SUMMARY
The underlying mechanisms by which prior immunity to dengue virus (DENV) affords cross-protection against the related flavivirus Zika virus (ZIKV) are poorly understood. Here, we examine the ability of DENV/ZIKV-cross-reactive CD4+ T cells to protect against versus exacerbate ZIKV infection by using a histocompatibility leukocyte antigen (HLA)-DRB1*0101 transgenic, interferon α/β receptor-deficient mouse model that supports robust DENV and ZIKV replication. By mapping the HLA-DRB1*0101-restricted T cell response, we identify DENV/ZIKV-cross-reactive CD4+ T cell epitopes that stimulate interferon gamma (IFNγ) and/or tumor necrosis factor (TNF) production. Vaccination of naive HLA-DRB1*0101 transgenic mice with these peptides induces a CD4+ T cell response sufficient to reduce tissue viral burden following ZIKV infection. Notably, this protective response requires IFNγ and/or TNF secretion but not anti-ZIKV immunoglobulin G (IgG) production. Thus, DENV/ZIKV-cross-reactive CD4+ T cells producing canonical Th1 cytokines can suppress ZIKV replication in an antibody-independent manner. These results may have important implications for increasing the efficacy and safety of DENV/ZIKV vaccines and for developing pan-flavivirus vaccines.
Graphical Abstract
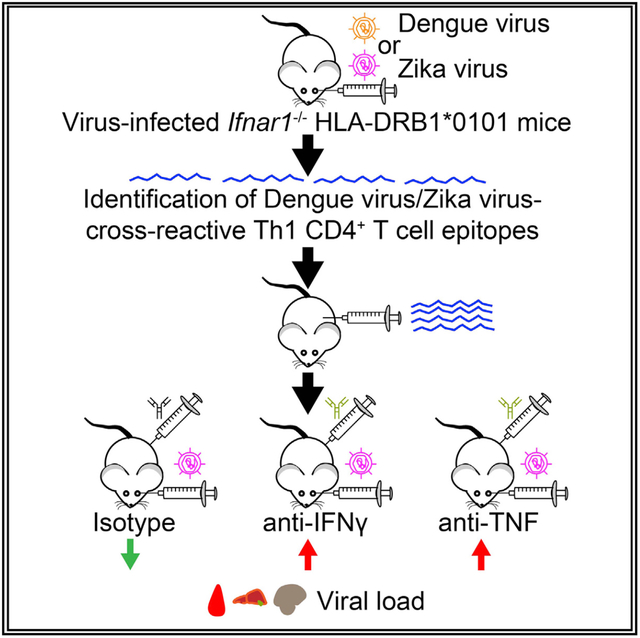
In Brief
Wen et al. show that dengue and Zika virus cross-reactive CD4+ T cells reduce Zika viral burden in interferon α/β receptor-deficient HLA-DRB1*0101 transgenic mice in an IFNγ- or TNF-dependent, antibody-independent manner.
INTRODUCTION
Zika virus (ZIKV) is a positive-sense, single-stranded, enveloped RNA virus of the Flavivirus genus, which includes the closely related dengue virus (DENV), Japanese encephalitis virus (JEV), West Nile virus (WNV), and yellow fever virus (YFV) (Choumet and Despres, 2015; Lazear and Diamond, 2016; Ngono and Shresta, 2018). ZIKV and DENV share similar amino acid sequences, with 43% overall homology and up to 68% identity for the non-structural proteins (Lazear and Diamond, 2016; Wen and Shresta, 2019). Additionally, ZIKV and DENV use the same vectors for transmission and have overlapping geographical ranges. Anti-DENV and anti-ZIKV immune responses have been shown to cross-react at the antibody (Ab) level (Castanha et al., 2017; Charles and Christofferson, 2016; Dejnirattisai et al., 2016; Kawiecki and Christofferson, 2016; Paul et al., 2016; Priyamvada et al., 2016; Swanstrom et al., 2016) and CD4+ and CD8+ T cell levels (Grifoni et al., 2017; Lim et al., 2018; Paquin-Proulx et al., 2017). These cross-reactive immune responses may contribute to both protection and pathogenesis during ZIKV and DENV infections (Ngono and Shresta, 2018). In particular, cross-reactive Abs produced during a primary infection with one DENV serotype can exacerbate, rather than protect, against secondary infection with a different DENV serotype (Katzelnick et al., 2017; Salje et al., 2018). This occurs through a process known as Ab-dependent enhancement (ADE) of infection and can lead to a potentially life-threatening infection with hemorrhagic fever and shock (known as severe dengue) (Halstead, 2007). Accordingly, studies using mouse models have shown that DENV/ZIKV-cross-reactive Abs play a dual role in mediating protection and pathogenesis during infection with DENV or ZIKV (Bardina et al., 2017; Fernandez et al., 2017; Fowler et al., 2018; Kam et al., 2017; Slon Campos et al., 2017). Although there is limited epidemiologic evidence demonstrating ZIKV-ADE in humans (Robbiani et al., 2019), three recent epidemiologic studies have demonstrated that prior DENV exposure provides cross-protection against ZIKV infection in humans (Gordon et al., 2019; Pedroso et al., 2019; Rodriguez-Barraquer et al., 2019). At present, the mechanisms responsible for the cross-protection in humans is poorly understood. Because natural infection and/or vaccination against these viruses could have either beneficial or disastrous consequences, it is crucial that we deepen our understanding of the mechanisms by which DENV/ZIKV-cross-reactive immunity can mediate these distinct outcomes.
A variety of mouse models have been used to investigate anti-DENV and anti-ZIKV T cell responses, including wild-type (WT) mice, mice deficient in the type I interferon (IFN) receptor on macrophages (LysMCre+Ifnar1−/−mice) or all cells (Ifnar1−/−mice), WT mice with Ab-mediated blockade of the type I IFN receptor, and—of particular importance for the human response—Ifnar1−/−histocompatibility leukocyte antigen (HLA) transgenic mice (Wen and Shresta, 2017; Wen and Shresta, 2019). Studies using these mouse models have demonstrated a critical role for CD8+ T cells in protecting against primary ZIKV infection (Elong Ngono et al., 2017; Huang et al., 2017) and for DENV-elicited CD8+ T cells in mediating cross-protection against subsequent ZIKV infection (Huang et al., 2017; Regla-Nava et al., 2018; Wen et al., 2017a, 2017b). Additionally, studies with Ifnar1−/−HLA-B*0702 and HLA-A*0101 transgenic mice, which express two of the most common HLA class I alleles in countries where ZIKV is endemic, showed that ZIKV-specific and DENV/ZIKV-cross-reactive CD8+ T cells from mice vaccinated with peptides can control the viral burden during subsequent ZIKV infection (Wen et al., 2017b). These findings suggest an important role for DENV-elicited CD8+ T cells in cross-protecting against ZIKV infection in humans.
More recent studies using mouse models have also identified a protective role for ZIKV-specific CD4+ T cells. In particular, ZIKV-specific CD4+ T cells are required for generation of a protective anti-ZIKV Ab response (Elong Ngono et al., 2019; Hassert et al., 2018; Lucas et al., 2018). A comparison of unmanipulated WT mice and anti-Ifnar1 Ab-treated WT mice demonstrated an important role for CD4+ T cell responses in controlling ZIKV infection in both mouse models (Liang et al., 2019). DENV/ ZIKV-cross-reactive CD4+ T cells have been described in HLA transgenic mice and in humans (Grifoni et al., 2017; Lim et al., 2018; Paquin-Proulx et al., 2017; Reynolds et al., 2018). Thus, CD4+ T cells primed by vaccination of HLA class II transgenic mice with purified ZIKV proteins show reactivity in vitro not only to the priming ZIKV peptides but also to variants of the same peptides present in the four DENV serotypes (DENV1–4), WNV, and YFV (Reynolds et al., 2018). Conversely, CD4+ T cells isolated from DENV-vaccinated individuals display cross-reactivity to ZIKV peptides in vitro (Grifoni et al., 2017; Lim et al., 2018; Paquin-Proulx et al., 2017). However, at present, the protective versus potentially pathogenic roles of DENV/ZIKV-cross-reactive CD4+ T cells are unknown.
In the present study, we investigated whether CD4+ T cells with cross-reactivity to HLA-class-II-restricted DENV2/ZIKV epitopes are protective versus pathogenic during ZIKV infection in the Ifnar1−/−HLA-DRB1*0101 transgenic mouse model. We identified a panel of ZIKV peptides predicted to bind to HLA-DRB1*0101 and then characterized the in vitro response to the peptides of CD4+ T cells from ZIKV- or DENV-infected HLA-DRB1*0101 mice. Of the 30 ZIKV peptides screened, 7 induced interferon gamma (IFNγ) and/or TNF production by ZIKV-primed CD4+ T cells and 4 induced IFNγ and/or TNF production by cross-reactive DENV2-primed CD4+ T cells. Vaccination of HLA-DRB1*0101 mice with DENV2/ZIKV-cross-reactive CD4+ T cell epitopes induced a protective response upon ZIKV infection, and viral control required IFNγ and/or TNF secretion but not anti-ZIKV immunoglobulin G (IgG). These findings suggest a mechanism by which prior DENV exposure cross-protects against ZIKV infection in humans and may, therefore, have important implications for vaccine development.
RESULTS
Identification of ZIKV-Derived HLA-DRB1*0101-Restricted CD4+ T Cell Epitopes
WT mice are highly resistant to DENV and ZIKV infection due to the inability of the viruses to inhibit the host IFN system (Aguirre et al., 2012; Ding et al., 2018; Grant et al., 2016; Yu et al., 2012). Because the antigenic load dictates the magnitude and quality of antiviral T cell responses (Earl et al., 2004; Panagioti et al., 2018; Salek-Ardakani et al., 2011), Ifnar1−/−mice have been widely used to investigate T cell responses to DENV and ZIKV (Ngono and Shresta, 2018; Tang et al., 2015). We previously showed that vaccination of Ifnar1−/−mice with DENV2 peptides elicited CD4+-T-cell-mediated protective immunity against reinfection with the DENV2 homotype (Yauch et al., 2010). HLA-DRB1*0101 is a commonly expressed HLA molecule in humans (Marsh et al., 1999), and human studies have confirmed the relevance of DENV-reactive CD4+ T cells identified using the HLA-DRB1*0101 transgenic mouse model (Ngono and Shresta, 2018; Weiskopf et al., 2011, 2014). Therefore, for the present study, we used the Ifnar1−/−HLA-DRB1*0101 mouse model. We selected a total of 30 ZIKV peptides from the top 2% of epitopes predicted to bind HLA-DRB1*0101 by the predictive database Immune Epitope Database and Analysis Resource (IEDB-AR) (Table 1). The 30 peptides were distributed in 9 ZIKV proteins: 3 in C, 2 in M, 4 in E, 1 in NS1, 6 in NS2A, 3 in NS3, 2 in NS4A, 7 in NS4B, and 2 in NS5. To test the reactivity of ZIKV-primed CD4+ T cells, Ifnar1−/−HLA-DRB1*0101 mice were infected with ZIKV SD001 strain for 7 days, and splenocytes were isolated, stimulated with the candidate epitopes for 6 h, and analyzed for production of canonical Th1 (IFNγ, TNF, and interleukin-2 [IL-2]), Th17 (IL-17), and Th2 (IL-4, IL-5) cytokines by intracellular staining and flow cytometry (ICS assay). This analysis identified eight peptides (C27–41, C53–67, C81–95, E134–148, E450–464, NS2A66–80, NS3601-NS4A12, and NS5222–236) as Th1 epitopes (Figure 1), of which NS5222–236 stimulated CD4+ T cells to produce IFNγ, TNF, and IL-2; and E134–148 stimulated CD4+ T cells to produce only IL-2; and the other six epitopes stimulated CD4+ T cells to produce IFNγ and/or TNF (Figure 1; Table 1). Notably, none of the 30 peptides tested induced CD4+ T cells to produce IL-17, IL-4, or IL-5 (Figure S1). These data indicate that the primary response to ZIKV infection in Ifnar1−/−HLA-DRB1*0101 transgenic mice is dominated by Th1 cytokine-secreting CD4+ T cells responding to eight peptides in ZIKV structural and non-structural proteins.
