Abstract
In mammalian cells, distinct H3K4 methylation states are created by deposition of methyl groups by multiple complexes of histone lysine methyltransferase 2 (KMT2) family proteins. For comprehensive analyses that directly compare the catalytic properties of all six human KMT2 complexes, we employed a biochemically defined system reconstituted with recombinant KMT2 core complexes (KMT2CoreCs) containing minimal components required for nucleosomal H3K4 methylation activity. We found that each KMT2CoreC generates distinct states and different levels of H3K4 methylation, and except for MLL3 all are stimulated by H2Bub. Notably, SET1BCoreC exhibited the strongest H3K4 methylation activity and, to our surprise, did not require H2B ubiquitylation (H2Bub); in contrast, H2Bub was required for the H3K4me2/3 activity of the paralog SET1ACoreC. We also found that WDR5, RbBP5, ASH2L and DPY30 are required for efficient H3K4 methyltransferase activities of all KMT2CoreCs except MLL3, which could produce H3K4me1 in the absence of WDR5. Importantly, deletion of the PHD2 domain of CFP1 led to complete loss of the H3K4me2/3 activities of SET1A/BCoreCs in the presence of H2Bub, indicating a critical role for this domain in the H2Bub-stimulated H3K4 methylation. Collectively, our results suggest that each KMT2 complex methylates H3K4 through distinct mechanisms in which individual subunits differentially participate.
INTRODUCTION
Histone H3 lysine 4 (H3K4) methylation plays an important role in many cellular processes, such as transcription and DNA replication, repair and recombination, that are all central to cell growth, differentiation and development (1–3). The association of impaired H3K4 methylation with various human diseases, including developmental defects and cancers (4–7), has motivated intense efforts to understand the precise roles of this histone modification in regulating cellular events.
H3K4 methylation can be present in three states: mono-methylated (H3K4me1), di-methylated (H3K4me2), and tri-methylated (H3K4me3). In mammalian cells, H3K4 methylation is mediated by at least six SET domain-containing KMT2 (lysine methyltransferase 2) family proteins—SET1A/KMT2F, SET1B/KMT2G, MLL1/KMT2A, MLL2/KMT2B, MLL3/KMT2C, and MLL4/KMT2D—each of which forms a multimeric complex (hereafter, KMT2 complex) with several subunits common to all complexes (e.g., WDR5, RbBP5, ASH2L, and DPY30) as well as some that are unique to each complex (7). The roles of individual KMT2 complexes in transcriptional regulation have been extensively studied. Preferential co-localization of SET1A with H3K4me3 at promoter regions of highly expressed genes supports a general regulatory role for the SET1A complex in transcriptional activation (8). MLL1 and MLL2 are reported to control hematopoietic development by inducing H3K4me3 at the promoter regions of Hox gene clusters through selective recruitment to these genes (9,10). For MLL2 specifically, it has been shown that MLL2-mediated H3K4me3 on bivalent promoters is critical for the precise differentiation of stem cells (11,12). In addition, MLL3 and MLL4 deposit H3K4me1 at enhancers involved in cell type-specific gene expression (13–15). Thus, despite having an identical target site on histone H3, these complexes are not redundant, marking methylations at different locations in their specific target genes and producing different methylation states, which in turn recruit distinct effector molecules (1,8).
Interestingly, early yeast genetic studies found that mono-ubiquitylation of histone H2B (H2Bub) at lysine 123 is a prerequisite for H3K4 methylation (16,17). Subsequent biochemical analyses using purified factors clearly demonstrated that H2Bub directly stimulates H3K4 methylation mediated by the yeast Set1 complex (18) and human SET1A complex (19). Recent studies have unveiled the mechanistic basis of this histone crosstalk in the yeast Set1 complex, showing that it involves H2Bub-induced conformational changes in the complex that support H3K4 methylation activity (20–22).
Compared with cellular functions of KMT2 proteins, which have been extensively investigated, our understanding of the catalytic properties of human KMT2 complexes, including their relative H3K4 methylation activities and associated H2Bub requirements for enzymatic activity, is limited. Here, using a biochemically defined system employing recombinant KMT2 complexes and chromatin templates, we performed comprehensive and comparative biochemical analyses to characterize H3K4 methyltransferase activities of human KMT2 complexes. Our studies revealed that the H3K4 methylation activities of all KMT2 complexes except MLL3 are stimulated by H2Bub. We further found that KMT2 complexes generate different levels of distinct H3K4 methylation states and have distinct subunit requirements for their catalytic activities, suggesting that distinct mechanistic pathways govern H3K4 methylation processes among KMT2 complexes.
MATERIALS AND METHODS
Construction of plasmids and baculoviruses, and purification of recombinant proteins and human KMT2 family complexes
For baculovirus-mediated expression, cDNAs were subcloned into pFASTBAC1 (Gibco-Invitrogen), with or without an epitope, and baculoviruses were generated according to the manufacturer's instructions (Gibco-Invitrogen). For reconstitution of complexes containing FLAG-tagged KMT2 family protein fragments, Sf9 cells were infected with combinations of baculoviruses, and complexes were affinity purified on M2 agarose (Sigma) as described (23). For plasmid transfection-mediated expression, cDNAs encoding N-terminally FLAG-tagged, full-length human KMT2 family proteins and cDNAs encoding untagged KMT2 family complex subunits were subcloned into pVLAD6 (Addgene) (24). HEK293E cells (∼1.5 × 106/ml) in suspension culture (300 ml) were transfected with combinations of expression plasmid (150 μg each) using the polyethylenimine (PEI; Sigma) method. After 60 h, cells were treated with 2 μM bortezomib (Millipore) and incubated for an additional 12 h. Cells were washed once with phosphate-buffered saline (PBS) and then with five packed-cell volumes (PCVs) of hypotonic buffer (10 mM Tris–HCl [pH 7.4], 10 mM KCl, and 1.5 mM MgCl2) each, after which they were resuspended in 2 PCVs of hypotonic buffer and incubated for 10 min. Swollen cells were disrupted with a Dounce homogenizer (pestle B, 15 strokes) and centrifuged at 3,900 rpm for 10 min. Nuclear pellets were resuspended in 10 ml extraction buffer (10 mM Tris–HCl [pH 7.9], 300 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, 1 mM PMSF, 2 mM DTT and protease inhibitor cocktail [Roche]), minced with a Dounce homogenizer (pestle B, 3 rounds of 10 strokes at 10 min intervals), and then centrifuged at 18,000 rpm for 20 min. Clarified extracts were incubated with 80 μl of M2 agarose for 4 h. After extensive washing with wash buffer (20 mM Tris–HCl [pH 7.9], 200 mM NaCl, 2 mM MgCl2, 0.2 mM EDTA, 15% glycerol, 0.1% NP40, 1 mM PMSF and 1 mM DTT), complexes were eluted with wash buffer containing 0.25 mg/ml FLAG peptide. The expression and purification of recombinant Xenopus histones, semi-synthetic H2Bub, histone octamers, NAP1 and the ACF complex were as described (23,25).
In vitro chromatin assembly
Procedures for chromatin assembly using the recombinant ACF/NAP1 system were as described (23). Briefly, 700 ng core histone octamer and 2.4 μg NAP1 in 55 μl HEG buffer (25 mM HEPES [pH 7.6], 0.1 mM EDTA and 10% glycerol) were incubated on ice for 30 min. After addition of 160 ng ACF complex and 700 ng p53ML plasmid (26), the reaction was adjusted to 25 mM HEPES [pH 7.6], 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol and 3.2 mM ATP in a final volume of 70 μl, and incubated at 27°C for 4 h.
In vitro histone methyltransferase assays
For free histone H3 methyltransferase assays, reactions containing 100 ng recombinant histone H3 and the indicated concentration of purified KMT2 complex in 20 μl reaction buffer (25 mM HEPES [pH 7.6], 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 10% glycerol) supplemented with 100 μM SAM (S-adenosyl methionine; NEB) were incubated at 30°C for 1 h. For recombinant chromatin methyltransferase assays, reactions containing 250 ng (histone amount) recombinant chromatin, the indicated concentration of purified KMT2 complex and 100 μM SAM were adjusted to 50 μl with HEG buffer, and incubated at 30°C for 1 h. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to immunoblot analyses.
Antibodies
Polyclonal anti-SET1A and anti-DPY30 antibodies were developed against purified histidine-tagged human SET1A fragment (amino acid residues 358–477) and human DPY-30 (full-length) proteins, respectively, and affinity purified (AbClon). Polyclonal anti-CFP1 and anti-WDR82 antibodies were obtained from the Roeder laboratory. Polyclonal anti-WDR5 antibody was obtained from the Allis laboratory. The following antibodies were obtained commercially: anti-SET1B (A302-281A), anti-MLL2 (A300-113A), anti-RbBP5 (A300-109A), anti-ASH2L (A300-489A), and anti-Menin (A300-105A) from Bethyl Laboratories; anti-WDR5 (ab22512), anti-H3 (ab1791), anti-H3K4me1 (ab8895), and anti-H3K4me2 (ab7766) from Abcam; anti-H3K4me3 (AM39159) from Active Motif; and anti-FLAG (A8592) from Sigma.
