Abstract
Background
Preferential accumulation of fat in the upper body (apple shape) is associated with higher risk of developing metabolic syndrome relative to lower body fat (pear shape). We previously discovered that chromatin openness partially defined the transcriptome of preadipocytes isolated from abdominal and gluteofemoral fat. However, the molecular mechanisms underlying interindividual variation in body shape are unknown.
Methods
Adipocyte fraction was isolated from abdominal and gluteofemoral fat biopsies of premenopausal women (age and body mass index matched) segregated initially only by their waist-to-hip ratio. We evaluated transcriptomic and chromatin accessibility using RNA sequencing and assay for transposase-accessible chromatin using sequencing (ATAC-seq) along with key clinical parameters.
Results
Our data showed that higher lower body fat mass was associated with better lipid profile and free fatty acid decrease after glucose administration. Lipid and glucose metabolic pathways genes were expressed at higher levels in gluteofemoral adipocyte fraction in pears, whereas genes associated with inflammation were higher both in abdominal and gluteofemoral apple adipocyte fraction. Gluteofemoral adipocyte chromatin from pear-shaped women contained a significantly higher number of differentially open ATAC-seq peaks relative to chromatin from the apple-shaped gluteofemoral adipocytes. In contrast, abdominal adipocyte chromatin openness showed few differences between apple- and pear-shaped women. We revealed a correlation between gene transcription and open chromatin at the proximity of the transcriptional start site of some of the differentially expressed genes.
Conclusions
Integration of data from all 3 approaches suggests that chromatin openness partially governs the transcriptome of gluteofemoral adipocytes and may be involved in the early metabolic syndrome predisposition associated with body shape.
Keywords: metabolic syndrome, fat distribution, adipocytes, gene expression, chromatin openness, gluteofemoral fat depot
Metabolic syndrome (MetS) is associated with a collection of several clinical phenotypes and is linked with increased risk of multiple chronic diseases, including type 2 diabetes (T2D), cancer, and cardiovascular disease [1, 2]. Central obesity, defined by an elevated waist circumference and often denoted as an “apple-shaped” body, is believed to be at the core of the syndrome [3, 4]. A major contributing factor is thought to be the presence of dysfunctional adipocytes, characterized by excess release of free fatty acids (FFAs) and inflammatory molecules. As a result, there is an ectopic accumulation of lipids in other major insulin target tissues (skeletal muscle, liver, and endothelial cells) that leads to global insulin resistance (IR) and systemic inflammation [5, 6]. Moreover, elevated FFA levels also inhibit insulin’s antilipolytic action on adipose tissue (AT), which will further increase the rate of FFA release into the circulation [7], resulting in a vicious cycle. According to this hypothesis, the visceral depot, which drains directly to the liver, plays a significant role and has been the focus of many studies. However, this depot accounts for only 2% to 3% of the total body fat in women [8] and contributes to no more than 15% of the total systemic FFA [7]. In contrast, the subcutaneous white adipose tissue (scWAT) is the largest fat depot in humans. The subcutaneous abdominal (ABD) fat is positively associated with risk factors of obesity-related complications [9] and has been implicated in the prevalence of MetS independently from the visceral fat mass [10, 11]. In contrast, individuals with lower-body obesity, characterized by preferential fat accumulation in the subcutaneous gluteofemoral (GF) depot and referred as a “pear-shaped” body, are at a lower risk for developing MetS [12, 13]. A recent integrative genomic analysis revealed 53 genomic loci strongly associated with IR, T2D, and coronary heart disease that were also related to lower hip circumference and reduced levels of gynoid and leg fat mass [14]. These correlation data suggest that an impaired GF fat expansion could be a significant determinant of diabetes and cardiovascular diseases [15].
Previous publications highlighted the differential tissue and cellular phenotypes existing between adipocytes isolated from subcutaneous ABD fat relative to lower-body fat depots [16, 17]; for example, adipocyte size, basal lipolysis, dietary fatty acid uptake, and insulin suppression of lipolysis vary according to the depot-specific origin of the fat analyzed. In addition, there are distinct gene expression signatures and differential CpG island methylation and histone marks in abdominal vs GF scWAT [9, 18-20], suggesting that there is an epigenomic code that underlies the known functional differences in scWAT depots. Still unexplored, however, are the molecular features and epigenetic signatures within the abdominal and GF-derived adipocyte that might differ between women with an “apple” vs “pear” body shape.
We hypothesized that differential chromatin openness plays an early role in AT deposition, before any appearance of metabolic complications, partially by regulating expression of body-shape–specific genes in the scWAT depots. To test this hypothesis, we isolated the adipocyte fraction from abdominal and GF scWAT biopsies obtained from groups of “apple” vs “pear” women segregated initially only by their waist-to-hip ratio (WHR) value. By using a combination of clinical assessments, RNA sequencing (RNA-seq) and assay for transposase-accessible chromatin using sequencing (ATAC-seq) technologies coupled with a comprehensive bioinformatic analysis, we unraveled the unique transcriptional signatures and open chromatin profiles of ABD- and GF-adipocyte fraction in apple- and pear-shaped women. Our studies reported here suggest that specific subcutaneous fat cells, identifiable by gene expression and chromatin openness, are part of distinct fat distribution and may account for early signs of MetS observed in otherwise healthy, apple-shaped, premenopausal women.
1. Materials and Methods
A. Clinical Study Design and Participant Details
Twenty-one healthy, eumenorrheic, premenopausal, weight-stable women age 20 to 40 years with a body mass index (BMI) between 24.8 and 33.9 kg/m2 were recruited in Orlando, Florida, using advertisements approved by an institutional review board. Participants were excluded if they reported a history of chronic disease (diabetes, heart or liver disease, high blood pressure, gastrointestinal disorder), recent weight loss or gain (> 3 kg over the past 12 weeks), had abnormal blood or urine values, or use of oral contraceptives or hormone replacement therapy. Participants were further categorized according to their WHR into 2 groups: apple-shaped (WHR > 0.85) and pear-shaped (WHR < 0.78). Detailed phenotyping of the participants included the following: anthropometric measures (weight, height, waist, hip, and thigh circumferences), fasting glucose, lipid, inflammation and hormone profiles, and body composition measured by dual‐energy x‐ray absorptiometry (DXA) using a GE Lunar iDXA whole‐body scanner. Volumetric measurement of fat and other organs (liver, kidneys, heart, brain, skeletal muscle, and intramuscular AT) was completed using a Philips Achieva 3T magnetic resonance imaging/multinuclear magnetic resonance spectroscopy. Resultant images were analyzed using Analyze software 11.0 (Biomedical Imaging Resource, Mayo Clinic).
AT biopsies were collected after overnight fasting with a 3-hole 2.5-mm liposuction cannula from the midabdomen approximately 5 to 8 cm lateral to the umbilicus. GF AT was collected 10 to 20 cm below the greater trochanter on the most lateral side of the upper thigh. Each sample was cleaned at bedside, a fraction of it was snap-frozen in liquid nitrogen, and the rest was immediately used for adipocytes isolation as described later. On a separate day, a standard 75 g frequently sampled intravenous glucose tolerance test (FSIVGTT) was performed as described in Parsons et al [21] and in Fig. 1B. Four times during the procedure, resting metabolic rate and respiratory quotient (RQ) were measured using a MAX‐II metabolic cart with a canopy attachment (AEI Technologies).
Figure 1.
Correlations between lower body fat mass and lipid parameters and frequently sampled intravenous glucose tolerance test (FSIVGTT data). A, Eleven apple- and 10 pear-shaped women were enrolled in the clinical trial. Correlations were established between the percentage of fat mass in the legs relative to total fat mass and circulating triglycerides levels, cholesterol/high-density lipoprotein (HDL), low-density lipoprotein (LDL)/HDL, percentage of liver fat determined by magnetic resonance spectroscopy, and visceral adipose tissue (VAT) mass estimated by dual‐energy X‐ray absorptiometry. B, FSIVGTT was performed and completed by 18 women in the initial group. The design of the test and the blood collection times are summarized at the arrows. The results of the minimal model method of Bergman are summarized in the tables under the graph. C-E, Respiratory quotient (RQ), glucose, and free fatty acid (FFA) levels during the FSIVGTT. C, RQ data were individually reported for each group and each time interval. *P less than .05 Wilcoxon test, RQ value compared to baseline value. D, Glucose concentrations for 5 hours after administration of glucose at time 0 and insulin at 20 minutes. For each time point, average data ± SD in each group were reported and plotted (red for apple and blue for pear). E, FFA concentrations (top graph) and percentage decrease of FFA (compared to baseline concentration—bottom graph) are represented. *P less than .05 repeated analysis of covariance (baseline as covariate).
B. Blood Analysis
Fasting blood samples were analyzed in clinical chemistry laboratory at either Adventhealth Orlando or onsite at the Translational Research Institute for Metabolism and Diabetes. Adiponectin was measured using the Human Adiponectin Kit (Meso Scale Discovery, catalog number K151BXC [22]) according to the manufacturer’s protocol.
