Abstract
The CRISPR–Cas12a is a class II, type V clustered regularly interspaced short palindromic repeat (CRISPR) system with both RNase and DNase activity. Compared to the CRISPR–Cas9 system, it recognizes T-rich PAM sequences and has the advantage of multiplex genomic editing. Here, in fission yeast Schizosaccharomyces pombe, we successfully implemented the CRISPR–Cas12a system for versatile genomic editing and manipulation. In addition to the rrk1 promoter, we used new pol II promoters from endogenous coding genes to express crRNA for Cas12a and obtained a much higher editing efficiency. This new design expands the promoter choices for potential applications in fission yeast and other organisms. In addition, we expressed a gRNA array using a strong constitutive pol II promoter. The array transcript is processed by Cas12a itself to release multiple mature crRNAs. With this construct, multiplex genomic editing of up to three loci was achieved from a single yeast transformation. We also built a CRISPR interference system using a DNase-dead Cas12a to significantly repress endogenous gene expression. Our study provides the first CRISPR-Cas12a toolkit for efficient and rapid genomic gene editing and regulation in fission yeast.
INTRODUCTION
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) was originally discovered as an adaptive immune system in many archaea and bacteria to detect and digest viral and other foreign invasive DNAs (1). This system was quickly deployed in human cells and many other organisms for precise genomic editing (2). CRISPR–Cas9, as a class II type II CRISPR–Cas system, was developed first and enjoys widespread use (3–5). The Cas9 endonuclease binds a precursor CRISPR RNA (crRNA) to form a functional complex for DNA editing at a specific locus defined by the protospacer adjacent motif (PAM) and guide RNA (gRNA) sequences. An Endonuclease Dead Cas9 (dCas9) was later engineered and subsequently modified by fusing the enzyme with transcriptional effectors to repress or activate the expression of specific target genes (6,7). Similar to CRISPR–Cas9, CRISPR–Cas12a (also called CRISPR-Cpf1) is a class II type V CRISPR–Cas system, but it recognizes a T-rich PAM upstream of the short guide RNA target sequence (8). CRISPR–Cas12a has several advantageous features. One key difference is that Cas12a has both endonuclease activity for DNA cleavage and RNase activity (9). As a result, Cas12a can process a precursor crRNA by itself by recognizing a short 19 bp specific direct repeat (DR) sequence and releasing the crRNA from the precursor transcript (10). With this feature, Cas12a was developed for multiplex genomic editing using a single construct, in which crRNA for various targets were co-assembled, separated by DRs, to form a crRNA array. This more compact crRNA structure (relative to Cas9) enables the design and synthesis of crRNA arrays bearing multiple tandem arrays consisting of DR/gRNA units. Another key difference is that DNA cleavage site of Cas12a leaves a sticky end at the distal end away from its PAM while Cas9 cleavage results in a blunt end close to the PAM locus (8). Consequently, the sticky end may affect the DNA repair preference of NHEJ, which precisely repairs such overhangs, at least in S. cerevisiae such that they remain targetable for subsequent rounds of Cas12a cleavage.
One limitation with CRISPR–Cas9 application is that the crRNA 5’ and 3’ ends require precise molecular definition (11). To solve this problem, a promoter transcribed by RNA polymerase III (pol III) is usually used, as pol III transcription starts at uniquely specified sites. In mammalian cells, crRNA is expressed from U6 snRNA promoter; transcription starts from a defined G residue and terminates at a poly-T stretch (consisting of 5–6 thymidines) (3). In Saccharomyces cerevisiae, the pol III tRNA promoter SNR52 is employed because the RNA precursor contains a leader RNA that is subsequently cleaved off at a defined location, yielding the appropriate mature 5’ end (12). The 3’ end is defined by the poly-T SUP4 terminator. Following the same concept, in fission yeast Sch. pombe, the pol II promoter of rrk1, encoding for K RNA from RNase O ribonucleoprotein, is used to express crRNA because its transcript also contains a cleavable leader RNA (11). This is the only promoter currently in use to enable expression of the Cas9 guides. The 3’ end is defined by a Hammerhead Ribozyme (HHR). SNR52 or rrk1 promoters may also be used to express the gRNA for CRISPR–Cas12a in Saccharomyces cerevisiae or Schizosaccharomyces pombe, respectively. But the implementation of CRISPR system in many other organisms is still restricted by the requirement for a proper crRNA promoter (13). Another problem is that the poly-T stretch has to be avoided in gRNA design because it terminates pol III transcription readily (14). This strongly limits gRNA selection, especially in the promoter regions and for long crRNA arrays with repetitive DR and multiple gRNAs. One solution for the CRISPR–Cas9 system is to flank the gRNA with cleavable RNA sequences (ribozymes and tRNAs) (15), or heterologous RNA processing sequences (Csy4) (16), which also enables multiplex genome editing using CRISPR–Cas9 system. But the constructs are complicated and their cloning remains cumbersome. In eukaryotes, most pol III transcripts are short non-coding RNAs while pol II promoters are employed to transcribe long endogenous protein coding genes. Combined with Cas12a's intrinsic crRNA processing ability, exploiting the large number of available pol II promoters to express single crRNAs or much longer crRNA arrays will provide more flexibility and perhaps open up additional applications.
By fusing DNase-dead Cas enzymes (dCas9 and dCas12a) with transcriptional effectors, CRISPR activation (CRISPRa) and CRISPR interference (CRISPRi) have been developed as efficient tools for specific gene expression regulation in various organisms (7,17). The dCas9–crRNA or dCas12a–crRNA complex interferes with transcription by impeding RNA polymerase binding and transcription elongation. Unlike Cas9, which recognizes downstream NGG as the PAM, Cas12a recognizes a T-rich PAM upstream of the protospacer. This feature makes dCas12a a better system for tunable transcription regulation by targeting transcription start sites (TSS), given that TSSs are typically T-rich (18–21). In bacteria, dCas12a alone shows highly efficient suppression activity for one or multiplex targets but has only modest effect with no significant repression in mammalian cells (20). To enhance repression activity, transcriptional regulators including Krüppel-associated box (22) and the EAR-repression domain (SRDX) were fused to dCas12a in human cells and plants, respectively (23,24). In S. cerevisiae, CRISPRi-dCas12a has not been reported but CRISPRi–dCas9 was tested in previous studies. dCas9 alone achieved high repression, which was further increased by fusing Mxi1, a mammalian transcriptional repressor domain to dCas9 (25,26). Mxi1 interacts with the histone deacetylase SIN3 in S. cerevisiae. In Sch. pombe, histone lysine H3 methyltransferase Clr4 has been used for gene silencing by introducing ectopic histone methylation (27,28). To our knowledge, the CRISPRi system has not been deployed in fission yeast. Whether dCas9 or dCas12a fused to the Clr4 enzymatic domain can introduce gene expression regulation remains unknown.
