Significance
Immunocompromised patients are at increased risk for multidrug-resistant infections, due to broad-spectrum antibiotic exposure and a host environment with limited innate defenses. This study explored how vancomycin-resistant Enterococcus faecium (VREfm), a pathogen endemic to many hospitals, underwent genomic and phenotypic changes during intestinal colonization and bloodstream infection of immunocompromised pediatric patients. We identified a mutation conferring bacterial growth in alternative sugars that arose de novo in two different patients and was also present in five other patients. We also characterized mutations in surface polysaccharide production associated with better adherence to surfaces and resistance to the innate immune factor lysozyme. These findings suggest that targeting carbohydrate availability and bacterial adherence may be worthwhile strategies to limit VREfm proliferation in immunocompromised hosts.
Keywords: vancomycin-resistant enterococci, pediatric infectious diseases, functional genomics
Abstract
Patients with hematological malignancies or undergoing hematopoietic stem cell transplantation are vulnerable to colonization and infection with multidrug-resistant organisms, including vancomycin-resistant Enterococcus faecium (VREfm). Over a 10-y period, we collected and sequenced the genomes of 110 VREfm isolates from gastrointestinal and blood cultures of 24 pediatric patients undergoing chemotherapy or hematopoietic stem cell transplantation for hematological malignancy at St. Jude Children’s Research Hospital. We used patient-specific reference genomes to identify variants that arose over time in subsequent gastrointestinal and blood isolates from each patient and analyzed these variants for insight into how VREfm adapted during colonization and bloodstream infection within each patient. Variants were enriched in genes involved in carbohydrate metabolism, and phenotypic analysis identified associated differences in carbohydrate utilization among isolates. In particular, a Y585C mutation in the sorbitol operon transcriptional regulator gutR was associated with increased bacterial growth in the presence of sorbitol. We also found differences in biofilm-formation capability between isolates and observed that increased biofilm formation correlated with mutations in the putative E. faecium capsular polysaccharide (cps) biosynthetic locus, with different mutations arising independently in distinct genetic backgrounds. Isolates with cps mutations showed improved survival following exposure to lysozyme, suggesting a possible reason for the selection of capsule-lacking bacteria. Finally, we observed mutations conferring increased tolerance of linezolid and daptomycin in patients who were treated with these antibiotics. Overall, this study documents known and previously undescribed ways that VREfm evolve during intestinal colonization and subsequent bloodstream infection in immunocompromised pediatric patients.
Enterococci are ancient and ubiquitous members of the healthy gastrointestinal tracts of a wide variety of land animals across the tree of life (1). In the antibiotic era, enterococci emerged as leading causes of drug-resistant and hospital-associated infections (2). Enterococci are intrinsically resistant to a large number of commonly used antibiotics, and they are particularly tolerant to stresses such as desiccation, starvation, and disinfectants (3–5). Additionally, genomic plasticity enables enterococci to readily acquire mobile genetic elements, and to share drug resistance genes with other pathogens, which further complicates infection treatment and control (1, 6, 7). Among the enterococci, Enterococcus faecalis and Enterococcus faecium are the most common causes of multidrug-resistant infection, including bacteremia, urinary tract infection, and surgical site infection (8, 9). The persistence of multidrug-resistant enterococci in the hospital environment poses a major threat to patient safety, and vancomycin-resistant E. faecium (VREfm) carrying VanA-type resistance currently pose the biggest threat in the United States (10–12).
Immunocompromised patients are prone to drug-resistant enterococcal infection, due to frequent use of broad-spectrum antibiotics in hospital environments where resistant lineages circulate (13). Patients undergoing chemotherapy and hematopoietic stem cell transplantation are at particular risk because of their compromised immune systems, including diminished innate immunity and permeable mucosal barriers, which facilitate colonization of the skin and intestinal tract and ultimately infection with multidrug-resistant organisms such as VREfm (14). The frequent use of central venous catheters in hospital settings creates an additional route for nosocomial pathogens to enter the bloodstream of patients during hospitalization. Whether they arrive in the blood via translocation through the intestinal mucosa or through an intravenous catheter, bacteria will experience distinct selective pressures in the bloodstream compared with those in the skin or in the intestinal tract, including altered nutrient availability, different host immune defenses, and variable concentrations of antibiotics (15). These conditions select for the outgrowth of variants better able to persist and proliferate in the bloodstream (16). A prior study in one immunocompromised pediatric patient found that VREfm adapted to conditions in the bloodstream by activation of the bacterial stringent response (17). For this patient, antibiotics alone were unable to effectively treat the infection, which resolved upon reconstitution of immune function with a granulocyte infusion. Whether VREfm adapt similarly in other patients, and how they evolve during intestinal colonization and bloodstream infection in immunocompromised hosts, remains to be determined.
Here, we studied VREfm intestinal colonization and bloodstream infection in 24 pediatric patients admitted over a 10-y period to the St. Jude Children’s Research Hospital for chemotherapy or hematopoietic stem cell transplantation. We used comparative genomic analyses to explore the genomic makeup of the VREfm isolates infecting patients over this time period. We then analyzed colonization and infection strain genomes from each patient to identify possible ways that the VREfm population within each patient adapted to persistently colonize the antibiotic-perturbed intestinal tract and to look for signals of bloodstream adaptation. Finally, we connected some of the genetic changes we observed to alterations in carbohydrate utilization, biofilm formation, and antibiotic tolerance.
Results
Overview of Sampled Patients and VREfm Population Structure.
A total of 110 VREfm isolates were collected from 24 pediatric patients over an ∼10-y period (Table 1, Fig. 1A, and Dataset S1). These included both stool or perianal swab (gastrointestinal) and blood isolates; participants were eligible if at least one VREfm isolate from blood and at least one isolate from preceding or concurrent gastrointestinal colonization were available. Length of sampling of patients ranged from 0 to 565 d, and 2 to 21 isolates were collected from each patient (Fig. 1A and Dataset S1). The genomes of all 110 isolates were sequenced on the Illumina platform (Dataset S1), and the earliest gastrointestinal isolate from each patient was also sequenced on the Oxford Nanopore MinION platform. Nanopore and Illumina sequencing reads were hybrid-assembled to generate a high-quality reference-genome assembly for each patient (Dataset S2).
Table 1.
Clinical characteristics of patients in the study cohort
| Characteristic | N | % |
| Sex | ||
| Female | 12 | 50 |
| Male | 12 | 50 |
| Race | ||
| Caucasian | 21 | 88 |
| African American | 2 | 8 |
| Other | 1 | 4 |
| Primary diagnosis | ||
| Acute lymphoblastic leukemia | 5 | 21 |
| Acute myeloid leukemia | 16 | 67 |
| Other | 3 | 13 |
| Hematopoietic cell transplantation | ||
| Allogeneic | 18 | 25 |
| None | 6 | 25 |
Age (median: 8.0 y [range: 0.6 to 21.4 y]) is reported as age at collection of first isolate. Race is self-reported race.
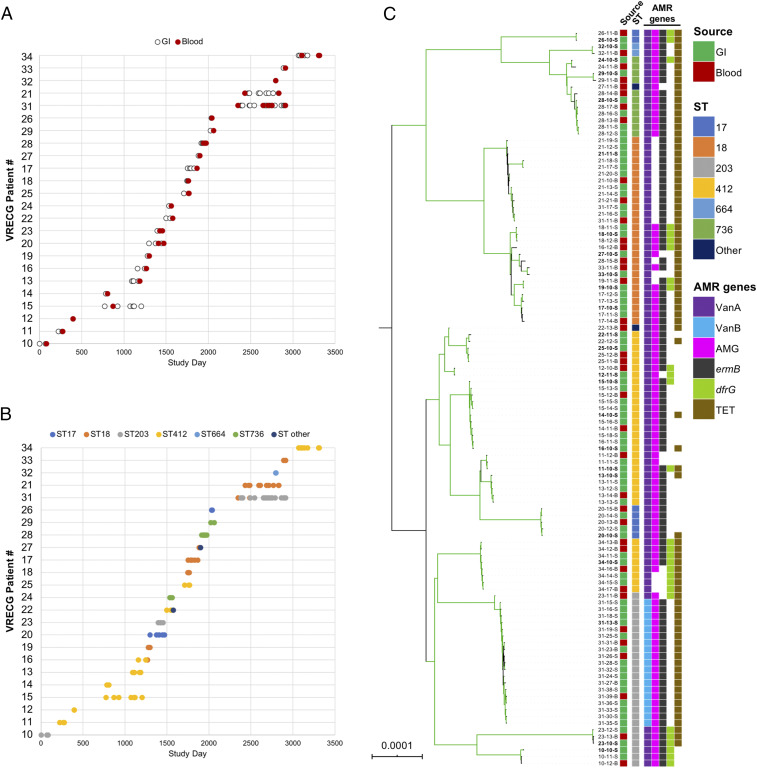
Fig. 1.
