Abstract
Aberrant KRAS signaling is a driver of many cancers and yet remains an elusive target for drug therapy. The nuclease hypersensitive element of the KRAS promoter has been reported to form secondary DNA structures called G-quadruplexes (G4s) which may play important roles in regulating KRAS expression, and has spurred interest in structural elucidation studies of the KRAS G-quadruplexes. Here, we report the first high-resolution crystal structure (1.6 Å) of a KRAS G-quadruplex as a 5′-head-to-head dimer with extensive poly-A π-stacking interactions observed across the dimer. Molecular dynamics simulations confirmed that the poly-A π-stacking interactions are also maintained in the G4 monomers. Docking and molecular dynamics simulations with two G4 ligands that display high stabilization of the KRAS G4 indicated the poly-A loop was a binding site for these ligands in addition to the 5′-G-tetrad. Given sequence and structural variability in the loop regions provide the opportunity for small-molecule targeting of specific G4s, we envisage this high-resolution crystal structure for the KRAS G-quadruplex will aid in the rational design of ligands to selectively target KRAS.
INTRODUCTION
The Kirsten sarcoma virus gene or KRAS encodes for KRAS, a small GTPase transducer protein involved in signal transduction (1). KRAS acts as a molecular switch inside the cell and is involved in the activation of several pathways such as MAPK, PI3K, PLCe and RalGDS (2). The MAPK and PI3K pathways in particular, activate cell proliferation and cell survival, which are two of the six hallmarks of cancer (3). In fact, aberrant KRAS signaling is a driver of many cancers, such as lung (4,5), prostate (6), colorectal (7,8), pancreatic (9) and breast cancer (10). Over the past three decades, much effort has gone into the development of small molecule based inhibitors targeting the KRAS protein, with only two compounds, AMG 510 and MRTX849, currently in phase I clinical trials (11–14). However, no drug has yet reached the market to date, highlighting the difficulty of this feat. KRAS binds its GDP/GTP substrate in high (picomolar) affinity, and it is perhaps the one reason why designing an inhibitor is difficult (14,15). Efforts have also gone into inhibiting the post-translational isoprenylation of KRAS that is required for attachment to the plasma membrane, but that too has been futile for the time being (14–16).
An alternative approach to targeting KRAS may still be through a small molecule, but instead of targeting the protein, one may choose to target the KRAS gene, in particular a G-quadruplex (G4) DNA in the promoter region (17). G4s are four-stranded DNA structures, found to be enriched in gene regulatory regions, such as the nuclease hypersensitive element (NHE) located upstream of transcription start sites (18). The stabilization of these G4 structures with small molecules has been shown to downregulate gene expression, even in the case of C-MYC, which was thought previously to be ‘undruggable’ (19,20).
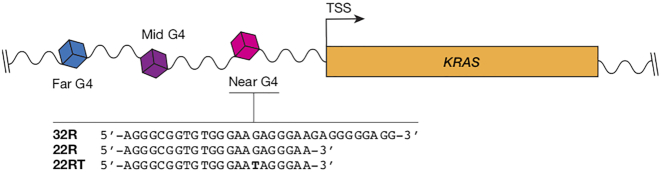
The promoter region of the KRAS gene contains three potential G4s (near, mid and far; Scheme 1) (21,22). The near KRAS G4 (namely 32R) is found ∼115 bp upstream of the transcription start site (TSS) and has been reported by circular dichroism (CD) and dimethyl sulfate (DMS) footprinting experiments to adopt a parallel G-quadruplex conformation with a thymidine bulge in one strand and a (1/1/11) looping topology (23,24). Xodo et al. reported that transcription factors such as MAZ (Myc associated zinc finger) and hnRNP A1, upregulate KRAS expression by binding to the near KRAS G4 (25). The same group reported that oxidation of guanine to 8-oxoguanine in the near KRAS G4 can elevate gene expression (26), similar to the findings of Burrows and co-workers (27). Recently, a publication has suggested that HMBG1 (high mobility group protein 1) binding the near KRAS G4 may play a role in KRAS regulation (28). These studies highlight that direct targeting of the near KRAS G4 could be a promising therapeutic strategy. To aid in the development of these targeted therapies, high-resolution structural studies of the near KRAS G4 are required.