Table 1.
Zika-Virus-Derived Potential HLA-DRB1*0101-Restricted Epitopes
| IEDB Prediction | % CD4+ | CD44+ | T Cells (ZIKV) | % CD4+ | CD44+ | T Cells (DENV2) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptidesa | Sequences | Percentile_Rank | INFγ+ | TNF+ | INFγ+TNF+ | IL-2+ | INFγ+ | TNF+ | INFγ+TNF+ | IL-2+ | |
| C27–41* | FGGLKRLPAGLLLGH | 0.25 | 0.19 | 0.12 | 0.10 | 0.08 | 0.11 | 0.11 | 0.09 | 0.06 | |
| C53–67 | FLRFTAIKPSLGLIN | 0.19 | 0.27 | 0.10 | 0.03 | 0.02 | 0.15 | 0.09 | 0.07 | 0.04 | |
| C81–95 | IKKFKKDLAAMLRII | 3.02 | 0.19 | 0.25 | 0.06 | 0.06 | 0.15 | 0.16 | 0.10 | 0.07 | |
| M39–53 | NPGFALAAAAIAWLL | 0.13 | 0.09 | 0.13 | 0.02 | 0.03 | 0.11 | 0.11 | 0.09 | 0.05 | |
| M63-E2 | YLVMILLIAPAYSIR | 0.96 | 0.07 | 0.11 | 0.02 | 0.01 | 0.08 | 0.14 | 0.07 | 0.03 | |
| E134-148*† | NLEYRIMLSVHGSQH | 2.27 | 0.06 | 0.13 | 0.02 | 0.19 | 0.11 | 0.19 | 0.07 | 0.46 | |
| E309–323 | TAAFTFTKIPAETLH | 1.58 | 0.10 | 0.08 | 0.02 | 0.02 | 0.09 | 0.09 | 0.08 | 0.06 | |
| E460-464 | GAAFKSLFGGMSWFS | 2.51 | 0.18 | 0.09 | 0.08 | 0.03 | 0.18 | 0.20 | 0.08 | 0.07 | |
| E492-NS11 | GGVLIFLSTAVSADV | 1.99 | 0.08 | 0.11 | 0.01 | 0.02 | 0.15 | 0.14 | 0.10 | 0.07 | |
| NS1206–220 | NDTWRLKRAHLIEMK | 2.73 | 0.06 | 0.09 | 0.02 | 0.02 | 0.14 | 0.14 | 0.08 | 0.05 | |
| NS2A18-32 | TTKIIISTSMAVLVA | 3.72 | 0.07 | 0.10 | 0.03 | 0.02 | 0.10 | 0.13 | 0.09 | 0.07 | |
| NS2A66-80*† | LALIAAFKVRPALLV | 1.81 | 0.17 | 0.11 | 0.06 | 0.03 | 0.39 | 0.20 | 0.34 | 0.20 | |
| NS2A122-136 | ALAWLAIRAMWPRT | 1.24 | 0.08 | 0.09 | 0.02 | 0.02 | 0.13 | 0.14 | 0.11 | 0.08 | |
| NS2A138–152 | NITLAILAALTPLAR | 2.51 | 0.07 | 0.09 | 0.02 | 0.02 | 0.11 | 0.12 | 0.07 | 0.06 | |
| NS2A167-181 | GGFMLLSLKGKGSVK | 2.27 | 0.07 | 0.07 | 0.03 | 0.03 | 0.10 | 0.12 | 0.09 | 0.07 | |
| NS2A184-198 | LPFVMALGLTAVRLV | 0.28 | 0.06 | 0.21 | 0.02 | 0.05 | 0.14 | 0.40 | 0.09 | 0.06 | |
| NS3126–140 | CGRVIGLYGNGWIK | 4.77 | 0.06 | 0.11 | 0.02 | 0.03 | 0.18 | 0.13 | 0.18 | 0.09 | |
| NS3199–213 | RLRTVILAPTRWAA | 1.81 | 0.11 | 0.11 | 0.04 | 0.03 | 0.14 | 0.12 | 0.11 | 0.06 | |
| NS36O1-NS4A12* | GAAFGVMEALGTLPG | 3.95 | 0.20 | 0.14 | 0.10 | 0.04 | 0.10 | 0.08 | 0.06 | 0.03 | |
| NS4A64-78 | GIFFVLMRNKGIGKM | 0.42 | 0.05 | 0.08 | 0.03 | 0.02 | 0.16 | 0.18 | 0.15 | 0.10 | |
| NS4A13O-144 | QMAIIIMVAVGLLGL | 0.96 | 0.10 | 0.12 | 0.02 | 0.03 | 0.17 | 0.17 | 0.10 | 0.06 | |
| NS4B40-54† | WAIYAALTTFITPAV | 3.95 | 0.09 | 0.20 | 0.05 | 0.03 | 0.22 | 0.37 | 0.10 | 0.06 | |
| NS4B64-78 | NYSLMAMATQAGVLF | 1.71 | 0.06 | 0.07 | 0.01 | 0.02 | 0.14 | 0.12 | 0.08 | 0.03 | |
| NS4B98-112 | IGCYSQLTPLTLIVA | 0.25 | 0.06 | 0.13 | 0.01 | 0.02 | 0.14 | 0.15 | 0.10 | 0.04 | |
| NS4B124-138 | IPGLQAAAARAAQKR | 1.06 | 0.05 | 0.08 | 0.02 | 0.02 | 0.11 | 0.12 | 0.04 | 0.04 | |
| NS4B170-184 | MGQVLLIAVAVSSAI | 2.05 | 0.06 | 0.11 | 0.04 | 0.02 | 0.14 | 0.15 | 0.07 | 0.07 | |
| NS4B227-241 | FRGSYLAGASLIYTV | 4.77 | 0.08 | 0.14 | 0.06 | 0.02 | 0.16 | 0.13 | 0.07 | 0.05 | |
| NS4B337-351 | GWSYYAATIRKVQEV | 1.53 | 0.07 | 0.12 | 0.03 | 0.02 | 0.12 | 0.12 | 0.11 | 0.06 | |
| NS5222–236*† | RAIWYMWLGARFLEF | 0.88 | 0.61 | 0.57 | 0.33 | 0.21 | 0.82 | 0.80 | 1.45 | 0.64 | |
| NS5519–533 | HRRDLRLMANAICSS | 1.9 | 0.10 | 0.11 | 0.04 | 0.01 | 0.19 | 0.15 | 0.13 | 0.06 | |
| Negative | no peptide | 0.06 | 0.14 | 0.03 | 0.03 | 0.11 | 0.12 | 0.09 | 0.07 | ||
| Positive | PMA/ionomycin | 35.69 | 8.14 | 18.28 | 9.40 | 32.13 | 6.21 | 40.02 | 19.54 | ||
The position of the peptides was determined according to the amino acid sequences of ZIKV strain FSS13025. The peptides denoted with an asterisk (*) were identified as ZIKV epitopes using the ICS assay. The peptides denoted with a dagger (†) were identified as DENV2/ZIKV-cross-reactive epitopes. The peptides underlined were used to immunize mice.
Figure 1. Cytokine Secretion Pattern of ZIKV-Peptide-Specific HLA-DRB1*0101-Restricted CD4+ T Cells.

Ifnar1−/−HLA-DRB1*0101 mice were infected retro-orbitally with 1 × 102 focus-forming unit (FFU) of ZIKV strain SD001 for 7 days. Splenocytes were isolated and stimulated in vitro for 6 h with the indicated ZIKV-derived peptides that were predicted to bind HLA-DRB1*0101, and the frequencies of cytokine-producing cells were detected using the ICS assay.
(A) Representative flow cytometry contour plots.
(B and C) Quantification of CD4+ T cells producing IFNγ, TNF, or IFNγ plus TNF (B) or IL-2 (C). Data represent the mean ± SEM of four independent experiments (n = 3–5 mice/experiment; total 16 mice). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-tailed Mann-Whitney test with a Benjamini-Hochberg (B-H) adjusted p value (FDR) < 10%. Negative and positive refer to cells incubated in medium alone or with phorbol-12-myristate-13-acetate (PMA) and ionomycin, respectively.
Identification of ZIKV-Derived CD4+ T Cell Epitopes That Stimulate DENV-Primed CD4+ T Cells
To determine the suitability of Ifnar1−/−HLA-DRB1*0101 mice for investigating the DENV/ZIKV-cross-reactive CD4+ T cell response, we assessed whether DENV-primed CD4+ T cells respond to cross-reactive ZIKV peptides in vitro. Splenocytes were isolated from Ifnar1−/−HLA-DRB1*0101 mice on day 7 after infection with the DENV2 S221 strain, stimulated in vitro for 6 h with the 30 ZIKV-derived candidate epitopes, and then analyzed for cytokine production by the ICS assay. Five ZIKV-derived CD4+ T cell epitopes elicited cross-reactive responses by DENV2-primed CD4+ T cells: NS2A184–198 and NS4B40–54 induced only TNF-producing cells; NS2A66–80 and NS5222–236 induced cells producing IFNγ plus TNF; and E134–148 and NS5222–236 induced cells producing IL-2, although their Benjamini-Hochberg adjusted p values (0.41 and 0.14, respectively) were higher than our target false discovery rate (FDR) of 10% (Figure 2; Table 1). Thus, in this mouse model, the non-structural proteins NS2A, NS4B, and NS5 in ZIKV contain epitopes that stimulate cross-reactive DENV-primed CD4+ T cells to produce the canonical Th1 cytokines IFNγ and TNF.
Figure 2. Cross-Reactivity of DENV2-Primed CD4+ T Cells to HLA-DRB1*0101-Binding ZIKV Peptides.

Ifnar1−/−HLA-DRB1*0101 mice were infected intraperitoneally with 2 × 103 FFU of DENV2 strain S221 for 7 days. Splenocytes were stimulated in vitro for 6 h with the indicated ZIKV-derived peptides that were predicted to bind HLA-DRB1*0101, and the frequencies of cytokine-producing cells were detected using the ICS assay.
(A) Representative flow cytometry contour plots.
(B and C) Quantification of CD4+ T cells producing IFNγ, TNF, or IFNγ plus TNF (B) or IL-2 (C). Data represent the mean ± SEM of three independent experiments (n = 3–4 mice/experiment; total 10 mice). **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-tailed Mann-Whitney test with a B-H adjusted p value (FDR) < 10%. Negative and positive refer to cells incubated in medium alone or with PMA and ionomycin, respectively.