Sharing more chromosome features with higher order eukaryotic cells, and being larger in size, which leads to better imaging compared to S. cerevisiae, Sch. pombe is preferred by some for research in chromatin dynamics, cell division, transposable elements and centromere and telomere regulation (29–33). Genomic engineering is more difficult in Sch. pombe with fewer available tools and lower recombination efficiency compared to S. cerevisiae, which is more widely studies in synthetic biology and metabolic engineering (34–39). Multiple edits can be introduced into the fission yeast genome by several rounds of CRISPR–Cas9 modification or crossing and meiotic recombination but this could take several weeks. Currently, CRISPRi using dCas12a has also not been demonstrated in either S. cerevisiae or Sch. pombe cells, although it has been tested in mammalian cells, plants and prokaryotic cells (20,23,24). To solve these problems, in this study, we successfully built the CRISPR–Cas12a system in Sch. pombe using FnCas12a from Francisella novicida. To achieve high editing efficiency with minimal yeast transformation efforts, the crRNA and FnCas12a enzyme were expressed from a single plasmid. To expand the crRNA promoter choices for potential application in yeast and other organisms, we tested several general pol II promoters from endogenous coding genes and achieved higher genomic editing efficiency. By expressing a crRNA array with a strong constitutive fba1 promoter, multiplex genomic editing was also achieved for up to three loci within a single yeast transformation step. Finally, using dCas12a, we achieved strong endogenous gene repression by impeding transcription, leading to an auxotrophic phenotype when deployed at ade6.
MATERIALS AND METHODS
Media
Sch. pombe strains were cultured in rich YES medium or chemically defined PMG medium supplied with the necessary supplements. PMG5 medium contains all five supplements, adenine, histidine, leucine, uracil, and lysine, at 225 mg/l, and drop-out media were prepared by leaving out one of the five supplements. In the PMG5 with low adenine medium, the adenine concentration was reduced to 22.5 mg/l. Top10 E. coli were grown in Luria Broth medium. In order to select bacteria with drug-resistant genes, carbenicillin (Sigma-Aldrich) or kanamycin (Sigma-Aldrich) were used at a final concentration of 75 or 50 mg/ml, respectively. Agar was added to 2% for preparation of solid media.
Plasmids
All plasmids constructed in this study are listed in Supplementary Table S1, including their Addgene accession number if applicable. The original plasmid with the FnCas12a codon optimized for human was obtained from Addgene (#103008) (40). Then, the FnCas12a CDS with nucleoplasmin nuclear localization signal (NLS) was assembled with adh1 promoter and CYC1 terminator by Golden Gate cloning. The whole cassette of Padh1-FnCas12a-TCYC1 was inserted into the empty vector with ura4 marker and ars1 replicating region. To build the initial crRNA expression module, the rrk1 promoter with direct repeat, NotI cutting site and hammerhead ribozyme (HHRz) were synthesized as a gBlock from IDT. To build the crRNA expression module with more pol II promoters from endogenous protein coding genes, we PCR amplified the promoter regions (around 1kb upstream from the start codon) and assembled them with direct repeats, a BsaI entry pad and TEF1 terminators using Golden Gate cloning. The resulting crRNA expression module was inserted into the plasmids with the FnCas12a cassette, to serve as the final entry vector.
To build the functional plasmid for genomic editing, a single gRNA or one gRNA array was inserted into the entry vector following similar strategies with previous reports (10). All gRNAs were designed using online tool CRISPOR (41). For the single gRNA, two complementary primers with gRNA sequence and BsaI adapter were ordered from IDT, annealed together, and then ligated to the entry vector that had been pre-digested with BsaI enzyme. For multiplex genomic editing, the gRNA array was ordered from IDT as a gBlock or multiple primers for annealing, then assembled into entry vectors with Golden Gate clone. A detailed protocol for gRNA clone is included as Supplementary Figure S1. All gRNA sequences used in this study are listed in Supplementary Table S2. The primers used to assemble the gRNAs into entry vectors are listed in Supplementary Table S3. The crRNA expression module sequence (Pfba1-crRNAarray-TTEF1) is in Supplementary Table S4.
For the plasmid with dCas12a, the D917A mutation was introduced into FnCas12a but the other components remain identical (8,42). To build the dCas12a fused with Clr4 DNA methyltransferase, the Clr4 enzymatic domain was PCR amplified from wild type Sch. pombe genomic DNA and assembled into the C-terminus of dCas12a. An additional SV40 NLS was fused to the N-terminus. To build the dCas12a fused with catalytic dead Clr4 domain, we used a two-step fusion PCR to introduce point mutations (G378S or G486D) into the Clr4 catalytic domain of dCas12a-Clr4 construct.
Yeast transformation and genomic editing
All fission yeast transformations were performed following a LiOAc transformation protocol (30). BP232 (h– ura4D-18) was used as the parent strain to test all the Cas12a genome editing efficiency. The cells were grown in PMG5 medium to an A600 between 0.45 and 0.5, and then harvested by centrifugation. The transformation efficiency was around 104 CFU/μg DNA. For genomic DNA editing, 2 μg of FnCas12a/crRNA plasmids were transformed, together with 2 μg linearized plasmid or PCR products as donor DNA if necessary. To compare editing efficiency of different gRNA sequences or the same gRNA driven by different promoters, we performed each group's experiments with replicates in parallel by using the same parent strain, transforming the same amount of plasmid and donor DNA. For multiplex genomic editing experiments, 2 μg donor DNA was used for each. To ensure efficient homologous recombination, usually 800bp homologous region was used on each side. Detailed homology lengths tested in this study are listed in Supplementary Table S5. After transformation, cells were spread on PMG5–Ura plates, and incubated at 30°C for 6–7 days to form single colonies. To check editing efficiency at ade6, cells were spread on PMG5–Ura plates with low adenine concentration (22.5 mg/l). For other auxotrophic markers, the plates with single colonies were replica-plating to the corresponding drop out media to check auxotrophy. Genomic editing was further confirmed by colony PCR and Sanger sequencing.
