Significance
TDP-43 is an RNA-binding protein closely associated with neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). We have previously shown that TDP-43 dysfunction leads to reduced dendritic complexity. In this study, we apply the recently developed genetic approach TRIBE (targets of RNA-binding proteins identified by editing) to identify the RNAs that are bound by TDP-43 in neurons. Of the many TDP-43 target RNAs identified, we observed that mRNAs encoding proteins related to the CREB signaling pathway are enriched. Next, we were able to rescue the dendritic complexity defect caused by TDP-43 dysfunction by restoring CREB signaling. Our study may provide pathways for therapeutic intervention in TDP-43−associated neurodegenerative diseases.
Keywords: TDP-43, TRIBE, CREB
Abstract
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are two related neurodegenerative diseases that present with similar TDP-43 pathology in patient tissue. TDP-43 is an RNA-binding protein which forms aggregates in neurons of ALS and FTD patients as well as in a subset of patients diagnosed with other neurodegenerative diseases. Despite our understanding that TDP-43 is essential for many aspects of RNA metabolism, it remains obscure how TDP-43 dysfunction contributes to neurodegeneration. Interestingly, altered neuronal dendritic morphology is a common theme among several neurological disorders and is thought to precede neurodegeneration. We previously found that both TDP-43 overexpression (OE) and knockdown (KD) result in reduced dendritic branching of cortical neurons. In this study, we used TRIBE (targets of RNA-binding proteins identified by editing) as an approach to identify signaling pathways that regulate dendritic branching downstream of TDP-43. We found that TDP-43 RNA targets are enriched for pathways that signal to the CREB transcription factor. We further found that TDP-43 dysfunction inhibits CREB activation and CREB transcriptional output, and restoring CREB signaling rescues defects in dendritic branching. Finally, we demonstrate, using RNA sequencing, that TDP-43 OE and KD cause similar changes in the abundance of specific messenger RNAs, consistent with their ability to produce similar morphological defects. Our data therefore provide a mechanism by which TDP-43 dysfunction interferes with dendritic branching, and may define pathways for therapeutic intervention in neurodegenerative diseases.
Amyotrophic lateral sclerosis (ALS) is a debilitating and rapidly progressing neurodegenerative disease that is often comorbid with frontotemporal dementia (FTD). A dominantly heritable form of ALS/FTD is caused by mutations in the gene TARDBP, which encodes for the TDP-43 protein (1, 2). TDP-43 is also a major component of pathological protein inclusions in more than 95% of sporadic ALS and ∼45% of FTD, suggesting that it is a central underlying factor in disease (3–5). TDP-43 is an RNA-binding protein (RBP) that shuttles between the nucleus and cytoplasm, and has been implicated in a variety of cellular functions, including DNA transcription, RNA splicing, microRNA processing, RNA transport, and translation (6, 7). Upon cellular insult, TDP-43 redistributes to the cytoplasm, where it associates with liquid stress granules, and can further irreversibly aggregate under pathological conditions (6, 8). Importantly, this TDP-43 cytoplasmic accumulation results in nuclear depletion of endogenous TDP-43, leading to the proposal that toxic gain of function of TDP-43 in the cytoplasm with concomitant TDP-43 loss of function in the nucleus contributes to the disease (8, 9).
We previously reported that either increased or decreased TDP-43 expression (modeling gain and loss of function, respectively) results in reduced dendritic outgrowth and complexity without an immediate effect on cell viability (10). In vivo, a major consequence of altered dendritic elaboration is disruption to overall neuronal circuit connectivity and cell-to-cell communication (11, 12), which has been suggested to be an underlying feature of the early stages of ALS and FTD (13–15). Importantly, the decreased dendritic branching observed upon TDP-43 overexpression (OE) is dependent on the RNA-binding ability of the protein (10). However, it remains unclear which TDP-43 RNA targets impinge on the many potential extrinsic and intrinsic signaling pathways that promote proper dendrite elaboration (16).
One obvious way forward is to identify TDP-43 RNA targets relevant to neurodegeneration (17–19). Using RNA immunoprecipitation followed by deep sequencing (RIP-seq) and cross-linking immunoprecipitation coupled with high-throughput sequencing (CLIP-seq and its variants), other groups independently generated lists of several thousand potential TDP-43 targets in the mammalian brain. Because of this large number and because RIP and CLIP have potential limitations (20), we applied an orthogonal approach to identify TDP-43 target RNAs.
TRIBE (targets of RNA-binding proteins identified by editing) is an antibody-independent method to identify RBP target messenger RNAs (mRNAs) (21, 22), which had only been previously applied to the Drosophila system. To identify putative TDP-43-bound target RNAs, we performed TRIBE using RNA obtained from primary cultures of mammalian neurons in which TDP-43 expression was altered. From the RNA target list generated, we identified multiple RNAs encoding various signaling pathway components, including those focused on signaling via the activity-regulated cAMP response element-binding protein (CREB) transcription factor (23).
Transcriptional programs such as those downstream of CREB activation control the elaboration of neuronal dendritic arbors (16). Upstream regulation of CREB activity is complex and involves multiple signal transduction networks, including the CaMK, MAPK, and PKA pathways (24–29). Some of these pathways contain TDP-43 targets, and we validated that CREB transcriptional activity is misregulated upon TDP-43 dysfunction, leading, in turn, to defects in dendrite morphogenesis. We were able to rescue dendrite elaboration in TDP-43−misregulated neurons by restoring CREB activity. Our RNA sequencing (RNA-seq) data also found that TDP-43 OE and knockdown (KD) result in similar mRNA abundance changes. This result is consistent with reduced dendritic branching observed upon TDP-43 OE or KD (10), suggesting that the mRNA changes may be downstream of the CREB pathway. Our data suggest that this pathway may play a role in TDP-43−related neurological diseases.
Results
TDP-43 RNA Targets Are Involved in Multiple Signaling Pathways That Regulate CREB.
TDP-43 is an RBP that has been suggested to interact with several thousand RNA transcripts in neuronal cells (17–19). RIP or CLIP methods were used in these studies, but these techniques have potential liabilities, for example, reassociation of RNAs into novel RBPs after cell lysis (30) and poor efficiency of cross-linking (31), respectively. These approaches also rely heavily on high-quality antibodies, which varied between these studies. These limitations make the identification of true target genes vs. false positives and false negatives difficult to assess. Consistent with this difficulty, the lists of putative target transcripts do not correlate well with each other, leading to considerable uncertainty about TDP-43 targets. TRIBE is independent of antibodies and an ideal orthogonal approach to identify TDP-43 targets.
The original TRIBE method takes advantage of the Drosophila RNA editing enzyme ADAR (referred to henceforth as dADAR), which endogenously interacts with specific structured RNAs and converts adenosine to inosine. Fusion of the ADAR catalytic domain (ADARcd) to an RBP of interest and expression in vivo directs the fusion protein to target RNAs of that RBP, on which the ADARcd then performs A-to-I editing up to several hundred base pairs from the RBP binding site (22). These editing events are detected as A-to-G mutations through standard RNA-seq and computational analysis (21, 22). To adapt the TRIBE method to mammalian systems, we used the catalytic domain of human ADAR2 (hADARcd), the human homolog of dADARcd.
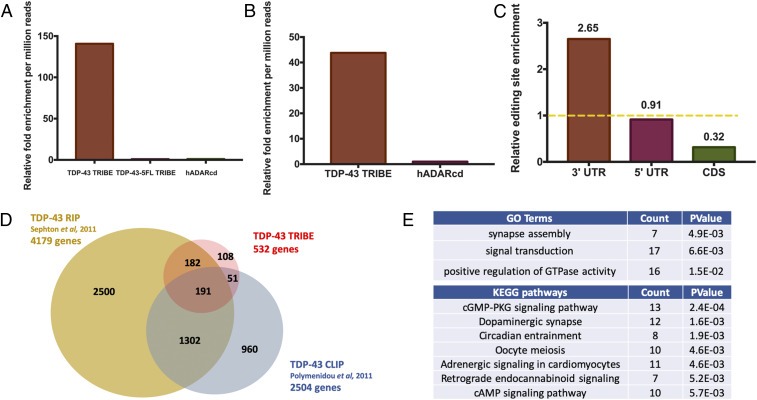
A pilot experiment was first performed in HEK293T cells to test the effectiveness of the TRIBE system in mammalian cells (32). Expression of the TDP-43 TRIBE fusion protein enhanced the number of editing sites by ∼150-fold (32) (Fig. 1A). Moreover, no above-background editing was detected in cells expressing TDP-43-5FL TRIBE, containing a non−RNA-binding mutant of TDP-43 (10), or the hADARcd alone (Fig. 1A). The data, not surprisingly, show that the efficiency and specificity of mammalian TRIBE is dependent on the RBP, as previously indicated in the Drosophila system (21, 22).