RESULTS
H2B ubiquitylation directly stimulates H3K4 methylation activities of human KMT2Core complexes
The large size of catalytic/scaffold KMT2 family proteins has been a major obstacle for purification of KMT2 complexes to homogeneity. In addition to a catalytic SET domain-containing KMT2 protein and the associating common subunits, WDR5, RbBP5, ASH2L and DPY30, collectively referred to a ‘WRAD’, each KMT2 family complex also has unique subunits that may differentially affect H3K4 methylation activities. Therefore, for comparative biochemical analyses of intrinsic enzyme activities of all six human KMT2 complexes, we sought to identify domains and subunits that are minimally required for H3K4 methyltransferase activities. In this context, we previously demonstrated that a partial yeast Set1 complex (C762 complex) composed of a Set1 fragment (residues 762–1080) encompassing the n-SET, SET and post-SET domains together with five associating subunits, Swd3 (a homolog of mammalian WDR5), Swd1 (RbBP5), Bre2 (ASH2L), Sdc1 (DPY30) and Spp1 (CFP1), exhibits robust H2B ubiquitylation-dependent H3K4 methylation activity toward chromatin substrates (18,21).
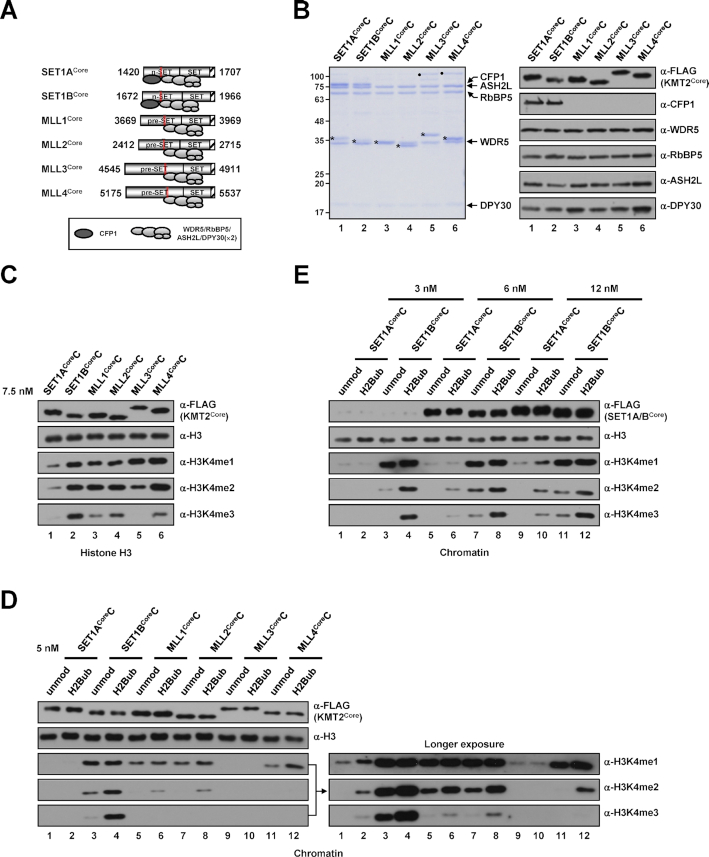
Accordingly, we reconstituted partial human KMT2 core complexes (hereafter, KMT2CoreCs) corresponding to the yeast C762 Set1 complex and affinity purified them from Sf9 insect cells infected with baculoviruses expressing FLAG-KMT2Core protein fragments (encompassing n-SET, SET and post-SET domains for SET1A/B, and pre-SET, SET and post-SET domains for MLL1/2/3/4) and five untagged subunits (WRAD and CFP1) (Figure 1A). All WRAD subunits were stably integrated into all purified complexes, but CFP1 was found only in SET1ACoreC and SET1BCoreC (Figure 1B), confirming SET1A- and SET1B-specific association of CFP1 (27,28) through the interaction with n-SET domains, similar to the case for the yeast Set1 complex (18).
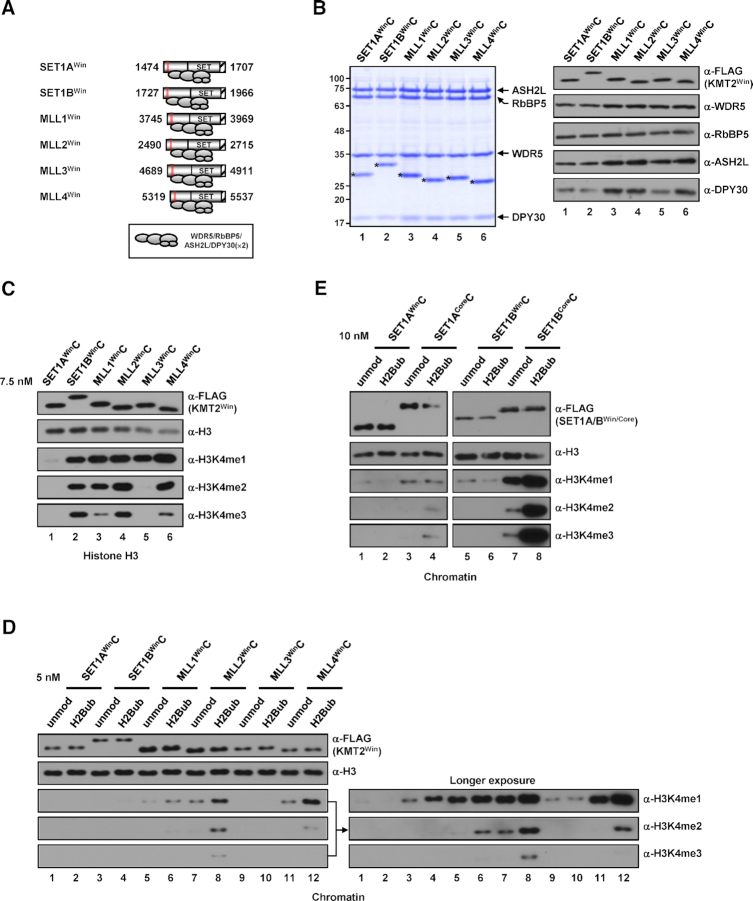
Figure 1.
H3K4 methylation activities of human KMT2Core complexes. (A) A schematic diagram of KMT2Core fragments used for the reconstitution of complexes and subunit associations deduced from (B). The n-SET, pre-SET, SET and post-SET (hatched box) domains and Win (red box) motif within each KMT2 protein are depicted. Numbers indicate amino acid residues of each protein. DPY30(×2) indicates the dimeric association of DPY30, which was established in recent structural analyses of KMT2 complexes (29,31,32). (B) SDS-PAGE/Coomassie blue staining (left) and immunoblot analyses (right) of purified KMT2Core complexes reconstituted with baculoviruses expressing FLAG-tagged KMT2Core protein (A), WDR5, RbBP5, ASH2L, DPY30 and CFP1. KMT2Core proteins are marked by asterisks. Note that an insect protein contaminant around 100 kDa (marked by dots) was found sometimes in the complexes prepared by single-step affinity purification and became more prominent when a large volume of samples (due to low concentration) was loaded. This protein could be removed by gel filtration and did not affect the H3K4 methylation activities of the affinity-purified complexes as demonstrated in several KMT2Core complexes (see Supplementary Figures). (C) Differential H3K4 methylation activities of KMT2Core complexes towards free histone H3 (H3) substrate. H3 was subjected to in vitro HMT assays with the indicated KMT2core complexes (7.5 nM). (D and E) Differential H3K4 methylation activities of KMT2core complexes towards chromatin substrates. Recombinant chromatin templates assembled with unmodified histone H2B (unmod)- or mono-ubiquitylated H2B at lysine 120 (H2Bub)-containing octamers were subjected to in vitro HMT assays with the indicated KMT2Core complexes at a concentration of 5 nM (D) or with the indicated concentrations of SET1ACore and SET1BCore complexes (E). H3K4 methylation status was monitored by immunoblotting with the indicated antibodies in this and other figures. Abbreviations: unmod, unmodified histone H2B; H2Bub, K120-ubiquitylated H2B; H3K4me1, K4-monomethylated H3; H3K4me2, K4-dimethylated H3; H3K4me3, K4-trimethylated H3.
In vitro histone methyltransferase (HMT) assays using free histone H3 (H3) as the substrate revealed that all purified KMT2CoreCs, except MLL3CoreC, are able to generate all three H3K4 methylation states; for MLL3CoreC, activity was restricted to H3K4me1 and H3K4me2 (Figure 1C). Notably, SET1BCoreC exhibited the strongest H3K4me3 activity (Figure 1C, lane 2), whereas the overall H3K4 methylation activity of SET1ACoreC was weakest among the tested complexes (Figure 1C, lane 1).
