Significance
Fungi play a key role in the global carbon cycle as the main decomposers of litter and wood. Although current climate models reflect limited functional variation in microbial groups, fungi differ vastly in their decomposing ability. Here, we examine which traits explain fungal-mediated wood decomposition. In a laboratory study of 34 fungal isolates, we found that decomposing ability varies along a spectrum from stress-tolerant, poorly decomposing fungi to fast-growing, competitive fungi that rapidly decompose wood. We observed similar patterns in a 5-y field experiment, in which communities of fast-growing fungi more rapidly decomposed logs in the forest. Finally, we show how linking decomposition rates to known spatial patterns in fungal traits could improve broad-scale predictions of wood decomposition by fungi.
Keywords: fungal traits, wood decomposition, carbon cycle, functional biogeography, decay rate
Abstract
As the primary decomposers of organic material in terrestrial ecosystems, fungi are critical agents of the global carbon cycle. Yet our ability to link fungal community composition to ecosystem functioning is constrained by a limited understanding of the factors accounting for different wood decomposition rates among fungi. Here we examine which traits best explain fungal decomposition ability by combining detailed trait-based assays on 34 saprotrophic fungi from across North America in the laboratory with a 5-y field study comprising 1,582 fungi isolated from 74 decomposing logs. Fungal growth rate (hyphal extension rate) was the strongest single predictor of fungal-mediated wood decomposition rate under laboratory conditions, and accounted for up to 27% of the in situ variation in decomposition in the field. At the individual level, decomposition rate was negatively correlated with moisture niche width (an indicator of drought stress tolerance) and with the production of nutrient-mineralizing extracellular enzymes. Together, these results suggest that decomposition rates strongly align with a dominance-tolerance life-history trade-off that was previously identified in these isolates, forming a spectrum from slow-growing, stress-tolerant fungi that are poor decomposers to fast-growing, highly competitive fungi with fast decomposition rates. Our study illustrates how an understanding of fungal trait variation could improve our predictive ability of the early and midstages of wood decay, to which our findings are most applicable. By mapping our results onto the biogeographic distribution of the dominance-tolerance trade-off across North America, we approximate broad-scale patterns in intrinsic fungal-mediated wood decomposition rates.
Fungi are a functionally critical component of terrestrial ecosystems because they govern the decomposition of organic material (1). The fungal community contributes to wood decay rates at least as much as local climate conditions (2), and thus represents a key driver of ecosystem function (3, 4). Accordingly, microbial processes are increasingly being incorporated into biogeochemical models of the global carbon cycle, which inform climate change forecasts (Earth System Models, ref. 5). Models traditionally used microbial biomass as a proxy of decomposer activity (3, 6), treating the microbial community as a single homogeneous group or a small number of functionally distinct pools (7). However, there is a growing appreciation that fungal taxa differ greatly in their decomposition ability (3, 8), leading to massive variation in decomposition across fungal communities (8–11). Understanding how decay rates vary with fungal community composition will be critical to making accurate forecasts of terrestrial carbon dynamics, as reflected in contemporary biogeochemical models of litter decomposition (12, 13). Improving these forecasts urgently requires a tangible, empirically tested link between fungal characteristics and their contribution to ecosystem function across landscapes (7, 14, 15).
Until recently, fungal ecologists have sought to comprehend the staggering functional diversity of fungi by using sequencing approaches to identify the taxonomic composition of entire communities (e.g., refs. 16 and 17), or by in-depth manipulative studies of up to a handful of species (e.g., refs. 9 and 18). These studies have contributed valuable insights into the dynamics of fungal systems, but neither approach permits a detailed exploration of fungal functional attributes across a broad range of taxa (19, 20). However, in recent years, the development of trait-based approaches has begun to transform our understanding of broad-scale functional patterns. By linking traits to ecosystem functioning, these approaches have been used in plant and animal ecology to infer the functioning of novel communities without prior knowledge of the taxa that are present (21, 22). Although the use of trait-based approaches in fungal ecology still lags behind that in plants and animals (20), recent studies have begun to reveal predictable patterns in fungal phenotypes at local (23, 24) and broad (25–28) spatial scales. For example, Maynard et al. (25) found that North American wood-decomposer fungi ranged from fast-growing, competitively dominant individuals to slow-growing stress-tolerant fungi, a trade-off that might underpin the spatial distribution of these organisms. The next critical step to comprehend the functional relevance of these community-level patterns is to link these key fungal traits to wood decomposition rates across a wide range of taxa (3, 20, 29). Such insight will allow us to translate physiological life-history trade-offs into predictions of ecosystem function.
Several hypotheses have been proposed to explain which fungal traits should predict fungal-mediated wood decomposition rates. At the phenotypic level, it has long been assumed that slow-growing fungi with a high hyphal density may decompose wood faster than thin fungi with a rapid outward extension rate (30). In contrast, the first studies incorporating the microbial community into decomposition models assumed that the rate of decay increases with the growth rate of the decomposers (3, 31, 32). At the genetic level, Treseder and Lennon (26) concluded that several functional genes regulating decomposition (specifically, breakdown of cellulose and lignin) were negatively associated with genes promoting stress tolerance. However, it remains unclear whether these genetic patterns relate to phenotypic trait expression (33). To test these hypothesized relationships between fungal traits and decomposition, empirical studies of a range of fungal taxa are needed that bridge the historical gap between the ecological focus of single-species studies and the greater taxonomic breath of community-level sequencing efforts.
Here, we explore which fungal characteristics predict wood decomposition rate across a range of common wood decomposer fungi. First, we use a database of 22 fungal traits previously measured in each of 34 wood rot fungi collected from a wide geographic range across North America (25, 34–36) to identify potential drivers of wood decomposition. Specifically, we measured the mass loss of wood blocks when colonized by each fungus to estimate a standardized wood decomposition rate, and examined which fungal traits in the database best explained the variation in wood decomposition across fungal isolates. Second, to evaluate the relevance of these trait measurements under complex natural conditions, we isolated 1,582 fungi from logs in a large field decomposition experiment (37) and tested whether these best-fitting traits likewise help us to explain the variation in mass loss of logs in situ. Finally, we combine our results with existing maps of fungal trait expression (25) to approximate the functional biogeography of fungal wood decomposition across North America.
To evaluate which characteristics of our fungi may predict wood decomposition, we considered a broad range of traits that influence different aspects of fungal ecology and physiology. A wide variety of trait definitions exist in the literature, ranging from physiological to performance-oriented properties of organisms (21, 23, 38, 39). Some can be linked directly to the expression of specific genes, while others are emergent properties that arise as a result of multiple genetic mechanisms. For the purposes of this study, we define a fungal trait as any characteristic of an individual fungus that can be measured under standardized growth conditions and compared across individuals. Specifically, we focused on three groups of traits for which previous work had demonstrated that they vary in consistent patterns across our fungal taxa (25). Hyphal extension rate and hyphal density reflect hyphal morphology and growth strategy, ecological performance traits (11 in total) relate to combative ability and tolerance of a range of temperature and moisture conditions (25), and finally, the production of oxidative and hydrolytic enzymes (9 traits) promotes nutrient acquisition from organic resources (26). All traits were measured directly on fungi growing in isolation under controlled laboratory conditions in previous work (25, 34–36), giving us a standardized estimate of potential trait expression. Likewise, the wood decomposition rate presented here for each fungus represents their intrinsic decay ability under standardized laboratory conditions.