Sampling overview and population structure of 110 VREfm from immunocompromised pediatric patients. (A) Date of sampling of gastrointestinal (GI) (white) and bloodstream (Blood) (red) VREfm isolates from 24 study patients. Patients are labeled according to their “VRECG” patient number. (B) Distribution of STs for all isolates in the sample set. (C) Single-copy core genome phylogeny of all isolates in the sample set. Isolates are named as XX-YY-S/B, where XX is the patient number, YY is the isolate identifier, S indicates GI isolates, and B indicates blood isolates. The RAxML phylogeny is based on a gapless alignment of SNPs in the core genome identified by Snippy, with SNPs due to recombination removed by ClonalFrameML. Branches colored green are supported by bootstrap values >90. Tips are annotated with isolate names, source, ST, Van operon type, and other drug resistance-associated (AMR) genes identified in each genome. Isolate names in bold mark the patient-specific reference genomes used to identify variants in subsequent isolates in each patient. AMG, aminoglycoside-resistance genes; TET, tetracycline-resistance genes.
VREfm isolates were found to belong to multilocus sequence type (ST) ST412 (n = 34), ST18 (n = 28), ST203 (n = 26), ST736 (n = 11), ST17 (n = 7), ST664 (n = 2), ST750 (n = 1), and a single locus variant of ST736 (n = 1) (Fig. 1B and Dataset S1). All isolates belonged to E. faecium clade A, and the most prevalent STs observed represent well-known multidrug-resistant and hospital-adapted lineages (18). Most patients maintained the same ST over time; however, isolates belonging to two different STs were isolated from five patients (Dataset S1). Nine of the 24 patients were colonized and infected with ST412 isolates, which dominated through the first 3 y of the study period and were then isolated periodically throughout the remainder of the study (Fig. 1B). ST412 reference genomes varied in size from 2.89 to 3.18 megabase pairs (Mb), many contained large chromosomal inversions, and they had variable plasmid contents (SI Appendix, Fig. S1 and Dataset S2). Analysis of antibiotic-resistance gene content in all study isolate genomes showed that all but one patient harbored VanA-encoding VREfm, and examination of closed reference genomes suggested that in all cases, VanA operons were carried on rep17 family plasmids of varying sizes (Dataset S2). Isolates from patient VRECG31 carried VanB integrated into the chromosome. These VanB-harboring isolates belonged to an ST203 lineage that was recovered from an immunocompromised pediatric patient in a prior study from the same institution (17). Finally, genes encoding resistance to aminoglycoside, macrolide, antifolate, and tetracycline antibiotics were also detected in isolates throughout the study but varied both between and within patients (Fig. 1C and Dataset S1).
Analysis of Variants that Arose during VREfm Intestinal Colonization and Bloodstream Infection.
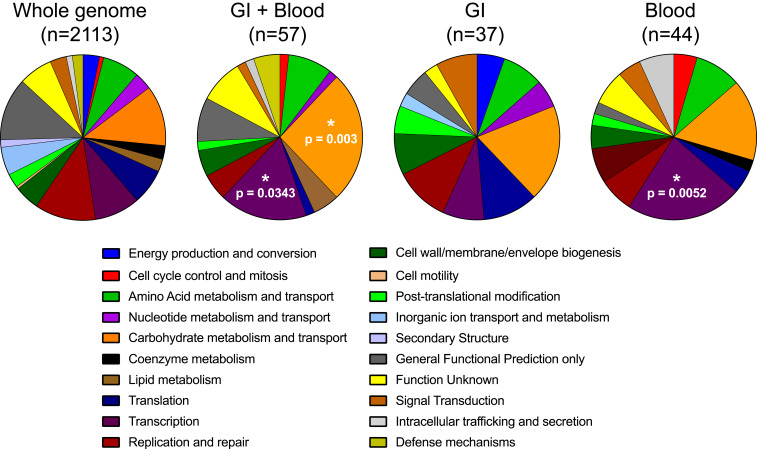
In order to understand the genetic factors that might have aided in persistent colonization and infection of patients, patient-specific reference genomes were employed to identify variants in the subsequent gastrointestinal and blood isolates of each patient. We focused on de novo chromosomal mutations in isolates from the same ST as the reference isolate derived from the same patient and identified 363 total variants with this approach (Dataset S3). Next, we investigated the functional classes of genes that were mutated in the subsequent gastrointestinal and blood isolates of each patient. We assigned Clusters of Orthologous Group (COG) functional categories to each of the genes in a representative VREfm genome and then compared the genome-wide COG category distribution with the categories of genes that were mutated only in gastrointestinal isolates, only in blood isolates, or in both gastrointestinal and blood isolates (Fig. 2). Compared with the genome-wide distribution, genes that were mutated in both gastrointestinal and blood isolates were enriched for “Carbohydrate metabolism and transport” (P = 0.003) and “Transcription” (P = 0.0343) categories. Mutated genes in these categories included those annotated as sugar (arabinose, lactose, sorbitol, and cellobiose) transporters, as well as transcriptional regulators of arabinose- and sorbitol-utilization pathways (Dataset S3). The “Transcription” category was also found to be enriched among the genes mutated only in blood isolates (P = 0.0052). Among these were genes annotated as PTS EIIA components, HTH-type transcriptional repressors or activators, rpoB/rpoC, and other transcriptional regulatory proteins (Dataset S3). These findings suggest that VREfm adapted during persistent intestinal colonization and bloodstream infection, in part, through transcriptional reprogramming, likely to optimize the use of distinct carbohydrates in the intestinal versus bloodstream environments.
Fig. 2.
COG functional category enrichment among mutated genes. COG categories were assigned to all genes in a representative genome in the dataset (Whole genome) or genes that were found to be mutated in both gastrointestinal and blood isolates (GI + Blood), only in GI isolates, or only in blood isolates. Distributions in each set were compared with the COG category distribution of the whole genome. P values were assessed with a Fisher’s exact test.
Differences in Carbohydrate Utilization between and within Patients.
Because we observed functional enrichment of variants in pathways related to carbohydrate utilization, we hypothesized that the VREfm isolates in our sample set might show growth differences in different carbon sources. We supplemented a complete and defined minimal medium with 10 different carbohydrates and screened all 110 isolates in our sample set for the ability to grow using each carbohydrate as a sole carbon source (SI Appendix, Fig. S2). Nearly all isolates grew readily in glucose, maltose, mannose, trehalose, and cellobiose. We observed modest differences in growth in ribose and fructose, while fewer isolates were able to grow in lactose, arabinose, or sorbitol. Growth in arabinose and sorbitol was variable between isolates from the same patient, suggesting that direct connections might exist between the genetic variants we observed and the ability of the isolates to utilize different carbon sources.
