Significance
Although recent advances in next-generation sequencing have facilitated the construction of whole genomes from hundreds of organisms, considerable barriers still restrict the functional understanding of the genes that they contain. An initial prerequisite for such an understanding is the availability of high-quality gene libraries (ORFeomes) amenable to high-throughput functional experiments. Here we develop a massively parallel next-generation sequencing method, PLATE-seq, and leverage the method to construct an ORFeome for rice, providing a toolkit for systematic functional study in an agricultural species. To demonstrate the utility of these resources, we present a map of rice protein–protein interactions.
Keywords: protein–protein interaction, rice, ORFeome, next-generation sequencing, networks
Abstract
Systematic mappings of protein interactome networks have provided invaluable functional information for numerous model organisms. Here we develop PCR-mediated Linkage of barcoded Adapters To nucleic acid Elements for sequencing (PLATE-seq) that serves as a general tool to rapidly sequence thousands of DNA elements. We validate its utility by generating the ORFeome for Oryza sativa covering 2,300 genes and constructing a high-quality protein–protein interactome map consisting of 322 interactions between 289 proteins, expanding the known interactions in rice by roughly 50%. Our work paves the way for high-throughput profiling of protein–protein interactions in a wide range of organisms.
The genomics revolution has democratized sequencing and structural annotation of genomes; however, assigning functions to predicted genes remains an important unsolved challenge. Identification of protein–protein interactions can help advance functional annotation in sequenced genomes. The first step in systematic, genome-wide mapping of protein–protein interactions involves the construction of a comprehensive set of high-quality open reading frames (ORFs). To generate such ORFeomes, tens of thousands of clones must be sequenced to ensure that only correct, full-length clones are retained. Traditionally, this is achieved through the labor-intensive and cost-prohibitive process of Sanger sequencing of each individual clone. Simple barcoding strategies that append a barcode at the beginning or end of each ORF are problematic because they require thousands of unique barcodes, provide extremely limited ORF coverage, and generate a high fraction of uninformative reads that contain no barcode (SI Appendix, Fig. S1A). A deep-well-pooling approach has recently been used to sequence ORFeome libraries (1, 2); however, these smart pooling approaches cannot accommodate the inclusion of homologous ORFs in one pool, rely on concrete prior knowledge regarding plate layout, and cannot detect potential cross-contamination between wells (SI Appendix, Fig. S1 C and D).
Here, we develop a massively parallel sequencing approach called PCR-mediated Linkage of barcoded Adapters To nucleic acid Elements for sequencing (PLATE-seq)—a broadly utilizable approach for rapid sequencing of thousands of DNA elements. We validate the utility of PLATE-seq by developing an ORFeome for rice and constructing a high-quality, experimentally validated protein–protein interactome map of this important monocot species. Despite being a staple food for over half of the world’s population, and an important model for monocot genomics, empirical annotations currently cover fewer than 5% of genes in the rice genome (3). Indeed, understanding the function of plant genes has become a major bottleneck for the field of plant biology as a whole.
The majority of plant functional studies have been carried out in the dicot model organism Arabidopsis thaliana (4, 5), and, to date, only limited characterization has been performed in monocot species. Combating this disparity in annotation, the development of a comprehensive, high-quality ORFeome for Oryza sativa would enable large-scale reverse-proteomics studies—including the systematic mapping of protein–protein interactions—and thus would significantly expand the functional genomics toolkit for plants.
A full-length complementary DNA (FLcDNA) clone library has previously been reported in O. sativa (6); however, such libraries are not suitable for high-throughput studies. Specifically, FLcDNA clones contain 5′ and 3′ untranslated regions and therefore are not amenable to C- and N-terminal tagging required for most functional studies (e.g., yeast two-hybrid [Y2H]). Furthermore, these clones were derived from pools of clones rather than single colonies—resulting in contamination of up to 80% of clones. While comprehensive Gateway-compatible ORFeomes amenable to high-throughput cloning and expression analysis have been extensively utilized in model organisms (2, 4, 7–10), that of the dicot species, A. thaliana, is currently the only ORFeome available for any plant. Although a handful of functional studies have been completed in rice by first cloning proteins of interest (11, 12), the lack of a unified ORFeome is a serious constraint to further advances.
In this study, we produce a fully sequenced, single-colony-derived, Gateway-cloning-compatible ORFeome for the monocot species, rice. Adapting a proven Y2H screening approach (13–15), we leverage the power of PLATE-seq and the ORFeome to systematically generate a high-quality rice protein–protein interaction network. Our work—while contributing a pipeline broadly useful for the biological community—expands the known map of the rice interactome, paves the way for future high-throughput rice biology studies, and provides a systematic characterization of a monocot genome, an internationally important crop species, and a model organism.
Results and Discussion
PLATE-Seq Achieves Robust Parallel Identification of ORFs Comparable to Sanger Sequencing.
