Significance
Pancreatic ductal adenocarcinoma (PDA) is one of the most lethal human malignancies, attributed in part to lineage infidelity downstream of deregulated lineage-specifying transcription factors (TFs). Here we define the biological effects of a poorly understood TF ZBED2 in the most aggressive subtype of PDA, defined by the expression of squamous lineage markers. Our study reveals two molecular functions of ZBED2 in PDA cells: An inhibitor of interferon response genes and a modifier of epithelial lineage programs. Both functions can be explained by the ability of ZBED2 to antagonize the functional output of interferon regulatory factor 1 (IRF1). Our study reinforces the concept of aberrant lineage identity in cancer and highlights an unexpected connection between interferon response pathways and squamous-subtype PDA.
Keywords: ZBED2, IRF1, interferon, lineage plasticity, pancreatic ductal adenocarcinoma
Abstract
Lineage plasticity is a prominent feature of pancreatic ductal adenocarcinoma (PDA) cells, which can occur via deregulation of lineage-specifying transcription factors. Here, we show that the zinc finger protein ZBED2 is aberrantly expressed in PDA and alters tumor cell identity in this disease. Unexpectedly, our epigenomic experiments reveal that ZBED2 is a sequence-specific transcriptional repressor of IFN-stimulated genes, which occurs through antagonism of IFN regulatory factor 1 (IRF1)-mediated transcriptional activation at cooccupied promoter elements. Consequently, ZBED2 attenuates the transcriptional output and growth arrest phenotypes downstream of IFN signaling in multiple PDA cell line models. We also found that ZBED2 is preferentially expressed in the squamous molecular subtype of human PDA, in association with inferior patient survival outcomes. Consistent with this observation, we show that ZBED2 can repress the pancreatic progenitor transcriptional program, enhance motility, and promote invasion in PDA cells. Collectively, our findings suggest that high ZBED2 expression is acquired during PDA progression to suppress the IFN response pathway and to promote lineage plasticity in this disease.
Cells are capable of adopting different identities along a phenotypic spectrum, a process referred to as lineage plasticity. While lineage plasticity is a critical feature of normal development and during wound-healing responses, it is also a powerful contributor to the pathogenesis of human cancer (1, 2). Through a multitude of genetic and nongenetic mechanisms, cancer cells gain access to diverse lineage and developmental transcriptional programs, resulting in heterogeneous cell identities emerging in a clonally derived tumor. The consequences of lineage plasticity in human cancer are far-reaching, but include a prominent role in the acquisition of metastatic traits and in the evasion of targeted therapy (1).
Pancreatic ductal adenocarcinoma (PDA) is an emerging paradigm of lineage plasticity in human cancer. While PDA is defined by its histopathological resemblance to ductal epithelial cells of the exocrine pancreas, studies in mice have shown that acinar cells can serve as a cell-of-origin for this disease, which transdifferentiate into the ductal fate following acquisition of a Kras activating mutation (3–5). At later stages of tumor development, aberrant up-regulation or silencing of master regulator transcription factors (TFs) in PDA can lead to reprogramming of ductal identity toward that of other cell lineages, including mesenchymal (6–8), foregut endodermal (9), or squamous epithelial fates (10–12). While each of these lineage transitions are capable of promoting disease progression in experimental systems, only the presence of squamous characteristics correlates with a shorter overall survival in human PDA patients (13, 14). For this reason, the identification of mechanisms that promote squamous transdifferentiation in PDA has become an active area of investigation in recent years (10–13, 15, 16).
The interferon (IFN) transcriptional response is a conserved pathway that protects organisms from infectious pathogens and malignancy (17, 18). IFN pathway activation occurs via autocrine or paracrine IFN signaling that can be triggered in response to the detection of foreign nucleic acids as well as ectopically located self-DNA (18, 19). Whereas almost all cell types can produce type I IFNs (e.g., IFN-α and IFN-β), type II IFN (i.e., IFN-γ) production is restricted to a subset of activated immune cells (20). IFN pathway activation promotes the transcriptional induction of hundreds of IFN-stimulated genes (ISGs), which encode diverse proteins with antiviral, antiproliferative, and immunostimulatory functions (21). The key TFs that promote ISG induction belong to the signal transducer and activator of transcription (STAT) and IFN regulatory factor (IRF) families, which can bind in an IFN-inducible manner at the promoters of ISGs (22, 23). In the classical pathway, phosphorylation of STATs downstream of IFN receptor activation triggers a rapid ISG response (23). This primary response includes the STAT-dependent transcriptional activation of several genes encoding IRFs, which subsequently drive an amplifier circuit resulting in sustained ISG induction (22). Within this complex transcriptional response, IRF1 is a critical positive regulator required for the full range of overlapping target gene activation following type I or type II IFN pathway activation (22). IRF1 is a broadly acting antiviral effector and exhibits tumor-suppressor functions in multiple cellular contexts (24, 25). With respect to PDA, prior studies have shown that IRF1 can promote a differentiated epithelial cell state and inhibit cell proliferation (26, 27).
The ZBED gene family encodes nine zinc finger-containing TFs in humans, which originated from a domesticated DNA transposase gene from an hAT transposable element (28). While lacking in transposase activity, human ZBED TFs instead retain their zinc finger domain to perform sequence-specific DNA binding and function as transcriptional regulators in a cell-type specific manner (29–31). Within this family, ZBED2 is one of the least-understood members, in part because of its recent evolution and lack of a mouse ortholog (28). A prior genomewide-association study identified ZBED2 as a candidate locus influencing risk of smoking-induced pancreatic cancer (32). More recently, ZBED2 was found to be highly expressed in the basal layer of the epidermis, where it plays a role in regulating keratinocyte differentiation (33). Another study identified ZBED2 as a marker of T cell exhaustion in human CD8 T cells, although the function of ZBED2 was not investigated in this context (34). We are unaware of any prior study characterizing a transcriptional function for ZBED2 or its role in cancer.
Here we identify ZBED2 as one of the most aberrantly up-regulated TFs in human PDA. This prompted our characterization of the transcriptional function of ZBED2, which we demonstrate to be a sequence-specific transcriptional repressor. We show that the repression targets of ZBED2 are highly enriched for genes within the IFN response pathway. By interacting with ISG promoters, ZBED2 blocks the transcriptional output and growth-arrest phenotypes caused by IRF1 activation downstream of IFN stimulation. We also provide evidence that ZBED2 is preferentially expressed in squamous-subtype PDA tumors and promotes loss of pancreatic progenitor cell identity in this context. Collectively, our findings suggest that aberrant ZBED2 expression in PDA cells blocks the IFN response and alters epithelial cell identity in this disease.
Results
Aberrant ZBED2 Expression in Pancreatic Ductal Adenocarcinoma Correlates with Inferior Patient Survival Outcomes.
In this study, we sought to identify novel TFs that deregulate cell identity in PDA. By evaluating previously published transcriptome data comparing normal human pancreas and PDA (35), we identified ZBED2 as among the most aberrantly up-regulated TF genes in tumor and metastatic lesions (Fig. 1 A and B). Notably, the fold-induction of ZBED2 was comparable to other aberrantly expressed TFs in PDA, FOXA1, and TP63, which have established roles in disease progression (Fig. 1A and Dataset S1) (9, 10). In two additional independent transcriptome studies of human PDA tumors (13, 36), we validated high levels of ZBED2 expression in 16 to 20% of primary patient samples (Fig. 1 C and D). We further corroborated the aberrant expression pattern of ZBED2 in ∼20% of established organoid cultures derived from human PDA tumors, while ZBED2 was expressed at low levels in organoids derived from normal pancreatic epithelial cells (Fig. 1E and Dataset S2) (37). Considering the limited number of prior studies of ZBED2, these observations prompted us to investigate the molecular function of ZBED2 in PDA.