To build yeast strains with synonymous mutations in the ade6 CDS, we first cloned the WT ade6 CDS with homology arms into a Blunt II-Topo vector via Zero Blunt Topo Cloning (ThermoFisher #450245) and introduced these point mutations by fusion PCR. To build the corresponding yeast strains, we started with the ade6Δ0 strain obtained from our first CRISPR editing experiments that targeted ade6+. Then, we transformed the DNA with synonymous mutations in ade6 and homology arms into the ade6Δ0 strain and selected on PMG5–Ade plates. These ‘synonymous’ strains were used to testCas12a PAM preference.
RESULTS
Expressing FnCas12a and crRNA for genomic editing in fission yeast
Many Cas12a (also called Cpf1) family proteins from different species have been applied for genomic editing. Among them, three Cpf1 variants are most widely used: FnCpf1 from Francisella novicida U112, AsCpf1 from Acidaminococcus sp. BV3L6 and LbCpf1 from Lachnospiraceae bacterium ND2006 (43). These variants have similar crRNA processing and genome editing capabilities and their direct repeat sequences also show high level of homology. In S. cerevisiae, FnCas12a (also called FnCpf1) has highly efficient genomic editing activity and low toxicity compared to AsCpf1 and LbCpf1 (40,43). Based on these properties, we selected FnCas12a to build the CRISPR–Cas12a system in fission yeast.
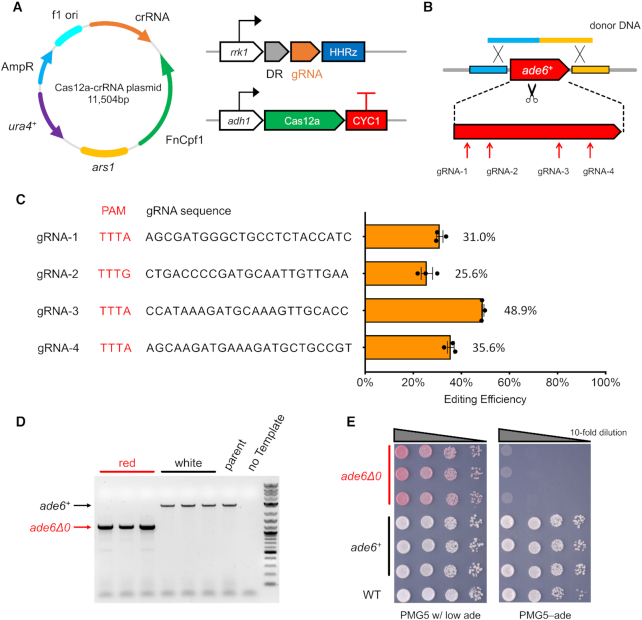
In order to achieve efficient genome editing and reduce the efforts of yeast transformation, we assembled the FnCas12a with crRNA together into one plasmid with the ura4 marker (Figure 1A). This one-plasmid strategy was shown to be more efficient for the CRISPR–Cas9 system in fission yeast (11). The FnCas12a was expressed using the constitutive adh1 promoter and the crRNA was initially expressed using the rrk1 promoter, followed by the Hammerhead Ribozyme at the 3’ end. This module was also used in the CRISPR–Cas9 system to ensure the maturation of crRNA with precise ends.
Figure 1.
Cas12a genome editing system. (A) FnCas12a and crRNA were expressed from a single plasmid with ura4 marker and ars1 replicating origin in fission yeast. As the first construct, FnCas12a and gRNA were expressed with adh1 and rrk1 promoter, respectively. (B) To test editing efficiency, we picked four gRNAs targeting the endogenous ade6+ CDS. One linear donor DNA was co-transformed to delete ade6 by homologous recombination. (C) The gRNA sequences used and their editing efficiency, with mean values ± S.E.M. The original colony counts are listed in Supplementary Table S6. (D) Colony PCR to check the ade6 deletion, in red colonies and white colonies from plates with PMG5–Ura w/ low adenine media. The 1 kb Plus DNA ladder from NEB (Catalog# N3200L) was used as the molecular weight standard. (E) Spot assay on plates to check adenine auxotroph.
As the first demonstration, we targeted ade6+ for deletion of the entire CDS by CRISPR–Cas12a (Figure 1B). Together with the Cas12a/gRNA plasmid, a linear donor DNA containing around 800bp homology at both ends was provided. From the ade6+ CDS region, four gRNAs with a PAM sequence of TTTV were selected and tested separately. For all of them, the ade6 CDS was successfully deleted at efficiencies of 25–50% (Figure 1C), resulting in a red colony color on plates with low adenine concentration (Supplementary Figure S2). In a control group transformed with the same amount of Cas12a/gRNA plasmid but not the donor DNA, fewer colonies appeared and none were red. To further confirm that these edits were designer deletions from homologous recombination (HR) with donor DNA, we randomly selected three red and another three white colonies from the gRNA-1 transformation and confirmed the deletion with colony PCR (Figure 1D). Shorter PCR products with the expected length, indicating the designer deletion, were obtained from all red colonies while the PCR products from all white colonies maintained the same length as wild-type colonies. We also performed a spot assay and found red colonies were auxotrophic for adenine while the white ones grew as well as wild type on plates with defined media without adenine (Figure 1E). Using Sanger sequencing, we found that in white colonies with no ade6 editing, the gRNA target sequence remained intact without any detectable mutations. This indicates that the DNA cleavage was mostly repaired by HR with donor DNA while a subset of colonies survived via resistance to Cas12a cleavage by an unknown mechanism. Similar results were also observed in the CRISPR–Cas9 system (11,44–46).