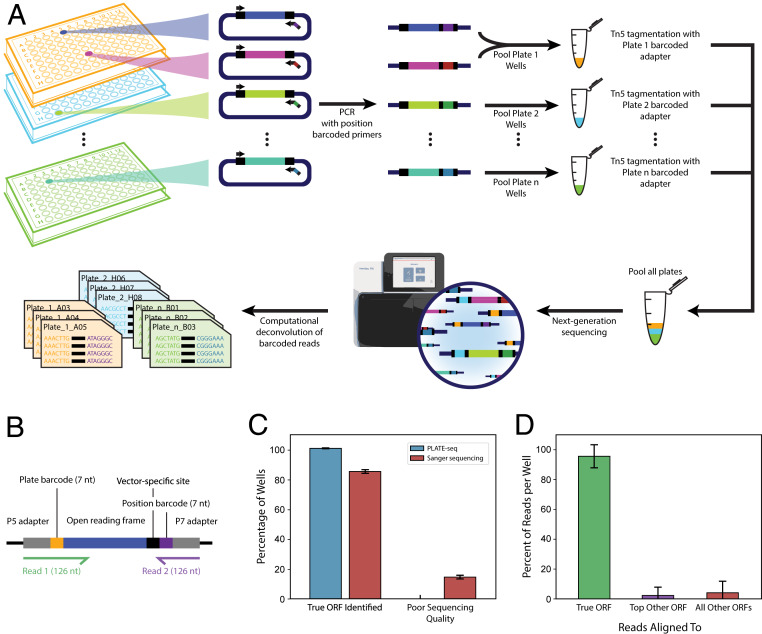
To facilitate the development of the rice and future ORFeomes, we developed PLATE-seq, a massively parallel barcoded sequencing approach to validate identities and locations within complex DNA libraries. Our PLATE-seq methodology (Fig. 1A) begins with a library of either single or pooled clones arrayed across a 96-well format. Individual PCR amplifications in each well append a unique position-specific barcode and the primary TruSeq sequencing adapter to the DNA product. Samples are then pooled together on a per-plate basis and tagmented by Tn5 transposase. The tagmentation reaction inserts a unique plate-specific barcode and the secondary TruSeq sequencing adapter at a random location within the ORF. Next, universal primers are used in a low-cycle PCR to enrich for clone fragments containing both position- and plate-specific barcodes (Fig. 1B and Dataset S1). Finally, amplicons from all plates are pooled together and subjected to massively parallel Illumina sequencing. The paired-end sequencing setup generates R2 reads just long enough to span the position-specific barcode and R1 reads maximized to span both the plate-specific barcode and the ORF sequence. Notably, the final low-cycle PCR enrichment ensures that all paired reads contain both barcodes and can be informatively mapped back to their exact source well. Moreover, because Tn5 tagmentation acts at a random position within the ORF, the fragments generated can theoretically span the entire ORF. However, in practice, cluster formation and amplification using current Illumina sequencers become inefficient for prohibitively large fragments. Therefore, we optimized our methods to provide coverage of roughly the last 800 to 1,000 bp of each ORF (SI Appendix, Fig. S1B).
Fig. 1.
A massively parallel approach to comprehensively index DNA libraries. (A) A schematic illustration of the PLATE-seq pipeline. (B) Barcoding design of PLATE-seq output products. (C) Fraction of ORFs in the human positive control plate that could be correctly identified by either PLATE-seq or Sanger sequencing (n = 94). Data are shown as ±SD. (D) Fraction of PLATE-seq reads mapping to true human positive control ORFs or other ORFs (n = 94). Data are shown as ±SD.
In order to benchmark the accuracy of PLATE-seq, we implemented the method on a test plate of 94 human ORF clones selected from the sequence-verified human ORFeome 8.1 library (2), demonstrating that PLATE-seq correctly identified the true ORF in 100% of the test cases (Fig. 1C). Determination of clone identity for each well was made by aligning the reads to the entire 8.1 reference library to calculate the fraction of reads in each well contributed by each ORF. Importantly, the vast majority of PLATE-seq reads aligned to the true ORF with only a minor fraction aligning to an incorrect ORF introduced through experimental contamination or alignment ambiguities (Fig. 1D). Moreover, the ability of PLATE-seq to detect these minor artifacts demonstrates that it possesses the resolution to discern the relative abundances of multiple clones in a pooled setup. By contrast, Sanger sequencing is ill-suited to handle a pooled setup or resolve contamination errors. Consequently, resequencing by Sanger was more prone to sequencing quality failures and was unable to fully reconfirm the identities of all control clones in one attempt (Fig. 1C).