Fig. 1.
Aberrant ZBED2 expression in pancreatic ductal adenocarcinoma correlates with inferior patient survival outcomes. (A) Expressed TFs ranked by mean log2 fold-change in primary PDA versus normal pancreas. Selected TFs are labeled along with their rank. (B) ZBED2 expression in normal pancreas and primary and metastatic PDA tumor samples. P value determined by one-way ANOVA. (C and D) ZBED2 expression across PDA patient samples. Scale bar indicates the standardized expression value. (E) ZBED2 expression in human organoids derived from normal pancreas or neoplastic PDA samples. (F and G) ZBED2 expression in normal tissues from the GTEx portal (F) or tumors from the TCGA Pan-Cancer Atlas (G). Arrow indicates pancreas tissue. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower-grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma. (H and I) Survival curves of patients stratified according to high or low ZBED2 expression. P value calculated using the log-rank (Mantel-Cox) test. (J) ZBED2 expression in cancer cell lines from the CCLE (44). Arrow indicates pancreas tissue. See also SI Appendix, Fig. S1.
To gain initial insight into ZBED2 function, we examined its expression across a diverse collection of normal and malignant human tissues using publicly available transcriptome data (38, 39). This analysis revealed a highly tissue-specific pattern of ZBED2 expression in normal tissues, with skin, lung, esophageal mucosa, and thyroid tissues expressing ZBED2 at the highest levels (Fig. 1F). In accord with the findings above, ZBED2 expression was not detected in the normal human pancreas (Fig. 1F). Transcriptome analysis of 32 tumor types profiled by the The Cancer Genome Atlas (TCGA) Pan-Cancer Atlas (39) revealed pervasive ZBED2 expression in numerous human cancer types (Fig. 1G). In specific malignancies, like thyroid carcinomas and squamous cell carcinomas (head and neck squamous cell carcinoma, lung squamous cell carcinoma, esophageal cancer), high ZBED2 levels can be explained by its expression pattern in the normal tissue counterpart (i.e., cell-of-origin) of these tumors. However, in other cancer types, like PDA and ovarian cancer, ZBED2 appears to be up-regulated in an aberrant manner (Fig. 1 F and G). In order to confirm that the ZBED2 expression observed in PDA tumors is attributable to the epithelial compartment, we analyzed single-cell RNA-sequencing (RNA-seq) data from 24 human PDA tumors and 11 normal pancreas tissue samples from a recently published study (40). This analysis revealed the most-abundant ZBED2 expression within the ductal cells of tumor samples and confirmed the absence of ZBED2 within normal pancreas tissues (SI Appendix, Fig. S1 A and B). Interestingly, we also observed ZBED2 expression in a subset of T cells within the tumor samples, which may represent T cells in an exhausted state (34). Indeed, analysis of RNA-seq data from three independent studies that have transcriptionally interrogated the exhausted T cell state (34, 41, 42) revealed ZBED2 as one of the most consistently up-regulated genes in this context (SI Appendix, Fig. S1C). We also found that patients with ZBED2high tumors have a significantly shorter overall survival than patients with ZBED2low tumors in the context of PDA, colorectal cancer, kidney cancer, glioblastoma/glioma, lung squamous cell carcinoma, and lung adenocarcinoma (Fig. 1 H and I and SI Appendix, Fig. S1D) (43). This highlights ZBED2 as a biomarker of disease aggressiveness across several tumor subtypes.
We further interrogated ZBED2 expression in 1,156 cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE) database (44) and identified high expression in ∼24% of cell lines (Fig. 1J). In accord with the TCGA analysis, ZBED2high cell lines were significantly enriched for lines derived from pancreatic, ovarian, intestinal, endometrial, and upper aerodigestive tract tumors (Fig. 1J and SI Appendix, Fig. S1E). In addition, ZBED2high cell lines were significantly enriched for genetic alterations in KRAS, TP53, CDKN2A, and SMAD4 (SI Appendix, Fig. S1 F and G), which are also the most recurrently mutated genes in PDA (45–47). Despite this high-level expression, analysis of CRISPR screening data from 559 cancer cell lines revealed that ZBED2 is not required to sustain cell growth in any cell line under standard tissue culture conditions (SI Appendix, Fig. S1H).
ZBED2 Inhibits the Expression of Genes in the IFN Response Pathway.
To understand the cancer-relevant function of ZBED2, we performed RNA-seq analysis following ZBED2 knockout or overexpression in PDA cell lines. Western blot analysis revealed high-level expression of endogenous ZBED2 in three of nine PDA cell lines examined (Fig. 2A). We used lentiviral CRISPR-Cas9 editing to target ZBED2 in the highest expressing PDA cell line (PANC0403) with two independent single-guide RNAs (sgRNAs) and performed RNA-seq analysis. An unbiased gene set enrichment analysis (GSEA) of the ZBED2 knockout RNA-seq data revealed induction of the IFN pathway as top-ranked gene signatures in this experiment (Fig. 2 B and C and Datasets S3 and S4). To complement this approach, we performed RNA-seq after overexpressing a ZBED2 cDNA in 15 different PDA cell lines, which likewise revealed consistent suppression of IFN pathway gene signatures as a top-ranked alteration (Fig. 2 B, D, and E, SI Appendix, Fig. S2A, and Datasets S3–S5). Of note, ZBED2 expression was also found to induce the epithelial-to-mesenchymal transition (EMT) signature in the majority of PDA cell lines tested (SI Appendix, Fig. S2A). Interestingly, we found that ZBED2 expression could be induced by TGF-β treatment and that the expression of ZBED2 augmented the induction of the EMT genes SNAI2, ZEB1, and TGFBI in response to TGF-β in certain cell contexts (SI Appendix, Fig. S2 B and C). We confirmed the down-regulation of the ISGs STAT2, CMPK2, and MX1 following ectopic ZBED2 expression by quantitative reverse-transcription–polymerase chain reaction (RT-qPCR) and Western blotting (Fig. 2F and SI Appendix, Fig. S2D). We also confirmed up-regulation of the ISGs STAT2 and CMPK2 in response to ZBED2 knockdown using short-hairpin RNA (shRNAs) as an alternative approach to CRISPR (SI Appendix, Fig. S2 E and F). Taken together, this transcriptome analysis implicates an inhibitory effect of ZBED2 on the IFN transcriptional response in PDA cells.
Fig. 2.
