or 24 and 23 nt [for (dT(45)3], respectively. The ligated strand is then annealed with oligo 6 (or oligo 6 and 7) and oligo 3, as illustrated in SI Appendix, Fig. S1. To prepare 351/dT(45)2-flap DNA substrates with different polarities, oligo 6 was replaced with oligo 8 or 9 including a dT3 flap at either the 3′- or 5′-end. Preparation of 351/dT(37+29+36) DNA substrate was carried out by a similar procedure as that for dT(45)3 but with replacing the annealed oligo 6 and 7 with oligos 10 to 13 containing one or two mismatches to oligo 5. All of the ssDNA were purified and verified by gel extraction. All oligo sequences are listed in SI Appendix, Table S1.
smTPM Assay.
The slide and streptavidin bead preparation were described previously (29, 48). The gap DNA substrates were specifically anchored onto the antidigoxigenin-coated surface with the distal end attached to a streptavidin-labeled bead (220-nm diameters, Bangs Laboratories). The surface was preblocked by BSA to prevent nonspecific bead sticking. All of the reaction mixtures were preincubated at 37 °C for 10 min before adding to the reaction chamber. The DNA tether images were acquired by an inverted microscope (Olympus IX71) as previously described (29, 48). The image acquisition rate is 30 Hz. Twenty microliters of reaction mixture containing recombinase was then added to initiate the assembly reaction. Typical TPM experiments were carried out at specified recombinase concentration, 1 mM nucleotide (ATP), 1 mM phosphoenolpyruvate, 4 units/mL pyruvate kinase, 30 mM Tris (pH 7.5), 50 mM KCl, 2.5 mM MgCl2, and 1 mM DTT. For the assembly experiment of mDMC1, mRAD51, or ScDmc1 at high K+ conditions, the KCl concentration was replaced by 150 mM.
The assembly of ScDmc1 on ssDNA containing short Rad51 filament patches was conducted as follows: 1) ScRad51 with AMP-PNP was introduced into the reaction chamber which was bound with 351/dT(37+29+36) DNA substrate. 2) After ScRad51 filament formation, free ScRad51 was washed way. 3) To remove the four oligos containing mismatch, excess complementary strands of the four oligos were added and incubated at 30 °C for 15 min. 4) Dissociated oligos were washed away at room temperature and then the substrate exposed 81 nt ssDNA, forming 351/ss(40+41)/Rad51 DNA. 5) ScDmc1 preincubated at 37 °C was introduced into a reaction chamber to form nucleoprotein filament on 351/ss(40+41)/Rad51 DNA.
Bead centroid position was determined by two-dimensional Gaussian fitting. Brownian motion (BM) amplitude is defined as the standard deviation (SD) from bead centroid position within 20 frames in sliding windows using a custom-made MATLAB program. The stage drift was corrected by subtracting the bead centroid position from the preadsorbed bead position. The solver function of Excel was used to fit the experimental time course for the minimum residuals with four output parameters: the initial BM amplitude, the final BM amplitude at the end of filament extension, the time point when the extension starts, and the time point at the end of extension.
Data Availability.
All other relevant data are described in SI Appendix or are available upon request.
Supplementary Material
Acknowledgments
This work was supported by National Taiwan University; by Academia Sinica; and by Ministry of Science and Technology Grants MOST 105-2314-B-002-073 and MOST 108-2321-B-002-054 (to P.C.) and Grant MOST 107-2113-M-002-010 (to H.-W. L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920368117/-/DCSupplemental.
