Significance
We identify a mechanism used by tumor cells to adhere to and migrate through a nanoporous, three-dimensional extracellular matrix characteristic of brain tissue. In this mechanism, cells engage hyaluronic acid, a key component within brain matrix, by assembling “microtentacles” that can extend tens of micrometers from the cell body and adhere to the matrix via the CD44 receptor. We also elucidate a molecular mechanism through which these structures can support motility based on the interplay of actin, microtubules, IQGAP1, and CLIP170. Analogous structures have previously been observed in circulating tumor cells and hypothesized to facilitate endothelial attachment and extravasation. Our studies strongly support this concept and further indicate that microtentacles can facilitate adhesion and invasion into tissue.
Keywords: glioblastoma, hyaluronic acid, extracellular matrix, mechanobiology, motility
Abstract
The structure and mechanics of many connective tissues are dictated by a collagen-rich extracellular matrix (ECM), where collagen fibers provide topological cues that direct cell migration. However, comparatively little is known about how cells navigate the hyaluronic acid (HA)-rich, nanoporous ECM of the brain, a problem with fundamental implications for development, inflammation, and tumor invasion. Here, we demonstrate that glioblastoma cells adhere to and invade HA-rich matrix using microtentacles (McTNs), which extend tens of micrometers from the cell body and are distinct from filopodia. We observe these structures in continuous culture models and primary patient-derived tumor cells, as well as in synthetic HA matrix and organotypic brain slices. High-magnification and superresolution imaging reveals McTNs are dynamic, CD44-coated tubular protrusions containing microtubules and actin filaments, which respectively drive McTN extension and retraction. Molecular mechanistic studies reveal that McTNs are stabilized by an interplay between microtubule-driven protrusion, actomyosin-driven retraction, and CD44-mediated adhesion, where adhesive and cytoskeletal components are mechanistically coupled by an IQGAP1–CLIP170 complex. McTNs represent a previously unappreciated mechanism through which cells engage nanoporous HA matrix and may represent an important molecular target in physiology and disease.
Tumor cell invasion is critically dependent on interactions with extracellular matrix (ECM) (1). Glioblastoma (GBM) is the most common and aggressive form of primary brain cancer, with outcomes improving only marginally over the last several decades (2, 3). GBMs are characterized by diffuse infiltration of tumor cells into surrounding healthy tissue, allowing cells to escape surgical resection and engage prosurvival microenvironmental cues that foster resistance to therapy (4). Targeting cell–ECM interactions in combination with conventional therapies therefore has potential to improve therapeutic outcome (5). The spatially heterogeneous composition and structure of brain ECM modulates invasion patterns (6). While the brain is generally richer in hyaluronic acid (HA) than other tissues, the intraparenchymal region is particularly HA-rich and generally lacking in fibrillar contact guidance cues associated with collagen, fibronectin, and laminin normally found in connective tissue (7). GBMs are more enriched in HA than lower-grade astrocytomas (8), and HA is a potent effector of aggressive invasion in GBM (9–11). Despite the clear functional significance of HA to GBM progression, the mechanisms by which cells invade HA-rich, nonfibrillar three-dimensional (3D) matrix are poorly understood.
CD44 is a transmembrane receptor for extracellular HA that plays a critical role in tumor progression and specifically GBM invasion (12). CD44 is highly overexpressed in GBM compared to healthy tissue and lower grade astrocytomas (13), and CD44 antibody blockade significantly reduces tumor size in rats with grafted C6 gliomas (14). Furthermore, knockdown (KD) of CD44 in human GBM tumors slows tumor growth and sensitizes tumors to cytotoxic agents (15). CD44 is also a marker of glioma stem cells (GSCs) (also known as tumor-initiating cells) and contributes to maintaining stemness (16). Our laboratory has shown that CD44 is necessary for adhesion and migration on engineered HA hydrogel-based matrix (17). The intracellular domain of CD44 interacts with the cytoskeleton through direct mechanical linkages or more indirectly through Src family kinase-based activation of Rho GTPase or focal adhesion kinase signaling (16, 18). Specifically, CD44 binds the actin cytoskeleton via ERM family proteins and the spectrin cytoskeleton via ankyrin proteins (18–20). IQGAP1 has also been reported to bind to CD44, which may provide an alternative mechanical linkage to the actin cytoskeleton (21). The relative roles of these binding interactions remain opaque and may be dependent on the cell type and microenvironment (22). Although CD44 is a recognized effector of cell invasion and several downstream binding partners have been identified, it remains unclear how the cytoskeleton coordinates to facilitate CD44-mediated invasion. Specifically, much remains to be discovered about the role of CD44 in protrusion formation and tension generation.
Here, we describe a mechanism through which GBM cells engage and invade HA-rich matrix through microtentacles (McTNs). McTNs engage HA via CD44 and are observed in both engineered HA scaffolds and tissue. Actin and microtubules often align in McTNs, undergo retrograde flow, and support tension generation against the ECM. Knockout (KO) of IQGAP1, a known cross-linker of actin and microtubules and regulator of microtubule growth, prevents cell adhesion and McTN formation and reduces migration speed on HA and HA–Arg-Gly-Asp (RGD). This work demonstrates that CD44-HA binding supports adhesion and migration through the formation of McTNs, which mediate adhesion and motility in HA-rich, nonfibrillar matrix.
Results
GBM Cell Adhesion and Migration on HA Are Associated with Extension of McTNs.
We had shown that CD44 is necessary for adhesion to HA in previous work, with CD44 KD or antibody blockade abrogating adhesion to HA-based hydrogels (17). However, the contributions of CD44 to force generation through the cytoskeleton remained unclear. To gain additional mechanistic insight into this result, we seeded both continuous GBM cells and GSCs on HA and performed phase and differential interference contrast (DIC) imaging. Cells exhibited partially rounded cell bodies surrounded by long, thin protrusions closely associated with the HA matrix visible at high magnification (Fig. 1A and SI Appendix, Fig. S1). The protrusions were positive for CD44 (Fig. 1B), with 65 ± 18% staining positive for both F-actin and tubulin in U-87 malignant glioma (MG) cells and 56 ± 17% in U-251 MG cells (Fig. 1B and SI Appendix, Fig. S2). Structured illumination microscopy (SIM) imaging revealed a fan of actin filaments supported by several microtubules interspersed throughout the protrusions in both continuous U-87 MG cells (Fig. 1C) and GSC-11 primary tumor-initiating cells (Fig. 1D). Acetylated microtubules were located in the center of the cell body and not in protrusions, suggesting that microtubules in protrusions are relatively dynamic (SI Appendix, Fig. S3A) (23). Tyrosinated microtubules, which regulate neuronal growth cone organization and are required for interaction with certain microtubule binding proteins, such as CLIP170, extended extensively into protrusions (SI Appendix, Fig. S3B) (24, 25). Detyrosinated tubulin, a marker of long-lived microtubules, was found interspersed along microtubules in protrusions, consistent with previous studies (SI Appendix, Fig. S3C) (26).
Fig. 1.
Cells interacting with HA matrix express actin- and microtubule-positive McTNs. (A) DIC imaging of live cells demonstrates that both continuous GBM cells and GSCs exhibit long, thin protrusions on HA. (Scale bar: 10 µm.) (B) SIM imaging of U-251 MG cells on HA demonstrates that protrusions are CD44-positive and frequently actin- and microtubule-positive. Dashed line box represents Inset area. Inset shows protrusions at cell periphery. (Scale bar: 10 µm.) (C) SIM imaging shows parallel actin filaments and microtubules in the core of protrusions of U-87 MG cells on HA. (Scale bar: 5 µm.) (D) GSC-11 cells express actin- and tubulin-positive protrusions. (Scale bar: 5 µm.) (E) CD44 shRNA KD U-251 MG cells seeded on HA or HA-RGD can form lamellipodia and large actin bundles at high RGD concentrations similar to the nontargeting control but are unable to spread or form protrusions on HA without RGD. (Scale bar: 20 µm.) (F) U-251 MG cells cultured for 6 d in 3D HA and 7 d in HA-RGD matrix. (Scale bar: 10 µm.) (G) GFP-tubulin and RFP-LifeAct–expressing U-251 MG cell embedded in 3D HA matrix and imaged by confocal microscopy. Dashed line box represents Inset area. Inset shows protrusions at cell periphery. (Scale bar: 10 µm.) (H) Schematic of ex vivo tissue culture slice model showing cells seeded onto brain tissue slices immersed in medium within a transmembrane insert. (I) GFP-tubulin and RFP-LifeAct–expressing U-251 MG cell after 1 d of culture in ex vivo tissue culture slice model as a Z-stack projection (12 slices, 1-µm spacing). (Scale bar: 10 µm.)
