Abstract
DNA double-strand breaks are repaired by end-joining or homologous recombination. A key-committing step of recombination is DNA end resection. In resection, phosphorylated CtIP first promotes the endonuclease of MRE11–RAD50–NBS1 (MRN). Subsequently, CtIP also stimulates the WRN/BLM–DNA2 pathway, coordinating thus both short and long-range resection. The structure of CtIP differs from its orthologues in yeast, as it contains a large internal unstructured region. Here, we conducted a domain analysis of CtIP to define the function of the internal region in DNA end resection. We found that residues 350–600 were entirely dispensable for resection in vitro. A mutant lacking these residues was unexpectedly more efficient than full-length CtIP in DNA end resection and homologous recombination in vivo, and consequently conferred resistance to lesions induced by the topoisomerase poison camptothecin, which require high MRN–CtIP-dependent resection activity for repair. This suggested that the internal CtIP region, further mapped to residues 550–600, may mediate a negative regulatory function to prevent over resection in vivo. The CtIP internal deletion mutant exhibited sensitivity to other DNA-damaging drugs, showing that upregulated resection may be instead toxic under different conditions. These experiments together identify a region within the central CtIP domain that negatively regulates DNA end resection.
INTRODUCTION
A DNA double-strand break (DSB) is an extremely toxic form of DNA damage. If repaired incorrectly, DSBs can generate mutations leading to cancer or cell death. There are two main pathways for DSB repair, homologous recombination (HR) and non-homologous end-joining (NHEJ) (1). HR utilizes an intact copy of DNA with a matching sequence, usually the sister chromatid, as a template to facilitate repair, thus reducing the likelihood of generating detrimental mutations. In order for HR to identify a homologous DNA sequence to use as a template, the broken DNA end must first be nucleolytically resected. DNA end resection occurs in a 5′ to 3′ direction, which exposes a 3′ single-stranded (ss) DNA overhang. This ssDNA tail can then invade homologous DNA template in a reaction catalyzed by RAD51, and prime DNA synthesis to restore missing DNA sequence in the vicinity of the break (1). The recombining DNA molecules are ultimately enzymatically processed to generate two intact DNA strands (1). Because a resected DSB can no longer be used for NHEJ, the initiation of end resection is the key committing step to determine if a DSB is repaired by NHEJ or HR. Unscheduled resection in the absence of sister chromatids may lead to recombination with a homologous chromosome or an ectopic template, leading to loss of heterozygosity or large translocations (2).
Multiple laboratories have demonstrated that DNA end resection can be a two-step process, particularly in mammalian cells (3,4). The first step is slow and involves limited resection of the 5′ DNA end, occurring up to 200–300 nucleotides away from the DSB. It is catalyzed by MRE11–RAD50–NBS1 (MRN) and CtIP in humans, and Mre11–Rad50–Xrs2 (MRX) and Sae2 in Saccharomyces cerevisiae (4–10). In this first step, phosphorylated CtIP/Sae2 promotes the endonuclease of MRE11 within the MRN/MRX complex to cleave the 5′-terminated DNA strand past protein blocks at sites internal to the DSBs, thus bypassing the end-bound proteins or secondary DNA structures (11,12). The second resection step is more processive and is facilitated by the EXO1 nuclease, or the DNA2 helicase-nuclease acting in conjunction with Bloom (BLM) or Werner (WRN) helicases in human cells (4,13–15). Components of the short-range resection pathways have additional structural functions to stimulate long-range resection (13,16–21). In particular, CtIP facilitates the DNA2-dependent resection pathway by promoting DNA unwinding by BLM, as well as the motor activity of DNA2 that accelerates the degradation of ssDNA unwound by BLM/WRN (16,17).
Because HR requires the use of a complementary template for repair, the timing of end resection is under a strict cell cycle control. Resection is limited to the S and G2 phases of the cell cycle when sister chromatids are available for use as template DNA (22). One of the key regulatory mechanisms to control end resection is the phosphorylation of CtIP and Sae2 (23,24). CtIP contains the key cyclin-dependent kinase (CDK) phosphorylation site (T847) that is analogous to the CDK site in Sae2 (S267), which are necessary for their function as co-factors to promote the MRN/MRX endonuclease, respectively (24). There are additional CDK phosphorylation sites in both Sae2 and CtIP, as well as in other DNA end resection factors with less-defined functions (25,26). It is generally thought that resection commits DSB repair to the recombination pathway and inhibits NHEJ. Recent work however identified that resection can be counteracted by a fill-in reaction mediated by the 53BP1–Shieldin–Polα pathway (27), suggesting that the DSB repair pathway choice may be more flexible than previously appreciated.
While the basic mechanism of DSB processing by MRN/X and CtIP/Sae2 is evolutionarily conserved, there are notable differences. Yeast Xrs2 is mostly dispensable for DNA end resection in yeast beyond mediating the nuclear import of MRX, and phosphorylation of Sae2 is sensed by the Rad50 subunit of the MRX complex (28–30). In contrast, NBS1 is comparatively much more important in human cells, where it is primarily responsible for sensing CtIP phosphorylation (26,31). Another dissimilarity is the size and domain structure between Sae2 and CtIP. Sae2 is a small protein consisting of 345 residues, which contains an N-terminal oligomerization domain and a C-terminal MRX regulatory domain bearing the key regulatory CDK phosphorylation site at S267. CtIP is instead a much larger protein, containing 897 residues (5). The very N-terminal domain of CtIP is related to that of Sae2 as it mediates tetramerization (32–34). Also, the very C-terminal region of CtIP bears limited sequence similarity with the C-terminal region of Sae2, is required to promote MRN and contains the T847 phosphorylation site (5,24). However, in contrast to Sae2, CtIP contains a large unstructured internal region (amino acids 165–790) with a poorly defined function in end resection. This internal region contains another CDK site (S327), which mediates interaction with BRCA1 (18,35,36). Additional CDK and ATM sites (including residues S233, T245, S276, T315, S347) facilitate interaction with NBS1 (26). Furthermore, the CtIP internal region (residues 690–740) facilitates the stimulation of the DNA2 translocase (16). Finally, the central region interacts with PCNA (residues 515–537) (37) and binds DNA (residues 509–557), however, it is unclear how important these interactions are for promoting end resection (38).
To define the role of CtIP in the regulation of end resection and to identify the key domains of CtIP important for this activity, we created a series of CtIP internal deletion mutants to test in a variety of cellular and biochemical assays. We found that the region between residues 350 and 600 containing the DNA and PCNA binding motifs is entirely dispensable for resection and for stimulation of the MRN endonuclease in vitro. The region was instead unexpectedly found to have a negative regulatory function, as a mutant lacking these residues was more efficient than wild type CtIP in resection assayed by single-strand annealing or quantitative PCR methods. Additional domain mapping identified a region between amino acids 550–600 of CtIP that mediates the negative regulation. Deletion of the entire internal domain between residues 165 and 790 of CtIP then dramatically reduced resection in vivo and in vitro, as it eliminated the interplay with NBS1 and DNA2. Together, this study helps clarify how the internal region of CtIP regulates DNA end resection to promote genome stability.
MATERIALS AND METHODS
Transfections and DSB reporter assays
For the DSB reporter assays, U2OS cells with the indicated reporters (SA-GFP, DR-GFP, EJ2-GFP, EJ5-GFP) (a kind gift from Jeremy Stark, City of Hope) (39) were transfected in 24-well plates. SA-GFP measures single strand annealing (SSA) by detecting the repair of an I-SceI-induced break in a 3′ fragment of green fluorescent protein (GFP) downstream of a 5′ fragment of GFP utilizing ∼200 nt of shared homology between the fragments, which generates a 2.7 kb deletion and expression of GFP. For the DR-GFP reporter assay, measuring homology-directed repair (HDR), the usage of the downstream internal GFP sequence (iGFP) as a template for repair of the I-SceI break replaces the I-SceI sequence with the complete GFP expression sequence and generates GFP-positive cells. The EJ2-GFP reporter assay measures microhomology-mediated end-joining (MMEJ) by detecting the repair of an I-SceI-induced DSB using the 8 nucleotides (nt) of flanking microhomology putting the GFP expression cassette in frame and generates GFP-positive cells. The EJ5-GFP reporter assay measures Distal-EJ as an indicator of the capacity for total end joining of the cell (a combination of both MMEJ and NHEJ). Repair of simultaneous I-SceI breaks using the distal ends deletes the puro gene and allows for expression of GFP. To deplete endogenous CtIP, a transfection mixture of 5 pmol siRNA (siCTRL, 5′-TGGTTTACATGTCGACTAA; siCtIP, 5′-GCTAAAACAGGAACGAATC; si53BP1, Dharmacon SMARTpool, 5′-GAAGGACGGAGTACTAATA, 5′-GCTATATCCTTGAAGATTT, 5′-GAGCTGGGAAGTATAAATT, 5′-GGACTCCAGTGTTGTCATT) and 1.8 μl RNAiMAX (Invitrogen) in 100 μl Opti-MEM (Gibco) was added to the wells, then 0.4 × 105 cells in 0.5 ml Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% FBS (Gibco) were seeded in each well and cultured for 20 h. Cells were then washed once and fed with 0.5 ml DMEM supplemented with 10% FBS and allowed to recover for 2 h. The cells were then transfected with 0.4 μg I-SceI (pCBASce) and 0.2 μg of either empty or siRNA-resistant CtIP variant expression vector (pcDNA3.1-derived) mixed with 1.8 μl Lipofectamine 2000 (Invitrogen) in 100 μl Opti-MEM, which was added to each well and incubated for 3 h. The cells were then washed 2× and fed with 1 ml DMEM supplemented with 10% FBS, 1% Pen Strep (Gibco) and 0.45 μg/μl plasmocin (Invivogen). After 3 days to allow for the repair of the I-SceI-catalyzed DSB, the cells were trypsinized, fixed in 1% formaldehyde and scored for the fraction of GFP-positive cells on a LSRFortessa flow cytometer (BD).
