Significance
Within nuclei, genomes are spatially partitioned into compartments of transcriptionally active and inactive chromatin. How compartments are formed and the functions these structures perform in gene regulation are not well understood. We show that heterochromatic histone modifications H3K9me1/me2/me3 play critical roles in establishing inactive Arm and active Center genome compartments in Caenorhabditis elegans via two distinct mechanisms. Anchoring of chromosome arms to the nuclear envelope via an H3K9me-binding protein promotes their cis association, thereby partitioning arms from center regions. An anchoring-independent mechanism compacts chromosome arms. Eliminating H3K9me modifications or disrupting perinuclear anchoring significantly weakens compartments. However, attenuation of compartments does not cause significant changes in gene expression. Our study demonstrates that chromatin modifications are important determinants of three-dimensional genome organization.
Keywords: genome compartments, histone modifications H3K9me1/me2/me3, chromosome compaction, perinuclear anchoring, gene expression
Abstract
Genomic regions preferentially associate with regions of similar transcriptional activity, partitioning genomes into active and inactive compartments within the nucleus. Here we explore mechanisms controlling genome compartment organization in Caenorhabditis elegans and investigate roles for compartments in regulating gene expression. Distal arms of C. elegans chromosomes, which are enriched for heterochromatic histone modifications H3K9me1/me2/me3, interact with each other both in cis and in trans, while interacting less frequently with central regions, leading to genome compartmentalization. Arms are anchored to the nuclear periphery via the nuclear envelope protein CEC-4, which binds to H3K9me. By performing genome-wide chromosome conformation capture experiments (Hi-C), we showed that eliminating H3K9me1/me2/me3 through mutations in the methyltransferase genes met-2 and set-25 significantly impaired formation of inactive Arm and active Center compartments. cec-4 mutations also impaired compartmentalization, but to a lesser extent. We found that H3K9me promotes compartmentalization through two distinct mechanisms: Perinuclear anchoring of chromosome arms via CEC-4 to promote their cis association, and an anchoring-independent mechanism that compacts individual chromosome arms. In both met-2 set-25 and cec-4 mutants, no dramatic changes in gene expression were found for genes that switched compartments or for genes that remained in their original compartment, suggesting that compartment strength does not dictate gene-expression levels. Furthermore, H3K9me, but not perinuclear anchoring, also contributes to formation of another prominent feature of chromosome organization, megabase-scale topologically associating domains on X established by the dosage compensation condensin complex. Our results demonstrate that H3K9me plays crucial roles in regulating genome organization at multiple levels.
Eukaryotic genomes are organized into a series of higher-order structures within the 3-dimensional (3D) space of the nucleus (1–3). Chromatin loops can connect promoters of genes to distal regulatory elements located tens to hundreds of kilobases away (4, 5). In metazoans, topologically associating domains (TADs) of ∼1 Mb confine chromatin interactions to distinct genomic neighborhoods (6–9). The functional roles of TADs remain obscure (9–11), but TAD disruption by chromosomal rearrangements can cause transcriptional misregulation due to ectopic enhancer–promoter contacts (12–14). At an even higher scale, genomic regions preferentially associate with regions of similar transcriptional activity on the same chromosome and on different chromosomes, partitioning mammalian genomes into transcriptionally active (A) and inactive (B) genome compartments (15). Deciphering the molecular mechanisms underlying these hierarchical structural features is crucial for understanding the organization and regulation of genome functions.
Although active and inactive genome compartments are among the most prominent features of chromosome organization, the mechanisms that establish them remain elusive. Most loops and TADs can be eliminated by abrogating binding of key architectural proteins, including the SMC complex cohesin and the zinc-finger protein CTCF (16–19). However, genome compartments are preserved and enhanced in the absence of TADs, revealing that compartments are created through different mechanisms (20). The clustering of active and repressive genomic regions into separate compartments has been proposed to enhance transcriptional regulation and stabilize different expression states. However, the inability to perturb genome compartments has hindered our understanding of the functions of these higher-order chromatin structures.
Genome compartments correlate strongly with chromatin states in diverse organisms (21). In mammals, euchromatic regions form the A compartment and are characterized by active transcription and histone modifications, such as H3K4me3 and H3K27Ac. In contrast, heterochromatic regions form the B compartment and are characterized by transcriptional repression and the histone H3 modifications H3K9me2, H3K9me3, and H3K27me3. Existing evidence suggests two possible ways that chromatin marks could contribute to the formation of genome compartments. First, regions with matching chromatin marks could associate with each other. In a recent simulation study, the compartment organization observed in mammalian cells was best explained by a model in which interactions among heterochromatic regions are energetically favorable (22). The heterochromatic binding protein HP1, which recognizes H3K9me and undergoes phase separation, could drive associations between heterochromatin regions (23, 24). Second, positioning of heterochromatin at the nuclear periphery could promote separation of the genome into compartments. Heterochromatin marks are known to position genomic regions to the nuclear periphery to promote their interactions and compaction (25–27). Thus, heterochromatin-associated histone modifications and proteins may play a prominent role in genome compartmentalization.
However, the extent to which heterochromatic marks contribute to genome organization in metazoans has been challenging to evaluate experimentally. Eliminating either H3K9me or H3K27me3 in mice causes lethality, thereby impeding assessment of their functions in genome organization (28–30). Although centromere-proximal H3K9me-enriched heterochromatin in the filamentous fungus Neurospora crassa exhibits reduced interactions with surrounding euchromatin, partitioning of euchromatin from heterochromatin is more severely disrupted by loss of subtelomeric H3K27me2/me3 than H3K9me1/me2/me3 (31, 32), making the role of H3K9me unclear.
Here we use Caenorhabditis elegans as a model to investigate the roles of H3K9me in genome organization. The C. elegans genome has a unique distribution of repressive and active chromatin. The distal arms of all five autosomes and the left arm of the X chromosome form the inactive compartment and are enriched for the heterochromatin marks H3K9me1/me2/me3 and H3K27me3 as well as for repetitive DNA elements (27, 33, 34). The heterochromatic chromosome arms preferentially associate with nuclear lamina via the nuclear envelope protein CEC-4, which binds to H3K9me (35, 36). In contrast, the central regions of all chromosomes form the active compartment and are depleted for repressive chromatin marks. The central regions also exhibit a low level of lamina association and are enriched for actively transcribed genes.
C. elegans embryos lacking all forms of H3K9me are viable and develop to adulthood under normal growth conditions. Hence the worm provides a unique opportunity to explore the functions of H3K9me in a metazoan. Eliminating H3K9me by disrupting the histone H3K9 methyltransferases does derepress repetitive DNA elements in germlines of adults and does cause accumulation of RNA:DNA hybrids at high growth temperatures, ultimately leading to genome instability and sterility (37–39).
In addition to genome compartment architecture, C. elegans chromosomes are organized into megabase-scale TADs. Hermaphrodite X chromosomes have a unique TAD organization that distinguishes them from autosomes and the male X. Prominent, regularly spaced TADs on X are created by the dosage compensation complex (DCC), a condensin complex that represses transcription by half to equalize X expression between sexes (XO males and XX hermaphrodites) (40). TAD boundaries are defined by sequence-specific recruitment elements on X (rex sites) that promote high-occupancy DCC binding (10, 40). TAD boundaries are strengthened by enrichment of H4K20me1 on X caused by the H4K20me2 demethylase activity of a DCC subunit (41, 42), but roles of other histone modifications in TAD architecture are not known.
In this study, we used genome-wide chromosome conformation capture experiments (Hi-C) to quantitatively assess the impact of H3K9me on C. elegans genome architecture. Comparison of genome-wide chromatin interaction profiles in wild-type embryos versus met-2 set-25 mutant embryos defective in H3K9 methyltransferases or cec-4 mutant embryos defective in perinuclear anchoring revealed that H3K9me promotes formation of genome compartments through two different mechanisms. H3K9me-mediated perinuclear anchoring via CEC-4 regulates cis-association of chromosome arms, while anchoring-independent functions of H3K9me control chromosome arm compaction. In addition, H3K9me but not perinuclear anchoring facilitates formation of DCC-dependent TADs on X.
Although loss of H3K9me or disruption of perinuclear anchoring led to significant weakening of compartmentalization, no significant changes in expression were found for genes that switched compartments, suggesting that the compartment in which a gene resides is not a major determinant of its expression level. Furthermore, increased compartment intermingling in mutants did not cause dramatic expression changes for genes that remained in their original compartments. Although genes that remained in the inactive Arm compartment exhibited an up-regulation in met-2 set-25 double and cec-4 single mutants relative to genes that remained in the active Center compartment, the expression changes were caused primarily by local repressive effects of H3K9me. Therefore, genome compartment strength does not dictate levels of gene expression. Finally, loss of H3K9me, but not disruption of perinuclear anchoring, led to weakening of TAD boundaries established by the DCC on hermaphrodite X chromosomes. Our findings demonstrate an intimate relationship between histone modifications and genome organization that broaden our understanding of the determinants and functions of 3D genome architecture.