ZBED2 inhibits the expression of genes in the IFN response pathway. (A) Western blot analysis for ZBED2 and IFN pathway components in human PDA cell lines. (B) GSEA following ZBED2 knockout in PANC0403 cells (Left) or ZBED2 cDNA expression in HPAFII cells (Right) versus their respective controls. Normalized enrichment score (NES) and family-wise error rate (FWER) P value were ranked and plotted for genes sets within MSigDB v7.0. Each gene set is depicted as a single dot. (C and D) GSEA plots evaluating the IFN response signature upon ZBED2 knockout in PANC0403 cells (C) or ZBED2 cDNA expression in HPAFII cells (D). (E) Gene-expression changes in HPAFII cells infected with ZBED2 cDNA versus empty vector control. ZBED2, STAT2, MX1, and CMPK2 are labeled along with their rank. (F) Western blot analysis for HSC70, ZBED2, STAT2, and MX1 following ZBED2 cDNA expression in HPAFII and PANC0403 cells. (G) ZBED2 expression versus the median expression value of IFN response genes across 15 PDA cell lines. (H) Median expression values of IFN response genes across all cancer cell lines within the CCLE database (Left) or in the indicated lineages stratified according to high or low ZBED2 expression. Each cell line is depicted as a single dot. **P < 0.01, ***P < 0.001 by Student’s t test. (I) Schematic representation of IFN signaling pathways. Modified from ref. 22. (J) Western blot analysis for HSC70, ZBED2, and the indicated IFN pathway components following 12-h stimulation with IFN-β or IFN-γ in AsPC1-ZBED2 cells or the empty vector. See also SI Appendix, Fig. S2.
Despite the inhibitory effect of ZBED2 on IFN pathway genes, our Western blot and RNA-seq analysis revealed that ZBED2high PDA lines tended to have a higher baseline level of IFN pathway activation, as judged by levels of STAT2 phosphorylation and ISG expression (Fig. 2 A and G). This high basal activity of the IFN pathway was not attributed to autocrine IFN production, since inactivating the IFN-α receptor (IFNAR1) did not suppress ISG levels (SI Appendix, Fig. S2 G and H). Moreover, the positive correlation between levels of ZBED2 and IFN pathway genes was observed across all 1,156 cell lines from the CCLE, encompassing PDA lines as well other diverse cancer types (Fig. 2H and SI Appendix, Fig. S2I). Additionally, we did not observe an up-regulation of ZBED2 following stimulation with IFN-β or IFN-γ, indicating that ZBED2 is not itself an ISG in this context (SI Appendix, Fig. S2J). Taken together, these findings suggest that ZBED2 expression may have been acquired in PDA cells to dampen a preexisting activation state of the IFN pathway.
We next validated the inhibitory effect of ZBED2 on the IFN pathway by performing Western blot analysis of IFN components following exposure of PDA cells to IFN-β or IFN-γ (Fig. 2 I and J). In these experiments, ZBED2 had no effect on STAT phosphorylation, which suggests that ZBED2 does not function in the signaling pathway downstream of receptor activation. In addition, this analysis revealed that specific ISG products were induced normally following IFN treatment (e.g., IRF1, IRF9), while others were attenuated by the presence of ZBED2 (STAT2, MX1, and CMPK2) (Fig. 2J). These findings suggest that ZBED2 functions as a repressor of a specific subset of ISGs.
Chromatin Immunoprecipitiation-Sequencing Analysis Implicates ZBED2 as a Sequence-Specific Repressor of ISG Promoters.
In order to investigate the function of ZBED2 at the molecular level, we performed chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) analysis of lentivirally expressed FLAG-ZBED2 in AsPC1 and SUIT2 PDA cell lines. We observed high concordance between ZBED2 occupancy in both cell lines, enabling us to define a set of 2,451 high-confidence binding sites (Fig. 3A, SI Appendix, Fig. S3A, and Dataset S6), which was highly biased toward promoter regions (Fig. 3B). Notably, genomic occupancy of ZBED2 was globally correlated with a repressive effect on transcription of neighboring genes when comparing ChIP-seq with the aforementioned RNA-seq analyses (Fig. 3C). This included several ISGs (e.g., CMPK2 and STAT2) that are both down-regulated and located near ZBED2 binding sites (Fig. 3C and Dataset S7). To further support a repressive function of ZBED2, we made use of GAL4 fusion proteins to artificially tether ZBED2 to a plasmid with luciferase expression downstream of a thymidine kinase promoter. Unlike the established activator protein IRF1, tethering ZBED2 to this promoter resulted in a significant decrease in luciferase expression (Fig. 3D). Moreover, using the GAL4 assay, we were able to map these repressive effects of ZBED2 to its N-terminal region and to its DNA binding domain (SI Appendix, Fig. S3 B–D). Together, these findings suggest that ZBED2 is a transcriptional repressor that occupies specific sites in the genome.
Fig. 3.
ChIP-seq analysis implicates ZBED2 as a sequence-specific repressor of ISG promoters. (A) Density plots showing FLAG-ZBED2 and H3K27ac enrichment surrounding a 2-kb interval centered on the summit of 2,451 high-confidence ZBED2 peaks in AsPC1 and SUIT2 cells, ranked by FLAG-ZBED2 peak intensity in AsPC1 cells. (B) Pie chart showing the distribution of FLAG-ZBED2 peaks. TSS, transcription start site; TTS, transcription termination site; UTR, untranslated region. (C) GSEA plot evaluating ZBED2 bound genes upon ZBED2 cDNA expression in HPAFII cells. Leading edge, IFN response genes are listed. (D) GAL4 fusion reporter assay testing full-length ZBED2 and IRF1 transactivation activity normalized to Renilla luciferase internal control. Mean + SEM is shown. n = 3. **P < 0.01, ***P < 0.001 by Student’s t test. (E) ZBED2 ChIP-seq–derived de novo motif logo, distribution, and E-value for the ZBED2 binding. (F) Metaprofile comparing ZBED2 occupancy in AsPC1 cells around the center of peaks with 0, 1, or ≥2 motif counts. (G) GSEA evaluating ZBED2 bound genes at promoter regions with 0, 1, or ≥2 motif counts upon ZBED2 cDNA expression in 15 PDA cell lines. (H) ZBED2 motif density frequency in the human genome. (I) Gene ontology (GO) analysis for genes annotated by HOMER to promoter regions with ≥2 ZBED2 motif counts. Terms are ranked by their significance (P value) and the most significant term is highlighted. (J) ChIP-seq profiles of FLAG-ZBED2 in AsPC1 cells and H3K27ac in HPAFII cells at the promoters of MX1, CMPK2, and STAT2. See also SI Appendix, Fig. S3.
Since other ZBED zinc fingers function as sequence-specific DNA binding domains (30, 31), we attempted to define a ZBED2 motif that correlates with its genomic occupancy. Using a de novo motif discovery analysis in the MEME-ChIP software (48), we derived an 8-nucleotide position weight matrix that closely correlated with ZBED2 enrichment observed by ChIP-seq (Fig. 3E). In addition, we found that the presence of two or more ZBED2 motifs correlated with strong genomic occupancy as well as with a stronger repressive effect on transcription than peaks displaying only a single ZBED2 motif (Fig. 3 F and G and SI Appendix, Fig. S3E). Notably, we found that less than 1% of all human promoters of protein-coding genes contain two or more ZBED2 motifs, and these promoters are strongly enriched for those driving expression of genes in the IFN pathway (Fig. 3 H and I, SI Appendix, Fig. S3F, and Dataset S8). MX1, CMPK2, and STAT2 are examples of ISGs that contain two ZBED2 motifs in their promoters (Fig. 3J). Collectively, these results suggest that ZBED2 functions as a sequence-specific DNA-binding repressor of select ISG promoters harboring multiple ZBED2 motifs.
Antagonistic Regulation of ISG Promoters by ZBED2 and IRF1.