Although our applications of PLATE-seq were limited to determination of the identity of the ORF(s) in each well, we note that the applications could be extended beyond these. If the reference sequences for the clones being sequenced were unknown, de novo sequence assembly could be applied to each set of reads after location deconvolution. To demonstrate this, we input reads from one of the wells of our human control plate into a contig assembly script. The pairwise alignment between our PLATE-seq–reconstructed sequence, the Sanger Sequencing result from the same clone, and the true sequence of the human ORF (BIRC7) is shown (SI Appendix, Fig. S2A). Our reconstructed sequence perfectly matched the true BIRC7 clone sequence; the one mismatch recapitulated a synonymous single-nucleotide polymorphism (SNP) that was reported for the BIRC7 clone when the human ORFeome 8.1 library was initially released (2). By contrast, the Sanger Sequencing result achieved only partial coverage and included many sequencing errors (SI Appendix, Fig. S2B). Although it may be possible through repeated trials to achieve Sanger Sequencing results of equal quality to our PLATE-seq reconstructed sequence, we emphasize that PLATE-seq sequence reconstruction can be applied simultaneously for hundreds of clones from one round of sequencing. Our ability to detect the C882T variant highlights further potential outside of de novo sequence reconstruction. By replacing the final step of PLATE-seq with a variant caller, it is possible to identify and uniquely assign SNPs among hundreds of copies of the same gene. To demonstrate this, we identified the same C882T variant in BIRC7 this time by aligning all genes to the known BIRC7 references and applying a variant caller to the read pileup (SI Appendix, Fig. S2D). We note that using PLATE-seq to call SNPs from a known reference would be more scalable than sequence reconstruction since there would be less need for robust sequencing depth.
A Draft Rice ORFeome Captures 2,300 Rice Genes Across a Diverse Functional Spectrum.
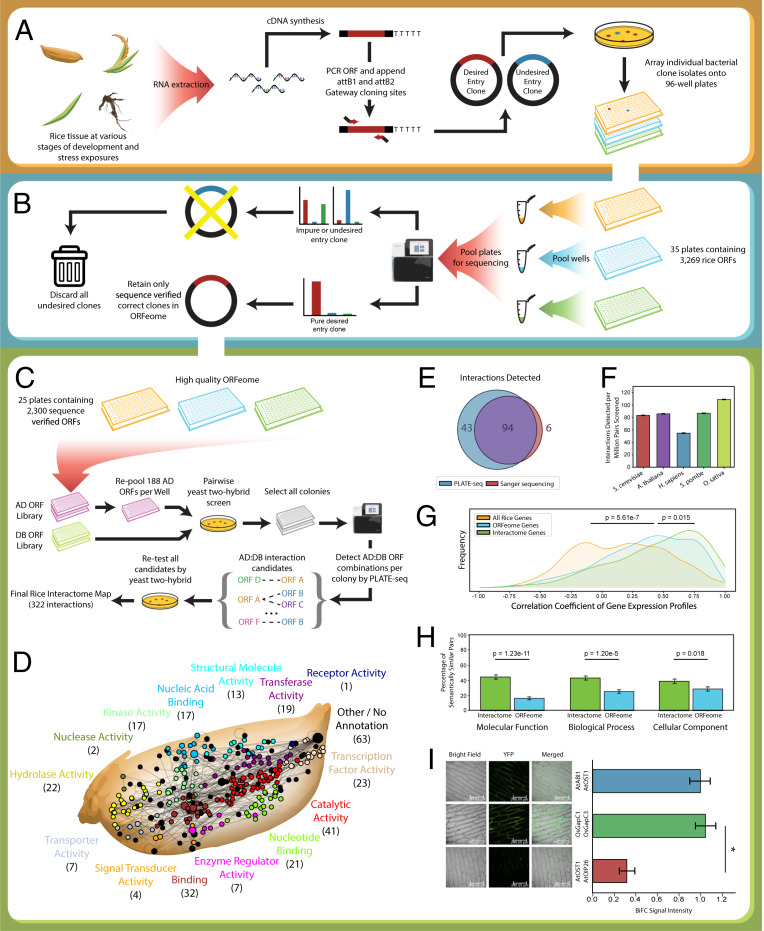
Having demonstrated PLATE-seq’s capacity for precise parallel determination of the identities and exact locations of clones within a complex library, we next sought to systematically construct and sequence-verify a first version of the rice ORFeome. To reduce the daunting scale of the full O. sativa genome, we initially used RiceNet (16, 17) to prioritize 3,269 genes predicted to be most closely associated with a seed set of 89 genes (Dataset S2) that had previously been linked to biotic or abiotic stress tolerance—either through experimental validation or association with validated stress tolerance genes (16, 18). To ensure maximum recovery of these genes, we designed primers for one representative ORF from each gene (Dataset S3) and amplified them using complementary DNA (cDNA) obtained from 40 combinations of developmental stages and stress exposures (Fig. 2A and Dataset S4). All cloning was carried out by Gateway recombination-based cloning to enable versatility for downstream cloning into various Gateway-compatible expression vectors for functional studies. To prevent contamination of the rice ORFeome with unwanted cloning by-products (e.g., PCR artifacts), we picked two single colonies per ORF, determined the true identity of every isolate by PLATE-seq, and eliminated any clones that did not align with the intended sequence (Fig. 2B). As a secondary check that only full-length clones were retained, approximate clone lengths were verified by gel electrophoresis (SI Appendix, Fig. S3).