Immunoblotting
For the immunoblot analysis, the cells were transfected as for the DSB reporter assays and lysed in RIPA buffer containing 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), and 1% protease inhibitor cocktail (Sigma, P8340). Cell lysates were incubated on ice for 20 min, and then cleared by centrifugation at 15 000 g at 4°C. Subsequent SDS-PAGE and Western blotting was performed using standard methods using the following antibodies and dilutions: anti-beta-Actin (Santa Cruz sc-47778, 1:1000), anti-CtIP (Active Motif 61141, 1:1000, or Santa Cruz sc-271339, 1:1000), anti-FLAG (Sigma F3165, 1:1000), anti-53BP1 (Novus Biologicals 7158, 1:1000) and anti-NBS1 (Novus Biologicals NB100-143, 1:1000).
Clonogenic survival assay
U2OS cells were transfected as described for the DSB reporter assays. Forty eight hours after transfection, 500 cells were seeded per well in a six-well plate, in triplicate, per each condition. Cells were allowed to adhere overnight in drug-free media and then treated with indicated concentrations of camptothecin (CPT, Sigma), olaparib (Axion Medchem), bleomycin (Sigma) and cisplatin (Cayman Chemical) for 1 h. The cells were then washed and fed with drug-free media and colonies were allowed to grow for 7–10 days. Colonies were fixed with 10% acetic acid and 10% methanol and stained with 0.5% crystal violet in 10% methanol. Colonies with more than ∼50 cells were then scored.
Chromatin-bound RPA assay
Chromatin-bound RPA was analyzed using a technique published previously (40). Briefly, cells were transfected as described for the DSB reporter assays and 48 h later treated with 1 μM CPT (Sigma) for 1 h to induce DNA lesions. Cells were then trypsinized, washed with cold PBS, and permeabilized with 0.2% Triton X-100 in PBS for 7 min. All wash steps were conducted by spinning cells in a desktop centrifuge at 660 g for 4 min. After permeabilization, cells were washed with 0.1% phosphate-buffered saline (PBS) and fixed with Cytofix/Cytoperm buffer (BD) and incubated at room temperature for 15 min in the dark. Cells were then washed with 0.1% BSA in PBS and incubated with RPA primary antibody (anti-RPA, Calbiochem NA19L, 1:200) in Perm/Wash buffer (BD) at room temperature for 1 h in the dark. Cells were then washed again with 0.1% BSA in PBS and incubated with a fluorescent secondary antibody (goat anti-mouse Alexa Fluor 647, Invitrogen, A21236, 1:200) in Perm/Wash buffer (BD) at room temperature for 30 min in the dark. Finally, cells were washed with 0.1% BSA in PBS and resuspended in 0.3 ml PBS with 0.02% sodium azide (Sigma), 250 μg/ml RNase A (Sigma, R4642), and 2 μg/ml DAPI (Sigma, D8417), and then incubated at 37°C for 30 min in the dark. The fraction of RPA positive cells was then scored on a LSRFortessa flow cytometer (BD) and quantified using FlowJo software (BD).
PCR-based end resection assay
DIvA (AsiSI-ER-U2OS) (41) cells were transfected on 10 cm plates (8 × 105 cells per plate) with siCTRL or siCtIP and complemented with empty vector, or vector expressing CtIP WT or CtIP Δ1 as described for the DSB reporter assays. Two days after transfection, cells were treated with 300 nM (Z)-4-Hydroxytamoxifen (Sigma), or mock treated for 4 h. For experiments with CDKi treatment, cells were mock-treated (DMSO) or treated with 25 μM Roscovitine (Sigma) for 24 h prior to induction of DSBs. Cells were then washed twice with PBS and collected in 1 ml PBS using a cell scraper. Cell were pelleted by centrifugation for 4 min at 420 g and genomic DNA was isolated using a DNAeasy kit (Qiagen) with RNAse A treatment during lysis. Then, 500 ng of genomic DNA was treated with 5 units of RNase H (New England BioLabs) for 15 min at 37°C and heat inactivated for 20 min at 65°C. After RNase H treatment, 150 ng of genomic DNA was digested with 16 units BanI (New England BioLabs), or mock digested overnight at 37°C, then heat inactivated for 20 min at 65°C. Finally, 10 ng of digested or mock digested genomic DNA was used in a 10 μl qPCR reaction (Quanta Bio PerfeCTa SYBR Green FastMix) with 300 nM primers flanking the BanI sites at 200 bp from the AsiSI site (forward primer: ACCATGAACGTGTTCCGAAT, reverse primer: GAGCTCCGCAAAGTTTCAAG). The qPCR reactions were run using MicroAmp Fast 96-well optical plates and adhesive film (Applied Biosystems) on a QuantStudio 3 Real-Time PCR System (Applied Biosystems) using the standard SYBR green program according to manufacturer's instructions. The percentage of ssDNA generated for each sample was calculated in Microsoft Excel using the following equation: % ssDNA = 1/((POWER(2,Ct digested sample – Ct non digested sample – 1) + 0.5)) × 100.
Cloning, expression and purification of recombinant proteins
Recombinant phosphorylated CtIP and MRN proteins were purified from Spodoptera frugiperda 9 (Sf9) cells using affinity chromatography (42). The wild type pCtIP expression vector was modified from the single MBP-tagged pFastBac1 vector used in our previous work (12), in order to include a second MBP tag on the N-terminus to create pFB–2xMBP–CtIP–10xHis. The pCtIP internal deletion mutants were created by PCR amplifying the desired regions from the original wild type CtIP vector and cloned into the pFB–2xMBP vector with NheI and XmaI restriction sites. The PCR primers used for generating the internal deletion mutants are listed in Supplementary Table S1. Large-scale preparation of the pCtIP proteins was carried out by seeding Sf9 cells at 0.5 × 106 cells/ml and infecting with baculovirus 16 h later. Cells were cultured for 48 h after infection and then incubated with 25 nM Okadaic acid (Calbiochem) for 4 h and 1 μM CPT (Sigma) for 1 h prior to collection of the cell pellet to promote phosphorylation. The cell pellets were stored at −80°C. The pCtIP cell pellets were then thawed and resuspended in ice-cold lysis buffer with 50 mM Tris–HCl pH 8.5, 1 mM dithiothreitol (DTT), 1:400 (v/v) protease inhibitor cocktail (Sigma, P8340), 30 μg/ml leupeptin (Merck), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM ethylenediaminetetraacetic (EDTA), 300 mM NaCl, 10% glycerol, 0.5% NP-40, 25 nM Okadaic acid, 15 mM Na4P2O7, 1 mM Na3VO4 and 20 mM NaF for 20 min at 4°C. The sample was sonicated with a probe sonicator (Branson SLPe) with 6 rounds of 1 s pulses at 70% intensity for 30 s each. The soluble extract was isolated by centrifugation at 50 000 g for 30 min at 4°C. To bind the MBP-tagged pCtIP proteins, the soluble extract was incubated with equilibrated amylose resin (New England Biolabs) for 1 h at 4°C under constant agitation. The amylose resin was then washed batch-wise four times by centrifugation at 2000 g for 2 min at 4°C with MBP wash buffer (50 mM Tris–HCl pH 7.5, 2 mM beta-mercaptoethanol, 250 mM NaCl, 10% glycerol, 1 mM PMSF, and 0.5% NP-40). The protein-bound amylose resin was transferred onto a 10 ml disposable column (ThermoFisher) and washed eight more times by gravity flow. The protein was eluted with MBP wash buffer supplemented with 10 mM maltose (Sigma). The eluate was incubated with a 1:5 ratio (w/w) of PreScission protease for 1 h at 4°C to cleave the 2xMBP tag. The cleaved protein was bound to equilibrated NiNTA resin (Qiagen) by incubating at 1 h at 4°C under constant agitation. The protein-bound NiNTA resin was transferred onto a 5 ml gravity column and washed 8 times with NiNTA wash buffer (50 mM Tris–HCl pH 7.5, 2 mM beta-mercaptoethanol, 150 mM NaCl, 10% glycerol, 1 mM PMSF and 20 mM imidazole) by gravity flow. The protein was eluted with NiNTA wash buffer supplemented with 300 mM imidazole and dialyzed (50 mM Tris–HCl pH 7.5, 2 mM beta-mercaptoethanol, 150 mM NaCl, 10% glycerol and 1 mM PMSF) to remove the imidazole before aliquoting, snap frozen with liquid nitrogen and stored at −80°C.