Testing PAM preference and additional target editing of CRISPR–Cas12a
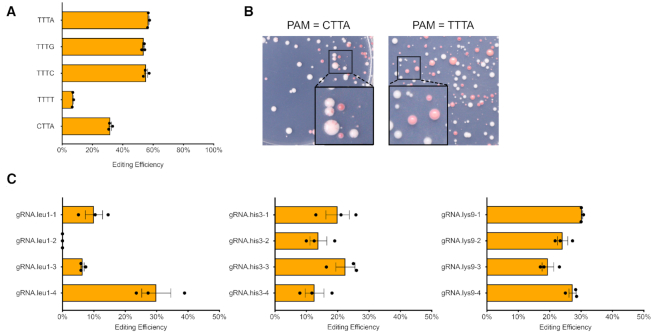
We also checked the PAM sequence preference of FnCas12a in fission yeast. Previously, the PAM for FnCas12a was defined as 5’-TTN-3’, which enables efficient DNA cleavage activity in vitro (8). But at least in S. cerevisiae, FnCas12a has a strong preference of 5’-TTTV-3’ (V = A, G or C) for genome editing in vivo (40). To check the PAM preference of FnCas12a in Sch. pombe, we modified the PAM sequence for the same gRNA target by introducing synonymous point mutations into the ade6+ CDS (Supplementary Figure S3). As a result, yeast strains with alternative PAMs have the same gRNA sequence and equally functional Ade6 protein. We observed that using PAM of TTTV, FnCas12a achieved relatively high editing efficiency, which was dropped dramatically to only around 7% using TTTT (Figure 2A). For PAM CTTA, the red colony ratio dropped by half, among which most colonies were actually chimeric resulting from inefficient genome editing (Figure 2B). This type of colony was not observed when the PAM sequence conformed to TTTV. In summary, a PAM sequence of TTTV seems to be preferred by FnCas12a for in vivo genome editing in fission yeast and this rule should be applied when gRNAs are designed.
Figure 2.
PAM preference for FnCas12a and genome editing at additional targets. (A) Yeast strains containing same gRNA targets but alternative PAMs with synonymous mutations were tested to measure the genome editing efficiency. (B) Transformants from parents strains with the PAM sequence CTTA compared to TTTA. (C) Cas12a editing efficiency targeting leu1, his3 and lys9, with mean values ± S.E.M. shown here. The original colony counts are listed in Supplementary Table S7.
To further test the CRISPR–Cas12a system, we selected additional auxotrophic markers leu1, his3 and lys9 to target for designer deletion by HR with donor DNA (Figure 2C). Four gRNAs were selected from the coding sequence (CDS) of each gene and tested for their genome editing efficiency. We consistently achieved efficiencies of 10–30%. Each ‘designer deletion’ was confirmed by replica plating and colony PCR (Supplementary Figures S4 and S5). One single gRNA out of four selected for leu1 deletion did not result in any activity. This result indicates that the CRISPR–Cas12a is a generally reliable genome editing system in fission yeast but the mechanism determining the efficiency of individual gRNAs remains unknown.
Expressing crRNA with endogenous pol II promoters
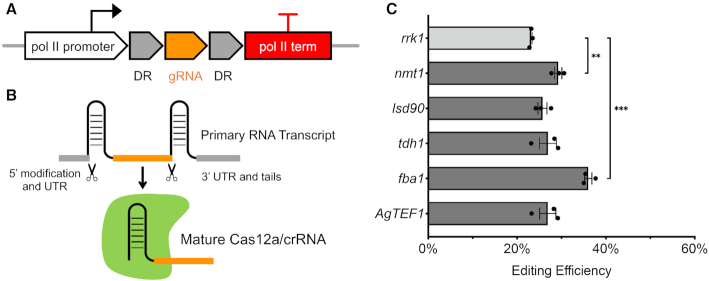
Compared to Cas9, one advantageous feature of Cas12a is its RNase activity, which enables Cas12a to process pre-crRNA by itself. With this feature, we tested several endogenous pol II promoters to express crRNAs designed to work with Cas12a. We designed a new module with two direct repeats flanking the gRNA sequence. Transcription is driven by strong endogenous pol II promoters (Figure 3A). After transcription, Cas12a recognizes the direct repeats in the primary transcript, releases the crRNA using its RNase activity, and then forms the mature Cas12a/crRNA ribonucleoprotein complex (Figure 3B). For these constructs, we selected the regulatable nmt1 promoter, three strong constitutive pol II promoters sourced from endogenous protein coding genes and the exogenous TEF1 promoter from fungus Ashbya gossypii, which is known to function well in S. cerevisiae. (47). Compared to the rrk1 promoter, nmt1 and fba1 are four and eight times stronger, respectively, based on their RNA molecular level during vegetative growth in minimal media (48). Using these promoters, the CRISPR–Cas12a system targeting ade6+ was functional at an even higher genome editing efficiency (Figure 3C). As the strongest promoter we tested, fba1 resulted in the highest editing efficiency, which was ∼30% higher than the original design employing rrk1. Using the regulatable nmt1 promoter, the editing efficiency was also higher. The crRNA expression was repressible by thiamine and as a result, the genomic editing efficiency was obviously reduced (Supplementary Figure S7). These results suggested that general endogenous and exogenous pol II promoters are able to drive the expression of crRNA, whose primary transcript is further processed by Cas12a to become the mature Cas12a/crRNA complex. Stronger expression also helped to increase genome editing efficiency presumably by making more crRNA available to form active Cas12a ribonucleoprotein particles.
Figure 3.
Expression of Cas12a crRNA using pol II promoters. (A) New constructs were designed using pol II promoters and terminators to express crRNA. (B) crRNA release from primary RNA transcripts. (C) New endogenous and exogenous pol II promoters tested and their editing efficiency targeting ade6+. The same gRNA sequence (gRNA-3) was used here. Error bars represent mean ± S.E.M. using three technical replicates. An unpaired t-test was used to assess the significance of higher efficiency using nmt1 or fba1 promoter compared to the rrk1 promoter (**P < 0.01, ***P < 0.001). The original colony counts were listed in Supplementary Table S8.