Scheme 1.
Schematic representation of the promoter G4s located in the NHE region of the KRAS gene.
The NMR solution structure of a truncated portion of the near KRAS G4, 22RT (a 22-nucleotide G16T mutant of the native 22R sequence, PDB: 5I2V), was previously reported by Salgado et al. to be a parallel G-quadruplex monomer with a thymidine bulge, two single-nucleotide propeller loops and a four-nucleotide loop (29). The G16T mutation was found to be more stable and had better resolved NMR peaks compared to the native 22R sequence.
Herein, we report the first crystal structure of KRAS-22RT and highlight unique features observed in the crystal structure which are absent in the previously reported NMR solution structure. We performed docking and molecular dynamics (MD) simulations with two multicarbazole ligands that demonstrate higher stabilization for the near KRAS G4 compared to other promoter G-quadruplexes (30), to provide further information on structural features that are important for small molecule binding to the near KRAS G4.
MATERIALS AND METHODS
All reagents for crystallization and circular dichroism (CD) were purchased from Merck or Sigma-Aldrich. Nuclease free water was purchased from Thermo Fisher (Australia). DNA oligos KRAS-22RT (5′-dAGGGCGGTGTGGGAATAGGGAA-3′) and the doubly 5-bromouracil modified sequence KRAS-22RBrU (5′-dAGGGCGGBrUGTGGGAABrUAGGGAA-3′) were purchased from Integrated DNA Technologies (Singapore) as a lyophilized solid with HPLC purification. CD was performed on KRAS-22RT and KRAS-22RBrU to determine topology in K+ buffer (100 mM KCl and 10 mM potassium cacodylate pH 6.5) prior to crystallization (Supplementary Figure S1).
Circular dichroism
G4 DNA was diluted to 4 μM in potassium cacodylate buffer (100 mM KCl and 10 mM potassium cacodylate pH 6.5), then annealed by heating to 95°C for 5 min and slowly cooled to room temperature overnight prior to CD analysis. CD was performed on a JASCO J-810 with Peltier temperature control (20°C), scanning from 220 to 320 nm, 100 nm/min and accumulation of three spectra (Supplementary Figure S1). Background and baseline were corrected for using a blank solution (potassium cacodylate buffer).
KRAS G4 crystallization
The DNA were resuspended in potassium cacodylate buffer (100 mM KCl and 10 mM potassium cacodylate pH 6.5) to a stock concentration of 3.0 mM. The DNA was further diluted to desired concentrations, checked with nanodrop and annealed by heating to 95°C for 5 min and allowed to slowly cool to room temperature overnight. Oligos were stored at 4°C after annealing until further use.
Conditions for crystal growth were screened in a sparse matrix 96-well sitting drop format screen using Natrix crystallization screens HR-116 and HR2-117 (Hampton Research), in 96-well Intelliplates (Hampton Research), and a Phenix liquid-handling robot (Arts Robbins Instruments, USA), and stored in a temperature-controlled room (25°C).
Fine screening of KRAS-22RT and KRAS-22RBrU crystals were produced in 24 well plates based on Natrix2 condition HR-117:35 (80 mM NaCl, 12 mM KCl, 20 mM MgCl•6H2O, 40 mM Na-cacodylate•3H2O pH 7.0, 35% v/v MPD and 12 mM spermine•4HCl) and 22RT or 22RBrU (at a concentration of 0.5 or 1.0 mM) in a drop ratio of 1:1 DNA:reservoir solution, then equilibrated against a 500 μl reservoir solution. Crystals were harvested in a nylon loop, flash frozen in liquid nitrogen (with ∼30% MPD as a cryoprotectant, if required). X-ray diffraction data were collected on an in-house Rigaku XtaLAB Synergy-S instrument, and at the MX1 (22RBrU) and MX2 (22RT) beamlines of the Australian Synchrotron (Victoria, Australia) (31).