MRN was purified as a complex by co-infecting Sf9 cells with three baculoviruses expressing the individual components of the complex (42). Sf9 cells were seeded at 0.5 × 106 cells/ml and infected with MRE11-His, RAD50-FLAG and NBS1 baculoviruses 16 h later. The infected cells were cultured for 52 h after infection before harvesting. Cell pellets were lysed with 50 mM Tris–HCl pH 7.5, 2 mM beta-mercaptoethanol, 1 mM EDTA, 1:400 (v/v) protease inhibitor cocktail (Sigma), 1 mM PMSF, 30 μg/ml leupeptin and 20 mM imidazole for 15 min at 4°C before adding glycerol and NaCl to a final concentration of 15% and 305 mM, respectively, and incubating for another 25 min at 4°C. The soluble extract was collected by centrifugation at 50 000 g for 30 min at 4°C. The soluble extract was then bound to equilibrated NiNTA resin (Qiagen) by incubating for 1 h at 4°C under constant agitation. The protein-bound resin was washed batch-wise four times by centrifugation at 2000 g for 2 min at 4°C with NiNTA wash buffer (50 mM Tris–HCl pH 7.5, 2 mM beta-mercaptoethanol, 300 mM NaCl, 10% glycerol, 1 mM PMSF and 20 mM imidazole), then transferred onto a 10 ml disposable column (ThermoFisher) and washed six times with NiNTA wash buffer by gravity flow. The protein was eluted with NiNTA wash buffer supplemented with 250 mM imidazole and diluted 1:2.5 with dilution buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10% glycerol and 1 mM PMSF) before binding to the equilibrated FLAG resin (Sigma) by incubating for 1 h at 4°C under constant agitation. The protein-bound FLAG resin was transferred onto a 5 ml gravity flow column (ThermoFisher) and washed four times with FLAG wash buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM PMSF and 1 mM beta-mercaptoethanol) by gravity flow. The protein was eluted with FLAG wash buffer supplemented with 150 ng/μl 3xFLAG peptide (ApexBio), aliquoted, snap frozen and stored at −80°C.
DNA2 was expressed in Spodoptera frugiperda 9 (Sf9) insect cells and purified by affinity chromatography using the N-terminal 6X his-tag and the C-terminal FLAG-tag (15). Human RPA was expressed in E. coli and purified using an ÄKTA pure (GE Healthcare) with HiTrap Blue HP, HiTrap Desalting and HiTrap Q chromatography columns (all GE Healthcare) (42).
Protein interaction assays
50 ml Sf9 cells per each binding reaction were infected with baculovirus expressing FLAG-NBS1. Cells were lysed 52 h later with 1.5 ml lysis buffer (50 mM Tris–HCl pH 7.5, 0.5 mM beta-mercaptoethanol, 1 mM EDTA, 1:400 (v/v) protease inhibitor cocktail [Sigma, P8340], 30 μg/ml leupeptin [Merck], 305 mM NaCl, 16% [v/v] glycerol) and FLAG-NBS1 (∼1.2 μg) was immobilized on 30 μl anti-FLAG M2 affinity gel (Sigma) as a bait protein. Next, in a 150 μl reaction, 80 nM pCtIP variants (prey proteins) were added and incubated in binding buffer (25 mM Tris–HCl pH 7.5, 0.5 mM DTT, 3 mM EDTA, 50 mM NaCl, 0.2 mg/ml BSA) for 1 h at 4°C with continuous rotation. Beads were washed four times with 300 μl wash buffer (50 mM Tris–HCl pH 7.5, 0.5 mM DTT, 3 mM EDTA, 80 mM NaCl, 0.1% NP-40) and proteins were eluted with wash buffer supplemented with 3× FLAG peptide (150 ng/μl, Sigma). Bound FLAG-NBS1 was detected by western blotting using rabbit anti-NBS1 antibody (Novus Biologicals, NB100-143, 1:1000) and pCtIP variants were detected by mouse anti-pCtIP (Santa Cruz, sc-271339, 1:1000). As a negative control, the prey proteins were incubated with resin without the bait protein.
Nuclease assays
Nuclease assays were conducted in 15 μl reactions with 25 mM Tris–HCl pH 7.5, 5 mM magnesium acetate, 1 mM manganese acetate, 1 mM DTT, 1 mM ATP, 0.25 mg/ml BSA (New England Biolabs), 1 mM phosphoenolpyruvate (Sigma), 80 U/ml pyruvate kinase (Sigma), and 1 nM oligonucleotide-based DNA substrate (in molecules) (43). To create the simulated protein blocks, 15 nM streptavidin (Sigma) was added to the biotinylated DNA substrate in nuclease buffer and incubated at 5 min at room temperature. The recombinant proteins were then added on ice and incubated at 37°C for 30 min. For the experiments with MR, the reactions were prepared as with MRN but incubated for 2 h (31). The following steps were the same for both MRN and MR reactions. Reactions were stopped by adding 0.5 μl of 0.5 M EDTA and 1 μl Proteinase K (19 mg/ml, Roche), and incubated at 50°C for 30 min. The samples were then mixed with 15 μl of loading dye (95% formamide, 20 mM EDTA, 0.1% bromophenol blue), boiled at 95°C for 4 min, and then separated on 15% polyacrylamide denaturing gels (7 M urea, 19:1 acrylamide-bisacrylamide, Bio-Rad). Resolved gels were fixed in 40% methanol, 10% acetic acid, and 5% glycerol for 30 min at room temperature before being dried on 3MM CHR paper (Whatman) (43). The dried gels were exposed on storage phosphor screens (GE Healthcare) and scanned with a Typhoon Phosphor Imager (GE Healthcare). Quantification of the nuclease products were carried out using ImageQuant software (GE Healthcare).
DNA2 nuclease assays were carried out in 15 μl reactions containing 25 mM Tris-acetate pH 7.5, 3 mM magnesium acetate, 1 mM ATP, 1 mM DTT, 0.1 mg/ml BSA (New England Biolabs), 1 mM phosphoenolpyruvate (PEP), 80 U/ml pyruvate kinase (Sigma) and 0.15 nM substrate (boiled λ DNA/HindIII fragments, in molecules). Reactions were incubated at 37°C as indicated. Human RPA (860 nM) was included to saturate all ssDNA. Reactions were stopped by adding 5 μl of 2% stop solution (150 mM EDTA, 2% sodium dodecyl sulfate, 30% glycerol, 0.1% bromophenol blue) and 1 μl of proteinase K (19 mg/ml, Roche) and incubated at 37°C for 10 min. Samples were analyzed by 1% agarose gel electrophoresis (16). Gels were dried on DE81 chromatography paper (Whatman), exposed to storage phosphor screens (GE Healthcare) and scanned by a Typhoon 9500 phosphorimager (GE Healthcare).
Preparation of DNA-based substrates
Oligonucleotides purified by polyacrylamide gel electrophoresis were purchased from Eurogentec. The 70 bp quadruple-blocked substrates were generated by labeling the 3′ end of PC210 (GTAAGTGCCGCGGTGCGGGTGCCAGGGCGTGCCCTTGGGCTCCCCGGGCGCGTACTCCACCTCATGCATC) and annealing to PC211 (GATGCATGAGGTGGAGTACGCGCCCGGGGAGCCCAAGGGCACGCCCTGGCACCCGCACCGCGGCACTTAC). The biotin labels were attached to internal thymidines (in bold). The labeling reaction was carried out using terminal deoxynucleotidyl transferase (New England Biolabs) and α-32P cordycepin 5′ triphosphate (Perkin Elmer) (43). For the DNA2 stimulation assays, HindIII digested λ DNA (New England Biolabs) was labeled at the 3′ end with [α-32P] dATP (Perkin Elmer) and the Klenow fragment of DNA polymerase I (New England Biolabs). Unincorporated nucleotides were removed with Micro Bio-Spin P-30 chromatography columns (Biorad). Subsequently, prior to each experiment, the substrate was heated at 95°C for 5 min to obtain ssDNA (16).
Propidium iodide staining for cell cycle analysis
Cells were transfected with siCTRL and empty vector as described for the DSB reporter assays two days prior to 24 h treatment with 25 μM Roscovitine (Sigma) or mock treated (DMSO). Cells were then trypsinized and washed twice with cold PBS. Cells were then fixed with 70% ethanol and incubated on ice for 1 h. Next, cells were centrifuged at 660 g for 4 min, the ethanol was removed and the cell pellet was resuspended in 500 μl PBS with 10 μg/ml RNAse A (Sigma) and 20 μg/ml propidium iodide (Sigma) and incubated at 30°C for 30 min. Samples were then analyzed on a LSRFortessa flow cytometer (BD) and quantified using FlowJo software (BD).