In our MEME analysis of ZBED2-enriched locations in the human genome, we noticed a strong correlation between ZBED2 occupancy and a motif recognized by the IRF family TFs (Fig. 4A and SI Appendix, Fig. S4A). RNA-seq analysis revealed variable expression of IRF family TFs in PDA cell lines; however, the levels of ZBED2 expression were closely correlated with that of IRF1 (Fig. 4B and SI Appendix, Fig. S4B). A significant positive correlation between ZBED2 and IRF1 expression was also observed across all 1,156 cell lines within the CCLE database (SI Appendix, Fig. S4C). We next performed ChIP-seq analysis of IRF1 in AsPC1 cells, which confirmed its overlap with ZBED2 occupancy (SI Appendix, Fig. S4D and Dataset S9). For further analysis, we defined a high-confidence set of IRF1/ZBED2 cooccupied sites (Fig. 4C and Dataset S10). Notably, the presence of IRF1 and ZBED2 cooccupancy was a feature that was highly enriched for ISG promoters, such as CMPK2 and STAT2 (Fig. 4 D and E and SI Appendix, Fig. S4 D–H).
Fig. 4.
Antagonistic regulation of ISG promoters by ZBED2 and IRF1. (A) CentriMo analysis for JASPAR motifs at ZBED2 binding sites. (B) ZBED2 and IRF1 expression across 15 human PDA cell lines. (C) Density plot showing IRF1 and FLAG-ZBED2 enrichment surrounding a 2-kb interval centered on the summit of 140 intersecting IRF1 and FLAG-ZBED2 sites in AsPC1 cells, ranked by IRF1 peak intensity. (D) ChIP-seq profiles of IRF1 and FLAG-ZBED2 in AsPC1 cells at the promoters of CMPK2 and STAT2. (E) GO analysis of genes annotated by HOMER to IRF1/ZBED2 sites. Terms are ranked by their significance (P value) and the most significant terms (−log10 P > 12) are shown. (F) RT-qPCR analysis of CMPK2 in AsPC1-empty and AsPC1-ZBED2 cells following IRF1 cDNA expression. Mean+SEM is shown. n = 3. **P < 0.01 by Student’s t test. (G and H) GSEA plots evaluating protein coding genes annotated by HOMER to IRF1/ZBED2 sites upon IRF1 (G) or ZBED2 (H) cDNA expression in AsPC1 cells. (I) Expression levels of protein coding genes annotated to IRF1/ZBED2 sites following 12-h treatment with 0.2 ng/μL of IFN-β, IFN-γ, or control. ****P < 0.0001, **P < 0.01 *P < 0.05 by one-way ANOVA. (J) IRF1 motif logo from the JASPAR database (Upper) and the ZBED2 motif logo (Lower). (K) ChIP-seq analysis showing the log2 fold-change in IRF1 occupancy at IRF1 binding sites in AsPC1-ZBED2 versus AsPC1-empty cells. (L) ChIP-seq profiles of FLAG-ZBED2 and IRF1 at the promoters of CMPK2 (Left) and STAT2 (Right) in AsPC1-empty or AsPC1-ZBED2 cells. ZBED2 and IRF1 motifs recovered by Find Individual Motif Occurrences (P < 0.001) are highlighted in purple and yellow, respectively. See also SI Appendix, Fig. S4.
Since ZBED2 is a transcriptional repressor and IRF1 is a transcriptional activator (Fig. 3D), we reasoned that these two TFs might function in an antagonistic manner to regulate ISG expression. Consistent with this hypothesis, RT-qPCR analysis showed that IRF1 led to potent activation of CMPK2, while this effect was ablated in the presence of ZBED2 (Fig. 4F). RNA-seq analysis further verified that IRF1 and ZBED2 function in an antagonistic manner to regulate the entire program of cooccupied sites (Fig. 4 G and H and SI Appendix, Fig. S4 I–L). The antagonistic activities of ZBED2 and IRF1 were also demonstrated at the promoter of STAT2 (Fig. 4D). In addition, a STAT2 knockout in PANC0403 cells revealed a strong correlation between genes regulated by STAT2 and IRF1 (SI Appendix, Fig. S4M). Moreover, the entire set of genes sustained by STAT2 were found to be antagonistically regulated by ZBED2 and IRF1 (SI Appendix, Fig. S4 N–P). Considering these observations, we next treated AsPC1-empty and AsPC1-ZBED2 cells with IFN-β, IFN-γ, or control for 12 h and performed RNA-seq analysis. This analysis revealed that genes associated with IRF1/ZBED2 cooccupied sites were significantly induced by IFN, and this effect was attenuated by the presence of ZBED2 (Fig. 4I and SI Appendix, Fig. S4Q). Taken together, these data indicate that ZBED2 colocalizes with IRF1 at a subset of ISG promoters that are potently modulated by IFN pathway activation.
We noticed that the IRF1 and ZBED2 motifs bear similarity with one another, with both factors able to recognize a GAAA sequence (Fig. 4J) (49). This led us to hypothesize that IRF1 and ZBED2 might bind to specific promoter sequences in a competitive manner, and this may contribute to the antagonistic function of these two TFs. To evaluate this, we performed IRF1 ChIP-seq in AsPC1 cells that either express or lack ZBED2. This analysis revealed a twofold reduction of IRF1 occupancy at ∼19% of IRF1/ZBED2 cobound sites, which represent less than 2% of all IRF1 binding sites in the genome (Fig. 4K and Dataset S11). This included sites at the promoters of ISGs STAT2 and CMPK2 (Fig. 4 K and L), which were consistently observed to be among the most strongly down-regulated genes upon ZBED2 expression (Figs. 2E and 3C). Notably, these two promoters both possess motifs with a common GAAA core that are capable of being recognized by both IRF1 and ZBED2 (Fig. 4L). Taken together, these findings reveal two distinct mechanisms by which ZBED2 can block the output of IRF1 at ISG promoters: Through an intrinsic repressor function and via competitive DNA binding at specific motifs.
To gain a deeper understanding of these antagonistic activities, we used a deep-learning approach to characterize ZBED2 and IRF1 binding sites. This analysis enabled us to identify IRF1 recognition motifs that were either “favorable” or “unfavorable” for ZBED2 binding, which highlighted the importance of a CGAAAC sequence for ZBED2 cooccupancy (SI Appendix, Fig. S4R). In order to distinguish the intrinsic repressive activities of ZBED2 from those associated with competition for IRF1 binding sites, we next analyzed our 140 IRF1/ZBED2 cobound sites and separated them into two categories based on our model-based predictions: 89 sites predicted to have directly overlapping IRF1/ZBED2 binding sites (overlapping sites), and 51 sites predicted to have binding sites at neighboring locations (nonoverlapping sites). Interestingly, upon ZBED2 expression, we observed comparable down-regulation of genes associated with both overlapping and nonoverlapping sites (SI Appendix, Fig. S4 S–W). This observation lends additional support that ZBED2 can down-regulate IRF1-activated genes through multiple mechanisms, including both competition for DNA binding and via an intrinsic repressive function.
ZBED2 Protects PDA Cells from IRF1- and IFN-γ–Induced Growth Arrest.