Results and Discussion
Fungal Traits Predicting Decomposition Rate.
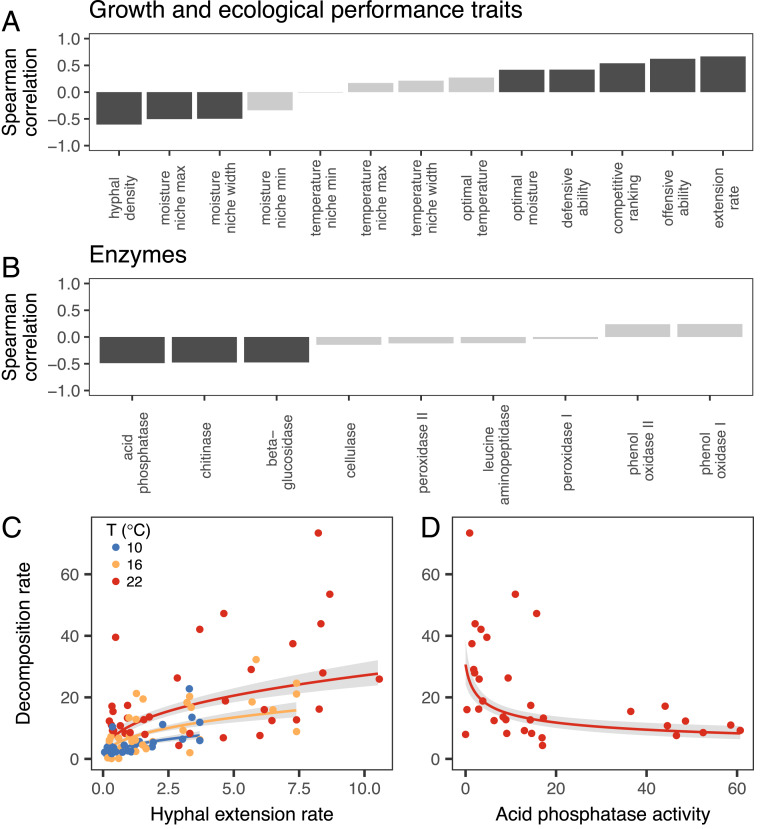
Of all 22 traits measured across our 34 fungal isolates, the strongest individual predictor of fungal-mediated wood decomposition was the hyphal extension rate of the fungal colony on 2% malt agar (ρ = 0.67; P < 0.001; Fig. 1A). Extension rate explained 19% of the variance in decomposition rate across our fungi (F1, 49.2 = 18.9; P < 0.001; semipartial R2 = 0.19; Fig. 1C) across three different temperatures (no significant interaction effect, F1, 69.6 = 0.07; P = 0.79). The positive relationship between extension rate and decomposition rate eventually leveled off for the fastest-growing isolates (slope = 0.39 ± 0.09 SEM on a log-log scale). In direct contrast to extension rate, hyphal density was negatively correlated with decomposition (ρ = −0.61; P < 0.001; Fig. 1A), which reflects the well-supported trade-off between colony density and extension rate (18). These results contradict the hypothesis that dense colonies should achieve higher intrinsic wood decomposition rates (30) and instead support models assuming that decomposition is positively related to growth or extension rate (31). Indeed, previous efforts to apply allometric scaling theory to fungi suggest that colonies with a higher extension rate capture and consume resources more rapidly and efficiently (40, 41). Our results provide strong empirical support for this hypothesis and suggest that hyphal extension rate could serve as an easily measurable proxy for the wood decay ability of fungi, as determined over a 4- to 5-mo period under standardized laboratory conditions.
Fig. 1.
Traits explaining wood decomposition across 34 fungal isolates under laboratory conditions. Decomposition was positively correlated with extension rate and combative ability, but negatively with hyphal density, moisture niche width, and the production of various enzymes (A and B). Bars represent Spearman’s rank correlation coefficients (ρ) between the geometric mean rate of decomposition (percentage mass loss over 122 d, measured at 10 °C, 16 °C, and 22 °C) and each trait, with dark shading indicating statistically significant correlations at α = 0.05. Extension rate (mm⋅day−1) was a strong predictor of decomposition across all three temperatures (C), and acid phosphatase production (D) had the strongest relationship with decomposition of all enzymes (measured at 22 °C). Points in C and D represent individual fungal isolates (each occurring 3 times in [C]), and lines and shading indicate model predictions ± SEM (full model details in Materials and Methods).
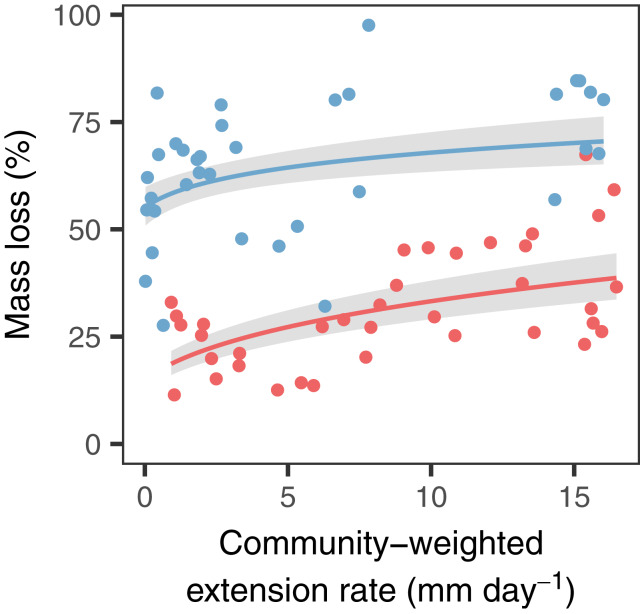
To evaluate the strength of the relationship between extension rate and decomposition under the complex natural conditions of forest ecosystems, we quantified hyphal extension rates at the community level in logs of 20 woody plant species that had been decomposing for 3 or 5 y in a landscape-scale field experiment in a temperate oak-hickory forest (37). Specifically, we cultivated 14 fungal isolates from each of 113 samples collected from 74 unique logs at the field site (1,582 isolated fungi in total) and estimated the intrinsic extension rate of each isolate when growing on agar. Consistent with the positive relationship between extension and decomposition rate found in our laboratory cultures, community-weighted hyphal extension rate also was a strong predictor of wood mass loss (representing cumulative decay) in the field (Fig. 2). That is, communities composed of fungi with high intrinsic hyphal extension rates were associated with more rapidly decomposing logs than those composed of slow-growing fungi. We found the strongest relationship between extension rate and wood mass loss after 3 y of decay (log-log scale, slope = 0.32 ± 0.04 SE; F1, 30.3 = 50.7; P < 0.001), with the slope of the relationship attenuating at the 5-y point (0.09 ± 0.04 SE; F1, 32.5 = 4.21; P = 0.048). As expected, based on species differences in the physical and chemical properties of wood (30, 42), tree species explained a large proportion of the variance in wood mass loss (57% and 31% after 3 and 5 y, respectively). Yet the intrinsic hyphal extension rate of the community explained an additional 27% (3 y) or 10% (5 y) of the variance in mass loss. This distinct relationship between community-weighted intrinsic growth rate in the laboratory and decomposition rate under field conditions after 3 y of decay provides one of the first tangible links between fungal community composition and wood decay, supporting growing calls to incorporate fungal community characteristics into broad-scale carbon cycling models (3, 4, 10, 14).