Colony-forming assay
Cells were transfected with the indicated siRNA and complementation vector as described for the DSB reporter assays. Two days after transfection, cells were seeded at low density on six-well plates with normal growth media and incubated for 7–10 days to allow colonies to form. Colonies were then fixed, stained and counted as described for the clonogenic survival assays. Results are shown as the plating efficiency (number of colonies/number of cells seeded).
RESULTS
The internal region of CtIP inhibits single-strand annealing in vivo
To determine the function of the internal CtIP region in regulating DNA end resection, we first created a pair of internal deletion mutants (Figure 1A). The first mutant (CtIP Δ1) lacked amino acids 350–600, which includes the PCNA and DNA binding sites but retains the CDK phosphorylation sites important for BRCA1 and NBS1 binding, as well as the region required for DNA2 stimulation (16,26,36). The second internal truncation mutant (CtIP Δ2) lacks residues 165–790, essentially fusing the N-terminal region important for tetramerization to the C-terminal region, creating a ‘Sae2-like’ CtIP. Finally, to further investigate the importance of the N-terminal oligomerization domain of CtIP, we also prepared the Sae2-like C-terminal region spanning amino acids 790–897 (CtIP Δ3).
Figure 1.
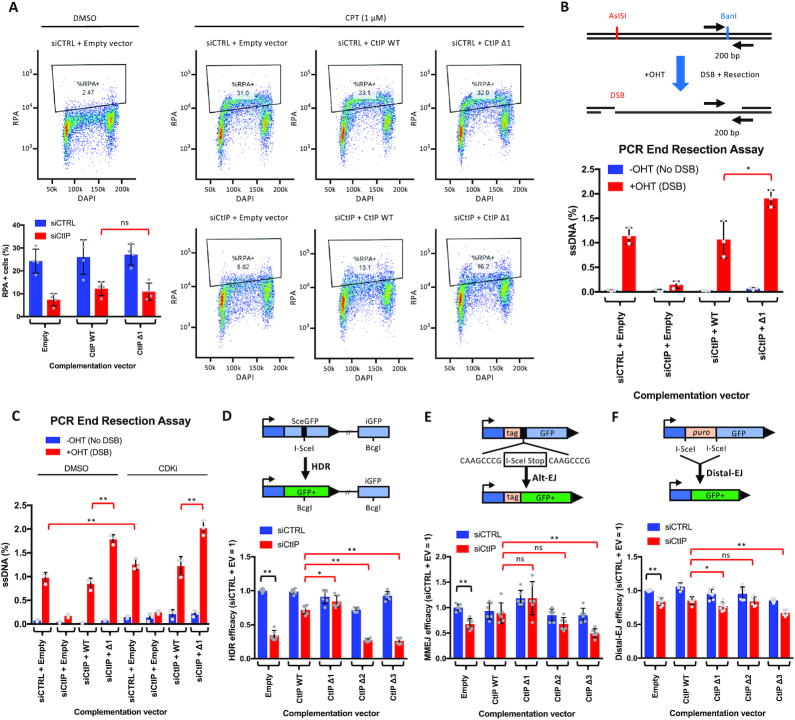
The internal region of CtIP inhibits single strand annealing. (A) Schematic of Sae2 and CtIP protein variants used in this study. Key domains and phosphorylation sites (P) are indicated. Missing amino acid regions in CtIP internal deletion mutants are indicated by a black line. (B) Schematic of the single strand annealing (SA-GFP DSB reporter) assay (top). The quantitation below shows the SSA efficacy after normalizing to siCTRL + empty vector (EV). Averages shown; n = 6; error bars, SD. Statistical denotations represent comparisons between samples indicated by brackets; ns (P > 0.05, not significant), **(P < 0.01), two-tailed t-test. (C) Western blot analysis showing depletion of endogenous CtIP by siCtIP (top left) and expression of the siRNA-resistant, FLAG-tagged CtIP (top right) using anti-CtIP antibody. The expression of the CtIP variants is shown using anti-FLAG antibody at the bottom. Beta-actin blots were used as loading controls.
We then tested how the CtIP truncations complement loss of endogenous CtIP using the single strand annealing (SSA) reporter assay in U2OS cells. In SSA, following an induced DSB, the ends are resected until two repeated matching sequences are found. The complementary sequences then anneal to each other and are further processed to result in a deletion (in this case 2.7 kb-long) of the region between the two repeats. As SSA is a mechanistically simple pathway independent of RAD51 and strand exchange, it is a reliable readout for DNA end resection activity in vivo (44). We depleted endogenous CtIP with siRNA (or mock-depleted with siCTRL) and simultaneously expressed siRNA-resistant FLAG-tagged CtIP variants in U2OS cells integrated with the SA-GFP DSB reporter (Figure 1B). Depletion of CtIP resulted in a dramatic reduction of SSA, which could be complemented by a vector expressing wild type CtIP, but not upon introduction of an empty vector (Figure 1B). Unexpectedly, complementation with CtIP Δ1 caused a ∼2-fold increase in SSA compared to complementation with wild type CtIP. The increase was apparent even when endogenous CtIP was not downregulated, indicating that the CtIP Δ1 fragment has a dominant effect to promote SSA (Figure 1B). One possible explanation for the observed increase in SSA after complementation with CtIP Δ1 was the increased expression level of CtIP Δ1 compared to wild type CtIP (Figure 1C). To address this, we varied the amount of transfected DNA during the complementation experiments. Even though the amount of expressed protein increased dramatically with the different DNA levels (Supplementary Figure S1A), we saw no significant change in SSA (Supplementary Figure S1B). Specifically, even when the amount of CtIP Δ1 was lower than that of wild type CtIP (Supplementary Figure S1A, compare lanes 4 and 5), the SSA was higher with CtIP Δ1, demonstrating that the observed difference is not due to various protein levels. We also note that the expression levels of all plasmid-encoded variants were higher than those of the endogenous CtIP protein (Figure 1C). The amino acid region between residues 350 and 600 has thus unexpectedly a negative regulatory function on SSA and possibly DNA end resection.
In contrast to CtIP Δ1, complementation of CtIP-depleted cells with CtIP Δ2 only minimally supported SSA. This is likely due to CtIP interactions with NBS1, BRCA1, and DNA2, which are disrupted in CtIP Δ2 (26,36,45), and an additional defect due to protein misfolding also cannot be excluded. In contrast, no SSA was observed upon complementation with CtIP Δ3 (Figure 1B). The importance of the N-terminal oligomerization domain was established previously (32,46). Collectively, these data indicate that the amino acid region lacking in CtIP Δ1 (350–600) negatively regulates SSA and potentially DNA end resection.
The internal region of CtIP is dispensable for stimulating MRN and DNA2 in vitro
Our observation that CtIP Δ1 is more efficient in SSA could mean that the variant is more efficient in stimulating the MRN and/or DNA2 nuclease directly, or the region may contain a post-translational modification or a binding site for an interaction partner in vivo that serves as a negative regulator of end resection indirectly. To distinguish between the two possibilities, we purified the CtIP variants (Figure 2A and Supplementary Figure S2A–D). A direct effect would be anticipated to result in elevated resection capacity in the reconstituted system. Wild type CtIP, as well as all its variants, were expressed in Sf9 cells in the presence of phosphatase inhibitors to preserve phosphorylation (12). Phosphorylation of CtIP is important for its capacity to stimulate the MRE11 endonuclease within the MRN complex, as well to stimulate the motor activity of DNA2 (12,16). The purified proteins were then used in reconstituted DNA end resection reactions with MRN and DNA2 nucleases in vitro.
Figure 2.
The internal region of CtIP is dispensable for stimulating MRN and DNA2 in vitro. (A) Purified pCtIP variants used in this study. The polyacrylamide gel was stained with Coomassie brilliant blue. (B) Representative endonuclease assays using recombinant MRN and the indicated pCtIP variants on 3′-end-labeled 70 bp dsDNA with all ends blocked with streptavidin. Reactions were incubated for 30 min at 37°C. Samples were separated on 15% denaturing polyacrylamide gels. (C) Quantitation of nuclease assays such as in (B). Averages shown; n = 3; error bars, SD. (D) Coimmunoprecipitation assays of the pCtIP variants and FLAG-NBS1. FLAG-NBS1 was immobilized on FLAG resin and incubated with the pCtIP variants. NBS1 interacts with wild type pCtIP and pCtIP Δ1 but not with pCtIP Δ2. Shown is a representative experiment. The lower bands in lanes 2 and 3 are truncation mutants of the recombinant pCtIP proteins. For input, see Supplementary Figure S2E. (E) Representative kinetic experiments showing degradation of ssDNA of different lengths by DNA2 alone or in the presence of different variants of pCtIP. All reactions contained 860 nM RPA.