Before and during the FSIVGTT, blood samples were collected on ice, centrifuged, aliquoted, and frozen at −80 °C until further analyses. Glucose was monitored during the procedure using a Nova StatStrip glucose meter (Nova Biomedical). FFAs were measured by the NEFA-HR (nonesterified fatty acid HR series) [2] assay (Fujifilm Wako Diagnostics) according to manufacturer’s protocol. Insulin and C-peptide were measured in undiluted human serum using the Human Insulin Kit (Meso Scale Discovery, catalog number K151BZC [23]) and the R-PLEX Human C-Peptide Antibody Set (Meso Scale Discovery, catalog number F213V [24]), respectively, according to the manufacturer’s protocol. The homeostasis model assessment of the IR index (HOMA‐IR) was calculated as follows: ([fasting insulin in mU/L] × [fasting glucose in mmol/L]/22.5). Data from the FSIVGTT were used to calculate the insulin sensitivity index, glucose effectiveness, acute insulin response to glucose, and the disposition index using the Minimal Model method of Bergman (MINMOD‐Millennium, R. Bergman) [25].
C. Isolation of Adipocytes
Ten participants (5 apples and 5 pears) of the initial cohort were randomly selected to perform adipocyte isolation and genome-wide analysis. Adipocytes were isolated by collagenase treatment of AT followed by low-speed centrifugation at 50 g for 2 minutes. Cells were washed twice with phosphate-buffered saline. A fraction of fat cell suspension was frozen and kept at –80 °C for future RNA analysis. Another fraction was immediately exploited to isolate chromatin as described as follows and to perform ATAC-seq.
D. RNA Sequencing
Total RNA was extracted from ABD- and GF-derived adipocytes using the GentleMACS Dissociator system (Milteny Biotec, GmbH) as described in Cirera [26].
Total RNA quality was assessed with the Agilent Bioanalyzer Nano (Agilent Technologies). A total of 1 μg of total RNA was used as starting material to construct RNA-seq libraries using the Illumina TruSeq Stranded Total RNA Library preparation kit (Illumina Inc) as per the instructions. First, total RNA was ribo-depleted to remove ribosomal from total RNA. The remaining nonribosomal RNA was fragmented using divalent cations at elevated temperature. Following fragmentation, first-strand complementary DNA was synthesized using random primers followed by second-strand synthesis using DNA polymerase I. Complementary DNA was ligated with index adapters for each sample followed by purification and enrichment by polymerase chain reaction (PCR) to create the final library. The quality and quantity of the libraries were analyzed using the Agilent Bioanalyzer and Kapa Biosystems quantitative PCR (Sigma-Aldrich). Multiplexed libraries were pooled, and single-end 50 base-pair sequencing was performed on one flow-cell of an Illumina HiSeq 2500.
E. RNA Sequencing Analysis
The RNA-seq samples’ quality was checked using the tool FastQC. After this step, a required trimming was executed reducing distortions on the “per base sequence content” at the beginning of the reads using Trimmomatic. The trimmed samples were aligned to the UCSC genome, assembly hg19, using STAR as described by Baruzzo and colleagues [27] in Supplementary Table 37. Reads were required to be uniquely mapped using Samtools for filtering, sorting, and indexing. RSeQC was used to assess the quality of each sample. The RNA sequencing abundance, raw read counts, and transcripts per million (TPM) values, were calculated with TPMCalculator using the default parameters. The differential gene expression analysis was executed using the Bioconductor packages Deseq2 and EdgeR. An adjusted P value of .05 and a logarithmic fold change of 1.5 was used to identified differentially expressed genes. All tools were executed using publicly available Common Workflows Language files and docker images with fixed tool versions [28].
The functional profiling of gene expression data was performed using DAVID (https://david.ncifcrf.gov/home.jsp). A P value threshold of .05 and enrichment score of 1 were used to identify KEGG biological pathways overrepresented in gene expression data.
F. Assay for Transposase-Accessible Chromatin Using Sequencing
ABD and GF adipocytes were used to isolate nuclei. Nuclei were used for tagmentation using Nextera DNA Library Preparation Kit (Illumina Inc). After tagmentation DNA was purified with a MinElute PCR Purification Kit (Qiagen). Tagmented DNA was amplified with a Kapa Hifi Hot Start Kit (Kapa Biosystems) using 16 PCR cycles. Amplified libraries were purified with Agencourt AMPure XP (Beckman Coulter). Fragment distribution of libraries was assessed with Agilent Bioanalyzer and libraries were sequenced on a HiSeq 2500 platform.
G. Assay for Transposase-Accessible Chromatin Using Sequencing Analysis
ATAC-seq data were analyzed as previously described [18] with the following modifications: From the BAM files, tag directories were created with the makeTagDirectory program of HOMER (v4.2), where the fragment length was set to 150 nucleotides [29]. Both peak calling and coverage files were created from the HOMER tag directories: “Peaks” or so-called nucleosome free regions were predicted with the findPeaks program by using the -style factor parameter.
Read distribution heat maps were generated by annotatePeaks of HOMER with -hist 50 and -ghist parameters and were normalized with the median value of the middle 50-bp bins.
Peaks were annotated by using the PeakAnnotator program to the nearest genes [30]. Genomic distribution of the peaks was defined also by the annotatePeaks.
H. DiffBind Analysis
Characterization of the differential open chromatin profile was performed with the DiffBind package in R (v3.4.4). Different consensus sets were generated based on the regions that were predicted from the ABD fat of each apple- or pear-shaped individuals and the same was performed for the GF fat of the apple- or pear-shaped individuals. The minOverlap parameter was set to the maximum 5 in all cases. Differential open chromatin regions that were used for the further analyses were determined according to a P value less than or equal to .05 or a P value less than or equal to .1 thresholds. Correlation heat maps with different P value settings were created using DiffBind. For volcano plots, significance and log fold-change values were also calculated by DiffBind.
I. Promoter Analysis
For promoter analysis, –1-kb to + 100-bp regions around transcriptional start site (TSSs) were used. Peaks overlapping with these extended regions were used to the further analysis. Reads per kilobase per million mapped reads (RPKM) values were calculated on the summit 100-bp regions according to the peak summits by using the coverageBed of BedTools program [31]. For box plots and RPKM-based heat maps, RPKM values were decile normalized before plotting. For this, a consensus set was generated with those peaks that could be predicted from at least 2 samples of interest, then RPKM values were calculated on the consensus set from each sample. The values, corresponding to the first deciles, were used to the division of the initial RPKM values.
J. Intergenic Analysis
DiffBind analysis was performed with the ATAC-seq signals calculated around a ± 100 kb genomic region relative to the TSSs of the body shape–specific genes. The minOverlap parameter was set to 3. Differential open chromatin regions were determined according to a P value of less than or equal to .05.
K. Motif Analysis
Motif enrichment analysis was carried out by the findMotifGenome of HOMER and was performed on the summit 200-bp regions of the differential open regions from which the promoter-TSS regions were excluded [29]. The targeted motif lengths were 8, 10, 10, and 14 bp. P values were calculated by comparing the enrichment within the target regions and that of a random set of regions (background) generated by HOMER.
For motif distribution plot, motif matrices were downloaded from the HOMER Motif Database and were mapped in 20-bp windows within 2-kb frames relative to the summits of the differential open regions using annotatePeaks with -mbed parameter.
L. Visualization
Read distribution and motif distribution heat maps were visualized by Java TreeView (v1.1.6r4). RPKM-based heat maps were plotted in R by using the pheatmap package and applying the row-wise scaling (rows were scaled to have mean zero and SD of one) to remove the scale effect of the variables.
Box plots and Volcano plots were plotted by using GraphPad Prism 8.
M. Immunohistochemistry Staining of Adipose Tissue
Samples fixed in Z-fix were dehydrated, paraffin embedded, and sectioned (thin sections of 5 µm). Immunohistochemical detection of CD68 (Atlas Antibodies catalog number AMAb90873 [32]) for macrophages and CD4 (Cell Marque Cat#104R-14 [33]) for T cells was performed with the avidin-biotin peroxidase method. The presence of CD68-positive cells was evaluated on the entire section of each participant and categorized into 3 groups: no staining, positive cells without specific organization, and positive cells organized in a crown-like structure (CLS).
N. Statistics
All statistical analyses on clinical data were performed in SAS (V9.4) and statistical significance was set at P less than .05.
We used a repeated-measures analysis of covariance with basal FFA as covariates (SAS PROC MIXED) to compare the mean levels of FFA between the pear-shaped and the apple-shaped women at different infusion phases. The model included shape, time, and shape by time interaction as fixed effects.
Unpaired t tests were used to compare apple vs pear chromatin openness. Paired t tests were used to compare ABD fat vs GF fat chromatin openness.
O. Data Availability
All sequencing data have been deposited to the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE143450.
P. Study Approval
All participants provided written informed consent. All procedures were performed under a research protocol approved by the Florida Hospital Institutional Review Board and was registered March 24 2016, at clinicaltrials.gov, NCT02728635 https://clinicaltrials.gov/ct2/show/NCT02728635.