Structure solution and refinement
X-ray data were reduced using XDS (32) and AIMLESS (33,34). Resolution cut-offs of 1.8 Å (22RBrU) and 1.6 Å (22RT) were chosen, using a combination of data completeness, half-dataset correlation coefficient, signal-to-noise ratio and merging R-factor as indicators (Supplementary Table S1). The crystal structure of 22RBrU was solved through molecular replacement using PHASER (35), in the CCP4 suite (36) with a search model consisting of the core G-tetrads extracted from the first model from the ensemble of the solution structure of KRAS G4 (PDB: 5I2V (29)). The model was manually completed in COOT (37) with iterative cycles of building/refinement in first REFMAC (38), and then PHENIX.REFINE (39). As the 22RT and 22RBrU data are isomorphous, the 22RT data was phased by rigid-body refinement with the 22RBrU structure, the model modified to reflect the 22RT sequence, and completed using the same protocol as for 22RBrU. Atomic coordinates and structure factors were deposited in the RCPB PDB with PDB ID of 6N65 and 6WCK. PyMOL was used for visualization and preparing figures (40).
Computational modeling
MD simulations were conducted on the KRAS-22RT G4 dimer and corresponding A17T mutant, each of the monomeric units of the KRAS-22RT G4, a A17T mutant monomeric unit and the NMR solution structure of the KRAS G4 (PDB ID: 5I2V). Functionalized multicarbazoles with an ethyl (2b) or propyl (2f) chain and a terminal pyrrolidine (Figure 7) were docked to the KRAS-22RT G4 dimer and corresponding A17T mutant, the monomeric unit for KRAS-22RT G4 and the A17T mutant monomeric unit using AutoDock Vina (41). The docking search space was defined as a box that included the entire G4 with 40 points included in each of the x- y- and z-directions (Supplementary Figure S2). The most stable docking pose with the dimer and monomer was simulated. By performing MD after docking the physical and chemical potential energy surface of the ligand binding to the quadruplex can be investigated as the system evolves with time to determine the minimum energy conformation of the system.
Figure 7.
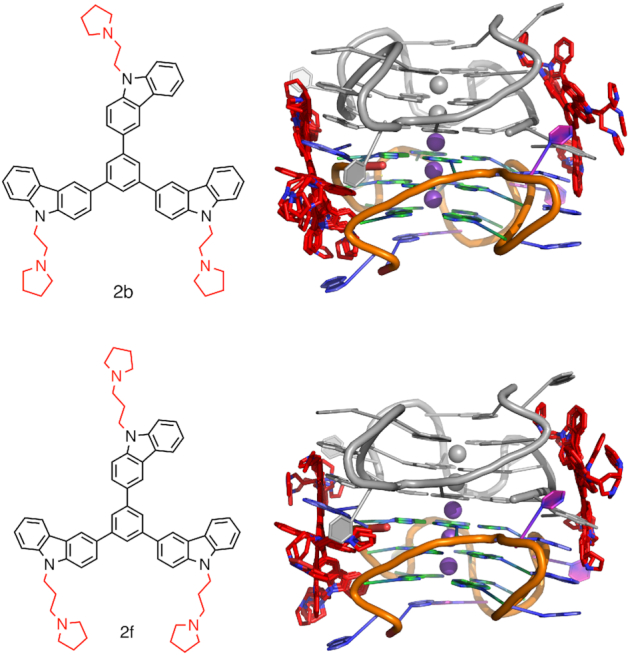
KRAS G4 dimer with 2b (top) or 2f (bottom) docked. Chain A coloured (A = blue, G = green, T = pink, C = yellow and K+ = purple), chain B in grey and the functionalized multicarbazoles in red.