Influence of DENV2 Immunity on the CD4+ T Cell Response to ZIKV Infection
Based on our previous finding that prior DENV immunity affords cross-protection against ZIKV (Regla-Nava et al., 2018; Wen et al., 2017a), we assessed the effect of pre-existing DENV2 immunity on the outcome of ZIKV infection in this mouse model. Naive Ifnar1−/−HLA-DRB1*0101 mice were inoculated with DENV2, followed by infection of the DENV2-immune or naive Ifnar1−/−HLA-DRB1*0101 mice with ZIKV 4 weeks later. ZIKV viral loads in the serum, liver, and brain were examined at 3 and 7 days post-infection (dpi). As expected, these experiments revealed near-complete suppression of ZIKV replication in DENV2-immune mice compared with naive mice (Figures 3A–3C and 3F). To investigate the influence of DENV2 immunity on the CD4+ T cell response, Ifnar1−/−HLA-DRB1*0101 mice or naive mice were primed with DENV2 and infected with ZIKV 4 weeks later, and splenocytes were isolated 3 or 7 dpi. Splenic CD4+ T cells were then evaluated for expression of IFNγ, TNF, or IFNγ plus TNF after in vitro stimulation with DENV2/ZIKV-cross-reactive peptides (NS2A66–80, NS4B40-54, and NS5222–236) and ZIKV-specific peptides (C27–41, C53–67, C81–95, E450–464, and NS3601-NS4A12). CD4+ T cells from naive mice harvested at 3 dpi showed no response to the peptides (Figure 3G), whereas cells harvested from DENV2-primed mice displayed a strong response to the DENV2/ZIKV-cross-reactive peptide NS5222–236, with expansion of CD4+ T cells producing IFNγ, TNF, and IFNγ plus TNF (Figure 3H). By 7 dpi, CD4+ T cells from both naive mice and DENV-immune mice showed responses to ZIKV-specific and DENV2/ZIKV-cross-reactive peptides (Figures 3I and 3J). These results show that the memory CD4+ T cell response elicited by prior DENV infection recognizes ZIKV in Ifnar1−/−HLA-DRB1*0101 mice.
Figure 3. Reduction in Viral Load in DENV-Immune Mice after ZIKV Infection.
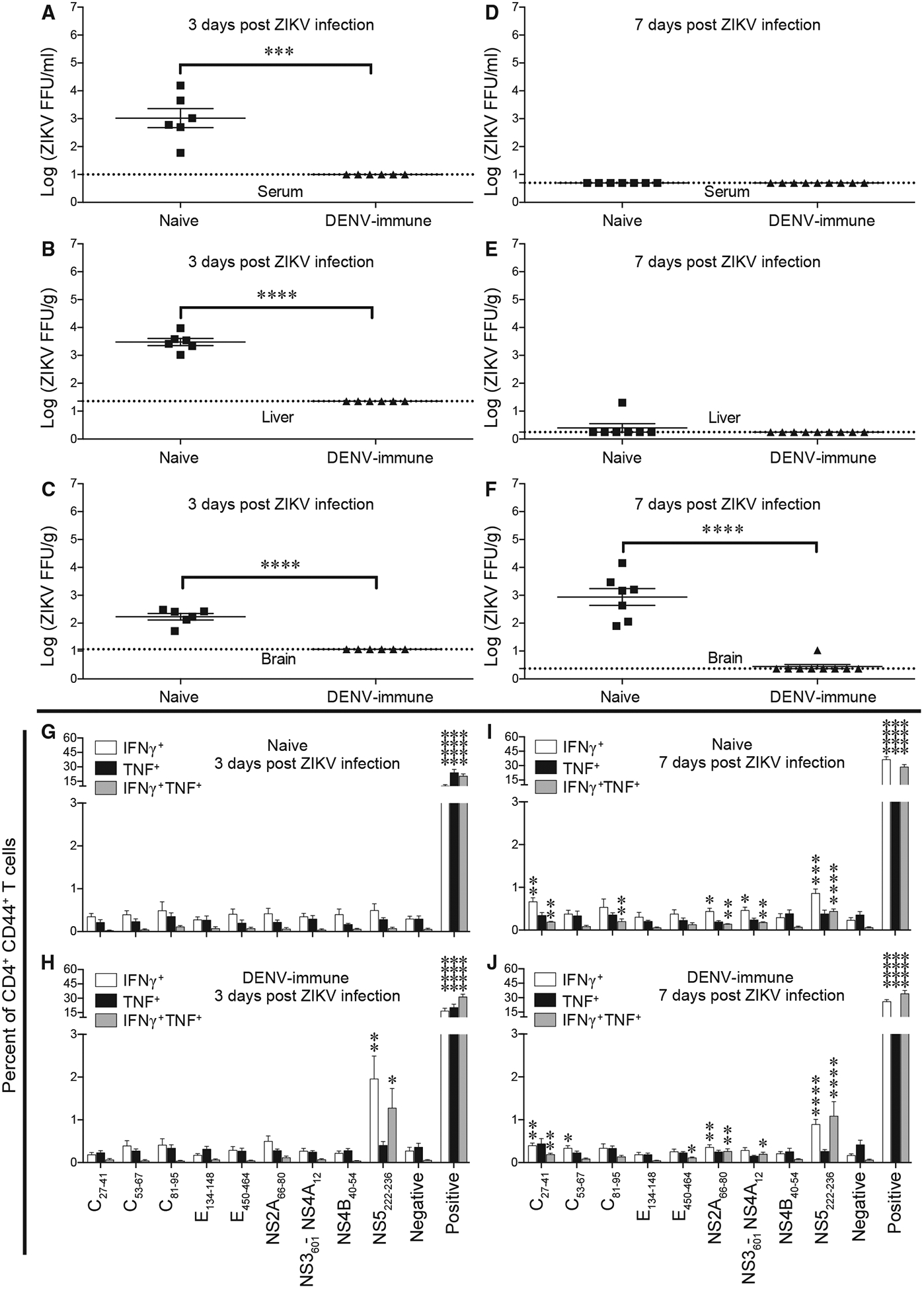
Ifnar1−/−HLA-DRB1*0101 mice were infected intraperitoneally with 2 × 103 FFU of DENV2 S221 for 4 weeks. Naive mice and DENV2-immune mice were infected retro-orbitally with 1 × 104 FFU of ZIKV strain SD001 for 3 (A–C and G–H) or 7 (D–F and I–J) days.
(A–F) Mouse blood (A and D), liver (B and E), and brain (C and F) were harvested, and levels of infectious ZIKV were determined using the FFA. Each point represents an individual mouse.
(G–J) Splenocytes were isolated from mice at 3 dpi (G and H) or 7 dpi (I and J) and stimulated in vitro for 6 h with the indicated ZIKV-specific and DENV2/ZIKV-cross-reactive peptides, and the frequencies of CD4+ T cells producing IFNγ, IFNγ plus TNF, or TNF were detected using the ICS assay. Negative and positive refer to cells incubated alone or with PMA and ionomycin, respectively. Data represent the mean ± SEM of two independent experiments (n = 3–5 mice/experiment).
(A–F) ***p < 0.001, ****p < 0.0001 by two-tailed Mann-Whitney test.
(G–J) *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-tailed Mann-Whitney test with a B-H adjusted p value (FDR) < 10%.
Protective Effect of Vaccination with ZIKV-Specific and DENV2/ZIKV-Cross-Reactive Epitopes in ZIKV-Infected Mice
Next, we asked whether the ZIKV-derived CD4+ T cell epitopes elicited a protective or pathogenic response during ZIKV infection. Ifnar1−/−HLA-DRB1*0101 mice were injected subcutaneously (s.c.) with adjuvant alone (mock vaccinated) or with a mixture of five ZIKV-derived peptides: two ZIKV specific (C27–41 and NS3601-NS4A12), and three DENV2/ZIKV cross-reactive (E134–148, NS2A66–80, and NS5222–236). Four weeks later, all mice were infected with ZIKV. At 3 dpi, blood, spleen, liver, and brain were harvested to determine viral titers by using a focus-forming assay (FFA). We found that infectious ZIKV titers were significantly lower in the serum, spleen, and brain of peptide-vaccinated mice than in mock-vaccinated mice (12-, 17-, and 12-fold, respectively), whereas no significant differences were observed between the liver viral titers in mock- and peptide-vaccinated mice (Figure S2). Thus, ZIKV-derived peptides (ZIKV specific and DENV2/ZIKV cross-reactive) elicit a protective response against subsequent ZIKV infection.
We then examined whether vaccination with DENV2/ZIKV-cross-reactive peptides alone could elicit a protective CD4+ T cell response upon ZIKV viral infection. We vaccinated Ifnar1−/−HLA-DRB1*0101 mice with the DENV2/ZIKV cross-reactive peptides E134–148, NS2A66–80, NS4B40–54, and NS5222–236 (targeting four different ZIKV proteins) or with four irrelevant (influenza hem-agglutinin derived) peptides and infected the mice with ZIKV 4 weeks later. At 3 or 7 dpi, the frequency of peptide-specific CD4+ T cells in the spleen was determined using the ICS assay, and ZIKV titers in the serum, liver, and brain were measured using the FFA. We found that the frequencies of peptide-specific IFNγ-, TNF-, and IFNγ plus TNF-producing CD4+ T cells were significantly higher in the DENV2/ZIKV-cross-reactive peptide-vaccinated mice than in the irrelevant-peptide-vaccinated mice at 3 dpi, whereas the response had waned by 7 dpi, at which time the frequencies of DENV2/ZIKV-cross-reactive CD4+ T cells were similar in the mice vaccinated with DENV2/ZIKV-cross-reactive peptides and irrelevant peptides (Figures 4A–4C). Because we previously showed that the development of a ZIKV-specific CD4+ T cell response develops later than 3 dpi (Elong Ngono etal., 2019), these results suggest that vaccination with DENV2/ZIKV-crossreactive peptides induced a memory CD4+ T cell response that expanded rapidly (within 3 days) upon ZIKV infection. Consistent with this finding, the level of infectious ZIKV was significantly lower in the serum, liver, and brain of mice vaccinated with DENV2/ZIKV-cross-reactive peptides than that of the mice vaccinated with irrelevant peptides at 3 dpi (7.6-, 3.6-, and 4.5-fold, respectively; Figures 4D–4F). At 7 dpi, the viral burden in the serum and liver of both mouse groups was completely cleared, likely because the primary T cell response to ZIKV developed by 7 dpi and contributed to ZIKVclearance;however, although the ZIKV burden in the brain of irrelevant-peptide-vaccinated mice remained high, it was significantly lower (21.7-fold) in DENV2/ZIKV-cross-reactive peptide-vaccinated mice (Figures 4D–4F).
Figure 4. Expansion of CD4+ T Cells and Suppression of Viral Burden in Mice Infected with ZIKV 4 Weeks after Vaccination with DENV2/ZIKV-Cross-Reactive Peptides.