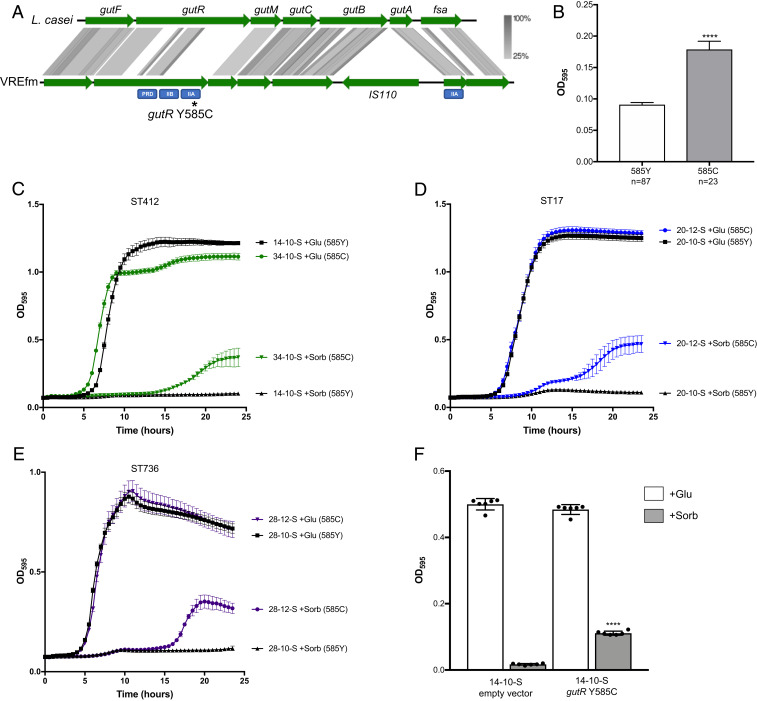
Bacterial growth in the presence of sorbitol was particularly variable, both between and within patients (SI Appendix, Fig. S2). Analysis of variants between closely related isolates that showed differential growth in sorbitol revealed a conserved mutation (Y585C) in a predicted licABCH/sorbitol operon regulator that was associated with growth in the presence of sorbitol (Fig. 3 A and B; P < 0.0001). The Y585C mutation arose in gastrointestinal isolates from two patients (VRECG20 and VRECG28) and was found in all gastrointestinal and blood isolates from an additional five patients (VRECG11, VRECG12, VRECG22, VRECG25, and VRECG34). The mutated regulator showed homology to the sorbitol utilization transcriptional activator gene gutR in Lactobacillus casei (Fig. 3A) (19), and we have adopted the same naming scheme here. When we compared the organization of the gut operon in VREfm with the gut operon in L. casei (19), we identified an IS110 family transposase insertion in between the gutB and gutA genes in all VREfm reference genomes (Fig. 3A). These genes are predicted to encode components of a sorbitol phosphotransferase system (PTS). The Y585C mutation in gutR resided in a predicted PTS IIA domain that appeared to be functionally similar to gutA. We performed kinetic growth assays on matched gutR wild-type (585Y) and mutant (585C) isolates belonging to three different STs in complete defined minimal medium supplemented with either glucose or sorbitol (Fig. 3 C–E). All isolate pairs grew similarly to one another in the presence of glucose, but wild-type isolates possessing the gutR 585Y wild-type allele were unable to grow in the presence of sorbitol. Isolates with the gutR mutant 585C allele, however, were able to grow in the presence of sorbitol (Fig. 3 C–E). To test this effect more directly, we introduced the gutR 585C allele into a wild-type isolate and observed that expression of the mutant allele was associated with growth in sorbitol (Fig. 3F). A search of E. faecium genome sequences deposited at the National Center for Biotechnology Information (NCBI) database revealed that ∼2.3% of deposited E. faecium genomes (28 of 1,227 deposited genomes containing the gene) also contained the gutR Y585C mutation. All 28 mutant isolates appeared to be from clinical sources (Dataset S4), suggesting that this mutation might be selected during the course of bacterial growth in hospitalized patients.
Fig. 3.
A Y585C mutation in the sorbitol operon regulator gutR is associated with VREfm growth in the presence of sorbitol. (A) Alignment of the gut sorbitol utilization operon in L. casei (Top) and VREfm (Bottom). Green arrows indicate coding sequences, and gray shading shows protein sequence homology ranging from 25 to 100%. Predicted functional domains in gutR and gutA are labeled below each gene in blue rectangles. The approximate position of the Y585C mutation in gutR is marked with an asterisk. (B) Association between the gutR Y585C mutation and growth in sorbitol among all 110 VREfm isolates in the dataset. Average bacterial growth ± SD as measured by OD595 after 24 h in a complete defined medium supplemented with sorbitol is shown. ****P < 0.0001 by two-tailed t test. (C–E) Kinetic growth curves of matched gutR wild-type and Y585C mutant isolate pairs in complete defined medium supplemented with glucose (+Glu) or sorbitol (+Sorb). Mean OD595 values ± SDs are plotted for at least three replicates of each isolate grown at 37 °C. The STs of each isolate pair are listed above the plots, and gutR alleles are listed to the right of the growth curves. (F) Expression of the gutR Y585C allele in a wild-type isolate confers growth in sorbitol. Average bacterial growth after 24 h at 37 °C is plotted ± SD for each isolate grown in a complete defined medium supplemented with glucose (+Glu, white bars) or sorbitol (+Sorb, black bars). ****P < 0.0001 by two-tailed t test comparing sorbitol growth versus empty vector.
Arabinose is a plant-derived sugar that has been explored as a food additive (20, 21). While most of the isolates in the sample set could grow on arabinose as a sole carbon source, all but two isolates from patient VRECG21 were unable to utilize this sugar (SI Appendix, Fig. S2). When we compared the closely related reference genome from patient VRECG19 (ST18, able to grow on arabinose) with the reference genome from patient VRECG21 (ST18, unable to grow), we identified a G305E mutation in the l-arabinose isomerase araA in patient VRECG21, which we hypothesized might prevent patient VRECG21 isolates from utilizing arabinose (SI Appendix, Fig. S3). A basic local alignment search tool search of the mutant allele in the NCBI nonredundant nucleotide collection revealed that the G305E mutation was unique to this study. Examination of genetic variants in 21-17-S and 21-18-S—the two isolates from patient VRECG21 that were able to grow on arabinose—revealed a nearly 100 kilobase pair chromosomal region of recombination, in which the ST18 nucleotide sequence appeared to have been replaced by sequence from an ST412 isolate (SI Appendix, Fig. S3). The recombined region included the l-arabinose isomerase gene and reverted the mutant 305 allele (305E) back to wild type (305G). Kinetic growth curves of patient VRECG21 isolates in the presence of glucose or arabinose confirmed the association between the l-arabinose isomerase G305E mutation and the inability to grow on arabinose (SI Appendix, Fig. S3).
Biofilm Formation Associated with Disruption of the Putative E. faecium Capsule Locus.
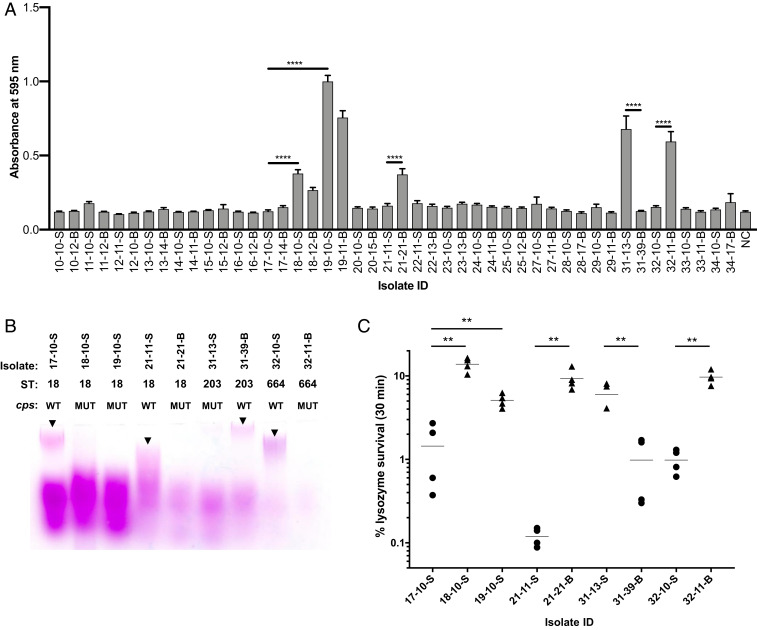
Because biofilm formation is considered to be an important contributor to bacterial colonization and persistence, we looked for differences in biofilm formation in a representative subset of our isolates. We quantified the ability of the earliest gastrointestinal and latest blood isolate from each patient to adhere to polystyrene plates using a standard in vitro crystal violet-based biofilm assay (22). Most of the isolates were unable to form robust biofilms; however, five patients yielded at least one isolate that could form either moderate or strong biofilms (Fig. 4A). Both gastrointestinal and blood isolates from patients VRECG18 and VRECG19 were able to form significantly more biofilm than the closely related isolates from patient VRECG17 (all ST18; P < 0.0001). The patient VRECG21 (ST18) and patient VRECG32 (ST664) blood isolates formed significantly more biofilm than their corresponding reference gastrointestinal isolates (P < 0.0001), and the patient VRECG31 (ST203) reference gastrointestinal isolate formed a much stronger biofilm than the last blood isolate from the same patient (P < 0.0001) (Fig. 4A). The same differences in biofilm formation were seen when biofilms were grown in fibronectin-coated wells (SI Appendix, Fig. S4), suggesting that isolates that form more biofilm might also be better able to attach to host surfaces.