Fig. 2.
The decomposition of logs also increases with the hyphal extension rate of the fungal community that colonized them. Community-weighted mean extension rate (n = 14 fungal isolates from the top and/or bottom of each log) is plotted against cumulative mass loss (n = 73 logs) after 3 y (red) or 5 y (blue) of wood decay in a forest ecosystem. Lines and shading represent general linear mixed model predictions ± SEM for each time period, with extension rate as a fixed effect and woody plant species as a random effect.
In our laboratory assays, fungal growth traits of the 34 isolates were not the only characteristics to predict intrinsic fungal-mediated wood decomposition. In particular, several physiological and biochemical traits correlated negatively with decomposition rate. Fungi that were tolerant of a wider range of moisture conditions (i.e., those with wide moisture niche widths) had lower decomposition rates (Fig. 1A), matching the genetic association between stress tolerance and wood decomposition ability found in previous work (26). In addition, the production of hydrolytic extracellular enzymes that release macronutrients from decaying organic material (acid phosphatase, P; chitinase, N; β-glucosidase, C; Fig. 1B) came at the cost of faster decomposition. Of all enzymes, acid phosphatase had the strongest negative relationship with decomposition rate (log-log scale, slope = −0.30 ± 0.10; F1, 30 = 9.66; P = 0.004; R2 = 0.31; Fig. 1D). The primary role of this enzyme is to convert organic phosphorus compounds into soluble inorganic forms, increasing phosphorus availability in the soil for both microorganisms and plants (43).
Broad-Scale Patterns in Fungal Decomposition.
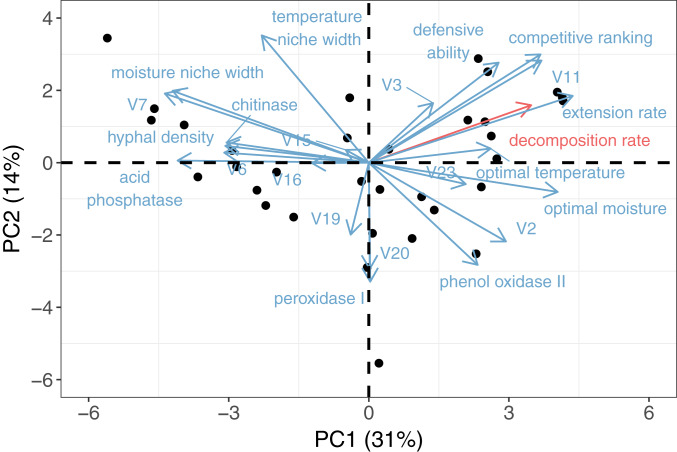
We have thus far considered each trait in our laboratory database in isolation, with our primary aim being to identify simple predictors of intrinsic decay rate across our fungal taxa. Together, these traits represent an apparent trade-off between competitive dominance and moisture stress tolerance in fungi, which was recently demonstrated by Maynard et al. (25) for the same trait dataset (a dataset that did not include decomposition rate). To evaluate the functional consequences of this physiological trade-off in terms of wood decay, we mapped intrinsic decomposition rate onto the trait space, using a principal component analysis (Fig. 3). We found that across our fungi, decomposition rate strongly aligns with the dominance-tolerance trade-off, parallel to extension rate (along axis PC1, which explains 31% of variation; Fig. 3). At one end of this spectrum, highly competitive fungi with high hyphal extension rates have high intrinsic rates of wood decomposition. At the other end of this spectrum, dense fungi that tolerate a wide range of moisture conditions and produce large quantities of N- and P-mineralizing enzymes were associated with slower rates of wood decomposition (Figs. 1 and 3).
Fig. 3.
Principal component analysis of decomposition, growth and ecological performance traits, and enzyme production across 34 fungal isolates (full variable names for V2, V3, etc., listed in SI Appendix, Table S1). The PC1 axis (explaining 31% of variation) represents a dominance-tolerance trade-off across taxa, ranging from fast-growing, highly competitive isolates to stress-tolerant isolates with wide moisture niches and high enzyme production. Decomposition rate (red, PC1 loading = 0.25) strongly aligns with extension rate and combative ability along this trade-off axis. Results therefore closely match a previous principal component analysis of the trait dataset only (25), with small differences due to the added variable of decomposition rate.
Throughout the fungal literature, a range of contrasting theories has been proposed to link fungal communities to ecosystem functioning (e.g., refs. 27 and 44–46). Our trait-based data support conceptual models (14, 29, 47, 48) and genetic studies (26) in suggesting that fungal-mediated decomposition rates might be governed by a fundamental trade-off between stress tolerance and competitive dominance. In his classic CSR framework of competitive (C), stress-tolerant (S), and ruderal (R) plant ecological strategies, Grime (49) originally proposed that the distinction between fast-growing competitors and slow-growing stress-tolerant individuals may also apply to fungi and relate to changes in decomposer ability through successional time. More recently, this framework has been expanded with the hypothesis that producing specific extracellular enzymes or cell damage repair compounds should be beneficial for fungi living in nutrient-limited or stressful environments (14, 44, 47). Indeed, genetic studies show a positive association between genes regulating acid phosphatase or chitinase production and genes promoting stress tolerance (26). Our results indicating that decomposition is negatively associated with production of nutrient-mineralizing enzymes and with moisture stress tolerance (Fig. 1 A, B, and D), while showing a positive relationship with combative ability (Fig. 1A) and extension rate (Figs. 1C and 2), support this framework at the phenotypic level.