Although the distribution of microtubules within protrusions and overall length varied by cell type, all cells expressed F-actin and microtubule-positive protrusions on HA. Furthermore, cells on HA generally lacked thick actin bundles and large lamellipodia typically observed on two-dimensional (2D), integrin-engaging substrates (27–29). The morphology of these protrusions, particularly their microtubule and actin architecture, seemed reminiscent of microtubule-positive McTNs that have been described in circulating breast cancer cells and implicated in endothelial docking and extravasation (30–33). Thus, we refer to these protrusions as McTNs. However, a key difference is that GBM cells associate with a solid-state ECM and rarely enter the circulatory system (34), suggesting drastically different functions for McTNs in each case.
To determine how McTNs might be affected by the presence of integrin ligands normally expected to promote lamellipodia, focal adhesions, and stress fibers, we investigated the effects of varying RGD concentration on cell morphology. With increasing RGD concentration, lamellipodia and large actin bundles increasingly dominated the phenotype in cells with both short hairpin RNA (shRNA) KD of CD44 and a nontargeting control (Fig. 1E). In contrast, only the control cells were able to spread and form McTNs on HA. Fibrin both contains integrin-adhesive sites and binds CD44, although the CD44–fibrin(ogen) interaction is weaker than CD44-HA (35, 36). Cells seeded on 2D fibrin gels also exhibited lamellipodia and large actin bundles, consistent with cells on HA-RGD (SI Appendix, Fig. S4). Integrin engagement is thus necessary for actin bundling and lamellipodia formation, and CD44-substrate binding alone is not sufficient to generate these morphologies.
When cells were encapsulated in HA matrix with a 3D topology characteristic of brain parenchyma, cells not only retained McTNs but lost the intervening fan-like regions of actin (Fig. 1F). Cells in both the 3D HA and 3D HA-RGD matrices formed protrusions similar in size, length, and frequency and over similar timescales (Fig. 1F). McTNs in 3D (∼1 wk) formed much more slowly than in 2D (∼minutes), consistent with a need for extensive matrix remodeling for McTN formation. Furthermore, the similarity in morphology between cells in 3D HA and 3D HA-RGD matrices suggests that McTNs may be more prominent in 3D even when integrin ligands are available. The protrusions exhibited by U-251 MG cells expressing GFP-tubulin and RFP-LifeAct in 3D HA were also actin- and tubulin-positive, with microtubules occupying the core of the protrusions and extending into the protrusion ends (Fig. 1G). We have previously demonstrated that U-87 MG cells invade HA-RGD by tunneling into the bulk matrix over the course of several weeks and that invasion is preceded by elaboration of long protrusions (37). We observe here that U-251 MG tumorspheres cultured in HA for 10 d exhibit protrusions into the matrix and initial tunneling (SI Appendix, Fig. S5A). These protrusions may be involved in matrix degradation as larger protrusions, and tunneling cells are only observed in regions where the HA matrix is degraded (SI Appendix, Fig. S5B). These observations also suggest a mechanism by which cells move through nanoporous HA matrix, which requires degradation to form a sufficiently large defect in the gel before cell migration can occur.
While 2D and 3D HA gels recapitulate some features of the brain ECM, such as dimensionality, bulk elastic modulus, and HA richness, this matrix does not fully capture the complexity of brain tissue. Ex vivo slice models have proven a valuable paradigm that retains the complexity of tissue while remaining amenable to high-resolution imaging (38, 39). We therefore seeded U-251 MG cells on ex vivo tissue culture slices, placed the slices on Boyden chamber (transwell) filters, and allowed the cells to invade for 24 h (Fig. 1H). SIM imaging of invaded cells revealed actin- and microtubule-positive McTNs of similar cytoskeletal composition to those observed in 2D and 3D culture (Fig. 1I). The consistency in cytoskeletal morphology between the HA platform and brain tissue culture supports the physiological significance of McTNs in driving adhesion and migration in brain tissue.
McTN Structure and Function Depend on CD44.
Given the abundance of the HA receptor CD44 on the McTN surface, we next investigated functional contributions of CD44 to McTN morphology and function. When we applied stochastic optical reconstruction microscopy (STORM) to image cells on HA, we observed that CD44 randomly distributed across the entire cell membrane, completely covering the periphery of McTNs (Fig. 2A). This is in contrast to previous reports of CD44 clustering into microdomains in other non-GBM cell types, which has been hypothesized to increase interactions between CD44 and certain membrane proteins (40). The broad distribution of CD44 even on membrane surfaces not in contact with the HA matrix is consistent with our earlier finding that CD44-mediated adhesion to HA is rapid (minutes) and driven by many weak contacts. This is in contrast to integrin/focal adhesion-based attachment, which requires much longer maturation times and is associated with fewer, stronger, and more discrete adhesive structures (17).
Fig. 2.
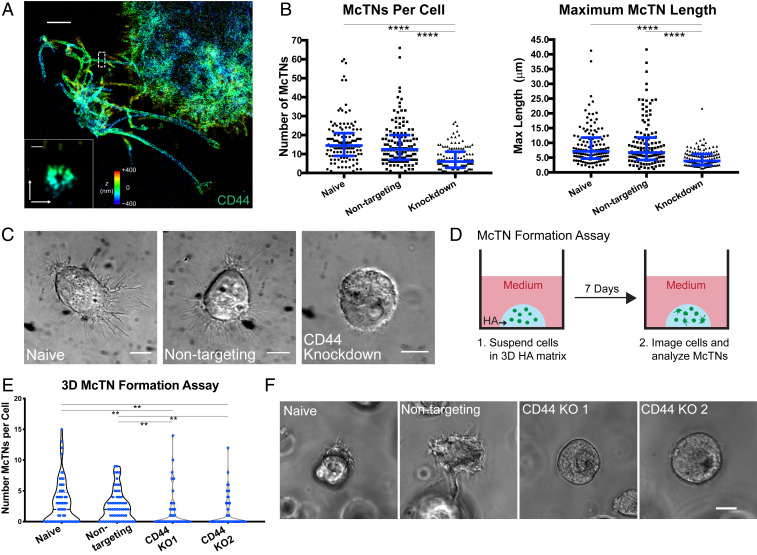
McTNs are CD44-dependent. (A) STORM imaging of CD44-covered McTNs at periphery of U-251 MG cell on HA matrix, with Inset showing transverse axis. Colors indicate z position. (Scale bar: A, 2 µm; Inset, 100 nm.) (B) shRNA KD of CD44 in U-251 MG cells on HA leads to a reduction of McTN number and length. n = 130 total cells from three independent experiments. ****P < 0.0001 by Kruskal–Wallis test followed by Dunn’s multiple comparisons test. Blue lines represent median with interquartile range. (C) DIC imaging of spread naive and nontargeting U-251 MG cells compared to rounded and poorly adhered shRNA CD44 KD cells. (Scale bars: 10 µm.) (D) Schematic of McTN formation assay in which cells are suspended in 3D HA, cultured for 7 d, and then imaged. (E) Number of McTNs formed per naive and nontargeting U-87 MG cells compared to CD44 KO controls. n = 60 total cells analyzed from three independent experiments. **P < 0.01 by Kruskal–Wallis test followed by Dunn’s multiple comparisons test. (F) Example phase images of cells from the high McTN expressing population in naive and nontargeting cells and the population expressing no McTNs in the CD44 KO cells. (Scale bar: 10 µm.)
We assessed the role of CD44 in McTN formation by counting the number of observable McTNs per cell from a population of randomly selected naive, nontargeting, and shRNA CD44 KD cells seeded on 2D HA. Using the same dataset, we measured the length of the longest McTN for each cell. CD44 KD cells expressed significantly fewer protrusions per cell (Fig. 2B). Similarly, CD44 KD cells generally did not form long protrusions. In particular, the population of cells expressing the most and longest McTNs was almost completely absent in CD44 KD cells. The KD cells remained rounded and extended only short protrusions, consistent with poor adhesion to the matrix (Fig. 2C).