Fig. 4.
Biofilm formation is associated with changes in surface polysaccharides and increased resistance to lysozyme. (A) Biofilm formation measured in the earliest gastrointestinal and latest bloodstream VREfm isolate from each patient in the dataset. Bars shown mean crystal violet absorbance values, and error bars show SEs of biological triplicate experiments, each with eight technical replicates. NC, negative control. ****P < 0.0001 by two-tailed t test. (B) Profiles of surface-associated polysaccharides extracted from biofilm-forming isolates harboring mutations in the cps capsule locus and matched wild-type controls. STs and cps genotypes are listed for each isolate (WT, wild type; MUT, mutant). Polysaccharides were separated on a polyacrylamide gel and were stained with Periodic acid-Schiff stain. Arrowheads point to high-molecular weight polysaccharide bands that are present in each WT isolate but absent in the corresponding MUT isolate. (C) Lysozyme survival among isolates shown in B. Percent survival following a 30-min exposure to 10 mg/mL or 50 mg/mL (isolates 32-10-S and 32-11-B) lysozyme is shown, with horizontal lines showing mean values of four replicate experiments. Data from isolates bearing wild-type cps alleles are plotted as circles, and data from isolates with cps mutations are plotted as triangles. **P < 0.005 by two-tailed t test. ID, identification number.
The biofilm-phenotype differences we observed prompted us to search for a genetic explanation. While no individual genes were consistently mutated or variably present in the genomes of isolates that could form biofilms, we did observe mutations in the putative E. faecium capsule locus (cps) (23), which appeared to be associated with increased biofilm formation in every case (Dataset S5). Every cps mutation was predicted to alter or eliminate the surface-associated capsular polysaccharide, suggesting that in contrast to our expectation, mutating the cps locus increased VREfm biofilm formation. To verify the nature of the mutations as loss-of-function, we extracted the cell surface-associated polysaccharides of cps mutants and closely related wild-type isolates using a protocol established for E. faecalis (24). Extracted polysaccharides were separated on a polyacrylamide gel and were stained with Periodic acid-Schiff stain (25). While the overall staining patterns we observed were different from those seen for E. faecalis, we nonetheless observed variations in polysaccharide profiles between wild-type and closely related cps mutant isolates (Fig. 4B). In particular, a high-molecular weight polysaccharide that we suspect corresponds to the E. faecium capsular polysaccharide was observed in the wild-type isolates but was greatly diminished or absent in cps mutants.
Lysozyme is an important antibacterial innate immune factor that hydrolyzes bacterial cell wall peptidoglycan (26). We tested whether biofilm-forming, cps mutant isolates showed altered susceptibility to lysozyme killing by measuring bacterial survival following exposure to a high concentration of the enzyme (Fig. 4C). Unexpectedly, all five cps mutant isolates showed significantly greater survival following lysozyme exposure compared with corresponding wild-type isolates (Fig. 4C, P < 0.005 in all cases). Lysozyme tolerance was also variable between patients; in particular, the ST664 isolates from patient VRECG32 were not killed upon exposure to 10 mg/mL lysozyme. For this reason, we used 50 mg/mL to test for killing differences in the isolates from this patient. We also tested the lysozyme susceptibility of isolates when they were grown in a biofilm and observed greater survival in the cps mutants compared with closely matched wild-type isolates (SI Appendix, Fig. S4). Overall, these data suggest that cps locus mutations might have arisen due to increased bacterial biofilm formation, increased protection against the antibacterial action of lysozyme (or a related antimicrobial factor), or both.
Antibiotic Treatment Selects for Resistance-Conferring Mutations in a Subset of Patients.
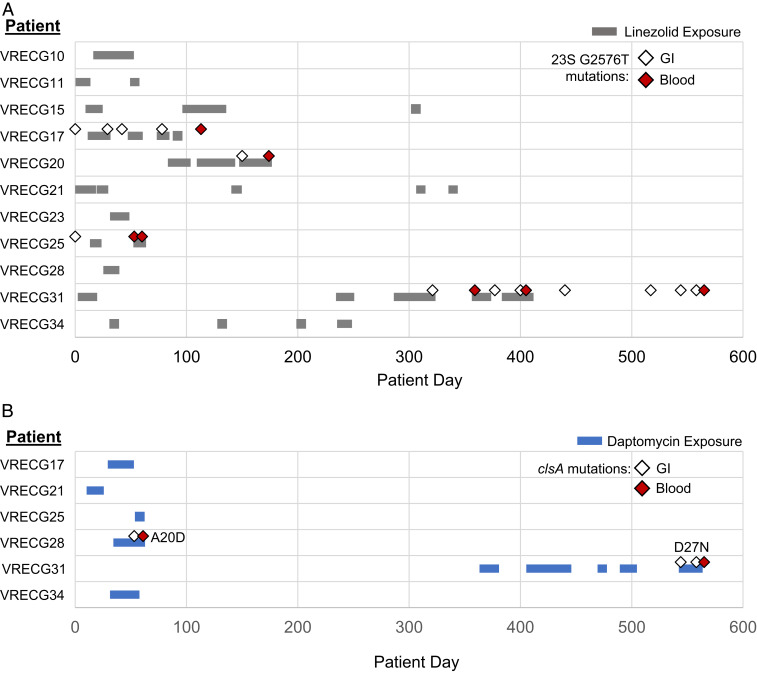
VREfm infections are often treated with second-line agents such as linezolid and daptomycin (27). Because some of the patients in this study received these agents, we looked for resistance-associated mutations in the VREfm isolate genomes from patients treated with these antibiotics. Linezolid resistance in enterococci has been predominantly associated with a G2576T mutation in the V domain of the 23S ribosomal RNA (28). Four of the 11 patients who were treated with linezolid during the study period were colonized or infected with isolates harboring the G2576T mutation in 1 or more copies of the 23S ribosomal RNA (rRNA) (Fig. 5A and Dataset S6). Two patients (VRECG17 and VRECG25) appear to have been initially colonized with isolates already carrying the G2576T mutation, while two other patients (VRECG20 and VRECG31) were initially colonized with wild-type isolates that developed de novo G2576T mutations following exposure to linezolid (Fig. 5A). The de novo occurrence of these mutations was confirmed by examining additional variants in the mutant isolates, which ruled out infection with a distinct strain (Dataset S3). In all four patients in whom resistance was detected, the G2576T mutation was first detected in gastrointestinal isolates, followed by detection in blood isolates. Additionally, the isolates bearing de novo G2576T mutations following linezolid exposure were isolated from the two patients with the greatest exposure to the drug (62 d in patient VRECG20 and 64 d in patient VRECG31). These findings suggest that linezolid pressure likely maintained the G2576T mutation in all isolates sampled from patients VRECG17 and VRECG25, and heavy drug exposure probably selected for the occurrence of the mutation in patients VRECG20 and VRECG31.
Fig. 5.
Antibiotic resistance-associated mutations arise in patients exposed to linezolid (A) and daptomycin (B). Colored bars show drug exposure in study patients who received linezolid (A, gray bars) or daptomycin (B, blue bars) therapy. Diamonds show the dates of isolation of VREfm isolates bearing resistance-associated mutations. White diamonds indicate gastrointestinal isolates; red diamonds indicate blood isolates. All resistant isolates in A encode a G2576T 23S rRNA mutation, while the resistant isolates in B encode two different mutations in the cardiolipin synthase clsA, which are listed in the figure.
Daptomycin resistance in enterococci often involves alterations in bacterial lipid metabolism (29, 30). The variant analysis we conducted showed that VREfm isolates from four patients in our study possessed mutations in various enzymes associated with lipid metabolism (Table 2 and Dataset S3). Blood isolates from three patients harbored mutations in the diacylglycerol kinases dagK (patient VRECG13) or dgkA (patients VRECG15 and VRECG28), compared with the reference gastrointestinal isolates from each patient. We also observed two different nonsynonymous single-nucleotide polymorphisms (SNPs) in the cardiolipin synthase clsA that arose de novo in isolates from patients VRECG28 and VRECG31 (Table 2 and Dataset S3). To investigate whether these variants were associated with altered daptomycin susceptibility, we determined the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of daptomycin against each mutant isolate compared with a corresponding wild-type isolate from the same patient. We observed that isolates 13-14-B (dagK P226A) and 15-12-B (dgkA G106E) were more susceptible to daptomycin than the reference, wild-type isolates from each patient (Table 2). In both cases, daptomycin MICs were twofold lower and MBCs were eightfold lower. Isolate 28-14-B possessed an insertion/deletion (indel) in dgkA, but this mutation did not appear to influence daptomycin susceptibility compared with the patient VRECG28 reference isolate (Table 2), perhaps because of the isolate’s genetic background (isolates from patient VRECG28 were all ST736, whereas isolates from patients VRECG13 and VRECG15 were ST412). In contrast, the gastrointestinal and blood isolates from patients VRECG28 and VRECG31 that carried SNPs in the cardiolipin synthase clsA all showed twofold increases in their daptomycin MICs and up to eightfold increases in their daptomycin MBCs compared with the patient-specific wild-type isolates (Table 2). Similar to the 23S rRNA G2576T mutation above, clsA mutations developed in the patients that had the greatest daptomycin exposure (21 d in patient VRECG28 and 60 d in patient VRECG31), arose de novo, and appeared first in gastrointestinal isolates and then in subsequent blood isolates (Fig. 5B).