Fig. 2.
A high-quality ORFeome and binary protein interactome in O. sativa. (A) A schematic illustration of the construction of the original rice ORF library. (B) The raw ORF library was sequence-verified by PLATE-seq, and only correct clones were retained. (C) Massively parallel Y2H screening was performed to detect putative Y2H-positive interactors, and each interaction was verified by pairwise retesting. (D) Network representation of the full rice interactome spanning 322 interactions across varied functional annotations. (E) Comparison of the successful Y2H-positive interactor detection rate across eight plates of putative interactors using either PLATE-seq or Sanger sequencing. (F) Comparison of the detection rates from previous high-throughput Y2H interactome screens to our rice interactome. Data are shown as ±SE. (G) Comparison of the distributions of gene coexpression between random rice gene pairs, random pairs sampled from our ORFeome, or our interactome pairs (n = 322). Coexpression is reported as Spearman rank correlation coefficients between gene expressions from 11 different rice tissue samples. Expression values were significantly more correlated among interactome pairs compared to random ORFeome pairs (P = 0.015 by two-sided Kolmogorov–Smirnov test). However, both interactome pairs and ORFeome pairs were significantly more often coexpressed than random genome pairs (P value = 5.61e−7 by two-sided Kolmogorov–Smirnov test). (H) Comparison of the fraction of detected interactions vs. random ORFeome pairs that share similar molecular function (MF), biological processes (BP), or cellular component (CC) gene ontology annotations. Similar GO annotation is defined as a semantic similarity score ≥ 0.75 as reported by GOssTo. Detected interactions were significantly more likely to be similarly annotated among all three classifications (MF: n = 236, P = 1.23e−11; BP: n = 254, P = 1.20e−5; CC: n = 206, P = 0.018; all tests are one-tailed Fisher’s exact test). Interactions lacking annotation for a specific GO term were excluded from each category. Data are shown as ±SE. (I) Representative BiFC confocal fluorescence images for the positive control (AtABI1-AtOST1), for the rice protein pair encoded by LOC_Os08g03290 (OsGapC1) and LOC_Os02g38920 (OsGapC3), and for negative control (AtOST1-AtOIP26). Average ratios of BiFC signals relative to the AtABI1-AtOST1–positive control. Data are shown as ±SD. Asterisk (*) denotes significance (P < 0.001) as ascertained by two-tailed t test.
In total, our final sequence-verified ORFeome provides one representative ORF clone for each of 2,300 rice genes sampled throughout the O. sativa genome (SI Appendix, Fig. S4A and Dataset S5). Analysis of the final composition of our ORFeome showed some evidence that the success rate of our cloning method was higher for certain genes. For example, although the distribution of gene lengths was similar to that of the entire O. sativa genome and prioritized gene set, it did show bias toward shorter genes (SI Appendix, Fig. S4B). Moreover, we observed disproportionate representation of highly expressed genes. Although this shift was introduced within our initial prioritized set of genes, it was further exacerbated in our final ORFeome (SI Appendix, Fig. S4C). These skews are consistent with known consequences of PCR amplification bias (19) and suggest that greater effort may be required in the future when cloning long or lowly expressed genes. Nonetheless, our ORFeome captures a wide diversity of biological processes broadly representative of the functional distributions over the entire O. sativa genome (SI Appendix, Fig. S4 D–F). We note that we do not observe extensive evidence of functional bias within our ORFeome despite the fact that the seed genes used for gene prioritization came from a specific functional study (18). High-scoring RiceNet predictions should accurately capture true functional associations with our seed set, and on their own may have contributed a functional bias. However, because we used a low confidence threshold when prioritizing genes to clone, a large number of less precise predictions may have counteracted this. The clear exception to this came from one of our seed genes, a 60S ribosomal protein L14 (LOC_Os02g40880), which contributed an enrichment for additional ribosomal proteins to our ORFeome (SI Appendix, Fig. S4E). Compared to the other seed genes, this ribosomal gene was a hub for functionally related true interactors that could be predicted with high confidence owing to the high degree of annotation transfer for this highly conserved complex available from other organisms. Thus, despite minor experimental limits, our fully validated rice ORFeome represents a wide and largely unbiased functional spectrum of the O. sativa genome.
A Systematic Yeast Two-Hybrid Screen Reveals 322 Rice Protein–Protein Interactions.