Because all fungi in our database are dominant species in the early to midstages of wood decay, our laboratory measurements are most informative about fungal dynamics during this phase of the decomposition process. Other studies suggest that fungal interactions may change at later stages of decay, affecting wood decomposition rates (reviewed in ref. 50). For example, Holmer and Stenlid (51) found that late-successional species outcompeted fungi from the earlier stages of wood decay in 6-mo laboratory trials on wood. It is possible that our fast-growing fungi with high intrinsic decay abilities best represent a ruderal, high-yield life history (sensu ref. 14) that is advantageous at early decay stages, while late-stage specialists could outcompete them by being able to access more complex substrates as wood decomposition progresses. This hypothesis is consistent with our result that oxidative enzymes necessary to break down lignin [biochemically the most complex step in wood decomposition (26)] were not significantly correlated with wood decay rates in our laboratory experiment (neutral and positive trends in Fig. 1B). Future studies could run fungal combat trials under less favorable nutrient conditions to test how competitive hierarchies change over the course of wood decay. Successional changes in the fungal community might also explain why the relationship between hyphal extension rate and decomposition rate leveled off between 3 and 5 y of decay in the field (refs. 42 and 50 and Fig. 2). In contrast to our laboratory study, field logs did reach the later stages of decay. The fungal community will likely have changed substantially over this time, although molecular data would have been necessary to confirm fungal identities genetically. As fungal communities become more complex during succession, higher investment into combat could reduce wood decay rates (52) and lessen the impact of fungal traits such as extension rate. Finally, changes in the relationship between extension rate and wood decay over time could also reflect the temporally variable nature of the wood decay process itself (53). Taking into account these constraints of our study system, we conclude that the dominance-tolerance trade-off (25) may play a key role in shaping the functional capacity of fungal communities, specifically in the early to midstages of wood decay.
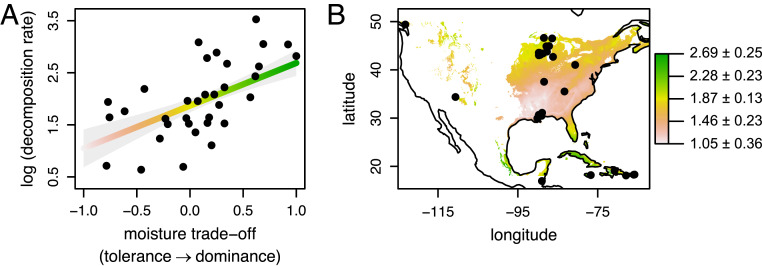
Our results directly build on previous findings obtained using these fungal isolates showing how the dominance-tolerance trade-off predicts their broad-scale biogeographic distributions (25). Here, we demonstrate that this same trade-off also governs intrinsic wood decay ability (slope = 0.82 ± 0.28; F1, 30 = 8.37; P = 0.007; R2 = 0.42; Fig. 4A), and thereby provides a link between community ecology and ecosystem function in wood decay fungi. By projecting this relationship onto the biogeographic distribution of the dominance-tolerance trade-off estimated previously (25), we can approximate the spatial variation of fungal-mediated wood decomposition rates across North American forests (Fig. 4B, map adapted from ref. 25). In essence, this map suggests that the slow-growing, stress-tolerant fungi that are more likely to exist in drier forests with high precipitation seasonality are likely to have poor intrinsic wood decay abilities. In contrast, the fast-growing, highly competitive fungi that are favored in more favorable environments are more likely to decompose wood more quickly, irrespective of the local microclimate. Thus, broad-scale environmental filters may select for fungal communities with certain traits that are in turn strongly linked to decomposition (54, 55). The direction of these indirect effects aligns with the direct effect of the climate on decomposition rate, which is higher in warm, moist environments for any given fungus (ref. 2 and Fig. 1C). Consequently, the biogeographic distribution of fungal traits may reinforce the climate-induced differences in wood decay rates, as fast-decomposing fungi exist in environments that are also conducive to high decomposer activity (as suggested by ref. 56).
Fig. 4.
The relationship between decomposition rate (percentage mass loss over 122 d, log-transformed) and the dominance/moisture tolerance trade-off (A), projected across North America (B). The moisture trade-off (x axis in A) is the difference between each isolate’s competitive ranking and their moisture niche width, both scaled to [0,1], as calculated by Maynard et al. (25). Points in A represent fungal isolates, the line and shading indicate predictions ± SEM from a general linear model (overall slope excluding species indicator effects of Armillaria gallica and Merulius tremellosus). Color shading in both A and B indicates decomposition rate, increasing along the fitted line in A. Thus, the map in B represents predicted decomposition rates, as projected onto the previously published spatial distribution of the moisture trade-off (25). Points in B indicate sampling locations of fungal isolates (n = 34). The sampling location of one isolate (−146.69, 60.73) falls outside the spatial extent of the map.
Notwithstanding these general patterns, it is widely acknowledged that local decomposition rates will be contingent upon microclimatic conditions, legacy effects, and nutrient quality (2), as observed in the deadwood decay field assays from which we cultured fungi (37). Although decay rates in the field will likely deviate substantially from the standardized rates estimated in the laboratory with pure isolates, the community-weighted means of hyphal extension rates for the field isolates helped explain observed wood decomposition in the field. Thus, our coupled laboratory and field approaches support the value of studying fungal activity in isolation under controlled laboratory conditions, in order to quantify and compare a large number of traits for a tractable number of fungi (57). The field experiment allowed us to demonstrate the validity of extrapolating our findings to field conditions, although our inference is constrained by the limited number of fungi we explored in depth and their unknown species identity.
We note that our results can partially be explained by phylogenetic relatedness among isolates (SI Appendix, Table S2), as seen in previous work (25). That is, fungal functional biogeography appears to be partly governed by spatial sorting of phylogenetic lineages, reflecting phylogenetic conservatism in physiological and functional traits that ultimately determine where a fungus can survive and its ability to decompose wood. Having identified the key traits predicting decomposition rates, future studies should explore habitat preference characteristics and their phylogenetic conservatism. Most importantly, the phylogenetic range should be expanded to include Ascomycota, which contain a great diversity of wood decomposer taxa (10, 42). Although the phylum was not represented in our laboratory trait database, it is likely that ascomycetes were present among the fungi isolated in the field experiment. Including a wider taxonomic range of fungi might further elucidate trade-off patterns between rapid wood decay and stress tolerance, given that fungal community composition has been associated with wood decay rates for phylogenetically diverse fungi that vary widely in enzyme production and competitive ability (9, 42, 48). By focusing on trait expression rather than the taxonomic identities of the field-based isolates, our study design does not permit us to identify the effect of community structure, composition, or genetic diversity on wood decomposition rates. Nevertheless, by taking a purely trait-based approach, our results demonstrate that community-weighted trait expression can provide meaningful insight into the functional capacity of wood decay fungi. Disentangling the relative importance of trait expression, genetic diversity, and community structure as drivers of fungal-mediated decomposition is a compelling future research question.
Conclusion and Future Directions.