2. Results
A. Clinical Characteristics of Study Participants
The characteristics of the study groups are presented in Table 1. Groups were matched both for BMI and age. By design, the average WHR was higher in the apple group than the pear group. This difference in ratio was mainly due to higher waist circumference in apples (P = .001), who also had higher android fat mass and visceral adipose tissue (VAT) mass (P = .01 and .01, respectively). The hip circumference, thigh circumference, and gynoid fat mass were indistinguishable between the groups (Table 1). We determined the average adipocyte diameter on histological section (Table 1). Importantly, among the 4 depots studied the biggest adipocytes were found in the GF depot of the pear-shaped women (74.8 µm). Previous studies showed that the GF depot have bigger adipocytes compared to the ABD depot. In our study this was confirmed only in pear-shaped women (ABD adipocytes = 64.2 µm, GF adipocytes = 74.8 µm); the apple-shaped women presented with similar adipocyte size in both depots (ABD adipocytes = 66.7 µm, GF adipocytes = 67.6 µm). We found an intriguing correlation between the size of the adipocyte in GF depot and the level of circulating potassium (Spearman correlation, P = .024, R = 0.50). Several studies have shown that higher potassium could alleviate obesity and MetS risk [34]; this topic is, however, still controversial and needs additional studies to confirm.
Table 1.
Clinical and biochemical characteristics of the study’s 2 groups of women
| Clinical parameters | Pear-shaped participants (n = 10) | Apple-shaped participants (n = 11) | P |
|---|---|---|---|
| Adiposity markers | |||
| BMI, kg/m2 | 27.4 ± 3.00 | 28.4 ± 3.57 | .54 |
| Weight, kg | 73 ± 9.3 | 76 ± 11 | .64 |
| Waist circumference, cm | 79.8 ± 6.16 | 93.5 ± 8.95 | .001 |
| Hip circumference, cm | 108.2 ± 7.9 | 106.4 ± 10.9 | .84 |
| Thigh circumference, cm | 60.1 ± 4.4 | 59.8 ± 5.1 | > .9 |
| Waist-to-hip ratio | 0.74 ± 0.04 | 0.88 ± 0.03 | < .001 |
| Total FM, kg | 28.2 ± 7.5 | 33.2 ± 9.2 | .25 |
| Total lean mass, kg | 43.1 ± 5.0 | 40.5 ± 3.4 | .22 |
| Leg lean mass, kg | 15.6 ± 2.1 | 14.2 ± 1.6 | .15 |
| FM, % | 39.1 ± 7.1 | 44.3 ± 6.3 | .11 |
| Android FM, kg | 1.7 ± 0.7 | 2.9 ± 1.0 | .01 |
| Gynoid FM, kg | 5.6 ± 1.2 | 5.9 ± 1.8 | > .9 |
| Fat leg/total FM, % | 45 ± 4.9 | 36 ± 4.8 | .001 |
| Circulating adiponectin, µg/mL | 12.3 ± 5.3 | 12.1 ± 5.4 | > .9 |
| VAT mass, g | 333 ± 258 | 731 ± 381 | .01 |
| Liver fat, % | 1.5 ± 1.0 | 2.5 ± 1.8 | .19 |
| IMAT, cm3 | 251 ± 91 | 289 ± 101 | > .9 |
| ABD_adipocyte diameter, µm | 64.2 ± 8.94 | 66.7 ± 8.10 | .52 |
| GF_adipocyte diameter, µm | 74.8 ± 6.59 | 67.6 ± 5.29 | .02 |
| Lipid profile | |||
| HDL, mg/dL | 67.8 ± 19.4 | 61.2 ± 17.0 | .77 |
| LDL, mg/dL | 96 ± 19.0 | 110 ± 34.7 | .54 |
| VLDL, mg/dL | 13.9 ± 6.47 | 17.8 ± 5.05 | .02 |
| TGL, mg/dL | 68.9 ± 31.8 | 89.0 ± 25.0 | .02 |
| Chol, mg/dL | 178 ± 28.2 | 189 ± 37.6 | .72 |
| Non-HDL chol, mg/dL | 110 ± 22.9 | 128 ± 36.4 | .32 |
| Chol/HDL | 2.73 ± 0.60 | 3.26 ± 0.88 | .15 |
| LDL/HDL | 1.48 ± 0.45 | 1.95 ± 0.77 | .12 |
| FFA, µmol/L | 567 ± 251 | 482 ± 145 | .40 |
| Glucose profile | |||
| Fasting glucose, mg/dL | 85.3 ± 7.30 | 91.4 ± 6.52 | .06 |
| Fasting insulin, µ(iU)/mL | 6.16 ± 3.67 | 7.93 ± 3.25 | .21 |
| HOMA-IR | 1.05 ± 0.66 | 1.56 ± 0.76 | .14 |
| HbA1c, % | 5.18 ± 0.41 | 5.17 ± 0.24 | .88 |
| C-peptide, ng/mL | 1.04 ± 0.46 | 1.30 ± 0.39 | .27 |
| Metabolic markers/hormones | |||
| Age, y | 33.4 ± 7.1 | 34.4 ± 6.4 | .90 |
| ΔRQ | 0.087 ± 0.089 | 0.055 ± 0.055 | .08 |
| RMR, kcal/min | 0.95 ± 0.10 | 1.00 ± 0.12 | > .9 |
| Testosterone, ng/dL | 28.5 ± 19.3 | 27.0 ± 14.2 | > .9 |
| CRP, mg/L | 3.22 ± 3.37 | 3.17 ± 1.63 | .56 |
Mean ± SD. Nonparametric Mann-Whitney test P less than or equal to .05.
Abbreviations: ABD, abdominal; BMI, body mass index; Chol, cholesterol; CRP, C-reactive protein; FFA, free fatty acids; FM, fat mass; GF, gluteofemoral; HbA1c, hemoglobin A1c; HDL, high-density lipoprotein; HOMA-IR, homeostatic model assessment of insulin resistance; IMAT, intramuscular adipose tissue; LDL, low-density lipoprotein; RMR, resting metabolic rate; RQ, respiratory quotient; TGL, triglycerides; VAT, visceral adipose tissue; VLDL, very low-density lipoprotein.
As expected, the apple-shaped women showed signs of dyslipidemia, reflected by significant higher very low-density lipoprotein and triglycerides (TGs) circulating levels (P = .02 for both, apples vs pears comparison), and trend to increase the ratios of low-density lipoprotein (LDL) to high-density lipoprotein (HDL) and cholesterol to HDL (P = .12, .15, respectively, apples vs pears comparison) (Table 1). They also presented with features associated with a trend toward IR including higher serum fasting glucose and HOMA-IR (P = .06, .14, respectively, apples vs pears comparison) (Table 1). In addition, apple-shaped participants tended to accumulate more lipid in the nonfat tissues such as liver and muscle (Table 1). We addressed the relative contribution of lower- and upper-body fat depot in the variation of lipid profiles by studying the correlations between the ratio leg fat divided by total fat mass (data measured by DXA) and the lipid parameters collected in the blood. The ratio fat leg/total fat mass correlated negatively with general markers of lipid metabolism, including circulating TGs, cholesterol/HDL, and LDL/HDL, as well as VAT mass and percentage of liver fat measured by magnetic resonance spectroscopy (Fig. 1A). These data confirmed that preferential fat accumulation in the lower body compared to the upper body decreased the risk of dyslipidemia and prevent accumulation of lipids in nonfat organs [35].
B. Increased Insulin Sensitivity and Decreased Free Fatty Acid Availability After Glucose Challenge Characterize Pear-Shaped Over Apple-Shaped Individuals
To directly compare insulin sensitivity between the 2 groups of women, we performed an intensive glucose tolerance test, whereby we repeatedly sampled circulating glucose and insulin over a 5-hour time window (FSIVGTT) with minimal model analysis (Fig. 1B). The pear-shaped individuals trended toward a greater insulin sensitivity index compared to the apple-shaped group (P = .10), with no difference detected in the glucose effectiveness (insulin-independent component), the acute insulin response to glucose, or the disposition index (product of insulin sensitivity and first-phase insulin secretion) (Fig. 1B).
IR is often associated with metabolic inflexibility, meaning impaired switching of substrate oxidation from fatty acids to glucose in response to insulin. The scWAT plays a role in these phenomena by being the major site of energy storage of the body [36]. WAT stores energy in the form of TGs when in positive energy balance and releases energy as FFAs when energy expenditure exceeds energy intake. To study the influence of differential fat distribution on metabolic flexibility variation, we first measured fasting RQ by indirect calorimetry in the 2 groups of women. Postabsorptive RQ was lower in pear-shaped individuals (limit of significance—Fig. 1C) consistent with a higher basal fat oxidation level. However, systemic RQ increased similarly after 40 minutes in both groups, as expected from the repeated glucose monitoring (Fig. 1D). Likewise, RQ similarly retreated to the fasting RQ level at the end of the FSIVGTT (Fig. 1C).
We next measured FFA levels in the blood collected during the frequently sampled IVGTT. After administering glucose, the decrease in circulating FFA during the first phase (10 to 20 minutes) was more prominent in pear-shaped compared to apple-shaped women (Fig. 1E). However, in the second phase (20 to 30 minutes), the insulin fully suppressed FFA release similarly in both groups (Fig. 1E), reflecting an equal late antilipolytic effect of insulin independently of body shape. Two mechanisms could explain the difference observed during the first phase of the test: an increase of FA esterification to sequester them as TGs in their lipid droplet and/or a better clearance of the FFA by other organs (skeletal muscle for example) via uptake and oxidation. Our RQ data suggest no difference in FFA clearance between the 2 groups of women, which suggests that differential FA accumulation in adipocytes contributes to the difference.