Multiplex genome editing using crRNA arrays
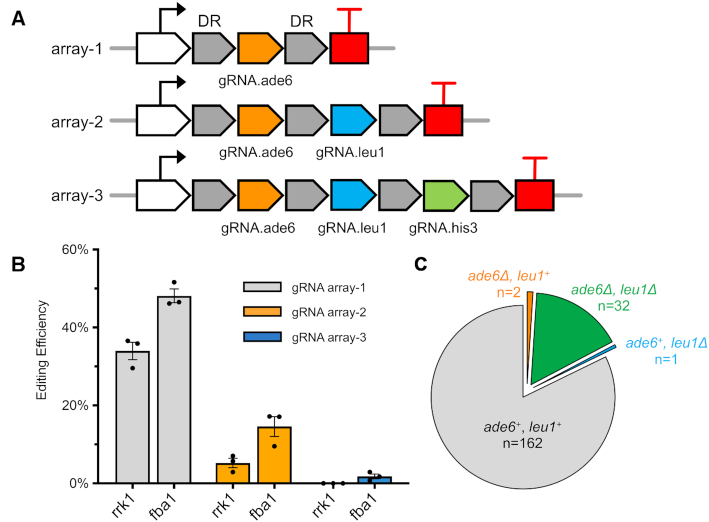
One major feature of Cas12a compared to Cas9 is its ‘multiplex’ genome editing activity, enabled by its RNase domain. To achieve this feature in fission yeast, we designed three gRNA arrays by flanking each gRNA with short direct repeats (19 bp) and tried to introduce designer deletions of ade6, leu1 and his3 simultaneously in a single yeast transformation step using linear donor DNAs corresponding to each auxotrophic marker (Figure 4A). Both the original rrk1 and strong fba1 promoters were tested to drive the expression of several crRNAs on a single transcript. Each gRNA was pre-tested for functionality in single editing experiments. For all crRNA arrays, CRISPR-Cas12a using the fba1 promoter achieved higher genome editing efficiency compared to the rrk1 promoter (Figure 4B). For double editing using crRNA array-2 driven by the fba1 promoter, we achieved an efficiency of 17% for double deletion (13 colonies out of 76) but for crRNA array-2 driven by the rrk1 promoter, we only identified 1 of 34 colonies with the double deletions. For triple edits, we successfully obtained 1 colony out of 40 that contained designer deletions of ade6, leu1 and his3, using the crRNA array-3 driven by the fba1 promoter. All these edits were designer deletions employing HR with donor DNA, as confirmed by colony PCR. This indicates that CRISPR–Cas12a can be used for multiplex genomic editing in fission yeast.
Figure 4.
Multiplex genome editing using crRNA arrays. (A) The crRNA arrays designed to express multiple crRNA on a single transcript. (B) Genome editing efficiency using rrk1 or fba1 promoter to drive the crRNA array expression, with their mean efficiency ± S.E.M. Gray: positive editing efficiency at ade6 alone using array-1. Orange: double positive editing efficiency at ade6 and leu1, using array-2. Blue: triple positive editing efficiency at ade6, leu1 and his3, using array-3. (C) The transformant count for ade6 or leu1 deletion from the group using array-2 driven by fba1 promoter.
The efficiency for triple genome editing is dramatically lower compared to double edits, making it harder to achieve in practice. One surprising observation in the double genome editing experiment is that editing of two of the targets, ade6 and leu1, was not independent (Figure 4C and Supplementary Figure S6). The majority of yeast transformants were divided into two groups: no genome edits at either target (n = 63, 82%) or genome edits at both targets (n = 13, 17%). This result suggests that a fraction of yeast cells somehow became resistant to CRISPR–Cas12a cleavage. This was also observed in our single edit experiments, in which colonies without any designer deletions still contain the complete gRNA target sequence without any mutagenesis. Interestingly, for single locus genome editing using CRISPR–Cas9, a portion of the transformants displayed very similar pattern of resistance (11,44–46). The mechanism remains unclear and controversial. Understanding how to reduce the resistance to CRISPR cleavage may help to increase the editing efficiency for both CRISPR–Cas9 and Cas12a. Moreover, this co-editing phenomenon was also used as a Co-CRISPR strategy in yeast, flies and human cells, to enrich for target modifications (49–51).
CRISPR interference to repress endogenous genes
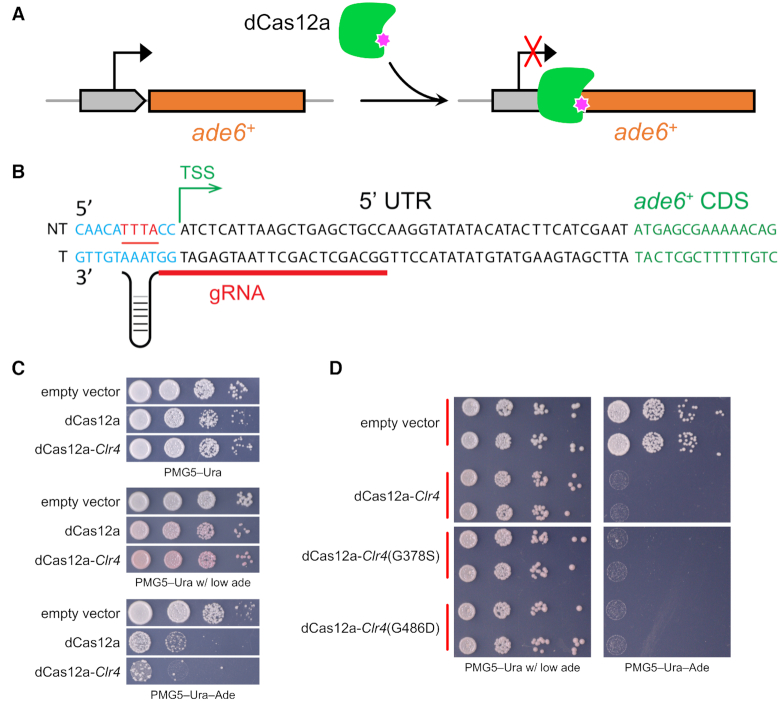
Transcriptional regulation using nuclease-inactive Cas9 or Cas12a has facilitated many genome engineering applications. Here, using a dCas12a, we developed a CRISPRi system to repress endogenous gene expression in fission yeast. The dCas12a and crRNA were expressed from a single plasmid, making cloning and yeast transformation extremely efficient (Figure 5A).
Figure 5.
CRISPR interference using dCas12a. (A) The dCas12a with D917A mutation was used to reduce expression of ade6+. (B) One gRNA proximal to TSS was used to repress the ade6 expression. (C) Spot assay to check CRISPRi repression using dCas12a alone or dCas12a-Clr4. (D) CRISPRi repression of ade6 using dCas12a fused to the Clr4 catalytic domain or catalytic dead mutants.