Fig. 1.
TDP-43 TRIBE reveals TDP-43 RNA targets involved in CREB signaling. (A) Quantification of the relative number of editing sites detected in HEK293T cells transfected with plasmids expressing TDP-43 TRIBE, TDP-43-5FL TRIBE, or hADARcd expressed under control of the CMV promoter (n = 2). All of the editing sites shown here and in all figures were present in both replicates. The y axis is the relative fold change compared to hADARcd and normalized by each sample’s sequencing depth. A requirement of 10 reads of 5% A-to-G editing is imposed to qualify as an editing site. (See Method section for details.) (B) Quantification of the relative number of editing sites detected in cultured rat cortical neurons transduced with lentivirus expressing TDP-43 TRIBE or hADARcd under control of the human synapsin promoter (n = 2). Normalization performed as in A. (C) Quantification of TDP-43 TRIBE editing sites enrichment in each mRNA region. Editing sites in 3′UTR, 5′UTR, or CDS were assigned based on RefSeq annotation, and enrichment was normalized to sequencing coverage. For example, 76% of all editing sites were located in 3′UTR, but 3′UTR only accounts for 29% of all sequencing depth, so it is 2.65-fold enriched. The yellow dashed line represents an enrichment score of 1. (D) Venn diagram demonstrating the 191 genes that overlap between three different studies, including ours, that sought to identify TDP-43 target RNAs. TDP-43 TRIBE identified 108 unique genes that were not identified previously by RIP-seq or CLIP-seq; 242 (45%) TRIBE targets were also identified by CLIP (Z test performed, P value < 0.0001) and 373 (70%) TRIBE targets were also identified by RIP (Z test performed, P value < 0.0001). (E) Top GO term and KEGG pathway hits with DAVID functional annotation analysis performed on TDP-43 TRIBE target list. The gene list was normalized to the background of the top 3,000 expressed genes in cultured rat cortical neurons (see Methods).
To identify RNA targets of TDP-43 in the central nervous system, we performed TDP-43 TRIBE in cultured rat cortical neurons (32), a frequently used system for studying the neurodegenerative pathology of TDP-43 (10, 33). We constructed lentiviral vectors encoding TDP-43 TRIBE or the hADARcd alone under the human synapsin 1 promoter (hSyn). After viral packaging, we transduced the viruses into cultured cortical neurons at 4 d in vitro (DIV). At 7 DIV, we harvested RNA, made mRNA-seq libraries, and performed high-throughput sequencing. Analysis of the sequence data with our computational pipeline (34) identified 532 genes, hosting 1,162 editing sites, as TDP-43 targets; this was ∼45-fold more editing sites than in the hADARcd control (Fig. 1 B and D). Analysis of the editing distribution within each mRNA region revealed a dramatic enrichment in 3′ untranslated region (UTR) (Fig. 1C), suggesting a strong preference of TDP-43 for 3′ UTR binding. This general 3′ UTR preference agrees with a previous publication (18) and has also been described for a handful of TDP-43 specific target mRNAs (19, 35, 36). The 3′ UTR binding also coincides with the proposed role of TDP-43 as an RNA stability and translational regulator (37, 38).
To examine how well the TDP-43 TRIBE target identification agrees with the RIP data (17) and CLIP data (19), we compared the target gene lists from the three datasets. Due to unavailability of the full gene list from ref. 19, we downloaded these raw data via the Gene Expression Omnibus (GEO) database and processed them using updated RNA-seq analysis standards, which reduced the number of target genes from 6,304 to 2,504.
TDP-43 TRIBE identified substantially fewer targets than the other two methods. This is most simply explained by a combination of a lower false-positive rate and higher false-negative rate with TRIBE. Another possibility is the absence of edited intronic sequences in our data, that is, nuclear targets, due to use of polyA RNA as our starting material (see Discussion). In any case, 191 high-confidence genes were identified in common between all three lists (Fig. 1D). This is a very large and highly significant fraction of the number of target genes identified by TRIBE (Fig. 1D; Z test performed, P < 0.001). The results, taken together, suggest that TDP-43 TRIBE is a viable method to identify bona fide RBP RNA targets in mammalian cells.
To gain insight into the biological processes potentially impacted by TDP-43, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) functional annotation tools for gene ontology term (GO term) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of TDP-43 TRIBE targets. From our TDP-43 TRIBE target list of 532 genes, both GO and KEGG generated lists of pathways and processes relevant to neuronal signaling and synapses (Fig. 1E and Dataset S1), consistent with our phenotypic observations (10). The results were similar when the 191 high-confidence genes were fed into the same analyses (Dataset S1). These findings also suggest that the shorter TDP-43 TRIBE target list likely reflects bona fide TDP-43 targets.
More specifically, the genes identified by TDP-43 TRIBE are enriched for GO terms involving dendritic branching and synapse assembly. These genes are also enriched in KEGG pathways that signal to the CREB transcription factor (Fig. 1E), which regulates dendrite elaboration (24–28). Notably, genes from TDP-43 TRIBE are enriched for pathways, including cAMP, Hippo, MAPK, Adrenergic signaling, and cGMP-PKG (Dataset S1, KEGG pathways for TRIBE targets). Activation of these signaling pathways results in CREB activation through phosphorylation at serine 133 (pCREB), which, together with recruited coactivators, activates downstream gene expression (39). Specifically, TDP-43 TRIBE targets include CaMKII, Calmodulin, MAPKs, PKA, and CREB2, suggesting that CREB transcriptional regulation might play an important role in TDP-43−dependent neuronal function.
TDP-43 Dysfunction Inhibits CREB Activation.
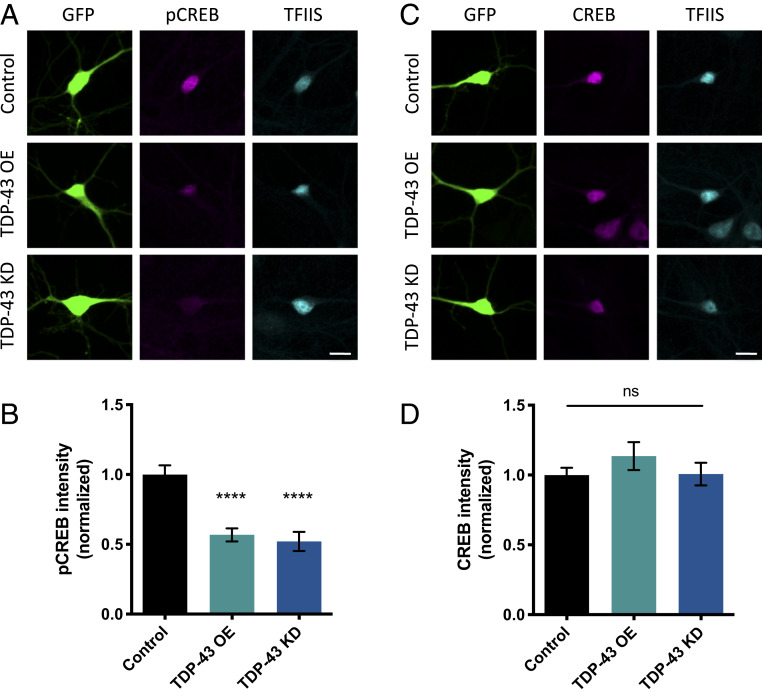
We next sought to determine whether TDP-43 dysfunction affects CREB activation in cultured mammalian neurons. We isolated cortical neurons from E18 rat pups, cocultured the neurons on a glial monolayer, and transfected them at 2 DIV with a vector expressing GFP and an empty vector control, GFP and TDP-43 (i.e., TDP-43 OE), or GFP and a short hairpin RNA (shRNA) targeting TDP-43 (i.e., TDP-43 KD). After an additional 5 DIV, during which time neurons mature and develop functional synaptic connections (40), neurons were fixed and immunostained using either an antibody that specifically recognizes pCREB or total CREB protein. The pCREB levels in these developing cultures could reflect CREB activation through a variety of signaling pathways, including growth factor signaling and calcium influx, in response to neuronal depolarization. Relative levels of pCREB and CREB were calculated by normalizing signal intensity to endogenous TFIIS, a ubiquitous transcription elongation factor (41).