The correlations established between the preferential fat accumulated in the leg and the lipid parameters and the fact that the 2 groups of women were segregated only by their WHR pinpointed the AT depots as a key player in the clinical differences described.
C. Gene Expression Comparison Across the 2 Body Shapes and the 2 Subcutaneous Fat Depots
Next, we evaluated a potential molecular basis for the differential fat accumulation observed between the 2 groups. We interrogated the gene expression signatures of collagenase-isolated ABD and GF-adipocytes from 5 apple- and 5 pear-shaped participants using RNA-seq. Their clinical characteristics are summarized in [28] and are very similar to the total of 21 apple- and pear-shaped women included in our study.
Differential analysis of gene expression between ABD and GF adipocyte fraction revealed 487 genes dysregulated in apple-shaped women and 74 genes dysregulated in pear-shaped women. In both groups, more genes are upregulated in GF compared to ABD cells (318 vs 169 in apples –52 vs 22 in pears [28]). The GF transcriptome seems to be more flexible or more sensitive to regulation than the ABD genes. We identified only 14 and 34 genes upregulated respectively in ABD and in GF fat cells simultaneously in apple- and pear-shaped women [28], which is not surprising based on the small number of differentially expressed genes in pear-shaped women. Consistent with our previous report and those of other groups [9, 20, 37], developmental genes (HOXA5, HOXA6, HOXB7 and HOXC10) and the lncRNA HOTAIR were found to be depot specific [28].
We next compared the gene expression profile between the adipocyte fraction from apple- and pear-shaped women. We named the genes upregulated in apple-shaped women “apple specific” and the genes up-regulated in pear-shaped women “pear specific.” A stringent analysis (cutoff fold change > 1.5 and P value < .05 according to 2 statistic models as described in section 2, “Materials and Methods”), revealed 95 body-shape specific genes in abdominal cells (Fig. 2A) and 4 times more (n = 428) body-shape–specific genes in GF cells (Fig. 2B). These data suggest that the GF cells seem to be more affected by the differential fat distribution. In both depots, around twice the number of genes were upregulated in apple- compared to pear-shaped individuals (Fig. 2A and 2B).
Figure 2.
Body shape-associated transcriptomic signatures of abdominal (ABD) and gluteofemoral (GF) fat depots. A and B, Heat map representation of the significantly changing genes (false discovery rate < 0.05) determined by RNA sequencing from the A, ABD and B, GF depots of 5 apple- and 5 pear-shaped women. The top signature genes of each group in each depot are listed. C, KEGG and GOTERM functional analysis of pear-specific gene set in GF depot. Chr indicates chromosome; FC, fold change pear/apple. When a gene was found in different pathways, all pathways are mentioned in order of P value.
D. Metabolic Pathways Are Upregulated in the Gluteofemoral Depot of Pear-Shaped Participants
We further analyzed the GF genes dysregulated in apple- compared to pear-shaped women’s isolated adipocytes and used a functional annotation tool to regroup them in biological pathways. Pathway enrichment analysis of the 123 genes preferentially expressed in pear-shaped women indicated that they regrouped in processes needed for adipocyte lipid storage, including cellular amino acid biosynthetic processes, fatty acid metabolism, general metabolic process, IR, and regulation of lipolysis (Fig. 2C). The genes involved in the 9 most significant pathways are listed in Fig. 2C with their corresponding fold change between pear- and apple-shaped women. Of particular interest, FASN and APOB, key regulators of fatty acid metabolism, showed high fold change between the 2 groups of participants (Fig. 2C). PCK1 (fold change = 2) is involved in lipid storage and fatty acid esterification. Its increase in pear-shaped GF cells is in favor of a greater fatty acid re-esterification in these women as suggested by our FSIGTT data. In addition, the ADRB3 gene, coding for the antilipolytic adrenergic receptor b3, was elevated in pear adipocytes (fold change = 10, Fig. 2C) suggesting a lower level of lipolysis in adipocytes isolated from apple-shaped women [38].
E. Inflammatory Pathways Are Upregulated in the Gluteofemoral Depot of Apple-Shaped Individuals
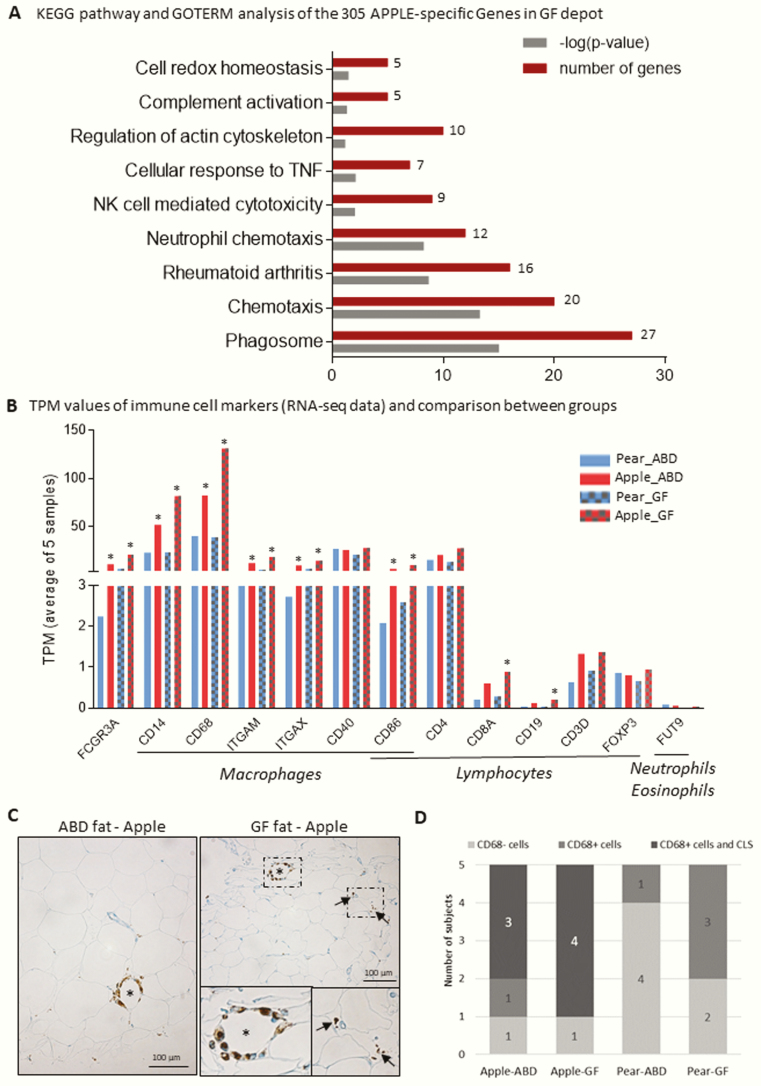
We identified 61 and 305 apple-selective genes in the ABD- and GF-derived cells, respectively (Fig. 2A and 2B). Pathway enrichment analysis of the 305 genes preferentially expressed in apple-shaped women in GF cells revealed that one-third of them were related to an inflammatory and immune response (Fig. 3A). Interestingly, a number of genes were involved in immune cell chemotaxis, suggesting an elevation of inflammatory cells inside the scWAT of apple- compared to pear-shaped women. Sixteen percent of the dysregulated genes in the apple GF depot were related to the extracellular matrix (including TIMP1 and MFAP5, 2 molecules involved in human AT inflammation and IR [39]), phagosome formation, and oxidative stress. These 3 pathways are associated with metabolic complications of obesity and may also be related to an increase of immune cells within apple-shaped women’s scWAT. Genes involved in inflammatory pathways also represented the most predominant signature in the apple ABD depot. However, a comparison of the ABD- and GF-enriched gene sets within the apple-shaped individuals revealed higher expression of inflammatory genes in the GF depot (data not shown).
Figure 3.
Inflammatory signature of abdominal and gluteofemoral adipocyte and fat depots of apple-shaped women. A, KEGG and GOTERM functional analysis of apple-specific gene set in gluteofemoral (GF) depot. B, Average of transcripts per million (TPM) values of immune cell markers in each group of participants and depots from the RNA-sequencing data. *P less than .05 nonparametric t test of pear vs apple individuals. C and D, Identification of CD68-positive cells in abdominal and GF white adipose tissue (WAT) section. C, Positive staining is revealed in apple women with DAB (3,3′-diaminobenzidine) system (brown staining). Nuclei were stained with hematoxylin (blue staining). The star shows positive cells organized in a crown-like structure (CLS) around an adipocyte; the arrows show positive cells without specific organization. D, A score was attributed to each individual as described in “Materials and Methods.” The histogram depicts the score repartition by participant in each group (5 women/group).
Some genes identified as apple specific in both depots were markers of macrophages, B cells, T cells, and other immune cells. Because the adipocyte fraction from an AT digestion could contain adherent stromal cells or macrophages, we cannot exclude the contribution of myeloid cells in the gene expression patterns observed. We looked closer at our RNA-seq data at some specific markers of these cell populations and reported their average TPM values in each subgroup (ABD and GF cells from apple- and pear-shaped participants—Fig. 3B). Gene expression of the general marker of myeloid cells (FCGR3A) was increased in apple-shaped women in both depots, however with relatively low average TPM values reported relative to the other immune function genes. There was a significant increase of macrophage marker gene expression in apple compared to pear individuals in both depots (CD14, CD68, ITGAM, and ITGAX). Two markers of lymphocytes (CD8A and CD19) were also reported to be significantly higher in apple participants, but only in GF cells (Fig. 3B). All together these data point out an infiltration by nonfat cells in the adipocyte fraction preparation that was more pronounced in both depots from apple-shaped women. However, the high TPM values described for CD14, CD68, CD40, and CD86 (Fig. 3B) and the recent publications showing these proteins are expressed in the adipocyte fraction of human AT [40-42] suggest that the increase in expression we observe here is directly from adipocytes.