It has been reported that for strong repression, dCas12a has a preference for crRNA targeting the template DNA strand (20,21). The opposite is true of dCas9, where the preference for crRNA is targeting the non-template DNA strand (7). To impede binding and movement of transcription machinery, gRNAs were usually selected to direct binding of dCas12a to the transcription starting site (TSS) (52). To further enhance repressive effects, we fused the enzymatic domain of Clr4 to dCas12a. The Clr4 protein is a histone lysine 9 methyltransferase, which contains two distinct domains: the N-terminal chromodomain, which binds to methylated histone H3K9, and the C-terminal enzymatic methyltransferase domain (residues 192–490) (53). We selected one gRNA targeting the template strand proximal to the TSS of ade6 (Figure 5B) and then tested the colony growth on defined media. The dCas12a enzyme alone showed obvious repression of ade6 expression, resulting in red colonies on plates with low adenine media and slow growth on plates without adenine (Figure 5C). With the help of Clr4, stronger repression was achieved, resulting in a deeper red color and poorer growth on plates with low adenine media or adenine drop-out media. In order to determine whether the improved CRISPRi efficiency benefits from Clr4 histone methyltransferase catalytic activity or by a ‘steric hindrance’ type mechanism, we introduced two mutations separately (G378S or G486D) into the Clr4 catalytic domain, which were previously shown to greatly reduce Clr4 methyltransferase activity (54). Their ade6 repression looks equally strong with no visible difference relative to the strain with the WT Clr4 enzymatic domain (Figure 5D). This indicates that steric hindrance from the added protein bulk may play the major role in enhancing CRISPRi repression, compared to dCas12a alone. We also observed a small number of colonies were able to grow on PMG5–Ura–Ade plates, suggesting that they may have acquired resistance to dCas12a CRISPRi. Taken together, these results demonstrate that dCas12a can be used efficiently to repress gene transcription at a target locus in Sch. pombe.
DISCUSSION
Sch. pombe is a unicellular organism whose genome structure resembles that of higher eukaryotes. But compared to budding yeast, genomic engineering is more challenging because of lower homologous recombination efficiency and a higher background of non-specific integration (32). Here, in our study, we successfully implemented the CRISPR–Cas12a system in fission yeast with editing efficiencies of 20–40% for a single target and 17% for two targets using the strong constitutive pol II fba1 promoter to drive crRNA designed to form deletion mutants ade6Δ0 and leu1Δ0. Our results demonstrate that the CRISPR–Cas12a system is an efficient means of genome editing, such as gene deletion or integration. Yeast cells with functional gRNA and Cas9 or Cas12a plasmids usually cannot survive with low background because of chromosome cleavage, unless they are repaired by homologous recombination with donor DNA (12,40,43). But fission yeast spends a majority of time in the G2 phase, leading to a more challenging ‘lifestyle’ for efficient genome editing (55). It was reported that synchronizing cells in G1 phase increased CRISPR–Cas9 editing efficiency (44).
The editing efficiency of CRISPR mainly depends on gRNA sequence design, genomic target accessibility and locations (56–58). But determining functional gRNAs with predictable editing efficiency remains a big challenge, especially for the CRISPR–Cas12a system. Algorithms to evaluate and predict CRISPR–Cas12a activity with gRNAs were reported (59,60). Large scale and high throughput screening of possible gRNA sequences in vivo, combined with a predictive algorithm and machine learning are still highly desired. In our study, we designed gRNAs using the online tool CRISPOR with TTTV as the PAM for FnCas12a but still ended up with one gRNA targeting at leu1 that was non-functional (41).
In our study, we showed that endogenous pol II promoters can be used to express crRNA for Cas12a. In our design, the 5’ and 3’ ends of crRNAs were defined by Cas12a RNase cutting at the DR locus, which is distinct from mature crRNA processing via RNA transcription or post-transcriptional modification. More than increasing CRISPR editing efficiency, this new strategy bypassed the requirements for pol III promoters or other complicated RNA processing-based designs. Since pol II promoters for coding genes exist in almost all model organisms, this new design significantly expands applications of the CRISPR–Cas12a system. Another advantage of pol II promoters is that they drive the expression of much longer crRNA arrays as a single transcript and allow the use of oligo-T stretches in gRNA designs while the transcripts from pol III promoters are usually short and terminate early at poly-T stretches (14). The idea of using endogenous pol II promoters, which seems to work in mammalian cells and Sch. pombe, to express crRNAs for Cas12a can also be tested in other organisms. In our study, the regulatable nmt1 promoter was also able to drive the crRNA expression, making it possible to build a CRISPR switch in fission yeast using thiamine (Supplementary Figure S7). In mammalian cells, using pol II promoters that temporally or spatially regulate Cas12a and crRNAs expression will enable many potential applications. Recently, the EF1a pol II promoter was shown to successfully express Cas12a and crRNAs on single transcripts in mammalian cells and achieved regulation of up to 25 endogenous genes (23). This system can also be applied into yeast systems easily.
CRISPR interference was also successfully implemented in fission yeast using DNase-dead dCas12a. To our knowledge, CRISPRi using dCas9 has not been tested in fission yeast but CRISPRi-dCas12a is preferable because its PAM (5’-TTTV-3’) exists more frequently proximal to TSS sites in promoter regions compared to the dCas9 PAM (5’-NGG-3’). It has been reported that dCas12a has a strong preference of gRNAs targeting at the template strand over non-template strand, proximal to the TSS locus for stronger repression (18,20,61). But without empirical testing, it is even more challenging to select the functional gRNAs to achieve the strongest CRISPRi repression. In this study, several gRNAs were also found to be unable to repress the ade6+ function by CRISPRi (Supplementary Figure S8). It seems that CRISPR interference with dCas12a has different gRNA sequence preferences as gRNAs that are functional for CRISPR cleavage may be non-functional for CRISPR interference (unpublished data). This is a significant limitation of the CRISPRi system. Fission yeast has a substantially different chromatin structure compared to S. cerevisiae and bacteria so the rules of gRNA design and targeting locus for CRISPRi may also be different. Therefore, further studies with more systematic gRNA screens are needed to reveal the general rules for CRISPRi in yeast.
In our study, we fused the catalytic methyltransferase domain of Clr4 to dCas12a in order to increase the magnitude of CRISPRi repression and create a novel DNA methyltransferase. Previously, in order to establish the ectopic heterochromatin in fission yeast, the Clr4 enzymatic domain was fused to the bacterial TetR protein, which facilitated its specific targeting to its cognate DNA binding sequence (10X tetO) which was pre-integrated (28). Our results suggest that the improved CRISPRi efficiency benefits mainly from ‘steric hindrance’ rather than enzymatic modification of chromatin. By using alternative promoters and fusion to other transcriptional effectors or enzymatic domains, dCas12a can be applied to more potential tissue- and organ-specific epigenetic modification, or gene expression pattern regulation.
DATA AVAILABILITY
All plasmids used in this study are listed in Supplementary Table S1 and are available upon request. The entry vectors for CRISPR genome editing with Cas12a were deposited to Addgene as accession number #132952 for pYZ221 and #132953 for pYZ673. The entry vectors for CRISPRi with dCas12a have been deposited to Addgene as #132954, #132955, #132956 for pYZ675, pYZ714 and pYZ694 respectively.