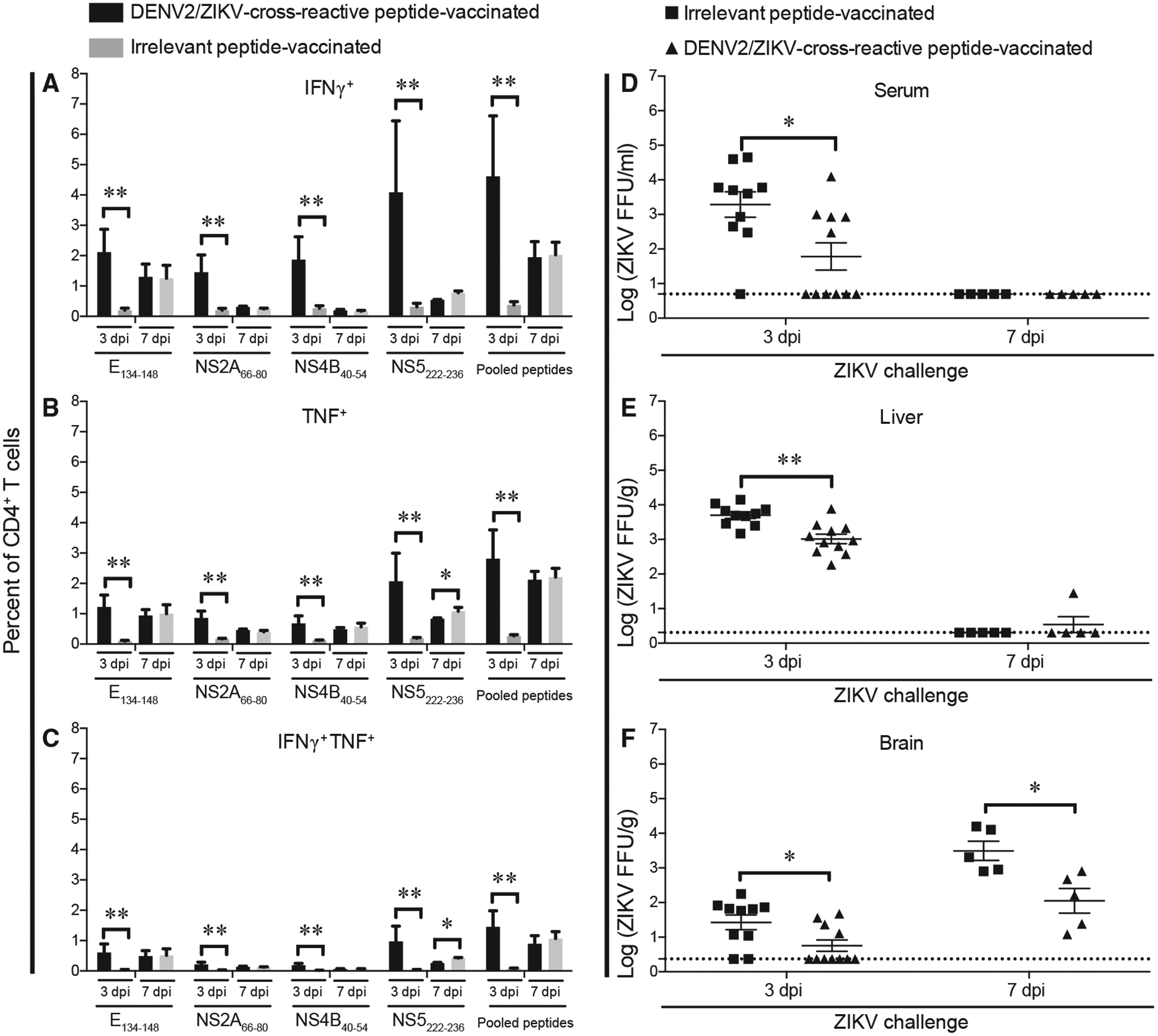
Ifnar1−/−HLA-DRB1*0101 mice were injected subcutaneously with four irrelevant peptides or four DENV2/ZIKV-cross-reactive peptides (E134–148, NS2A66–80, NS4B40–54, and NS5222–236) and boosted in the same manner 2 weeks later. Two weeks after boosting, all mice were infected by retro-orbital injection with 1 × 104 FFU of ZIKV strain SD001. Three or 7 days later, mice were sacrificed.
(A–C) Splenocytes were isolated and stimulated for 6 h in vitro with the indicated DENV2/ZIKV-cross-reactive peptides, and the frequencies of CD4+ T cells producing IFNγ (A), TNF (B), or IFNγ plus TNF (C) were detected using the ICS assay.
(D–F) Blood (D), liver (E), and brain (F) were harvested, and levels of infectious ZIKV were determined using the FFA. Data represent the mean ± SEM of two independent experiments (n = 3–6 mice/experiment). *p < 0.05, **p < 0.01, ***p < 0.001 by two-tailed Mann-Whitney test.
To determine whether the ZIKV-protective response could be elicited in mice vaccinated for a longer period than 4 weeks, we also examined the CD4+ T cell response and viral burden in mice infected with ZIKV at 6 weeks after peptide vaccination. Notably, the results were comparable to those obtained at 4 weeks post-vaccination. Thus, significantly higher frequencies of cytokine-producing CD4+ T cells were detected in the spleens from mice vaccinated with DENV2/ZIKV-cross-reactive peptides than from mice vaccinated with irrelevant peptides at 3 dpi (Figures S3A–S3C), and ZIKV infection was effectively controlled, with viral titers in the liver reduced by 4.8-fold in the DENV2/ZIKV-cross-reactive peptide-vaccinated compared with the irrelevant-peptide-vaccinated mice (Figures S3D–S3F). We note that ZIKV was not detectable in the serum samples from either mouse group at 3 or 7 dpi. This may be because the 11-week-old (6-week-immunized) mice are more resistant to ZIKV infection than 9-week-old (4-week-immunized) mice.
Collectively, these results demonstrate that vaccination with DENV2/ZIKV-cross-reactive CD4+ T cell epitopes can induce a memory CD4+ T cell response that expands after ZIKV infection and contribute to the reduction of viral load at 4 and 6 weeks post-vaccination.
Protection Induced by Vaccination with DENV2/ZIKV-Cross-Reactive CD4+ T Cell Epitopes Is Mediated by IFNγ and/or TNF in the Absence of Antibodies
To confirm that protection conferred by 4-week vaccination with DENV2/ZIKV-cross-reactive epitopes was mediated by CD4+ T cells and the canonical Th1 cytokines, we depleted the vaccinated mice of CD4+ T cells, IFNγ, or TNF. Ifnar1−/−HLA-DRB1*0101 mice were vaccinated with four DENV2/ZIKV-cross-reactive peptides (as described above), and 4 weeks later, they were treated with a CD4+-T-cell-depleting Ab or isotype control Ab before (days −3 and −1) and after (day +1) ZIKV infection. Blood and tissues were then harvested and evaluated for infectious ZIKV titers at 3 dpi. Although the serum ZIKV titers were not significantly different between the two mouse groups (Figure 5A), ZIKV titers were markedly higher in the spleen, liver, and brain of CD4+-T-cell-depleted mice than isotype control Ab-treated mice (5.7-, 12.2-, and 1.5-fold, respectively; Figures 5B–5D), confirming that CD4+ T cells elicited by the DENV2/ZIKV-cross-reactive peptides contributed to protection against ZIKV infection.
Figure 5. Reduction in Protective Immunity against ZIKV Infection in Mice Vaccinated with DENV2/ZIKV-Cross-Reactive Peptides and Depleted of CD4+ T Cells.
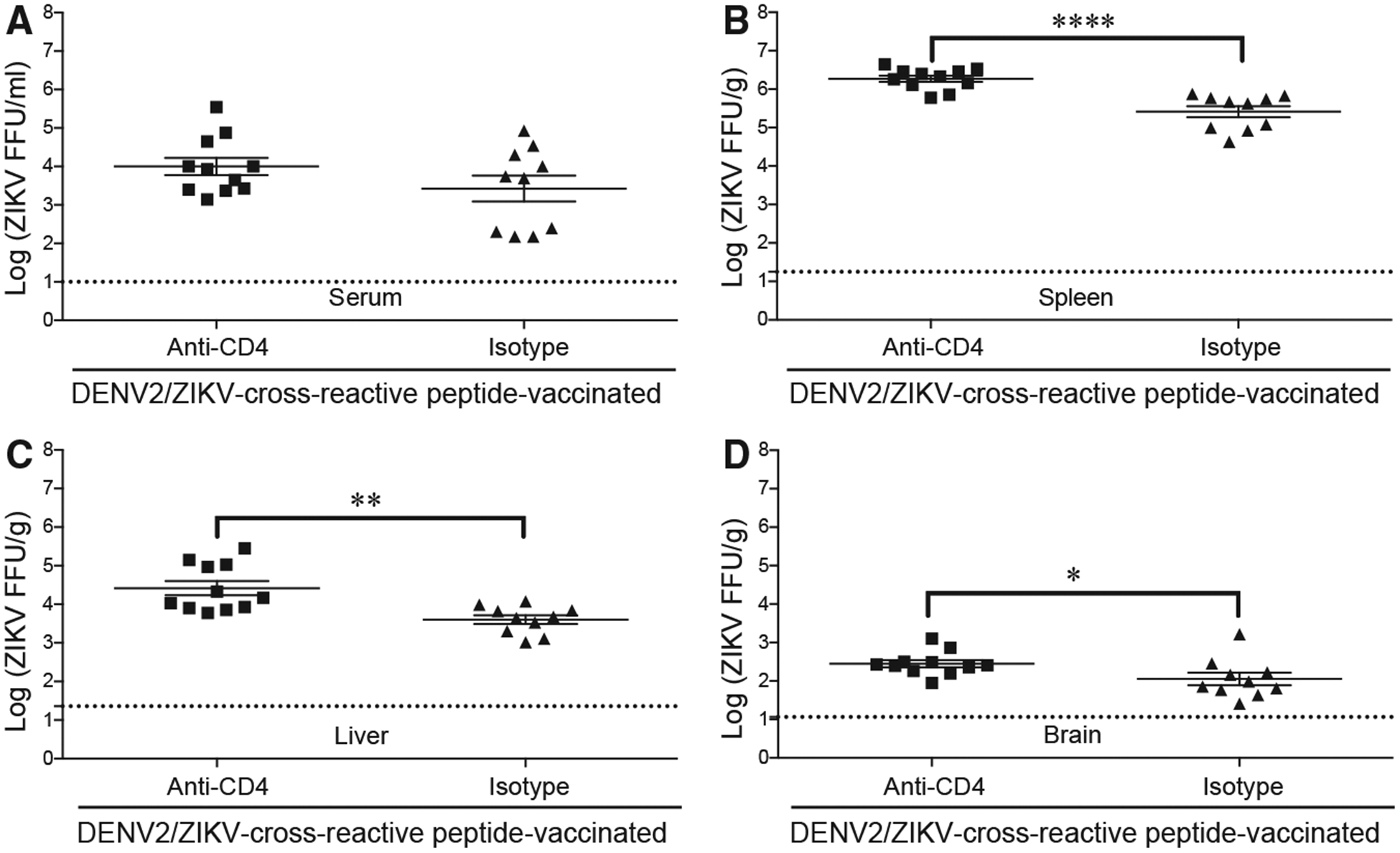
Ifnar1−/− HLA-DRB1*0101 mice were injected subcutaneously with four DENV2/ZIKV-crossreactive peptides (E134–148, NS2A66–80, NS4B40–54, and NS5222–236) and boosted in the same manner 2 weeks later. Two weeks after boosting, all mice were infected by retro-orbital injection with 1 × 104 FFU of ZIKV strain SD001. Mice were injected intraperitoneally with isotype control Ab or anti-CD4 Ab days 3 and 1 before and day 1 after ZIKV infection. At 3 dpi, levels of infectious ZIKV in serum (A), spleen (B), liver (C), and brain (D) were determined using the FFA. Each point represents an individual mouse. Data represent the mean ± SEM of two independent experiments (n = 5–6 mice/experiment). *p < 0.05, **p < 0.01, ****p < 0.0001 by two-tailed Mann-Whitney test.