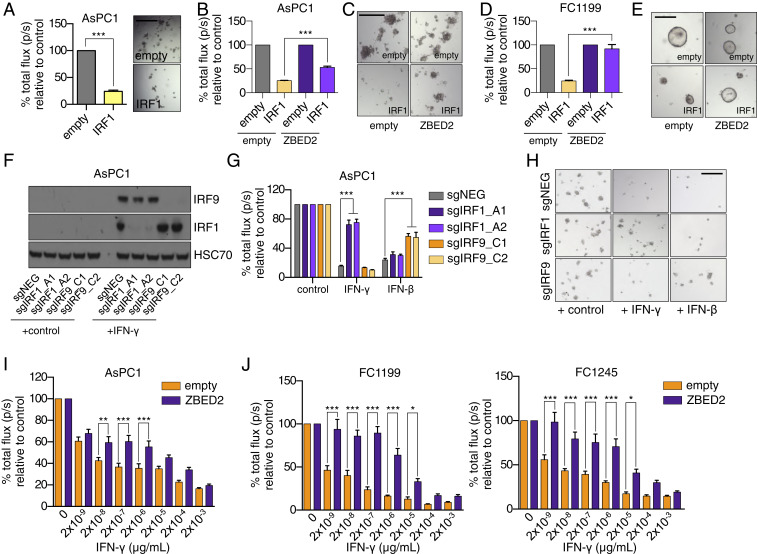
In addition to coordinating an antiviral cellular response, IRF1 leads to a powerful growth arrest when activated in tumor cells (50–52). Consistent with this observation, we found that forced IRF1 expression in several human and murine PDA cell line contexts resulted in a significant growth arrest (Fig. 5A and SI Appendix, Fig. S5A). However, if we repeated this experiment in the presence of ZBED2, the IRF1-induced growth arrest was prevented (Fig. 5 B–E). To verify this result in the setting of a more physiological context, we used exogenous IFN-γ to promote IRF1 activation in PDA cells. Notably, in PDA cell lines, IRF1 knockout only blocked IFN-γ–induced growth arrest but not growth arrest caused by IFN-β, which instead relied on IRF9 (Fig. 5 F–H and SI Appendix, Fig. S5 B–D). Given that the growth-inhibition effects of IFN-γ stimulation are at least partially mediated through IRF1 induction, we reasoned that ZBED2 expression might confer a protective effect in this context. Consistent with this hypothesis, ZBED2 expression was sufficient to protect PDA cells from the effects of IFN-γ–mediated growth arrest, which was observed in AsPC1 cells, as well as in three murine PDA cell lines derived from the KPC (Kras+/LSL-G12D; Trp53+/LSL-R172H; Pdx1-Cre) mouse model (Fig. 5 I and J and SI Appendix, Fig. S5E) (53, 54). Analysis of single-cell RNA-seq data (40) revealed IFNB1-expressing ductal tumor cells and IFNG-expressing T cells as possible sources of IFN production in human PDA tumors (SI Appendix, Fig. S5 F–H). Taken together, these data suggest that ZBED2 can antagonize both the transcriptional and phenotypic consequences of IRF1 activation in PDA cells.
Fig. 5.
ZBED2 protects PDA cells from IRF1- and IFN-γ–induced growth arrest. (A) Luciferase-based quantification of cell viability of AsPC1 cells grown in Matrigel on day 7 postinfection with IRF1 cDNA or the empty vector. Representative bright-field images (Right) are shown. (Scale bar, 200 μm.) ***P < 0.001 by Student’s t test. (B) Luciferase-based quantification of cell viability of AsPC1-empty and AsPC1-ZBED2 cells following coexpression of IRF1 cDNA or empty vector for 7 d. ***P < 0.001 by Student’s t test. (C) Representative bright-field images from B. (Scale bar, 500 μm.) (D) Luciferase-based quantification of cell viability of KPC-derived FC1199 cells expressing ZBED2 or the empty vector following coexpression of IRF1 cDNA or empty vector for 7 d. (E) Representative bright-field images from D. (Scale bar, 200 μm.) ***P < 0.001 by Student’s t test. (F–H) AsPC1 cells infected with sgRNAs targeting IRF1, IRF9, or a control sgRNA (sgNEG) were plated in Matrigel and grown for 7 d in the presence of 20 ng/μL of IFN-γ, IFN-β, or control. Representative Western blots following overnight stimulation with 20 ng/μL of IFN-γ (F), luciferase-based quantification (G), and bright-field images on day 7 (H) are shown. ***P < 0.001 by Student’s t test. (Scale bar, 500 μm.) (I and J) AsPC1 cells (I) or the indicated KPC cell lines (J) were infected with the ZBED2 cDNA or an empty vector and grown in Matrigel with the increasing concentrations of IFN-γ. Bar charts show luciferase-based quantification on day 7. Mean + SEM is shown. n = 3. For I and J, ***P < 0.001, **P < 0.01, *P < 0.05 by one-way ANOVA. See also SI Appendix, Fig. S5.
ZBED2 Represses Pancreatic Progenitor Lineage Identity in PDA.
Prior transcriptome analyses of tumor samples have revealed two major molecular subtypes of PDA: A “pancreatic progenitor” and a “squamous” subtype (13, 35, 36, 55). The pancreatic progenitor subtype is also known as classical PDA, and expresses endodermal TFs (e.g., GATA6, HNF4, FOXA) at high levels. In contrast, the squamous subtype (also known as basal-like) silences the pancreatic progenitor gene signature and instead expresses markers of the squamous lineage (e.g., KRT5, TP63) in association with inferior patient survival outcomes (10, 13). We found in three independent human tumor transcriptome datasets (13, 35, 36), ZBED2 expression was highly associated with squamous subtype PDA tumors (Fig. 6 A–F and Dataset S12). This prompted us to investigate whether ZBED2 has a causal role in influencing the pancreatic progenitor or squamous transcriptional signatures in PDA cell lines. Using the aforementioned RNA-seq analysis, we found that the pancreatic progenitor signature was significantly repressed by ZBED2 expression in 13 of 15 PDA cell lines examined (Fig. 6 G and H). The two exceptions were MIAPaca2 and PANC1 cells, which do not express pancreatic progenitor signature genes (10). The squamous signature was more variably affected by ZBED2 expression (SI Appendix, Fig. S6A). In accord with previous findings (27), we found that IRF1 activates the pancreatic progenitor signature, which is consistent with its antagonism with ZBED2 (Fig. 6I). We noticed that one of the IRF1/ZBED2 cooccupied sites was found at the promoter of the GATA6 gene, which encodes an established marker of the pancreatic progenitor transcriptional program (Fig. 6 J and K) (36, 55–57). Consistent with their antagonistic activities, we found ZBED2 or IRF1 expression resulted in the repression or activation of GATA6 in this context, respectively (Fig. 6L and SI Appendix, Fig. S6 B and C). Of note, of the 15 PDA cell lines used in this study, we observed the lowest levels of GATA6 and IRF1 in MIAPaca2 and PANC1 cells (SI Appendix, Fig. S6 D and E).
Fig. 6.