Supplementary Material
ACKNOWLEDGEMENTS
Y.Z. and J.D.B conceived the ideas. Y.Z. performed all the experiments and collected the data. Y.Z. and J.D.B prepared the manuscript. We thank Neta Agmon from Boeke lab for helpful discussions about CRISPR–Cas12a designs, Fei Li and Qianhua Dong from NYU Department of Biology for helpful discussions about CRISPR interference. We also thank Stephanie Lauer, Julie Trolle and Hala Iqbal, together with Y.Z. as the ‘Tuesday bay’ members at Boeke lab for their help during manuscript preparation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NSF [MCB-1616111 to J.D.B.]. Funding for open access charge: NSF [MCB-1616111 to J.D.B.].
Conflict of interest statement. J.D.B. is a founder and director of the following: Neochromosome, Inc., the Center of Excellence for Engineering Biology, and CDI Labs, Inc. and serves on the Scientific Advisory Board of the following: Sangamo, Inc., Modern Meadow, Inc., and Sample6, Inc.
REFERENCES
- 1. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P.. CRISPR provides acquired resistance against viruses in prokaryotes. Science (New York, N.Y.). 2007; 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 2. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.). 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al.. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.). 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science (New York, N.Y.). 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR–Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dominguez A.A., Lim W.A., Qi L.S.. Beyond editing: repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016; 17:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. et al.. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y., Fedorova I., Nakane T., Makarova K.S., Koonin E.V. et al.. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016; 165:949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zetsche B., Heidenreich M., Mohanraju P., Fedorova I., Kneppers J., DeGennaro E.M., Winblad N., Choudhury S.R., Abudayyeh O.O., Gootenberg J.S. et al.. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 2017; 35:31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jacobs J.Z., Ciccaglione K.M., Tournier V., Zaratiegui M.. Implementation of the CRISPR–Cas9 system in fission yeast. Nat. Commun. 2014; 5:5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DiCarlo J.E., Norville J.E., Mali P., Rios X., Aach J., Church G.M.. Genome engineering in Saccharomyces cerevisiae using CRISPR–Cas systems. Nucleic Acids Res. 2013; 41:4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raschmanova H., Weninger A., Glieder A., Kovar K., Vogl T.. Implementing CRISPR–Cas technologies in conventional and non-conventional yeasts: Current state and future prospects. Biotechnol. Adv. 2018; 36:641–665. [DOI] [PubMed] [Google Scholar]
- 14. Nielsen S., Yuzenkova Y., Zenkin N.. Mechanism of eukaryotic RNA polymerase III transcription termination. Science (New York, N.Y.). 2013; 340:1577–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ryan O.W., Skerker J.M., Maurer M.J., Li X., Tsai J.C., Poddar S., Lee M.E., DeLoache W., Dueber J.E., Arkin A.P. et al.. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014; 3:e03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferreira R., Skrekas C., Nielsen J., David F.. Multiplexed CRISPR/Cas9 genome editing and gene regulation using Csy4 in Saccharomyces cerevisiae. ACS Synth. Biol. 2018; 7:10–15. [DOI] [PubMed] [Google Scholar]
- 17. Wu W.Y., Lebbink J.H.G., Kanaar R., Geijsen N., van der Oost J.. Genome editing by natural and engineered CRISPR-associated nucleases. Nat. Chem. Biol. 2018; 14:642–651. [DOI] [PubMed] [Google Scholar]
- 18. Kim S.K., Kim H., Ahn W.C., Park K.H., Woo E.J., Lee D.H., Lee S.G.. Efficient transcriptional gene repression by type V-A CRISPR-Cpf1 from Eubacterium eligens. ACS Synth. Biol. 2017; 6:1273–1282. [DOI] [PubMed] [Google Scholar]
- 19. Tak Y.E., Kleinstiver B.P., Nuñez J.K., Hsu J.Y., Horng J.E., Gong J., Weissman J.S., Joung J.K.. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat. Methods. 2017; 14:1163–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X., Wang J., Cheng Q., Zheng X., Zhao G., Wang J.. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017; 3:17018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miao C., Zhao H., Qian L., Lou C.. Systematically investigating the key features of the DNase deactivated Cpf1 for tunable transcription regulation in prokaryotic cells. Synth. Syst. Biotechnol. 2019; 4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zimmer M., Schiebner T., Krabusch M., Wolf K.. Nucleotide sequence of the unassigned reading frame urf a in the mitochondrial genome of three Schizosaccharomyces pombe strains. Nucleic Acids Res. 1990; 18:6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Campa C.C., Weisbach N.R., Santinha A.J., Incarnato D., Platt R.J.. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods. 2019; 16:887–893. [DOI] [PubMed] [Google Scholar]
- 24. Tang X., Lowder L.G., Zhang T., Malzahn A.A., Zheng X., Voytas D.F., Zhong Z., Chen Y., Ren Q., Li Q. et al.. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants. 2017; 3:17018. [DOI] [PubMed] [Google Scholar]
- 25. Smith J.D., Suresh S., Schlecht U., Wu M., Wagih O., Peltz G., Davis R.W., Steinmetz L.M., Parts L., St Onge R.P.. Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biol. 2016; 17:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. et al.. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013; 154:442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kagansky A., Folco H.D., Almeida R., Pidoux A.L., Boukaba A., Simmer F., Urano T., Hamilton G.L., Allshire R.C.. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science (New York, N.Y.). 2009; 324:1716–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ragunathan K., Jih G., Moazed D.. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science (New York, N.Y.). 2015; 348:1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keeney J.B., Boeke J.D.. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994; 136:849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Forsburg S.L., Rhind N.. Basic methods for fission yeast. Yeast (Chichester, England). 2006; 23:173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fantes P.A., Hoffman C.S.. A brief history of schizosaccharomyces pombe research: a perspective over the past 70 years. Genetics. 2016; 203:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffman C.S., Wood V., Fantes P.A.. An ancient yeast for young geneticists: a primer on the schizosaccharomyces pombe model system. Genetics. 2015; 201:403–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feng G., Leem Y.E., Levin H.L.. Transposon integration enhances expression of stress response genes. Nucleic Acids Res. 2013; 41:775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao Y., Boeke J.D.. Construction of designer selectable marker deletions with a CRISPR–Cas9 toolbox in Schizosaccharomyces pombe and new design of common entry vectors. G3 (Bethesda, Md.). 2018; 8:789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mitchell L.A., Wang A., Stracquadanio G., Kuang Z., Wang X., Yang K., Richardson S., Martin J.A., Zhao Y., Walker R. et al.. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science (New York, N.Y.). 2017; 355:eaaf4831. [DOI] [PubMed] [Google Scholar]