We next tested the H3K4 methylation activity of complexes towards more physiologically relevant chromatin templates containing either unmodified histone H2B (H2B) or mono-ubiquitylated H2B at lysine 120 (H2Bub) (25). Consistent with its weak activity towards free H3, SET1ACoreC showed weak H3K4 methylation activity that was detectable in immunoblots only after prolonged exposure (Figure 1D, lanes 1 and 2) or in reactions with a high concentration of the complex (Supplementary Figure S1A). More importantly, SET1ACoreC–generated H3K4me2/3 exhibited H2Bub dependence. Consistent with its strong activity towards free H3, SET1BCoreC generated all three H3K4 methylation states at the highest levels among the tested complexes (Figure 1D, lanes 3 and 4). In vitro HMT assays using the complexes further purified by Superose 6 size exclusion chromatography also revealed that SET1BCoreC exhibited much stronger H3K4 methylation activities than SET1ACoreC (Supplementary Figure S2). Although H2Bub increased the H3K4 methylation activity of SET1BCoreC at all tested concentrations (Supplementary Figure S1B), we found that, in clear contrast to SET1ACoreC (Figure 1E), SET1BCoreC generated H3K4 methylation even in the absence of H2Bub. This suggests that, despite having a domain and subunit compositions similar to that of the SET1A complex and yeast Set1 complex, the SET1B complex is able to bypass the requirement of H2Bub for H3K4 methylation because of its strong intrinsic methyltransferase activity. We also found that both MLL1CoreC and MLL2CoreC were able to generate all three H3K4 methylation states, activities that were stimulated, albeit modestly, by H2Bub (Figure 1D, lanes 5–8, Supplementary Figure S1C and D). The chromatin H3K4 methylation activity of MLL3CoreC was very weak compared with that of other complexes and was restricted to H3K4me1 (Figure 1D, lanes 9 and 10, Supplementary Figure S1E). H2Bub-mediated stimulation of MLL3CoreC activity was not clearly observed, but this could be because the intrinsic H3K4me1 activity of MLL3CoreC is too weak to allow possible H2Bub stimulatory effects to be monitored by immunoblot analysis. This interpretation is in line with the results of a previous study using an alternative luminescent method that showed that H2Bub stimulated the H3K4me1 activity of a partial MLL3 complex (29). Lastly, MLL4CoreC exhibited mainly H3K4me1/2 activities that were stimulated by H2Bub; however, the stimulatory effect of H2Bub was modest, requiring longer exposure of immunoblots (Figure 1D, lanes 11 and 12) or reactions with a higher concentration of the complex (Supplementary Figure S1F) to be detectable. Collectively, our biochemical analyses demonstrate that all KMT2Core complexes have distinctive H3K4 methyltransferase properties, with different strengths of activity and methylation species produced, but they were stimulated to varying degrees by the presence of H2Bub on the chromatin. Thus, these observations suggest that a common component(s) in KMT2 family complexes might be responsible for their sensitivity to H2Bub-stimulated H3K4 methylation (see Discussion).
Human SET1ACore and SET1BCore complexes exhibit distinctive subunit requirements for H3K4 methylation activities
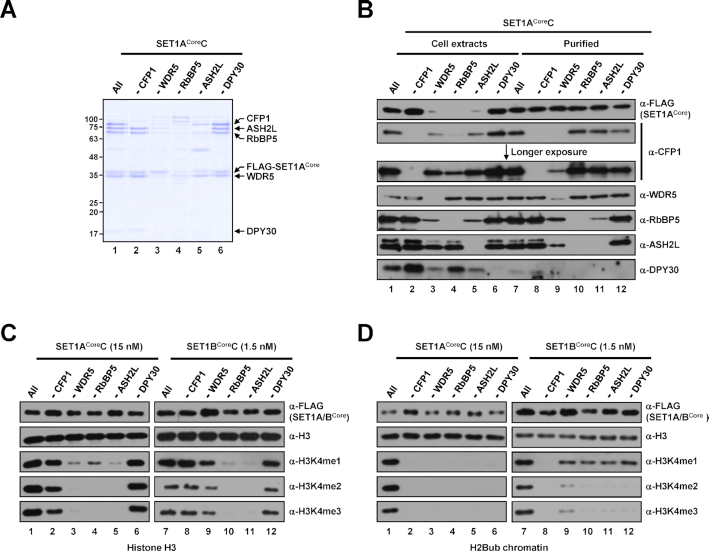
Given our ability to reconstitute catalytically active KMT2Core complexes, we next sought to establish the requirements of individual subunits within each KMT2CoreC for H3K4 methylation activity. To this end, we first reconstituted and affinity-purified SET1ACore and SET1BCore complexes lacking individual subunits. Yields of purified complexes obtained in the absence of WDR5, RbBP5 or ASH2L were significantly lower, suggesting contributions of these subunits to the efficient formation and stability of SET1ACoreC (Figure 2A, lanes 3–5) and SET1BCoreC (Supplementary Figure S3A). This conclusion is further supported by the much lower expression of scaffold SET1ACore and SET1BCore proteins as well as some subunits in Sf9 cells infected with baculoviruses lacking WDR5, RbBP5 or ASH2L (Figure 2B, lanes 3–5, Supplementary Figure S3B).
Figure 2.
Subunit requirements for H3K4 methylation activities of human SET1ACore and SET1BCore complexes. (A) SDS-PAGE/Coomassie blue staining of purified SET1ACore complexes reconstituted with baculoviruses in the absence of the indicated subunits. Sample loadings were normalized to FLAG-KMT2Core proteins in this and other figures. Note that because of inefficient complex formation (∼10–20-fold lower concentration compared with the intact complex), 5- and 2.5-fold lower amounts (maximum loading volumes) of RbBP5 and ASH2L-deficient complexes, respectively, relative to others were used for SDS-PAGE. (B) Immunoblot analyses of whole-cell extracts of Sf9 cells infected with baculoviruses in the absence of the indicated subunits and SET1ACore complexes purified from each cell extract. Note that equal volumes of cell extracts were loaded (lanes 1–6), and the loading of purified complexes was normalized to FLAG-SET1ACore protein (lanes 7–12). The slightly fast migrating band in anti-WDR5 immunoblot (lane 9) is considered an insect WDR5 homolog in this and other figures. The upper bands observed in anti-ASH2L immunoblot are considered a post-translationally modified form(s) of ASH2L in this and other figures. (C and D) H3 (C) and H2Bub chromatin (D) substrates were subjected to in vitro HMT assays with the indicated subunit-lacking SET1ACore (left) or SET1BCore (right) complexes. Note that because of large differences in intrinsic H3K4 methyltransferase activities (Figure 1), different concentrations of SET1ACore (15 nM) and SET1BCore complexes (1.5 nM) were used for assays.
Immunoblots of purified complexes, analyzed densitometrically and normalized to FLAG-SET1ACore protein level, led us to draw a number of conclusions regarding SET1ACoreC integrity that are also generalizable to all other KMT2Core complexes (Figures 3B and 4B, Supplementary Figure S3). First, omission of RbBP5 resulted in the concomitant disappearance of ASH2L (and DPY30, see below) (Figure 2B, lane 10), indicating RbBP5-dependent association of ASH2L with KMT2 complexes and a pivotal role of RbBP5 for stable complex formation (see below). This is consistent with a previous similar reconstitution analysis of the core MLL1 complex (30) and is further supported by recent structural studies that revealed interaction networks of RbBP5, ASH2L and the SET domain of MLL1 or MLL3 (29,31,32). Second, exclusion of ASH2L caused the simultaneous disappearance of DPY30, whereas ASH2L was retained in the complex in the absence of DPY30 (Figure 2B, lanes 11 and 12). These results differ from the case of the yeast Set1 complex, in which Bre2 and Swd1 are jointly required for their concurrent residence within the complex (18,33), and thus suggest differential subunit interaction networks between yeast and human H3K4 methyltransferase complexes.
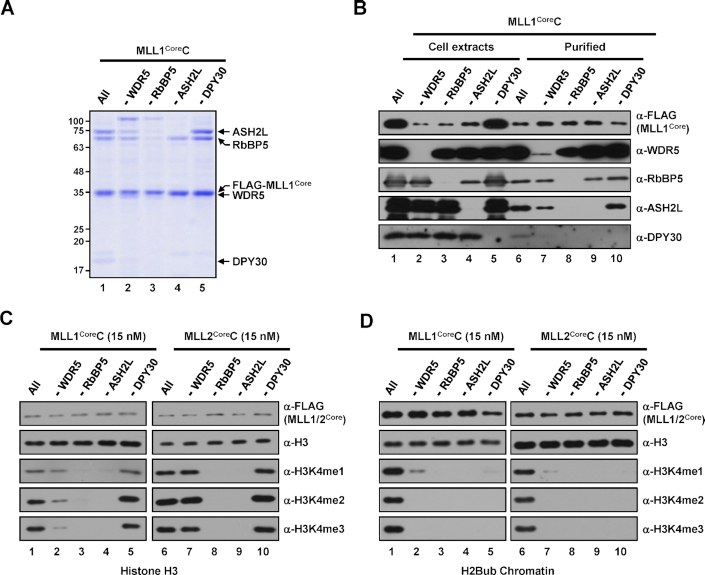
Figure 3.