MD simulations were performed using AMBER16 (42) with the AMBER BSC1 force field (43). The functionalized multicarbazoles were assigned AMBER BSC1 atom types supplemented with GAFF using ANTECHAMBER (44). Partial charges for the lesions were developed using RESP charge fitting based on a B3LYP/6-31G(d,p) structure and the HF/6-31G(d) electrostatic potential. The quadruplexes were solvated in a TIP4PEW octahedral box such that the minimum distance between the box edge and quadruplex is 10 Å. KCl was added to neutralize the system at a final concentration of 0.1 M.
Systems were minimized in a step-wise procedure with minimization of (i) water and counter ions, (ii) DNA hydrogen atoms, (iii) DNA and (iv) the entire system. Each step of minimization included 2500 steps of steepest descent and 2500 steps of conjugate gradient minimization, while applying a 50 kcal/(mol Å2) restraint on all other atoms. The systems were then heated from 0 to 310 K in six steps, with each 10 ps simulation having an increased temperature (i.e. 10, 60, 120, 180, 210, 260 and 310 K). Heating was performed with a 1 fs time step, 10 kcal/(mol Å2) restraint on the solute and the Langevin thermostat (γ = 1.0). The systems were then equilibrated over five 20 ps simulations while reducing the constraint on the DNA (i.e. 20, 15, 10, 5 and 1 kcal/(mol Å2)). Equilibration was performed at 310 K with a 2 fs time step. Finally, unconstrained production simulations of 1 μs were performed in triplicate for each system. Throughout all simulations the pressure was maintained at 1 bar using isotropic position scaling and the Berendsen barostat (τP = 2.0 ps), SHAKE, the periodic boundary condition, a nonbonded cutoff of 10 Å, and long-range electrostatic interactions were treated with the Particle Mesh Ewald (PME) method.
Analysis was performed on frames spaced by 0.1 ns using the cpptraj module of AMBER16 (38). Representative structure for systems were selected by clustering using the hierarchical agglomerative algorithm (ε = 3.0). Only clusters with occupancies >10% have been reported in the figures. Visualization of the simulations was performed using VMD (45) and figures were created using both VMD and PyMOL (46). Molecular Mechanics/Generalized Born Surface Area (MMGBSA) pairwise energies were calculated for the interactions between the functionalized multicarbazoles and the quadruplex.
Native DNA PAGE
KRAS-22RT (AGGGCGGTGTGGGAATAGGGAA) and a KRAS-22RT mutant oligo (AGAGCAGTGTGAGAATAAGGAA) (IDT, Singapore), were reconstituted in ddH2O to a concentration of 10 mM. The ladder was prepared from repeating dT oligos (IDT, Singapore) reconstituted in ddH2O to a concentration of 1 mM, and mixed in ddH2O with final concentrations shown in parentheses: dT60 (21 μM), dT50 (25 μM), dT40 (62 μM), dT30 (41 μM), dT22 (56 μM), dT15 (82 μM).
For the PAGE gel KRAS-22RT and KRAS-22RT mutant oligos were further diluted in buffer (10 mM lithium cacodylate pH 7.4, 150 mM KCl) to final concentrations of 1000, 316 and 100 μM in the absence and presence of compound 2f in the following oligo:compound ratios (μM): 1000:1000, 316:316, 100:100, 316:1000, 316:316, 316:100. The resulting solutions were annealed at 95°C for 5 min, and incubated at 4°C overnight. Samples were then further diluted in the same buffer to normalize the DNA concentration to 100 μM and 10 μl of this solution were mixed with 5 μl of glucose 50% (w/v) prior to loading in wells.