First, we sought to investigate if the increase in SSA when complementing with CtIP Δ1 was due to a more efficient stimulation of the MRE11 endonuclease activity. To this point, phosphorylated CtIP variants (pCtIP) were incubated with recombinant MRN in a previously-established nuclease assay designed to detect DNA cleavage near protein-blocked DNA ends (Figure 2B) (11). The assay utilizes oligonucleotide-based dsDNA with a radioactive phosphate at the 3′ terminus of one DNA end and biotin–streptavidin tags on all four DNA ends to simulate a protein-blocked DSB, and to prevent exonucleolytic DNA degradation that is CtIP-independent (12). Using limiting CtIP variant concentrations, we observed that both wild type pCtIP and the pCtIP Δ1 mutant stimulated the endonuclease of MRN to a similar extent (Figure 2B,C). In contrast, much lower endonuclease activity was detected with the CtIP Δ2 variant. While this defect can be due to protein unfolding, we hypothesize that it can also be because of disrupted physical interaction between the FHA/BRCT domains of NBS1 and the phosphorylated residues in the CtIP region 233–347, which is missing in pCtIP Δ2 (Figure 1A) (26,31). To confirm this, we immobilized FLAG-tagged NBS1 on anti-FLAG resin, and incubated with the CtIP variants. While the interaction of NBS1 with pCtIP Δ1 was comparable to that with full-length CtIP, the physical interaction of NBS1 with pCtIP Δ2 was abrogated (Figure 2D and Supplementary Figure S2E). Accordingly, upon longer incubation, residual DNA cleavage by MR (without NBS1) was stimulated similarly by wild type pCtIP, as well as by pCtIP Δ1 and pCtIP Δ2 variants (Supplementary Figure S2F,G). These experiments are in agreement with the weak, NBS1-independent, resection observed previously (31). We conclude that the reduced MRE11-dependent endonuclease activity we observed with pCtIP Δ2 is due to abrogated physical and functional interaction of the pCtIP Δ2 variant with NBS1.
Next, we compared the capacity of full-length pCtIP and pCtIP Δ1 polypeptides to promote the nuclease of DNA2. To this point, we used an assay that monitors the degradation of ssDNA by wild type DNA2 (16,47). The use of ssDNA bypasses the requirement of the BLM/WRN helicase, and is thus a simple readout of the effect of pCtIP on DNA2. The residues 350–600 lie outside of the region of CtIP required to promote DNA2 (16). To compare resection efficacy quantitatively, we next performed kinetic experiments with DNA2 and either wild type pCtIP or pCtIP Δ1. The experiments clearly indicated that pCtIP Δ1 was just as efficient as wild type pCtIP in stimulating DNA2 (Figure 2E) (16). No nuclease activity has been detected in this assay with pCtIP alone (12,16). Collectively, as the pCtIP Δ1 variant was comparable to wild type pCtIP in promoting MRE11- and DNA2-dependent DNA degradation in vitro, our data suggested that the internal truncation does not result in a structural change in CtIP that would make its function as a co-factor of the MRE11 and DNA2 nucleases more efficient. Rather, the data suggest that the region missing in CtIP Δ1 may have an indirect negative regulatory function to prevent extensive resection in vivo.
The internal region of CtIP inhibits resection and is dispensable for other aspects of DSB repair
To further investigate the role of the internal CtIP region between residues 350 and 600 in DNA end resection, we examined the accumulation of chromatin-bound replication protein A (RPA) after induction of DSBs upon treatment with the DNA topoisomerase I (TopoI) inhibitor, camptothecin (CPT). RPA is a ssDNA binding protein that binds to the ssDNA tails generated by end resection and is used as an indicator of cellular end resection proficiency. Chromatin-bound RPA was then scored by flow cytometry (40). In contrast to the SSA assay, we observed no statistical difference in chromatin-bound RPA levels between cells complemented with wild type CtIP and CtIP Δ1 (Figure 3A). However, this assay based on detection of RPA-foci is a low-resolution method that does not yield sufficient quantitative information about the extent of resected DNA (48). To this point, we used an RT-PCR based assay that allows accurate quantitation of the fraction of resected DNA at a defined position away from an endonuclease-induced DSB (41). In contrast to the RPA-foci formation, the high-resolution of the PCR-based method provides better quantitative data (Figure 3B). We observed that the depletion of CtIP resulted in a dramatic decrease of DNA end resection, which could be rescued by expressing wild type CtIP. Importantly, the siRNA-resistant CtIP Δ1 variant rescued the resection defect even better than wild type CtIP (Figure 3B), in agreement with our data from the SSA reporter assay.
Figure 3.
The internal region of CtIP inhibits DNA end resection. (A) Flow cytometry-based DNA end resection assay showing chromatin-bound RPA after CPT treatment. Shown are the representative flow cytometry plots of U2OS cells treated with siCTRL or siCtIP and complemented with the indicated construct. Quantitation is shown at bottom left; averages shown; n = 4; error bars, SD. Statistical denotations represent comparison to siCtIP + CtIP WT; ns (P > 0.05, not significant), two-tailed t-test. (B) Schematic of RT-PCR based DNA end resection assay in DivA cells. DSBs at AsiSI sites were induced with 4-hydroxytamoxifen (OHT). DNA end resection was then monitored at a site 200 bp away from the DSB. Arrows indicate the position of the PCR primers. Below are the quantification of the RT-PCR based DNA end resection assay. Averages shown; n = 3; error bars, SD. Statistical denotations represent comparison to siCtIP + CtIP WT + OHT; * (P < 0.05) two-tailed t-test. (C) Quantification of the RT-PCR based DNA end resection assays after CDK inhibition. Averages shown; n = 3; error bars, SD. Statistical denotations represent comparison between siCTRL + empty vector DMSO versus CDKi treatment or CtIP WT versus CtIP Δ1 treated cells; **(P < 0.01), two-tailed t-test. (D) Homology directed repair (HDR) efficacy estimated by the DR-GFP DSB reporter assay in cells where CtIP was not depleted (siCTRL) or upon CtIP depletion (siCtIP) and complementation with the indicated siRNA-resistant construct. The values are presented as relative to siCTRL with empty vector. Averages shown; n = 6; error bars, SD. Statistical denotations represent comparison to siCTIP + CtIP WT; * (P < 0.05), **(P < 0.01), two-tailed t-test. (E) Microhomology-mediated end-joining (MMEJ) efficacy estimated by the EJ2-GFP DSB reporter assay in cells where CtIP was not depleted (siCTRL) or upon CtIP depletion (siCtIP) and complementation with the indicated siRNA-resistant construct. The values are presented as relative to siCTRL with empty vector. Averages shown; n = 6; error bars, SD. Statistical denotations represent comparison to siCTIP + CtIP WT; ns (P > 0.05, not significant), **(P < 0.01), two-tailed t-test. (F) Distal-EJ efficacy estimated by the EJ5-GFP DSB reporter assay in cells where CtIP was not depleted (siCTRL) or upon CtIP depletion (siCtIP) and complementation with the indicated siRNA-resistant construct. The values are presented as relative to siCTRL with empty vector. Averages shown; n ≥ 6; error bars, SD. Statistical denotations represent comparison to siCTIP + CtIP WT; ns (P > 0.05, not significant), * (P < 0.05), **(P < 0.01), two-tailed t-test.
Due to the cell cycle-dependent regulatory control that restricts DNA end resection to the S and G2 phases (23,24,49), we set out to determine if the elevated levels of DNA end resection in CtIP Δ1 result from uninhibited resection in G1. To address this, we analyzed DNA end resection levels after treatment with the CDK inhibitor Roscovitine. Roscovitine caused cells to accumulate in G2 (Supplementary Figure S3A). As anticipated, we observed a corresponding increase in end resection in cells treated with siCTRL complemented with an empty vector after CDKi treatment (Figure 3C). Importantly, we also saw a proportional increase in DNA end resection in cells complemented with both full-length CtIP and CtIP Δ1 upon CDKi treatment (Figure 3C). These results indicate that the elevated levels of end resection in CtIP Δ1 do not result from mis-regulated resection in G1.
SSA generally requires extensive resection to reveal repetitive DNA sequences on both ends of the DSB that can mediate annealing. However, homology-directed repair (HDR) and microhomology-mediated end-joining (MMEJ, also termed Alt-EJ) also require the use of homologous sequences and thus end resection, although to a different extent (1). Accordingly, CtIP has been shown to be important for both of these repair pathways (5,39). In contrast, NHEJ, which does not require homology for repair, proceeds independently of CtIP (1). To evaluate the role of the central region of CtIP on pathway choice in DSB repair, we used cell-based DSB reporter assays DR-GFP, EJ2-GFP and EJ5-GFP to measure HDR, MMEJ and distal end-joining (EJ) (a combination of both MMEJ and NHEJ, which is used as an indicator of the total end-joining capacity of a cell), respectively (39) (Figure 3D-F). We observed a decrease in HDR and MMEJ after the depletion of endogenous CtIP (siCTRL vs siCtIP after complementation with EV) (Figure 3D and E), as reported previously (39). In contrast, we observed no major decrease in distal-EJ after CtIP depletion (Figure 3F), as expected. Complementation with wild type CtIP partially restored HDR and MMEJ levels. In HDR, the CtIP Δ1 variant restored repair to a level higher than wild type CtIP, while in MMEJ the small increase was not statistically significant (Figure 3D and E). Neither CtIP Δ2 nor CtIP Δ3 managed to effectively restore HDR or MMEJ back to endogenous levels (Figure 3D and E). Conversely, there was a small but statistically significant decrease in distal-EJ after complementation with CtIP Δ1, compared to CtIP WT (Figure 3F). This decrease in distal-EJ indicates a shift in the balance between HDR and EJ towards HDR when CtIP Δ1 was present. Collectively, these data indicate that the internal region of CtIP is required for a regulatory control mechanism that prevents over-resection that can be detected by the SSA and quantitative RT-PCR-based assays. The elevated resection in CtIP Δ1 cells then affects the pathway choice in DSB repair.