ZBED2 represses pancreatic progenitor lineage identity in PDA. (A–C) TF expression in molecular subtypes of PDA. TFs are ranked by their mean log2 fold-change in Basal-like vs. classic (A and C) or bquamous vs. progenitor (B) patient samples from the indicated studies. (D–F) ZBED2 expression in PDA patient samples stratified according to molecular subtype. Each dot represents one patient sample. P value was calculated using Student’s t test. (G) GSEA evaluating the progenitor PDA Identity signature upon ZBED2 cDNA expression in 15 PDA cell lines. (H and I) GSEA plots evaluating the progenitor-PDA Identity signature following expression of ZBED2 (H) or IRF1 (I) in AsPC1 cells. (J) Overlap of progenitor PDA identity genes with protein coding genes associated with IRF1/ZBED2 sites. (K) ChIP-seq profiles of IRF1 and FLAG-ZBED2 at the promoter of GATA6 in AsPC1 cells. (L) Overlap of leading edge (LE) genes associated with IRF1/ZBED2 sites that are repressed by ZBED2 or activated by IRF1 in AsPC1 cells. (M) Quantification of colony size in Matrigel assays. Means + SEM are shown. n = 3. Representative images at day 7 are shown (Right). (N) Quantification of scratch assays at the indicated time points postseeding and representative images (Right). (Scale bar, 500 μm.) Means + SEM are shown. n = 3. ***P < 0.001, **P < 0.01 *P < 0.05 by Student’s t test; ns, not significant. See also SI Appendix, Fig. S6.
In order to determine the impact of GATA6 expression on the pancreatic progenitor signature, we first performed RNA-seq analysis following GATA6 knockout in PATU8988S cells, the cell line that expresses GATA6 at the highest levels (SI Appendix, Fig. S6D). Notably, a GATA6 knockout resulted in a potent repression of the pancreatic progenitor signature (SI Appendix, Fig. S6 F and G). Next, to investigate whether the down-regulation of GATA6 was a requirement for ZBED2-mediated repression of the progenitor transcriptional program, we performed RNA-seq analysis following the expression of ZBED2 or GATA6 cDNAs alone, or in combination, in SUIT2 PDA cells (SI Appendix, Fig. S6 H–J). As expected, ZBED2 expression alone resulted in repression of the pancreatic progenitor signature. In contrast, GATA6 expression was sufficient to potently activate the pancreatic progenitor signature, and this was true whether GATA6 was expressed alone or in the presence of ZBED2 (SI Appendix, Fig. S6 H–J). Finally, by intersecting our GATA6 knockout and overexpression datasets, we defined a set of genes that were potently regulated by GATA6 (SI Appendix, Fig. S6K and Dataset S13). Consistent with the antagonistic activities of ZBED2 and IRF1 at the promoter of GATA6, we observed potent repression and activation of GATA6-regulated genes in response to ZBED2 and IRF1 expression, respectively (SI Appendix, Fig. S6 L–N).
Consistent with the results above, patient tumor samples with high levels of ZBED2 expression were much more likely to present with poorly differentiated tumors compared to those patients with low ZBED2 expression (SI Appendix, Fig. S6 O and P). Moreover, we found that ZBED2 expression in PDA cells was sufficient to promote loss of epithelial integrity and increased invasiveness when cells were plated in Matrigel (Fig. 6M), as well as enhanced migratory behavior in scratch assays (Fig. 6N). These observations are consistent with our findings that ZBED2 expression can repress GATA6 and induce an EMT transcriptional program. Collectively, these findings are in support of ZBED2 regulating the lineage identity of PDA cells and can account for the biased expression of ZBED2 in squamous subtype PDA tumors.
Discussion
Here, we have shown that a previously uncharacterized zinc finger protein ZBED2 is aberrantly expressed in a diverse collection of human tumors in a manner that correlates with poor clinical outcomes. Using PDA cells as an experimental system, we have defined two major molecular functions of ZBED2 that may underpin its role in promoting cancer: As an antagonist of the IFN response and as a modulator of epithelial cell identity. In addition, we provide evidence that both of these functions of ZBED2 can be accounted for by antagonism of IRF1-mediated transcriptional activation.
A key mechanistic advance in our study is in revealing the DNA sequence motif recognized by ZBED2, which we show is disproportionately enriched at ISG promoters in juxtaposition with IRF1 motifs. At these sites, we have shown that ZBED2 can suppress transcription through multiple mechanisms, including an intrinsic transcriptional repressor function as well as the ability to compete with IRF1 for DNA binding. While both mechanisms are likely to contribute to the suppression of IRF1 transcriptional output, we note that eviction of IRF1 by ZBED2 only occurs at a small fraction of cobound promoters. Taken together, these observations suggest that the evolved function of ZBED2 in mammalian biology is as a tissue-specific attenuator of the IFN response. The tissues that express ZBED2 at the highest levels include thyroid, esophagus, lung, and skin, which are organs that are often exposed to pathogens and may employ ZBED2 to define thresholds for IFN responsiveness. Notably, inflammation of the thyroid is one of the most common side effects of IFN-α therapy in humans (58), which leads us to speculate that ZBED2 may have evolved to protect vulnerable tissues from the damaging effects of chronic IFN stimulation. Other ZBED TFs have also been implicated in the regulation of immune responses. For example, ZBED1 has been described as a viral restriction factor that negatively regulates viral growth (59). In addition, a recent study in plants identified a ZBED protein as a novel regulator of immunity (60). Thus, the function of ZBED2 defined in our study may have its origins in the early evolutionary functions of ZBED proteins as important regulators of immune responses.
We have defined ZBED2 as an attenuator of IFN responses through its ability to antagonize the transcriptional output of IRF1. A paradoxical observation revealed from our studies is that PDA cells that have acquired high levels of ZBED2 also tend to display higher levels of IRF1 expression and IFN pathway activation. We attribute this to ZBED2 functioning as a quantitative dampener of the IFN transcriptional response during the initiation and progression of PDA. Importantly, IRF1 is a validated tumor suppressor (50–52, 61) and IFN pathways have a well-established role in the regulation of antitumor immunity (62). Thus, antagonism of IRF1 provides a plausible explanation for why high ZBED2 expression is selected for during tumorigenesis. Less clear are the initial triggers of IRF1/IFN pathway activation in PDA. A recent study demonstrated that cancer cells exhibit high ISG expression due to chronic IFN signaling sustained by the STING adaptor protein, which is activated by various intracellular nucleic acid sensors (63, 64). Thus, elevated ISG expression in cancer cells could be attributed to the accumulation of cytosolic double-stranded DNA, which may arise from the disruption of micronuclei as a result of DNA damage, chromosome mis-segregation, and chromothripsis (65, 66). Such features of high genomic instability have been strongly implicated in the development and progression of PDA (46, 67). However, sustained activation of ISG expression via the STING pathway was shown to be dependent on chronic production of type I IFNs (63). In our analysis of human PDA cell lines, we observed high basal levels of ISG expression in PDA cells in a manner that is independent of type I IFNs. While the triggers of the ISG pathway in PDA await further characterization, our results strongly suggest that the function of ZBED2 in this context is to block the antitumor effects of IRF1 within the IFN pathway. It is also noteworthy that, despite direct repression of STAT2 and the type I IFN transcriptional response, ZBED2 was unable to suppress the effects of IFN-β–mediated growth arrest in the PDA cell lines tested. While the mechanisms of IFN-β–mediated growth arrest of PDA cell lines was not extensively investigated in this study, it is clear that they occur independent of IRF1 and instead rely more on IRF9. These data suggest that the functional antagonism demonstrated for ZBED2 and IRF1 may not be a feature that is equally shared across the IRF transcription factor family. Notably, immune checkpoint therapy requires an intact IFN pathway for effective tumor clearance (68), and this approach has been found to be ineffective in PDA when compared to other tumor subtypes. While several factors are likely contributing to immune evasion in PDA (69), we speculate that high ZBED2 expression could be a mechanism by which tumor cells evade cytotoxic T cells.