We observed a significant decrease in pCREB signal in neurons that overexpress TDP-43 compared to controls (Fig. 2 A and B). Similarly, pCREB intensity is decreased in neurons in which TDP-43 was knocked down (Fig. 2 A and B). These results are consistent with our previous observation that dendritic branching is decreased by either TDP-43 OE or TDP-43 KD (10). Total CREB levels remained constant in both TDP-43 OE and TDP-43 KD conditions compared to control (Fig. 2 C and D), suggesting that TDP-43 dysfunction specifically inhibits CREB activation without affecting overall CREB protein abundance.
Fig. 2.
CREB phosphorylation is decreased by TDP-43 KD and OE. (A) Representative images of 7 DIV cultured cortical neurons transfected with plasmids expressing GFP and either empty vector control, TDP-43 OE, or an shRNA targeting TDP-43 for KD immunostained with an antibody that recognizes phosphorylated CREB at Ser133 (pCREB). (Scale bar for all images: 10 µm.) (B) Quantification of average fluorescence intensity for pCREB immunostaining normalized to the fluorescence intensity of the same neuron immunostained with an antibody that recognizes TFIIS. Control, n = 68; TDP-43 OE, n = 69; TDP-43 KD, n = 69. (C) Representative images of 7 DIV cultured cortical neurons transfected with plasmids expressing GFP and either empty vector control, TDP-43, or an shRNA targeting TDP-43 for KD immunostained with an antibody that recognizes total CREB protein. (Scale bar for all images: 10 µm.) (D) Quantification of average fluorescence intensity for CREB immunostaining normalized to the fluorescence intensity of the same neuron immunostained with an antibody that recognizes TFIIS. ns = not significant. Control, n = 70; TDP-43 OE, n = 68; TDP-43 KD, n = 65. ****P < 0.0001, one-way ANOVA, error bars represent SEM.
CREB Transcriptional Output Is Suppressed by TDP-43 Dysfunction.
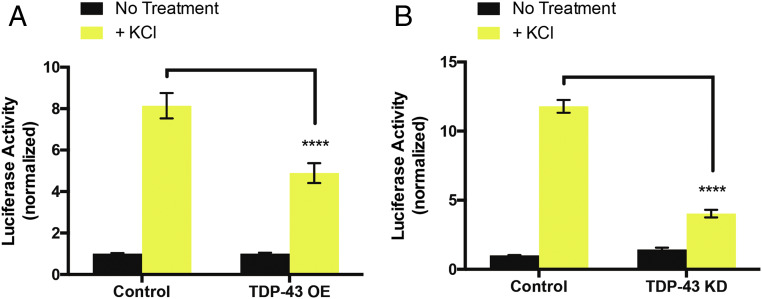
One likely consequence of TDP-43−dependent decrease in CREB phosphorylation is suppression of CREB-dependent transcriptional activity. Therefore, we investigated whether TDP-43 dysfunction interfered with activity-dependent CREB transcriptional output using a dual luciferase assay to quantify CREB transcriptional activity. To this end, we transfected primary cultures of rat cortical neurons at 2 DIV with either an empty vector control, a plasmid expressing TDP-43 (TDP-43 OE), or an shRNA targeting TDP-43 (TDP-43 KD) along with a reporter construct containing four CREB response elements (4x CRE) upstream of a firefly luciferase gene and a control reporter expressing Renilla luciferase from the thymidine kinase promoter. At 6 DIV, all cultures were treated overnight with 1 µM of the sodium channel blocker tetrodotoxin to suppress spontaneous activity, and therefore CREB activation, in the cultures. At 7 DIV, a subset of control, TDP-43 OE, and TDP-43 KD neuronal cultures were treated with 55 mM KCl for 6 h to mimic neuronal depolarization and induce CREB activation (26); this protocol causes calcium- and CREB-dependent transcription from the 4x CRE luciferase reporter (26). After 6 h, cell lysates were harvested, and firefly and Renilla luminescence was quantified for each condition. We observed a significant decrease in KCl-dependent CREB transcriptional output upon either TDP-43 OE or TDP-43 KD (Fig. 3), indicating that TDP-43 dysfunction indeed inhibits CREB activation and suppresses CREB transcriptional activity. Further, these results imply that TDP-43 dysregulation might specifically abrogate CREB activation through voltage gated Ca2+ channels, consistent with our TRIBE results.
Fig. 3.
KCl-dependent CREB transcriptional activity is suppressed by TDP-43 dysfunction. Normalized luciferase activity (firefly luciferase luminescence/Renilla luciferase luminescence) from cell lysates of cortical neurons transfected with an empty vector control, (A) TDP-43 OE, or (B) an shRNA targeting TDP-43 for KD. Yellow bars represent conditions where a subset of cortical neurons were exposed to 55 mM KCl for 6 h, and black bars represent cortical neurons that were not stimulated with KCl. To account for changes in transfection efficiency, firefly was normalized to Renilla for each condition. Each condition was then normalized to the “No treatment” control condition for each experiment. Each experiment was performed in triplicate and repeated at least three times. Data are presented as the mean of normalized luciferase activity. Two-way ANOVA with Tukey’s test. ****P < 0.0001. Error bars represent SEM.
Activation of CaMK Pathways Restores Dendritic Branching Defects Induced by TDP-43 Dysfunction.