As noted earlier, McTN formation is much slower in 3D than in 2D HA matrix (days vs. hours), which strongly implies McTN formation within 3D nanoporous HA matrix requires cells to degrade or otherwise remodel the surrounding matrix. To explore the functional importance of CD44 while accounting for these radically different kinetics, we designed a McTN formation assay in which cells were cultured in 3D HA for a week and then imaged within a single plane, and any visible McTNs were counted (Fig. 2D). During this extended period, we were concerned that the intrinsic heterogeneities associated with shRNA-mediated KD could allow selective pressures to artifactually enrich the population in cells with high CD44 expression. To avoid this confounding issue, we applied CRISPR-based gene editing to generate CD44 KO U-87 MG cells, for which we validated the lack of CD44 function through an inability to adhere to the bare HA matrix (SI Appendix, Fig. S6). In CD44 KO cells, we observed between 0 and ∼15 McTNs per cell, with most cells having fewer than five McTNs within the imaging plane (Fig. 2E). While this does not account for protrusions extending in the z-direction, the results are overall consistent with 2D data (Fig. 2B). Again, we observed a subpopulation of cells with more numerous McTNs in controls that was not present in CD44 KO cells (Fig. 2E). Furthermore, a larger proportion of CD44 KO cells expressed no McTNs. Cells expressing no McTNs were completely rounded and did not appear to productively engage the surrounding matrix (Fig. 2F). The finding that some CD44 KO cells are able to express McTNs in the 3D matrix may result from several alternative mechanisms that emerge in the absence of CD44-HA binding. First, other hyaladherins, such as RHAMM, not strongly bound in 2D may engage more tightly in the 3D matrix and compensate for CD44. Another possibility is that cells cultured in 3D HA over the course of a week secrete matrix components that engage adhesive receptors. Collectively, these data strongly indicate that McTN formation is CD44-dependent.
McTNs Uniquely Depend on Microtubules over Actin, Cdc42, and Myosin X.
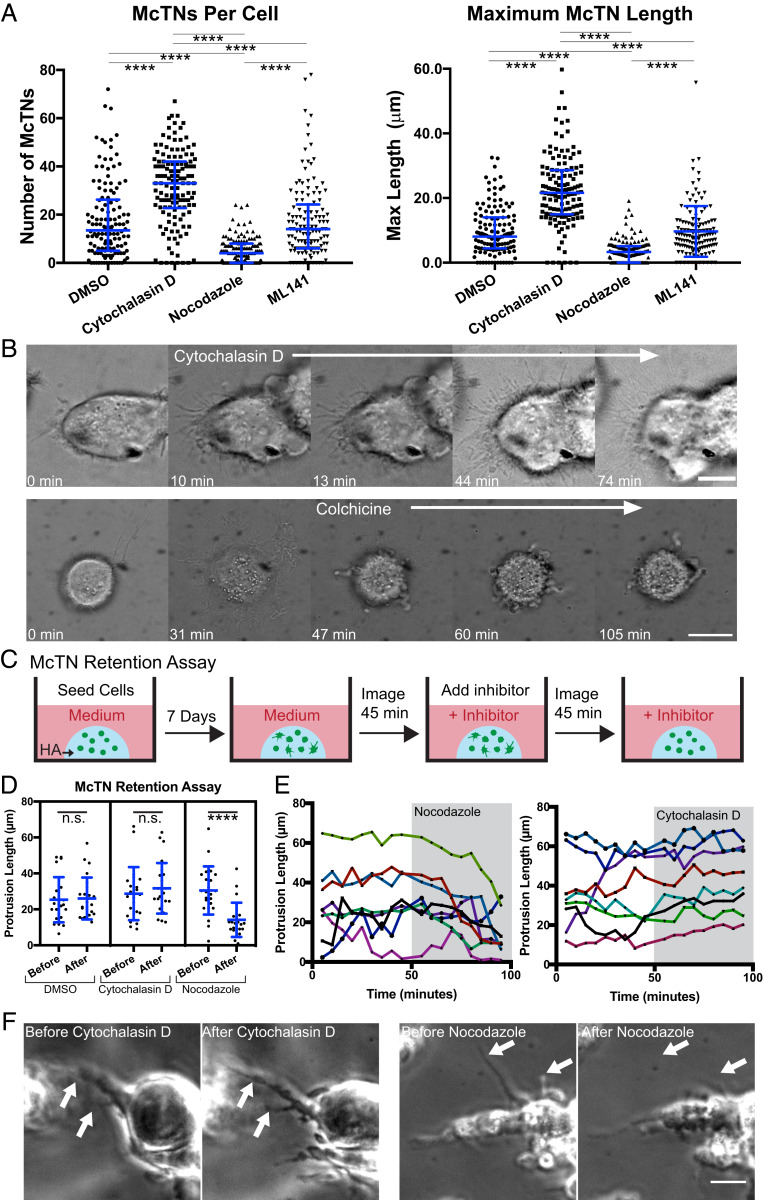
We next examined the extent to which McTNs depend on actin and microtubules. Cytochalasin D-mediated inhibition of actin polymerization increased both the number and length of McTNs, whereas nocodazole-based inhibition of microtubule polymerization reduced both the number of McTNs and the maximum protrusion length (Fig. 3A). Addition of cytochalasin D to CD44 knockout cells, however, did not generate significant growth of McTNs, indicating the necessity of CD44-HA adhesion to reinforce McTN formation and extension (SI Appendix, Fig. S7). The few, short McTNs that formed were likely microtubules that began to protrude but could not stabilize and grow. Notably, McTN number and length were insensitive to the Cdc42 inhibitor ML141 (Fig. 3A), implying that these structures are not filopodia (41). DIC imaging of cells during inhibitor treatment revealed that previously existing McTNs not only persisted but continued to grow after cytochalasin D treatment and were accompanied by induction of new McTNs (Fig. 3B). In contrast, colchicine treatment produced an initial collapse of most McTNs, followed by slow collapse of the remaining McTNs (Fig. 3B). Addition of cytochalasin D and nocodazole did not reduce localization of CD44 to the cell membrane (SI Appendix, Fig. S8). Thus, McTNs are functionally distinct from filopodia, with McTN formation and protrusion along the matrix promoted by microtubule polymerization and restricted by actin polymerization.
Fig. 3.
McTNs are distinct from filopodia and are stabilized by a balance between microtubules and actin. (A) Number of McTNs per U-251 MG cell and maximum McTN length per cell. n = 130 total cells analyzed from three independent experiments, ****P < 0.0001 by Kruskal–Wallis test followed by Dunn’s multiple comparisons test. Blue lines represent median with interquartile range. (B) DIC imaging of U-251 MG cells shows McTNs increase in length and number with cytochalasin D treatment while McTNs collapse with colchicine treatment. (Scale bars: 10 µm.) (C) Schematic showing McTN formation assay. Cells are seeded in 3D HA and cultured for 7 d. Cells are imaged for at least 45 min, an inhibitor is added, and then cells are again imaged for 45 min while McTN lengths are tracked. (D) Length of tracked McTNs at 45 min before inhibitor addition compared to 45 min after inhibitor addition in U-87 MG cells. n = 24 McTNs from 12 cells analyzed. ****P < 0.001, with lengths before and after each inhibitor compared by a paired t test. Blue lines represent mean and SD. n.s., not significant. (E) Lengths of example individual protrusions tracked before and after inhibitor treatment. Each color represents a single McTN. (F) Examples of tracked protrusions (white arrows) 45 min before inhibitor addition and 45 min after inhibitor addition. (Scale bar: 20 µm.)
To explore these regulatory concepts in 3D, we modified our McTN formation assay (Fig. 2D) to quantify McTN retention. Specifically, cells were cultured for 1 wk to allow for McTNs to grow and then imaged for 45 min prior to inhibitor treatment and 45 min after treatment (Fig. 3C). We hypothesized that if microtubules were truly critical to McTN retention, inhibition of microtubules would result in McTN collapse, but inhibition of actin would not. Comparing the lengths of individual McTNs 45 min prior to inhibitor treatment and 45 min after inhibitor treatment showed that dimethyl sulfoxide (DMSO) and cytochalasin D treatment did not significantly affect protrusion length, but nocodazole treatment resulted in decreased McTN length (Fig. 3D). Observation of individual protrusions suggested that while McTNs collapsed gradually after nocodazole treatment, McTNs were largely unaffected by cytochalasin D treatment (Fig. 3 E and F). The lack of McTN growth in response to microtubule disruption is consistent with a model in which McTN extension requires degradation and remodeling of the HA matrix, which would be expected to occur over days. Together, these results suggest a similar microtubule dependence of McTNs in 3D matrix as observed on 2D matrix.