Table 2.
Daptomycin MICs and MBCs for VREfm isolates with mutations in lipid metabolism genes
| Isolate | ST | Source | Genotype | Daptomycin MIC (µg/mL) | Daptomycin MBC (µg/mL) |
| 13-10-S | 412 | GI | Wild type | 2 | 16 |
| 13-14-B | 412 | Blood | dagK(P226A) | 1 | 2 |
| 15-10-S | 412 | GI | Wild type | 2 | 16 |
| 15-12-B | 412 | Blood | dgkA (G106E) | 1 | 2 |
| 28-10-S | 736 | GI | Wild type | 4 | 8 to 16 |
| 28-14-B | 736 | Blood | dgkA (indel) | 4 | 8 to 16 |
| 28-16-S | 736 | GI | clsA (A20D) | 8 | 32 |
| 28-17-B | 736 | Blood | clsA (A20D) | 8 | 32 to 64 |
| 31-13-S | 203 | GI | Wild type | 4 | 8 to 16 |
| 31-36-S | 203 | GI | clsA (D27N) | 8 | 32 |
| 31-38-S | 203 | GI | clsA (D27N) | 8 | 32 |
| 31-39-B | 203 | Blood | clsA (D27N) | 8 | 16 |
GI, gastrointestinal.
Discussion
VREfm are leading hospital-acquired pathogens that readily colonize and infect patients undergoing hematopoietic stem cell transplantation and chemotherapy (31). Here, we used comparative and functional genomics to identify and characterize signals of VREfm adaptation during gastrointestinal tract colonization and bloodstream infection in immunocompromised pediatric hosts. We identified trends in the adaptation of VREfm across different genetic backgrounds and connected these genetic changes to clinically relevant phenotypic differences. Rather than observing one type of variation that caused a large phenotypic effect, we instead found evidence for multiple different variants, each associated with phenotypic differences. In particular, we uncovered convergent evolution in carbohydrate utilization and surface-associated polysaccharide production, with mutations in the latter causing increased biofilm formation and resistance to lysozyme. We also documented the occurrence of known linezolid and daptomycin resistance-associated mutations and found that these mutations emerged in patients who received prolonged therapy with either agent.
The bacterial isolates we studied all belong to the hospital-adapted clade of E. faecium (18), and the overall population structure resembles other VREfm populations collected from hospitalized patients (27, 32). The large number of ST412 isolates collected here is somewhat notable, as ST412 is a relatively new VREfm lineage that has been isolated from hospitals in North America, South America, and the Caribbean islands (33, 34). We identified large-scale rearrangements in the genomes of ST412 isolates from five patients, some of which resulted in chromosomal inversions larger than 1 Mb in length (SI Appendix, Fig. S1). Such rearrangements have been previously identified in the genomes of other hospital-associated E. faecium (35, 36); whether and how these rearrangements might contribute to survival in the hospital environment remains to be determined.
Intestinal colonization of VREfm often precedes clinical infection, but the factors contributing to bacterial translocation from the intestine to other body sites are not well understood (31). It is clear, however, that the risk of enterococcal bloodstream infection is directly tied to the abundance of enterococci in the intestinal tract (37). The enrichment for mutations in carbohydrate metabolism enzymes and transcriptional regulators that we observed in both gastrointestinal and blood isolates could promote enterococcal intestinal domination and thereby contribute to bloodstream infection. Alternately, these mutations might enhance the ability of the bacteria to actively translocate into the bloodstream from the intestinal tract and/or to adapt to life in blood. Recent studies of VREfm adaptation to the human intestinal tract have also identified changes in carbohydrate metabolism that occur during prolonged colonization (38, 39), but clear signals of common adaptations across distinct genetic lineages have not been previously described. Future studies of larger datasets that incorporate metabolic modeling approaches could aid in the identification of causative mutations that affect carbohydrate utilization.
One common adaptation related to carbohydrate utilization that we observed in our dataset was the independent occurrence of a Y585C mutation in the sorbitol operon transcriptional regulator gutR that occurred across different STs, suggesting convergent evolution of this mutation in independent VREfm genetic lineages. The gut operon was previously found to be enriched among hospital-adapted E. faecium isolates compared with commensal isolates (18); however, in all 24 patient-specific reference genomes in our study, and in additional publicly available clinical E. faecium genomes that we analyzed, the operon is interrupted by the insertion of an IS110 family transposase in between gutB and gutA. The gutA gene encodes a PTS IIA component predicted to aid in transporting sorbitol into the bacterial cell (19). The orientation of the IS110 insertion in the genomes of hospital-adapted VREfm might interfere with gutA transcription or translation, which could explain why most isolates in our study were unable to grow with sorbitol as a sole carbon source. The Y585C mutation we identified in gutR occurred in a PTS IIA domain and was associated with bacterial growth on sorbitol. These data are consistent with a working model whereby the gutR Y585C mutation compensates for the lack of PTS IIA domain activity due to gutA inactivation by the IS110 transposase. We suspect that the Y585C mutation alters the structure and/or activity of gutR; however, this hypothesis remains to be formally tested. It does seem likely, however, that the VREfm we isolated would have been exposed to sorbitol in the intestinal tracts of the patients in this study; sorbitol has been increasingly used as a dietary sugar substitute and is also administered as an osmotic laxative to relieve chemotherapy-induced constipation (40). We observed de novo occurrence of the gutR Y585C mutation in two patients, while all of the isolates from five additional patients were found to also have the same mutation. We suspect that sorbitol exposure could have exerted selection in both cases, either by driving the gutR Y585C mutation from low to high frequency or by maintaining the mutation at a high frequency in a population where it was initially present. The fact that we found additional publicly available VREfm genomes harboring the same gutR Y585C mutation suggests that sorbitol ingested by or administered to hospitalized patients might be driving the occurrence of this mutation more generally; this will be a focus of our future work.
The ability to form biofilms is an important feature employed by a wide variety of bacterial pathogens. When we tested our VREfm isolates for their ability to form biofilms in vitro, we found that one or more isolates from five different patients formed significant biofilms. To identify a genetic explanation for this finding, we first looked for changes in genes that are known to contribute to biofilm formation in E. faecium, including the biofilm-associated pilus ebpABC, enterococcal surface protein esp, antibiotic stress and response regulator asrR, collagen adhesion protein acm, and the autolysin atlA (41). None of these genes was differentially present or mutated in the biofilm-forming isolates in our study. Comparisons using whole-genome alignment of closely related biofilm forming and nonforming isolates, however, revealed large deletions in the putative capsular polysaccharide (cps) locus, and further analysis uncovered distinct mutations in the cps locus in all biofilm-forming isolates. Very little is currently known about the capsular polysaccharide in E. faecium beyond the identification of a putative capsule biosynthetic locus, which is variable between STs (23). We hypothesize that the cps locus mutations we identified cause alterations in cell surface-associated polysaccharide production, which increase biofilm formation and might contribute to improved colonization or persistence of the VREfm isolates harboring these mutations. Biofilm formation increases the ability of bacteria to infect and cause disease in a multifactorial manner through better adhesion to host tissues and indwelling devices like catheters, greater resistance to phagocytosis, and increased tolerance to antibiotics (22, 42). The fact that increased biofilm formation was not consistently associated with only gastrointestinal or blood isolates further suggests that multiple pressures might select for this phenotype. While the patients from whom these bacteria were isolated were all immunocompromised and likely depleted for phagocytic cells, we think the ability to better adhere and/or tolerate antibiotic stress could have selected for cps locus mutations in these isolates. Since the composition of enterococcal cell surface-associated polysaccharides has been shown to impact bacterial sensitivity to lysozyme (43), we also tested whether our capsule locus mutants showed altered susceptibility to lysozyme. While susceptibility to lysozyme varied between isolates and across STs, all five cps mutant isolates we tested were more resistant to lysozyme killing compared with closely related wild-type isolates, suggesting that the ability to tolerate constitutive innate immune system defenses such as lysozyme could have also played a role in selecting for capsule-lacking, biofilm-forming bacteria (44).