Histopathological and transcriptome studies have shown that a subset of aggressive PDA tumors undergo transdifferentiation into the squamous lineage (13, 70–72). In addition to the aberrant up-regulation of squamous lineage markers, a key attribute of these tumors is the silencing of genes associated with pancreatic progenitor cell identity. A key finding in our study is that ZBED2 is selectively up-regulated in squamous-subtype PDA tumors, and in this context represses the pancreatic progenitor transcriptional program. Notably, we also observe high ZBED2 expression in normal squamous epithelial tissues, such as the skin, lung, and esophageal mucosa, suggesting a normal function for ZBED2 in the regulation of squamous epithelial cells. Interestingly, single-cell RNA-seq analysis recently identified ZBED2 as a key TF in the regulation of human keratinocyte differentiation (33). ZBED2 was predicted to promote the basal state and experimental depletion of ZBED2 was shown to induce keratinocyte differentiation, demonstrated by the up-regulation of KRT10 (33). It is noteworthy that IRF family members have also been ascribed functional roles in regulation of the skin epidermis. For example, both IRF2 and IRF6 are necessary for regulating proliferation and terminal differentiation of keratinocytes within the epidermis (73–76). Thus, it is possible that the ability of ZBED2 to modulate the transcriptional output of IRF TFs may extend to its ability to regulate stem cell function and cell identity within normal squamous epithelial tissues.
Similar to ZBED2, prior studies have implicated TP63 and GLI2 as additional determinants of squamous cell identity in PDA (10, 11, 13, 15). Our analysis has shown that ZBED2 and TP63 are among the most aberrantly expressed TFs in the squamous subtype of PDA; however, these two TFs are not expressed in a mutually exclusive manner. These findings suggest the aberrant expression of TP63, GLI2, or ZBED2 offer distinct mechanisms by which pancreatic cancer cells with a ductal identity adopt squamous features during the pathogenesis and progression of this disease. An important area of future investigation will be to determine whether such factors can function in a collaborative manner to regulate squamous identity.
An important feature of squamous subtype tumors is their poorly differentiated histopathological features and an exceptionally poor prognosis (13, 35, 36, 55, 77, 78). Interestingly, a recent study identified IRF1 as one of several candidate TFs responsible for preserving differentiation and epithelial identity in human PDA (27). In this study, IRF1 was shown to associate with “low-grade” enhancers controlling grade-specific transcriptional programs, which is in accord with our functional studies of IRF1. Given the dual role of IRF1 as a tumor suppressor and as a regulator of cellular differentiation in PDA cells, we speculate that ZBED2-mediated dampening of the IFN response may support PDA cell growth at early stages of tumor progression. This may also explain, at least in part, the positive correlation that is observed between ZBED2 and IRF1 expression in PDA cell lines, which runs counterintuitive to the repressor function of ZBED2 on IRF1 transcriptional output. We have shown that a key IRF1/ZBED2 cooccupied target gene is GATA6, which is overexpressed and amplified in the pancreatic progenitor subtype of PDA (55, 57, 79). Indeed, GATA6 expression is altered by multiple mechanisms in pancreatic tumors and was found to be a robust biomarker for PDA subtypes (36, 78). We have now established GATA6 as a master regulator of the pancreatic progenitor subtype of PDA, strongly implicating the regulation of the GATA6 promoter as a critical region of ZBED2-mediated control of PDA cell identity. Thus, our study further reinforces how cell identity in PDA is defined by antagonism among TFs important for maintaining the epithelial identity of this lineage.
Materials and Methods
CRISPR-Based Targeting.
To generate cell lines in which ZBED2, IRF1, IRF9, STAT2, GATA6, or IFNAR1 had been stably knocked out, PDA cells expressing Cas9 in the LentiV-Cas9-puro vector (Addgene #108100) were infected in a pooled fashion with domain-targeting sgRNAs or a control sgRNA (sgNEG) in the LRG2.1_Neo vector (addgene #125593). sgRNA targeting sequences can be found in Dataset S14.
shRNA-Based Targeting.
shRNAs targeting ZBED2 or control were cloned into the miR30-based retroviral shRNA expression vector LENC (LTR-miR30-shRNA-PGK-neo-mCherry) (#111163) (80). Two days postinfection with shRNAs, transduced cells were selected with 1 mg/mL of G418 for 5 d prior to RNA extraction. shRNA sequences can be found in Dataset S14.
Data and Software Availability.
ChIP-seq and RNA-seq data reported in this paper is available via the Gene Expression Omnibus (GEO) accession no. GSE141607. Previously deposited ChIP-seq data (10) can be found in the GEO, accession no. GSE115463. PDA patient microarray data (35) can be found in the GEO, accession no. GSE71729.
Supplementary Material
Acknowledgments
The authors thank the Cold Spring Harbor Cancer Center Support Grant shared resources, Bioinformatics Shared Resource, and Next Generation Sequencing Core Facility. We also thank Kenneth Olive and Carlos Maurer (Columbia University) for critical discussions; and Noah Dukler (Cold Spring Harbor Laboratory) for phylogenetic analysis of ZBED family members. T.D.D.S. was supported by a grant from the State of New York, Contract C150158. C.R.V. was supported by Pershing Square Sohn Cancer Research Alliance, the Cold Spring Harbor Laboratory and Northwell Health Affiliation, the National Cancer Institute 5P01CA013106-Project 4 and 1RO1CA229699, the Thompson Family Foundation, the Simons Foundation, and a Career Development Award from the Pancreatic Cancer Action Network–American Association for Cancer Research 16-20-25-VAKO.
Footnotes
Competing interest statement: C.R.V. has received funding from Boehringer-Ingelheim and is an advisor to KSQ Therapeutics.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE141607).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921484117/-/DCSupplemental.