Results
H3K9me Regulates Genome Compartments in C. elegans.
Our previous Hi-C results from mixed-stage embryos revealed that the two arms of all C. elegans autosomes preferentially associate with each other, both in cis and in trans, while the central regions of autosomes preferentially associate with each other in trans (40). The left arm of chromosome X associates with autosomal arms in trans, and the remainder of X associates with the central regions of autosomes. This segregation of large active Center and inactive Arm chromatin regions partitions the genome into two compartments, comparable to the active A and inactive B compartments observed in flies and mammals, thereby providing the opportunity to dissect the molecular basis of genome compartmentalization.
Further analysis of our genome-wide Hi-C data in wild-type C. elegans embryos (10, 40) revealed architectural differences among the different chromosomes (Fig. 1A). The two arms of the three smaller autosomes (chromosomes I, II, and III) interact in cis at a higher frequency than expected for loci at that distance (Fig. 1 A and D), while the arms of the larger autosomes (chromosomes IV and V), which are separated by longer central regions, do not exhibit elevated cis interactions (Fig. 1 A and J). The right arm of chromosome V exhibits a unique topology. The ∼3-Mb region at the distal end of chromosome V forms a well-defined subdomain (ArmD subdomain) that separates from the remainder of the right arm. In contrast, the more central ∼3-Mb region of the right arm (ArmC subdomain) does not exhibit strong self-association. Both ArmD and ArmC subdomains interact strongly with the center regions adjacent to the Arm–Center junction (Fig. 1J and SI Appendix, Fig. S1G).
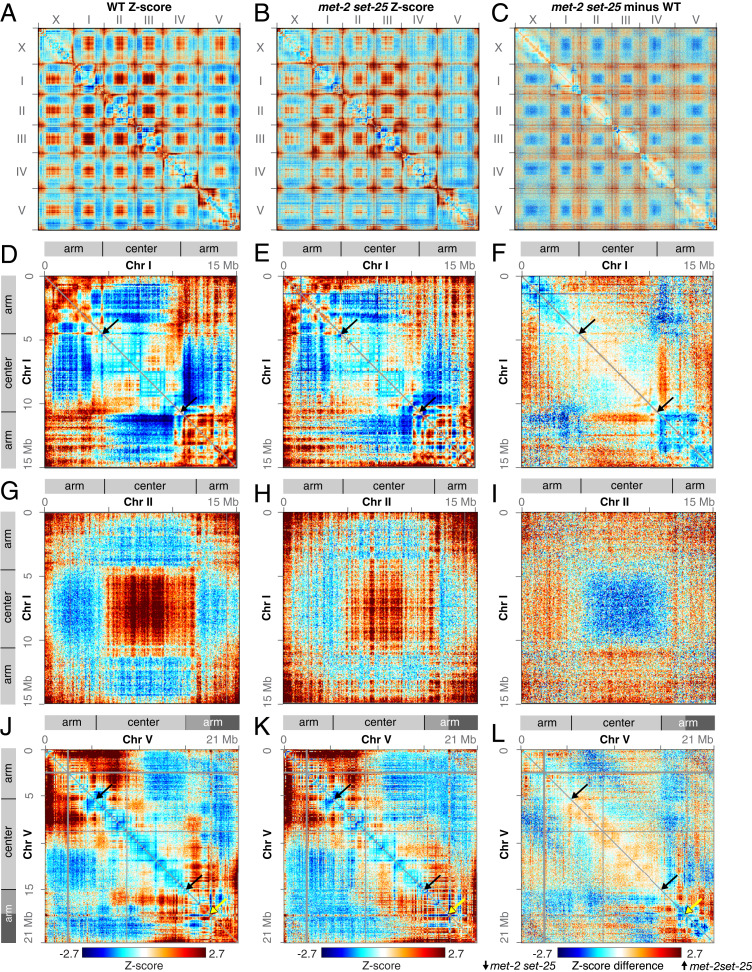
Fig. 1.
Elimination of H3K9me weakens genome compartments. (A and B) Whole-genome heatmaps show Hi-C Z-scores for the X chromosome and autosomes I–V in wild-type and met-2 set-25 mutant embryos. Red indicates higher interactions than expected for loci at a given distance, and blue indicates lower interactions than expected. The plaid pattern in interchromosomal interactions reflects compartment formation (high interactions between chromosome centers in trans and between chromosome arms in trans). (C) A Z-score subtraction heatmap shows increased (red) interactions between arms and centers and decreased (blue) interactions between centers and between arms in met-2 set-25 mutant compared to wild-type embryos. (D–F) Chromosome I heatmaps show Hi-C Z-scores in wild-type and met-2 set-25 mutant embryos and their difference. Diagrams delineate the Arm and Center compartments based on principal component analysis. Black arrows mark borders between Arm and Center domains. (G–I) Z-score heatmaps show trans interactions between chromosomes I and II in wild-type and met-2 set-25 mutant embryos and their difference. (J–L) The Z-score heatmaps show that the right arm of chromosome V consists of two subdomains (ArmC, mid gray, and ArmD, dark gray) demarcated by yellow arrows. Interactions between the two subdomains are low in wild-type but increase in met-2 set-25 mutant embryos. Black arrows mark borders between Arm and Center domains. All heatmaps are binned at 50 kb.
To assess the role of H3K9me in C. elegans genome organization, we performed in situ Hi-C in embryos lacking functional MET-2 and SET-25 methyltransferases, the major enzymes that deposit H3K9me1/me2 and H3K9me3, respectively. The met-2 set-25 mutant embryos have no detectable H3K9me of any form (27, 37). Comparison of genomic distance-normalized Hi-C interaction frequencies (Z-scores) between met-2 set-25 mutant and wild-type embryos (Fig. 1 A–C and SI Appendix, Fig. S1 A and B) revealed several obvious changes in chromatin interactions that reflect a loss of compartmentalization in the absence of H3K9me. First, in the double mutant, the two distal arms of chromosomes I, II, and III interact less frequently in cis (Fig. 1 D–F and SI Appendix, Fig. S1 D and E). In contrast, arms of chromosomes IV and V, which exhibit less prominent cis–arm association in wild-type embryos, are less affected in met-2 set-25 mutants (Fig. 1 J–L and SI Appendix, Fig. S1 G and H). Second, interactions among central regions of all chromosomes decrease significantly in the mutants (Fig. 1 A–C and G–I). Third, cis interactions between chromosome arms and central regions on chromosomes I, II, and III increase significantly, as do trans Arm–Center interactions among all chromosomes (Fig. 1 D–I and SI Appendix, Fig. S1 D and E). On the right side of chromosome V, the ArmD subdomain exhibits higher cis-interactions with both the central region and the ArmC subdomain (Fig. 1 J–L and SI Appendix, Fig. S1 G and H). Altogether, these changes demonstrate that H3K9 methylation plays a critical role in regulating the formation or maintenance of genome compartments in C. elegans.
H3K9me-Mediated Nuclear Peripheral Anchoring Facilitates Compartment Formation.