F. Presence of Macrophages Organized in a Crown-Like Structure in the Subcutaneous Adipose Tissue of Apple-Shaped Women
We wanted to further assess myeloid cell infiltration within the AT of the 10 individuals we previously studied for the RNA-seq analysis. Their AT sections were stained with CD68 and CD4 (markers respectively of macrophages and lymphocytes identified in our RNA-seq data with high read count numbers—Fig. 3B) and observed by histology. We did not detect CD4-positive T cells in any depots or participants, suggesting no or very little scWAT infiltration by T lymphocytes in both groups of women. In contrast, CD68 staining revealed the presence of macrophages in the scWAT of the individuals tested. Remarkably, the macrophages observed in the adipose samples of apple-shaped women were found in a CLS pattern surrounding adipocytes (Fig. 3C). Interestingly, no CLS were visualized in the pear-shaped women’s scWAT, whereas 7 of the 10 apple women’s sections presented those specific structures (Fig. 3D).
G. Distinct Chromatin Accessibility Between Apple- and Pear-Shaped Subjects Is Observed in Gluteofemoral Fat
Changes in chromatin structure have been associated with alterations in gene expression linked to obesity, T2D, MetS, and cardiovascular diseases (reviewed in Carson and Lawson [43]). Notably, hyperglycemia and excess dietary fat consumption alter DNA methylation and chromatin accessibility around genes involved in glucose regulation and fat metabolism [44]. Comparative ATAC-seq profiling has been developed to identify differentially open chromatin regions [45]. Here we applied this technology to the ABD- and GF-derived adipocyte fraction samples in our 5-member apple- and 5-member pear-shaped groups to compare their genome-wide chromatin accessibility and to assess the link between body-shape–specific chromatin availability, differential gene expression, and putative metabolic risk factors. We performed the analysis independently in ABD and GF depots. All the quality control analyses for the ATAC-seq samples showed they were comparable and yielded similar overall read densities suggesting all samples were of comparable technical quality [28].
To identify differentially open chromatin regions, we determined the consensus peak set in our adipocyte fraction samples for each depot and each group of women. To avoid the person-to-person variability within apple- and pear-shaped participant groups, only the common open chromatin regions were considered in all 5 individuals. Apple-shaped women have 11 301 and 12 404 peaks in the ABD and GF depot, respectively, whereas pear-shaped women have 11 054 and 20 114 peaks in the ABD and GF depot, respectively. We used these consensus sets to determine the differentially accessible genomic regions. Differential analysis of the ATAC-seq data and hierarchical clustering revealed a strong correlation between individual apple and pear samples in the GF depot, but there was not a clear separation between the 2 body types for the peaks from the ABD depot [28]. This finding suggests there is a distinct difference in chromatin organization in lower-body adipocytes but not in ABD adipocytes. Interestingly, we found a strong correlation between the number of consensus peaks and the average adipocyte size per group (R2 = 0.95).
By using a P less than .05 cutoff, we identified more open chromatin regions in pear-shaped individuals compared to apple-shaped individuals in both depots (Fig. 4A and 4B). Consistent with our previous RNA-seq data, an extremely small number of ATAC-seq peaks were identified as body shape-specific in the ABD cells (n = 19 in total) (Fig. 4A), whereas 159 apple-specific and 2296 pear-specific open chromatin regions were observed in the GF cells (Fig. 4B). This finding suggests there is a distinct body composition–related difference in chromatin organization in lower-body adipocytes but not in ABD adipocytes.
Figure 4.
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) analysis revealed apple- and pear-specific open chromatin regions in the gluteofemoral (GF) depot rather than in the abdominal (ABD) depot. A and B, Volcano plot representation of apple- (highlighted in red) and pear-specific (highlighted in blue) open chromatin regions in adipocyte fraction isolated from A, ABD and B, GF depot (P < .05). C and D, HOMER was used to annotate each identified open chromatin region to the closest gene using the hg19 reference genome. Open chromatin regions that could be annotated to differentially expressed genes (DEGs) are represented on the volcano: Open chromatin regions linked to apple- and pear-specific DEGs are highlighted in red and blue respectively in C, ABD and D, GF depots. Open chromatin regions that could not be linked to DEGs are represented in gray. E, Integrative Genomic Viewer (IGV) snapshot of ATAC-seq coverage in the GF depot representing 10 differentially opened chromatin regions (P < .05) between apple- and pear-shaped individuals associated with DEGs (false discovery rate < 0.05). The more accessible chromatin regions in apples are highlighted in red; more accessible regions in pears are highlighted in blue.
Genome-wide distribution of ATAC-seq peaks revealed that a large majority of the open chromatin regions localized to the promoter-TSS (60% in the abdominal depot and 44% in the GF depot) [28]; however, the body-shape–specific peaks in the GF depot rather localized in intronic and intergenic regions [28]. It suggests that the direct regulation of the genes through the promoter elements are less variable between the investigated depots; they are regulated more likely by body-shape–specific enhancer elements driven possibly by a different set of coregulatory proteins.
H. Chromatin Accessibility Associates With Gene Expression
To examine the correlation between depot-specific open chromatin regions from the ATAC-seq analysis with depot-specific gene expression signatures, we integrated our ATAC-seq and RNA-seq data sets. First, we interrogated the full list of body-shape–specific open chromatin regions annotated to their closest gene (using PeakAnnotator). As shown in Fig. 4C for the ABD depot and Fig. 4D for the GF depot, only a small fraction of the body-shape–specific open chromatin regions was annotated to differentially expressed genes. These observations suggest that body-shape–specific chromatin structure is weakly correlated with the expression of their nearest neighboring genes. Interestingly, 191 pear-specific open chromatin regions were annotated to apple-specific genes (Fig. 4D). In other words, those regions were more open in the pear-shaped chromatin but were associated with lower transcription. These ATAC-seq peaks may represent sites of transcriptional repressor binding, or they may regulate genes that are located far from these sites. Fig. 4E depicts some examples of open-region chromatin detected by ATAC-seq at the proximity of 10 differentially expressed genes between apple- and pear-shaped participants.
To further understand how chromatin openness correlates with differential gene expression in our GF samples, we focused on the ATAC-seq signals at the proximal promoters (1000 bp upstream and 100 bp downstream from the TSSs) of the differentially expressed genes. We determined the normalized read counts on the promoter regions of the 305 apple-specific genes. This analysis revealed significantly higher chromatin accessibility in the promoters of the apple-specific genes in the chromatin isolated from apple women compared to the chromatin originating from the pear women (Fig. 5A heat map and top graph). Conversely, read counts were higher within the promoters of the 123 pear-specific genes in the pear-women chromatin compared to the apple-women chromatin (Fig. 5A, heat map and bottom graph). To investigate the relationship between chromatin openness and gene expression in a larger genomic region, we decided to extend our analysis around the TSSs of the depot-specific genes (±100 kb of TSS). Around the apple-specific genes, we identified 107 apple-dominant open chromatin regions (linked to 84 genes) (Fig. 5B top graph). On the other hand, 96 pear-dominant open chromatin regions were observed (owned by 52 pear-specific genes) (Fig. 5B bottom graph). Surprisingly, an additional 70 pear-dominant open chromatin regions (linked to 54 genes) were related to apple-specific gene expression (Fig. 5B top graph), potentially revealing sites of transcriptional repressor binding of apple-specific genes in pear-shaped women in the GF depot.
Figure 5.
Integration of RNA sequencing and assay for transposase-accessible chromatin using sequencing (ATAC-seq) data. A, Promoter analysis of differentially expressed genes (DEGs) in gluteofemoral (GF) from apple- (305 genes) and pear-shaped (123 genes) participants based on ATAC-seq signals represented as heat map and boxplots, RPKM indicates reads per kilobase per million). ***P less than .001 and *P less than .05 unpaired t test. B, Volcano plot representation of differentially open intergenic regulatory regions that could be linked to apple- and pear-specific differentially expressed genes, respectively. Apple-specific open chromatin regions are highlighted in red, whereas pear-specific open chromatin regions are highlighted in blue.
To compare the chromatin accessibility between ABD and GF chromatin, we integrated and performed bioinformatic analyses on RNA-seq and ATAC-seq data from adipocyte fractions isolated from both depots. Correlation analysis of the ATAC-seq data revealed a segregation by anatomical location in the 2 groups of women [28]. Notably, there was a higher number of ATAC-seq peaks in GF compared to the ABD depot [28]. We then compared the normalized read counts at the proximity of the depot-specific gene promoters. We found that the ABD chromatin was more open in the promoter regions of ABD-specific genes, and the GF chromatin was more opened in the promoter regions of the GF-specific genes [28].