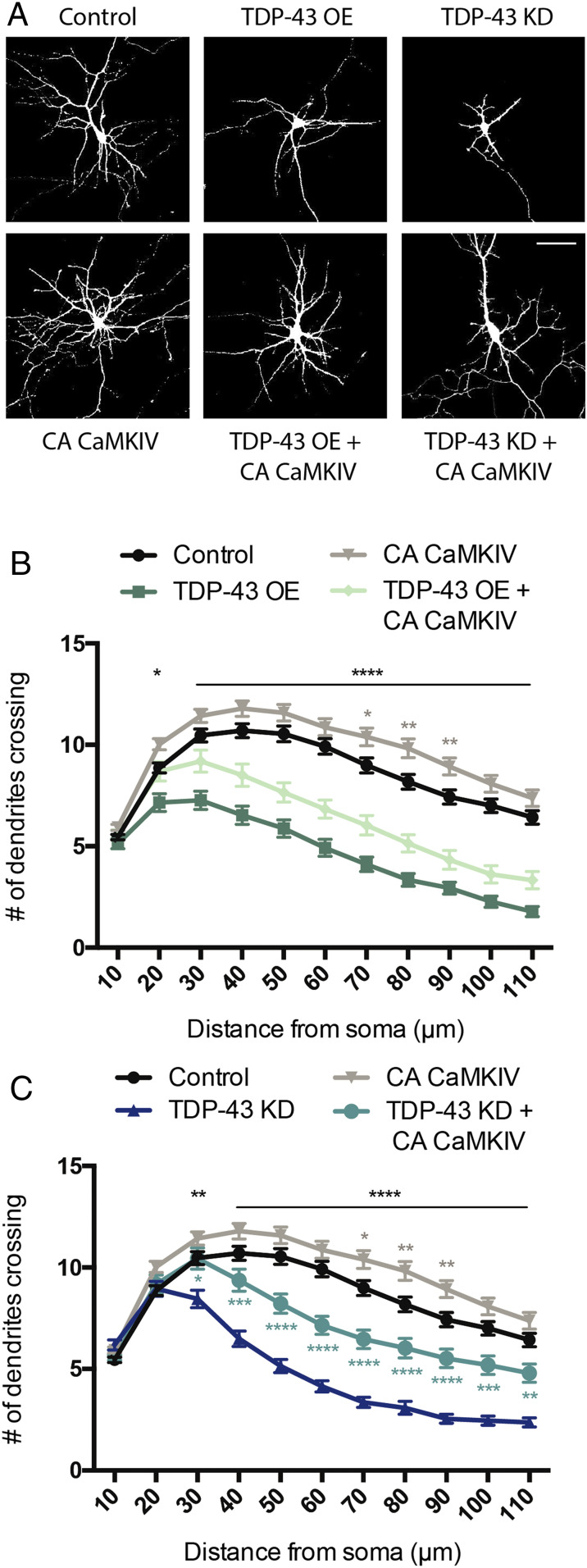
Calcium/calmodulin-dependent protein kinase IV (CaMKIV) stimulates dendritic growth via CREB phosphorylation at Ser133 and subsequent activation (24, 42). We therefore asked whether expression of a constitutively active form of CaMKIV (CaMKIV CA) (43, 44) would restore dendritic branching under conditions in which TDP-43 expression is increased or decreased. CaMKIV was made constitutively active by forcing nuclear localization via addition of a nuclear localization signal to the N terminus (45); expression of this construct in hippocampal neurons has previously been shown to increase dendritic complexity (42). Hippocampal neurons isolated from E18 rat pups were cocultured on a glial monolayer and transfected at 2 DIV with GFP and either an empty vector control or a plasmid expressing CaMKIV CA alone or in combination with appropriate constructs to achieve TDP-43 OE or TDP-43 KD (as described above). At 5 DIV, these neurons were fixed, imaged, and analyzed for dendritic complexity by Sholl analysis (46). Expression of CaMKIV CA alone increased dendritic complexity relative to control, consistent with previous reports (24, 42). We also observed decreased dendritic branching in both TDP-43 OE and TDP-43 KD conditions similar to previous reports (Fig. 4) (10, 47, 48).
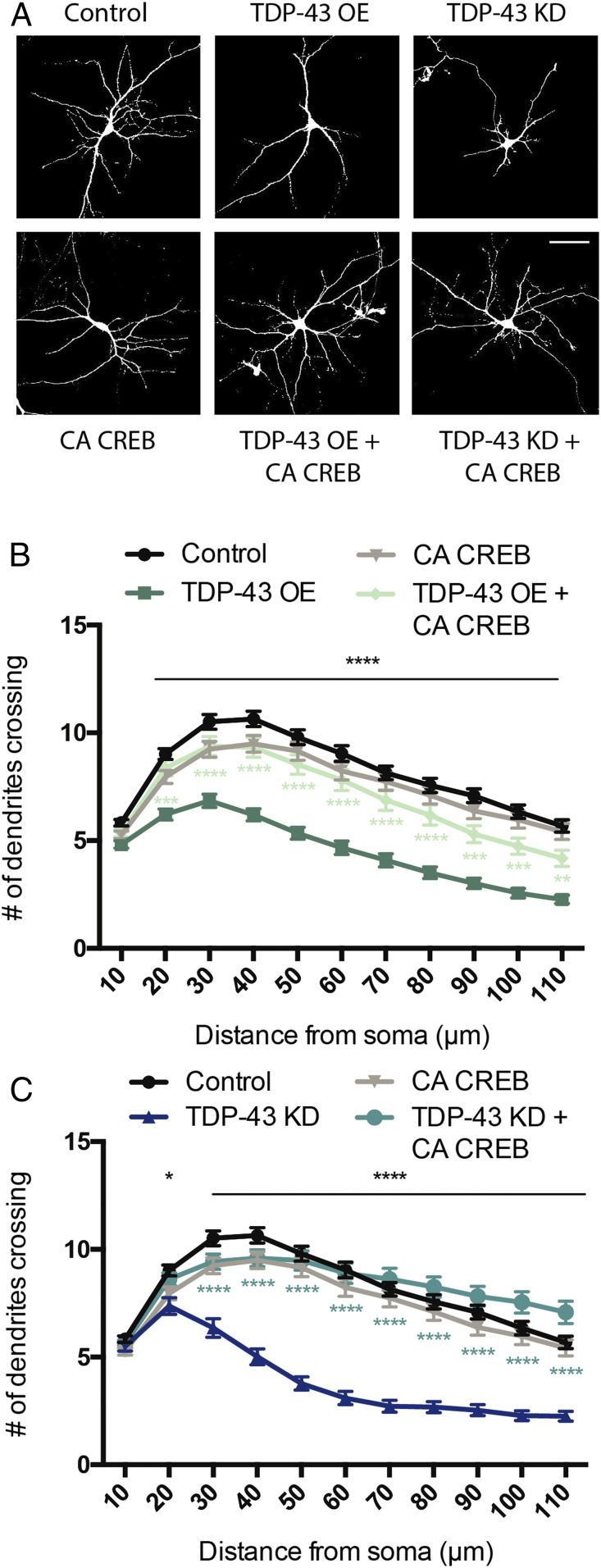
Fig. 4.
Constitutively active CaMKIV partially restores dendritic branching defects induced by TDP-43 dysfunction. (A) Representative images of 7 DIV cultured hippocampal neurons transfected with plasmids expressing GFP and either empty vector control, TDP-43 OE, or an shRNA targeting TDP-43 for KD (Upper). A similar set of experiments were performed in neurons cotransfected with CA CaMKIV (Lower). (Scale bar for all panels: 50 µm.) (B) Quantification of dendritic branching via Sholl analysis for each condition (Control, n = 164; TDP-43 OE = 64; CA CaMKIV = 157; TDP-43 OE + CA CAMKIV = 55). (C) Quantification of dendritic branching via Sholl analysis for each condition. Control and CA CaMKIV traces from B are replotted for comparison (Control, n = 164; TDP-43 KD = 82; CA CaMKIV = 157; TDP-43 KD + CA CaMKIV = 73). Two-way ANOVA with Tukey’s test. Black asterisks, control vs. TDP-43 OE or TDP-43 KD; gray asterisks, control vs. CA CaMKIV; turquoise asterisks, TDP-43 KD vs. TDP-43 KD + CA CaMKIV; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Error bars represent SEM.
We observed that expression of CaMKIV CA in neurons in which TDP-43 has been knocked down partially restored the reduction in dendritic branching (Fig. 4 A and C). In contrast, we found that expression of CaMKIV CA in neurons overexpressing TDP-43 did not have a significant effect on branching (Fig. 4 A and B). The fact that boosting CaMK signaling compensates for TDP-43 KD, but not TDP-43 OE, suggests that there are notable differences in upstream signaling pathways that are impacted by these manipulations.
As described above, our TRIBE experiment identified potential TDP-43 targets in multiple signaling pathways culminating in CREB activation. Since our observations indicated that both loss and gain of function of TDP-43 in cultured neurons results in a decrease in CREB activation and subsequent transcriptional output (Figs. 2 and 3), we sought to determine whether restoring CREB activity directly could rescue the defective dendritic morphology in both these conditions. To this end, we employed a CREB protein that was rendered constitutively active by fusing the VP16 transactivation domain from the Herpes Simplex Virus to full length CREB (CA CREB; ref. 26). Like the experiments described above with CaMKIV, hippocampal neurons were transfected at 2 DIV and fixed for morphological analysis at 5 DIV. Interestingly, the data show that expression of CA CREB in the context of both TDP-43 OE and TDP-43 KD restored dendritic complexity to near-normal levels (Fig. 5). Expression of CA CREB alone did not affect dendritic branching (Fig. 5), consistent with previous reports (24). These results indicate that changes in dendritic morphology resulting from dysfunction of TDP-43 can be rescued by restoring CREB transcriptional output.