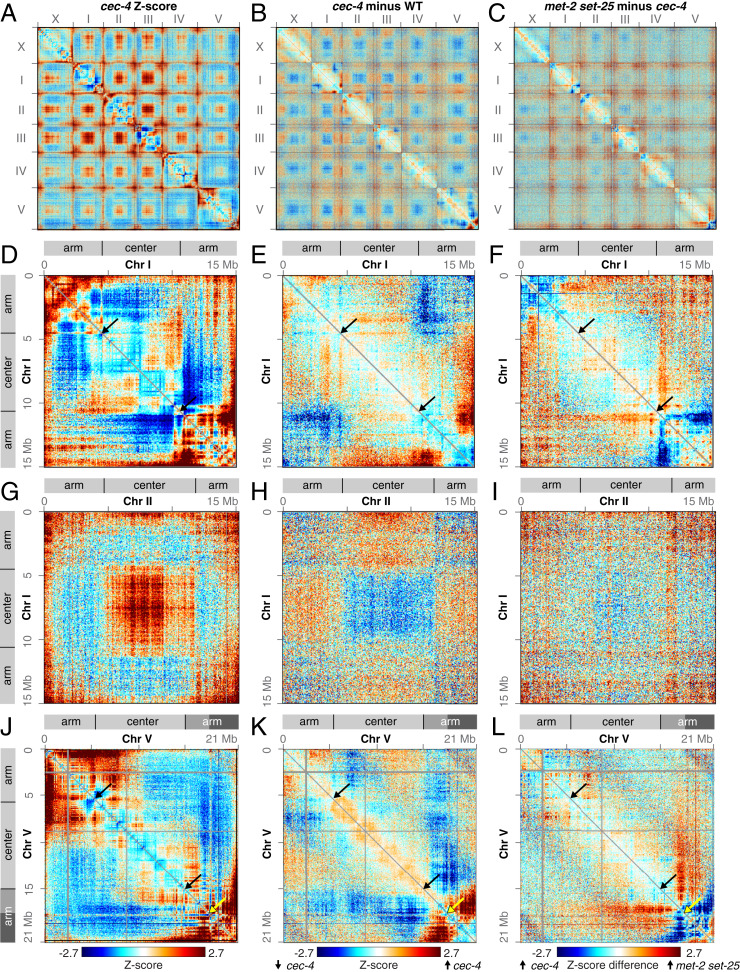
Disrupting perinuclear anchoring of chromosome arms while maintaining H3K9 methylation revealed that perinuclear anchoring is required for full separation of Arm and Center compartments. In C. elegans, H3K9me-enriched chromatin is anchored to the nuclear periphery by the chromodomain protein CEC-4, which localizes to the nuclear envelope and binds to all three forms of H3K9me. Prior studies showed that disrupting CEC-4 caused delocalization of chromosome arms from the nuclear periphery to the same extent as disrupting the H3K9 methyltransferases (36). To assess whether H3K9me’s role in compartmentalization is due to perinuclear anchoring of chromatin, we performed Hi-C in cec-4 mutant embryos (Fig, 2 A, D, G, and J). Comparison between Z-score heatmaps of cec-4 mutant and wild-type embryos revealed distinct changes in chromatin interactions among all chromosomes in a fashion similar to those in met-2 set-25 mutant embryos (Fig. 2 B, E, H, and K and SI Appendix, Fig. S1 C, F, and I). Thus, perinuclear anchoring via CEC-4 appears to facilitate genome compartmentalization (Fig. 2 A–L and SI Appendix, Fig. S1 A, C, F, and I).
Fig. 2.
Perinuclear anchoring of arms contributes to formation of genome compartments. (A) Whole-genome heatmap shows Hi-C Z-scores in cec-4 mutant embryos, which lack anchoring of the H3K9me-enriched arms to the nuclear periphery. (B) Whole-genome Z-score subtraction heatmap shows increased (red) and decreased (blue) interactions in cec-4 mutant compared to wild-type embryos. (C) Whole-genome Z-score subtraction heatmap shows increased (red) and decreased (blue) interactions in met-2 set-25 compared to cec-4 mutant embryos. (D–F) Chromosome I heatmaps show Hi-C Z-scores in cec-4 mutant embryos and Z-scores in cec-4 mutant compared to wild-type and met-2 set-25 mutant embryos. Diagrams delineate the Arm and Center compartments based on principal component analysis. (G–I) The Z-score heatmaps show trans interactions between chromosomes I and II in cec-4 mutant embryos and cec-4 mutant compared to wild-type and met-2 set-25 mutant embryos. (J–L) The Z-score heatmaps show that the right end of chromosome V strongly interacts with the rest of the right arm in cec-4 mutant embryos. These interactions are greatly diminished in met-2 set-25 mutant embryos. Black arrows mark borders between Arm and Center domains. Yellow arrows demarcate the boundary between the ArmC (mid gray) and ArmD (dark gray) subdomains. All heatmaps are binned at 50 kb.
However, disruption of compartment organization in cec-4 mutants is less substantial than that in met-2 set-25 mutants, as indicated by differences in chromatin interaction patterns between the two mutant strains (Fig. 2 C, F, I, and L). We quantified the strength of genome compartments in cec-4 and met-2 set-25 mutants using principal component analysis of Hi-C interactions. The first principal component (PC1), which is highly correlated with nuclear lamina association, defines the genome compartments in wild-type embryos. Genomic 50-kb bins with positive first eigenvector values are in the Center compartment, and bins with negative first eigenvector values are in the Arm compartment. In the cec-4 and met-2 set-25 mutants, PC1 still delineated Arm and Center compartments, and nearly 90% of the genomic bins remained in their original compartment (Fig. 3 and SI Appendix, Fig. S2 A–D). However, while PC1 explained 89% of the genome-wide variation in Hi-C interactions in wild-type embryos, it only explained 69% of the variation in cec-4 embryos and 63% in met-2 set-25 embryos, indicating the compartments were weakened. Hence, perinuclear anchoring activity of H3K9me accounts for a large proportion, but not all of its functions in genome compartmentalization.
Fig. 3.
Loci that switch compartments are near compartment borders. (A and B) Plots show eigenvector1 values for chromosomes V and X in wild-type, cec-4, and met-2 set-25 mutant embryos. Positive values define the Center compartment and negative values define the Arm compartment. Some loci near compartment borders switch compartments. Black horizontal lines mark examples of TADs that switch compartments as a unit. (C) Eigenvector1 plots show that the same TAD on the left side of X that switches compartments in cec-4 and met-2 set-25 mutants also switches compartments in a DCC mutant (Middle). In 8rexΔ (Bottom) the TAD exhibits an intermediate pattern, with eigenvector values closer to 0. The left vertical line for chromosome X is rex-32, which is deleted in 8rex∆.
In addition to causing increased mixing between compartments, loss of perinuclear anchoring led to increased interactions within the right arm of chromosome V, specifically between the ArmC and ArmD subdomains (Fig. 2K and SI Appendix, Fig. S1I). Interactions between ArmC and ArmD were not further increased in met-2 set-25 mutant embryos (Fig. 2L and SI Appendix, Fig. S1H). Thus, perinuclear anchoring not only promotes partitioning of active and inactive chromatin, it also promotes partitioning of two domains with similar chromatin marks.
Loci that Switch Compartments Reside Near Compartment Borders.
In both cec-4 and met-2 set-25 mutants, the ∼10% of loci that switched compartments tended to be near compartment borders (Fig. 3 A and B and SI Appendix, Fig. S2 A–D). In some cases, a TAD adjacent to a compartment border switched compartments as a unit (Fig. 3 A and B). For example, a TAD on the left side of chromosome V switched from the Center compartment to the Arm component (Fig. 3A). In addition, a TAD within the Arm compartment on the left side of chromosome X moved into the Center compartment in both met-2 set-25 and cec-4 mutants, shifting the compartment border (Fig. 3B). Furthermore, a broad region consisting of multiple TADs on the right side of chromosome X switched from the Center compartment to the Arm compartment in met-2 set-25 mutants, greatly expanding the Arm compartment on X (Fig. 3B).
Because the topology of X is regulated by a DCC, which resembles condensin, we asked whether the DCC also regulates compartment organization on X. We examined compartment structure after depleting a DCC subunit (SDC-2) that is essential for DCC binding to X. Loss of SDC-2 causes elevated X expression, decompaction of X, and disruption of TAD structure. DNA corresponding to a TAD on the left of X in wild-type embryos switched from the Arm to the Center compartment, as in met-2 set-25 mutants (Fig. 3C). Moreover, disrupting the TAD structure by deleting only the eight DCC binding sites (8rexΔ) at DCC-dependent TAD boundaries caused an intermediate change in the left Arm–Center border in which the eigenvector values approached 0, indicating the domain does not associate strongly with either the Arm or Center compartment (Fig. 3C). Together, these results indicate that compartment switching by depleting SDC-2 is not solely the consequence of disrupting DCC-dependent TAD boundaries; it also requires loss of DCC activity. Since DCC disruption does not affect the distribution of H3K9me3 (43), both DCC-dependent and H3K9me3-dependent mechanisms must regulate compartment organization on X.
Perinuclear Anchoring of H3K9me and Anchoring-Independent Functions of H3K9me Differentially Regulate Genome Organization.