I. Motif Analysis of Pear- and Apple-Specific Open Chromatin Regions in Gluteofemoral Fat
We next performed de novo motif analyses on the shape-specific regulatory elements identified in GF cells (2296 and 159 peaks).
On one hand, the apple-specific sites showed significant enrichment for the activator protein transcription factor, which is known to be involved in early adipocyte commitment.
On the other hand, pear-specific ATAC-seq peaks exhibited strong enrichment for C/EBP, CTCF, NF1, EBF, STAT, USF, and NR half sites (Fig. 6A). All these factors are known to contribute to adipogenesis or adipocyte lipid accumulation (Fig. 6A for detailed references). Interestingly, the NR half sites are associated with nuclear receptor binding, which are key regulators of metabolism and required for adipocyte differentiation. Additionally, they are important for regulation of insulin sensitivity, lipogenesis, and adipocyte survival and function [46]. The expression of all the members of the nuclear receptor superfamily in the apple- and pear-shaped women are listed in [28]. Out of the nuclear receptors, the half-site—which is enriched within the 2296 regions—can be bound by almost any of them. Interestingly peroxisome proliferator-activated receptor γ showed the highest expression both in ABD and GF cells [28].
Figure 6.
Motif analysis of differential open regions in gluteofemoral (GF) fat depot. A, Motif analysis on the 159 apple-specific open chromatin regions and 2296 pear-specific open chromatin regions in GF fat depot. Enriched motif matrices are presented along with the rank in the analysis, the P value, the percentages of each motif found in the target (Target%), background (Bg%) genomic regions, the reference revealing the implication of the transcriptional factor associated with the motif in adipogenesis and triglycerides storage. B, Read distribution plot of assay for transposase-accessible chromatin using sequencing signals for apple- and pear-specific open chromatin regions in adipocytes isolated from GF depot (P < .05). Left panel represents around ± 200 bp of the summit of the peak. Right panel, Motif distribution plot of the enriched transcription factor motifs from the table described in A. Pear-specific open chromatin regions are sorted into 4 groups containing an equal number of genomic regions based on the fold-change difference they show starting from the smallest (group I) to the highest (group IV). The number of transcription factor motifs in each group are presented as a heat map.
We were intrigued that several transcription factor motifs showed enrichment in the pear-specific open chromatin regions. Therefore, we looked closer at these regions, hoping to be able to predict/define the relative importance of these motifs in the establishment of open chromatin. First, we ordered these genomic regions by the magnitude of peak intensity compared to the corresponding region in apple chromatin (fold change). Second, we split the peaks into 4 equal groups (quartiles), then we analyzed the significantly enriched motifs in each quartile (Fig. 6B). The adipogenic transcription factor motifs (C/EBP, NF1, EBF, and STAT), usually associated with enhancer regions, showed a higher enrichment in the open chromatin regions with the highest read density (third and fourth quartiles) (Fig. 6B, heat map), suggesting that the corresponding transcription factors are important in regulating chromatin openness. CCCTC binding factor (CTCF) motifs were preferentially found in the less open chromatin regions, or the first quartile (Fig. 6B, heat map). Given the role of CTCF in regulating higher-order chromatin structure, these results indicate that the 3-dimensional chromatin architecture of adipocytes might be also affected. This observation merits further investigation.
3. Discussion
Much has been learned about the metabolic profiles of apple- and pear-shaped body types and the biology of their abdominal WAT (subcutaneous and visceral). However, the mechanisms that lead to different fat distribution remain unclear and the differential molecular characteristics of the upper- and lower-body adipocytes have not been thoroughly investigated. In this study, we used anthropometric and DXA measurements to segregate 2 different groups of relatively healthy premenopausal women characterized by similar BMI but with clear separation of “apple” vs “pear” fat deposition.
Although the women included in our study are relatively young and we excluded any individuals with T2D or impaired fasting glucose, the pear-shaped women already demonstrated, in comparison to apple-shaped women, better metabolic profiles reflected by a healthier insulin response, reduced dyslipidemia markers, and lower circulating FFA after a glucose challenge. Notably, the pear-shaped women also have a higher RQ compared to apple-shaped women (Fig. 1C—P = .11). This difference could be explained by the higher leg lean mass (Table 1—P = .15) observed in the pear-shaped women. As it was previously reported, preferential accumulation of fat in the leg observed in the pear-shaped women negatively correlates with circulating lipid levels [36]. Importantly, since both groups showed similar resting metabolic rates, the differences observed suggest there is a primary adipocyte biology dysregulation in apple-shaped women rather than a defect in peripheral organ metabolism. Moreover, the percentage of lower fat is negatively correlated to ectopic fat accumulation, suggesting its protective role against unhealthy lipid apparition in other organs.
It is noteworthy that some of the mentioned and some additional comparisons provided in [28] trended toward a more unhealthy profile in the apple group, but they did not reach the statistical threshold of .05, probably because of the limited sample size and the age of the cohort. Notably, our inclusion and exclusion criteria ensured that all the women in our study were in general good health, of a relatively young age, and before overt MetS features develop. It should also be noted that an independent influence of VAT on lipid and glucose metabolism could not be excluded, because the VAT mass was higher in apple-shaped women [28]. Additionally, elevated circulating adiponectin, described in women with higher fat leg mass [47, 48] is potentially related to better insulin sensitivity. However, there was no significant difference in circulating FFA or adiponectin between the 2 groups in our study [28]. Therefore, the effects observed here occur before differences in FFA and adiponectin emerge and are thus independent of systemic FFA and adiponectin levels.
Transcriptomic and ATAC-seq analyses of the adipocyte fraction derived from the ABD and GF fat depots of apple- and pear-shaped participants allowed us to pinpoint the main differences between these groups at the gene expression and chromatin structure level. Genes involved in fatty acid metabolism and regulation of lipolysis are upregulated in GF adipocytes isolated from apple- compared to pear-shaped women. The gene expression signatures identified suggest a greater FA sequestering previously observed in pear- compared to apple-shaped individuals [17, 49] and recently in GF depot compared to ABD depot [49]. The higher variation of FFA in pear-shaped women after a glucose challenge supports our gene expression data (see Table 1 and Fig. 2 for example higher expression of the ADRB3 gene, also observed upregulated in obese individuals). Moreover, the GF adipocytes were reported to be bigger in pear-shaped women compared to the apple-shaped women (Table 1), confirming a more metabolic active cell in the pear GF depot (higher TG storage and higher lipolytic rate under stimulation). A recent study demonstrated that a small fraction of systemic FFA was stored back into subcutaneous fat without going through the very low-density lipoprotein–TG pathway [50]. This mechanism has not been studied in the context of fat distribution and merits further investigation.
We showed that apple adiposity is associated with an increase in inflammatory markers both in ABD and GF tissues. Among these markers, numerous chemokines were found, which could potentially lead to increased inflammatory cell infiltration in the fat depots of apple-shaped women. We indeed observed an accumulation of macrophages organized in CLS, specifically in the scWAT of apple-shaped women. CLS were described 13 years ago in mice [51], where they were associated with obesity and IR in rodent models. However, the appearance of CLS is rarer and much more variable in human obese AT [52], and their role is still unclear. Activated macrophages in AT are thought to be involved in the clearance of dead adipocytes. In this case the macrophages are loaded with lipids, making it difficult to separate them from the adipocyte fraction. The increase in gene expression signature for phagosome pathways in apple-shaped women is consistent with the presence of macrophages organized in CLS in the adipocyte fraction isolated from apple-shaped women. Additionally, elevated expression of SLC37A2, which codes for a macrophage-specific putative sugar transporter [53], in adipocyte fraction from both depots of apple-shaped women is consistent with the presence of active macrophages in their scWAT. We identified the macrophages with a general marker (CD68); the use of additional markers will be necessary to define the level of activation of these macrophages and explore their function. In the actual study, we cannot exclude the contribution of macrophages in the molecular signature of the 2 groups of women. However, a number of markers used to identify macrophages are also expressed by adipocytes. Notably, CD86 have been shown to be expressed by large adipocytes and play the role of antigen-presenting cell to activate T cells in obese AT [54]. In the future RNA-seq and ATAC-seq single-cell analysis would help to distinguish between immune cells’ and adipocytes’ molecular signature. Altogether these data suggest that the scWAT of apple-shaped women displays characteristics associated with low-grade inflammation, which could contribute to the latter increased IR state of the apple adipocyte.
The hypothesis that CLS are markers of increased inflammation and metabolic disease is based on data in rodent models but, as mentioned earlier, studies in humans have failed to consistently reveal an association between CLS in human obese and IR AT. This discordance suggests the appearance of CLS may not be a conserved feature of adipose inflammation and metabolic disease. However, we consistently observed CLS in all apple-derived adipose samples along with increased inflammatory gene markers and clinical signs of early metabolic disease. In contrast, CLS were absent from all pear samples. This striking difference provides evidence that the model for the association of metabolic disease with CLS and increased macrophage inflammation is likely also relevant in humans.