Subunit requirements for H3K4 methylation activities of human MLL1Core and MLL2Core complexes. (A) SDS-PAGE/Coomassie blue staining of purified MLL1Core complexes lacking the indicated subunits. (B) Whole-cell extracts and purified MLL1Core complexes prepared from Sf9 cells infected with baculoviruses in the absence of the indicated subunits were subjected to immunoblot analyses as in Figure 2B. (C and D) H3 (C) and H2Bub chromatin (D) substrates were subjected to in vitro HMT assays with subunit-lacking MLL1Core (left) or MLL2Core (right) complexes (15 nM).
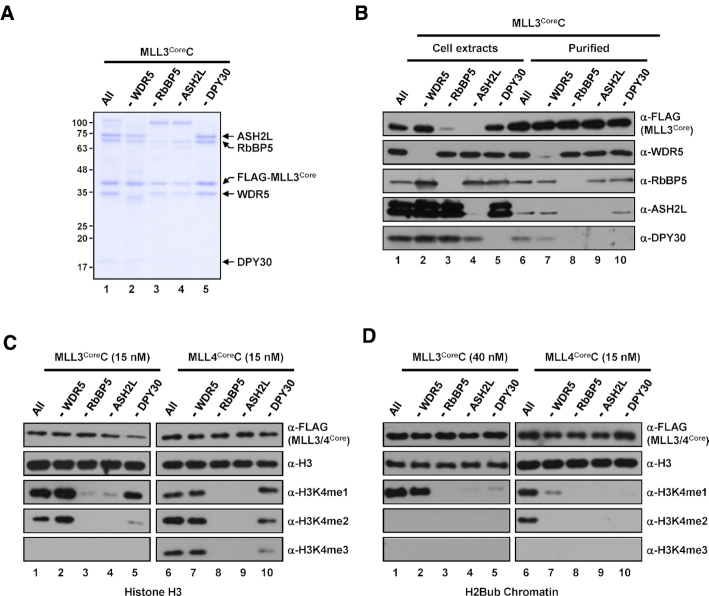
Figure 4.
Subunit requirements for H3K4 methylation activities of human MLL3Core and MLL4Core complexes. (A) SDS-PAGE/Coomassie blue staining of purified MLL3Core complexes lacking the indicated subunits. (B) Whole-cell extracts and purified MLL3Core complexes prepared from Sf9 cells infected with baculoviruses in the absence of the indicated subunits were subjected to immunoblot analyses as in Figure 2B. (C and D) H3 (C) and H2Bub chromatin (D) substrates were subjected to in vitro HMT assays with the indicated concentrations of subunit-lacking MLL3Core (left) and MLL4Core (right) complexes. Note that because of weak H3K4 methylation activity (Figure 1), a much higher concentration of MLL3Core complex (40 nM) than MLL4Core complex (15 nM) was used for chromatin HMT assays.
In vitro HMT assays performed on individual subunit-lacking complexes using free H3 as the substrate revealed several features in common for both SET1ACore and SET1BCore complexes. First, CFP1- or DPY30-deficient complexes exhibited H3K4 methylation activities comparable to that of the intact complex (Figure 2C, lanes 2 and 6 versus lane 1, lanes 8 and 12 versus 7). Second, omission of RbBP5 and ASH2L resulted in significantly decreased H3K4me1 activity and no H3Kme2/3 activities (Figure 2C, lanes 4, 5, 10 and 11), indicating the critical roles of these subunits for the intrinsic H3K4 methyltransferase activities of SET1A/BCoreCs; the importance of these subunits for H3K4 methylation activities also extended to all other KMT2CoreCs (Figures 3C and 4C). Interestingly, and in contrast to the shared contributions of CFP1, DPY30, RbBP5 and ASH2L subunits to the activities of SET1ACoreC and SET1BCoreC, described above, a WDR5 deficiency had different effects on SET1ACoreC and SET1BCoreC. Specifically, WDR5-lacking SET1ACoreC generated markedly decreased levels of H3K4me1 and failed to generate H3K4me2/3 (Figure 2C, lane 3), whereas WDR5-deficient SET1BCoreC exhibited only slightly decreased H3K4 methylation activity relative to the intact complex (Figure 2C, lane 9 versus lane 7). These findings suggest differential contributions of WDR5 to the catalytic activities of SET1A and SET1B complexes.
We next tested the contributions of each subunit to H3K4 methylation activities towards an H2Bub-containing chromatin template. Consistent with previous biochemical analyses of the purified human SET1A complex (19,34), both intact SET1ACoreC and SET1BCoreC were able to generate all H3K4 methylation states (Figure 2D, lanes 1 and 7). Although several complexes were shown to have intrinsic H3K4 methyltransferase activity towards H3, all SET1ACoreCs lacking any individual subunit completely lacked H3K4 methylation activity (Figure 2D, lanes 2–6). Unlike the case for SET1ACoreC, all SET1BCoreCs lacking any of WRAD subunits generated moderately and markedly weakened levels of H3K4me1 and H3K4me2/3, respectively (Figure 2D, lanes 9–12). However, not all subunit effects differed between SET1ACoreC and SET1BCoreC. As was the case for SET1ACoreC, CFP1-depleted SET1BCoreC showed a complete loss of H3K4 methylation activity (Figure 2D, lane 8), indicating the chromatin substrate-specific critical role of the CFP1 subunit, as has also been demonstrated for Spp1 (a yeast CFP1 homolog) in the yeast Set1 complex (18). In relation to this, we found that omission of WDR5 during complex reconstitution significantly decreased the level of CFP1 in SET1ACoreC (Figure 2B, lane 9), but not in SET1BCoreC (Supplementary Figure S3B) Therefore, based on the essential role of CFP1 for the nucleosomal H3K4 methylation activities of SET1ACoreC and SET1BCoreC, the loss of H3K4 methylation activity observed in WDR5-deficient SET1ACoreC could be explained, at least in part, by the disappearance of CFP1 in the complex. Thus, our in vitro system revealed differential subunit requirements for chromatin H3K4 methylation activities between SET1A and SET1B complexes.
Notably, we observed that, albeit little, an insect WDR5 homolog is present in purified SET1A/BCoreCs reconstituted in the absence of human WDR5 (Figure 2B, lane 9, Supplementary Figure S3B). These SET1A/BCoreCs showed significantly diminished H3K4 methylation activities relative to the intact complexes, indicating that the insect homolog does not fully replace the function of human WDR5 in the complexes. In relation to this and in support of our observations that WDR5 is required for the catalytic activities of SET1A/B complexes, a recent biochemical analysis that employed a partial MLL1 complex reconstituted with bacterially expressed subunits also demonstrated that omission of WDR5 largely decreased the H3K4 methylation activity of the complex (29).
All WRAD subunits are required for nucleosomal H3K4 methylation activities of human MLL1Core and MLL2Core complexes
We next tested subunit requirements for the H3K4 methylation activities of purified MLL1CoreC (Figure 3A) and MLL2CoreC (Supplementary Figure S3C), of which subunit compositions were examined by immunoblot analyses (Figure 3B, Supplementary Figure S3D). In in vitro HMT assays with free H3, intact MLL1CoreC and MLL2CoreC containing all WRAD subunits were able to generate all H3K4 methylation states. As was the case for SET1BCoreC, RbBP5 and ASH2L were found to be essential for H3K4 methylation activity, whereas WDR5 and DPY30 were dispensable (Figure 3C). Consistent with previous biochemical analyses of purified human MLL1 and MLL2 complexes (35,36), both intact MLL1CoreC and MLL2CoreC were able to generate all three H3K4 methylation states (Figure 3D, lanes 1 and 6) with similar activities (Figure 1D) towards H2Bub-containing chromatin templates. We further found that, with the exception of very weak residual H3K4me1 activity in the specific absence of WDR5, exclusion of WRAD subunits led to a complete loss of H3K4 methylation activities (Figure 3D, lanes 2–5 and 7–10), demonstrating the essential roles of all WRAD subunits for nucleosomal H3K4 methylation by MLL1 and MLL2 complexes.
To confirm stable complex formations, all MLL2CoreCs reconstituted without a single subunit were further analyzed by size exclusion chromatography (Supplementary Figure S4). We found that MLL2CoreCs reconstituted in the absence of WDR5, ASH2L or DPY30 were shown to form stable complexes. In contrast, RbBP5-deficient complex retained only MLL2Core fragment and WDR5, and was diluted out during gel filtration, indicating a critical role of RbBP5 for stable complex formation. This is consistent with recent structural studies that revealed extensive contacts of RbBP5 with other subunits, which are important for stable complex formation (29,31,32). Therefore, lack of the H3K4 methylation activity of RbBP5-deficient complexes is considered to be attributed by the absence of multiple catalytic subunits.