Our study reveals key traits predicting fungal-mediated wood decomposition, a critical driver of the global carbon cycle (3). Specifically, intrinsic decay ability under standardized laboratory conditions can be predicted from simple information about hyphal extension rate, as faster-growing, more competitive fungi have higher decomposition rates than slower-growing, stress-tolerant fungi. This close association between decomposition rates and fungal life-history strategies allows us to translate previously documented physiological trade-offs (25) into spatial patterns of intrinsic functional capacity. That these apparent trade-offs in extension rate and decomposition ability could be discerned within the complexity of the field environment, with hyperdiverse natural communities and variable environmental conditions, highlights the predictive strength of the mechanism. Because the traits predicting decomposition are associated with broad-scale biogeographic distributions of wood-decomposer fungi, they provide unique insight into their functional biogeography.
We hope that our results motivate broad-scale efforts to validate these patterns using fungal isolates sampled across the globe from a wide variety of environments. Future studies should include taxonomically diverse fungi from all stages of the decay process, to further elucidate how the relationships between fungal traits and decomposition rates change over time. If the patterns we observed among our North American fungi hold across taxa and ecosystems, then this research may prove to be a useful step toward the meaningful incorporation of fungal processes into global biogeochemical models. For example, it is expected that we can currently account for ∼50% of spatial variation in wood decomposition rates by considering the extrinsic drivers (climate and plant traits) of soil organic matter turnover (2, 7). Our analysis suggests that accounting for the intrinsic variation in wood decay ability of fungi in the field (10) might ultimately enhance our ability to predict broad-scale variation in wood decomposition rates to a great degree (in our field study, by up to 10% to 27%). The next key steps in this regard will be to incorporate continuous fungal trait variation into spatially explicit models of wood decay, analogous to recent models of litter decomposition (13, 58). Because each additional biotic layer of a biogeochemical model can introduce new model uncertainty (59), field validation of model outputs (14) and model sensitivity analyses (7) will be needed to determine the optimal level of ecological detail at which the functional biogeography of fungi should be represented.
Materials and Methods
Fungal Isolates.
Our database comprises 34 saprotrophic basidiomycete fungi from 20 unique species (SI Appendix, Fig. S1 and Table S3), previously described in refs. 25, 34, 35, and 36. All fungal isolates were obtained from the US Forest Service culture collection at the Center for Forest Mycology Research (Madison, WI). They were collected from fruiting bodies on dead wood in mixed-hardwood forests across North America and stored in liquid nitrogen without serial transfer. All species are dominant decomposers during the early to midstages of wood decay, from newly fallen logs to the point where cellulose and labile carbon compounds have largely been decomposed and the wood has started to disintegrate (60, 61). Thus, our fungi cover a wide taxonomic and geographic range, but have similar ecological roles (34), allowing us to examine general patterns in fungal traits and decomposition rate.
Laboratory Trait Measurements.
Experimental design.
All laboratory trait data in our database have been described in previous work (25, 34–36). We used these data to find the best predictors of wood decomposition and summarize the general methodology of the trait measurements here (traits listed in SI Appendix, Table S1). To evaluate fungal characteristics in a standardized environment, all traits were measured on single isolates growing on artificial media. Because these systems are structurally different from all other substrates, they provide a nonbiased arena to quantify trait variation for comparison across species (29, 57). Fungi were grown in deep-well, 10-cm-diameter Petri dishes with 2% malt extract agar, sealed with Petri-Seal (full experimental protocol in ref. 34). All trait measurements were carried out at 22 °C and −0.5 MPa moisture potential, unless otherwise noted. This design provides near-optimal growth conditions for all fungal isolates in our study and resembles the nutrient conditions of decaying wood.
Physiological growth.
To characterize the growth of each isolate, we quantified its hyphal extension rate and hyphal density (presented in ref. 34). In brief, we inoculated the center of five replicate Petri dishes (plates) per isolate with a 5-mm-diameter plug of sample culture. Plates were incubated for 2 wk or until the growing isolate reached the edge of the plate. Hyphal extension rate was quantified as the linear extension rate in millimeters per day. Hyphal density was measured using a similar design with media covered by a layer of cellophane, which allowed us to remove and weigh the mycelium (as described in ref. 34, following refs. 62 and 63). We quantified hyphal density as micrograms dry mass per cubic centimer at 1 cm from the edge of the growing front.
Temperature and moisture niche.
Physiological response curves have previously been described for our fungi in ref. 25. Skew-normal distribution models were fit to the extension rate of each isolate across a temperature (10 °C to 40 °C, five replicates per isolate) and moisture (water potentials −0.5 to −4.5 MPa, three replicates) gradient, with extension rate measured at six values along each gradient. To quantify temperature and moisture niche traits of each isolate based on these response curves (full methods in ref. 25, based on ref. 23), we derived the niche width, minimum, and maximum (marking the range of conditions that supports at least half the maximum extension rate of the fungus), as well as the optimal temperature and moisture conditions (where maximum extension rate is reached).
Combative ability.
To quantify the ability of our fungal isolates to displace other isolates in direct combat, we conducted pairwise competition trials with all 20 species, excluding intraspecific interactions (presented in ref. 34, methods following ref. 64). We inoculated plates with two straight lines composed of three plugs per isolate, facing each other at 2 cm from the center of the plate. Inoculation time was adjusted to known differences in extension rate, such that both species formed a hyphal front across half the plate 1 cm apart at the start of the trial. Plates were incubated for up to 8 wk, until one fungus completely displaced the other (competitive exclusion), or when no displacement was observed for 3 wk (deadlock). Based on the outcome of 615 unique trials, we quantified three traits at the species level. “Competitive ranking” is the position of a species in the overall competitive hierarchy among our fungi, calculated using the Elo ranking system (34, 65). “Offensive ability” is the average extension rate of a fungus when displacing its competitor, divided by its extension rate in monoculture. Finally, “defensive ability” is the average rate at which a fungus is overgrown, divided by the competitor’s extension rate in monoculture (25).
Enzyme production.
For each fungal isolate, we quantified the production of five hydrolytic and four oxidative enzymes (described in ref. 34, following refs. 66 and 67). We studied the enzymes acid phosphatase (substrate: phosphate), N-acetyl-β-glucosaminidase (a chitinase, substrate: N-acetyl-β-d-glucosaminide), β-glucosidase (substrate: β-d-glucopyranoside), cellobiohydrolase (a cellulase, substrate: β-d-cellobioside), leucine aminopeptidase (substrate: l-leucine), two peroxidases (substrates 0.3% hydrogen peroxide and tetramethylbenzidine, referred to as peroxidase I and II, respectively), and two phenol oxidases [substrates L-3,4-dihydroxyphenylalanine (l-DOPA) and 2,2'-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid) (ABTS), referred to as phenol oxidase I and II]. Fungal isolates were cultured over a period of 7 d, after which enzyme activity was measured per unit biomass for 4 plugs of agar (diameter 7 mm) sampled 1 cm behind the growing front of each isolate. Hydrolytic enzymes activity was assayed using fluorescence methods, and oxidative enzyme activity using absorbance methods (following ref. 68).
Laboratory Decomposition Experiment.