Fig. 5.
Constitutively active CREB restores dendritic branching defects induced by TDP-43 dysfunction. (A) Representative images of 7 DIV cultured hippocampal neurons transfected with plasmids expressing GFP and either empty vector control, TDP-43 OE, or an shRNA targeting TDP-43 for KD (Upper). A similar set of experiments were performed in neurons cotransfected with CA CREB (Lower). (Scale barfor all panels: 50 µm.) (B) Quantification of dendritic branching via Sholl analysis for each condition (Control, n = 119; TDP-43 OE = 102; CA CREB = 111; TDP-43 OE + CA CREB = 70). (C) Quantification of dendritic branching via Sholl analysis for each condition. Control and CA CREB traces from B are replotted for comparison (Control, n = 119; TDP-43 KD = 71; CA CREB = 111; TDP-43 KD + CA CREB = 91). Two-way ANOVA with Tukey’s test. Black asterisks, control vs. TDP-43 OE or TDP-43 KD; light green asterisks, TDP-43 OE vs. TDP-43 OE + CA CREB; turquoise asterisks, TDP-43 KD vs. TDP-43 KD + CA CREB; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Error bars represent SEM.
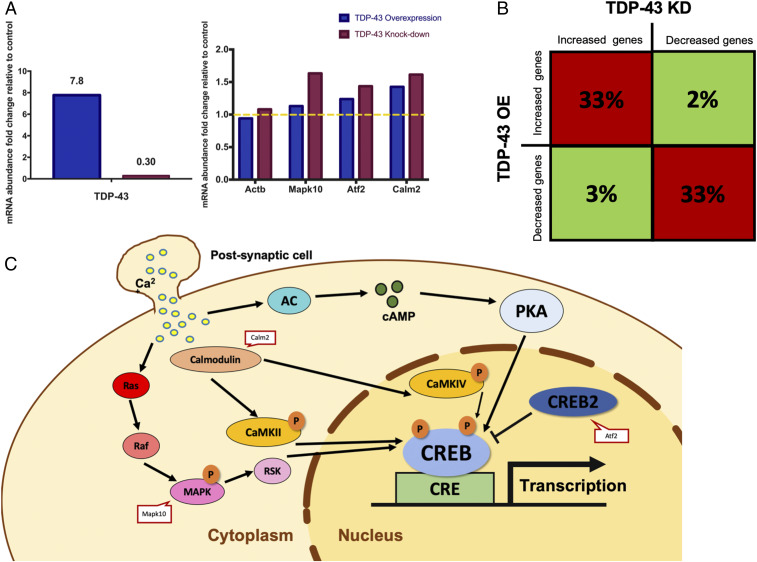
Finally, we sought to understand how TDP-43 dysfunction might affect levels of specific RNA transcripts, including those identified by TRIBE. We performed RNA-seq experiments using RNA isolated from the N2A mouse neuroblastoma cell line transfected with plasmids which cause TDP-43 OE or KD ; this neuronal cell line was chosen for its ease of culturing and high transfection efficiency with plasmid DNA. RNA-seq analysis revealed that TDP-43 RNA levels were strongly increased or decreased, indicating successful OE or KD, respectively (Fig. 6 A, Left). In contrast, transcriptome-wide RNA abundance changes were modest in both cases (Dataset S2), suggesting that the overall phenotypic effects of either TDP-43 OE or KD are caused by combinatorial changes in the expression of multiple genes.
Fig. 6.
Both TDP-43 OE and KD in cultured N2A cells cause similar changes in gene expression. (A) Bar graphs reflecting abundance of mRNAs in N2A cells after TDP-43 OE/KD relative to wild-type N2A cells transfected with empty vector. The y axis shows the fold change of the abundance of each mRNA relative to the vector control condition (FPKM). Left indicates the efficiency of TDP-43 OE/KD. Right includes several TDP-43 TRIBE hits related to CREB signaling pathway and the Actin-b gene as a negative control. Yellow dashed line represents no change in mRNA abundance. (B) The 1,000 genes that were most up-regulated or down-regulated with TDP-43 OE or KD were selected out of the 8,225 genes expressed (FPKM > 3) in N2A cells (also Dataset S2). Significantly more genes are misregulated in the same direction (i.e., increased or decreased) with TDP-43 OE and KD (33%, red blocks, Z test performed, P < 0.00001); few genes are misregulated in the opposite directions (2 or 3%, green blocks, Z test performed, P < 0.00001). The remaining 65% of the top 1,000 genes for a given condition (e.g., OE) are not in the top 1,000 genes for the other condition (e.g., KD). (C) Model of TDP-43 TRIBE hits related to CREB signaling pathway.
Despite the modest changes in expression levels, these data provide two important insights into the relationship between TDP-43 TRIBE targets and the cellular consequences of TDP-43 dysfunction. First, we found that several CREB-related, TDP-43 TRIBE target transcripts (Mapk10, Atf2, and Calm2) were more abundant in this dataset after both TDP-43 OE and KD (Fig. 6 A, Right and C). Second, many more of the top 1,000 up-regulated or down-regulated transcripts changed in the same direction in response to OE and KD, whereas many fewer transcripts changed in opposite directions (Fig. 6B). This result is consistent with the idea that TDP-43 OE sequesters TDP-43 into nonfunctional aggregates or granules, thus causing similar gene expression responses to TDP-43 KD. Further, these observations parallel and may help explain how both TDP-43 OE and KD decrease dendritic branching (10).
Discussion
TDP-43 is a key regulator of RNA metabolism and the central component in ALS and FTD pathology. Our previous data demonstrated that TDP-43 dysfunction disrupts normal dendritic morphology, and this effect is dependent on the RNA-binding ability of TDP-43 (10). In this paper, we applied the recently developed TRIBE method to a mammalian system and identified neuronal signaling pathways as targets of TDP-43. Based on the identity of the affected signaling pathways, we hypothesized and validated that CREB transcriptional activity is an important regulator of TDP-43−dependent changes in dendritic morphology. Activation of CREB alleviated the defects in dendritic morphology caused by TDP-43 dysfunction, thus validating our use of TRIBE to identify TDP-43 targets relevant to cellular function. In addition, TRIBE was recently performed for other mammalian RBPs (49, 50), indicating that TRIBE will have wide applicability.
Previous studies reported that TDP-43 associates with several thousand nascent and mature RNA transcripts in neurons (17–19). One group performed TDP-43 RIP-seq from cultured cortical neurons from rats, similar to our culture system (17), whereas the other used adult mouse brain to perform TDP-43 CLIP-seq (19). The overlap between our datasets strongly suggests that TDP-43 function is highly conserved in neurons.
Our TRIBE approach generated a shorter list of candidate targets than both of these other studies in addition to a high level of consistency between replicates (>50% of all editing sites). The discrepancy in the number of potential targets between these different techniques may be due, in part, to the fact that the RIP and CLIP studies assayed total TDP-43 RNA including pre-mRNA. Indeed, 2/3 of the CLIP targets have only intronic signal, and subtracting nuclear RNA further reduces the number of candidate CLIP TDP-43 target mRNAs to 842 (19), comparable to the number of candidate TRIBE TDP-43 target mRNAs identified here. We used polyA RNA in this study to specifically focus on mRNA targets, but we note that, in the future, TDP-43 TRIBE might be applied to nascent RNA targets (21, 51), potentially identifying new intronic targets of TDP-43 that may regulate cryptic splicing (52).
TRIBE target genes that are also identified with other methods are less likely to be false positives and may reflect RNAs bound at high affinity by the RBP (22, 34). By comparison, TRIBE is an antibody-independent method, and RIP and CLIP targets may include false positives due to intrinsic antibody issues (49). The shorter TRIBE gene list may also reflect more false negatives, due perhaps to inefficiencies of TRIBE as previously discussed (22).