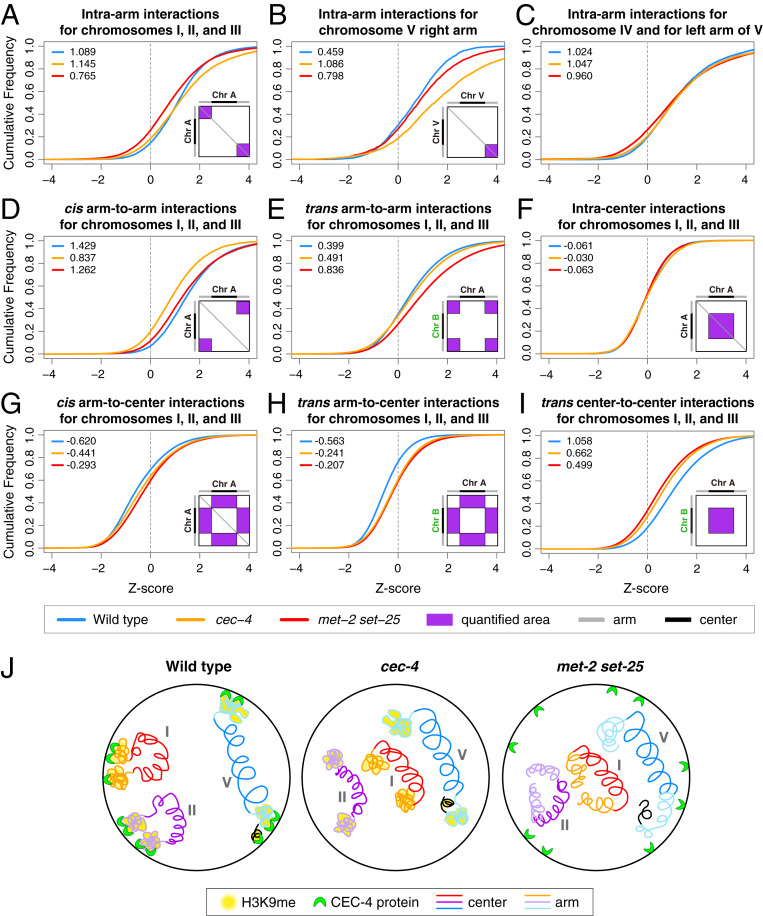
Our Hi-C results indicate that H3K9me regulates genome compartmentalization through two mechanisms: The CEC-4–dependent perinuclear anchoring of chromosome arms and a CEC-4–independent mechanism. To further delineate how the two mechanisms influence genome compartmentalization, we quantified the changes for different categories of chromatin interactions in both met-2 set-25 and cec-4 mutants and illustrated these changes in interaction strength using cumulative frequency plots.
Quantification of interactions within chromosome arms revealed that H3K9me promotes compartmentalization, in part, by facilitating compaction of chromosome arms through a mechanism that is independent of perinuclear anchoring. Loss of H3K9me in met-2 set-25 mutants resulted in a dramatic decrease in interactions within each chromosome arm (intra-arm) for the smaller and more compartmentalized chromosomes I, II, and III (median intra-arm Z-score 1.09 for wild-type and 0.76 for met2 set-25) (Fig. 4A), indicating decompaction of chromosome arms. In contrast, the left arm of chromosome V and both arms of chromosome IV exhibited only a slight decrease in intra-arm interactions in met-2 set-25 mutants (Fig. 4C), and the right arm of chromosome V exhibited a slight increase (Fig. 4B). In cec-4 mutants intra-arm interactions for different chromosome arms were either maintained or elevated (Fig. 4 A–C). Thus, anchoring-independent functions of H3K9me, but not CEC-4–dependent perinuclear anchoring, compact chromosome arms.
Fig. 4.
Perinuclear anchoring and anchoring-independent functions of H3K9me in genome organization. (A–I) Cumulative frequency plots illustrate shifts in distribution of Hi-C Z-scores and changes in Z-score median values for different categories of interactions on chromosomes of wild-type, met-2 set-25, and cec-4 embryos. Diagrams within the cumulative plots illustrate the quantified areas (purple) on the Z-score heatmaps for each interaction category. Schematics on the top and left of the diagrams depict chromosome arms (gray) and central regions (black). The median Z-score for each genotype is in the upper left corner of each plot. (A–C) In met-2 set-25 but not cec-4 mutants, the distribution of Z-scores for interactions within arms of chromosomes I, II, and III shifts leftward relative to that in wild-type embryos, indicating reduced interactions (A). Interactions within the right arm (B) but not left arm (C) of chromosome V increase dramatically in cec-4 mutants. (D) Cis interactions between arms of chromosomes I, II, and III decrease more dramatically in cec-4 than met-2 set-25 mutants. (E) Trans interactions between arms of different chromosomes increase dramatically in met-2 set-25 mutants. (F) Within the Center compartment, interactions within centers of chromosomes I, II, and III show no clear changes, while trans interactions between centers of different chromosomes dramatically decrease in both mutants (I). (G) cis and (H) trans interactions between arms and centers of chromosomes I, II, and III exhibit clear increases in both mutants, indicating increased intermingling between compartments. (J) The cartoons present a simplified view of how H3K9me likely controls compartmentalization through two mechanisms. For smaller autosomes I, II, and III, CEC-4-mediated perinuclear anchoring of chromosome arms brings the arms together. For chromosome V, perinuclear anchoring does not cause increased cis-association between arms, but limits interactions between the most distal region of the right arm (black) and the rest of the arm. In cec-4 mutants, the right end of V loops back internally. Independently of anchoring, H3K9me promotes compaction of arms. Compartment formation was not eliminated entirely in met-2 set-25 mutants, implying that other mechanisms contribute to compartmentalization (see Discussion).
The increases in intra-arm interactions upon disruption of CEC-4 (Fig. 4 A–C) can be attributed to increased flexibility of chromosome ends, as indicated by the more frequent interactions between the chromosome ends adjacent to the telomeres and the remainder of the chromosome arms (SI Appendix, Fig. S2 E–G). The phenomenon was most prominent at the right end of chromosome V, where the ArmD subdomain clearly gained interactions with the ArmC subdomain in cec-4 mutants (Fig. 2 B, J, and K and SI Appendix, Fig. S2G). This inward looping topology was inhibited by the elimination of H3K9me in met-2 set-25 mutants (Fig. 2L and SI Appendix, Fig. S2G).
Perinuclear anchoring of the two distal arms for the three smaller autosomes promoted long-range cis–arm interactions. In cec-4 mutants, the cis arm-to-arm interactions on chromosomes I, II, and III decreased dramatically (median Z-score 1.43 for wild type and 0.84 for cec-4) (Fig. 4D). Chromosomes IV and V, which do not show enriched cis arm-to-arm interactions in wild-type embryos, were affected to a smaller extent (SI Appendix, Fig. S3A). Furthermore, interactions among chromosome arms of different autosomes (trans arm-to-arm) increased slightly in cec-4 mutants (Fig. 4E and SI Appendix, Fig. S3B). Thus, for chromosomes I, II, and III, perinuclear anchoring brings the two arms of the same chromosome into closer proximity while spatially constraining each chromosome within its own territory, creating a configuration that favors cis arm-to-arm interactions over trans arm-to-arm interactions.
Both cis arm-to-arm and trans arm-to-arm interactions increased in met-2 set-25 mutants compared to cec-4 mutants (Fig. 4 D and E and SI Appendix, Fig. S3 A and B), suggesting that decompaction of chromosome arms increases their chances of contacting other arms over long distance. Taken together, these results indicate that CEC-4–dependent perinuclear anchoring, but not anchoring-independent functions of H3K9me, promotes interactions in cis between the two arms of the three smaller autosomes to facilitate the formation of the Arm compartment.
Perinuclear anchoring of chromosome arms also dramatically affects interactions between chromosome arms and central regions. In cec-4 mutants, arms of the three smaller autosomes interacted more frequently with the central region of the same chromosome (cis arm-to-center) (Fig. 4G). However, interactions between the right arm and center of chromosome V do not increase, likely due to the strong engagement between the right end of chromosome V with the rest of the chromosome arm (Fig. 2K and SI Appendix, Fig. S3D). In addition, loss of spatial constraints for chromosome arms in cec-4 mutants enabled the arms of all chromosomes to gain interactions with central regions of different chromosomes (trans arm-to-center) (Fig. 4H and SI Appendix, Fig. S3E). Neither cis nor trans arm-to-center interactions were further increased upon elimination of H3K9me (Fig. 4 G and H and SI Appendix, Fig. S3 D and E), suggesting that perinuclear anchoring acts as the main inhibitor of intermingling between Arm and Center compartments.
Interactions within the central regions of all autosomes (intra-center) were minimally affected in both cec-4 and met-2 set-25 mutants, consistent with the paucity of H3K9me in the central regions (Fig. 4F and SI Appendix, Fig. S3C). However, interactions among central regions of different chromosomes (trans center-to-center) decreased in both cec-4 and met-2 set-25 mutants (Fig. 4I and SI Appendix, Fig. S3F), suggesting that the Center compartment can be influenced indirectly by loss of perinuclear anchoring of chromosome arms.
Compared to autosomes, compartmentalization of the dosage-compensated X chromosomes is less influenced by H3K9me. Trans-association between the central region of X and central regions of autosomes decreased in both cec-4 and met-2 set-25 mutants (SI Appendix, Fig. S3L), indicating some influence of H3K9me on X compartmentalization. However, changes in most other categories of interactions were minimal (SI Appendix, Fig. S3 G–K). These results, combined with the finding that DCC disruption alters the Arm–Center border on the left side of X (Fig. 3C), reinforce the view that both H3K9me-dependent and DCC-dependent mechanisms are required to achieve proper X compartment organization.