VREfm are challenging to treat because there are few available antibiotics that can effectively target them, with linezolid and daptomycin being the preferred options (27). Prior studies have shown that both linezolid- and daptomycin-resistant enterococci can emerge during the course of therapy and can also be isolated in the absence of prior exposure to these antibiotics (28, 45, 46). Four patients in our study were colonized and infected with linezolid-resistant VREfm isolates, all of which harbored G2576T mutations in one or more copies of the 23S rRNA. No additional 23S rRNA mutations were detected, nor did we detect the presence of other linezolid resistance-associated genes such as cfr, cfr(B), optrA, and poxtA (28). In two patients, resistance mutations emerged during linezolid therapy, and in the other two cases, mutations were found to be preexisting before therapy was initiated. These latter two patients may have received antibiotic therapy earlier in their treatment, and perhaps their linezolid exposure during this study period helped maintain the presence of the G2576T mutation in their colonizing and infecting bacterial populations. We also observed multiple mutations that affected daptomycin susceptibility, including two independent mutations in the cardiolipin synthase clsA that were both associated with increased resistance to daptomycin killing. Both of the mutations we identified have been observed in other studies (47–49). We also identified several other mutations in genes presumably involved in phospholipid metabolism; mutations in the diacylglycerol kinases dagK and dgkA were identified only in blood isolates and were associated with increased sensitivity to daptomycin in a ST-dependent manner. A prior study found the emergence of a dagK mutation during in vitro selection for increased daptomycin resistance (50), but the selective pressure(s) that caused the emergence of these mutations in our study remains to be explored.
This study is unique in examining isolates from dozens of patients, and in using patient-specific reference genomes for a comprehensive comparative analysis, which allowed us to uncover signals of VREfm adaptation to the intestinal tract and bloodstream. The study also has several limitations. Isolates were collected as they were available, and patients were not systematically sampled at defined intervals. Additionally, a single isolate from each sampling time point was sequenced and analyzed as representative of the colonizing or infecting population at that time point, but other cooccurring clones, if they existed, were not sampled. Finally, the relevance of the mutations we identified should be explored further using isogenic wild-type and mutant strains to determine the molecular mechanisms connecting genotypes to phenotypes and to assess how these mutations contribute to colonization or infection in relevant animal models.
In summary, this study demonstrates how VREfm evolve during colonization of the antibiotic-perturbed intestinal tract as well as infection of the bloodstream in immunocompromised pediatric patients. Further investigation is needed to examine this evolution in other patient populations and to better understand how it contributes to enterococcal pathogenesis. Our hope is that these findings can ultimately be leveraged to develop more effective strategies for the treatment and control of enterococcal infections in vulnerable patient populations.
Materials and Methods
Collection of Isolates and Growth Conditions.
Bacterial isolates used in this study are summarized in SI Appendix. Patients are referenced throughout the text according to their “VRECG” patient number, and isolates are named as XX-YY-S/B, where XX is the patient identifier, YY is the isolate ID, S indicates gastrointestinal (GI) isolates, and B indicates blood isolates. Isolates were collected as part of routine clinical care and were identified retrospectively for inclusion in this study. This study was approved by the Institutional Review Board of the St. Jude Children’s Research Hospital (XPD17-062). Clinical data were collected retrospectively from patient medical records and from pharmacy and pathology databases. VREfm isolates were routinely cultured in either brain heart infusion (BHI) broth or Mueller Hilton broth (MHB) at 37 °C with agitation at 250 rpm.
Genome Sequencing and Analysis.
Genomic DNA from 110 VREfm gastrointestinal (n = 72) or blood (n = 38) isolates was extracted using a DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD), next-generation sequencing libraries were prepared with a Nextera XT kit (Illumina, San Diego, CA), and libraries were sequenced on an Illumina HiSeq using 2 × 100 bp paired-end reads. Genomic DNA from the earliest gastrointestinal isolate of each patient was also sequenced with long-read technology using a MinION device (Oxford Nanopore Technologies, Oxford, United Kingdom), and a hybrid reference assembly for each patient was generated with Unicycler (51). Reference assemblies were annotated with prokka (52) and were compared with one another with Roary (53). Phylogenetic analyses were conducted using Snippy v4.4.5 (https://github.com/tseemann/snippy), ClonalFrameML (54), and RAxML (55). STs, antimicrobial-resistance genes, and plasmid replicons were determined using online tools provided by the Center for Genomic Epidemiology (https://cge.cbs.dtu.dk/). Reference-assembly locus tags were customized using the orthogroups generated by Roary, so that locus tags were consistent across different reference genomes. Variants between isolates were then identified using BreSEq (56) and were manually filtered to remove variants on plasmids and those that were suspected to be due to recombination. Whole-chromosome alignments were generated in Geneious v11.1.5 (Biomatters, Auckland, New Zealand) using Mauve or with EasyFig (57). The G2576T mutant allele of the 23S rRNA associated with linezolid resistance was identified using LRE-Finder (28). Illumina read data for each isolate have been submitted to the Sequence Read Archive with accession numbers listed in SI Appendix. Hybrid assembled reference genomes have been submitted to NCBI under BioProject PRJNA575852.
COG Analysis of Mutated Genes.
The COG database (58) was used to group mutated genes together into categories with related function. The 18-10-S chromosome was used to determine the COG distribution of all genes in a representative VREfm isolate chromosome; chromosomal genes were annotated using the BASys bacterial annotation system tool (59), which assigned COG numbers and categories to 1,909/2,659 annotated genes. Some of the 1,909 genes with COG numbers were assigned to multiple categories; for distribution analysis, these were counted once in each category to yield a total of 2,113 categorized genes. The subset of genes that were mutated in both gastrointestinal and blood isolates, or only in gastrointestinal or blood isolates, were also analyzed for their COG category distributions. Genes that were found to have the same mutation in multiple isolates from the same patient were only counted once, while genes that were independently mutated in different patients were counted once per patient.
Carbohydrate-Utilization Assays.
A minimal and chemically defined medium (CDM) was used to assess differences in carbohydrate utilization among all 110 isolates in the sample set (60). Isolates were first arrayed into 96-well deep-well plates by inoculating each isolate into 1 mL of BHI broth and growing overnight at 37 °C. Then 500 µL of glycerol was added to each well, and plates were stored at −80 °C. Frozen cultures from deep-well plates were reinoculated into 96-well plates containing 200 µL of BHI per well with a 96-head pin-replicator and were incubated overnight at 37 °C. Bacteria were then pinned into 200 µL of CDM alone or supplemented with 0.5% arabinose, cellobiose, fructose, glucose, galactose, lactose, maltose, ribose, sorbitol, or trehalose. After 24 h of static incubation at 37 °C, the optical density at 595 nm (OD595) was measured in each well using a Synergy H1 microplate reader (Biotek, Winooski, VT). Growth of each isolate was quantified by first subtracting the OD595 of blank (CDM with no added sugar) wells and then calculating the percentage of maximal growth for each carbohydrate. All isolates were tested for growth on each carbohydrate in triplicate. Kinetic growth assays were conducted by diluting overnight cultures of selected isolates 1:100 into CDM plus 0.5% glucose, sorbitol, or arabinose, and growth was then monitored for 24 h at 37 °C by measuring OD595 every 30 min.
Cloning and Expression of gutR Y585C.