Differential subunit requirements of human MLL3Core and MLL4Core complexes for nucleosomal H3K4 methylation activity
We then examined subunit requirements of purified MLL3CoreC (Figure 4A) and MLL4CoreC (Supplementary Figure S3E) for H3K4 methylation activity. Unlike the complexes described above, we found that protein expression level of scaffold MLL3CoreC was not decreased in the absence of WDR5 (Figure 4B, lane 2 versus lane 1), which allowed us to obtain a purified WDR5-deficient complex at a concentration comparable to that of the intact complex. This also suggests a distinct contribution of WDR5 to the assembly of the MLL3 complex, possibly leading to different effects of WDR5 on H3K4 methylation activity compared with that in other complexes (see below). In vitro HMT assays revealed that the intrinsic methyltransferase activity of MLL3CoreC on free H3 was restricted to H3K4me1/2, and required RbBP5 and ASH2L, but not WDR5 and DPY30 (Figure 4C, lanes 1–5). Interestingly, we found that WDR5-deficient MLL3CoreC exhibited slightly increased H3K4me1/2 activities relative to the intact complex (Figure 4C, lane 2 versus lane 1). This is consistent with a previous similar HMT assay that employed a partial MLL3 complex (equivalent to our MLL3WinC, see below) and H3 peptides as the substrates (37). In accordance with the role of MLL3 as a major H3K4 mono-methyltransferase around enhancer regions (38), chromatin methylation activity of MLL3CoreC was restricted to H3K4me1 (Figure 4D, lane 1). This activity was almost completely lost in the absence of RbBP5, ASH2L and DPY30 but, importantly was not diminished by omission of WDR5 (Figure 4D, lanes 2–5). In a related observation, a recent study showed that omission of WDR5 from a partial MLL3 complex (equivalent to our MLL3WinC) reconstituted with bacterially expressed proteins rather increased H3K4me1 activity of the complex (29). These findings reflect the clearly different pattern of MLL3CoreC activity compared with that of MLL1/2CoreCs (Figure 3D) and further suggest differences in structural organization between MLL3 and other KMT2 complexes (29).
H3K4 methylation activity of purified MLL4CoreC towards free H3 was dependent on RbBP5 and ASH2L (Figure 4C, lanes 6–10). As was the case for MLL1/2CoreCs, MLL4CoreC also required all WRAD subunits for chromatin methylation activities but retained weak H3K4me1 activity in the absence of WDR5 (Figure 4D, lanes 6–10). In addition, similar to the cases for SET1A/BCoreCs and MLL2CoreC, MLL3/4CoreCs further purified by gel filtration exhibited indistinguishable H3K4 methylation activities to those of the affinity-purified complexes (Supplementary Figure S5).
The n-SET domain is essential for nucleosomal H3K4 methylation activities of SET1ACore and SET1BCore complexes
So far, we have examined H3K4 methylation activities of KMT2Core complexes that contain partial KMT2 fragments harboring entire n-SET or pre-SET domains at their N-terminus (Figure 1A). However, all previous biochemical and structural analyses have employed complexes containing partial KMT2 fragments encompassing a region beginning near the WDR5-interaction (Win) motif residing within their n-SET or pre-SET domains because these complexes are minimally sufficient to exert H3K4 methylation activity (29,32,37,39). In a related observation, the SET domain of the yeast Set1 complex alone was shown to be sufficient to form a stable complex with all WRAD homologs (18,21,39); however, we failed to purify stable complex using the SET domain from any human KMT2 protein and WRAD subunits (data not shown). Collectively, these results indicate that, in addition to the SET domain, the Win motif located in the N-terminus is also required for stable complex formation, as previously reported (40,41).
To test H3K4 methylation activities of complexes containing KMT2 protein fragments that are minimally sufficient to form complexes with WRAD subunits, we reconstituted partial human KMT2 Win complexes (hereafter, KMT2WinCs) and affinity purified them from Sf9 insect cells infected with baculoviruses expressing FLAG-KMT2Win protein fragments (encompassing the Win motif, SET and post-SET domains) (37) and untagged WRAD and CFP1 (Figure 5A). From the standpoint of complex integrity, we found that all WRAD subunits were stably integrated into all purified complexes, but CFP1 was absent (Figure 5B). The absence of SET1A/B complex-specific CFP1 (Figure 1B) in SET1A/BWinCs suggests that the missing N-terminal region of the n-SET domain is required for retention of CFP1 in SET1A and SET1B complexes. All KMT2WinCs exhibited H3K4 methylation activities towards free H3, exhibiting relative catalytic strengths and product methylation states similar to those of KMT2CoreCs (compare Figure 5C and 1C). These findings confirm that KMT2Win protein fragments are sufficient to confer intrinsic catalytic activities to the complexes. Strikingly different results were obtained in in vitro HMT assays using chromatin templates. All MLL1/2/3/4WinCs exhibited H3K4 methylation activities (Figure 5D, lanes 5–12), with patterns of H2Bub-stimulatory methylation states generally similar to those of MLL1/2/3/4CoreCs. Importantly, however, both SET1AWinC and SET1BWinC showed a complete loss of H3K4me2/3 activities, even in the presence of H2Bub (Figure 5D, lanes 1–4), despite the fact that SET1BCoreC showed the strongest activity among all KMT2CoreCs. A direct comparison of H3K4 methylation activities further confirmed that SET1A/BWinCs, in contrast to SET1A/BCoreCs, do not possess H2Bub-dependent nucleosomal H3K4 methylation activity (Figure 5E). These results indicate that n-SET domains, likely in conjunction with associating CFP1 subunits (see below), play a critical role in regulating the catalytic activities of SET1A and SET1B complexes.
Figure 5.
H3K4 methylation activities of human KMT2Win complexes. (A) A schematic diagram of KMT2Win fragments used for the reconstitution of complexes and subunit associations deduced from (B). The SET and post-SET (hatched box) domain and Win (red box) motif within each KMT2 protein are depicted. Numbers indicate amino acid residues of each protein. Note that all proteins contain truncated n-SET or pre-SET domains. (B) SDS-PAGE/Coomassie blue staining (left) and immunoblot analyses (right) of purified KMT2Win complexes reconstituted with baculoviruses expressing FLAG-tagged KMT2Win protein (A), WDR5, RbBP5, ASH2L, DPY30 and CFP1. KMT2Win proteins are marked by asterisks. (C) Differential H3K4 methylation activities of KMT2Win complexes towards H3 substrate. H3 was subjected to in vitro HMT assays with the indicated KMT2Win complexes (7.5 nM). (D) Differential H3K4 methylation activities of KMT2Win complexes towards chromatin substrates. Unmodified or H2Bub chromatin substrates were subjected to in vitro HMT assays with the indicated KMT2Win complexes (5 nM). (E) Comparison of H3K4 methylation activities of SET1A/BWin and SET1A/BCore complexes. Unmodified and H2Bub chromatin substrates were subjected to in vitro HMT assays with SET1AWin and SET1ACore complexes (left) and SET1BWin and SET1BCore complexes (right), present at a final concentration of 10 nM.
The PHD2 domain within CFP1 is essential for H2B ubiquitylation-dependent H3K4me2 and H3K4me3 activities of SET1ACore and SET1BCore complexes
The disappearance of CFP1 in SET1A/BWinCs caused by n-SET domain truncation raises the possibility that the observed loss of H3K4 methylation activities of these complexes is attributable to the lack of CFP1. In this same vein, we showed above that CFP1-deficient SET1A/BCoreCs are completely inactive with respect to nucleosomal H3K4 methylation (Figure 2D), establishing a critical role of CFP1 in catalytic activity that is specific to SET1A/B complexes.