Because proteins function primarily by physically interacting with each other (20–22), protein interactome networks provide a crucial resource to discover functions associated with protein-coding genes. To date, these interactome maps have been pivotal in uncovering functional relationships between proteins in a wide variety of organisms (5, 15, 23, 24). Although several resources have applied homology-based annotation transfer to predict O. sativa protein–protein interactions (16, 17, 25–27), a large-scale experimental survey is yet to materialize. A tandem affinity purification method has been employed to detect rice kinase complex associations (11, 28), and a few Y2H studies have probed other specific functional subcomponents of the interactome (12, 18, 29). However, these Y2H studies have relied on cDNA libraries that generally produce lower-quality Y2H interactome mapping compared to sequence-verified, full-length ORFeome clone libraries (14). In order to provide a large-scale, experimentally validated rice interactome map, we tested all pairwise combinations of Y2H-amenable proteins encoded by our rice ORFeome (1,671 × 1,671 ∼2.7 million protein–protein pairs tested) using the same high-throughput Y2H assay that we previously used to generate the budding yeast, human, and fission yeast interactome networks (14, 15, 30) (Fig. 2C). Previous high-throughput Y2H screening-sequencing approaches identified interaction candidates by screening 188 activating domain (AD) ORFs against one DNA binding domain (DB) ORF at a time but were subject to a bottleneck because each positive colony must be sequenced individually to determine the AD interactors (13–15). Leveraging PLATE-seq we were able to uncover all protein-pair interaction candidates from our Y2H screen in one sequencing step and, subsequently, validate them by pairwise Y2H retest. Our full workflow resulted in a high-quality rice protein interactome network consisting of 322 high-quality interactions between 289 rice proteins (Fig. 2D and Dataset S6) across a wide span of molecular processes and cellular localizations (SI Appendix, Fig. S4 D–F). Notably, sequencing by PLATE-seq boasted higher identification of truly interacting protein pairs when compared to Sanger sequencing (Fig. 2E), and our overall detection rate was comparable to previous Y2H interactome screens (Fig. 2F and SI Appendix, Fig. S5).
To support the biological relevance of our network, we analyzed the quality of our rice interactome map based on the functional relationship between interacting proteins compared to noninteracting proteins. For a physiologically relevant protein–protein interaction to occur, the corresponding genes must be expressed under similar spatiotemporal conditions. We demonstrate that interacting genes exhibit higher coexpression compared to random gene pairs selected from our ORFeome or the entire rice genome (Fig. 2G). However, we do note that gene coexpression is one of the features used in the RiceNet predictions that we used to prioritize genes for inclusion in our first draft ORFeome. As a consequence of this selection, all pairs within the ORFeome were already significantly coexpressed. We additionally note that our ORFeome captures a high proportion of highly conserved genes (SI Appendix, Fig. S6), potentially also as a consequence of RiceNet prioritization since such genes may borrow evidence from homologs in other organisms. While these caveats must be considered when interpreting our analyses, the high conservation rate among interacting genes highlights the broad applicability of our interactome for high-confidence annotation transfer to other plant species. Because biological pathways involve protein–protein interactions, we also expect interacting protein pairs to be enriched in similar functional annotations. We show that, compared to a random sampling of protein pairs from our ORFeome, our interactome map contains a significantly higher proportion of similarly annotated protein pairs across all classifications of gene ontology (GO) terms (Fig. 2H). Finally, to demonstrate the robustness and accuracy of our Y2H approach, we validated a subset of our interactions through an orthogonal assay. We performed bimolecular fluorescence complementation (BiFC) on a random subset of seven interactions. Six of the seven interactions (85.7%) were robustly recapitulated (Dataset S7). As a representative example, we confirmed an interaction between two predicted glyceraldehyde-3-phosphate dehydrogenases, LOC_Os08g03290 (OsGapC1) and LOC_Os02g38920 (OsGapC3) (Fig. 2I and SI Appendix, Fig. S7).
Our Rice Interactome Map Increases the Current Literature Interactome Map by 50% and Uncovers Conserved Interactions.