Altogether, our analysis of chromatin interactions revealed two tiers of genome organization imposed by different functions of H3K9me (Fig. 4J). The presence of H3K9me on chromosome arms leads to arm compaction independently of anchoring. The binding of H3K9me by CEC-4 anchors chromosome arms to the nuclear periphery greatly promotes the cis-association of arms in the shorter chromosomes I, II, and III, and prevents the intermingling of inactive Arm and active Center genomic regions. Perinuclear anchoring of the right arm of chromosome V also limits interactions between the end of the chromosome (ArmD subdomain) and the rest of the arm. The collective effects of H3K9me result in effective compartmentalization of the C. elegans genome.
Genes that Switch Compartments Do Not Show Significant Changes in Expression.
Attenuation of Arm and Center compartments in met-2 set-25 and cec-4 mutants offered a unique opportunity to assess the role of genome compartmentalization in the regulation of transcription. In addition, our finding that cec-4 mutants showed weakened genome compartmentalization, while retaining H3K9me levels allowed us to separate the global effect of genome compartmentalization from the local repressive effect of H3K9me.
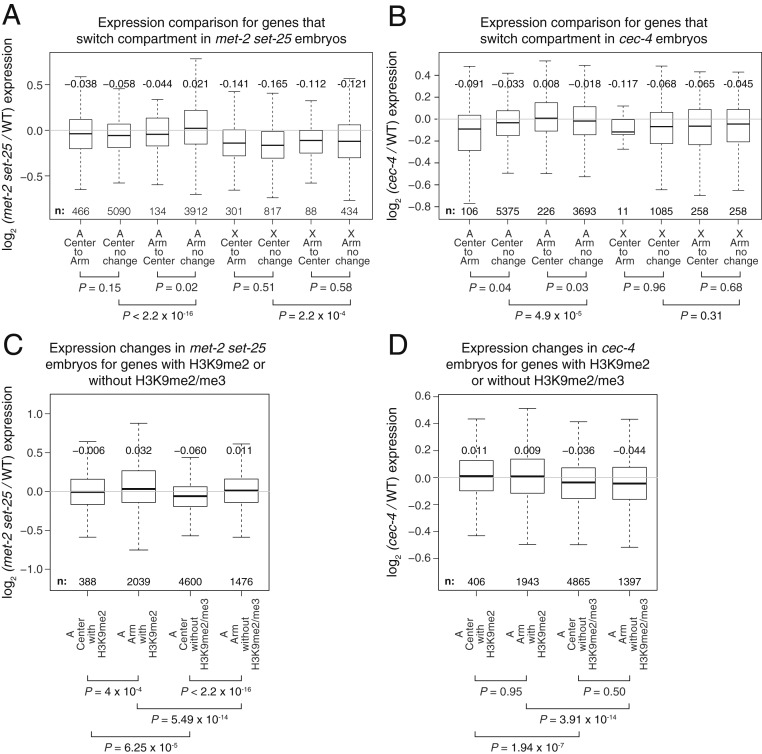
We first explored whether the expression levels of genes that switched compartments changed to reflect their new compartment. Our analysis of published gene-expression datasets from met-2 set-25 and cec-4 young mutant embryos (36) revealed that only 3 of the 600 expressed autosomal genes that switched compartments in met-2 set-25 mutants and none of the 332 autosomal genes that switched compartments in cec-4 mutants showed significant (adjusted P < 0.05) changes in expression.
Although only rare individual genes exhibited a significant change in expression upon switching compartments, we tested the possibility that an entire set of genes that relocated into the active or repressive compartment might have significant changes in expression compared to those that remained in their original compartment. In met-2 set-25 mutants, expression of the autosomal genes that switched from the repressive Arm compartment to the active Center compartment decreased slightly compared to genes that remained in the Arm compartment (Fig. 5A) (P = 0.02 for 134 genes, two-tailed t test), a trend opposite to that expected, while in cec-4 mutants expression of the genes that switched from the Arm to the Center compartment increased only slightly (Fig. 5B) (P = 0.03 for 226 genes). Autosomal genes that switched from the Center to the Arm compartment in met-2 set-25 mutants had no significant changes in expression (Fig. 5A) (P = 0.15 for 466 genes) compared to those that remained in the Center. In cec-4 mutants, autosomal genes that switched from the Center to the Arm compartment had only a slight decrease in expression (Fig. 5B) (P = 0.04 for 106 genes). Therefore, genes that switch between active and repressive compartments do not assume the transcriptional status of their new compartment.
Fig. 5.
Weakening of compartments does not cause significant changes in gene expression. (A and B) Box plot shows expression changes in met-2 set-25 or cec-4 mutant versus wild-type embryos for genes in Center and Arm compartments and genes that switch compartments on autosomes (A) and X. For each comparison, the median fold-change and number of genes included are listed. Genes with >1 fragment per kilobase of transcript per million mapped reads (FPKM) are included. Gene expression data are from ref. 36. (C and D) Box plots show early embryo expression changes in met-2 set-25 or cec-4 mutants for autosomal genes in Arm or Center compartments with H3K9me2 peaks (including those that also have H3K9me3 peaks) in the gene body or promoter, and with neither H3K9me2 nor H3K9me3 peaks. Genes with >1 FPKM that remain in the original compartment in mutant embryos are included. X-linked genes are excluded from analysis because of the small number of H3K9me2/me3 peaks. H3K9me2/me3 ChIP-seq data are from ref. 37.
The converse was also found: The group of genes with significant changes in expression was not enriched for genes that switched compartments. Of 132 genes with significant expression changes in met-2 set-25 mutants (adjusted P < 0.05), none switched compartments. Only one gene had a significant expression change in cec-4 mutants, and it did not switch compartments.
We also observed the same trends for X-linked genes, which we considered separately because they are subject to two different modes of chromosome-wide regulation. First, in both sexes genes on X are silenced in the germline, and this silencing is lost during early embryonic development. Second, starting in early embryonic development of hermaphrodites, genes on X are repressed twofold by the dosage-compensation process. As on autosomes, genes on X that switched between compartments in embryos of either mutant did not show changes in expression (Figs. 3B and 5 A and B) (P > 0.5).
Analysis of published gene-expression datasets from L1 larvae (44) revealed results similar to those observed in embryos. In general, autosomal and X-linked genes that switched between compartments in either of the L1 mutants were not significantly misregulated compared to genes that remained in the original compartments (SI Appendix, Fig. S4 G and H). An exception occurred for the 176 autosomal genes that switched from the active Center compartment to the repressive Arm compartment in cec-4 mutants. They exhibited significant up-regulation, contrary to the expectation if the compartment dictated the level of gene expression. The L1 gene-expression data further corroborate the conclusion that the compartment in which a gene resides is not a major determinant of its expression level in C. elegans.
Relative Elevation in Arm Gene Expression Is Caused Primarily by Local Effects of H3K9me Rather than Compartment Intermingling.
Using gene-expression data from young embryos, we assessed how expression of the genes that remained in their original compartment (90% of total expressed genes) was affected by loss of H3K9me2/me3 deposition and by loss of perinuclear anchoring. In met-2 set-25 mutants, autosomal genes remaining in the Arm compartment were significantly more up-regulated than genes remaining in the Center compartment (Fig. 5A) (P < 2.2 × 10−16, two-tailed t test). In cec-4 mutants, autosomal genes in the Arm compartment were also relatively elevated compared to genes in the Center compartment (Fig. 5B) (P = 5 × 10−5), although the differences were much smaller. Transcription of repetitive sequences was affected by loss of H3K9me (37, 38), but the inability to map repetitive reads uniquely prevented attribution of these changes to repeats in Arm versus Center compartments.
To understand the mechanisms underlying the relative elevation in Arm gene expression in mutants, we asked whether the elevation resulted from 1) loss of heterochromatic marks having local effects on genes with H3K9me peaks in wild-type embryos (45, 46), which are over-represented in Arm compartments (SI Appendix, Fig. S5 A–C), or from 2) increased compartment intermingling causing increased transcription throughout the Arm compartment. If the elevation of Arm gene expression was caused predominantly by impairment of a local H3K9me repressive function, we would expect genes associated with H3K9me in wild-type embryos, but not genes without H3K9me, to be up-regulated in mutants regardless of the compartment in which they resided. In contrast, if the increased compartment intermingling influenced gene expression, genes in the Arm compartment would be consistently more up-regulated than genes in the Center compartment, even when not associated with H3K9me. As shown below, the relative up-regulation of autosomal genes in Arm vs. Center compartments in mutants is generally better explained by a local repressive effect of H3K9me.
We categorized genes by their H3K9me2/me3 levels in wild-type embryos using chromatin immunoprecipitation sequencing (ChIP-seq) datasets (37) and then examined each category of genes for changes in expression (Fig. 5 C and D). Autosomal genes with H3K9me2 peaks in their promoter or gene body in wild-type embryos were significantly more elevated in expression in mutants than genes not associated with H3K9me2/me3 peaks (Fig. 5 C and D) for both Arm (P < 10−13, met-2 set-25 and P < 10−13, cec-4, two-tailed t test) and Center (P < 10−4, met-2 set-25 and P < 10−6, cec-4) compartments. These results indicate that H3K9me has a local repressive function in both Arm and Center compartments.