The gutR sequence was cloned into the pMSP3535 vector (61) using Gibson assembly. Briefly, the insert sequence was amplified with PCR from genomic DNA of the 34-10-S isolate using primers 5′-ATA AGG AGG CAC TCA AAA TGA CGC TAG TAA ACA GAT GG-3′ and 5′-CTC GCA TGC GAA TTC CTG CAG TCA TTC ATG AAG TCT CTC TC-3′, and the vector was amplified with PCR using primers 5′-TGC AGG AAT TCG CAT GCG-3′ and 5′-CAT TTT GAG TGC CTC CTT ATA ATT TAT TTTG-3′. The use of the above vector primers allowed for placement of the gutR coding sequence directly adjacent to the nisin-inducible promoter. Amplified insert and vector were purified using a PCR Purification Kit (Qiagen), and Gibson assembly was conducted using a HiFi DNA Assembly Cloning Kit (New England Biolabs). The Gibson product was transformed into NEB 5-alpha competent E. coli, and transformants were selected on Lysogeny broth agar containing 300 µg/mL erythromycin. To transform into E. faecium, the 14-10-S isolate was first grown in BHI media at 42 °C and was passaged by 1:1,000 dilution daily for 5 d. After passaging, single colonies were screened to identify a clone that was no longer resistant to erythromycin. The gutR Y585C-encoding vector or pMSP3535 empty vector was then transformed into the erythromycin-susceptible 14-10-S derivative E. faecium strain, and transformants were selected on BHI agar containing 15 µg/mL erythromycin. Transformants were confirmed with PCR using primers specific to the pMSP3535 plasmid backbone.
To test whether gutR Y585C expression conferred bacterial growth in the presence of sorbitol, transformants were grown without shaking in 200 µL of complete defined minimal media supplemented with 15 µg/mL erythromycin, 25 ng/mL nisin, and 0.5% glucose or sorbitol for 24 h at 37 °C. Wells were resuspended to disperse clumping that might have occurred during the incubation, and then the OD595 was measured in each well using a Synergy H1 microplate reader. Growth of each isolate was quantified by subtracting the OD595 of blank (media with no added sugar) wells. Six replicate wells were measured for each transformant.
Biofilm Assays.
Microtiter plate-based biofilm assays were performed as previously described (62). Briefly, overnight cultures of the earliest gastrointestinal and latest blood VREfm isolate from each patient were diluted 100-fold into BHI broth supplemented with 0.25% glucose; 200 µL of each culture was plated into 8 replicate wells of a 96-well untreated polystyrene microtiter plate, and plates were incubated for 24 h at 37 °C under static conditions. Plates were washed three times with 250 µL 1× phosphate-buffered saline (PBS) to remove unattached cells, and then wells were stained with 200 µL of 0.1% crystal violet (CV) in water. After incubation for 30 min at 4 °C, stained wells were washed twice with 250 µL of 1× PBS to remove excess stain. Plates were dried, and then 250 µL of 4:1 ethanol:acetone was added to each well to solubilize the CV-stained biofilm. After incubation for 45 min at room temperature, the absorbance of the dissolved CV was measured at 595 nm using a Synergy H1 microplate reader (Biotek). To assess biofilm formation on fibronectin, the 96-well polystyrene microtiter plates were first coated with 200 µL of 5 µg/mL bovine plasma fibronectin (Alfa Aesar, Haverhill, MA), and plates were incubated overnight at 37 °C. The next day, the fibronectin was removed, and the biofilm assay was performed as described above.
Capsular Polysaccharide Extraction and Analysis.
Capsular polysaccharides were isolated from VREfm isolates with cps locus mutations and closely related wild-type isolates following methods developed previously for E. faecalis (24), with minor modifications. Briefly, overnight VREfm cultures were diluted 100-fold in BHI broth supplemented with 0.25% glucose and were grown until they reached an OD595 of 0.6 to 0.8. Cells were pelleted, washed, and resuspended in lysis buffer—25% sucrose, 10 mM tris(hydroxymethyl)aminomethane (Tris), 0.05% sodium azide—and were then treated with lysozyme (1 mg/mL) and mutanolysin (10 U/mL) for 16 h with gentle rocking at 37 °C. After pelleting the cells by centrifugation, supernatants were collected and treated with RNase (100 µg/mL), DNase (10 U/mL), MgCl2 (2.5 mM), and CaCl2 (0.5 mM) for 4 h at 37 °C. This was followed by treatment with pronase (50 µg/mL) for 16 h at 37 °C. Afterward, 500 μL of chloroform was added and mixed to obtain phase separation, and the aqueous phase was preserved. Carbohydrates were precipitated by adding 900 μL of 100% ethanol and incubating overnight at −80 °C and were subsequently dried until excess ethanol evaporated. Pellets were resuspended in 80 µL of distilled water, and 12 μL aliquots were mixed with 6× carbohydrate loading dye (0.02% bromophenol blue, 2M sucrose) and 1× Tris-glycine running buffer, prior to being loaded onto NuPAGE 7% Tris-Acetate Protein Gels (Thermo Fisher Scientific, Waltham, MA). After electrophoresis at 120 V for 1 h, gels were stained with Pierce Glycoprotein Staining Kit (Thermo Fisher Scientific), according to the manufacturer’s protocol.
Lysozyme-Sensitivity Assays.
To test bacterial sensitivity to lysozyme, 500 μL of overnight cultures grown in BHI broth at 37 °C supplemented with 0.25% glucose were centrifuged, and pellets were washed once with Tris-ethylenediaminetetraacetic acid (TE) buffer. Bacteria were then resuspended in 500 μL of TE buffer containing 10 mg/mL or 50 mg/mL (for isolates 32-10-S and 32-11-B) lysozyme (Millipore Sigma, Burlington, MA). Colony forming units (CFUs) per milliliter were quantified before lysozyme addition and following a 30-min incubation at 37 °C with lysozyme, and the percentage survival was calculated by dividing CFUs per milliliter after 30 min by the initial CFUs per milliliter. To assess lysozyme sensitivity of isolates grown in biofilms, biofilms were grown in 96-well untreated polystyrene microtiter plates as described above. After incubation for 24 h at 37 °C, culture media were removed and wells were washed three times with 250 μL of 1× PBS to remove unattached cells. Then, 200 μL of either TE buffer or TE containing 10 mg/mL lysozyme was added to each well, and the plate was incubated at 37 °C for 30 min under static conditions. Wells were resuspended by pipetting to disrupt and homogenize cells, and the contents of each well were serially diluted 10-fold in 1× PBS. CFUs per milliliter were determined for each well using track dilution (63), and percentage survival was calculated by dividing CFUs per milliliter in the treated wells by the untreated wells.
Antimicrobial-Susceptibility Testing.
Daptomycin MIC assays were carried out by broth microdilution (64), in MHB containing 50 μg/mL CaCl2 (MHBc). Overnight cultures of VREfm were diluted to an OD595 of 0.1 and were further diluted 1:1,000 into fresh MHBc broth; 100 μL of this culture was then transferred to 96-well plates containing 100 μL of MHBc with serial twofold dilutions of daptomycin, ranging in concentration from 0 to 128 μg/mL. Plates were incubated for 24 h at 37 °C under static conditions, and growth in each well was analyzed by both visual inspection and by OD595 measurement. MBCs were determined by resuspending each well from the MIC assay plate and then spotting 5 μL of each well onto BHI agar plates and incubating overnight at 37 °C. MBCs were recorded as the lowest concentration of daptomycin with no growth from 5 μL following 24 h of drug exposure.
Statistical Analyses.
COG functional category enrichment was calculated with Fisher’s exact test. Differences in sorbitol growth, biofilm formation, and lysozyme sensitivity between isolates were assessed with a two-tailed t test.
Data Availability.
Genome sequence data have been deposited in the Sequence Read Archive or GenBank with accession information listed in SI Appendix. Bacterial strains used in the study are available upon request.
Supplementary Material
Acknowledgments
We thank all members of the J.W.R. and D.V.T laboratories for helpful discussions throughout the preparation of this manuscript. We also thank Matthew Culyba for assistance with molecular cloning. This work was supported by Public Health Service Grants AI083214 (to M.S.G.), AI124302 and AI110618 (to J.W.R.), and EY028222 (to D.V.T.) and by the University of Pittsburgh Department of Medicine. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Illumina read data for each isolate have been submitted to the Sequence Read Archive with accession numbers listed in SI Appendix, Table S1. Hybrid assembled reference genomes have been submitted to NCBI under BioProject PRJNA575852.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917130117/-/DCSupplemental.