Finally, we compared our reported interactions to the previous literature. Using a curated set of high-quality binary protein–protein interactions (31) compiled from seven primary interaction databases—BioGRID (32), Molecular Interaction Database (33), iRefWeb (34), Database of Interacting Proteins (35), IntAct (36), Human Protein Reference Database (37, 38), Munich Information Center for Protein Sequences (39), and the Protein Data Bank (40, 41)—we uncovered 237 interactions in O. sativa. We supplemented this set with an additional 372 interactions from a high-throughput Y2H rice-kinase interactome screen (12) for a total of 609 previously reported interactions. Notably, our additions to the rice interactome map cover a unique search space; among literature interactions only seven were recapitulated by our screen and only about 5% could have theoretically been recapitulated from our ORFeome using an 80% sequence identify cutoff (SI Appendix, Fig. S8 A and B). Two of our interactions—one between Elongation factor 1 delta (LOC_Os07g42300) and Elongation factor 1 beta (LOC_Os07g46750), another between a DUF851 domain-containing protein (LOC_Os04g49660) and serine/threonine protein kinases OSK4 (LOC_Os08g37800)—showed near exact sequence identity to a previously reported interaction. Thus, our network greatly expands the known rice protein interactome. We repeat this analysis for the interactomes from four additional organisms (SI Appendix, Fig. S8 C–F). For the distantly related organisms—yeast, human, and Escherichia coli—we note that, for nearly all rice interactions where both interacting proteins had a homolog in the other organism, a homologous interaction was in fact reported, potentially suggesting sampling from a core interactome the functionality of which is tightly conserved across species. In A. thaliana, by contrast, although more conserved interactions were detected, there was a larger discrepancy between the number of rice interactions that could have been detected using Arabidopsis homologs and the number homologous interactions between those homologs that actually were reported. This may indicate some interaction rewiring between the fringe components of the interactomes of rice and Arabidopsis. However, we emphasize that at the time our rice interactome map is not large enough to allow any statistically meaningful interpretation of the interactome conservation between species.
To explore the implications of conserved interactions further, we conducted a manual literature search of the top 20 most highly coexpressed interacting genes (Spearman's rank correlation coefficient [SCC] ≥ 0.8), which yielded evidence supporting the existence of heteromeric protein complexes for the majority of interactions (Dataset S8), providing further confirmation that our method identifies robust protein interactions. Among these, our interactome map shows a physical interaction between the RAD23 DNA repair protein (LOC_Os02g08300) and a component of the 26S proteasome assembly (LOC_Os03g13970). Previous studies in humans and Arabidopsis have demonstrated that the ubiquitin receptor RAD23 serves as a link between the nucleotide excision repair and 26S proteasomal degradation pathways (42–44). We further found that the proteasomal protein LOC_Os03g13970 interacts with a glutaredoxin family protein (LOC_Os04g17050) and a ubiquitin-conjugating enzyme (LOC_Os08g28680). Glutaredoxin proteins have previously been suggested as candidates for modulating the gate of the 26S proteasomal channel through deglutathionylation of the 20S proteosomal subunit in bacteria and eukaryotes (45, 46). We also detected an interaction between the cytosolic and plastidic versions of fructose-1,6-bisphosphatase responsible for catalyzing the reaction from fructose-1,6-bisphosphate to fructose 6-phosphate during gluconeogenesis and the Calvin cycle in the cytosol and chloroplast, respectively. A previous study in pea (Pisum sativum) demonstrated that these proteins colocalize in the nucleus but do not experimentally probe the interaction (47). Although the functional consequences of this interaction need to be further characterized, these findings suggest that this interaction may be conserved between monocots and dicots, highlighting the potential utility of such interactome networks for understanding evolutionary relationships among interacting proteins.
Conclusion
Overall, our work presents a systematic, experimentally validated advance in the functional annotation of the rice genome. The importance of and effort toward characterization of plant genomes including those of key agricultural species has continued to grow over the years. A recent study has applied a mass spectrometry approach to identify protein complex assemblies broadly conserved throughout the Viridiplantae clade to which O. sativa belongs (48). Our annotations alongside others in the literature have critical applications for both basic and translational research that aims to improve the productivity, nutritional value, and climate resilience of this important crop species. Moreover, O. sativa is now the first monocot, first agriculturally relevant organism—and indeed the only plant outside of A. thaliana—with an ORFeome amenable to high-throughput functional characterization. Thus, our ORFeome and interactome map provide a vital resource to help bridge the ∼150-million-year evolutionary gap separating A. thaliana from monocot species, including major crop staples such as maize, sorghum, wheat, or barley. We recognize that the work presented here is limited to interrogating a subset of the ORFeome. The genome of O. sativa is predicted to encode a staggering 30,000 to 50,000 genes (49), dwarfing the number of genes in humans and Arabidopsis (50, 51). Our reported rice ORFeome currently spans less than 10% of the complete O. sativa genome, and as noted above, likely oversamples the most highly expressed genes that are easiest to clone. Moreover, despite matching the recall rate of previous interactomes, and increasing the currently known rice interactome by about 50%, our Y2H screen to date has likely captured less than 1% of the roughly 100,000 protein interactions expected to occur within the proteome as a whole (SI Appendix, Fig. S5A). Nonetheless, our PLATE-seq strategy is massively parallel and highly scalable and thus constitutes a vital tool that will accelerate future high-throughput functional biology studies aimed at filling these gaps.
Materials and Methods
PLATE-Seq.