Repression occurs primarily for genes that have H3K9me2 in wild-type embryos but not for genes that have only H3K9me3 and no H3K9me2 (SI Appendix, Fig. S5 D and E), indicating a more critical role for H3K9me2 in repression. This finding is consistent with results of others examining repression of germline genes in embryonic somatic cells (46).
In cec-4 mutants, the local repressive effects of H3K9me are sufficient to explain the up-regulation of genes. Autosomal genes with H3K9me2/me3 peaks showed similar increases in expression regardless of the compartment (P = 0.95), and genes without H3K9me2/3 showed similar decreases in expression regardless of the compartment (P = 0.50) (Fig. 5D). Therefore, increased compartment intermingling does not account for the relative up-regulation of Arm genes in cec-4 mutants.
In met-2 set-25 mutants, compartment-wide effects in addition to local repressive H3K9me effects contribute to the increase in Arm gene expression. Autosomal Arm genes in met-2 set-25 mutants, unlike in cec-4 mutants, were consistently elevated compared to Center genes, whether or not they had H3K9me2/me3 peaks in wild-type embryos (P < 10−3 for all Center to Arm comparisons) (Fig. 5C). This compartment-wide effect cannot be attributed to increased intermingling between compartments because met-2 set-25 mutants show a similar level of interactions between Arm and Center compartments as cec-4 mutants (Fig. 4 G and H). Instead, the Arm up-regulation could result from long-range effects caused by decompaction of chromosome arms, which is observed in met-2 set-25 but not cec-4 mutants.
H3K9me, but Not Perinuclear Anchoring, Modulates DCC-Dependent TAD Organization on X Chromosomes.
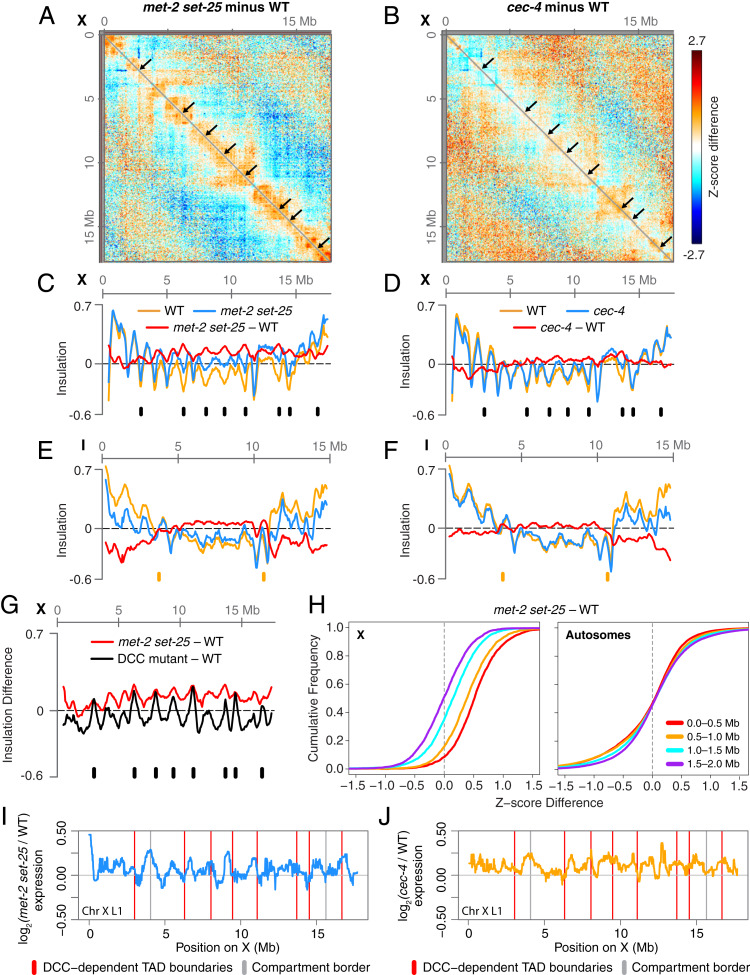
In addition to its role in compartment formation, H3K9me influences the formation of TAD boundaries on X chromosomes. Using a previously described, insulation-score approach (Fig. 6) (40), we found that the eight DCC-dependent TAD boundaries on X, but not the DCC-independent boundaries, were significantly weakened in met-2 set-25 mutants (Fig. 6 A and C and SI Appendix, Figs. S1 J and K and S6 E and F). Boundaries that were completely eliminated in DCC mutant embryos were only weakened in met-2 set-25 mutants (Fig. 6G), suggesting that H3K9me is necessary to reinforce but not create the boundaries.
Fig. 6.
Loss of H3K9me weakens DCC-dependent TAD boundaries on X. (A and B) X chromosome heatmaps binned at 50 kb show increased (red) or decreased (blue) Hi-C Z-scores in met-2 set-25 or cec-4 mutant compared to wild-type embryos. Black arrows mark DCC-dependent TAD boundaries. (C–F) Insulation plots show insulation scores across chromosome X or I in met-2 set-25 or cec-4 mutant (blue) and wild-type embryos (orange) and the insulation score difference between genotypes (red). Insulation profiles were generated by aggregating the interactions within a 500-kb window across every genomic interval. Local minima correspond to TAD boundaries, and the insulation differences between the minima at the boundaries and the maxima in the neighboring regions indicate the strength of the boundaries. Black ticks mark the position of DCC-dependent TAD boundaries. Orange ticks mark the position of borders between Arm and Center compartments. (G) X chromosome insulation plot shows the difference in insulation scores between either met-2 set-25 mutant or the DCC mutant sdc-2(y93, RNAi) and wild-type embryos. TAD boundaries lost in the DCC mutant (black ticks) are weakened to a lesser extent in met-2 set-25. DCC mutant data are from ref. 10. (H) Cumulative plots show Z-score differences between met-2 set-25 and wild-type chromosomes at different length scales. On X, interactions within a 1.5-Mb increase in met-2 set-25 compared to wild-type embryos, while autosomal interactions are unchanged. (I and J) Median gene expression changes in met-2 set-25 (blue) or cec-4 (yellow) mutant L1s versus wild-type L1s in a 200-kb sliding window across chromosome X. Boundaries between Arm and Center compartments are marked with gray vertical lines. DCC-dependent TAD boundaries on X are marked with red vertical lines. Gene expression data come from ref. 44.
In contrast, neither DCC-dependent nor DCC-independent TAD boundaries on X were significantly changed in cec-4 mutants (Fig. 6 B and D and SI Appendix, Figs. S1L and S6 A–F). Therefore, our results suggest that the presence of H3K9me on X, but not the perinuclear anchoring activity of H3K9me, plays a significant role in regulating DCC-dependent TAD boundaries on X.
In addition to loss of DCC-dependent TAD boundaries, DCC-mediated interactions between strong rex sites were lost in met-2 set-25 mutants but not in cec-4 mutants. The interactions among the 25 highest DCC-occupied rex sites were significantly reduced in met-2 set-25 mutant versus wild-type embryos, but not in cec-4 mutant embryos (SI Appendix, Fig. S6G), further illustrating the interplay between the DCC and H3K9me, but not perinuclear anchoring, in shaping X-chromosome structure.
Although DCC-mediated TAD boundaries and rex–rex interactions require H3K9me, another DCC-mediated structure is retained in met-2 set-25 mutants. The DCC promotes interactions between loci within 0.1 to 1 Mb, resulting in increased interactions on X compared to autosomes at that length scale (10). Unlike a mutant in which the DCC fails to assemble onto X, interactions within 1 Mb on X were unchanged in cec-4 mutants (SI Appendix, Fig. S6I) and increased in met-2 set-25 mutants (Fig. 6H).
The weakening of DCC-dependent TAD boundaries and rex–rex interactions in met-2 set-25 mutants is not due to loss of DCC binding. Previous cytological studies showed neither met-2 set-25 nor cec-4 mutations prevented DCC binding to X (44). We further explored DCC occupancy by performing ChIP-seq for the DCC component SDC-3 (SI Appendix, Fig. S7) and also analyzed available ChIP-seq datasets (43) (SI Appendix, Fig. S7 C and D). Consistent with cytological studies, our DCC binding analyses show no significant changes in DCC occupancy at rex sites in either mutant (SI Appendix, Fig. S7), indicating that H3K9me modulates TAD boundary formation without affecting DCC binding to rex sites. We found no notable enrichment of H3K9me at or near the rex sites in published ChIP-seq datasets (34) (SI Appendix, Fig. S6H). Thus, H3K9me is not required for proper loading of the DCC. To regulate TADs, either a low level of H3K9me may be required at rex sites to facilitate TAD boundary formation, or H3K9me and its binding proteins could act distantly from rex sites to control TAD formation.