References
- 1.Van Tyne D., Gilmore M. S., Friend turned foe: Evolution of enterococcal virulence and antibiotic resistance. Annu. Rev. Microbiol. 68, 337–356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias C. A., Murray B. E., The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pidot S. J., et al. , Increasing tolerance of hospital Enterococcus faecium to handwash alcohols. Sci. Transl. Med. 10, eaar6115 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Wendt C., Wiesenthal B., Dietz E., Rüden H., Survival of vancomycin-resistant and vancomycin-susceptible enterococci on dry surfaces. J. Clin. Microbiol. 36, 3734–3736 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebreton F., et al. , Tracing the enterococci from Paleozoic origins to the hospital. Cell 169, 849–861.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kos V. N., et al. , Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with Methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio 3, e00112-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilmore M. S., Lebreton F., van Schaik W., Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr. Opin. Microbiol. 16, 10–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher K., Phillips C., The ecology, epidemiology and virulence of Enterococcus. Microbiology 155, 1749–1757 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Tien B. Y. Q., et al. , Enterococcus faecalis promotes innate immune suppression and polymicrobial catheter-associated urinary tract infection. Infect. Immun. 85, e00378-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peleg A. Y., et al. , Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 7, e28316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faron M. L., Ledeboer N. A., Buchan B. W., Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant enterococcus in the health care setting. J. Clin. Microbiol. 54, 2436–2447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Driscoll T., Crank C. W., Vancomycin-resistant enterococcal infections: Epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 8, 217–230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ubeda C., et al. , Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120, 4332–4341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balian C., Garcia M., Ward J., A retrospective analysis of bloodstream infections in pediatric allogeneic stem cell transplant recipients: The role of central venous catheters and mucosal barrier injury. J. Pediatr. Oncol. Nurs. 35, 210–217 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Van Tyne D., Gilmore M. S., Raising the alarmone: Within-host evolution of antibiotic-tolerant Enterococcus faecium. MBio 8, e00066-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Tyne D., et al. , Impact of antibiotic treatment and host innate immune pressure on enterococcal adaptation in the human bloodstream. Sci. Transl. Med. 11, eaat8418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honsa E. S., et al. , RelA mutant Enterococcus faecium with multiantibiotic tolerance arising in an immunocompromised host. MBio 8, e02124-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebreton F., et al. , Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 4, e00534-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alcántara C., et al. , Regulation of Lactobacillus casei sorbitol utilization genes requires DNA-binding transcriptional activator GutR and the conserved protein GutM. Appl. Environ. Microbiol. 74, 5731–5740 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotake T., Yamanashi Y., Imaizumi C., Tsumuraya Y., Metabolism of L-arabinose in plants. J. Plant Res. 129, 781–792 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krog-Mikkelsen I., et al. , The effects of L-arabinose on intestinal sucrase activity: Dose-response studies in vitro and in humans. Am. J. Clin. Nutr. 94, 472–478 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Hashem Y. A., Amin H. M., Essam T. M., Yassin A. S., Aziz R. K., Biofilm formation in enterococci: Genotype-phenotype correlations and inhibition by vancomycin. Sci. Rep. 7, 5733 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer K. L., et al. , Comparative genomics of enterococci: Variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio 3, e00318-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thurlow L. R., Thomas V. C., Hancock L. E., Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 191, 6203–6210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiss-Liebisch S., et al. , Secondary cell wall polymers of Enterococcus faecalis are critical for resistance to complement activation via mannose-binding lectin. J. Biol. Chem. 287, 37769–37777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ragland S. A., Criss A. K., From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 13, e1006512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee T., Pang S., Abraham S., Coombs G. W., Antimicrobial-resistant CC17 Enterococcus faecium: The past, the present and the future. J. Glob. Antimicrob. Resist. 16, 36–47 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Hasman H., et al. , LRE-Finder, a Web tool for detection of the 23S rRNA mutations and the optrA, cfr, cfr(B) and poxtA genes encoding linezolid resistance in enterococci from whole-genome sequences. J. Antimicrob. Chemother. 74, 1473–1476 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Tran T. T., et al. , Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio 4, e00281-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller W. R., et al. , LiaR-independent pathways to daptomycin resistance in Enterococcus faecalis reveal a multilayer defense against cell envelope antibiotics. Mol. Microbiol. 111, 811–824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubin K. A., et al. , Diversification and evolution of vancomycin-resistant Enterococcus faecium during intestinal domination. Infect. Immun. 87, e00102-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willems R. J. L., et al. , Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11, 821–828 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panesso D., et al. , Molecular epidemiology of vancomycin-resistant Enterococcus faecium: A prospective, multicenter study in South American hospitals. J. Clin. Microbiol. 48, 1562–1569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akpaka P. E., et al. , Genetic characteristics and molecular epidemiology of vancomycin-resistant Enterococci isolates from Caribbean countries. PLoS One 12, e0185920 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam M. M. C., et al. , Comparative analysis of the first complete Enterococcus faecium genome. J. Bacteriol. 194, 2334–2341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arredondo-Alonso S., et al. , Genomes of a major nosocomial pathogen Enterococcus faecium are shaped by adaptive evolution of the chromosome and plasmidome. bioRxiv:10.1101/530725 (28 January 2019).
- 37.Taur Y., et al. , Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 55, 905–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayjanov J. R., et al. , Enterococcus faecium genome dynamics during long-term asymptomatic patient gut colonization. Microb. Genom. 5, e000277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Didelot X., Walker A. S., Peto T. E., Crook D. W., Wilson D. J., Within-host evolution of bacterial pathogens. Nat. Rev. Microbiol. 14, 150–162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McQuade R. M., Stojanovska V., Abalo R., Bornstein J. C., Nurgali K., Chemotherapy-induced constipation and diarrhea: Pathophysiology, current and emerging treatments. Front. Pharmacol. 7, 414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ch’ng J.-H., Chong K. K. L., Lam L. N., Wong J. J., Kline K. A., Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 17, 82–94 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Dunny G. M., Hancock L. E., Shankar N., “Enterococcal biofilm structure and role in colonization and disease” in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection Boston, (NCBI Bookshelf, 2014). [Google Scholar]
- 43.Smith R. E., et al. , Decoration of the enterococcal polysaccharide antigen EPA is essential for virulence, cell surface charge and interaction with effectors of the innate immune system. PLoS Pathog. 15, e1007730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hébert L., et al. , Enterococcus faecalis constitutes an unusual bacterial model in lysozyme resistance. Infect. Immun. 75, 5390–5398 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahim S., et al. , Linezolid-resistant, vancomycin-resistant Enterococcus faecium infection in patients without prior exposure to linezolid. Clin. Infect. Dis. 36, E146–E148 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Arias C. A., et al. , Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365, 892–900 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lellek H., et al. , Emergence of daptomycin non-susceptibility in colonizing vancomycin-resistant Enterococcus faecium isolates during daptomycin therapy. Int. J. Med. Microbiol. 305, 902–909 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Prater A. G., et al. , Environment shapes the accessible daptomycin resistance mechanisms in Enterococcus faecium. Antimicrob. Agents Chemother. 63, e00790-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G., et al. , Evolution and mutations predisposing to daptomycin resistance in vancomycin-resistant Enterococcus faecium ST736 strains. PLoS One 13, e0209785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mello S. S., et al. , A mutation in the glycosyltransferase gene lafB causes daptomycin hypersusceptibility in Enterococcus faecium. J. Antimicrob. Chemother. 75, 36–45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wick R. R., Judd L. M., Gorrie C. L., Holt K. E., Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Page A. J., et al. , Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Didelot X., Wilson D. J., ClonalFrameML: Efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 11, e1004041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deatherage D. E., Barrick J. E., Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan M. J., Petty N. K., Beatson S. A., Easyfig: A genome comparison visualizer. Bioinformatics 27, 1009–1010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V., The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Domselaar G. H., et al. , BASys: A web server for automated bacterial genome annotation. Nucleic Acids Res. 33, W455–W459 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang G., Mills D. A., Block D. E., Development of chemically defined media supporting high-cell-density growth of lactococci, enterococci, and streptococci. Appl. Environ. Microbiol. 75, 1080–1087 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bryan E. M., Bae T., Kleerebezem M., Dunny G. M., Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44, 183–190 (2000). [DOI] [PubMed] [Google Scholar]
- 62.O’Toole G. A., Microtiter dish biofilm formation assay. J. Vis. Exp., 2437 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jett B. D., Hatter K. L., Huycke M. M., Gilmore M. S., Simplified agar plate method for quantifying viable bacteria. Biotechniques 23, 648–650 (1997). [DOI] [PubMed] [Google Scholar]
- 64.Wiegand I., Hilpert K., Hancock R. E. W., Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequence data have been deposited in the Sequence Read Archive or GenBank with accession information listed in SI Appendix. Bacterial strains used in the study are available upon request.