In addition, there were 108 genes identified by TDP-43 TRIBE only (Fig. 1D). Because of our stringent requirement for reproducibility (the exact editing sites must be identified in both replicates), most of these genes are also unlikely to be false positives and may therefore reflect bona fide TDP-43 targets that escaped identification with the other methods. Notably, some of these genes, for example, MAPK8 and MAPK8IP1, are components of the top hit KEGG signaling pathways. In any case, the shorter candidate TRIBE list is certainly advantageous for target validation purposes.
This shorter TRIBE list also contrasts with the many thousands of target genes identified in previous TRIBE experiments with the RBP Hrp48 in Drosophila cultured S2 cells and in Drosophila neurons (22). This suggests that a general inefficiency of the TRIBE technique is unlikely to be a major explanation for the relatively small number of TDP-43 targets. Although the method could be less efficient in mammalian cells compared to Drosophila, Hrp48 is probably a more promiscuous RBP compared to TDP-43. Perhaps the most likely explanation is that the expression systems used in all previous Drosophila experiments employed substantial OE compared to endogenous protein levels and also compared to the lentiviral expression of the TDP-43−hADARcd fusion protein used in this study. To this point, our TRIBE data revealed that the level of TDP-43−hADARcd RNA was very similar to that of endogenous rat TDP-43 RNA.
We and others have shown that reduced dendritic branching results from TDP-43 dysfunction (10, 47, 48). Changes in dendritic arbors occur in a variety of neurological disorders, and morphological changes are known to precede neurodegeneration (13–15). Alterations in dendritic complexity may disrupt neuronal connectivity and cell-to-cell communication, and ultimately cause detrimental effects to cell health and survival. We were therefore curious whether the 532 TDP-43 targets identified by TRIBE functioned in specific signaling pathways that might impact dendritic morphology.
CREB is an activity-dependent transcription factor with a well-established role in regulating dendritic complexity (24–28), and DAVID GO term and KEGG pathway analyses revealed that the TDP-43 TRIBE target genes are significantly enriched for signaling pathways that converge on CREB activation. Importantly, CREB signaling and gene expression have previously been shown to be down-regulated in several neurodegenerative diseases (53, 54). Taken together with the 3′UTR preference of the TDP-43 TRIBE signals, our data suggest that TDP-43 posttranscriptional regulation of discrete targets is upstream of CREB function, which contributes to the dendritic branching defects of the pathologic state.
Our data demonstrate that coexpression of CA CREB with TDP-43 OE or TDP-43 KD fully rescued dendritic branching. In a complementary pathway, loss of TDP-43 in primary hippocampal neurons reduces surface expression of the growth factor receptor ErbB4 and impairs downstream signaling, also resulting in decreased dendritic complexity (48). In addition, dendritic branching can be restored in TDP-43–depleted neurons by overexpressing ErbB4 (48). Interestingly, our GO term analyses of TRIBE hits also identified enriched processes that establish neuronal communication at chemical synapses: synapse assembly and dendritic spine morphogenesis (Fig. 1). It is therefore possible that TDP-43 dysfunction disrupts dendritic morphogenesis processes directly, as well as indirectly, through signaling pathways that are engaged by synaptic transmission and, in turn, impact CREB activation. This may explain why CaMKIV CA expression can only partially rescue the dendritic complexity phenotype. Taken together, these data suggest that multiple signaling pathways that influence dendritic branching are affected by TDP-43 dysfunction.
We observed that the abundance of a significant portion of TDP-43 TRIBE targets (including the CREB-related targets Mapk10, Atf2, and Calm2) increased after both TDP-43 OE and KD in N2A cells. This finding supports our hypothesis that TDP-43 targets identified by TRIBE are dysregulated upon changes in TDP-43 levels. Furthermore, the mRNA abundance of many genes changed in the same direction upon TDP-43 OE or KD, consistent with the similar morphological defects in dendritic branching observed in TDP-43 OE/KD neurons. This finding supports the well-established idea that -overexpression of TDP-43 causes a loss-of-function phenotype, likely by sequestering TDP-43 in the cytoplasm in a nonfunctional state (8, 9). Further, the large number of RNAs with abundance changes is likely beyond the direct effect of TDP-43 dysfunction and, for example, could be secondary to misregulated signaling pathways like CREB.
In conclusion, our findings reveal a role for TDP-43 in the regulation of dendritic morphology via RNA targets that converge on dendritic branching, at least in part, by modulation of CREB transcriptional activity. Given that reduced dendritic complexity may precede neurodegeneration (14), a delay between the change in morphology and neuronal function serves as a potential window for therapeutic intervention. Although many early ALS studies focused on the morphological changes that happen at the distal axon in the peripheral nervous system (55), our data support a model that alterations of the dendritic arbor in the central nervous system may also contribute to disease pathogenesis. Indeed, ALS patient neurons display reduced dendrite morphologies in both upper and lower motor neurons which may lead to neurodegeneration (56, 57). Our work therefore suggests that reestablishing nervous system connectivity could be a therapeutic target in ALS/FTD.
Data Availability.
Both raw and processed next-generation RNA-seq data generated from this study were deposited at National Center for Biotechnology Information GEO database with accession number GSE147432.
Methods
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. All animal procedures were approved by the Brandeis University Institutional Animal Care and Usage Committee, and all experiments were performed in accordance with relevant guidelines and regulations.
Plasmids.
The pCMV vector was used to create pCMV-TRIBE constructs for HEK293T cell expression. The hTDP-43-hADARcd-E488Q, hTDP-43-5FL-hADARcd-E488Q, or hADARcd-E488Q was inserted into the vector by Gibson Assembly. The pCMV-TRIBE plasmids were individually transfected into HEK293T cells by standard calcium phosphate transfection. The pCMV-EGFP was cotransfected to monitor transfection efficiency.
The pLV-hSyn-RFP was a gift from Edward Callaway (Salk Institute for Biological Studies, La Jolla, CA) (Addgene plasmid #22909). The pLV-hSyn-RFP plasmid was linearized by cutting it with BamHI. A myc-hTDP-43-P2A cassette with complementary overhangs was synthesized using BioXP Custom cloning (Synthetic Genomics Inc.) and cloned in frame by Gibson Assembly. For TDP-43 TRIBE plasmids cloning, pLV-hSyn-RFP were cut with PmeI and AgeI. The hTDP-43-hADARcd-E488Q or hADARcd-E488Q fragment with complementary overhang was inserted into the digested backbone by Gibson Assembly. RFP remains in frame and serves as a marker for transfection efficiency.
Cell Culture and Transfection.
Twelve-millimeter glass coverslips were coated with poly-D-lysine (20 μg/mL) and laminin (3.4 μg/mL) in 24 well plates for 1 h to 24 h. Then the coverslips were washed three times with deionized H2O and two times with Neurobasal medium. Dissociated hippocampal neurons from E18 rat embryos were cultured on an astrocyte feeder layer, as described previously (58), at a density of 80,000 neurons per well (for Sholl analysis). Luciferase assays were performed with dissociated cortical neurons from E18 rat embryos without an astrocyte feeder layer. Cultured cortical neurons were plated at a density of 500,000 neurons per well. Both hippocampal and cortical cultures were grown in Neurobasal medium supplemented with B27 (Thermo Fisher) at 37 °C.
For Sholl analysis, neurons were transfected on DIV 2 by the calcium phosphate method (59), which transfects a low percentage of cells, allowing the genetically manipulated neuron to grow in an environment surrounded by unaffected or wild-type cells. This method is also ideal to visualize the dendritic morphology of individual cells in a dense population. For all experimental conditions, unless noted otherwise, neurons were cotransfected with pCMV-GFP plasmid at 500 ng per well to visualize neuronal morphology. Along with pCMV-GFP, neurons were either transfected with pCMV-TDP-43 (TDP-43 OE) at 500 ng per well or shRNAs expressed from pSuper targeting endogenous TDP-43 at 33 ng per well (TDP-43 KD). For coexpression experiments with constitutive active forms of CaMKIV and CREB, plasmids expressing these genes were transfected at 100 ng per well either alone or with TDP-43 OE or TDP-43 KD.