References
- 1.Yuan S., Norgard R. J., Stanger B. Z., Cellular plasticity in cancer. Cancer Discov. 9, 837–851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merrell A. J., Stanger B. Z., Adult cell plasticity in vivo: De-differentiation and transdifferentiation are back in style. Nat. Rev. Mol. Cell Biol. 17, 413–425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grippo P. J., Nowlin P. S., Demeure M. J., Longnecker D. S., Sandgren E. P., Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 63, 2016–2019 (2003). [PubMed] [Google Scholar]
- 4.Kopp J. L., et al. , Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22, 737–750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De La O J. P., et al. , Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc. Natl. Acad. Sci. U.S.A. 105, 18907–18912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhim A. D., et al. , EMT and dissemination precede pancreatic tumor formation. Cell 148, 349–361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krebs A. M., et al. , The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 19, 518–529 (2017). [DOI] [PubMed] [Google Scholar]
- 8.David C. J., et al. , TGF-β tumor suppression through a lethal EMT. Cell 164, 1015–1030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roe J.-S., et al. , Enhancer reprogramming promotes pancreatic cancer metastasis. Cell 170, 875–888.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Somerville T. D. D., et al. , TP63-Mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal adenocarcinoma. Cell Rep. 25, 1741–1755.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamdan F. H., Johnsen S. A., DeltaNp63-dependent super enhancers define molecular identity in pancreatic cancer by an interconnected transcription factor network. Proc. Natl. Acad. Sci. U.S.A. 115, E12343–E12352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andricovich J., et al. , Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell 33, 512–526.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey P., et al. ; Australian Pancreatic Cancer Genome Initiative , Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Collisson E. A., Bailey P., Chang D. K., Biankin A. V., Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Adams C. R., et al. , Transcriptional control of subtype switching ensures adaptation and growth of pancreatic cancer. eLife 8, e45313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Candido J. B., et al. , CSF1R+ macrophages sustain pancreatic tumor growth through T cell suppression and maintenance of key gene programs that define the squamous subtype. Cell Rep. 23, 1448–1460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zitvogel L., Galluzzi L., Kepp O., Smyth M. J., Kroemer G., Type I interferons in anticancer immunity. Nat. Rev. Immunol. 15, 405–414 (2015). [DOI] [PubMed] [Google Scholar]
- 18.McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A., Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motwani M., Pesiridis S., Fitzgerald K. A., DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Pestka S., Krause C. D., Walter M. R., Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202, 8–32 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Schneider W. M., Chevillotte M. D., Rice C. M., Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michalska A., Blaszczyk K., Wesoly J., Bluyssen H. A. R., A positive feedback amplifier circuit that regulates interferon (IFN)-Stimulated gene expression and controls type I and type II IFN responses. Front. Immunol. 9, 1135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platanias L. C., Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Schoggins J. W., et al. , A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alsamman K., El-Masry O. S., Interferon regulatory factor 1 inactivation in human cancer. Biosci. Rep. 38, BSR20171672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai T., et al. , The roles of interferon regulatory factors 1 and 2 in the progression of human pancreatic cancer. Pancreas 43, 909–916 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Diaferia G. R., et al. , Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J. 35, 595–617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayward A., Ghazal A., Andersson G., Andersson L., Jern P., ZBED evolution: Repeated utilization of DNA transposons as regulators of diverse host functions. PLoS One 8, e59940 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita D., et al. , hDREF regulates cell proliferation and expression of ribosomal protein genes. Mol. Cell. Biol. 27, 2003–2013 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mokhonov V. V., Theendakara V. P., Gribanova Y. E., Ahmedli N. B., Farber D. B., Sequence-specific binding of recombinant Zbed4 to DNA: Insights into Zbed4 participation in gene transcription and its association with other proteins. PLoS One 7, e35317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markljung E., et al. , ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 7, e1000256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang H., et al. , Axonal guidance signaling pathway interacting with smoking in modifying the risk of pancreatic cancer: A gene- and pathway-based interaction analysis of GWAS data. Carcinogenesis 35, 1039–1045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finnegan A., et al. , Single-cell transcriptomics reveals spatial and temporal turnover of keratinocyte differentiation regulators. Front. Genet. 10, 775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., et al. , Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 176, 775–789.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moffitt R. A., et al. , Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 47, 1168–1178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network , Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiriac H., et al. , Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 8, 1112–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GTEx Consortium , Human genomics. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoadley K. A., et al. ; Cancer Genome Atlas Network , Cell-of-Origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173, 291–304.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng J., et al. , Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 29, 725–738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo X., et al. , Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24, 978–985 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Zheng C., et al. , Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342–1356.e16 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Smith J. C., Sheltzer J. M., Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. eLife 7, e39217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghandi M., et al. , Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biankin A. V., et al. ; Australian Pancreatic Cancer Genome Initiative , Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waddell N., et al. ; Australian Pancreatic Cancer Genome Initiative , Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones S., et al. , Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Machanick P., Bailey T. L., MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Escalante C. R., Yie J., Thanos D., Aggarwal A. K., Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature 391, 103–106 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Tanaka N., et al. , Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell 77, 829–839 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Kirchhoff S., Schaper F., Hauser H., Interferon regulatory factor 1 (IRF-1) mediates cell growth inhibition by transactivation of downstream target genes. Nucleic Acids Res. 21, 2881–2889 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harada H., et al. , Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science 259, 971–974 (1993). [DOI] [PubMed] [Google Scholar]
- 53.Engle D. D., et al. , The glycan CA19-9 promotes pancreatitis and pancreatic cancer in mice. Science 364, 1156–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hingorani S. R., et al. , Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Collisson E. A., et al. , Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Decker K., Goldman D. C. L., Grasch C. L., Sussel L., Gata6 is an important regulator of mouse pancreas development. Dev. Biol. 298, 415–429 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu B., Luo M., Lakkur S., Lucito R., Iacobuzio-Donahue C. A., Frequent genomic copy number gain and overexpression of GATA-6 in pancreatic carcinoma. Cancer Biol. Ther. 7, 1593–1601 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Tomer Y., Blackard J. T., Akeno N., Interferon alpha treatment and thyroid dysfunction. Endocrinol. Metab. Clin. North Am. 36, 1051–1066, x–xi (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Radko S., et al. , Adenovirus E1A targets the DREF nuclear factor to regulate virus gene expression, DNA replication, and growth. J. Virol. 88, 13469–13481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kroj T., Chanclud E., Michel-Romiti C., Grand X., Morel J.-B., Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210, 618–626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nozawa H., et al. , Loss of transcription factor IRF-1 affects tumor susceptibility in mice carrying the Ha-ras transgene or nullizygosity for p53. Genes Dev. 13, 1240–1245 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parker B. S., Rautela J., Hertzog P. J., Antitumour actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer 16, 131–144 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Liu H., et al. , Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med. 25, 95–102 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Ng K. W., Marshall E. A., Bell J. C., Lam W. L., cGAS-STING and cancer: Dichotomous roles in tumor immunity and development. Trends Immunol. 39, 44–54 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Mackenzie K. J., et al. , cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang C.-Z., et al. , Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Notta F., et al. , A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 538, 378–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Minn A. J., Wherry E. J., Combination cancer therapies with immune checkpoint blockade: Convergence on interferon signaling. Cell 165, 272–275 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Joyce J. A., Fearon D. T., T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Ishikawa O., et al. , Adenosquamous carcinoma of the pancreas: A clinicopathologic study and report of three cases. Cancer 46, 1192–1196 (1980). [DOI] [PubMed] [Google Scholar]
- 71.Morohoshi T., Held G., Klöppel G., Exocrine pancreatic tumours and their histological classification. A study based on 167 autopsy and 97 surgical cases. Histopathology 7, 645–661 (1983). [DOI] [PubMed] [Google Scholar]
- 72.Motojima K., Tomioka T., Kohara N., Tsunoda T., Kanematsu T., Immunohistochemical characteristics of adenosquamous carcinoma of the pancreas. J. Surg. Oncol. 49, 58–62 (1992). [DOI] [PubMed] [Google Scholar]
- 73.Ingraham C. R., et al. , Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat. Genet. 38, 1335–1340 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mercado N., et al. , IRF2 is a master regulator of human keratinocyte stem cell fate. Nat. Commun. 10, 4676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hida S., et al. , CD8(+) T cell-mediated skin disease in mice lacking IRF-2, the transcriptional attenuator of interferon-α/β signaling. Immunity 13, 643–655 (2000). [DOI] [PubMed] [Google Scholar]
- 76.Oberbeck N., et al. , The RIPK4-IRF6 signalling axis safeguards epidermal differentiation and barrier function. Nature 574, 249–253 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Maurer C., et al. , Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut 68, 1034–1043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aung K. L., et al. , Genomics-driven precision medicine for advanced pancreatic cancer: Early results from the COMPASS trial. Clin. Cancer Res. 24, 1344–1354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martinelli P., et al. , GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut 66, 1665–1676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fellmann C., et al. , An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5, 1704–1713 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.