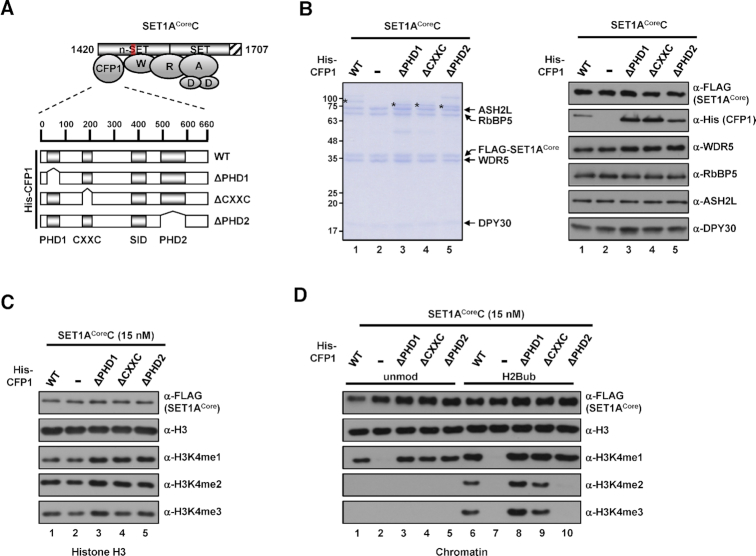
CFP1 contains two evolutionarily conserved PHDs (plant homeodomains), a SID (Set1-interacting domain), and a CXXC domain with demonstrated individual functions in binding to methylated histones (42), SET1A/B proteins (43), and unmethylated CpG islands around promoter regions (44,45), respectively. To determine the domain within CFP1 that is involved in the H3K4 methylation activity of the SET1A complex, we reconstituted SET1ACore protein-based complexes and affinity purified them from Sf9 cells infected with baculoviruses expressing FLAG-SET1ACore protein, untagged WRAD, and His-tagged wild-type (WT) or mutant CFP1 (Figure 6A). (Note that the complex containing ΔSID-CFP1 was not included because this protein was not expressed in soluble form in Sf9 cells). All subunits were found to be stably integrated into the complex (Figure 6B). In tests designed to determine the contribution of each domain within CFP1 to intrinsic H3K4 methyltransferase activity towards free H3, all complexes exhibited comparable levels of all H3K4 methylation states (Figure 6C). In in vitro HMT assays using chromatin templates, complexes containing ΔPHD1-CFP1 or ΔCXXC-CFP1 generated all three H3K4 methylation states at levels comparable to those observed with the complete SET1ACoreC in the presence of H2Bub (Figure 6D, lanes 8 and 9 versus lane 6). Importantly, we further found that the complex reconstituted with ΔPHD2-CFP1 was capable of producing H3K4me1, but completely failed to generate H3K4me2/3 (Figure 6D, lane 10). Notably, because the ΔPHD2 complex showed intrinsic H3K4me1 activity (Figure 6D, lane 5), these results strongly suggest that the PHD2 domain plays a critical role in H2Bub-dependent H3K4me2/3 activities. In addition, the same series of experiments performed with SET1BCoreC (Supplementary Figure S6) collectively indicate a critical role for CFP1 PHD2 in the H2Bub-dependent H3K4 methylation activities of SET1A and SET1B complexes. Given that WRAD subunits alone are insufficient and that CFP1 is additionally required for SET1A/BCoreCs to exhibit efficient H3K4 methylation activities, these results suggest that CFP1 works together with WRAD to establish H3K4 methylation-competent SET1A/B complexes, as proposed for the yeast Set1 complex (20,21). In addition, the SET1A/BCoreC-specific requirement of CFP1 for catalytic activity strongly suggests that the H3K4 methylation mechanism is somewhat different for SET1A/B and MLL1/2/3/4 complexes (see Discussion).
Figure 6.
Requirement of CFP1 domains for H3K4 methylation activities of the human SET1ACore complex. (A) A schematic diagram of SET1ACore protein-based complexes containing wild-type (WT) or mutant CFP1. PHD1, CXXC, SID, and PHD2 domains are depicted. Physical interactions between subunits were deduced from previous structural studies of human MLL1 and MLL3 complexes (29,31,32). Abbreviations: W, WDR5; R, RbBP5; A, ASH2L; D, DPY30. (B) SDS-PAGE/Coomassie blue staining (left) and immunoblot analyses (right) of purified SET1ACore protein-based complexes reconstituted with baculoviruses expressing FLAG-tagged SET1ACore protein, WDR5, RbBP5, ASH2L, DPY30 and His-tagged WT or mutant CFP1. His-CFP1 polypeptides are marked by asterisks. (C and D) Requirement of the PHD2 domain of CFP1 for H2Bub-dependent H3K4 methylation activities of the SET1ACore complex. H3 (C) and unmodified or H2Bub chromatin substrates (D) were subjected to in vitro HMT assays with the indicated SET1ACore protein-based complexes (15 nM).
Comparisons of H3K4 methylation activities of full-length KMT2-containing complexes
To directly compare intrinsic H3K4 methyltransferase activities, we have deployed biochemical analyses using partial KMT2 complexes containing KMT2 protein fragments and the subunits that are minimally required for catalytic activity. However, it is still important to analyze biochemical properties of complete KMT2 complexes to understand their precise in vivo functions. Although several previous studies have reported H3K4 methylation activities of individual KMT2 complexes (34–36,46), there have been no comprehensive biochemical characterizations and comparisons of intact KMT2 complexes prepared using identical conditions. On a related note, we tried to prepare recombinant complexes containing full-length KMT2 protein and all subunits of the complex using a baculovirus expression system, but these approaches have so far proved unsuccessful (data not shown). Instead, we found that we could successfully reconstitute SET1A, SET1B and MLL2 complexes and affinity purify them from HEK293E cells transfected with combinations of plasmids expressing FLAG-tagged KMT2 proteins and subunits that constitute each complex (7) (Figure 7A and B). (Note that the same approach was not successful for the MLL1 complex, and we did not attempt to purify MLL3/4 complexes owing to difficulties in preparing plasmids expressing full-length MLL3 or MLL4.)
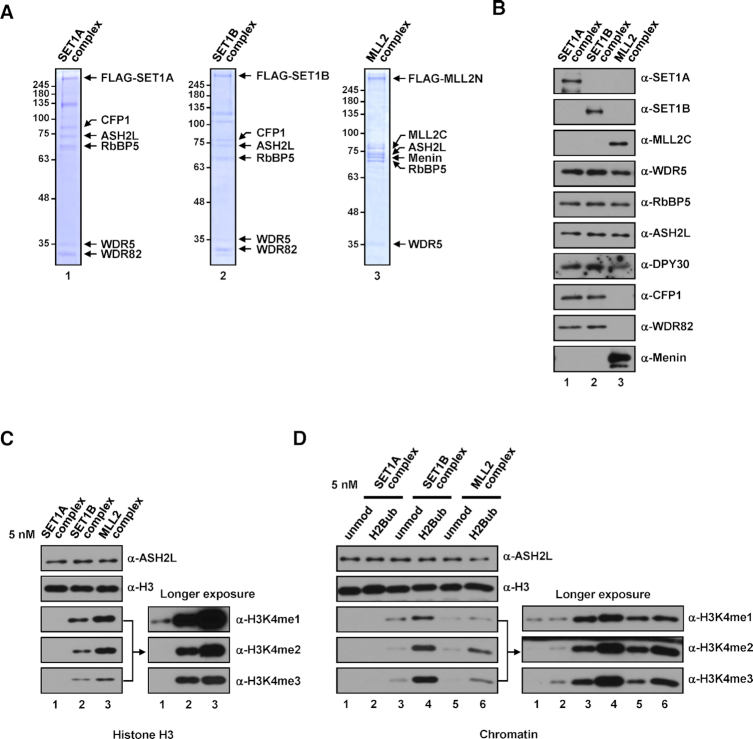
Figure 7.
H3K4 methylation activities of human SET1A, SET1B, and MLL2 complexes. (A and B) SDS-PAGE/Coomassie blue staining (A) and immunoblot analyses (B) of SET1A, SET1B, and MLL2 complexes purified from HEK293E cells transfected with combinations of plasmids expressing subunits that constitute each complex. MLL2N and MLL2C, N-terminal and C-terminal parts of MLL2, respectively, resulting from Taspase 1 cleavage (54). (C and D) Differential H3K4 methylation activities of SET1A, SET1B, and MLL2 complexes. H3 (C) and unmodified or H2Bub chromatin substrates (D) were subjected to in vitro HMT assays with the indicated complexes (5 nM).
In tests of intrinsic enzymatic activities, all purified complete complexes generated H3K4 methylation to different degrees. Similar to the case for KMT2CoreCs (Figure 1C), SET1B and MLL2 complexes exhibited much stronger H3K4 methylation activities towards free H3 than did the SET1A complex (Figure 7C). Likewise, we also found that all complete complexes were able to generate all H3K4 methylation states and, importantly, generated these states at increased levels in the presence of nucleosomal H2Bub (Figure 7D). The rank order of the strengths of enzyme activities were SET1B > MLL2 > SET1A complexes, which is the same order as that obtained with KMT2CoreCs (Figure 1D). These results validate our comparative biochemical analyses using KMT2Core complexes for recapitulating H3K4 methylation activities of complete KMT2 complexes.
DISCUSSION
The presence of multiple KMT2 family complexes and many other cellular factors that directly and indirectly affect H3K4 methylation makes it difficult to address how each KMT2 complex contributes individually to the levels and states of H3K4 methylation in cells. To directly assess the catalytic properties of all six human KMT2 complexes, we used biochemically defined in vitro HMT assays employing purified KMT2 complexes and recombinant chromatin substrates. Our studies provide a number of valuable insights regarding the nucleosomal H3K4 methylation activities of human KMT2 complexes: (i) H2Bub stimulates H3K4 methylation activities of KMT2 complexes; (ii) H2Bub is strictly required for H3K4me2/3 activities of the SET1A complex, but is dispensable for SET1B complex activities; (iii) the SET1B complex exhibits the strongest H3K4 methylation activity among KMT2 complexes; (iv) the CFP1 PHD2 domain plays a critical role in the catalytic activities of SET1A and SET1B complexes; (v) MLL1 and MLL2 complexes can generate comparable levels of all H3K4 methylation states; (vi) the H3K4 methylation activity of MLL3 is restricted to H3K4me1, whereas MLL4 can produce both H3K4me1 and H3K4me2; (vii) in contrast to the case for other complexes, WDR5 is dispensable for the H3K4me1 activity of the MLL3 complex. Taken together, these results provide fundamental insights into nucleosomal H3K4 methylation activities of human KMT2 complexes.