For luciferase assays, neurons were transfected on DIV 2 by the calcium phosphate method with either an empty vector control, TDP-43 OE, or TDP-43 KD at the same concentrations as mentioned above. In addition, each condition was cotransfected with reporter constructs containing the firefly luciferase gene downstream of concatemerized CREB binding sites at 500 ng per well. A separate reporter construct expressing Renilla luciferase being driven by the thymidine kinase promoter or the EF-1α promoter was transfected at 100 ng per well to serve as an internal control for normalizing transfection efficiency. On DIV 6, neurons were treated with 1 μM TTX to block sodium channels. On DIV 7, 55 mM KCl was added to a subset of neurons from each condition. After 6 h of KCl treatment, neurons were washed with 1× phosphate-buffered saline (PBS) and lysed with passive lysis buffer (Promega), and luciferase levels were measured using a dual luciferase assay. Firefly luciferase was measured by adding 100 μL of 75 mM Hepes, pH 8.0, 5 mM MgSO4, 20 mM dithiothreitol, 100 mM (ethylenedinitrilo)tetraacetic acid (EDTA), and 530 μM adenosine 5′-triphosphate, 0.5 mM d-luciferin, and 0.5 mM CoA. Renilla luciferase was measured using 100 μL of 25 mM Na4PPi, 10 mM NaOAc, 15 mM EDTA, 0.5 M Na2SO4, 1 M NaCl, and 0.1 mM Coelenterazine, pH 5.0. All conditions were repeated at least three times in triplicate and normalized to Renilla luciferase and then to the untreated control condition.
For TDP-43 TRIBE experiments, dissociated cortical neurons from E18 rat embryos were cultured in a six-well plate at 4 million neurons per well and grown in glia-conditioned Neurobasal medium supplemented with B27 at 37 °C. Neurons were transduced at DIV 5 with TDP-43 TRIBE virus (packaged by Vigene Biosciences, Inc.), and RNA was harvested 3 d later on DIV 7 (21, 34, 60).
For the RNA-seq experiment, N2A cells were cultured in a six-well plate at 250,000 cells per well and maintained at 37 °C and 5% CO2. Cells were cultured in Dulbecco’s modified Eagle’s medium (GE Healthcare) supplemented with 10% fetal bovine serum (GE Healthcare), 100 units per mL penicillin, 100 units per mL streptomycin, and 2 mm glutamine. When the cells reached ∼70% confluence, 2,500 ng of plasmid DNA (GFP along with an empty vector control, TDP-43 OE, or TDP-43 KD) was transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol.
RNA-seq Library Preparation and Sequencing.
Transfected HEK293T cells or neurons were harvested 3 d after transfection in TRIzolTM reagent (Thermo Fisher Scientific, 15596026). RNA was extracted as recommended by TRIzol product manual, dissolved in Milli-Q H2O, and stored in a −80 °C freezer. One to two micrograms of total RNA was poly(A) bead selected and used to prepare for mRNA-seq library with the Nextera DNA Flex Library Rapid Directional qRNA-Seq Kit (NEXTFLEX kit).
RNA from N2A cells was harvested 2 d after transfection using RNeasy isolation kit (Qiagen) and reverse transcribed using Random Primer Mix (New England BioLabs) following manufacturer’s protocol. Sequencing libraries were generated using illumina Nextera XT DNA library kit with 1-ng complementary DNA input.
All library product was quantified with Agilent 4200 TapeStation System and normalized to 2 nM for sequencing purpose. Libraries were pooled and then pair-end sequenced for 75 base pairs by NextSeq 500 System by Illumina (NextSeq High Output Kit v2, 75 cycles). Each library had ∼50 million pair-end reads.
RNA-seq Data Analysis.
RNA-seq data were analyzed as described in ref. 34, and scripts used are available on GitHub (https://github.com/rosbashlab/HyperTRIBE). Hg38 genome build was used for HEK293T cell data, and rn6 genome was used for rat neuron data. PCR duplication was removed using Unique Molecular Identifier (UMI) tools (61) which removes UMIs included in the NEXTFLEX kit. Sequencing data of RNA from wild-type HEK293T cells or untransfected rat neurons were used as the reference for calling RNA editing events. The criteria for RNA editing events were that 1) the nucleotide is covered by a minimum of 11 reads in each replicate, 2) more than 80% of wild-type RNA reads at this nucleotide are A with zero G (use the reverse complement if annotated gene is in the reverse strand), 3) a minimum of 5% G is observed at this site in mRNA (or C for the reverse strand), and 4) editing sites are present in both replicates. Distribution of editing sites in each mRNA region was determined using bedtools, intersecting editing sites with RefSeq annotated coding sequence (CDS), 5′UTR, and 3′UTR. Distribution of the transcriptome in these regions was calculated using RSeQC (http://rseqc.sourceforge.net/).
Gene expression profiles were generated by using cufflinks, and ranked by fragments per kilobase of transcript per million mapped reads (FPKM) value. The top 3,000 genes from RNA-seq of wild-type rat neurons were used as the background list for DAVID GO analysis.
Immunostaining.
Immunostaining was performed as described in ref. 10. The following antibodies were used: Anti-Phospho-CREB-Ser133 (87G3) (Rabbit mAb # 9198) from Cell Signaling Technologies (used at 1:500), Anti-CREB (Cat. No. 06-863) from Millipore Sigma (used at 1:100), and Anti-TFIIS (611204) from BD Transduction Laboratories (used at 1:275). Secondary antibodies were conjugated to Cy-3 or Cy-5 (1:500, Jackson ImmunoResearch Laboratories).
Neurons were imaged as described (10) using identical acquisition settings for laser power, exposure time, and detector gain across conditions. Data were acquired and analyzed in a blinded manner.
Image Analysis.
To quantify pCREB levels, sum projections were generated from individual Z stacks using ImageJ. In ImageJ, using GFP as a marker for transfected cells, a region of interest was drawn around the soma, and fluorescence intensity was measured in that area in the pCREB channel. Then, TFIIS fluorescence intensity was measured using the same region of interest. pCREB intensity was normalized to TFIIS intensity for every cell. The same procedure was used for the CREB antibody.
Analysis of Neuronal Morphology.
Transfected neurons were fixed at DIV 7 with 4% paraformaldehyde + 4% sucrose solution in PBS for 8 min at room temperature and washed three times with 1× PBS. An Olympus Fluoview 300 confocal microscope using a 20× oil objective (numerical aperture 0.85) was used to image GFP-transfected cells (5 to 10 steps at 1-μm optical sectioning). All images were analyzed in ImageJ using Sholl analysis where maximum intensity projections were generated from individual Z stacks. To quantify dendritic complexity, 11 concentric circles (10-μm intervals) centered at the soma were overlaid on the image, and the number of dendritic crossings was counted for each 10-µm interval. For each experimental condition, a total of 10 to 30 neurons were analyzed from two to three coverslips, and each experiment was independently repeated at least twice as a biological replicate. Imaging and analysis were performed in a blinded manner, and unblinded only after analysis.
Supplementary Material
Acknowledgments
We would like to thank members of the S.P., A.A.R., and M.R. labs for helpful comments and suggestions throughout the project. This work was supported by a Blazeman Foundation for ALS Research Postdoctoral Fellowship (M.D.) and by NIH grants R01NS103967 (A.R.), R01DA037721 (M.R), R01AG052465 (M.R.), and R01NS065856 (S.P.).
Footnotes
Competing interest statement: W.X. and M.R. declare that a PCT patent application (PCT patent application no. PCT/US2016/054525) has been filed on the TRIBE technique.
Data deposition: Both raw and processed next generation RNA sequencing data generated from this study were deposited at National Center for Biotechnology Information Gene Expression Omnibus database with accession number GSE147432.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917038117/-/DCSupplemental.