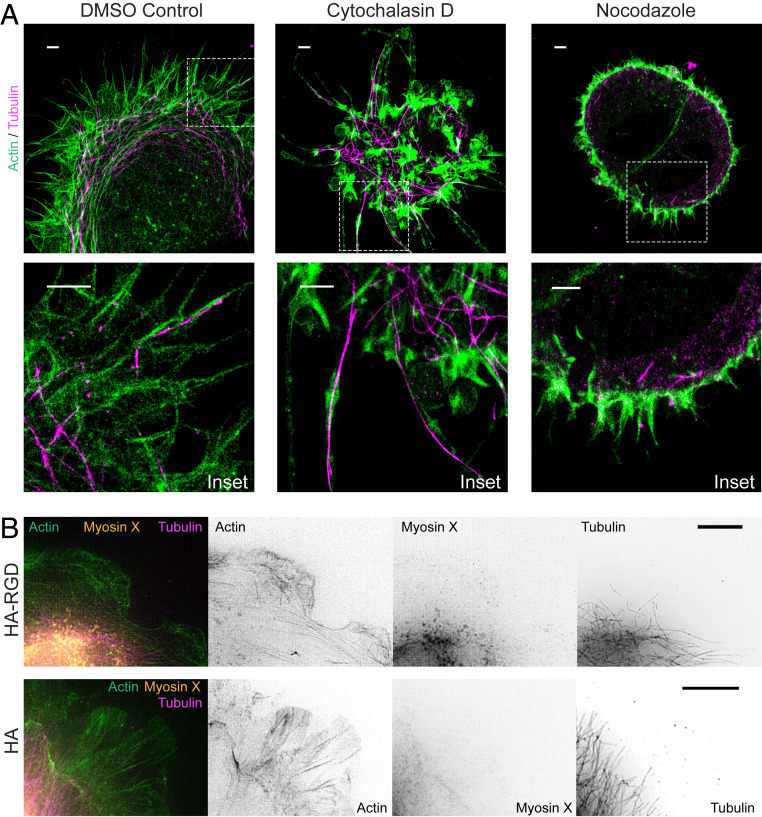
To gain additional insight into McTN ultrastructure, we revisited STORM to investigate cytoskeletal organization in these structures (Fig. 4A). In DMSO-treated controls, cell protrusions were composed of linear actin filaments that extended to the tips of some protrusions with interspersed and coaligned microtubules. Notably, cells on HA matrix lacked large actin bundles or lamellipodia. After cytochalasin D treatment, filamentous actin organization was not discernible, and microtubules dominated the core of long protrusions. Nocodazole treatment disrupted nearly all microtubule structures, with some short microtubule fragments present at the periphery. Nocodazole-treated cells expressed short actin filaments around the periphery. As a whole, nocodazole-treated cells were rounded and did not appear to interact with the HA matrix, consistent with our previous results. We also used SIM to examine the localization of myosin X, a recognized filopodial tip marker thought to contribute to actin polymerization and traction force generation (42). While these cells did not strongly express myosin X at the tips of filopodia even when seeded on HA-RGD, some myosin X was present in the lamellipodia of cells on HA-RGD (Fig. 4B). In contrast, cells on HA matrix expressed almost no myosin X in protrusions. Together, these results underscore mechanistic differences between filopodia and McTNs.
Fig. 4.
Organization of microtubules and actin within McTNs. (A) STORM imaging of U-251 MG cells treated with DMSO as a control shows actin filaments extending around the cell periphery with microtubules interspersed and extending to the ends of McTN tips. After cytochalasin D treatment, most protrusions contain microtubules at the core. Cells treated with nocodazole adhere poorly to HA, remain rounded, and express only short, actin-positive protrusions at the periphery. Dashed line boxes represent Inset area. Insets show protrusions at cell periphery. (Scale bars: 2 µm.) (B) U-251 MG cells on bare HA do not express myosin X at the cell periphery. Cells on HA-RGD have some myosin X at the leading edge compared to cells on HA. (Scale bars: 5 µm.)
McTNs Are Dynamic and Participate in Cell Adhesion and Motility.
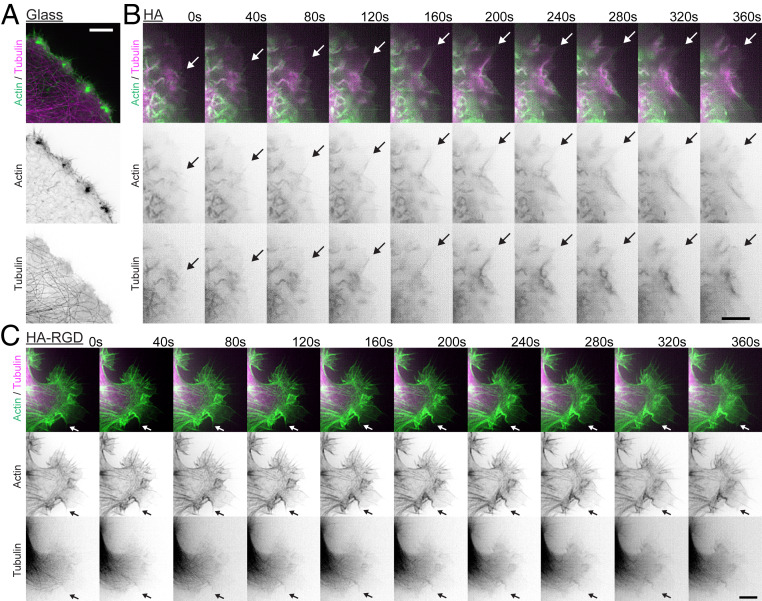
To gain deeper mechanistic insight into the dynamic interplay of microtubules and actin in McTNs, we applied live-cell SIM imaging to cells on HA matrix expressing GFP-tubulin and RFP-LifeAct. While control cells on glass exhibited more traditional migratory structures in which microtubules and actin were not strongly colocalized and short filopodial structures decorated the leading edge, cells on HA displayed a different morphology (Fig. 5 A and B). Cells on bare HA matrix exhibited strong colocalization of actin filaments and microtubules in linear protrusions. Furthermore, colocalized actin filaments and microtubules underwent retrograde flow at the leading edge of the cell at qualitatively similar rates (Fig. 5B and Movie S1). Cells on HA-RGD rarely showed colocalization of actin and microtubules, with actin filaments again outlining the cell periphery (Fig. 5C and Movie S2). Retrograde flow on HA-RGD also appeared to be much slower than on bare HA matrix (Movie S2).
Fig. 5.
Microtubules and actin align and undergo coordinated retrograde flow in McTNs. (A) On glass, U-251 MG cells express short actin filaments. (Scale bar: 5 µm.) (B) Microtubules and actin grow in parallel and undergo retrograde flow in U-251 MG cells on HA. Arrow shows an example microtubule and actin microfilament in parallel. (Scale bar: 5 µm.) (C) Microtubule and actin are less strongly colocalized in U-251 MG cells on HA-RGD compared to HA. The arrow shows an example where a microtubule is growing in parallel with actin. (Scale bar: 5 µm.)
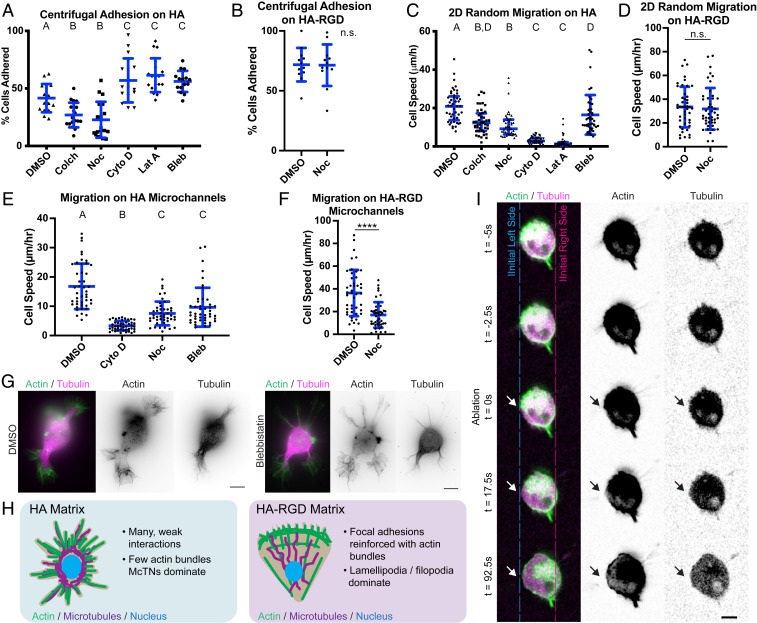
To investigate the role of McTNs in cell adhesion, mechanics, and motility, we pharmacologically manipulated McTN dynamics while probing these processes in several different in vitro paradigms. Using a centrifugal adhesion assay, we found that disruption of microtubules reduced adhesion while disruption of actin polymerization increased adhesion in both U-87 MG and U-251 MG cells (Fig. 6A and SI Appendix, Fig. S9A). These results were consistent with our previous observations that nocodazole-treated cells are rounded on HA matrix while cytochalasin D-treated cells express more and longer McTNs. In contrast, disruption of microtubule polymerization did not affect adhesion to HA-RGD matrix (Fig. 6B). Disruption of either microtubules or actin decreased 2D random migration on HA, with actin disruption resulting in a more extreme reduction of motility (Fig. 6C and SI Appendix, Fig. S9B). However, disruption of microtubules did not affect 2D migration on HA-RGD, despite affecting cell morphology and spreading (Fig. 6D and SI Appendix, Fig. S10A). This implies that cells on HA as opposed to HA-RGD treated with microtubule polymerization inhibitors do not adhere strongly enough to generate traction forces necessary for migration. The loss of microtubule-mediated signaling may also or alternatively reduce F-actin assembly, thereby reducing the area of adhesion and number of adhesive bonds engaged with the surface. Conversely, cells treated with actin polymerization inhibitors can adhere strongly but cannot generate sufficient actomyosin-based traction to support motility. Moderate inhibition of myosin II-mediated contractility by blebbistatin resulted in an increase in HA adhesion and decrease of migration speed on 2D HA substrates (Fig. 6 A and C). These results mirrored trends observed with actin polymerization inhibitors although to a lesser extreme. To characterize migration in environments characteristic of confined geometries found in tissue, we seeded cells onto HA or HA-RGD microchannels. We saw a similar relative effect of inhibitors on migration of cells in HA microchannels as on 2D matrix (Fig. 6E). Microtubule disruption slowed migration of cells on HA-RGD microchannels (Fig. 6F) but did not prevent cells from attaching and spreading to some degree in HA-RGD channels (SI Appendix, Fig. S10B). SIM imaging revealed that blebbistatin treatment resulted in longer, thinner McTNs with more prominent microtubule-based structures than with DMSO alone (Fig. 6G). Microtubule-driven protrusion or microtubule-based signaling and CD44-mediated adhesion thus seem to balance actomyosin-driven contractility in McTN-based motility (Fig. 6H). In contrast, integrin-RGD binding strengthens adhesion and reinforces actin bundles, which reduces the need for mechanical balance by microtubule polymerization or signaling.