In contrast to the strong effects that met-2 set-25 mutations have on DCC-dependent X TADs, almost no effects were found on autosomal TADs (Fig. 6 E and F and SI Appendix, Fig. S6 A–C). In both met-2 set-25 and cec-4 mutants, insulation profiles on autosomes were shifted, reflecting a change in chromatin compaction (SI Appendix, Fig. S2 J and K), but TAD boundary strength was unchanged (SI Appendix, Fig. S6 B and C) and TAD boundaries between Arm and Center regions were not weakened (SI Appendix, Fig. S6D). Our combined results reveal a role for H3K9me in regulating the formation of DCC-dependent TAD organization on X but not TAD organization on autosomes.
The up-regulation of X-linked genes in H3K9me mutants is not correlated with TAD organization. We plotted fold-changes of gene expression in embryos and in L1 larvae along the genomic coordinates and found no discernible patterns of expression changes for X-linked (Fig. 6 I and J and SI Appendix, Fig. S4 C, D, G, and H) or autosomal genes (SI Appendix, Fig. S4 A, B, E, and F) relative to TADs in met-2 set-25 or cec-4 mutants. While only met-2 set-25 mutant embryos exhibit substantial weakening of DCC-dependent TAD boundaries on X, both met-2 set-25 and cec-4 mutant L1 larvae (SI Appendix, Fig. S4 G and H) exhibit a similar magnitude of up-regulation along X. These results corroborate our earlier discovery that X-chromosome–wide gene repression is not coupled to DCC-driven TAD boundary formation (10).
Discussion
Our work has elucidated mechanisms that organize eukaryotic genomes into large-scale active and inactive chromatin compartments. Although patterns of histone modifications have been used to predict features of 3D genome architecture (47, 48), the roles played in genome organization by specific chromatin modifications, such as the heterochromatic modification of H3K9me, have been challenging to assess because of their essential roles in development. Here we show that H3K9me regulates genome organization in C. elegans by controlling the formation of transcriptionally active and inactive genome compartments on all chromosomes and by regulating the condensin-driven formation of TADs on X chromosomes.
Our analysis of chromatin interaction patterns caused by eliminating H3K9me revealed that this prominent heterochromatin mark regulates compartment formation in C. elegans through two distinct mechanisms. First, H3K9me promotes partitioning of inactive Arm from active Center compartments by regulating the radial positioning of chromosome arms. We showed that the perinuclear anchoring of H3K9me-enriched chromosome arms via chromodomain protein CEC-4 establishes a distinct chromosome configuration for the smaller autosomes by bringing the two distal arms of each chromosome into proximity. The larger autosomes IV and V do not exhibit such a CEC-4–dependent cis arm association, suggesting the size of central regions may present a topological barrier for the two arms to interact. However, for chromosome V, CEC-4–dependent perinuclear anchoring does modulate chromosome arm conformation by constraining interactions between subdomains within the right arm. Anchoring of the ArmD subdomain obstructs interactions between ArmD and the remainder of the arm, resulting in a more extended right arm conformation. For all autosomes, the spatial confinement of chromosome arms to the nuclear periphery promotes the separation between heterochromatic arms and euchromatic central regions, while strengthening interactions among central regions and thereby enhancing genome compartmentalization.
A recent cytological study of chromosome configuration suggested that CEC-4–dependent perinuclear anchoring influences genome organization differently in early-stage C. elegans embryos versus later-stage embryos in our study (49). Prior to gastrulation, perinuclear anchoring of chromosome arms led to an extended chromosome configuration, with low interactions between the two distal arms. Upon gastrulation, cis associations between arms increased, and a conventional Arm/Center compartment configuration emerged, as seen in our Hi-C studies. Changes that occur during embryogenesis may underlie the differential effects caused by CEC-4 at different stages of development, including the maturation of constitutive heterochromatin, extended duration of cell cycle, and decrease in nuclear volume.
The second way H3K9me facilitates compartment formation is by promoting intra-arm chromatin interactions and thereby increasing compaction of chromosome arms. This process does not require CEC-4–dependent perinuclear anchoring. Clustering and compaction of heterochromatin in Drosophila is achieved by HP1α, the major binding protein of H3K9me, which forms phase-separated condensates in vitro and in vivo (23, 24). Such phase separation of heterochromatin could underlie aspects of genome compartmentalization in C. elegans.
Organization of the genome into Arm and Center compartments was not entirely eliminated in the absence H3K9me (Fig. 1B and SI Appendix, Fig. S1B). Thus, H3K9me-independent mechanisms also contribute to the formation of genome compartments in C. elegans. In the case of X, we showed that the DCC also contributes to defining compartment borders. In N. crassa, the heterochromatin mark, H3K27me2/me3, promotes intra- and interchromosomal interactions between subtelomeric heterochromatin (32), supporting the idea that multiple heterochromatin marks may collectively drive the association of heterochromatin regions. In Drosophila, interactions between actively transcribed genes that form the active A compartment are reduced upon inhibition of transcriptional elongation, suggesting that RNA polymerase II or other factors that bind active chromatin also contribute to compartment formation (50).
The functional significance of genome compartmentalization in the regulation of transcription had not been explicitly defined. In mouse and Drosophila cells, disruption of nuclear lamina results in increased intermingling between active and inactive compartments, detachment of a subset of lamina-associated domains from the nuclear periphery, and up-regulation of genes within lamina-associated domains (51, 52). However, lamina disruption also affects the chromatin association of many transcription regulators, such as histone deacetylases (53). Whether the observed gene-expression changes are caused by overall changes in genome architecture or instead the loss of chromatin-associated factors has not been resolved in these cases. Furthermore, many genomic regions switch between A and B compartments during differentiation of mammalian stem cells (54, 55). A subset of genes that switch compartments exhibit changes in expression that match the identity of the new compartment, although the correlation is modest. Mechanisms underlying these changes in expression are not known.
In this study, while the defects in H3K9 methylation or H3K9me-dependent perinuclear anchoring significantly weakened genome compartmentalization, they did not cause dramatic changes in gene expression in either active or inactive genomic compartments. With rare exception (3 of 932), genes that switched compartments did not show significant changes in expression. However, mild changes in expression were found for an entire set of genes that remained in its original compartment, with Arm genes exhibiting a relative up-regulation compared to Center genes. These changes in gene expression are attributable primarily to the impairment of a local H3K9me-repressive function, rather than to increased intermingling of inactive Arm and active Center compartments. In our experiments, eliminating H3K9me does not abolish compartments entirely and causes only 10% of the genome to transition between compartments. Hence, the possibility remains that complete disruption of genome compartments could cause more dramatic changes in gene expression.
In line with our results, the widespread transcriptional response triggered by heat shock was not accompanied by changes in A/B compartments in human or Drosophila cells (56), suggesting that compartment changes are not required for gene-expression changes. These findings support the view that the transcriptional status of a compartment, either active or inactive, does not play a deterministic role in setting the level of gene expression.
Although eliminating H3K9me in C. elegans causes minimal changes in expression of nonrepetitive genes in somatic cells, it does lead to significant derepression of repetitive elements in both the germline and soma, resulting in genome instability (37). A critical role for H3K9me in silencing repetitive elements and immobilizing transposons has also been observed in fission yeast, flies, and mammals (57–59). Whether the role of H3K9me in promoting genome compartmentalization has functional relevance for its conserved role in ensuring genome stability and genome defense is yet to be determined.
In addition to discovering functions for H3K9me in driving genome compartmentalization, we observed that H3K9me plays an unexpected role in regulating TAD organization. The C. elegans DCC, which includes a condensin subcomplex, induces formation of prominent TADs on X chromosomes. Current data suggest the DCC drives TAD formation through a loop-extrusion mechanism (10), as has been proposed for TAD formation in mammals (3, 20, 60, 61). Accordingly, the condensin subunits serve as the loop extruder whose extrusion activity is halted when the entire DCC binds to its highest-occupancy binding sites on X. The observed weakening of DCC-dependent TAD boundaries and decreased interactions between TAD boundaries in the absence of H3K9me are consistent with impaired functioning of the extrusion barriers. Curiously, although this study of H3K9me and our previous study of H4K20me (41) showed that the strength of DCC-dependent TAD boundaries is dependent on specific histone modifications, we found that H3K9me and H4K20me are enriched on chromosome arms or across X, respectively, rather than simply being enriched at the strong DCC-binding sites (rex sites) that define TAD boundaries. These findings, together with reports that TAD boundaries are disrupted in mammals by DNA methylation at specific CTCF binding sites that serve as loop extrusion barriers (62), suggest that TAD organization is subject to intricate regulation through both DNA and histone modifications.