Additional internal deletion mutants of CtIP identify amino acids 550–600 as the key domain in regulating SSA
To more precisely identify the region of CtIP responsible for the inhibition of SSA in the wild type protein, we next generated a series of internal deletion mutants by sequentially adding 50 amino acids to either side of the region deleted in the CtIP Δ1 mutant (Figure 4A). After complementing the loss of endogenous CtIP in U2OS cells with the SA-GFP reporter with each of these new CtIP internal deletion mutants, we observed increased SSA with deletion mutants where amino acids were added back from the N-terminus (mutants Δ4, Δ5 and Δ6), compared to wild type CtIP. In contrast, the increase in SSA was abrogated when amino acids were added back from the C-terminus (mutants Δ7, Δ8 and Δ9, Figure 4B and Supplementary Figure S4A). Specifically, the CtIP Δ7 variant, missing residues 350–550, did not show the increased SSA as CtIP Δ1, missing residues 350–600. There was no additional difference in SSA after further addition of amino acids to the C-terminal side of the central domain (compare CtIP Δ7 with Δ8 and Δ9, Figure 4A and B), indicating the region between amino acids 550–600 of CtIP contains the negative regulatory domain for SSA.
Figure 4.
The CtIP domain limiting SSA maps to amino acid region 550–600. (A) Schematic of additional CtIP internal deletion mutants, with the deleted amino acid regions indicated by a horizontal black line. For CtIP Δ1-Sgs1, the 250 amino acids deleted in CtIP Δ1 were replaced with an unstructured region of S. cerevisiae Sgs1 of the same length (residues 90–340 of Sgs1) indicated by the striped box. The symbols on the right depict the SSA efficacy relative to wild type CtIP (++, strongly elevated; + elevated; –, reduced). (B) Quantitation of SSA efficacy using SA-GFP reporter assay. CtIP was either not depleted (siCTRL) or depleted (siCtIP) and complemented with the indicated siRNA-resistant internal deletion construct. The values are presented as relative to siCTRL with empty vector. The dotted line indicates the average SSA efficiacy of siCtIP + CtIP WT experiments for comparison. Averages shown; n = 4; error bars, SD. Statistical denotations represent comparison to siCTIP + CtIP WT; ns (P > 0.05, not significant), * (P < 0.05), **(P < 0.01), two-tailed t-test. (C) Quantification of RT-PCR based DNA end resection assays in cells complemented with CtIP Δ10, wild type CtIP and CtIP Δ1. Averages shown; n = 3; error bars, SD. **(P < 0.01), two-tailed t-test. (D) Representative nuclease assays showing the proficiency of recombinant pCtIP Δ7 construct to promote the endonuclease of MRN. Compare to Figure 2, panel B. (E) Representative kinetic experiment showing comparable ssDNA degradation by DNA2 in complex with pCtIP Δ7 and pCtIP Δ1. For DNA degradation without pCtIP, see Figure 2, panel E (lanes 3–9). All reactions contained 860 nM RPA.
To further support this conclusion, we next expressed an internal deletion mutant of CtIP lacking only amino acids 550–600 (CtIP Δ10, Figure 4A, Supplementary Figure S4C). This mutant showed an increase in SSA compared to CtIP WT (Figure 4B), as well as elevated end resection as measured by the RT-PCR assay (Figure 4C), supporting the conclusion that the negative regulatory domain of CtIP lies between amino acids 550–600. While there were differences between the expression levels of the individual truncation constructs, they did not correlate with the capacity of the variants to promote SSA (Figure 4B and Supplementary Figure S4A). Additionally, we also generated a CtIP variant where the amino acids deleted from CtIP Δ1 were replaced with a non-specific unstructured stretch of residues of the same length (sequence taken from S. cerevisiae Sgs1, residues 90–340, CtIP Δ1-Sgs1) (Figure 4A). No human protein is expected to specifically bind this unique yeast protein sequence. Complementation with the CtIP Δ1-Sgs1 variant caused a similar increase in SSA as the CtIP Δ1 mutant (Figure 4B). This indicated that both the deletion and the replacement of the region including residues 550–600 with a non-specific sequence abrogates the negative effect of this sequence on SSA.
Importantly, as the CtIP Δ7 variant was less efficient than CtIP Δ1 to promote SSA, we investigated its capacity to promote MRN and DNA2 in vitro. pCtIP Δ7 was fully proficient in stimulating the MRE11 endonuclease within the MRN complex (Figure 4D and Supplementary Figure S4B), as well as the DNA2 nuclease (Figure 4E) in vitro. Therefore, wild type pCtIP, pCtIP Δ1 and pCtIP Δ7 were indistinguishable from each other in their capacities to promote the MRE11 and DNA2 nucleases in vitro. This showed that the presence of the 550–600 amino acid region of CtIP does not result in structural alterations that directly affect the interplay of CtIP with the resection nucleases. Rather, these data suggest that the 550–600 region within CtIP appears to function specifically as a negative regulator of resection and SSA in vivo.
The regulatory domain in CtIP functions independently of 53BP1 and BRCA1
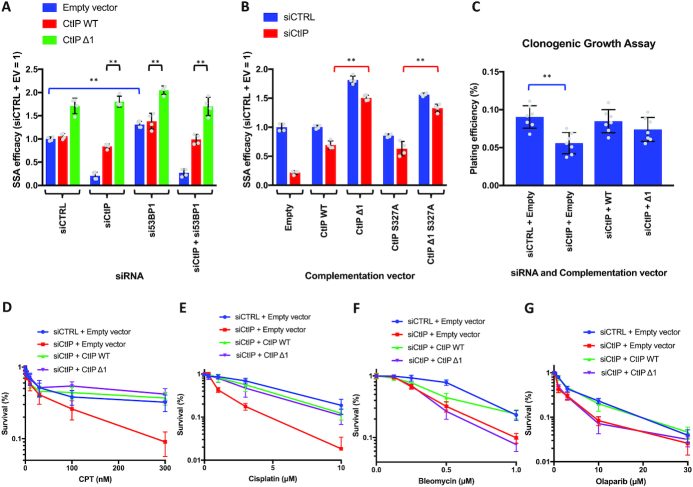
Due to the important role of the balance between 53BP1 and BRCA1 factors on regulating DNA end resection (50), we wanted to determine if the regulatory domain we identified in CtIP is functionally linked to either of these proteins. 53BP1 with its effectors such as the Shieldin complex has been identified as pro EJ factor that negative regulates or counteracts DNA end resection (27,51–53). Therefore, we reasoned that CtIP Δ1 mutant might have escaped the control by 53BP1. To test this, we used siRNAs to deplete 53BP1, CtIP or both, and complemented the loss of endogenous CtIP with CtIP WT and CtIP Δ1 variants. We then analyzed the efficacy of SSA. As anticipated, SSA increased after the depletion of 53BP1. However, CtIP Δ1, compared to wild type CtIP, promoted SSA to a higher level also in the absence of 53BP1. Therefore, the regulatory domain missing in CtIP Δ1 restricts resection by a mechanism that is independent of 53BP1 (Figure 5A, Supplementary Figure S5A). Conversely, BRCA1 has been shown to be a factor important for promoting end resection (48,54,55). BRCA1 has been shown to interact with CtIP upon phosphorylation at S327 of CtIP (26,35,36,45). Upon introduction of the non-phosphorylatable CtIP S327A mutation into both full-length CtIP and CtIP Δ1 backgrounds, we monitored the ability of the CtIP variants to complement the loss of CtIP in the SSA reporter assay. We observed a similar increase in DNA end resection comparing full-length and CtIP Δ1 even in the presence of the CtIP S327A mutation (Figure 5B, Supplementary Figure S4C). Collectively, these results indicate that the end resection phenotype observed in CtIP Δ1 cells cannot be explained by a mis-regulation of the BRCA1-53BP1 axis.
Figure 5.