Fig. 6.
McTNs facilitate cell adhesion and motility on HA matrix. (A) Centrifugal adhesion assay for U-87 MG cells interacting with HA. n = 15–18 total gels analyzed from three independent experiments. A, B, and C represent statistical families with a significant difference of P < 0.05 by ANOVA followed with Tukey–Kramer multiple comparisons test. Blue lines represent mean and SD. Inhibitors include colchicine (Colch), nocodazole (Noc), cytochalasin D (Cyto D), latrunculin A (Lat A), and blebbistatin (Bleb). (B) Centrifugal adhesion assay for U-87 MG cells interacting with HA-RGD. n = 12 total gels analyzed from three independent experiments, with no significant difference by Student’s t test. Blue lines represent mean and SD. (C) Random 2D migration speeds of U-87 MG cells on HA. n = 45 total cells analyzed from three independent experiments. A, B, C, and D represent statistical families with a significant difference of P < 0.05 by ANOVA followed with Tukey–Kramer multiple comparisons test. Blue lines represent mean and SD. (D) Random 2D migration speeds of U-87 MG cells on HA-RGD. n = 45 total cells analyzed from three independent experiments. No significant difference detected by Student’s t test. Blue lines represent mean and SD. (E) The 1D random migration of U-87 MG cells on microchannels in HA. n = 45 total cells analyzed from three independent experiments. A, B, and C represent statistical families with a significant difference of P < 0.05 by ANOVA followed with Tukey–Kramer multiple comparisons test. Blue lines represent mean and SD. (F) The 1D random migration of U-87 MG cells on microchannels composed of HA-RGD. n = 45 total cells analyzed from three independent experiments. ****P < 0.0001 by Student’s t test. (G) SIM imaging of U-87 MG cells treated with blebbistatin compared with cells treated with the vehicle control DMSO. (Scale bars: 10 µm.) (H) Schematic of cell morphology on HA compared to HA-RGD. Cells on HA exhibit McTNs. (I) Laser ablation of McTN in U-251 MG cell expressing GFP-tubulin and RFP-LifeAct. An arrow indicates an ablation site. (Scale bar: 10 µm.)
Based on this model, we hypothesized that McTNs must bear tension. Laser ablation of an McTN extending from a U-251 MG cell on HA directly verified this hypothesis (Fig. 6I and Movie S3) After ablation, the microtubule component was no longer visible, presumably due to rapid microtubule depolymerization induced by the ablation (43). The cell body shifted away from the site of laser ablation, implying that the ablated McTN had been maintaining a tensile force between the cell body and the HA matrix.
McTN Formation Requires IQGAP1.
The coupling of cytoskeletal forces to cell traction and motility in integrin-based adhesion has traditionally been framed in terms of a motor-clutch model (44, 45). In this paradigm, actin polymerization advances the leading edge and establishes matrix adhesions, which act as clutches that transmit myosin-based centripetal forces to the matrix to allow forward translocation of the cell. Based on our SIM imaging revealing close coupling between actin and microtubule dynamics, we hypothesized that an analogous motor-clutch mechanism may be at play in McTNs, with McTNs acting as the protrusive element. Such a model would require specific proteins to couple microtubules, actin, and CD44. The IQGAP1/CLIP170 complex is a natural candidate in this regard. IQGAP1 has previously been shown to complex with the microtubule-binding protein CLIP170 where it can participate in microtubule capture to membrane-localized Rac1 and Cdc42 in the leading edge of fibroblasts (46). Cross-linking of actin and microtubules via IQGAP1 and CLIP170 has also been implicated in neuronal dendrite and axonal growth cone extension (47). IQGAP1-positive protrusions have been identified in brain tissue culture (48), and IQGAP1 has previously been suggested as a biomarker for aggressive GBM (49). Given that IQGAP1 can also bind to CD44 and is important for HA binding by CD44 (21), we asked whether IQGAP1, CLIP170, and CD44 collectively contribute to McTN formation, cell adhesion, and cell motility.
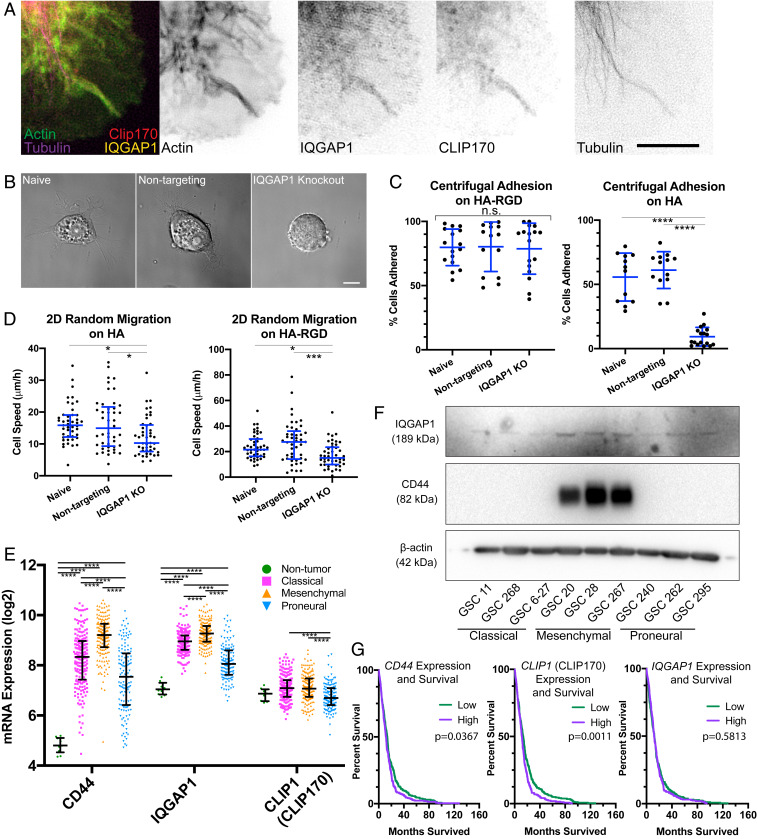
SIM imaging of cells on HA revealed colocalization of IQGAP1, CLIP170, actin, and microtubules in McTNs (Fig. 7A). IQGAP1 strongly colocalized to actin both in cells on HA and HA-RGD matrices where it is available for complexation with CLIP170 (SI Appendix, Fig. S11 A and B). Protrusions positive for actin and microtubules were also positive for IQGAP1 and CLIP170, with 71 ± 18% of protrusions positive for all four components (SI Appendix, Fig. S11C). To test functional contributions of IQGAP1 to McTN formation, we used CRISPR/Cas9 gene editing to knock out IQGAP1 in U-87 MG cells (SI Appendix, Fig. S11D). Fewer IQGAP1 KO cells formed long McTNs (>10 µm) than controls (Fig. 7B and SI Appendix, Fig. S11E). IQGAP1 KO cells also exhibited more blebbing than controls, perhaps indicating a more contractile state (SI Appendix, Fig. S11 F and G) (50). IQGAP1 KO cells still spread on HA-RGD, albeit with a more circular, less polarized morphology (SI Appendix, Fig. S12), possibly reflecting IQGAP1’s association with actin and Rho GTPases (51). Consistent with these morphological observations, IQGAP1 KO had no effect on cell adhesion to the HA-RGD matrix but dramatically reduced adhesion to the HA matrix (Fig. 7C). The random 2D migration speed of IQGAP1 KO cells was moderately reduced both on HA and HA-RGD (Fig. 7D). Thus, RGD-integrin binding is largely preserved in the absence of IQGAP1, but CD44-mediated binding to the HA-rich matrix is largely abrogated.