To examine the contribution of IFNγ and/or TNF to the protection against ZIKV infection, we performed the same experiments as for CD4+ T cell depletion, except that mice were instead treated with neutralizing Abs against either IFNγ or TNF before (days 3 and 1) and after (day +1) ZIKV infection. DENV2/ZIKV-cross-reactive peptide immunization elicited IFNγ- and/ or TNF-producing CD4+ T cell response to individual peptide stimulation (Figures 6A–6C). Compared with the mice treated with the isotype control Ab, infectious ZIKV titers were significantly higher in the serum and brain of mice treated with the neutralizing anti-TNF Ab (6.6- and 35-fold, respectively) or anti-IFNγ Ab (59.2- and 26.4-fold, respectively) (Figures 6D and 6F), whereas ZIKV titers in the liver were significantly higher in mice depleted of IFNγ (82.1-fold) but not of TNF activity (Figure 6E). The same experiment performed in mice vaccinated with DENV2/ZIKV-cross-reactive peptides for 6 weeks before ZIKV infection confirmed that IFNγ was required for the reduction of ZIKV viral load in serum, liver, and brain (Figures S3G–S3I). No significant role for TNF was observed in this experiment, although the levels of infectious ZIKV trended to be higher in the liver and brain, but not the serum, of mice with TNF blockade than mice treated with the isotype control mAb. These results indicate that both IFNγ and TNF contribute to the cross-reactive peptide vaccine-induced protection against ZIKV depending on the tissue and vaccination regimen.
Figure 6. Anti-ZIKV Immune Response in Mice Depleted of IFNγ or TNF and Infected with ZIKV 4 Weeks after Vaccination with DENV2/ZIKV-Cross-Reactive Peptides.

Ifnar1−/−HLA-DRB1*0101 mice were injected subcutaneously with adjuvant alone (mock) or four irrelevant peptides or four DENV2/ZIKV-cross-reactive peptides (E134–148, NS2A66–80, NS4B40–54, and NS5222–236) and boosted in the same manner 2 weeks later. Two weeks after boosting, mice were infected by retro-orbital injection with 1 × 104 FFU of ZIKV strain SD001. Three days later, mice were sacrificed and blood and tissues were harvested.
(A–C) Splenocytes were stimulated for 6 h in vitro with the indicated DENV2/ZIKV-cross-reactive peptides, and the frequencies of CD4+ T cells producing IFNγ (A), TNF (B), and IFNγ plus TNF (C) were detected using the ICS assay.
(D–F) Mice were injected intraperitoneally with a neutralizing Ab against TNF or IFNγ or an isotype control Ab on days 3 and 1 before and 1 day after ZIKV infection. Levels of infectious ZIKV in the serum, liver, and brain at 3 dpi were determined using the FFA. Each point represents an individual mouse.
(G) Sera were tested for the presence of ZIKV E-reactive IgG by ELISA. Data represent the mean ± SEM of two independent experiments, with a total of 10 mice per group (A–C) or with 10 mice (8 females, 2 males) for a-TNF, 8 mice (2 females, 6 males) for α-IFNγ, 8 mice (1 female, 7 males) for isotype, or 8 mice (8 males) for mock (D–F) groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by two-tailed Mann-Whitney test (A–C) or Kruskal-Wallis one-way ANOVA with Dunn’s correction (D–F).
Finally, we assessed whether an anti-ZIKV Ab response contributed to the protection against ZIKV infection in mice vaccinated with DENV2/ZIKV-cross-reactive peptides. Ifnar1−/−HLA-DRB1*0101 mice were vaccinated with four DENV2/ZIKV-cross-reactive peptides, infected with ZIKV, and bled at 3 and 7 dpi for analysis of serum ZIKV E-reactive IgG titers. At 3 dpi, anti-ZIKV IgG titers were undetectable in mice vaccinated with either DENV2/ZIKV-cross-reactive peptides or irrelevant control peptides, whereas at 7 dpi, a low titer of anti-ZIKV IgG was detected in both mouse groups. Notably, however, there was no significant difference between the IgG titers in irrelevant-peptide-vaccinated and DENV2/ZIKV-cross-reactive peptide-vaccinated mice (Figure 6G). Thus, an anti-ZIKV IgG response had not developed in the vaccinated mice at 3 dpi, indicating that Abs do not contribute to the cross-reactive peptide vaccine-induced protection seen at 3 dpi.
Taken together, the results of this study demonstrate that protection against ZIKV after vaccination with DENV2/ZIKV-crossreactive epitopes is mediated by CD4+ T cells and IFNγ- and/or TNF in an Ab-independent manner.
DISCUSSION
Recent epidemiologic studies have shown that prior DENV exposure provides protection against subsequent ZIKV infection in humans (Gordon et al., 2019; Pedroso et al., 2019; Rodriguez-Barraquer et al., 2019), but the mechanisms responsible for this cross-protection have yet to be defined. Evidence from mouse models supports roles for both Abs and CD8+ T cells as mediators of cross-protective immunity (Fernandez et al., 2017; Huang et al., 2017; Kam et al., 2017; Regla-Nava et al., 2018; Wen et al., 2017a, 2017b), and more recent studies support a protective role for CD4+ T cells in primary and secondary ZIKV infection (Elong Ngono et al., 2019; Hassert et al., 2018; Liang et al., 2019; Lucas et al., 2018; Pardy et al., 2017). However, the pathogenic versus protective functions of DENV-elicited CD4+ T cells during ZIKV infection have not previously been investigated. Using Ifnar1−/−HLA-DRB1*0101 transgenic mice, we identified DENV2/ZIKV cross-reactive CD4+ T cell epitopes and showed that vaccination with the peptides induced a CD4+ T cell response that protected against ZIKV infection in a manner dependent on CD4+ T cells and IFNγ and/or TNF but not on anti-ZIKV Abs. Thus, in addition to Abs and CD8+ T cells, the results of the present study establish that CD4+ T cells and the canonical Th1 effector cytokines act in an Ab-independent manner to confer cross-protective immunity against ZIKV in mice. These results have important implications for ZIKV infections in humans previously exposed to DENV.
Our previous finding that the same epitopes and immunodominance patterns are detected in Ifnar1−/−HLA transgenic mice and humans (Elong Ngono et al., 2016; Weiskopf et al., 2011, 2013, 2014, 2015b) validate the use of the Ifnar1−/−HLA-DRB1*0101 transgenic mouse model to evaluate human-relevant DENV/ZIKV-cross-reactive CD4+ T cell responses against ZIKV infection. One of the ZIKV-specific epitopes identified here (C27–41) shares a 13-amino acid sequence with a 15-mer (SPFGGLKRLPAGLLL) identified as a CD4+ T cell epitope in humans by Koblischke et al. (2018), who examined the IL-2 response of CD8+-T-cell-depleted peripheral blood mononuclear cells from ZIKV-infected individuals. The E134–148 peptide, which we demonstrated here induces IL-2-producing CD4+ T cells, shares a 14-amino acid sequence with another human CD4+ T cell epitope (ENLEYRIMLSVHGSQ) reported in the same human study (Koblischke et al., 2018). Given that C27–41 and E134–148 were identified based on their predicted binding to HLA-DRB1*0101, it seems likely that the two epitopes identified by Koblischke et al. (2018) are also HLA-DRB1*0101 restricted.
Our results demonstrating that the DENV2/ZIKV-cross-reactive CD4+ T cell response expanded early (day 3) after ZIKV infection and remained in the later phase of the response (day 7) is in agreement with data showing that ZIKV cross-reactive CD4+ T cells are immunodominant and rapidly activated in DENV-immune individuals (Grifoni et al., 2017). This finding is also consistent with studies on the responses of cross-reactive CD8+ T cells during sequential infections with DENV and ZIKV (Grifoni et al., 2017; Wen et al., 2017b) or with heterologous DENV serotypes (Weiskopf et al., 2014) in humans and Ifnar1−/−HLA transgenic mice. Thus, prior DENV infection shapes both CD4+ and CD8+ T cell responses during subsequent ZIKV infection in humans and Ifnar1−/−HLA transgenic mice. As DENV and ZIKV infection in Ifnar1−/−HLA transgenic mice evoke T cell responses that are similar to those in humans with respect to timing, antigen specificity, and immunodominance patterns, Ifnar1−/−HLA transgenic mouse models represent a valuable model for examining key features of human-relevant T cell responses in various DENV and ZIKV infection and vaccination settings.
Consistent with publications reporting cross-reactivity of pre-existing DENV-elicited CD4+ T cell responses to ZIKV (Grifoni et al., 2017; Lim et al., 2018; Paquin-Proulx et al., 2017; Reynolds et al., 2018), we identified five HLA-DRB1*0101-restricted ZIKV epitopes (E134–148, NS2A66–80, NS2A184–198, NS4B40–54, and NS5222–236) that were cross-reactive on DENV-primed CD4+ T cells. The five DENV2/ZIKV-cross-reactive epitopes identified here share 33%–100% homology with the corresponding DENV2 sequences and 27%–100%, 33%– 93%, and 33%–100% homology with variants in DENV1, DENV3, and DENV4, respectively (Table S1). Notably, NS5222–236 is a conserved T cell epitope among multiple flaviviruses; the identical sequence is present in DENV1, 2, and 4 and YFV, and the homologous sequences in DENV3 and JEV differ by one and two residues, respectively. Similarly, we previously identified an HLA-B*0702-restricted CD8+ T cell epitope that is conserved among many flaviviruses, including ZIKV, DENV1–4, WNV, JEV, Usutu virus, Murray Valley encephalitis virus, and Kunjin virus (Wen et al., 2017b). The identification of conserved CD4+ T cell and CD8+ T cell epitopes among flaviviruses supports the feasibility of developing flavivirus vaccines that elicit both CD4+- and CD8+-T-cell-mediated cross-protective immunity.
In line with studies suggesting a protective role for serotype-cross-reactive CD4+ T cells in DENV immunity (Weiskopf et al., 2015a, 2016), the present study provides evidence in support of a role for CD4+ T cells in conferring cross-protection against ZIKV in DENV2-immune mice. CD4+ T cells from DENV2/ZIKV-cross-reactive peptide-vaccinated mice produced IFNγ and TNF upon cognate peptide restimulation in vitro, and depletion of CD4+ T cells and neutralization of IFNγ and/or TNF disrupted the protection induced by peptide vaccination in mice. Thus, the protective roles of CD4+ T cells elicited by peptide vaccination are likely to be mediated by ZIKV-induced secretion of IFNγ and/or TNF. Taken together with studies identifying important roles for CD8+ T cells in cross-protecting against DENV (de Alwis et al., 2016; Elong Ngono et al., 2016; Weiskopf et al., 2013, 2015b; Zellweger et al., 2015) and ZIKV infections (Huang et al., 2017; Regla-Nava et al., 2018; Wen et al., 2017a, 2017b), our results suggest that a pan-flavivirus vaccine that induces CD8+ T cell responses and canonical Th1-cytokine-producing CD4+ T cell responses may not only be broadly effective against both DENV and ZIKV but also avoid ADE. This is a crucial point because the DENV- and ZIKV-specific vaccines that are currently licensed or in clinical trials aim to elicit Ab responses and may, at least in theory, cause ADE if the vaccine-induced Ab response is inefficient or wanes.
In summary, our results demonstrate that vaccination with DENV2/ZIKV-cross-reactive peptides elicits a Th1 CD4+ T cell effector response that promotes protection against ZIKV infection in an IFNγ/TNF-dependent, Ab-independent manner. These findings suggest that the cross-reactive CD4+ T cell response is one mechanism by which humans with prior DENV exposure are protected against ZIKV. Inclusion of cross-reactive CD8+ T cell and CD4+ Th1 cell epitopes could not only enhance the efficacy of ZIKV-specific vaccines but also pave the way for the development of a pan-flavivirus vaccine that simultaneously protects against both DENV and ZIKV and minimizes ADE.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sujan Shresta (sujan@lji.org).