Interestingly, in a previous study that did not stratify for WHR, a comparison of the ABD scWAT gene expression between never-obese women and obese women showed an upregulation of inflammatory signaling pathways in the obese group compared to the lean group [55]. A total of 277 of the 428 genes differentially expressed between apples and pears women in the GF depot (64%) were also dysregulated between lean and obese women. Interestingly, the genes upregulated in pears were also upregulated in lean and those upregulated in apples were upregulated in obese women. Similar observations were made for the body-shape differentially expressed genes in ABD depot. The adipocytes isolated from pear-shaped women seem to represent a population of healthy cells similar to the ones found in lean individuals, whereas the apple-adipocyte transcriptome is similar to the transcriptome signature of unhealthy obese fat cells. The apple-shaped participants showed the highest heterogeneity between the 2 scWAT (6 times more differentially expressed genes in apple- compared to pear-shaped women), maybe resulting in a more drastic change in their specific function. It also can be the consequence of higher environmental modifications in the scWAT of apple women; for example, the observation of CLS macrophages could play a significant role in adipocyte transcriptome regulation.
Our study brings novel insights on an apparent epigenomic origin of differences between apple- vs pear-shaped women. The main differences in gene expression and chromatin openness between apple- and pear-shaped women were observed in the GF cells, suggesting that lower-body fat may represent the primary depot responding to fat accumulation or environmental changes. The apples clearly showed an excess both of upper-body adipose as well as extra-adipose organ fat accumulation. Our data suggest that a primary difference in GF depot biology can directly or indirectly influence the accumulation of lipids in other fat depots and/or organs. Additional studies, for example, using reverse transcriptase-quantitative PCR or Western blot, will be necessary to validate the differential gene expression observed and further explore the contribution of specific genes in fat distribution.
We attempted to correlate the gene expression signature of the isolated adipocytes with their chromatin openness. Our first approach, limited to the closest annotated genes, revealed a weak correlation between the transcriptome and the chromatin structure of the adipocytes, although matching with other reports [56]. Analysis of wider chromatin regions around the TSS of the differentially expressed genes highlighted a better correlation between gene expression and chromatin openness (in the GF depot, 107 more open chromatin regions at a proximity of 305 apple-specific genes and 96 more open chromatin regions at a proximity of 123 pear-specific genes). A very recent study highlighted that tissue-specific genes were highly expressed and had only medium to low promoter accessibility [57]. Therefore, open chromatin at the promoter region may not be the most significant way to define scWAT depot body-shape transcriptomic signature.
The presence of more differentially open regions in the chromatin of pear-shaped individuals, closed to apple-up regulated genes (70 regions, Fig. 5B) suggests this tissue may be positioned in a latent or “poised” state. This hypothesis can be tested with a more comprehensive analysis of histone modifications that distinguish “poised” vs “active” enhancers. This speculation is for now supported by 2 results: 1) A total of 139 pear-specific open chromatin regions were annotated to apple-specific genes (Fig. 4D). In other words, those regions were more open in the pear-shaped women but were associated with lower transcription. These ATAC-seq peaks may represent sites of active transcriptional repressor binding. They may also regulate genes that are located far from these sites. Current studies are exploring this possibility. 2) Further analysis identified several motifs of key transcription factors involved in adipocyte metabolism and fat storage within the GF-specific ATAC-seq peaks. These observations are consistent with the ancestral role of the GF depot in women during pregnancy and lactation, phases during which a sufficient amount of body fat accumulation is needed for the developing baby and robust mechanisms to store sufficient lipids released for the increased energy required for lactation [58].
Our results are also consistent with an early defect in the capacity for lipid storage of apple adipocytes characterized by inflammation, potentially leading to the inability to store excess fat within the GF depot. The excess energy may be then diverted to extra-GF sites including the AB subcutaneous and visceral adipose depots, and nonfat organs such as the liver and muscle. In this framework, the gene expression signature observed in apple-shaped women could be the potential consequences of a more condensed chromatin structure that is incapable of responding to signals associated with adipocyte expansion and lipid storage, which could lead to an environment that is permissive for inflammation and the appearance of general IR later. The role of epigenetic control on adipocytes inflammation merits further investigation.
4. Conclusions
In conclusion, our results suggest that chromatin programming of the molecular state of GF fat may have deterministic roles in fat distribution and in the overall predisposition of an individual to metabolic complications. Future studies should aim to assess the transcriptomic and chromatin accessibility profiles of a larger cohort and include more advanced stages of apple/pear divergence and in a more dynamic fashion.
Acknowledgments
We thank the study volunteers for their participation and the Translational Research Institute for Metabolism and Diabetes clinical research staff for their contributions. We are grateful to Heather Cornell and Chris Bock respectively for the imaging and calorimetric data collection and interpretation, Elvis Alvarez Carnero for assistance in calorimetric data interpretation, John Marchica for performing the RNA and DNA sequencing and Roberto Vera Alvarez for performing the RNA-seq analysis, and Alexey Eroshkin for the DNA methylation array analysis and interpretation. We thank Laura Tao for the realization of the transcriptional heat map and Seema Sernovitz for her editing support.
Financial Support: This work was supported by the National Institutes of Health (Grant Number R01DK107009-02); the Hungarian Scientific Research Fund (Grant project code NKFIH K-129166); MOLMEDEX FUN-OMICS (Grant GINOP-2.3.3-15-2016-00007); the Debrecen Venture Catapult Program (Grant EFOP-3.6.1-16-2016-00022); the University of Debrecen Medical Faculty through the Bridging Fund system; and internal research funding provided by the Department of Biochemistry and Molecular Biology (to B.L.B. and D.B.). B.L.B. was a Szodoray Fellow of the University of Debrecen, Faculty of Medicine, and an alumni of the Magyary Zoltan fellowship supported by the TAMOP 4.2.4.A/2-11-1-2012-0001 grant. These funds were implemented through the New Hungary Development Plan cofinanced by the European Social Fund and the European Regional Development Fund. D.B. is a recipient of the New National Excellence Program grant by the Ministry of Human Capacities (ÚNKP-18-3-III-DE-253).
Clinical Trial Information: This trial was registered March 24, 2016, at https://clinicaltrials.gov/ct2/show/NCT02728635.
Glossary
Abbreviations
- ATAC-seq
assay for transposase-accessible chromatin using sequencing
- CLS
crown-like structure
- CTCF
CCCTC binding factor
- DEG
differentially expressed gene
- FFAs
free fatty acids
- GF
gluteofemoral
- HOMA-IR
homeostatic model assessment of insulin resistance
- IR
insulin resistant
- MetS
metabolic syndrome
- MRI
magnetic resonance imaging
- RQ
respiratory quotient
- scWAT
subcutaneous white adipose tissue
- T2D
type 2 diabetes
- TGs
triglycerides
- TPM
transcripts per million
- TSS
transcriptional start site
- VAT
visceral adipose tissue
- WAT
white adipose tissue;
- WHR
, waist-to-hip ratio
Additional Information
Disclosure Summary: The authors have nothing to disclose.
Data availability: The supplemental data have been deposited to the Dryad Digital Repository 2017 (reference 28), January 6, 2020. http://doi.org/10.5061/dryad0.7sqv9s4pb.
References
- 1. Ben-Shmuel S, Rostoker R, Scheinman EJ, LeRoith D. Metabolic syndrome, type 2 diabetes, and cancer: epidemiology and potential mechanisms. Handb Exp Pharmacol. 2016;233:355-372. [DOI] [PubMed] [Google Scholar]
- 2. Cornier MA, Dabelea D, Hernandez TL, et al. The metabolic syndrome. Endocr Rev. 2008;29(7):777-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Björntorp P. Metabolic implications of body fat distribution. Diabetes Care. 1991;14(12):1132-1143. [DOI] [PubMed] [Google Scholar]
- 4. Ritchie SA, Connell JM. The link between abdominal obesity, metabolic syndrome and cardiovascular disease. Nutr Metab Cardiovasc Dis. 2007;17(4):319-326. [DOI] [PubMed] [Google Scholar]
- 5. Gastaldelli A, Gaggini M, DeFronzo RA. Role of adipose tissue insulin resistance in the natural history of type 2 diabetes: results from the San Antonio Metabolism Study. Diabetes. 2017;66(4):815-822. [DOI] [PubMed] [Google Scholar]
- 6. Suiter C, Singha SK, Khalili R, Shariat-Madar Z. Free fatty acids: circulating contributors of metabolic syndrome. Cardiovasc Hematol Agents Med Chem. 2018;16(1):20-34. [DOI] [PubMed] [Google Scholar]
- 7. Jensen MD, Haymond MW, Rizza RA, Cryer PE, Miles JM. Influence of body fat distribution on free fatty acid metabolism in obesity. J Clin Invest. 1989;83(4):1168-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miazgowski T, Kucharski R, Sołtysiak M, Taszarek A, Miazgowski B, Widecka K. Visceral fat reference values derived from healthy European men and women aged 20-30 years using GE Healthcare dual-energy x-ray absorptiometry. PLoS One. 2017;12(7):e0180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pinnick KE, Nicholson G, Manolopoulos KN, et al. ; MolPAGE Consortium Distinct developmental profile of lower-body adipose tissue defines resistance against obesity-associated metabolic complications. Diabetes. 2014;63(11):3785-3797. [DOI] [PubMed] [Google Scholar]
- 10. Goodpaster BH, Thaete FL, Simoneau JA, Kelley DE. Subcutaneous abdominal fat and thigh muscle composition predict insulin sensitivity independently of visceral fat. Diabetes. 1997;46(10):1579-1585. [DOI] [PubMed] [Google Scholar]
- 11. Patel P, Abate N. Role of subcutaneous adipose tissue in the pathogenesis of insulin resistance. J Obes. 2013;2013:489187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karpe F, Pinnick KE. Biology of upper-body and lower-body adipose tissue—link to whole-body phenotypes. Nat Rev Endocrinol. 2015;11(2):90-100. [DOI] [PubMed] [Google Scholar]
- 13. Tchkonia T, Thomou T, Zhu Y, et al. Mechanisms and metabolic implications of regional differences among fat depots. Cell Metab. 2013;17(5):644-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lotta LA, Gulati P, Day FR, et al. ; EPIC-InterAct Consortium; Cambridge FPLD1 Consortium Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet. 2017;49(1):17-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crewe C, An YA, Scherer PE. The ominous triad of adipose tissue dysfunction: inflammation, fibrosis, and impaired angiogenesis. J Clin Invest. 2017;127(1):74-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fried SK, Lee MJ, Karastergiou K. Shaping fat distribution: new insights into the molecular determinants of depot- and sex-dependent adipose biology. Obesity (Silver Spring). 2015;23(7):1345-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jensen MD, Sarr MG, Dumesic DA, Southorn PA, Levine JA. Regional uptake of meal fatty acids in humans. Am J Physiol Endocrinol Metab. 2003;285(6):E1282-E1288. [DOI] [PubMed] [Google Scholar]
- 18. Divoux A, Sandor K, Bojcsuk D, et al. Differential open chromatin profile and transcriptomic signature define depot-specific human subcutaneous preadipocytes: primary outcomes. Clin Epigenetics. 2018;10(1):148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gehrke S, Brueckner B, Schepky A, et al. Epigenetic regulation of depot-specific gene expression in adipose tissue. PLoS One. 2013;8(12):e82516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karastergiou K, Fried SK, Xie H, et al. Distinct developmental signatures of human abdominal and gluteal subcutaneous adipose tissue depots. J Clin Endocrinol Metab. 2013;98(1):362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parsons SA, Jones KP, Yi F, et al. A novel clinical approach to evaluating changes in fat oxidation in healthy, overnight-fasted subjects. Transl Med Commun. 2016;1(6). [Google Scholar]
- 22. RRID:AB_ 2819056 https://scicrunch.org/resolver/AB_2819056.