Fig. 7.
McTNs are mechanically coupled to the cytoskeleton and ECM through CD44, IQGAP1, and CLIP170. (A) SIM of U-87 MG cell on HA demonstrating that IQGAP1, CLIP170, tubulin, and actin are colocalized in McTNs. (Scale bar: 5 µm.) (B) DIC imaging demonstrates that IQGAP1 KO cells remain rounded on the HA matrix compared to naive and nontargeting controls. (Scale bar: 10 µm.) (C) IQGAP1 KO reduces adhesion to HA but not HA-RGD matrix. n = 13–17 total gels analyzed from three independent experiments. ****P < 0.001 by one-way ANOVA followed by Tukey–Kramer multiple comparisons test. Blue lines represent mean and SD. (D) IQGAP1 KO reduces 2D migration speed on both HA and the HA-RGD matrix. n = 45 total cells analyzed from three independent experiments. *P < 0.05; ***P < 0.001, by one-way ANOVA followed by Tukey–Kramer multiple comparisons test. Blue lines represent mean and SD. (E) Plot of log2 transformed mRNA levels obtained from TCGA of CD44, IQGAP1, and CLIP1 (CLIP170) by subtype. Black lines represent median and interquartile range. n = 10 nontumor, 199 classical, 166 mesenchymal, and 137 proneural samples from independent patients analyzed. ****P < 0.001 by one-way ANOVA followed by Tukey–Kramer’s multiple comparisons test using log2 transformed mRNA expression levels and compared between subtypes for each gene. (F) Western blot of IQGAP1, CD44, and β-actin in patient-derived GSC cells indicates variable yet ubiquitous IQGAP1 expression, but high CD44 expression only in cells of the mesenchymal subtype. Blot was first probed for CD44 and β-actin, followed by stripping and probing for IQGAP1 (G) Kaplan–Meier survival correlating with high or low levels of CD44, CLIP1 (CLIP170), and IQGAP1 mRNA with median expression level used as cutoff. n = 262 patients in the high expression group, and n = 263 patients in the low expression group. P values determined by log-rank Mantel–Cox comparison.
To more deeply explore the disease relevance of these findings, we analyzed The Cancer Genome Atlas (TCGA) to explore associations between IQGAP1, CLIP1 (CLIP170), and CD44 gene expression and GBM. CD44 and IQGAP1 gene expression was strongly correlated (Pearson’s product-moment coefficient of 0.61). Both CD44 and IQGAP1 are highly overexpressed in GBM (Fig. 7E). Furthermore, IQGAP1, CLIP1 (CLIP170), and CD44 show GBM subtype-specific expression patterns, with the highest expression levels in the most aggressive mesenchymal subtype and the lowest expression levels in the least aggressive proneural subtype. We also probed the CD44 and IQGAP1 status in a panel of GSCs which have been previously characterized by subtype (52) (Fig. 7F). Consistent with results from TCGA, we observed a dramatic up-regulation in CD44 as well as an up-regulation of IQGAP1 in the mesenchymal subtype compared to the proneural and classical subtypes. All of the cell lines expressed levels of the standard isoform of CD44 (CD44s) detectable by Western blotting, except GSC 268 and GSC 6-27, and isoform expression of CD44 was not observed (Fig. 7F and SI Appendix, Figs. S6A and S13). While low IQGAP1 expression in histological samples has been associated with longer patient survival relative to samples with high expression (49), no correlation was observed between gene expression and survival using median expression as a cutoff for survival analysis (Fig. 7G). CD44 and CLIP1 (CLIP170) gene expression correlated with patient survival (Fig. 7G). The modest survival difference correlated with high compared to low CD44 expression is consistent with previous findings that an intermediate level of CD44 exhibits the strongest correlation with poor patient survival (39).
Discussion
Here, we investigated molecular and biophysical mechanisms through which GBM cells adhere to HA matrix. We find that GBM cells interact with HA via McTNs, which are long, thin protrusions rich in the HA receptor CD44, which we show is necessary for cell adhesion and McTN stability. Motility assays and laser ablation indicate that McTNs are stabilized by a balance of actomyosin-driven contraction, microtubule-driven protrusion, and CD44-mediated adhesion. Mechanical coupling of actin and microtubules with McTNs appears to be reinforced by a complex of IQGAP1 and CLIP170, and loss of IQGAP1 specifically disrupts adhesion and motility on HA.
McTNs, which can be functionally defined as microtubule-positive protrusions that elongate following actin depolymerization, have been observed in circulating tumor cells (CTCs), where they are believed to facilitate endothelial adhesion prior to extravasation (30–32). However, to our knowledge, these structures have not been previously appreciated in the context of 2D or 3D migration in or on a solid-state matrix. It is therefore informative to compare McTNs to canonically described migratory processes. McTNs share some morphological similarities with filopodia, such as the high aspect ratio and presence of actin filaments (53). However, the lack of Cdc42 dependence and myosin X positivity, together with the dependence on CD44 and microtubules for formation and elongation, suggests that McTNs are structurally and functionally distinct from filopodia. McTNs in 3D show some similarities with pseudopodia, although pseudopodia are usually observed in a context of integrin adhesion in a fibrillar matrix rather than a nanoporous matrix lacking integrin ligands (54, 55). Nonetheless, additional characterization and comparison of molecular composition and dynamics are necessary to establish the degree of similarity between these protrusions and develop a more rigorous operational definition. Our findings may also offer mechanistic insight into GBM progression in vivo. Micronemes, microtubes, and other long, protrusive structures have been described in GBMs yet their molecular characteristics and mechanisms of formation remain incompletely elucidated. In particular, McTNs bear several key similarities to recently reported membrane microtubes, which are postulated to connect cells into a multicellular network that facilitates exchange of resistance factors (56). Key similarities are the inclusion of actin and microtubules, lack of myosin X, and similar morphology. While it is not clear that McTNs are functionally related to microtubes, our results and model may offer mechanistic insights into how microtubes and related processes arise in GBM.
These data also provide insights into how CD44 supports cell motility. Our HA platform enables us to isolate and investigate the role of CD44-HA binding in driving invasion. Importantly, CD44 is sufficient to drive the formation of tension-bearing protrusions that enable motility in the complete absence of integrin ligands. We have previously demonstrated that CD44-HA–based adhesion occurs more rapidly than integrin-based adhesion (17), and imaging in this study suggests CD44 covers the membrane rather than localizing to specific binding regions analogous to integrins in focal adhesions. Furthermore, CD44-mediated adhesion alone does not support strong actin bundling or the formation of large lamellipodia. These findings suggest that CD44 binding is characterized by many rapid, weak interactions with the ECM, whereas integrin binding is characterized by fewer, more slowly mechanically reinforced interactions. These two systems give rise to fundamentally different cytoskeletal architectures. Microtubule polymerization, through mechanical reinforcement and/or modulation of tubulin-mediated signaling, reduces retrograde actin flow or up-regulates F-actin assembly, which balances actomyosin-based contractility (57, 58). With the lack of reinforced, localized cell–matrix interactions in the context of CD44-HA binding and low integrin binding found in cells in HA-rich matrix, more internal protrusive forces or signaling cues from microtubules may be needed to balance actomyosin contractility (59). Consistent with this idea, microtubule-based motility is increasingly recognized as key to migration through tightly confined channels or soft 3D matrix in which focal adhesion formation may be restricted (59, 60). Furthermore, GBM cells have been reported to migrate despite treatment with pharmacological inhibitors of actin polymerization (61). This may also explain why we observed McTNs in both 3D HA and HA-RGD but not in 2D HA and HA-RGD, with much greater prominence in 3D than in 2D.
Several studies have identified IQGAP1 as an important marker of GBM invasion, but the mechanisms underlying the expression of this biomarker remain poorly understood (48, 49, 51). In particular, IQGAP1-positive processes protruding from GBM cells have been identified in brain tissue culture slices (48). Our work suggests that IQGAP1 may be contributing directly to invasive McTN machinery through the coupling of actin and microtubules in complex with CLIP170 and possibly CD44. Nonetheless, it is important to acknowledge that CD44, IQGAP1, and microtubules can all trigger other types of intracellular signals which likely contribute to migratory phenotypes. Microtubule disruption releases monomeric tubulin potentially along with actin regulatory factors, which could in turn promote actin polymerization and contractility (57, 62, 63). CD44, IQGAP1, and microtubules all up-regulate and depend to some degree on Rac1 signaling (55, 64). It is possible that disrupting one of these components also reduces Rac1 signaling, which in turn impedes McTN growth. While the nature of the relationship between IQGAP1, CLIP170, CD44, actin, and microtubules remains to be further elucidated, our study suggests strong interplay that is particularly important in the context of low-integrin, high-CD44 engagement compared to high-integrin engagement.