In brief, plasmid(s) from individual wells of 96-well plates were amplified by PCR using a plasmid-specific forward primer and a reverse primer consisting of a position-specific barcode and TruSEq. 3′ sequencing adapter. Amplicons derived from the same 96-well plate were pooled and purified by PCR purification columns (Qiagen). Each amplicon pool was subject to Tn5 tagmentation to fragment the amplicons and append adapters consisting of a plate-specific barcode and TruSEq. 5′ sequencing adapter. Tagmented DNA was purified by PCR purification columns (Qiagen) and pooled across all 96-well plates. These pools were then subjected to low-cycle PCR both to extend the TruSeq end adapters with sequences compatible for binding to the Illumina flowcell and to enrich for only DNA fragments consisting of TruSeq adapter sequences on both ends of the plate-specific and position-specific barcodes. The primers used can be found in Dataset S1. Details related to PLATE-seq are available in SI Appendix, Supplementary Methods.
Construction of the O. sativa ORFeome.
O. sativa RNA was isolated from a wide range of rice plant tissue (e.g., leaf, stem, nodes, roots) at different developmental stages and under various stress conditions (e.g., light, dark, cold stress, salt stress, drought stress) as detailed in Dataset S4 using the primers listed in Dataset S3. The 3,269 genes to clone were selected using RiceNet (16, 17) to prioritize genes most associated with a seed set of 89 genes listed in Dataset S2. After cDNA synthesis and cloning into Gateway entry vector pDONR223, clones were transformed into bacterial carriers. All clones were validated by isolation of single bacteria and sequencing by PLATE-seq.
Generating the Rice Interactome.
We screened all possible pairs (∼2.7 million) of 1,671 O. sativa ORFs for interaction by high-throughput yeast Y2H. Initial interaction screening was performed by testing one DB ORF against minipools composed of 188 AD ORFs at a time. All Y2H-positive colonies were collected for sequencing. Pairs of ORFs encoding putative Y2H-positive interactors were identified by PLATE-seq and validated by pairwise Y2H retest. Additional details regarding the Y2H screen can be found in SI Appendix, Supplementary Methods.
BiFC.
BiFC assays were performed by onion infiltration using Agrobacterium harboring Gateway-compatible pSAT expression clones. All interactions were tested in all possible pairwise combinations with respect to N-terminal or C-terminal tags to minimize the possibility of not detecting true interactions due to hindrance from tags. BiFC signal was normalized to the negative control. If one combination showed BiFC signal significantly above negative control, the interaction was considered positive. The positive control used for comparison is the AtABI1-AtOST1 strong interaction and the negative control used is AtOST1-AtOIP26 as previously reported (52). All samples were observed by a Leica TCS-SP5 microscope (Leica Microsystems), and quantification was performed using ImageJ (version 1.51n). Reported values are a ratio of the absolute fluorescence measured for each sample relative to the positive control. Statistical analyses were performed using two-tailed t test in reference to the negative control.
Data Availability.
All datasets of direct relevance to the paper have been included alongside the publication as Datasets S1–S8. In the event any additional raw data not included here is requested, we will make that data available whenever possible.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (NSF #1661380 to H.Y.; NSF #1639075 to H.Y. and S.R.M.; NSF #1444511 to S.R.M.; NSF #0520253 to J.R.E.; NSF #0703905 to M.V., D.E.H., and J.R.E.; and NSF #1237975 to P.C.R.), the National Institute of General Medical Sciences (R01 GM124559 to H.Y.; R01 GM125639 to H.Y.; and R01 GM122968 to P.C.R.), the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK115398 to H.Y.), the National Institute of Food and Agriculture (2013-67013-21379 to S.R.M.), the Deutsche Forschungsgemeinschaft (Award #411255989 to L.H.K.), the Foundation for Food and Agriculture Research (534683 to P.C.R.), the European Research Council’s Horizon 2020 Research and Innovation Programme (648420 to P.F.-B.), and the Bio-Oriented Technology Research Advancement Institution (to S.K.). J.R.E. is an Investigator of the Howard Hughes Medical Institute. Positive and negative control clones used for BiFC experiments were generous gifts from Julian I. Schroeder. We thank Horacio Caniza and Alberto Paccanaro for useful discussions about the use and interpretation of semantic similarity scores and in particular Horacio Caniza for assistance and feedback in running the GOssTO software to calculate these semantic similarity scores.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918068117/-/DCSupplemental.