- 23. RRID:AB_2819057. https://scicrunch.org/resolver/AB_2819057.
- 24. RRID:AB_2819058. https://scicrunch.org/resolver/AB_2819058.
- 25. Pacini G, Bergman RN. MINMOD: a computer program to calculate insulin sensitivity and pancreatic responsivity from the frequently sampled intravenous glucose tolerance test. Comput Methods Programs Biomed. 1986;23(2):113-122. [DOI] [PubMed] [Google Scholar]
- 26. Cirera S. Highly efficient method for isolation of total RNA from adipose tissue. BMC Res Notes. 2013;6:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baruzzo G, Hayer KE, Kim EJ, Di Camillo B, FitzGerald GA, Grant GR. Simulation-based comprehensive benchmarking of RNA-seq aligners. Nat Methods. 2017;14(2):135-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dryad Digital Repository 2017 DJ Fat distribution in women associates with depot-specific transcriptomics signatures and chromatin structure. January 6, 2020. https://datadryad.org/stash/dataset/doi:10.5061/dryad.7sqv9s4pb. [DOI] [PMC free article] [PubMed]
- 29. Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics. 2010;11:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. RRID:AB_1516770 https://scicrunch.org/resolver/AB_1516770.
- 33. RRID:AB_2665705 https://scicrunch.org/resolver/AB_2665705.
- 34. Cai X, Li X, Fan W, et al. Potassium and obesity/metabolic syndrome: a systematic review and meta-analysis of the epidemiological evidence. Nutrients. 2016;8(4):183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Snijder MB, Visser M, Dekker JM, et al. ; Health ABC Study Low subcutaneous thigh fat is a risk factor for unfavourable glucose and lipid levels, independently of high abdominal fat. The Health ABC Study. Diabetologia. 2005;48(2):301-308. [DOI] [PubMed] [Google Scholar]
- 36. Sparks LM, Pasarica M, Sereda O, et al. Effect of adipose tissue on the sexual dimorphism in metabolic flexibility. Metabolism. 2009;58(11):1564-1571. [DOI] [PubMed] [Google Scholar]
- 37. Divoux A, Karastergiou K, Xie H, et al. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity (Silver Spring). 2014;22(8):1781-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kasher-Meron M, Youn DY, Zong H, Pessin JE. Lipolysis defect in white adipose tissue and rapid weight regain. Am J Physiol Endocrinol Metab. 2019;317(2):E185-E193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vaittinen M, Kolehmainen M, Rydén M, et al. MFAP5 is related to obesity-associated adipose tissue and extracellular matrix remodeling and inflammation. Obesity (Silver Spring). 2015;23(7):1371-1378. [DOI] [PubMed] [Google Scholar]
- 40. Khazen W, M’bika JP, Tomkiewicz C, et al. Expression of macrophage-selective markers in human and rodent adipocytes. FEBS Lett. 2005;579(25):5631-5634. [DOI] [PubMed] [Google Scholar]
- 41. Poggi M, Jager J, Paulmyer-Lacroix O, et al. The inflammatory receptor CD40 is expressed on human adipocytes: contribution to crosstalk between lymphocytes and adipocytes. Diabetologia. 2009;52(6):1152-1163. [DOI] [PubMed] [Google Scholar]
- 42. Deng T, Lyon CJ, Minze LJ, et al. Class II major histocompatibility complex plays an essential role in obesity-induced adipose inflammation. Cell Metab. 2013;17(3):411-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carson C, Lawson HA. Epigenetics of metabolic syndrome. Physiol Genomics. 2018;50(11):947-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leung A, Parks BW, Du J, et al. Open chromatin profiling in mice livers reveals unique chromatin variations induced by high fat diet. J Biol Chem. 2014;289(34):23557-23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bogacka I, Xie H, Bray GA, Smith SR. The effect of pioglitazone on peroxisome proliferator-activated receptor-gamma target genes related to lipid storage in vivo. Diabetes Care. 2004;27(7):1660-1667. [DOI] [PubMed] [Google Scholar]
- 47. Manolopoulos KN, Karpe F, Frayn KN. Gluteofemoral body fat as a determinant of metabolic health. Int J Obes (Lond). 2010;34(6):949-959. [DOI] [PubMed] [Google Scholar]
- 48. Shadid S, Stehouwer CD, Jensen MD. Diet/Exercise versus pioglitazone: effects of insulin sensitization with decreasing or increasing fat mass on adipokines and inflammatory markers. J Clin Endocrinol Metab. 2006;91(9):3418-3425. [DOI] [PubMed] [Google Scholar]
- 49. Raajendiran A, Ooi G, Bayliss J, et al. Identification of metabolically distinct adipocyte progenitor cells in human adipose tissues. Cell Rep. 2019;27(5):1528-1540.e7. [DOI] [PubMed] [Google Scholar]
- 50. Shadid S, Koutsari C, Jensen MD. Direct free fatty acid uptake into human adipocytes in vivo: relation to body fat distribution. Diabetes. 2007;56(5):1369-1375. [DOI] [PubMed] [Google Scholar]
- 51. Cinti S, Mitchell G, Barbatelli G, et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res. 2005;46(11):2347-2355. [DOI] [PubMed] [Google Scholar]
- 52. Cancello R, Tordjman J, Poitou C, et al. Increased infiltration of macrophages in omental adipose tissue is associated with marked hepatic lesions in morbid human obesity. Diabetes. 2006;55(6):1554-1561. [DOI] [PubMed] [Google Scholar]
- 53. Kim JY, Tillison K, Zhou S, Wu Y, Smas CM. The major facilitator superfamily member Slc37a2 is a novel macrophage-specific gene selectively expressed in obese white adipose tissue. Am J Physiol Endocrinol Metab. 2007;293(1):E110-E120. [DOI] [PubMed] [Google Scholar]
- 54. Xiao L, Yang X, Lin Y, et al. Large adipocytes function as antigen-presenting cells to activate CD4(+) T cells via upregulating MHCII in obesity. Int J Obes (Lond). 2016;40(1):112-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arner P, Sinha I, Thorell A, Rydén M, Dahlman-Wright K, Dahlman I. The epigenetic signature of subcutaneous fat cells is linked to altered expression of genes implicated in lipid metabolism in obese women. Clin Epigenetics. 2015;7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dechassa ML, Tryndyak V, de Conti A, Xiao W, Beland FA, Pogribny IP. Identification of chromatin-accessible domains in non-alcoholic steatohepatitis-derived hepatocellular carcinoma. Mol Carcinog. 2018;57(8):978-987. [DOI] [PubMed] [Google Scholar]
- 57. Starks RR, Biswas A, Jain A, Tuteja G. Combined analysis of dissimilar promoter accessibility and gene expression profiles identifies tissue-specific genes and actively repressed networks. Epigenetics Chromatin. 2019;12(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lassek WD, Gaulin SJ. Brief communication: menarche is related to fat distribution. Am J Phys Anthropol. 2007;133(4):1147-1151. [DOI] [PubMed] [Google Scholar]