References
- 1.San Filippo J., Sung P., Klein H., Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Crickard J. B., Greene E. C., The biochemistry of early meiotic recombination intermediates. Cell Cycle 17, 2520–2530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masson J.-Y., West S. C., The Rad51 and Dmc1 recombinases: A non-identical twin relationship. Trends Biochem. Sci. 26, 131–136 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Lin Z., Kong H., Nei M., Ma H., Origins and evolution of the recA/RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer. Proc. Natl. Acad. Sci. U.S.A. 103, 10328–10333 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Story R. M., Bishop D. K., Kleckner N., Steitz T. A., Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science 259, 1892–1896 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Lee J. Y., et al. , DNA RECOMBINATION. Base triplet stepping by the Rad51/RecA family of recombinases. Science 349, 977–981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi Z., et al. , DNA sequence alignment by microhomology sampling during homologous recombination. Cell 160, 856–869 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop D. K., Park D., Xu L., Kleckner N., DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69, 439–456 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Sung P., Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265, 1241–1243 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Sehorn M. G., Sigurdsson S., Bussen W., Unger V. M., Sung P., Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature 429, 433–437 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Steinfeld J. B., et al. , Defining the influence of Rad51 and Dmc1 lineage-specific amino acids on genetic recombination. Genes Dev. 33, 1191–1207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lao J. P., et al. , Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 9, e1003978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J. Y., et al. , Sequence imperfections and base triplet recognition by the Rad51/RecA family of recombinases. J. Biol. Chem. 292, 11125–11135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callender T. L., et al. , Mek1 down regulates Rad51 activity during yeast meiosis by phosphorylation of Hed1. PLoS Genet. 12, e1006226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busygina V., et al. , Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 22, 786–795 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsubouchi H., Roeder G. S., Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 20, 1766–1775 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crickard J. B., et al. , Regulation of Hed1 and Rad54 binding during maturation of the meiosis-specific presynaptic complex. EMBO J. 37, e98728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cloud V., Chan Y. L., Grubb J., Budke B., Bishop D. K., Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337, 1222–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinohara M., Gasior S. L., Bishop D. K., Shinohara A., Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. U.S.A. 97, 10814–10819 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurzbauer M.-T., Uanschou C., Chen D., Schlögelhofer P., The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell 24, 2058–2070 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown M. S., Grubb J., Zhang A., Rust M. J., Bishop D. K., Small Rad51 and Dmc1 complexes often co-occupy both ends of a meiotic DNA double strand break. PLoS Genet. 11, e1005653 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crickard J. B., Kaniecki K., Kwon Y., Sung P., Greene E. C., Spontaneous self-segregation of Rad51 and Dmc1 DNA recombinases within mixed recombinase filaments. J. Biol. Chem. 293, 4191–4200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop D. K., RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79, 1081–1092 (1994). [DOI] [PubMed] [Google Scholar]
- 24.Shinohara A., Gasior S., Ogawa T., Kleckner N., Bishop D. K., Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2, 615–629 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Sauvageau S., et al. , Fission yeast rad51 and dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol. Cell. Biol. 25, 4377–4387 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holthausen J. T., Wyman C., Kanaar R., Regulation of DNA strand exchange in homologous recombination. DNA Repair 9, 1264–1272 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Morrison C., et al. , The essential functions of human Rad51 are independent of ATP hydrolysis. Mol. Cell. Biol. 19, 6891–6897 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C.-H., et al. , Swi5-Sfr1 stimulates Rad51 recombinase filament assembly by modulating Rad51 dissociation. Proc. Natl. Acad. Sci. U.S.A. 115, E10059–E10068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu H.-Y., Lu C.-H., Li H.-W., RecA-SSB interaction modulates RecA nucleoprotein filament formation on SSB-wrapped DNA. Sci. Rep. 7, 11876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pugh B. F., Cox M. M., General mechanism for RecA protein binding to duplex DNA. J. Mol. Biol. 203, 479–493 (1988). [DOI] [PubMed] [Google Scholar]
- 31.Lu Y.-W., et al. , Using single-molecule approaches to study archaeal DNA-binding protein Alba1. Biochemistry 52, 7714–7722 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Piechura J. R., et al. , Biochemical characterization of RecA variants that contribute to extreme resistance to ionizing radiation. DNA Repair (Amst.) 26, 30–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu H. F., Ngo K. V., Chitteni-Pattu S., Cox M. M., Li H. W., Investigating Deinococcus radiodurans RecA protein filament formation on double-stranded DNA by a real-time single-molecule approach. Biochemistry 50, 8270–8280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung P., Klein H., Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7, 739–750 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Ogawa T., Yu X., Shinohara A., Egelman E. H., Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259, 1896–1899 (1993). [DOI] [PubMed] [Google Scholar]
- 36.Conway A. B., et al. , Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 11, 791–796 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Sheridan S. D., et al. , A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 36, 4057–4066 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hilario J., Amitani I., Baskin R. J., Kowalczykowski S. C., Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl. Acad. Sci. U.S.A. 106, 361–368 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keeney S., Giroux C. N., Kleckner N., Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Heyer W.-D., Ehmsen K. T., Liu J., Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y., Caron P., Legube G., Paull T. T., Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 42, e19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimonkar A. V., et al. , BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niu H., et al. , Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467, 108–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cejka P., et al. , DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467, 112–116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Busygina V., et al. , Functional attributes of the Saccharomyces cerevisiae meiotic recombinase Dmc1. DNA Repair (Amst.) 12, 707–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang H. Y., et al. , Functional relationship of ATP hydrolysis, presynaptic filament stability, and homologous DNA pairing activity of the human meiotic recombinase DMC1. J. Biol. Chem. 290, 19863–19873 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai S.-P., et al. , Rad51 presynaptic filament stabilization function of the mouse Swi5-Sfr1 heterodimeric complex. Nucleic Acids Res. 40, 6558–6569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung C., Li H.-W., Direct observation of RecBCD helicase as single-stranded DNA translocases. J. Am. Chem. Soc. 135, 8920–8925 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other relevant data are described in SI Appendix or are available upon request.