Decomposition measurements in the laboratory at 10 °C, 16 °C, and 22 °C are newly presented in this study. To obtain a standardized estimate of the decomposition rate for each fungal isolate, we quantified their ability to decompose blocks of maple wood (Acer spp.) in the laboratory. The experiment (full methodological details provided in ref. 34) was conducted on similar plates as the trait assay. Preliminary experiments showed that maple wood supports the growth of all fungal taxa in our study. Thus, fungi could obtain carbon from both the wood and the medium. We inoculated the center of each plate with a 5-mm plug of sample culture. Three sterilized wood blocks (10 mm × 10 mm × 5 mm) were placed 15 mm from the plug and equidistant from each other, secured between two squares of stainless-steel micro mesh. Plates were sealed with Parafilm and incubated for 14 to 18 wk. To measure decomposition at different temperatures for each fungal isolate, we incubated plates at 10 °C, 16 °C, and 22 °C, with six replicate wood blocks (distributed over two plates) per isolate and temperature. After incubation, any fungal residues were carefully scraped from the surface of each block with a razor blade. Wood blocks were dried at 40 °C to constant mass. We quantified decomposition rate as the average mass loss (percentage dry weight) of the six replicate blocks over a period of 122 d. To examine the relationship between extension rate and decomposition, the extension rate of each isolate was measured at 10 °C, 16 °C, and 22 °C, as previously described. Hyphal extension rate at 22 °C was highly consistent (r = 0.95; P < 0.001) with our earlier measurements at this temperature (34).
Field Decomposition Experiment.
To evaluate whether intrinsic extension rate measured under laboratory conditions could predict wood decomposition by natural communities of fungi in the field, we sampled the fungal community from wood logs in a 7-y decomposition study in a temperate deciduous oak-hickory forest at Tyson Research Center in Missouri (37). We established common garden decay sites with logs (22 cm long and 5 to 9 cm ∅) from 21 woody species widely spread across seed plant families, deployed in two cohorts: one beginning in 2009 and the another in 2011 (SI Appendix, Table S5). We present here data from 2014, when we harvested subsamples for analyzing fungal communities from one upland and two lowland sites within the forest. Sawdust was sampled to 2.5-cm depth from eight locations across the log surfaces that had been closest to and furthest from the soil and preserved for cultivation of fungi (full details of sample preparation in refs. 37 and 53). The eight sawdust subsamples from each side of a log were pooled as top and bottom samples and shipped to the laboratory. Samples that appeared contaminated upon arrival or following cultivation were discarded. We cultivated 14 fungal isolates from each of the 113 remaining top or bottom samples (now comprising 74 unique logs from 20 woody species), and measured the extension rate of each isolate (1,582 in total) on agar as previously described. Although it is impossible to capture the entire fungal community inside the logs (species diversity is high and not all species can be cultivated), the 14 isolated fungi per log top or bottom were likely the most abundant variants contributing most to fungal biomass. Although we detected great variation in morphologies and growth rates across isolates from the same logs, suggesting that there were multiple different species present, we were not able to confirm this genetically. We calculated the community-weighted hyphal extension rate of the 74 unique logs by calculating the average extension rate across all 14 fungal isolates in a sample and taking the average of top and bottom samples from the same log where available (SI Appendix, Table S5). We used the mass loss (percentage) of each log as a measure of cumulative decay over a 3- or 5-y period, noting that our samples represent a snapshot of a decay process that may change through time (53).
Statistical Analysis.
All statistical analyses were conducted in R v3.5.1 (69). As a general measure of decomposition under standardized conditions, we used the geometric mean of decomposition rate at 10 °C, 16 °C, and 22 °C measured for each fungal isolate, unless otherwise noted (decomposition rate at each temperature was first averaged across six replicates per isolate). Spearman’s ρ was used to compute pairwise rank correlations between each trait and decomposition rate. We used standard principal component analysis to explore the position of decomposition within the entire trait space.
To further explore the relationship between decomposition rate and extension rate (both in the laboratory and in the field) and between decomposition rate and acid phosphatase activity, we first log-transformed all these rates and analyzed the relationships between them using general linear (mixed) models. The slope of the relationship at a log-log scale corresponds to the exponent of a power-law relationship at a linear scale, which was used for graphical representation. Where necessary due to very small data values, we used an offset of +1 for the log-transformation to satisfy assumptions of normality. To account for species identity effects in our laboratory data, we included a species indicator variable for each species with more than one isolate. Only significant species indicators (α = 0.05, two-sided) were retained in the model, with the exception of the indicator for Armillaria gallica, which was always retained because of the high number of isolates for this species. Plotted regression lines show the predicted mean regression trend, with the indicators set to zero.
To test whether the relationship between decomposition and extension rate was consistent across the three temperatures, we fitted a mixed-effects model with extension rate, temperature, and their interaction as fixed effects (in addition to the species indicators) and fungal isolate as a random effect (lme4 package, ref. 70). Main and interaction effects were evaluated using Type II Wald F tests with Kenward-Roger degrees of freedom (df) (car package, ref. 71). We used a similar approach to analyze the field decomposition data, with extension rate, years decayed (three or five) and their interaction as fixed effects and plant species as a random effect. Because the interaction was significant, we then fitted separate models for each decay period. R2-values for the mixed models were estimated following Nakagawa and Schielzeth (72), using the packages r2glmm (73) and MuMIn (74). Finally, we analyzed the relationship between acid phosphatase activity and decomposition rate at 22 °C, the temperature that enzyme activity was measured at. We fitted a linear model and evaluated the effects of acid phosphatase activity and species indicators using Type II F-tests (71).
Data for growth and ecological performance traits and enzymes are available in ref. 25. Decomposition and extension rate data from the laboratory can be accessed in SI Appendix, Tables S3 and S4, and field data in ref. 75 and SI Appendix, Table S5.
Mapping Decomposition Across North America.
To map predicted rates of decomposition across North America, we quantified the relationship between decomposition rate and the dominance-tolerance trade-off across our fungi and visualized the result on a previously published map of this trade-off. In brief, Maynard et al. (25) defined a metric of the dominance-tolerance trade-off for moisture, calculated as the difference between each isolate’s competitive ranking and their moisture niche width, both scaled to [0,1]. Therefore, the trade-off metric ranged from −1 to 1, with 1 representing high competitive dominance/low moisture tolerance and −1 the reverse. To explore whether climate might predict the distribution of this trade-off, climate data for the sampling location of each fungal isolate were obtained from the WorldClim Global Climate Data database (76). The relationship between the moisture trade-off and key climate variables was analyzed using general linear models with species indicator variables, which yielded regression coefficients that were used to estimate moisture trade-off values across a raster map of North America (25). Here, we quantified the relationship between decomposition rate (log-transformed) and this moisture trade-off using similar linear models. We then projected this linear relationship onto the previously published map of the moisture trade-off, such that color scaling now reflects predicted decomposition rates across North America.