- 36. Dymond J.S., Richardson S.M., Coombes C.E., Babatz T., Muller H., Annaluru N., Blake W.J., Schwerzmann J.W., Dai J., Lindstrom D.L. et al.. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011; 477:471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Annaluru N., Muller H., Mitchell L.A., Ramalingam S., Stracquadanio G., Richardson S.M., Dymond J.S., Kuang Z., Scheifele L.Z., Cooper E.M. et al.. Total synthesis of a functional designer eukaryotic chromosome. Science (New York, N.Y.). 2014; 344:55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Richardson S.M., Mitchell L.A., Stracquadanio G., Yang K., Dymond J.S., DiCarlo J.E., Lee D., Huang C.L., Chandrasegaran S., Cai Y. et al.. Design of a synthetic yeast genome. Science (New York, N.Y.). 2017; 355:1040–1044. [DOI] [PubMed] [Google Scholar]
- 39. Tanaka T., Yamada R., Ogino C., Kondo A.. Recent developments in yeast cell surface display toward extended applications in biotechnology. Appl. Microbiol. Biotechnol. 2012; 95:577–591. [DOI] [PubMed] [Google Scholar]
- 40. Swiat M.A., Dashko S., den Ridder M., Wijsman M., van der Oost J., Daran J.M., Daran-Lapujade P.. FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res. 2017; 45:12585–12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haeussler M., Schonig K., Eckert H., Eschstruth A., Mianne J., Renaud J.B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J. et al.. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016; 17:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fonfara I., Richter H., Bratovič M., Le Rhun A., Charpentier E.. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016; 532:517. [DOI] [PubMed] [Google Scholar]
- 43. Verwaal R., Buiting-Wiessenhaan N., Dalhuijsen S., Roubos J.A.. CRISPR/Cpf1 enables fast and simple genome editing of Saccharomyces cerevisiae. Yeast (Chichester, England). 2018; 35:201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rodriguez-Lopez M., Cotobal C., Fernandez-Sanchez O., Borbaran Bravo N., Oktriani R., Abendroth H., Uka D., Hoti M., Wang J., Zaratiegui M. et al.. A CRISPR/Cas9-based method and primer design tool for seamless genome editing in fission yeast. Wellcome Open Res. 2016; 1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang X.-R., He J.-B., Wang Y.-Z., Du L.-L.. A Cloning-Free method for CRISPR/Cas9-Mediated genome editing in fission Yeast. G3 (Bethesda, Md.). 2018; 8:2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hayashi A., Tanaka K.. Short-Homology-Mediated CRISPR/Cas9-Based method for genome editing in fission Yeast. G3 (Bethesda, Md.). 2019; 9:1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Verma H.K., Shukla P., Alfatah M., Khare A.K., Upadhyay U., Ganesan K., Singh J.. High level constitutive expression of luciferase reporter by lsd90 promoter in fission yeast. PLoS One. 2014; 9:e101201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., Bahler J.. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012; 151:671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Després P.C., Dubé A.K., Nielly-Thibault L., Yachie N., Landry C.R.. Double selection enhances the efficiency of Target-AID and Cas9-Based genome editing in Yeast. G3: Genes Genomes Genetics. 2018; 8:3163–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kane N.S., Vora M., Varre K.J., Padgett R.W.. Efficient screening of CRISPR/Cas9-induced events in Drosophila using a co-CRISPR strategy. G3: Genes Genomes Genetics. 2017; 7:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Agudelo D., Duringer A., Bozoyan L., Huard C.C., Carter S., Loehr J., Synodinou D., Drouin M., Salsman J., Dellaire G.. Marker-free coselection for CRISPR-driven genome editing in human cells. Nat. Methods. 2017; 14:615. [DOI] [PubMed] [Google Scholar]
- 52. Jensen M.K. Design principles for nuclease-deficient CRISPR-based transcriptional regulators. FEMS Yeast Res. 2018; 18:foy039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Min J., Zhang X., Cheng X., Grewal S.I., Xu R.M.. Structure of the SET domain histone lysine methyltransferase Clr4. Nat. Struct. Biol. 2002; 9:828–832. [DOI] [PubMed] [Google Scholar]
- 54. Nakayama J.-I., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I.. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science (New York, N.Y.). 2001; 292:110–113. [DOI] [PubMed] [Google Scholar]
- 55. Gomez E.B., Forsburg S.L.. Analysis of the fission yeast Schizosaccharomyces pombe cell cycle. Methods Mol. Biol. 2004; 241:93–111. [DOI] [PubMed] [Google Scholar]
- 56. Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E.. Rational design of highly active sgRNAs for CRISPR–Cas9-mediated gene inactivation. Nat. Biotechnol. 2014; 32:1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al.. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gisler S., Goncalves J.P., Akhtar W., de Jong J., Pindyurin A.V., Wessels L.F.A., van Lohuizen M.. Multiplexed Cas9 targeting reveals genomic location effects and gRNA-based staggered breaks influencing mutation efficiency. Nat. Commun. 2019; 10:1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim H.K., Song M., Lee J., Menon A.V., Jung S., Kang Y.M., Choi J.W., Woo E., Koh H.C., Nam J.W. et al.. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods. 2017; 14:153–159. [DOI] [PubMed] [Google Scholar]
- 60. Kim H.K., Min S., Song M., Jung S., Choi J.W., Kim Y., Lee S., Yoon S., Kim H.H.. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat. Biotechnol. 2018; 36:239–241. [DOI] [PubMed] [Google Scholar]
- 61. Liu Y., Han J., Chen Z., Wu H., Dong H., Nie G.. Engineering cell signaling using tunable CRISPR-Cpf1-based transcription factors. Nat. Commun. 2017; 8:2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All plasmids used in this study are listed in Supplementary Table S1 and are available upon request. The entry vectors for CRISPR genome editing with Cas12a were deposited to Addgene as accession number #132952 for pYZ221 and #132953 for pYZ673. The entry vectors for CRISPRi with dCas12a have been deposited to Addgene as #132954, #132955, #132956 for pYZ675, pYZ714 and pYZ694 respectively.