This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Ifnar1−/−HLA-DRB1*0101 mice have been described in our previous study of anti-DENV responses (Weiskopf et al., 2011). Mice were bred under specific pathogen-free conditions with a 12 hour on/off light cycle at the La Jolla Institute for Immunology. Mouse experiments were approved by the Institutional Animal Care and Use Committee (protocol no. AP028-SS1–0615). Sample sizes were estimated based on experiments in similar studies, and the experiments were not randomized or blinded.
Cell Lines
Baby hamster kidney (BHK)-21 cells (ATCC, CCL-10) were grown in MEM-α medium containing 10% FBS and 1 3 penicillin/streptomycin. Growing cells were allowed to proliferate until 90% confluency and then split for propagation.
Virus Strains
ZIKV strain SD001 was isolated from the urine of a ZIKV-infected individual who traveled to Venezuela during the 2016 ZIKV epidemic. PCR sequencing showed that ZIKV SD001 belongs to the Asian lineage and is phylogenetically related to ZIKV isolates circulating in South American countries (Carlin et al., 2018). The mouse-adapted DENV2 strain S221 is a triple-plaque-purified clone derived from DENV2 D2S10 (Yauch et al., 2009). Both ZIKV and DENV2 were grown in C6/36 mosquito cells, and viral titers were measured using an FFA with the baby hamster kidney (BHK)-21 cell line as described below.
METHOD DETAILS
Peptide prediction and synthesis
The online software Immune Epitope Database and Analysis Resource (IEDB-AR) (http://www.iedb.org) (Vita et al., 2019) was used to predict HLA-DRB1*0101-binding peptides from ZIKV strain FSS13025 (Cambodia, 2010; Asian lineage). Thirty predicted epitope candidates were synthesized by Synthetic Biomolecules as crude peptides (> 75% purity) for use in in vitro experiments. Two ZIKV-specific (C27–41 and NS3601-NS4A12), four DENV2/ZIKV-cross-reactive (E134–148, NS2A66–80, NS4B40–54, and NS5222–236) (ZIKV sequence numbering; Table 1), and four irrelevant peptides (influenza hemagglutinin-derived HLA-DRB1*0101-restricted epitopes: H1221–235 [SRYSKKFKPEIAIRP], H1225–239 [KKFKPEIAIRPKVRD], H1305–319 [TSLPFQNIHPITIGK], and H1441–455 [ELLVLLENERTLDYH]) (DiPiazza et al., 2017) were synthesized at high purity (> 99%) for use in vitro and in mouse vaccination experiments. All peptides were dissolved in dimethyl sulfoxide (DMSO) at a concentration of 40 mg/ml and were stored at −20°C.
ZIKV infection of mice and peptide screening
For the 30-peptide epitope screening, 5-week-old female or male mice were infected retro-orbitally (r.o.) with either 1 × 102 focus-forming units (FFU) of ZIKV SD001 or 2 × 103 FFU of DENV2 S221 in 200 μL of 10% fetal bovine serum in phosphate-buffered saline (FBS/PBS). At 7 dpi, spleens were harvested and single-cell splenocyte suspensions were prepared. A total of 1 × 106 splenocytes was plated in each well of a 96-well U-bottom plate and stimulated with individual peptides (20 μg crude peptide/well) for 6 h, with brefeldin A added for the final 5 h (GolgiPlug, BD Biosciences). Positive and negative controls were splenocytes stimulated with phor-bol-12-myristate-13-acetate (PMA, 0.1 μg/ml) and ionomycin (1 μg/ml) (Thermo Fisher Scientific) or incubated with medium alone, respectively. Cells were harvested, washed, and processed for the ICS assay as described below.
ZIKV infection of DENV2-immune mice
Five-week-old female or male mice were inoculated intraperitoneally (i.p.) with 2 × 103 FFU of DENV2 S221. Four weeks later, the mice were infected r.o. with 1 × 104 FFU of ZIKV SD001, and on day 3 or 7 after infection, all mice were sacrificed and blood samples were collected for serum preparation. After cardiac perfusion with PBS, the spleen, liver, and brain were harvested. Splenocytes were stimulated in vitro with 20 μg crude peptide (C27–41, C53–67, C81–95, E450–464 or NS3601-NS4A12) or 1 μg purified peptide (E134–148, NS2A66–80, NS4B40-54 or NS5222–236) per well. Positive and negative controls were included in all experiments. Cells were processed for the ICS assay as described below. ZIKV viral titers in serum and tissues were measured using the FFA as described below.
ZIKV infection of peptide-vaccinated mice
Several protocols were employed. Mixtures of five ZIKV-specific and DENV2/ZIKV-cross-reactive peptides (C27–41, E134–148, NS2A66–80, NS3601-NS4A12, and NS5222–236; 50 μg of each peptide/mouse) were emulsified in complete Freund’s adjuvant and injected subcutaneously (s.c.) into 5-week-old female or male mice. Mock-vaccinated mice received the adjuvants in DMSO without peptides. Two weeks later, the mice were boosted by injection of the same peptides in incomplete Freund’s adjuvant. Two weeks after the boost, mice were infected r.o. with 1 × 104 FFU of ZIKV SD001. Three days later, all mice were sacrificed and blood, spleen, liver, and brain were harvested. ZIKV viral titers in serum and tissues were measured using the FFA as described below.
The four DENV2/ZIKV-cross-reactive peptides (E134–148, NS2A66–80, NS4B40–54, and NS5222–236; 50 μg of each peptide/mouse) were emulsified in complete Freund’s adjuvant and injected s.c. into 5-week-old female or male mice. Mock-vaccinated mice received the adjuvants in DMSO without peptides, and the irrelevant peptide-vaccinated mice received the adjuvants with four influenza-derived peptides (H1221–235, H1225–239, H1305–319, and H1441–455; 50 μg of each peptide/mouse). Two weeks later, the mice were boosted by injection of the same peptides in incomplete Freund’s adjuvant. Two or 4 weeks after the boost, mice were treated by one of three protocols as follows. (i) Mice were infected r.o. with 1 × 104 FFU of ZIKV SD001. Three or 7 days later, all mice were sacrificed and blood, spleen, liver, and brain were harvested. Splenocytes were stimulated in vitro with individual or pooled DENV2/ZIKV-crossreactive peptides (E134–148, NS2A66–80, NS4B40–54, and NS5222–236) (1 μg purified peptide/well) and processed for the ICS assay as described below. ZIKV-reactive IgG in serum was measured using a capture ELISA assay, and ZIKV viral titers in serum and tissues were measured using the FFA as described below. (ii) Mice were infected r.o. with 1 × 104 FFU of ZIKV SD001 and injected i.p. with 250 μg of a CD4+ T cell-depleting Ab (clone GK1.5, Bio X Cell) or isotype control Ab (LTF-2, Bio X Cell) on days 3 and 1 before and 1 day after ZIKV infection. Three days later, all mice were sacrificed and blood, spleen, liver, and brain were harvested. ZIKV viral titers in serum and tissues were measured using the FFA. (iii) Mice were infected r.o. with 1 × 104 FFU of ZIKV SD001 and injected i.p. with 100 μg of neutralizing anti-TNF monoclonal Ab (mAb; clone XT3.11, Bio X Cell), anti-IFNγ mAb (clone XMG1.2, Thermo Fisher Scientific), or isotype control mAb (clone HPRN, Bio X Cell) on days 3 and 1 before and 1 day after ZIKV infection. At 3 dpi, all mice were sacrificed and blood, spleen, liver, and brain were harvested. ZIKV viral titers in serum and tissues were measured using the FFA.
ICS assay
After incubation of splenocytes with peptides or PMA/ionomycin, cells were harvested, washed, and incubated with Fc Block (CD16/CD32 mAb 2.4G2; BD Biosciences), stained with fixable Live/Dead blue viability stain (Thermo Fisher Scientific), and then stained with the following Abs: PerCP-Cy5.5-conjugated anti-CD3 mAb (clone 145–2C11, Tonbo Biosciences), APC-eFluor780-conjugated anti-CD4 mAb (clone GK1.5, Thermo Fisher Scientific), Brilliant Violet (BV) 785-conjugated anti-CD44 mAb (clone IM7, BioLegend), Alexa Fluor 700-conjugated anti-CD62L mAb (clone MEL-14, Thermo Fisher Scientific), PE-conjugated anti-CD11a (clone M17/4, BioLegend), BV605-conjugated anti-CD49d (clone R1–2, BD Biosciences), PE-conjugated anti-CD25 mAb (clone PC61, BioLegend), PE-conjugated or biotin-conjugated anti-CD185 mAb (CXCR5, clone SPRC15; Thermo Fisher Scientific), BV 605-conjugated anti-CD279 mAb (PD1, clone 29F.1A12; BioLegend), and/or BV 421-conjugated streptavidin (#405225, BioLegend). Cells were then fixed and permeabilized using Cytofix/Cytoperm solution (#554722, BD Biosciences), followed by staining with FITC-conjugated anti-IFNγ mAb (clone XMG1.2, Tonbo Biosciences), Alexa Fluor 700- or APC-conjugated anti-TNF mAb (clone MP6-XT22, Thermo Fisher Scientific), PE-, BV 421-, or BV 711-conjugated anti-IL-2 mAb (clone JES6–5H4, BioLegend), APC-conjugated anti-IL-4 mAb (clone 11B11, BioLegend), PE-conjugated anti-IL-5 mAb (clone TRFK5, Thermo Fisher Scientific), BV510-conjugated anti-IL-17A mAb (clone TC11–18H10.1, BioLegend), and/or Alexa Fluor 700-conjugated anti-FoxP3 mAb (clone FJK-16S, Thermo Fisher Scientific). Data were collected using an LSR Fortessa flow cytometer (BD Biosciences) and analyzed using FlowJo software X 10.0.7 (Tree Star).
ELISA
To quantify ZIKV-reactive IgG, 96-well high-affinity ELISA plates (Costar) were coated with ZIKV E protein (1 mg/ml ZIKVSU-ENV, The Native Antigen) in 100 μl coating buffer (0.1 M NaHCO3) overnight at 4°C and then blocked for 1 h at room temperature (RT) with 5% Blocker Casein in PBS (Thermo Fisher Scientific). Mouse serum samples were serially diluted three-fold from 1:30 to 1:65,610 in 1% bovine serum albumin (BSA)/PBS and added to the coated wells. As a positive control, 10 μg of the pan-flavivirus envelope protein-specific mAb 4G2 (Absolute Antibody) was diluted in 1% BSA/PBS and titrated three-fold in the same manner as for the sera. After 1.5 h incubation at RT, the wells were washed with washing buffer (0.05% Tween 20 in PBS) and then incubated with HRP-conjugated goat anti-mouse IgG (1:5000 in 1% BSA/PBS) for 1.5 h at RT. TMB chromogen solution (Thermo Fisher Scientific) was added to the wells, the reaction was stopped by addition of 2N sulfuric acid, and the absorbance at 450 nm was read on a Spectramax M2E microplate reader (Molecular Devices). The ZIKV-specific IgG endpoint titers were calculated as the reciprocal of the highest serum dilution that gave a reading twice the cutoff absorbance of the negative control (1% BSA/PBS).