Previous biochemical analyses using purified KMT2 complexes prepared by bacterial (37) or baculovirus (39) expression systems also reported their differential H3K4 methylation activities towards H3 peptide (37) or non-ubiquitylated recombinant nucleosome (39). It is important to note that these analyses employed complexes containing partial KMT2 fragments encompassing a region beginning near the Win motif, which are equivalent to KMT2WinCs in our study (Figure 5). In contrast, we used KMT2Core fragments that contain entire n-SET or pre-SET domains at their N-terminus for complex reconstitutions (i.e. KMT2CoreCs), which allowed incorporation of CFP1 specifically into SET1A and SET1B complexes (Figure 1). This allowed us to uniquely demonstrate a critical role of the CFP1 subunit in the H3K4 methylation activities of SET1A and SET1B complexes towards physiologically relevant H2Bub-containing chromatin substrates (Figures 2 and 6), which was not addressed by previous studies.
H3K4 methylation activities of human KMT2 complexes towards chromatin templates
Our ability to reconstitute KMT2 complexes lacking specific subunits allowed us to directly assess the contribution of individual subunits to H3K4 methylation activities towards free H3 and chromatin templates. For all KMT2 complexes, we found that all WRAD subunits are required for full H3K4 methylation activities towards chromatin (except WDR5 for MLL3 complex), but only RbBP5 and ASH2L are essential for methylation of free H3. These different subunit requirements reflect the fact that extensive subunit interactions with DNA, as well as new binding surface created by the nucleosome structure, are critical for nucleosomal H3K4 methylation (20,22,29,31,32). They also highlight the merits of our biochemical analyses using physiologically relevant chromatin substrates rather than free histone substrates for proper characterization of chromatin-modifying enzymes.
The large differences in enzymatic activities observed for SET1ACoreC and SET1BCoreC (Figure 1E) indicate that KMT2 protein itself determines the catalytic power of the complex. SET1BCoreC exhibited reduced, but still significant, H3K4 methylation activities in the absence of H2Bub, even at much lower concentrations than SET1ACoreC. This suggests that the Set1 protein underwent divergent evolution to produce one weak descendant (SET1A) and one strong descendant (SET1B). Structural studies detailing how SET1A and SET1B proteins differentially contribute to catalytic power are warranted.
In support of our finding that SET1A complex has a weak catalytic activity, it was reported that the removal of the SET1A SET domain in mouse V6.5 embryonic stem cells did not diminish global H3K4 methylation level; instead, only a subset of genes shows decreased H3K4me3 levels (47). Consistent with this, shRNA-mediated depletion of SET1A was shown to have no effect on global H3K4 methylation levels in cancer cell lines (48,49). In addition, in a test for the relative contribution of SET1A and SET1B to cellular H3K4 methylation, we further found that ectopic expression of SET1B resulted in more significant increases in global H3K4me2/3 levels than that of SET1A (Supplementary Figure S7). In contrast to above results, another study has reported that inducible knockout of SET1A led to a significant reduction in global H3K4 methylation levels in mouse R1 embryonic stem cells and embryos, whereas that of SET1B had no effect (50), suggesting a principal role of SET1A in the maintenance of global H3K4 methylation levels in their study. However, in relation to the latter observation, it is worth to note that the HeLa cell nucleus contains about 130-fold more SET1A than SET1B as demonstrated by quantitative mass spectrometric analysis (51). Therefore, SET1A could have a more significant effect on H3K4 methylation levels in some cells that contain SET1A as a dominant population although its intrinsic catalytic activity is much weaker than SET1B. Collectively, these suggest that, in addition to intrinsic enzyme activities, several features of KMT2 proteins, including their relative amounts in cells and subcellular localization that could vary according to cell types and developmental stages (51,52) determine cellular H3K4 methylation levels.
A notable difference among KMT2Core proteins is that SET1 and MLL homologs have n-SET and pre-SET domains, respectively. To test whether these domains affected the catalytic properties of other groups of homologs, we performed domain-swapping experiments (Supplementary Figure S8). We found that fusion of the SET1B n-SET domain to the MLL3 or MLL4 SET and post-SET domains resulted in inclusion of CFP1 in purified complexes that exhibited significantly enhanced H3K4 methylation activities. Importantly, however, the methylation states remained unchanged. These observations suggest that the amino acid sequence within each SET domain itself determines the methylation product states of the complex. Support for this is provided by the previous demonstration that unique amino acid sequences in the SET domain of MLL3 specify its H3Kme1-restricted activity (39,53).
H2B ubiquitylation stimulates H3K4 methylation activities of human KMT2 complexes
Because of similarities in domain architecture and subunit composition between SET1A/B complexes and the yeast Set1 complex (7), we initially expected that only SET1A and SET1B complexes would exhibit H2Bub-dependent H3K4 methylation activities. Surprisingly, however, we found that all KMT2 complexes (except MLL3, see above) exhibited H2Bub-stimulated H3K4 methylation activities, suggesting that a component(s) common to all complexes might be responsible for this trans-tail stimulatory effect. In support of this suggestion, a recent study demonstrated that the WD40 domain of RbBP5 directly binds to H2Bub, leading to more extensive and stabilized binding of MLL1 or MLL3 to nucleosomes and thus enhanced catalytic activities of the complexes (29). Because RbBP5 is present in all complexes, it might contribute to H2Bub-mediated enhanced H3K4 methylation activities through the same mechanism.
In addition, a very recent structural study on the yeast Set1 complex proposed another mechanism to explain how H2Bub activates H3K4 methylation (20,22). According to this model, the arginine-rich RXXXRR motif that we previously demonstrated critical for the H2Bub-dependent H3K4 methylation activity of the Set1 complex (18) makes extensive contacts with H2Bub as well as the acidic patch of histones H2A and H2B; thus, increased nucleosome interaction potentiates H3K4 methylation activity. Because the RXXXRR motif is conserved only in human SET1A/B but not in MLL1/2/3/4, this mechanism could be specific to SET1A and SET1B complexes. In this context, the greater enhancement of H3K4 methylation activities by H2Bub observed in SET1A/BCoreCs compared with MLL1/2/4CoreCs (Supplementary Figure S1) could be explained by cooperative contributions of the RbBP5 subunit and the RXXXRR motif in SET1A/B. In relation to this, we also found that mutations in the RXXXRR motif resulted in complete loss of the H3K4 methylation activities of SET1A/BCoreCs (Supplementary Figure S9), suggesting an evolutionarily conserved role of the RXXXRR motif in the regulation of H3K4 methylation activities of SET1A and SET1B complexes.
In a related observation, we found that SET1AWinC exert no H3K4me2/3 activities, even in the presence of H2Bub (Figure 5D), despite the fact that these complexes contain both RbBP5 and the RXXXRR motif. Instead, we found that inclusion of the n-SET domain and CFP1 (i.e. SET1ACoreC) converts the complexes into an H3K4me2/3-active form in the presence of H2Bub (Figure 5E). In contrast to other domain-defective complexes, the CFP1 PHD2 domain-deficient SET1ACoreC showed no H3K4me2/3 activities in the presence of H2Bub, but did exhibit intrinsic H3K4me1 activity (Figure 6D). These results strongly suggest that CFP1 acts through its PHD2 to participate in H2Bub-induced H3K4me2/3 activities of SET1A complex. Additional structural analyses of CFP1-containing complexes will be needed to detail the contribution of CFP1 to this process.
In the current study, using a biochemically defined system reconstituted with recombinant human KMT2 complexes and recombinant chromatin substrates, we found that each KMT2 complex generates distinct H3K4 methylation states and different levels of methylation, suggesting mechanistic differences in H3K4 methylation processes among KMT2 complexes. This detailed catalytic characterization will be invaluable for guiding and interpreting high-resolution structural studies of KMT2 complexes that have recently begun to emerge, to identify roles of KMT2 complexes in transcriptional regulation, and for future epigenetic drug development efforts related to H3K4 methylation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Robert G. Roeder for anti-CFP1 and anti-WDR82 antibodies; Dr. C. David Allis for anti-WDR5 antibody; and Dr. Daeyoup Lee for valuable discussion.