In conclusion, we have demonstrated that chromatin modifications function as important determinants of 3D genome architecture, particularly for the formation of genome compartments and TADs. Our studies open new directions for understanding the mechanisms underlying 3D genome architecture in metazoans and the consequent regulation of genome functions.
Materials and Methods
Detailed materials and methods are available in SI Appendix, SI Methods and Methods.
Strains.
C. elegans strains used in this study include: N2 Bristol strain (wild-type), RB2301 cec-4(ok3124) IV, and GW0638 met-2(n4256) set-25(n5021) III. All strains were maintained at 20 °C on NGM plates seeded with OP50 grown in LB.
Hi-C and Data Analysis.
The in situ Hi-C protocol was performed on mixed-stage mutant embryos of genotypes cec-4 (two biological replicates) and met-2 set-25 (three biological replicates), as described previously (41). Hi-C data analyses were performed using scripts derived from hiclib and cworld packages (40, 41) (SI Appendix). Comparisons were made to our Hi-C data (10) from wild-type embryos (GSM3680067 in the National Center for Biotechnology Information Gene Expression Omnibus [GEO]: accession no. GSE128568).
ChIP-Seq and Data Analysis.
SDC-3 ChIP-seq was performed on mixed-stage mutant embryos of genotypes met-2 set-25 (two biological replicates) and cec-4 (one biological replicate) as described in SI Appendix. SDC3 ChIP-seq datasets from this study, and from published SDC-3 ChIP-seq datasets downloaded from GEO (accession no. GSE122639) (43), were analyzed as in ref. 10. Previously published early-embryo H3K9me2 and H3K9me3 ChIP-seq datasets (accession no. SRP080806), L1 larvae H3K9me2 ChIP-seq datasets (accession no. GSE126884), and early-embryo H3K9me1, H3K9me2, and H3K9me3 ChIP-chip datasets (accession nos. GSE22720, GSE22740, and GSE22746) were downloaded from the Sequence Read Archive (SRA) and GEO and reanalyzed (SI Appendix).
RNA-Seq Data Analysis.
RNA-seq data from N2, cec-4, and met-2 set-25 early embryos (accession no. GSE74134) (36) and L1 larvae (accession no. GSE79597) (44) were downloaded from the GEO and analyzed using the DESeq2 package in R (SI Appendix).
Data Availability.
Hi-C and ChIP-seq datasets from this study are available through GEO accession no. GSE144253 (63).
Supplementary Material
Acknowledgments
We thank D. Stalford for graphic design and B.J.M. laboratory members for discussions. This work used the Vincent J. Coates Genomics Sequencing Laboratory at University of California, Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. Q.B. is supported by The Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (TP2018044), the Shanghai Pujiang Program (18PJ1406800), and the National Natural Science Foundation of China (31801056 and 31970585). B.J.M. is an investigator of the Howard Hughes Medical Institute and received funding via NIH Grants R01GM30702 and R35GM131845.
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE144253).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002068117/-/DCSupplemental.
References
- 1.Bickmore W. A., van Steensel B., Genome architecture: Domain organization of interphase chromosomes. Cell 152, 1270–1284 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Bonev B., Cavalli G., Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Mirny L. A., Imakaev M., Abdennur N., Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol. 58, 142–152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanyal A., Lajoie B. R., Jain G., Dekker J., The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao S. S. P., et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon J. R., et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nora E. P., et al. , Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robson M. I., Ringel A. R., Mundlos S., Regulatory landscaping: How enhancer-promoter communication is sculpted in 3D. Mol. Cell 74, 1110–1122 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Beagan J. A., Phillips-Cremins J. E., On the existence and functionality of topologically associating domains. Nat. Genet. 52, 8–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson E. C., et al. , X chromosome domain architecture regulates Caenorhabditis elegans lifespan but not dosage compensation. Dev. Cell 51, 192–207.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williamson I., et al. , Developmentally regulated Shh expression is robust to TAD perturbations. Development 146, dev179523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupiáñez D. G., et al. , Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke M., et al. , Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Despang A., et al. , Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Genet. 51, 1263–1271 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Lieberman-Aiden E., et al. , Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nora E. P., et al. , Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao S. S. P., et al. , Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarzer W., et al. , Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wutz G., et al. , Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nuebler J., Fudenberg G., Imakaev M., Abdennur N., Mirny L. A., Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl. Acad. Sci. U.S.A. 115, E6697–E6706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowley M. J., et al. , Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67, 837–852.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falk M., et al. , Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570, 395–399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larson A. G., et al. , Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strom A. R., et al. , Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bian Q., Khanna N., Alvikas J., Belmont A. S., β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J. Cell Biol. 203, 767–783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harr J. C., et al. , Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol. 208, 33–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Towbin B. D., et al. , Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150, 934–947 (2012). [DOI] [PubMed] [Google Scholar]
- 28.O’Carroll D., et al. , The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21, 4330–4336 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters A. H. F. M., et al. , Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Tachibana M., et al. , G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galazka J. M., et al. , Neurospora chromosomes are organized by blocks of importin alpha-dependent heterochromatin that are largely independent of H3K9me3. Genome Res. 26, 1069–1080 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klocko A. D., et al. , Normal chromosome conformation depends on subtelomeric facultative heterochromatin in Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 113, 15048–15053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahringer J., Gasser S. M., Repressive chromatin in Caenorhabditis elegans: Establishment, composition, and function. Genetics 208, 491–511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T., et al. , Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 21, 227–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikegami K., Egelhofer T. A., Strome S., Lieb J. D., Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 11, R120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez-Sandoval A., et al. , Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell 163, 1333–1347 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Zeller P., et al. , Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48, 1385–1395 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Padeken J., et al. , Synergistic lethality between BRCA1 and H3K9me2 loss reflects satellite derepression. Genes Dev. 33, 436–451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaney C. E., et al. , Heterochromatic foci and transcriptional repression by an unstructured MET-2/SETDB1 co-factor LIN-65. J. Cell Biol. 218, 820–838 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crane E., et al. , Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brejc K., et al. , Dynamic control of X chromosome conformation and repression by a histone H4K20 demethylase. Cell 171, 85–102.e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bian Q., Anderson E. C., Brejc K., Meyer B. J., Dynamic control of chromosome topology and gene expression by a chromatin modification. Cold Spring Harb. Symp. Quant. Biol. 82, 279–291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Street L. A., et al. , Binding of an X-specific condensin correlates with a reduction in active histone modifications at gene regulatory elements. Genetics 212, 729–742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snyder M. J., et al. , Anchoring of heterochromatin to the nuclear lamina reinforces dosage compensation-mediated gene repression. PLoS Genet. 12, e1006341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snowden A. W., Gregory P. D., Case C. C., Pabo C. O., Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr. Biol. 12, 2159–2166 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Rechtsteiner A., et al. , Repression of germline genes in Caenorhabditis elegans somatic tissues by H3K9 dimethylation of their promoters. Genetics 212, 125–140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di Pierro M., Cheng R. R., Lieberman Aiden E., Wolynes P. G., Onuchic J. N., De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture. Proc. Natl. Acad. Sci. U.S.A. 114, 12126–12131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi Y., Zhang B., Predicting three-dimensional genome organization with chromatin states. PLOS Comput. Biol. 15, e1007024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawh A. N., et al. , Lamina-dependent stretching and unconventional chromosome compartments in early C. elegans embryos. Mol. Cell 78, 96–111.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowley M. J., et al. , Condensin II counteracts cohesin and RNA polymerase II in the establishment of 3D chromatin organization. Cell Rep. 26, 2890–2903.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng X., et al. , Lamins organize the global three-dimensional genome from the nuclear periphery. Mol. Cell 71, 802–815.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulianov S. V., et al. , Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat. Commun. 10, 1176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barton L. J., Soshnev A. A., Geyer P. K., Networking in the nucleus: A spotlight on LEM-domain proteins. Curr. Opin. Cell Biol. 34, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dixon J. R., et al. , Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonev B., et al. , Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557–572.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ray J., et al. , Chromatin conformation remains stable upon extensive transcriptional changes driven by heat shock. Proc. Natl. Acad. Sci. U.S.A. 116, 19431–19439 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cam H. P., et al. , Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37, 809–819 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Wood J. G., et al. , Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 113, 11277–11282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bulut-Karslioglu A., et al. , Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell 55, 277–290 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Fudenberg G., et al. , formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanborn A. L., et al. , Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 112, E6456–E6465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flavahan W. A., et al. , Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meyer B. J., et al. , Histone H3K9 methylation promotes formation of genome compartments in C. elegans via chromosome compaction and perinuclear anchoring. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144253. Deposited 25 January 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Hi-C and ChIP-seq datasets from this study are available through GEO accession no. GSE144253 (63).