The regulatory domain functions independently of 53BP1 and BRCA1 and provides resistance to DNA damaging agents that require resection for repair. (A) Quantitation of SSA efficacy using SA-GFP reporter assay. CtIP and 53BP1 were either not depleted (siCTRL) or depleted (siCtIP, and/or si53BP1) and complemented with the indicated siRNA-resistant CtIP construct. The values are presented as relative to siCTRL with empty vector. Averages shown; n = 4; error bars, SD. Statistical denotations represent comparison to siCTRL + empty vector or CtIP WT vs CtIP Δ1; **(P < 0.01), two-tailed t-test. (B) Quantitation of SSA efficacy using SA-GFP reporter assay. CtIP was either not depleted (siCTRL) or depleted (siCtIP) and complemented with the indicated siRNA-resistant CtIP construct. The values are presented as relative to siCTRL with empty vector. Averages shown; n = 4; error bars, SD. Statistical denotations represent comparison to siCtIP + WT; ns (P > 0.05, not significant), two-tailed t-test. (C) Quantification of cell growth assays. Cells were transfected as for the DSB reporter assays and then seeded at low density to allow for colonies to form. Results are shown as plating efficiency. Averages shown; n = 7; error bars, SD. Statistical denotations represent comparison to siCTRL + empty vector; **(P < 0.01), two-tailed t-test. (D) Quantitation of cell survival upon CPT treatment in cells where CtIP was not depleted (siCTRL) or upon CtIP depletion (siCtIP) and complementation with the indicated construct. The values are presented as relative to cells not treated with CPT. Averages shown; n = 3; error bars, SD. (E) Assay as in D, but upon cisplatin treatment. Averages shown; n = 3; error bars, SD. (F) Assay as in D, but upon bleomycin treatment. Averages shown; n = 3; error bars, SD. (G) Assay as in D, but upon olaparib treatment. Averages shown; n = 3; error bars, SD.
Increased resection in CtIP Δ1 cells promotes resistance to camptothecin but not other genotoxic drugs
To investigate the biological significance of the control mechanism conferred by the internal CtIP domain, we first evaluated whether expressing the CtIP Δ1 mutant affects cell viability in the absence of DNA damage. To this point, we monitored colony-forming capacity of cells complemented with CtIP WT and CtIP Δ1. Our results show a clear decrease in plating efficiency upon transient depletion of CtIP, which was rescued by complementation with wild type CtIP (Figure 5C). Interestingly, CtIP Δ1 did not provide any advantage over wild type CtIP, in fact there was a slight decrease in plating efficiency of CtIP Δ1 cells compared to CtIP WT, which was however not statistically significant (Figure 5C).
Next, we tested cellular sensitivity to various DNA-damaging drugs. CtIP-deficient cells are in particular sensitive to topoisomerase inhibitors, which result in DSBs blocked with covalently attached topoisomerase DNA cleavage complexes (5,56,57). The protein blocks can be removed by MRN and CtIP (56,57). To this point, we treated transiently complemented U2OS cells with increasing concentrations of the DNA topoisomerase I inhibitor camptothecin (CPT) and monitored survival (Figure 5D). We observed a strong decrease in cell survival in CPT-treated cells after the depletion of endogenous CtIP (siCtIP + empty vector). The expression of CtIP Δ1 could efficiently rescue this sensitivity, even more so than wild type CtIP (Figure 5D). In contrast, we did not observe such as resistance in CtIP Δ1 cells upon treatment with other classes of DNA damaging drugs (Figure 5E–G). In particular, CtIP Δ1 could not complement cells treated with bleomycin or olaparib; in these cases, wild type CtIP complemented the cellular sensitivity very efficiently (Figure 5F, G). This indicated that when a high level of DNA end resection activity is required such as to remove covalent protein blocks from DSBs, the upregulated DNA end resection in CtIP Δ1 efficiently promotes cellular resistance. In response to other type of DNA damage however, the mis-regulated activity of CtIP Δ1 is detrimental.
DISCUSSION
Resection at DSB sites is needed to reveal 3′-tailed ssDNA of sufficient length to localize matching template DNA, which is usually the sister chromatid and rarely the homologous chromosome in vegetative cells. In cases when homologous DNA is not found, recombination may take place between ectopic loci with homeologous sequences, potentially leading to DNA translocations (2,44). The likelihood of such events may increase with the length of the resected DNA. Similarly, extended DNA end resection may lead to SSA, leading to deletions between two repetitive DNA sequences (1,2). Therefore, cells have developed regulatory mechanisms that place resection under CDK control, which allows it to occur only in the S/G2 phases of the cell cycle when sister chromatids are available (22). CtIP and its phosphorylation is the key target of such a regulatory mechanism, as it controls key nucleases involved in resection including MRE11 and DNA2 (12,24). Resection is further regulated by 53BP1 and its effectors that inhibit resection or promote fill-in DNA synthesis (27,51–53), and by BRCA1 that facilitates resection by a yet undefined mechanism (50). Many additional factors that regulate resection both positively and negatively have been identified, although the underlying molecular mechanisms are not always understood (26,58–60). The proper balance between the pro and anti-resection signals is required for optimal maintenance of genome stability.
Here we show that CtIP, the key pro-resection factor in human cells, contains a domain that has an unexpected negative regulatory function in resection. We revealed that deletion or replacement of a stretch of residues between amino acids 550 and 600 from the internal region of CtIP leads to an unanticipated increase of SSA and DNA end resection in cellular assays. The same internally truncated CtIP is indistinguishable from the full-length protein in its capacity to promote MRN and DNA2 nucleases in vitro, suggesting that the truncation does not result in a structural alteration of CtIP that leads to a gain-of-function. Rather, we propose that the internal region of CtIP might serve as a binding site for a negative regulatory factor, which may affect CtIP either by direct interaction or post-translational modifications. We also performed mass spectrometry analysis to identify potential interacting partners that might be responsible for this negative regulation, however our experiments failed to reveal promising candidates. Therefore, the interaction might be transient, as would be anticipated, e.g. for kinases and phosphatases. More work is needed to understand the underlying mechanism leading to increased resection in the CtIP Δ1 and related mutants. Interestingly, the increased resection observed in cells expressing internally truncated CtIP confers resistance to camptothecin that likely requires high MRN-CtIP resection activity to remove stalled DNA topoisomerase I from the ends of DNA breaks, as shown for topoisomerase inhibitors in general (56,57). In contrast, the same CtIP variant was incapable to support survival upon treatment with other DNA-damaging drugs. These results underpin the need for optimal balance of DNA end resection. Less than physiological resection activity results in decreased survival both under unperturbed conditions and upon DNA damage. Higher than normal resection can be advantageous under specific conditions such as in response to camptothecin, but this comes at the cost of elevated SSA that is very mutagenic, and impaired response to other types of DNA damage.
One of the most exciting breakthroughs in biomedical technology in the last few years has been the development of CRISPR-Cas9 for gene editing. This technique is very efficient in generating gene knockouts based on inaccurate repair by NHEJ. However, the efficacy of homology-driven repair aimed to introduce specific DNA sequence alterations is much lower and limiting in particular for applications where only low amounts of cells are available and no selection markers can be used. To this point, it has been proposed to decrease the efficacy of NHEJ by specific inhibitors to shift the repair balance and thus promote HR-driven events (61). Furthermore, another promising strategy is to fuse Cas9 with HR factors. This has been already demonstrated for Cas9-CtIP fusions, which increased the efficacy of homology-directed repair ∼2-fold (62,63). Interestingly, the N-terminal oligomerization domain containing residues 1–270 was as efficient as the full-length CtIP protein (63). While the underlying mechanism is not fully clear, it is possible that the CtIP fragment retains its capacity to interact with endogenous full-length CtIP, which targets functional CtIP to the DSB sites and mediates the stimulatory function. Our experiments demonstrated that CtIP Δ1 was more efficient than wild type in promoting DNA resection, and thus such a fragment fused to Cas9 may promote homology-driven repair even further, in particular as we observed that the CtIP Δ1 fragment increased SSA in a dominant manner. This strategy may increase the success rate for Cas9-mediated genome engineering.