Focus-forming assay
BHK-21 cells (2 × 105/well) were plated in 24-well culture plates and incubated at 37°C in a CO2 incubator overnight. Mouse spleen, liver, and brain were homogenized using TissueLyser II (QIAGEN) and centrifuged at 6000 rpm for 10 min. Aliquots of the supernatants (100 μl) were serially diluted 10-fold in medium, added to the BHK-21 cells, and incubated at 37°C for 1 h. The viral supernatant was aspirated, and a pre-warmed solution of 1% carboxymethyl cellulose medium was added to each well. After 2.5 days incubation, the cells were fixed with 4% paraformaldehyde solution for 30 min at RT, washed with PBS, permeabilized with 1% Triton X-100 for 20 min at RT, and washed again with PBS. Plates were blocked with 10% FBS/PBS for 40 min at RT and incubated with the pan-flavivirus envelope protein-specific mAb 4G2 (1 μg/ml) for 1 h at RT. Plates were washed with PBS and incubated with horseradish peroxidase-conjugated goat anti-mouse IgG mAb (1:1000 dilution) for 1.5 h at RT. Finally, the plates were washed with PBS and developed with TrueBlue peroxidase substrate for 20 min at RT. Foci were counted and the viral titers were expressed as FFU/ml serum or FFU/g tissue.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics and graphs
Statistics and graphs were prepared using Prism software version 6.0 (GraphPad Software) and are expressed as the mean ± standard error (s.e.m.). Statistically significant differences between two groups were determined using the non-parametric Mann– Whitney test followed by Benjamini-Hochberg (B-H) correction in R studio (v3.6.1), fixing the false discovery rate (FDR) at 10% or Kruskal–Wallis one-way ANOVA with Dunn’s correction for multiple comparisons. p < 0.05 was considered significant. Further details on statistical analysis are listed in the figure legends.
DATA AND CODE AVAILABILITY
This study did not generate any unique datasets or code.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-mouse CD16/CD32 (clone 2.4G2) | 3D Biosciences | Cat#553141; RRID:AB_394656 |
| Rat monoclonal anti-mouse CD49d (clone R1–2), Brilliant Violet 605 | 3D Biosciences | Cat#740341; RRID:AB_2740074 |
| Rat monoclonal anti-mouse CD3 (clone 145–2C11), PerCP-Cy5.5 | Tonbo Biosciences | Cat#65–0031; RRID:AB_2621872 |
| Rat monoclonal anti-mouse IFNg (clone XMG1.2), FITC | Tonbo Biosciences | Cat#35–7311; RRID:AB_2621724 |
| Rat monoclonal anti-mouse CD44 (clone IM7), Brilliant Violet 785 | BioLegend | Cat#103041; RRID:AB_11218802 |
| Rat monoclonal anti-mouse CD11a (clone M17/4), PE | BioLegend | Cat#101107; RRID:AB_312780 |
| Rat monoclonal anti-mouse CD25 (clone PC61), PE | BioLegend | Cat#102008; RRID:AB_312857 |
| Rat monoclonal anti-mouse IL-2 (clone JES6–5H4), PE | BioLegend | Cat#503807; RRID:AB_315301 |
| Rat monoclonal anti-mouse IL-2 (clone JES6–5H4), Brilliant Violet 421 | BioLegend | Cat#503825; RRID:AB_10895901 |
| Rat monoclonal anti-mouse IL-2 (clone JES6–5H4), Brilliant Violet 711 | BioLegend | Cat#503837; RRID:AB_2564225 |
| Rat monoclonal anti-mouse IL-4 (clone 11B11), APC | BioLegend | Cat#504105; RRID:AB_315319 |
| Rat monoclonal anti-mouse CD279 (clone 29F.1A12), Brilliant Violet 605 | BioLegend | Cat#135219; RRID:AB_11125371 |
| Rat monoclonal anti-mouse IL-17A (clone TC11–18H10.1), Brilliant Violet 510 | BioLegend | Cat#506933; RRID:AB_2562668 |
| Rat monoclonal anti-mouse CD185 (CXCR5) (clone SPRCL5), PE | Thermo Fisher Scientific | Cat#12–7185-82; RRID:AB_11217882 |
| Rat monoclonal anti-mouse CD185 (CXCR5) (clone SPRC15), biotin | Thermo Fisher Scientific | Cat#13–7185-82; RRID:AB_2572800 |
| Rat monoclonal anti-mouse CD4 (clone GK1.5), APC-eFluor780 | Thermo Fisher Scientific | Cat#47–0041-82; RRID:AB_11218896 |
| Rat monoclonal anti-mouse CD62L (clone MEL-14), Alexa Fluor 700 | Thermo Fisher Scientific | Cat#56–0621-82; RRID:AB_494003 |
| Rat monoclonal anti-mouse TNF (clone MP6-XT22), Alexa Fluor 700 | Thermo Fisher Scientific | Cat#56–7349-42; RRID:AB_10671335 |
| Rat monoclonal anti-mouse TNF (clone MP6-XT22), APC | Thermo Fisher Scientific | Cat#17–7321-82; RRID:AB_469508 |
| Rat monoclonal anti-mouse IL-5 (clone TRFK5), PE | Thermo Fisher Scientific | Cat#12–7052-82; RRID:AB_763587 |
| Rat monoclonal anti-mouse FoxP3 (clone FJK-16 s), Alexa Fluor 700 | Thermo Fisher Scientific | Cat#56–5773-82; RRID:AB_1210557 |
| Rat monoclonal anti-mouse IFNg (clone XMG1.2), neutralizing antibody | Thermo Fisher Scientific | Cat#16–7311-81; RRID:AB_469242 |
| Rat monoclonal anti-mouse CD4 (clone GK1.5), CD4+ T cell-depleting antibody | Bio X Cell | Cat#BP0003–1; RRID:AB_1107636 |
| Rat isotype control monoclonal antibody (clone LTF-2) | Bio X Cell | Cat#BP0090; RRID:AB_1107780 |
| Rat monoclonal anti-mouse TNF (clone XT3.11), neutralizing antibody | Bio X Cell | Cat#BP0058; RRID:AB_1107764 |
| Isotype control monoclonal antibody (clone HPRN) | Bio X Cell | N/A |
| Pan-flavivirus envelope protein-specific monoclonal antibody 4G2 (clone D1–4G2–4-15) | Absolute Antibody | Cat#Ab00230–2.0; RRID:AB_2715504 |
| Goat polyclonal anti-mouse IgG, horseradish peroxidase | Sigma-Aldrich | Cat#A0168; RRID:AB_257867 |
| Bacterial and Virus Strains | ||
| ZIKV strain SD001 | Carlin et al., 2018 | N/A |
| DENV2 strain S221 | Yauch et al., 2009 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Live/Dead blue viability stain | Thermo Fisher Scientific | Cat#50-112-1524 |
| Cell Stimulation Cocktail (containing phorbol-12-myristate-13-acetate(PMA)/ionomycin) | Thermo Fisher Scientific | Cat#00-4970-93 |
| TMB chromogen solution | Thermo Fisher Scientific | Cat#00-202-3 |
| Brilliant Violet 421-conjugated streptavidin | BioLegend | Cat#405225 |
| Cytofix/Cytoperm solution | BD Biosciences | Cat#554722 |
| Protein transport inhibitor (containing Brefeldin A) | BD Biosciences | Cat#555029 |
| Carboxymethyl cellulose | Sigma-Aldrich | Cat#9004-32-4 |
| TrueBlue peroxidase substrate | KPL | Cat#5510-0030 |
| ZIKV E protein | The Native Antigen | N/A |
| Experimental Models: Cell Lines | ||
| Baby hamster kidney (BHK)-21 cells | ATCC | CCL-10 |
| Mouse: Ifnar1−/− HLA-DRB1*0101 mice | Weiskopf et al., 2011 | N/A |
| Software and Algorithms | ||
| Immune Epitope Database and Analysis Resource (IEDB-AR) | Vita et al., 2019 | http://www.iedb.org/home_v3.php |
Highlights.
Vaccination with DENV/ZIKV-cross-reactive peptides elicits a Th1 CD4+ T cell response
Peptide vaccine-elicited cross-reactive CD4+ T cells protect against ZIKV infection
Cross-reactive CD4+ T-cell-mediated protection is dependent on IFNγ or TNF
Cross-reactive CD4+ T-cell-mediated protection occurs in the absence of antibodies
ACKNOWLEDGMENTS
This work was supported by the grants from the National Institutes of Health (AI116813, AI140063, and NS106387 to S.S.), the National Natural Science Foundation of China (31870159 to J.W.), and the Zhejiang Provincial Natural Science Foundation (LY17C010004 to J.W.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107566.
DECLARATION OF INTERESTS
The La Jolla Institute for Immunology has a pending USA patent application, 16/537,447, covering methods and compositions utilizing the epitopes described in this manuscript for the treatment and prevention of flaviviruses, with S.S. listed as the inventor.