References
- 1.Sreedharan J. et al., TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovacs G. G. et al., TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov. Disord. 24, 1843–1847 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai T. et al., TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Neumann M. et al., Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Ling S. C., Polymenidou M., Cleveland D. W., Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasad A., Bharathi V., Sivalingam V., Girdhar A., Patel B. K., Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 12, 25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nussbacher J. K., Tabet R., Yeo G. W., Lagier-Tourenne C., Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron 102, 294–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birsa N., Bentham M. P., Fratta P., Cytoplasmic functions of TDP-43 and FUS and their role in ALS. Semin. Cell Dev. Biol. 99, 193–201 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Xu Z. S., Does a loss of TDP-43 function cause neurodegeneration? Mol. Neurodegener. 7, 27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herzog J. J., Deshpande M., Shapiro L., Rodal A. A., Paradis S., TDP-43 misexpression causes defects in dendritic growth. Sci. Rep. 7, 15656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart G. J., Spruston N., Dendritic integration: 60 years of progress. Nat. Neurosci. 18, 1713–1721 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Lefebvre J. L., Sanes J. R., Kay J. N., Development of dendritic form and function. Annu. Rev. Cell Dev. Biol. 31, 741–777 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Kulkarni V. A., Firestein B. L., The dendritic tree and brain disorders. Mol. Cell. Neurosci. 50, 10–20 (2012). [DOI] [PubMed] [Google Scholar]
- 14.López-Doménech G. et al., Loss of dendritic complexity precedes neurodegeneration in a mouse model with disrupted mitochondrial distribution in mature dendrites. Cell Rep. 17, 317–327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kweon J. H., Kim S., Lee S. B., The cellular basis of dendrite pathology in neurodegenerative diseases. BMB Rep. 50, 5–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong X., Shen K., Bülow H. E., Intrinsic and extrinsic mechanisms of dendritic morphogenesis. Annu. Rev. Physiol. 77, 271–300 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Sephton C. F. et al., Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 286, 1204–1215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tollervey J. R. et al., Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polymenidou M. et al., Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler E. C., Van Nostrand E. L., Yeo G. W., Advances and challenges in the detection of transcriptome-wide protein-RNA interactions. Wiley Interdiscip. Rev. RNA 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon A. C. et al., TRIBE: Hijacking an RNA-editing enzyme to identify cell-specific targets of RNA-binding proteins. Cell 165, 742–753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu W., Rahman R., Rosbash M., Mechanistic implications of enhanced editing by a HyperTRIBE RNA-binding protein. RNA 24, 173–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberini C. M., Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 89, 121–145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redmond L., Kashani A. H., Ghosh A., Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron 34, 999–1010 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Watson F. L. et al., Neurotrophins use the Erk5 pathway to mediate a retrograde survival response. Nat. Neurosci. 4, 981–988 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Tao X., Finkbeiner S., Arnold D. B., Shaywitz A. J., Greenberg M. E., Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20, 709–726 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Tan Y. et al., FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15, 4629–4642 (1996). [PMC free article] [PubMed] [Google Scholar]
- 28.Alboni S. et al., Stress induces altered CRE/CREB pathway activity and BDNF expression in the hippocampus of glucocorticoid receptor-impaired mice. Neuropharmacology 60, 1337–1346 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Cohen S., Greenberg M. E., Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu. Rev. Cell Dev. Biol. 24, 183–209 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mili S., Steitz J. A., Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA 10, 1692–1694 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darnell R. B., HITS-CLIP: Panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip. Rev. RNA 1, 266–286 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herzog J., Xu W., Rosbash M., Paradis S., TDP-43 dysfunction restricts dendritic complexity by inhibiting CREB activation and altering gene expression. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147432. Deposited 23 March 2020. [DOI] [PMC free article] [PubMed]
- 33.Baskaran P., Shaw C., Guthrie S., TDP-43 causes neurotoxicity and cytoskeletal dysfunction in primary cortical neurons. PLoS One 13, e0196528 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Rahman R., Xu W., Jin H., Rosbash M., Identification of RNA-binding protein targets with HyperTRIBE. Nat. Protoc. 13, 1829–1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y., Arslan P. E., Won A., Yip C. M., Chakrabartty A., Binding of TDP-43 to the 3’UTR of its cognate mRNA enhances its solubility. Biochemistry 53, 5885–5894 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Fukushima M., Hosoda N., Chifu K., Hoshino S. I., TDP-43 accelerates deadenylation of target mRNAs by recruiting Caf1 deadenylase. FEBS Lett. 593, 277–287 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Tank E. M. et al., Abnormal RNA stability in amyotrophic lateral sclerosis. Nat. Commun. 9, 2845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neelagandan N. et al., TDP-43 enhances translation of specific mRNAs linked to neurodegenerative disease. Nucleic Acids Res. 47, 341–361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaywitz A. J., Greenberg M. E., CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68, 821–861 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Fletcher T. L., De Camilli P., Banker G., Synaptogenesis in hippocampal cultures: Evidence indicating that axons and dendrites become competent to form synapses at different stages of neuronal development. J. Neurosci. 14, 6695–6706 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wind M., Reines D., Transcription elongation factor SII. BioEssays 22, 327–336 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghiretti A. E., Kenny K., Marr M. T. 2nd, Paradis S., CaMKII-dependent phosphorylation of the GTPase Rem2 is required to restrict dendritic complexity. J. Neurosci. 33, 6504–6515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wayman G. A. et al., Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron 50, 897–909 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Wayman G. A., Lee Y. S., Tokumitsu H., Silva A. J., Soderling T. R., Calmodulin-kinases: Modulators of neuronal development and plasticity. Neuron 59, 914–931 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt J. M., Wayman G. A., Nozaki N., Soderling T. R., Calcium activation of ERK mediated by calmodulin kinase I. J. Biol. Chem. 279, 24064–24072 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Sholl D. A., Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 87, 387–406 (1953). [PMC free article] [PubMed] [Google Scholar]
- 47.Majumder P. et al., TDP-43 regulates the mammalian spinogenesis through translational repression of Rac1. Acta Neuropathol. 124, 231–245 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Schwenk B. M. et al., TDP-43 loss of function inhibits endosomal trafficking and alters trophic signaling in neurons. EMBO J. 35, 2350–2370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biswas J., Rahman R., Gupta V., Rosbash M., Singer R. H., MS2-TRIBE evaluates protein-RNA interactions and nuclear organization of transcription by RNA editing. bioRxiv:829606 (4 November 2019). [DOI] [PMC free article] [PubMed]
- 50.Jin H., et al. , TRIBE editing reveals specific mRNA targets of eIF4E-BP in Drosophila and in mammals. bioRxiv:2020.02.24.962852 (25 February 2020). [DOI] [PMC free article] [PubMed]
- 51.Menet J. S., Rodriguez J., Abruzzi K. C., Rosbash M., Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife 1, e00011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ling J. P., Pletnikova O., Troncoso J. C., Wong P. C., TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349, 650–655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mantamadiotis T. et al., Disruption of CREB function in brain leads to neurodegeneration. Nat. Genet. 31, 47–54 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Saura C. A., Valero J., The role of CREB signaling in Alzheimer’s disease and other cognitive disorders. Rev. Neurosci. 22, 153–169 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Chou S. M., Norris F. H., Amyotrophic lateral sclerosis: Lower motor neuron disease spreading to upper motor neurons. Muscle Nerve 16, 864–869 (1993). [DOI] [PubMed] [Google Scholar]
- 56.Genç B. et al., Apical dendrite degeneration, a novel cellular pathology for Betz cells in ALS. Sci. Rep. 7, 41765 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takeda T. et al., Dendritic retraction, but not atrophy, is consistent in amyotrophic lateral sclerosis-comparison between Onuf’s neurons and other sacral motor neurons. Acta Neuropathol. Commun. 2, 11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghiretti A. E., Paradis S., The GTPase Rem2 regulates synapse development and dendritic morphology. Dev. Neurobiol. 71, 374–389 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia Z., Dudek H., Miranti C. K., Greenberg M. E., Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J. Neurosci. 16, 5425–5436 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez J., Menet J. S., Rosbash M., Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol. Cell 47, 27–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith T., Heger A., Sudbery I., UMI-tools: Modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Both raw and processed next-generation RNA-seq data generated from this study were deposited at National Center for Biotechnology Information GEO database with accession number GSE147432.