Notes
Present address: Jeong-Heon Lee, Division of Experimental Pathology and Laboratory Medicine, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN 55905, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Samsung Science and Technology Foundation [SSTF-BA1702-13 to J.K.]; National Institutes of Health [R37 GM086868, P01 CA196539 to T.W.M., P01DK068055 to J.H.L., 1ZIADK075003 to K.G.]. Funding for open access charge: Samsung Science and Technology Foundation [SSTF-BA1702-13].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hyun K., Jeon J., Park K., Kim J.. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017; 49:e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li B., Carey M., Workman J.L.. The role of chromatin during transcription. Cell. 2007; 128:707–719. [DOI] [PubMed] [Google Scholar]
- 3. Rivera C., Gurard-Levin Z.A., Almouzni G., Loyola A.. Histone lysine methylation and chromatin replication. Biochim. Biophys. Acta. 2014; 1839:1433–1439. [DOI] [PubMed] [Google Scholar]
- 4. Chi P., Allis C.D., Wang G.G.. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer. 2010; 10:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collins B.E., Greer C.B., Coleman B.C., Sweatt J.D.. Histone H3 lysine K4 methylation and its role in learning and memory. Epigenet. Chromatin. 2019; 12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rao R.C., Dou Y.. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer. 2015; 15:334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012; 81:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crump N.T., Milne T.A.. Why are so many MLL lysine methyltransferases required for normal mammalian development. Cell. Mol. Life Sci. 2019; 76:2885–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang W., Ernst P.. Distinct functions of histone H3, lysine 4 methyltransferases in normal and malignant hematopoiesis. Curr. Opin. Hematol. 2017; 24:322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang W., Ernst P.. SET/MLL family proteins in hematopoiesis and leukemia. Int. J. Hematol. 2017; 105:7–16. [DOI] [PubMed] [Google Scholar]
- 11. Harikumar A., Meshorer E.. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 2015; 16:1609–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang Y., Dominguez P.M., Melnick A.M.. The many layers of epigenetic dysfunction in B-cell lymphomas. Curr. Opin. Hematol. 2016; 23:377–384. [DOI] [PubMed] [Google Scholar]
- 13. Froimchuk E., Jang Y., Ge K.. Histone H3 lysine 4 methyltransferase KMT2D. Gene. 2017; 627:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang C., Lee J.E., Lai B., Macfarlan T.S., Xu S., Zhuang L., Liu C., Peng W., Ge K.. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:11871–11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee J.E., Wang C., Xu S., Cho Y.W., Wang L., Feng X., Baldridge A., Sartorelli V., Zhuang L., Peng W., Ge K.. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013; 2:e01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dover J., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A.. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002; 277:28368–28371. [DOI] [PubMed] [Google Scholar]
- 17. Sun Z.W., Allis C.D.. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002; 418:104–108. [DOI] [PubMed] [Google Scholar]
- 18. Kim J., Kim J.A., McGinty R.K., Nguyen U.T., Muir T.W., Allis C.D., Roeder R.G.. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol. Cell. 2013; 49:1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim J., Guermah M., McGinty R.K., Lee J.S., Tang Z., Milne T.A., Shilatifard A., Muir T.W., Roeder R.G.. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009; 137:459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu P.L., Shi H., Leonen C., Kang J., Chatterjee C., Zheng N.. Structural basis of H2B ubiquitination-dependent H3K4 methylation by COMPASS. Mol. Cell. 2019; 76:712–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeon J., McGinty R.K., Muir T.W., Kim J.A., Kim J.. Crosstalk among Set1 complex subunits involved in H2B ubiquitylation-dependent H3K4 methylation. Nucleic Acids Res. 2018; 46:11129–11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Worden E.J., Zhang X., Wolberger C.. Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome. eLife. 2020; 9:e53199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim J., Roeder R.G.. Nucleosomal H2B ubiquitylation with purified factors. Methods. 2011; 54:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dukkipati A., Park H.H., Waghray D., Fischer S., Garcia K.C.. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr. Purif. 2008; 62:160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McGinty R.K., Kohn M., Chatterjee C., Chiang K.P., Pratt M.R., Muir T.W.. Structure-activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem. Biol. 2009; 4:958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. An W., Kim J., Roeder R.G.. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004; 117:735–748. [DOI] [PubMed] [Google Scholar]
- 27. Lee J.H., Skalnik D.G.. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 2005; 280:41725–41731. [DOI] [PubMed] [Google Scholar]
- 28. Lee J.H., Tate C.M., You J.S., Skalnik D.G.. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 2007; 282:13419–13428. [DOI] [PubMed] [Google Scholar]
- 29. Xue H., Yao T., Cao M., Zhu G., Li Y., Yuan G., Chen Y., Lei M., Huang J.. Structural basis of nucleosome recognition and modification by MLL methyltransferases. Nature. 2019; 573:445–449. [DOI] [PubMed] [Google Scholar]
- 30. Dou Y., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G.. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006; 13:713–719. [DOI] [PubMed] [Google Scholar]
- 31. Park S.H., Ayoub A., Lee Y.T., Xu J., Kim H., Zhang W., Zhang B., Sha L., An S., Zhang Y. et al.. Cryo-EM structure of the human MLL1 core complex to the nucleosome. Nat. Commun. 2019; 10:5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y., Han J., Zhang Y., Cao F., Liu Z., Li S., Wu J., Hu C., Wang Y., Shuai J. et al.. Structural basis for activity regulation of MLL family methyltransferases. Nature. 2016; 530:447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. South P.F., Fingerman I.M., Mersman D.P., Du H.N., Briggs S.D.. A conserved interaction between the SDI domain of Bre2 and the Dpy-30 domain of Sdc1 is required for histone methylation and gene expression. J. Biol. Chem. 2010; 285:595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang Z., Chen W.Y., Shimada M., Nguyen U.T., Kim J., Sun X.J., Sengoku T., McGinty R.K., Fernandez J.P., Muir T.W. et al.. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013; 154:297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dou Y., Milne T.A., Tackett A.J., Smith E.R., Fukuda A., Wysocka J., Allis C.D., Chait B.T., Hess J.L., Roeder R.G.. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005; 121:873–885. [DOI] [PubMed] [Google Scholar]
- 36. Jiang H., Lu X., Shimada M., Dou Y., Tang Z., Roeder R.G.. Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95. Nat. Struct. Mol. Biol. 2013; 20:1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shinsky S.A., Monteith K.E., Viggiano S., Cosgrove M.S.. Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation. J. Biol. Chem. 2015; 290:6361–6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu D., Gao X., Morgan M.A., Herz H.M., Smith E.R., Shilatifard A.. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 2013; 33:4745–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hsu P.L., Li H., Lau H.T., Leonen C., Dhall A., Ong S.E., Chatterjee C., Zheng N.. Crystal structure of the COMPASS H3K4 methyltransferase catalytic module. Cell. 2018; 174:1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patel A., Vought V.E., Dharmarajan V., Cosgrove M.S.. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J. Biol. Chem. 2008; 283:32162–32175. [DOI] [PubMed] [Google Scholar]
- 41. Song J.J., Kingston R.E.. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 2008; 283:35258–35264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eberl H.C., Spruijt C.G., Kelstrup C.D., Vermeulen M., Mann M.. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell. 2013; 49:368–378. [DOI] [PubMed] [Google Scholar]
- 43. Butler J.S., Lee J.H., Skalnik D.G.. CFP1 interacts with DNMT1 independently of association with the Setd1 Histone H3K4 methyltransferase complexes. DNA Cell Biol. 2008; 27:533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brown D.A., Di Cerbo V., Feldmann A., Ahn J., Ito S., Blackledge N.P., Nakayama M., McClellan M., Dimitrova E., Turberfield A.H. et al.. The SET1 complex selects actively transcribed target genes via multivalent interaction with CpG island chromatin. Cell Rep. 2017; 20:2313–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Voo K.S., Carlone D.L., Jacobsen B.M., Flodin A., Skalnik D.G.. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell. Biol. 2000; 20:2108–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cho Y.W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G.R., Copeland T.D., Kalkum M. et al.. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007; 282:20395–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sze C.C., Cao K., Collings C.K., Marshall S.A., Rendleman E.J., Ozark P.A., Chen F.X., Morgan M.A., Wang L., Shilatifard A.. Histone H3K4 methylation-dependent and -independent functions of Set1A/COMPASS in embryonic stem cell self-renewal and differentiation. Genes Dev. 2017; 31:1732–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salz T., Deng C., Pampo C., Siemann D., Qiu Y., Brown K., Huang S.. Histone methyltransferase hSETD1A is a novel regulator of metastasis in breast cancer. Mol. Cancer Res. 2015; 13:461–469. [DOI] [PubMed] [Google Scholar]
- 49. Tajima K., Yae T., Javaid S., Tam O., Comaills V., Morris R., Wittner B.S., Liu M., Engstrom A., Takahashi F. et al.. SETD1A modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat. Commun. 2015; 6:8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bledau A.S., Schmidt K., Neumann K., Hill U., Ciotta G., Gupta A., Torres D.C., Fu J., Kranz A., Stewart A.F. et al.. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development. 2014; 141:1022–1035. [DOI] [PubMed] [Google Scholar]
- 51. van Nuland R., Smits A.H., Pallaki P., Jansen P.W., Vermeulen M., Timmers H.T.. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol. Cell. Biol. 2013; 33:2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang L., Collings C.K., Zhao Z., Cozzolino K.A., Ma Q., Liang K., Marshall S.A., Sze C.C., Hashizume R., Savas J.N. et al.. A cytoplasmic COMPASS is necessary for cell survival and triple-negative breast cancer pathogenesis by regulating metabolism. Genes Dev. 2017; 31:2056–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weirich S., Kudithipudi S., Kycia I., Jeltsch A.. Somatic cancer mutations in the MLL3-SET domain alter the catalytic properties of the enzyme. Clin. Epigenet. 2015; 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takeda S., Chen D.Y., Westergard T.D., Fisher J.K., Rubens J.A., Sasagawa S., Kan J.T., Korsmeyer S.J., Cheng E.H., Hsieh J.J.. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 2006; 20:2397–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.