Supplementary Material
Acknowledgments
We thank S. Thomas, A. Neupane, E. Karlsen-Ayala and C. Delavaux for laboratory assistance, and M. Walton, D. Young, A. Milo and K. Dunham for their help with deployment, harvest and sampling of the field experiment. The Tyson Research Center provided logistical support. Finally, we thank L. Boddy for critical discussions on this topic. This study was funded by grants from DOB Ecology, Plant for the Planet, the Yale Climate and Energy Institute, British Ecological Society, and Marie Skłodowska-Curie Actions Fellowship (to T.W.C.), the Yale Institute for Biospheric Studies (to D.S.M.), the US National Science Foundation (Grants DEB-1021098 and DEB-1457614 to M.A.B., T.W.C., and D.S.M.; DEB-1302797 to A.E.Z.), the Swiss National Science Foundation (Grant P2EZP3_178481 to N.L.), and the US Forest Service.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909166117/-/DCSupplemental.
References
- 1.Baldrian P., Forest microbiome: Diversity, complexity and dynamics. FEMS Microbiol. Rev. 41, 109–130 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Bradford M. A., et al. , Climate fails to predict wood decomposition at regional scales. Nat. Clim. Chang. 4, 625–630 (2014). [Google Scholar]
- 3.McGuire K. L., Treseder K. K., Microbial communities and their relevance for ecosystem models: Decomposition as a case study. Soil Biol. Biochem. 42, 529–535 (2010). [Google Scholar]
- 4.Cavicchioli R., et al. , Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 17, 569–586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wieder W. R., Bonan G. B., Allison S. D., Global soil carbon projections are improved by modelling microbial processes. Nat. Clim. Chang. 3, 909–912 (2013). [Google Scholar]
- 6.Manzoni S., Porporato A., Soil carbon and nitrogen mineralization: Theory and models across scales. Soil Biol. Biochem. 41, 1355–1379 (2009). [Google Scholar]
- 7.Crowther T. W., et al. , The global soil community and its influence on biogeochemistry. Science 365, eaav0550 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Dickie I. A., Fukami T., Wilkie J. P., Allen R. B., Buchanan P. K., Do assembly history effects attenuate from species to ecosystem properties? A field test with wood-inhabiting fungi. Ecol. Lett. 15, 133–141 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Fukami T., et al. , Assembly history dictates ecosystem functioning: Evidence from wood decomposer communities. Ecol. Lett. 13, 675–684 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Van der Wal A., Ottosson E., De Boer W., Neglected role of fungal community composition in explaining variation in wood decay rates. Ecology 96, 124–133 (2015). [DOI] [PubMed] [Google Scholar]
- 11.van der Wal A., Klein Gunnewiek P. J. A., Cornelissen J. H. C., Crowther T. W., de Boer W., Patterns of natural fungal community assembly during initial decay of coniferous and broadleaf tree logs. Ecosphere 7, e01393 (2016). [Google Scholar]
- 12.Wieder W. R., Grandy A. S., Kallenbach C. M., Taylor P. G., Bonan G. B., Representing life in the Earth system with soil microbial functional traits in the MIMICS model. Geosci. Model Dev. 8, 1789–1808 (2015). [Google Scholar]
- 13.Allison S. D., Goulden M. L., Consequences of drought tolerance traits for microbial decomposition in the DEMENT model. Soil Biol. Biochem. 107, 104–113 (2017). [Google Scholar]
- 14.Malik A. A., et al. , Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J. 14, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall E. K., et al. , Understanding how microbiomes influence the systems they inhabit. Nat. Microbiol. 3, 977–982 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Kumar T. K. A., et al. , An ontology of fungal subcellular traits. Am. J. Bot. 98, 1504–1510 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Tedersoo L., et al. , Fungal biogeography. Global diversity and geography of soil fungi. Science 346, 1256688 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Boddy L., Saprotrophic cord-forming fungi: Warfare strategies and other ecological aspects. Mycol. Res. 97, 641–655 (1993). [Google Scholar]
- 19.Green J. L., Bohannan B. J. M., Whitaker R. J., Microbial biogeography: From taxonomy to traits. Science 320, 1039–1043 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Aguilar-Trigueros C. A., et al. , Branching out: Towards a trait-based understanding of fungal ecology. Fungal Biol. Rev. 29, 34–41 (2015). [Google Scholar]
- 21.McGill B. J., Enquist B. J., Weiher E., Westoby M., Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21, 178–185 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Violle C., Reich P. B., Pacala S. W., Enquist B. J., Kattge J., The emergence and promise of functional biogeography. Proc. Natl. Acad. Sci. U.S.A. 111, 13690–13696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lennon J. T., Aanderud Z. T., Lehmkuhl B. K., Schoolmaster D. R. Jr, Mapping the niche space of soil microorganisms using taxonomy and traits. Ecology 93, 1867–1879 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Chagnon P.-L., Bradley R. L., Maherali H., Klironomos J. N., A trait-based framework to understand life history of mycorrhizal fungi. Trends Plant Sci. 18, 484–491 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Maynard D. S., et al. , Consistent trade-offs in fungal trait expression across broad spatial scales. Nat. Microbiol. 4, 846–853 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Treseder K. K., Lennon J. T., Fungal traits that drive ecosystem dynamics on land. Microbiol. Mol. Biol. Rev. 79, 243–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talbot J. M., et al. , Endemism and functional convergence across the North American soil mycobiome. Proc. Natl. Acad. Sci. U.S.A. 111, 6341–6346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordén J., Penttilä R., Siitonen J., Tomppo E., Ovaskainen O., Specialist species of wood-inhabiting fungi struggle while generalists thrive in fragmented boreal forests. J. Ecol. 101, 701–712 (2013). [Google Scholar]
- 29.Crowther T. W., et al. , Untangling the fungal niche: The trait-based approach. Front. Microbiol. 5, 579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boddy L., Fungal community ecology and wood decomposition processes in angiosperms: From standing tree to complete decay of coarse woody debris. Ecol. Bull. 49, 43–56 (2001). [Google Scholar]
- 31.Parnas H., Model for decomposition of organic material by microorganisms. Soil Biol. Biochem. 7, 161–169 (1975). [Google Scholar]
- 32.Walse C., Berg B., Sverdrup H., Review and synthesis of experimental data on organic matter decomposition with respect to the effect of temperature, moisture, and acidity. Environ. Rev. 6, 25–40 (1998). [Google Scholar]
- 33.Louis B. P., et al. , Soil C and N models that integrate microbial diversity. Environ. Chem. Lett. 14, 331–344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maynard D. S., et al. , Diversity begets diversity in competition for space. Nat. Ecol. Evol. 1, 0156 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Maynard D. S., Crowther T. W., Bradford M. A., Fungal interactions reduce carbon use efficiency. Ecol. Lett. 20, 1034–1042 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Maynard D. S., Crowther T. W., Bradford M. A., Competitive network determines the direction of the diversity-function relationship. Proc. Natl. Acad. Sci. U.S.A. 114, 11464–11469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zanne A. E., et al. , A deteriorating state of affairs: How endogenous and exogenous factors determine plant decay rates. J. Ecol. 103, 1421–1431 (2015). [Google Scholar]