DATA AVAILABILITY
The entire primary dataset is available online at Dryad repository: https://doi.org/10.5061/dryad.msbcc2fvd.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jeremy Stark (City of Hope), Pavel Janscak (University of Zurich), Tanya Paull (UT Austin), Gaelle Legube (CBI Toulouse) and Silvia Monticelli (IRB Bellinzona) for cell lines, plasmids and reagents. We thank members of the Cejka laboratory (S. Halder, G. Reginato, E. Cannavo, A. Acharya, A. Sanchez, V. Kissling) for comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [31003A_175444 to P.C.]; European Research Council [681630 to P.C.]; European Molecular Biology Organization [ALTF 1560-2015 to S.M.H.]; the Institute for Research in Biomedicine is supported by the Helmut Horten Foundation. Funding for open access charge: SNSF.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ranjha L., Howard S.M., Cejka P.. Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma. 2018; 127:187–214. [DOI] [PubMed] [Google Scholar]
- 2. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daley J.M., Niu H., Miller A.S., Sung P.. Biochemical mechanism of DSB end resection and its regulation. DNA Repair (Amst.). 2015; 32:66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G.. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008; 134:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P.. Human CtIP promotes DNA end resection. Nature. 2007; 450:509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gravel S., Chapman J.R., Magill C., Jackson S.P.. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008; 22:2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mimitou E.P., Symington L.S.. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008; 455:770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J.L., Kowalczykowski S.C.. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010; 467:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Niu H., Chung W.H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L. et al.. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010; 467:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Usui T., Ohta T., Oshiumi H., Tomizawa J., Ogawa H., Ogawa T.. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998; 95:705–716. [DOI] [PubMed] [Google Scholar]
- 11. Cannavo E., Cejka P.. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014; 514:122–125. [DOI] [PubMed] [Google Scholar]
- 12. Anand R., Ranjha L., Cannavo E., Cejka P.. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 2016; 64:940–950. [DOI] [PubMed] [Google Scholar]
- 13. Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., Kowalczykowski S.C.. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011; 25:350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sturzenegger A., Burdova K., Kanagaraj R., Levikova M., Pinto C., Cejka P., Janscak P.. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 2014; 289:27314–27326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pinto C., Kasaciunaite K., Seidel R., Cejka P.. Human DNA2 possesses a cryptic DNA unwinding activity that functionally integrates with BLM or WRN helicases. Elife. 2016; 5:e18574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ceppi I., Howard S.M., Kasaciunaite K., Pinto C., Anand R., Seidel R., Cejka P.. CtIP promotes the motor activity of DNA2 to accelerate long-range DNA end resection. Proc. Natl. Acad. Sci. U.S.A. 2020; doi:10.1073/pnas.2001165117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daley J.M., Jimenez-Sainz J., Wang W., Miller A.S., Xue X., Nguyen K.A., Jensen R.B., Sung P.. Enhancement of BLM-DNA2-Mediated Long-Range DNA End Resection by CtIP. Cell Rep. 2017; 21:324–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoa N.N., Kobayashi J., Omura M., Hirakawa M., Yang S.H., Komatsu K., Paull T.T., Takeda S., Sasanuma H.. BRCA1 and CtIP are both required to recruit dna2 at double-strand breaks in homologous recombination. PLoS One. 2015; 10:e0124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peterson S.E., Li Y., Wu-Baer F., Chait B.T., Baer R., Yan H., Gottesman M.E., Gautier J.. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol. Cell. 2013; 49:657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nicolette M.L., Lee K., Guo Z., Rani M., Chow J.M., Lee S.E., Paull T.T.. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2010; 17:1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cannavo E., Cejka P., Kowalczykowski S.C.. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E1661–E1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M. et al.. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004; 431:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huertas P., Cortes-Ledesma F., Sartori A.A., Aguilera A., Jackson S.P.. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008; 455:689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huertas P., Jackson S.P.. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009; 284:9558–9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ferretti L.P., Lafranchi L., Sartori A.A.. Controlling DNA-end resection: a new task for CDKs. Front. Genet. 2013; 4:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang H., Shi L.Z., Wong C.C., Han X., Hwang P.Y., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W. et al.. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013; 9:e1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mirman Z., Lottersberger F., Takai H., Kibe T., Gong Y., Takai K., Bianchi A., Zimmermann M., Durocher D., de Lange T.. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature. 2018; 560:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oh J., Al-Zain A., Cannavo E., Cejka P., Symington L.S.. Xrs2 dependent and independent functions of the Mre11-Rad50 complex. Mol. Cell. 2016; 64:405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cannavo E., Johnson D., Andres S.N., Kissling V.M., Reinert J.K., Garcia V., Erie D.A., Hess D., Thoma N.H., Enchev R.I. et al.. Regulatory control of DNA end resection by Sae2 phosphorylation. Nat. Commun. 2018; 9:4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang W., Daley J.M., Kwon Y., Krasner D.S., Sung P.. Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev. 2017; 31:2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anand R., Jasrotia A., Bundschuh D., Howard S.M., Ranjha L., Stucki M., Cejka P.. NBS1 promotes the endonuclease activity of the MRE11-RAD50 complex by sensing CtIP phosphorylation. EMBO J. 2019; 38:e101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davies O.R., Forment J.V., Sun M., Belotserkovskaya R., Coates J., Galanty Y., Demir M., Morton C.R., Rzechorzek N.J., Jackson S.P. et al.. CtIP tetramer assembly is required for DNA-end resection and repair. Nat. Struct. Mol. Biol. 2015; 22:150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andres S.N., Appel C.D., Westmoreland J.W., Williams J.S., Nguyen Y., Robertson P.D., Resnick M.A., Williams R.S.. Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat. Struct. Mol. Biol. 2015; 22:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilkinson O.J., Martin-Gonzalez A., Kang H., Northall S.J., Wigley D.B., Moreno-Herrero F., Dillingham M.S.. CtIP forms a tetrameric dumbbell-shaped particle which bridges complex DNA end structures for double-strand break repair. Elife. 2019; 8:e42129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu X., Chen J.. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004; 24:9478–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen L., Nievera C.J., Lee A.Y., Wu X.. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008; 283:7713–7720. [DOI] [PubMed] [Google Scholar]
- 37. Gu B., Chen P.L.. Expression of PCNA-binding domain of CtIP, a motif required for CtIP localization at DNA replication foci, causes DNA damage and activation of DNA damage checkpoint. Cell Cycle. 2009; 8:1409–1420. [DOI] [PubMed] [Google Scholar]
- 38. You Z., Shi L.Z., Zhu Q., Wu P., Zhang Y.W., Basilio A., Tonnu N., Verma I.M., Berns M.W., Hunter T.. CtIP links DNA double-strand break sensing to resection. Mol. Cell. 2009; 36:954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bennardo N., Cheng A., Huang N., Stark J.M.. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008; 4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Forment J.V., Walker R.V., Jackson S.P.. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry A. 2012; 81:922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou Y., Caron P., Legube G., Paull T.T.. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014; 42:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Anand R., Pinto C., Cejka P.. Methods to study DNA end resection I: recombinant protein purification. Methods Enzymol. 2018; 600:25–66. [DOI] [PubMed] [Google Scholar]
- 43. Pinto C., Anand R., Cejka P.. Methods to study DNA end resection II: biochemical reconstitution assays. Methods Enzymol. 2018; 600:67–106. [DOI] [PubMed] [Google Scholar]
- 44. Bhargava R., Onyango D.O., Stark J.M.. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 2016; 32:566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yun M.H., Hiom K.. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009; 459:460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yuan J., Chen J.. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J. Biol. Chem. 2009; 284:31746–31752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Levikova M., Pinto C., Cejka P.. The motor activity of DNA2 functions as an ssDNA translocase to promote DNA end resection. Genes Dev. 2017; 31:493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cruz-Garcia A., Lopez-Saavedra A., Huertas P.. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014; 9:451–459. [DOI] [PubMed] [Google Scholar]
- 49. Orthwein A., Noordermeer S.M., Wilson M.D., Landry S., Enchev R.I., Sherker A., Munro M., Pinder J., Salsman J., Dellaire G. et al.. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015; 528:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50. Bunting S.F., Callen E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L. et al.. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010; 141:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dev H., Chiang T.W., Lescale C., de Krijger I., Martin A.G., Pilger D., Coates J., Sczaniecka-Clift M., Wei W., Ostermaier M. et al.. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 2018; 20:954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gupta R., Somyajit K., Narita T., Maskey E., Stanlie A., Kremer M., Typas D., Lammers M., Mailand N., Nussenzweig A. et al.. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell. 2018; 173:972–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noordermeer S.M., Adam S., Setiaputra D., Barazas M., Pettitt S.J., Ling A.K., Olivieri M., Alvarez-Quilon A., Moatti N., Zimmermann M. et al.. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018; 560:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aparicio T., Gautier J.. BRCA1-CtIP interaction in the repair of DNA double-strand breaks. Mol Cell Oncol. 2016; 3:e1169343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aparicio T., Baer R., Gottesman M., Gautier J.. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J. Cell Biol. 2016; 212:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hoa N.N., Shimizu T., Zhou Z.W., Wang Z.Q., Deshpande R.A., Paull T.T., Akter S., Tsuda M., Furuta R., Tsusui K. et al.. Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol. Cell. 2016; 64:580–592. [DOI] [PubMed] [Google Scholar]
- 57. Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E. et al.. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLos Genet. 2010; 6:e1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jimeno S., Fernandez-Avila M.J., Cruz-Garcia A., Cepeda-Garcia C., Gomez-Cabello D., Huertas P.. Neddylation inhibits CtIP-mediated resection and regulates DNA double strand break repair pathway choice. Nucleic Acids Res. 2015; 43:987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lopez-Saavedra A., Gomez-Cabello D., Dominguez-Sanchez M.S., Mejias-Navarro F., Fernandez-Avila M.J., Dinant C., Martinez-Macias M.I., Bartek J., Huertas P.. A genome-wide screening uncovers the role of CCAR2 as an antagonist of DNA end resection. Nat. Commun. 2016; 7:12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gong Y., Handa N., Kowalczykowski S.C., de Lange T.. PHF11 promotes DSB resection, ATR signaling, and HR. Genes Dev. 2017; 31:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L.. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015; 33:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tran N.T., Bashir S., Li X., Rossius J., Chu V.T., Rajewsky K., Kuhn R.. Enhancement of precise gene editing by the association of Cas9 with homologous recombination factors. Front Genet. 2019; 10:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Charpentier M., Khedher A.H.Y., Menoret S., Brion A., Lamribet K., Dardillac E., Boix C., Perrouault L., Tesson L., Geny S. et al.. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Commun. 2018; 9:1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The entire primary dataset is available online at Dryad repository: https://doi.org/10.5061/dryad.msbcc2fvd.