While targeting CD44 is effective in reducing tumor invasion in animal models (14, 15), initial clinical studies of antibodies targeting one isoform of CD44 in breast or squamous cell carcinoma failed due to high toxicity from nonspecific interactions with CD44 in other organs, such as skin (65, 66). Our findings highlight the need to understand the role of CD44–cytoskeletal interactions to elucidate more specific targets. These findings also underscore compositional and mechanistic differences in protrusions forming in microenvironments lacking nonfibrillar guidance cues and rich in HA. McTN function should be further studied to better understand the relevance of these structures in disease progression, such as how McTNs participate in matrix remodeling or whether they facilitate cell–cell communication. Given the morphological similarities between McTNs and protrusions involved in invasion and cell–cell communication in vivo (38, 56), establishment of functional similarities could generate new mechanistic insight and therapeutic targets difficult to elucidate in animal models. Finally, understanding how McTN expression relates to molecular subtype and tumor recurrence would focus efforts to target McTN-based motility in patients who would most benefit.
Methods
Cell Lines.
U-251 MG and U-87 MG human GBM cells were obtained from the University of California, Berkeley Tissue Culture Facility, which sources its cultures directly from the ATCC. GSC-11, GSC-268, GSC6-27, GSC-28, GSC-267, GSC-240, GSC-262, GSC-20, and GSC-295 cells were kindly provided from Joseph McCarty and Erik Sulman, MD Anderson Cancer Center, Houston, TX. L0 cells were originally obtained from Brent Reynolds, University of Florida, Gainesville, FL, and maintained as described in previous publications (67–69).
Ex Vivo Mouse Brain Model.
Samples were derived from mice that were cared for by the University of California, Berkeley Office for Animal Care and Use, and all experiments were approved by Berkeley’s Institutional Animal Care and Use Committee. Brain slice culture was performed following institutional and national regulations in accordance with the American Association for Accreditation of Laboratory Animal Care using a previously established method (70). Mouse brain tissue (female, 18 mo old) was cut into 1-mm-thick slices immediately after transcardial saline perfusion and extraction. Brain slices were cultured in 10% (vol/vol) serum medium in the upper chamber of a Boyden (transwell) chamber on top of a 5-µm pore size polyester membrane (Corning) with 10% (vol/vol) serum medium in the lower chamber. To each slice, ∼1,500 U-251 MG cells expressing GFP-tubulin and RFP-LifeAct in 50 µL of medium were seeded onto the center of the slice, and invasion was tracked using confocal microscopy. After culture, the whole slice was fixed using 4% (wt/vol) paraformaldehyde in PBS for 1 h, placed on a glass-bottom dish, and immediately imaged by SIM.
Laser Ablation.
For laser ablation studies, an upright Olympus BX51WI microscope (Olympus Corporation) equipped with swept field confocal technology (Bruker) and a Ti:Sapphire 2-photon Chameleon Ultra II laser (Coherent) was used. The two-photon laser was set to 770 nm, and ablation was performed using three 20-ms pulses. Live-cell imaging was performed using an Olympus LUMPlanFL N 60×/1.0 water dipping objective. Cells were kept at 37 °C using a stage-top sample heater (Warner Instruments). Images were captured using an electron multiplying charge-coupled device (EM-CCD) camera (Photometrics). The following emission filters were used: Quad FF-01-446/523/600/677-25 (Semrock) and 525/50 ET525/50 (Chroma). PrairieView Software (v. 5.3 U3, Bruker) was used to acquire images.
STORM Imaging.
The 3D-STORM imaging (71, 72) was carried out on a home-built setup using a Nikon CFI Plan Apo λ 100× oil immersion objective (numerical aperture 1.45), as described previously (33, 73). Briefly, the sample was mounted with an imaging buffer consisting of 5% (wt/vol) glucose, 100 mM cysteamine, 0.8 mg/mL glucose oxidase, and 40 µg/mL catalase in 1 M Tris⋅HCI (pH 7.5). Dye molecules were photoswitched to the dark state and imaged using either 647- or 560-nm lasers; these lasers were passed through an acousto-optic tunable filter and introduced through an optical fiber into the back focal plane of the microscope and onto the sample at ∼2 kW⋅cm−2. A translation stage shifted the laser beams toward the edge of the objective so that light reached the sample at incident angles slightly smaller than the critical angle of the glass–water interface. Single-molecule emission was recorded with an Andor iXon Ultra 897 EM-CCD camera at a frame rate of 110 Hz, for a total of ∼80,000 frames per image. For 3D-STORM, a cylindrical lens of focal length 1 m was inserted into the imaging path to introduce astigmatism (72). The raw STORM data were analyzed according to previously described methods (71, 72). Two-color imaging was performed via sequential imaging of targets labeled by Alexa Fluor 647 and CF568.
Additional methods and details can be found in SI Appendix.
Data Availability.
All data discussed in the paper are included in this published article and SI Appendix.
Supplementary Material
Acknowledgments
Confocal images were acquired at the California Institute for Regenerative Medicine/QB3 Shared Stem Cell Facility at the University of California, Berkeley (UC Berkeley), which provided the Olympus BX51WI microscope with swept field confocal technology. We thank Dr. Mary West for assistance and training. Structured illumination microscopy was conducted at the UC Berkeley Biological Imaging Facility, which was supported in part by the NIH S10 program under Award 1S10OD018136-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank Dr. Steve Ruzin and Dr. Denise Schichnes for their technical assistance and training. We also thank the following groups and individuals: Dr. Joseph McCarty and Dr. Erik Sulman (MD Anderson Cancer Center) for providing the GSC patient-derived cells; Dr. David Schaffer for sharing equipment for Western blot and immunohistochemistry analysis; Hector Nolla for technical assistance with flow cytometry; and the Northwest Animal Care Facility. Finally, we gratefully acknowledge financial support from the following sources: NIH Grants F31CA228317 (to K.J.W.); F31GM119329 (to S.L.); R21CA174573, R21EB016359, R01CA227136, and R01GM122375 (to S.K.); the European Union’s Horizon 2020 Research and Innovation Programme under Marie Sklodowska-Curie Grant Agreement 752097 (to J.D.C.); and the W. M. Keck Foundation (S.K.). K.X. is a Chan Zuckerberg Biohub Investigator and acknowledges support from NSF Grant CHE-1554717.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914294117/-/DCSupplemental.