References
- 1.Salehi-Ashtiani K., et al. , Isoform discovery by targeted cloning, ‘deep-well’ pooling and parallel sequencing. Nat. Methods 5, 597–600 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X., et al. , A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakai H., et al. , Rice annotation project database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 54, e6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong W., et al. , Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 135, 773–782 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arabidopsis Interactome Mapping Consortium , Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice Full-Length cDNA Consortium ; et al. , Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301, 376–379 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Reboul J., et al. , C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34, 35–41 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Rual J. F., et al. , Human ORFeome version 1.1: A platform for reverse proteomics. Genome Res. 14, 2128–2135 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelperin D. M., et al. , Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816–2826 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuyama A., et al. , ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24, 841–847 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Rohila J. S., et al. , Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J. 46, 1–13 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Ding X., et al. , A rice kinase-protein interaction map. Plant Physiol. 149, 1478–1492 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rual J. F., et al. , Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Yu H., et al. , High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vo T. V., et al. , A proteome-wide fission yeast interactome reveals network evolution principles from yeasts to human. Cell 164, 310–323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee I., et al. , Genetic dissection of the biotic stress response using a genome-scale gene network for rice. Proc. Natl. Acad. Sci. U.S.A. 108, 18548–18553 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee T., et al. , RiceNet v2: An improved network prioritization server for rice genes. Nucleic Acids Res. 43, W122–W127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo Y. S., et al. , Towards establishment of a rice stress response interactome. PLoS Genet. 7, e1002020 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krehenwinkel H., et al. , Estimating and mitigating amplification bias in qualitative and quantitative arthropod metabarcoding. Sci. Rep. 7, 17668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vidal M., A unifying view of 21st century systems biology. FEBS Lett. 583, 3891–3894 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Robinson C. V., Sali A., Baumeister W., The molecular sociology of the cell. Nature 450, 973–982 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Barabási A. L., Gulbahce N., Loscalzo J., Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 12, 56–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stelzl U., et al. , A human protein-protein interaction network: A resource for annotating the proteome. Cell 122, 957–968 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Vidal M., Cusick M. E., Barabási A. L., Interactome networks and human disease. Cell 144, 986–998 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snel B., Lehmann G., Bork P., Huynen M. A., STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28, 3442–3444 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D., et al. , STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu H., Zhu P., Jiao Y., Meng Y., Chen M., PRIN: A predicted rice interactome network. BMC Bioinformatics 12, 161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohila J. S., Chen M., Cerny R., Fromm M. E., Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38, 172–181 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Cooper B., et al. , A network of rice genes associated with stress response and seed development. Proc. Natl. Acad. Sci. U.S.A. 100, 4945–4950 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das J., et al. , Cross-species protein interactome mapping reveals species-specific wiring of stress response pathways. Sci. Signal. 6, ra38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das J., Yu H., HINT: High-quality protein interactomes and their applications in understanding human disease. BMC Syst. Biol. 6, 92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark C., et al. , BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Licata L., et al. , MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 40, D857–D861 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner B., et al. , iRefWeb: interactive analysis of consolidated protein interaction data and their supporting evidence. Database (Oxford) 2010, baq023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xenarios I., et al. , DIP: The database of interacting proteins. Nucleic Acids Res. 28, 289–291 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orchard S., et al. , The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, D358–D363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peri S., et al. , Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 13, 2363–2371 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keshava Prasad T. S., et al. , Human protein reference database: 2009 update. Nucleic Acids Res. 37, D767–D772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pagel P., et al. , The MIPS mammalian protein-protein interaction database. Bioinformatics 21, 832–834 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Berman H. M., et al. , The protein data bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berman H., Henrick K., Nakamura H., Announcing the worldwide protein data bank. Nat. Struct. Biol. 10, 980 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Schauber C., et al. , Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391, 715–718 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Dantuma N. P., Heinen C., Hoogstraten D., The ubiquitin receptor Rad23: At the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst.) 8, 449–460 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Farmer L. M., et al. , The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 22, 124–142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silva G. M., et al. , Role of glutaredoxin 2 and cytosolic thioredoxins in cysteinyl-based redox modification of the 20S proteasome. FEBS J. 275, 2942–2955 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Demasi M., et al. , Redox regulation of the proteasome via S-glutathionylation. Redox Biol. 2, 44–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson L. E., Yousefzai R., Ringenberg M. R., Carol A. A., Both chloroplastic and cytosolic fructose bisphosphatase isozymes are present in the pea leaf nucleus. Plant Sci. 166, 721–730 (2004). [Google Scholar]
- 48.McWhite C. D., et al. , A pan-plant protein complex map reveals deep conservation and novel assemblies. Cell, 10.1016/j.cell.2020.02.049. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goff S. A., et al. , A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Venter J. C., et al. , The sequence of the human genome. Science 291, 1304–1351 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Arabidopsis Genome Initiative , Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Waadt R., et al. , Identification of open stomata1-interacting proteins reveals interactions with sucrose non-fermenting1-related protein Kinases2 and with type 2A protein phosphatases that function in abscisic acid responses. Plant Physiol. 169, 760–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets of direct relevance to the paper have been included alongside the publication as Datasets S1–S8. In the event any additional raw data not included here is requested, we will make that data available whenever possible.