REFERENCES
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, et al. (2012). DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 8, e1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D, et al. (2017). Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 356, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin AF, Wen J, Vizcarra EA, McCauley M, Chaillon A, Akrami K, Kim C, Ngono AE, Lara-Marquez ML, Smith DM, et al. (2018). A longitudinal systems immunologic investigation of acute Zika virus infection in an individual infected while traveling to Caracas, Venezuela. PLoS Negl. Trop. Dis 12, e0007053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanha PMS, Nascimento EJM, Braga C, Cordeiro MT, de Carvalho OV, de Mendonça LR, Azevedo EAN, França RFO, Dhalia R, and Marques ETA (2017). Dengue Virus-Specific Antibodies Enhance Brazilian Zika Virus Infection. J. Infect. Dis 215, 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles AS, and Christofferson RC (2016). Utility of a Dengue-Derived Monoclonal Antibody to Enhance Zika Infection In Vitro. PLoS Curr. 8, ecurrents.outbreaks.4ab8bc87c945eb41cd8a49e127082620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choumet V, and Despres P (2015). Dengue and other flavivirus infections. Rev. Sci. Tech 34, 473–478, 467–472. [PubMed] [Google Scholar]
- de Alwis R, Bangs DJ, Angelo MA, Cerpas C, Fernando A, Sidney J, Peters B, Gresh L, Balmaseda A, de Silva AD, et al. (2016). Immunodominant Dengue Virus-Specific CD8+ T Cell Responses Are Associated with a Memory PD-1+ Phenotype. J. Virol 90, 4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, et al. (2016). Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol 17, 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Gaska JM, Douam F, Wei L, Kim D, Balev M, Heller B, and Ploss A (2018). Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc. Natl. Acad. Sci. USA 115, E6310–E6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPiazza A, Richards K, Poulton N, and Sant AJ (2017). Avian and Human Seasonal Influenza Hemagglutinin Proteins Elicit CD4 T Cell Responses That Are Comparable in Epitope Abundance and Diversity. Clin. Vaccine Immunol 24, e00548–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Americo JL, Wyatt LS, Eller LA, Whitbeck JC, Cohen GH, Eisenberg RJ, Hartmann CJ, Jackson DL, Kulesh DA, et al. (2004). Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature 428, 182–185. [DOI] [PubMed] [Google Scholar]
- Elong Ngono A, Chen HW, Tang WW, Joo Y, King K, Weiskopf D, Sidney J, Sette A, and Shresta S (2016). Protective Role of Cross-Reactive CD8 T Cells Against Dengue Virus Infection. EBioMedicine 13, 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elong Ngono A, Vizcarra EA, Tang WW, Sheets N, Joo Y, Kim K, Gorman MJ, Diamond MS, and Shresta S (2017). Mapping and Role of the CD8+ T Cell Response During Primary Zika Virus Infection in Mice . Cell Host Microbe 21, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elong Ngono A, Young MP, Bunz M, Xu Z, Hattakam S, Vizcarra E, Regla-Nava JA, Tang WW, Yamabhai M, Wen J, and Shresta S (2019). CD4+ T cells promote humoral immunity and viral control during Zika virus infection. PLoS Pathog. 15, e1007474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez E, Dejnirattisai W, Cao B, Scheaffer SM, Supasa P, Wongwiwat W, Esakky P, Drury A, Mongkolsapaya J, Moley KH, et al. (2017). Human antibodies to the dengue virus E-dimer epitope have therapeutic activity against Zika virus infection. Nat. Immunol 18, 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler AM, Tang WW, Young MP, Mamidi A, Viramontes KM, McCauley MD, Carlin AF, Schooley RT, Swanstrom J, Baric RS, et al. (2018). Maternally Acquired Zika Antibodies Enhance Dengue Disease Severity in Mice. Cell Host Microbe 24, 743–750.e745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A, Gresh L, Ojeda S, Katzelnick LC, Sanchez N, Mercado JC, Chowell G, Lopez B, Elizondo D, Coloma J, et al. (2019). Prior dengue virus infection and risk of Zika: A pediatric cohort in Nicaragua. PLoS Med. 16, e1002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sánchez-Seco MP, Evans MJ, Best SM, and García-Sastre A (2016). Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe 19, 882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Pham J, Sidney J, O’Rourke PH, Paul S, Peters B, Martini SR, de Silva AD, Ricciardi MJ, Magnani DM, et al. (2017). Prior Dengue virus exposure shapes T cell immunity to Zika virus in humans. J. Virol 91, e01469–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB (2007). Dengue. Lancet 370, 1644–1652. [DOI] [PubMed] [Google Scholar]
- Hassert M, Wolf KJ, Schwetye KE, DiPaolo RJ, Brien JD, and Pinto AK (2018). CD4+T cells mediate protection against Zika associated severe disease in a mouse model of infection. PLoS Pathog. 14, e1007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Li S, Zhang Y, Han X, Jia B, Liu H, Liu D, Tan S, Wang Q, Bi Y, et al. (2017). CD8+ T Cell Immune Response in Immunocompetent Mice during Zika Virus Infection. J. Virol 91, e00900–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam YW, Lee CY, Teo TH, Howland SW, Amrun SN, Lum FM, See P, Kng NQ, Huber RG, Xu MH, et al. (2017). Cross-reactive dengue human monoclonal antibody prevents severe pathologies and death from Zika virus infections. JCI Insight 2, 92428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, Balmaseda A, and Harris E (2017). Antibody-dependent enhancement of severe dengue disease in humans. Science 358, 929–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawiecki AB, and Christofferson RC (2016). Zika Virus-Induced Antibody Response Enhances Dengue Virus Serotype 2 Replication In Vitro. J. Infect. Dis 214, 1357–1360. [DOI] [PubMed] [Google Scholar]
- Koblischke M, Stiasny K, Aberle SW, Malafa S, Tsouchnikas G, Schwaiger J, Kundi M, Heinz FX, and Aberle JH (2018). Structural Influence on the Dominance of Virus-Specific CD4 T Cell Epitopes in Zika Virus Infection. Front. Immunol 9, 1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, and Diamond MS (2016). Zika Virus: New Clinical Syndromes and Its Emergence in the Western Hemisphere. J. Virol 90, 4864–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Tang J, Liu Z, Liu Y, Huang Y, Xu Y, Hao P, Yin Z, Zhong J, Ye L, et al. (2019). ZIKV infection induces robust Th1-like Tfh cell and long-term protective antibody responses in immunocompetent mice. Nat. Commun 10, 3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MQ, Kumaran EAP, Tan HC, Lye DC, Leo YS, Ooi EE, MacAry PA, Bertoletti A, and Rivino L (2018). Cross-Reactivity and Anti-viral Function of Dengue Capsid and NS3-Specific Memory T Cells Toward Zika Virus. Front. Immunol 9, 2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas CGO, Kitoko JZ, Ferreira FM, Suzart VG, Papa MP, Coelho SVA, Cavazzoni CB, Paula-Neto HA, Olsen PC, Iwasaki A, et al. (2018). Critical role of CD4+ T cells and IFNγ signaling in antibody-mediated resistance to Zika virus infection. Nat. Commun 9, 3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh SGE, Parham P, and Barber LD (1999). The HLA FactsBook (Academic Press; ). [Google Scholar]
- Ngono AE, and Shresta S (2018). Immune Response to Dengue and Zika. Annu. Rev. Immunol 36, 279–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagioti E, Klenerman P, Lee LN, van der Burg SH, and Arens R (2018). Features of Effective T Cell-Inducing Vaccines against Chronic Viral Infections. Front. Immunol 9, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin-Proulx D, Leal FE, Terrassani Silveira CG, Maestri A, Brockmeyer C, Kitchen SM, Cabido VD, Kallas EG, and Nixon DF (2017). T-cell Responses in Individuals Infected with Zika Virus and in Those Vaccinated Against Dengue Virus. Pathog. Immun 2, 274–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardy RD, Rajah MM, Condotta SA, Taylor NG, Sagan SM, and Richer MJ (2017). Analysis of the T Cell Response to Zika Virus and Identification of a Novel CD8+ T Cell Epitope in Immunocompetent Mice. PLoS Pathog. 13, e1006184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul LM, Carlin ER, Jenkins MM, Tan AL, Barcellona CM, Nicholson CO, Michael SF, and Isern S (2016). Dengue virus antibodies enhance Zika virus infection. Clin. Transl. Immunology 5, e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroso C, Fischer C, Feldmann M, Sarno M, Luz E, Moreira-Soto A, Cabral R, Netto EM, Brites C, Kummerer €BM, and Drexler JF (2019). Cross-Protection of Dengue Virus Infection against Congenital Zika Syndrome, Northeastern Brazil. Emerg. Infect. Dis 25, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, et al. (2016). Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc. Natl. Acad. Sci. USA 113, 7852–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regla-Nava JA, Elong Ngono A, Viramontes KM, Huynh AT, Wang YT, Nguyen AT, Salgado R, Mamidi A, Kim K, Diamond MS, and Shresta S (2018). Cross-reactive Dengue virus-specific CD8+ T cells protect against Zika virus during pregnancy. Nat. Commun 9, 3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CJ, Suleyman OM, Ortega-Prieto AM, Skelton JK, Bonnesoeur P, Blohm A, Carregaro V, Silva JS, James EA, Maillère B, et al. (2018). T cell immunity to Zika virus targets immunodominant epitopes that show cross-reactivity with other Flaviviruses. Sci. Rep 8, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Olsen PC, Costa F, Wang Q, Oliveira TY, Nery N Jr., Aromolaran A, do Rosário MS, Sacramento GA, Cruz JS, et al. (2019). Risk of Zika microcephaly correlates with features of maternal antibodies. J. Exp. Med 216, 2302–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Barraquer I, Costa F, Nascimento EJM, Nery N, Castanha PMS, Sacramento GA, Cruz J, Carvalho M, De Olivera D, Hagan JE, et al. (2019). Impact of preexisting dengue immunity on Zika virus emergence in a dengue endemic region. Science 363, 607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salek-Ardakani S, Flynn R, Arens R, Yagita H, Smith GL, Borst J, Schoenberger SP, and Croft M (2011). The TNFR family members OX40 and CD27 link viral virulence to protective T cell vaccines in mice. J. Clin. Invest 121, 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje H, Cummings DAT, Rodriguez-Barraquer I, Katzelnick LC, Lessler J, Klungthong C, Thaisomboonsuk B, Nisalak A, Weg A, Ellison D, et al. (2018). Reconstruction of antibody dynamics and infection histories to evaluate dengue risk. Nature 557, 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slon Campos JL, Poggianella M, Marchese S, Mossenta M, Rana J, Arnoldi F, Bestagno M, and Burrone OR (2017). DNA-immunisation with dengue virus E protein domains I/II, but not domain III, enhances Zika, West Nile and Yellow Fever virus infection. PLoS One 12, e0181734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom JA, Plante JA, Plante KS, Young EF, McGowan E, Gallichotte EN, Widman DG, Heise MT, de Silva AM, and Baric RS (2016). Dengue Virus Envelope Dimer Epitope Monoclonal Antibodies Isolated from Dengue Patients Are Protective against Zika Virus. MBio 7, e01123–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WW, Grewal R, and Shresta S (2015). Influence of antibodies and T cells on dengue disease outcome: insights from interferon receptor-deficient mouse models. Curr. Opin. Virol 13, 61–66. [DOI] [PubMed] [Google Scholar]
- Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, and Peters B (2019). The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 47, D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Yauch LE, Angelo MA, John DV, Greenbaum JA, Sidney J, Kolla RV, De Silva AD, de Silva AM, Grey H, et al. (2011). Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. J. Immunol 187, 4268–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, Broadwater A, Kolla RV, De Silva AD, de Silva AM, et al. (2013). Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA 110, E2046–E2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Angelo MA, Sidney J, Peters B, Shresta S, and Sette A (2014). Immunodominance changes as a function of the infecting dengue virus serotype and primary versus secondary infection. J. Virol 88, 11383–11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, de Silva AM, Crotty S, Peters B, and Sette A (2015a). Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc. Natl. Acad. Sci. USA 112, E4256–E4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Cerpas C, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, Sanches FP, Silvera CG, Costa PR, et al. (2015b). Human CD8+ T-Cell Responses Against the 4 Dengue Virus Serotypes Are Associated With Distinct Patterns of Protein Targets. J. Infect. Dis 212, 1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Angelo MA, Grifoni A, O’Rourke PH, Sidney J, Paul S, De Silva AD, Phillips E, Mallal S, Premawansa S, et al. (2016). HLA-DRB1 Alleles Are Associated With Different Magnitudes of Dengue Virus-Specific CD4+ T-Cell Responses. J. Infect. Dis 214, 1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, and Shresta S (2017). T Cell Immunity to Zika and Dengue Viral Infections. J. Interferon Cytokine Res 37, 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, and Shresta S (2019). Antigenic cross-reactivity between Zika and dengue viruses: is it time to develop a universal vaccine? Curr. Opin. Immunol 59, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Elong Ngono A, Regla-Nava JA, Kim K, Gorman MJ, Diamond MS, and Shresta S (2017a). Dengue virus-reactive CD8+ T cells mediate cross-protection against subsequent Zika virus challenge. Nat. Commun 8, 1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Tang WW, Sheets N, Ellison J, Sette A, Kim K, and Shresta S (2017b). Identification of Zika virus epitopes reveals immunodominant and protective roles for dengue virus cross-reactive CD8+ T cells. Nat. Microbiol 2, 17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, Prestwood TR, Sette A, and Shresta S (2009). A protective role for dengue virus-specific CD8+ T cells. J. Immunol 182, 4865–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch LE, Prestwood TR, May MM, Morar MM, Zellweger RM, Peters B, Sette A, and Shresta S (2010). CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J. Immunol 185, 5405–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, and Lin YL (2012). Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 8, e1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger RM, Tang WW, Eddy WE, King K, Sanchez MC, and Shresta S (2015). CD8+ T Cells Can Mediate Short-Term Protection against Heterotypic Dengue Virus Reinfection in Mice. J. Virol 89, 6494–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.