References
- 1.Bissell M. J., Radisky D., Putting tumours in context. Nat. Rev. Cancer 1, 46–54 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom Q. T., et al. , CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol. 17 (suppl. 4), iv1–iv62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R., et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups ; National Cancer Institute of Canada Clinical Trials Group , Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10, 459–466 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Giese A., Bjerkvig R., Berens M. E., Westphal M., Cost of migration: Invasion of malignant gliomas and implications for treatment. J. Clin. Oncol. 21, 1624–1636 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Carbonell W. S., DeLay M., Jahangiri A., Park C. C., Aghi M. K., β1 integrin targeting potentiates antiangiogenic therapy and inhibits the growth of bevacizumab-resistant glioblastoma. Cancer Res. 73, 3145–3154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gladson C. L., The extracellular matrix of gliomas: Modulation of cell function. J. Neuropathol. Exp. Neurol. 58, 1029–1040 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Bellail A. C., Hunter S. B., Brat D. J., Tan C., Van Meir E. G., Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int. J. Biochem. Cell Biol. 36, 1046–1069 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Delpech B., et al. , Hyaluronan and hyaluronectin in the extracellular matrix of human brain tumour stroma. Eur. J. Cancer 29A, 1012–1017 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Ward J. A., Huang L., Guo H., Ghatak S., Toole B. P., Perturbation of hyaluronan interactions inhibits malignant properties of glioma cells. Am. J. Pathol. 162, 1403–1409 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedron S., Becka E., Harley B. A. C. C., Regulation of glioma cell phenotype in 3D matrices by hyaluronic acid. Biomaterials 34, 7408–7417 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Park J. B., Kwak H.-J., Lee S.-H., Role of hyaluronan in glioma invasion. Cell Adhes. Migr. 2, 202–207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toole B. P., Hyaluronan-CD44 interactions in cancer: Paradoxes and possibilities. Clin. Cancer Res. 15, 7462–7468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida T., Matsuda Y., Naito Z., Ishiwata T., CD44 in human glioma correlates with histopathological grade and cell migration. Pathol. Int. 62, 463–470 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Breyer R., et al. , Disruption of intracerebral progression of C6 rat glioblastoma by in vivo treatment with anti-CD44 monoclonal antibody. J. Neurosurg. 92, 140–149 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Xu Y., Stamenkovic I., Yu Q., CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 70, 2455–2464 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zöller M., CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 11, 254–267 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Kim Y., Kumar S., CD44-mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility. Mol. Cancer Res. 12, 1416–1429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponta H., Sherman L., Herrlich P. A., CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 4, 33–45 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Clucas J., Valderrama F., ERM proteins in cancer progression. J. Cell Sci. 127, 267–275 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Zhu D., Bourguignon L. Y. W., The ankyrin-binding domain of CD44s is involved in regulating hyaluronic acid-mediated functions and prostate tumor cell transformation. Cell Motil. Cytoskeleton 39, 209–222 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Bourguignon L. Y. W., Gilad E., Rothman K., Peyrollier K., Hyaluronan-CD44 interaction with IQGAP1 promotes Cdc42 and ERK signaling, leading to actin binding, Elk-1/estrogen receptor transcriptional activation, and ovarian cancer progression. J. Biol. Chem. 280, 11961–11972 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Wolf K. J., Kumar S., Hyaluronic acid: Incorporating the bio into the material. ACS Biomater. Sci. Eng. 5, 3753–3765 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wloga D., Joachimiak E., Fabczak H., Tubulin post-translational modifications and microtubule dynamics. Int. J. Mol. Sci. 18, E2207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bieling P., et al. , CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 183, 1223–1233 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcos S., et al. , Tubulin tyrosination is required for the proper organization and pathfinding of the growth cone. PLoS One 4, e5405 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y., Brady S. T., Post-translational modifications of tubulin: Pathways to functional diversity of microtubules. Trends Cell Biol. 25, 125–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson M. F., Sahai E., The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis 26, 273–287 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Arnold M., et al. , Activation of integrin function by nanopatterned adhesive interfaces. ChemPhysChem 5, 383–388 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Rottner K., Stradal T. E., Actin dynamics and turnover in cell motility. Curr. Opin. Cell Biol. 23, 569–578 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Whipple R. A., Cheung A. M., Martin S. S., Detyrosinated microtubule protrusions in suspended mammary epithelial cells promote reattachment. Exp. Cell Res. 313, 1326–1336 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whipple R. A., et al. , Vimentin filaments support extension of tubulin-based microtentacles in detached breast tumor cells. Cancer Res. 68, 5678–5688 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matrone M. A., et al. , Metastatic breast tumors express increased tau, which promotes microtentacle formation and the reattachment of detached breast tumor cells. Oncogene 29, 3217–3227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Killilea A. N., et al. , Cytoskeletal organization in microtentacles. Exp. Cell Res. 357, 291–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernstein J. J., Woodard C. A., Glioblastoma cells do not intravasate into blood vessels. Neurosurgery 36, 124–132, discussion 132 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Adams R. A., Passino M., Sachs B. D., Nuriel T., Akassoglou K., Fibrin mechanisms and functions in nervous system pathology. Mol. Interv. 4, 163–176 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Raman P. S., Alves C. S., Wirtz D., Konstantopoulos K., Distinct kinetic and molecular requirements govern CD44 binding to hyaluronan versus fibrin(ogen). Biophys. J. 103, 415–423 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf K. J., Lee S., Kumar S., A 3D topographical model of parenchymal infiltration and perivascular invasion in glioblastoma. APL Bioeng. 2, 031903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirata E., et al. , In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J. Cell Sci. 125, 858–868 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Klank R. L., et al. , Biphasic dependence of glioma survival and cell migration on CD44 expression level. Cell Rep. 18, 23–31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ilangumaran S., Briol A., Hoessli D. C., CD44 selectively associates with active Src family protein tyrosine kinases Lck and Fyn in glycosphingolipid-rich plasma membrane domains of human peripheral blood lymphocytes. Blood 91, 3901–3908 (1998). [PubMed] [Google Scholar]
- 41.Heasman S. J., Ridley A. J., Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690–701 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Sousa A. D., Cheney R. E., Myosin-X: A molecular motor at the cell’s fingertips. Trends Cell Biol. 15, 533–539 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Heisterkamp A., et al. , Pulse energy dependence of subcellular dissection by femtosecond laser pulses. Opt. Express 13, 3690–3696 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Chan C. E., Odde D. J., Traction dynamics of filopodia on compliant substrates. Science 322, 1687–1691 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Mitchison T., Kirschner M., Cytoskeletal dynamics and nerve growth. Neuron 1, 761–772 (1988). [DOI] [PubMed] [Google Scholar]
- 46.Fukata M., et al. , Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 109, 873–885 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Coles C. H., Bradke F., Coordinating neuronal actin-microtubule dynamics. Curr. Biol. 25, R677–R691 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Rotoli D., et al. , IQGAP1 in podosomes/invadosomes is involved in the progression of glioblastoma multiforme depending on the tumor status. Int. J. Mol. Sci. 18, 150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald K. L., et al. , IQGAP1 and IGFBP2: Valuable biomarkers for determining prognosis in glioma patients. J. Neuropathol. Exp. Neurol. 66, 405–417 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Paluch E. K., Raz E., The role and regulation of blebs in cell migration. Curr. Opin. Cell Biol. 25, 582–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noritake J., Watanabe T., Sato K., Wang S., Kaibuchi K., IQGAP1: A key regulator of adhesion and migration. J. Cell Sci. 118, 2085–2092 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Bhat K. P. L., et al. , Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 24, 331–346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy D. A., Courtneidge S. A., The ‘ins’ and ‘outs’ of podosomes and invadopodia: Characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413–426 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bard J. B., Hay E. D., The behavior of fibroblasts from the developing avian cornea. Morphology and movement in situ and in vitro. J. Cell Biol. 67, 400–418 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouchet B. P., Akhmanova A., Microtubules in 3D cell motility. J. Cell Sci. 130, 39–50 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Osswald M., et al. , Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Prahl L. S., et al. , Microtubule-based control of motor-clutch system mechanics in glioma cell migration. Cell Rep. 25, 2591–2604.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stamenović D., Mijailovich S. M., Tolić-Nørrelykke I. M., Chen J., Wang N., Cell prestress. II. Contribution of microtubules. Am. J. Physiol. Cell Physiol. 282, C617–C624 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Balzer E. M., et al. , Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 26, 4045–4056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rhee S., Jiang H., Ho C.-H., Grinnell F., Microtubule function in fibroblast spreading is modulated according to the tension state of cell-matrix interactions. Proc. Natl. Acad. Sci. U.S.A. 104, 5425–5430 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Panopoulos A., Howell M., Fotedar R., Margolis R. L., Glioblastoma motility occurs in the absence of actin polymer. Mol. Biol. Cell 22, 2212–2220 (2011).21551075 [Google Scholar]
- 62.Heck J. N., et al. , Microtubules regulate GEF-H1 in response to extracellular matrix stiffness. Mol. Biol. Cell 23, 2583–2592 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng I. K., et al. , GEF-H1 over-expression in hepatocellular carcinoma promotes cell motility via activation of RhoA signalling. J. Pathol. 228, 575–585 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Marhaba R., Zöller M., CD44 in cancer progression: Adhesion, migration and growth regulation. J. Mol. Histol. 35, 211–231 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Tijink B. M., et al. , A phase I dose escalation study with anti-CD44v6 bivatuzumab mertansine in patients with incurable squamous cell carcinoma of the head and neck or esophagus. Clin. Cancer Res. 12, 6064–6072 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Rupp U., et al. , Safety and pharmacokinetics of bivatuzumab mertansine in patients with CD44v6-positive metastatic breast cancer: Final results of a phase I study. Anticancer Drugs 18, 477–485 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Deleyrolle L. P., et al. , Evidence for label-retaining tumour-initiating cells in human glioblastoma. Brain 134, 1331–1343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong S. Y., et al. , Constitutive activation of myosin-dependent contractility sensitizes glioma tumor-initiating cells to mechanical inputs and reduces tissue invasion. Cancer Res. 75, 1113–1122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin J. G., et al. , Linking invasive motility to protein expression in single tumor cells. Lab Chip 18, 371–384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jung S., et al. , Brain tumor invasion model system using organotypic brain-slice culture as an alternative to in vivo model. J. Cancer Res. Clin. Oncol. 128, 469–476 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rust M. J., Bates M., Zhuang X., Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang B., Wang W., Bates M., Zhuang X., Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wojcik M., Hauser M., Li W., Moon S., Xu K., Graphene-enabled electron microscopy and correlated super-resolution microscopy of wet cells. Nat. Commun. 6, 7384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper are included in this published article and SI Appendix.