- 38.Violle C., et al. , Let the concept of trait be functional! Oikos 116, 882–892 (2007). [Google Scholar]
- 39.Dawson S. K., et al. , Handbook for the measurement of macrofungal functional traits: A start with basidiomycete wood fungi. Funct. Ecol. 33, 372–387 (2019). [Google Scholar]
- 40.Aguilar-Trigueros C. A., Rillig M. C., Crowther T. W., Applying allometric theory to fungi. ISME J. 11, 2175–2180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crowther T. W., Bradford M. A., Thermal acclimation in widespread heterotrophic soil microbes. Ecol. Lett. 16, 469–477 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Hoppe B., et al. , Linking molecular deadwood-inhabiting fungal diversity and community dynamics to ecosystem functions and processes in Central European forests. Fungal Divers. 77, 367–379 (2016). [Google Scholar]
- 43.Della Mónica I. F., Godoy M. S., Godeas A. M., Scervino J. M., Fungal extracellular phosphatases: Their role in P cycling under different pH and P sources availability. J. Appl. Microbiol. 124, 155–165 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Boddy L., Heilmann-Clausen J., “Basidiomycete community development in temperate angiosperm wood” in British Mycological Society Symposia Series, Boddy L., Frankland J. C., van West P., Eds. (Elsevier, 2008), pp. 211–237. [Google Scholar]
- 45.Boddy L., Interspecific combative interactions between wood-decaying basidiomycetes. FEMS Microbiol. Ecol. 31, 185–194 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Henningsson B., Physiology and Decay Activity of the Birch Conk Fungus Polyporus betulinus (Bull.) Fr (Skogshögskolan, 1965). [Google Scholar]
- 47.Krause S., et al. , Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 5, 251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heilmann-Clausen J., A gradient analysis of communities of macrofungi and slime moulds on decaying beech logs. Mycol. Res. 105, 575–596 (2001). [Google Scholar]
- 49.Grime J. P., Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am. Nat. 111, 1169–1194 (1977). [Google Scholar]
- 50.Hiscox J., O’Leary J., Boddy L., Fungus wars: Basidiomycete battles in wood decay. Stud. Mycol. 89, 117–124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holmer L., Stenlid J., Competitive hierarchies of wood decomposing basidiomycetes in artificial systems based on variable inoculum sizes. Oikos 79, 77–84 (1997). [Google Scholar]
- 52.Toljander Y. K., Lindahl B. D., Holmer L., Högberg N. O. S., Environmental fluctuations facilitate species co-existence and increase decomposition in communities of wood decay fungi. Oecologia 148, 625–631 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Oberle B., et al. , Accurate forest projections require long-term wood decay experiments because plant trait effects change through time. Glob. Change Biol. 26, 864–875 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Jacobs J. M., Work T. T., Linking deadwood-associated beetles and fungi with wood decomposition rates in managed black spruce forests. Can. J. For. Res. 42, 1477–1490 (2012). [Google Scholar]
- 55.Rubenstein M. A., Crowther T. W., Maynard D. S., Schilling J. S., Bradford M. A., Decoupling direct and indirect effects of temperature on decomposition. Soil Biol. Biochem. 112, 110–116 (2017). [Google Scholar]
- 56.Heilmann-Clausen J., et al. , Communities of wood-inhabiting bryophytes and fungi on dead beech logs in Europe–Reflecting substrate quality or shaped by climate and forest conditions? J. Biogeogr. 41, 2269–2282 (2014). [Google Scholar]
- 57.Crowther T. W., Boddy L., Maynard D. S., The use of artificial media in fungal ecology. Fungal Ecol. 32, 87–91 (2017). [Google Scholar]
- 58.Kaiser C., Franklin O., Richter A., Dieckmann U., Social dynamics within decomposer communities lead to nitrogen retention and organic matter build-up in soils. Nat. Commun. 6, 8960 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi Z., Crowell S., Luo Y., Moore B. 3rd, Model structures amplify uncertainty in predicted soil carbon responses to climate change. Nat. Commun. 9, 2171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laiho R., Prescott C. E., Decay and nutrient dynamics of coarse woody debris in northern coniferous forests: A synthesis. Can. J. For. Res. 34, 763–777 (2004). [Google Scholar]
- 61.Rajala T., Peltoniemi M., Pennanen T., Mäkipää R., Fungal community dynamics in relation to substrate quality of decaying Norway spruce ( Picea abies [L.] Karst.) logs in boreal forests. FEMS Microbiol. Ecol. 81, 494–505 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Nesci A., Etcheverry M., Magan N., Osmotic and matric potential effects on growth, sugar alcohol and sugar accumulation by Aspergillus section Flavi strains from Argentina. J. Appl. Microbiol. 96, 965–972 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Ritchie F., McQuilken M. P., Bain R. A., Effects of water potential on mycelial growth, sclerotial production, and germination of Rhizoctonia solani from potato. Mycol. Res. 110, 725–733 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Magan N., Lacey J., Effect of water activity, temperature and substrate on interactions between field and storage fungi. Trans. Br. Mycol. Soc. 82, 83–93 (1984). [Google Scholar]
- 65.Elo A. E., The Rating of Chessplayers, Past and Present (Arco, 1978). [Google Scholar]
- 66.Baldrian P., et al. , Production of extracellular enzymes and degradation of biopolymers by saprotrophic microfungi from the upper layers of forest soil. Plant Soil 338, 111–125 (2011). [Google Scholar]
- 67.Žifčáková L., Dobiášová P., Kolářová Z., Koukol O., Baldrian P., Enzyme activities of fungi associated with Picea abies needles. Fungal Ecol. 4, 427–436 (2011). [Google Scholar]
- 68.Crowther T. W., et al. , Biotic interactions mediate soil microbial feedbacks to climate change. Proc. Natl. Acad. Sci. U.S.A. 112, 7033–7038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2018). [Google Scholar]
- 70.Bates D., Maechler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 71.Fox J., Weisberg S., An R Companion to Applied Regression, (Sage, Thousand Oaks, ed. 3, 2011). [Google Scholar]
- 72.Nakagawa S., Schielzeth H., A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 4, 133–142 (2013). [Google Scholar]
- 73.Jaeger B., R2glmm: Computes R Squared for Mixed (Multilevel) Models (Version 0.1.2, 2017).
- 74.Bartoń K., MuMIn: Multi-Model Inference (Version 1.43.6, 2018).
- 75.Oberle B., “Data and Code for, “Accurate forest projections require long-term wood decay experiments because plant trait effects change through time.” Available at http://ncf.sobek.ufl.edu/AA00026436. Deposited 10 March 2019” (New College of Florida Institutional Data Repository, 2019). [DOI] [PubMed]
- 76.Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G., Jarvis A., Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.