Two acrylamide gels (15%) were cast using 4.368 ml ddH2O, 4.50 ml acrylamide/bisacrylamide solution 19:1 40% (w/v), 1200 μl 10× TBE, 1800 μl KCl 1 M, 120 μl ammonium persulfate 10% (w/v), and 12 μl TEMED in a Bio-Rad Mini-PROTEAN Tetra cell apparatus (gel size 8.3 × 7.3 cm, thickness 1 mm). The running buffer consisted of 1× TBE + 150 mM KCl (1000 ml). Gels were run at 65 V, 90 mA, 4°C for 3.5 h using a mixture of bromophenol blue (runs at ∼22 nt) and xylene cyanol (∼60 nt) to follow progress. After completion, gels were transferred to plastic wrap, placed on a fluorescent silica TLC plate and illuminated manually from above at 254 nm with a handheld UV lamp. Images were captured using a Bio-Rad ChemiDoc XRS+ (no illumination, 1 × 1 binning, 0.015 s exposure).
RESULTS AND DISCUSSION
The truncated 22nt KRAS promoter G4 crystallizes as a dimer with head-to-head stacking
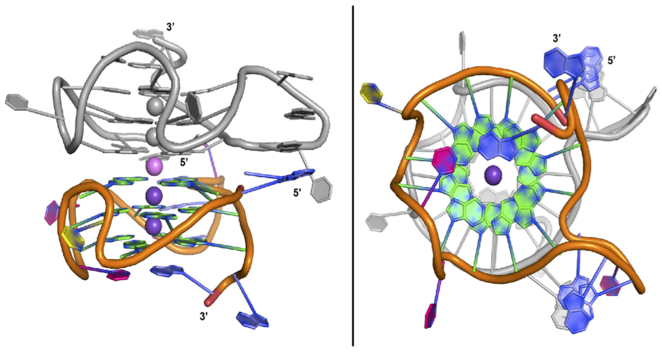
The KRAS-22RT G4 crystallized in space group P1211 as a 5′-head-to-head stacked dimer in the asymmetric unit, which differs from the previously reported monomeric NMR solution structure (29) (Figure 1 and Supplementary Table S1). In parallel, the 5-bromouracil derivative (22RBrU) was also crystallized with a view to use anomalous scattering phasing where it was required to solve the structure of 22RT (Supplementary Figure S3 and Supplementary Table S1). Following successful molecular replacement, the 22RBrU data was used to confirm the sequence register of the structure, by location of the bromine atoms in an anomalous difference electron density map (Supplementary Figure S4). There was little variation between the 22RBrU and 22RT crystal structures (RMSD = 0.12 Å, dimer), with small difference in the orientation of T16 versus BrU16, which is expected due to the higher B-factor of BrU residues (Supplementary Figure S5). MD simulations were performed to gain a greater understanding of the stability of the dimeric G4 structure. A backbone RMSD of < 4 Å was maintained across all simulations, indicating the stability of the G4 conformation (Supplementary Figure S6).
Figure 1.
Crystal structure of KRAS-22RT G-quadruplex. Side view (left) and top view (right). Chain A colored (A = blue, G = green, T = pink and C = yellow, potassium cations = dark purple) and chain B in grey. Potassium cations can be observed in the central channel, sandwiched between two G-tetrads. The potassium ion at the dimer interface is coloured in light purple.
A total of five potassium ions were found in the central channel of the dimeric G4 formed in the asymmetric unit, with each individual G4 containing two potassium ions positioned between the stacked G-tetrads and an additional potassium ion located at the dimer interface (Figure 1). In the crystal structures, the distances from each potassium ion is ∼3.3 Å and for guanines participating in G-tetrad formation the K+ to O6 guanine oxygen distances ranged from 2.7 to 3.0 Å. Upon simulation, the distance between each potassium ion is slightly elongated (3.6 ± 0.2 Å), while the K+···O6(G) distances remain consistent with the crystal structure geometry (2.7 ± 0.2 Å, Supplementary Table S2). Additionally, the overall geometry about the K+ ions is consistent with that of previously reported G4 crystal structures (47–53). Based on the G4 folding conventions of da Silva (54), the KRAS-22RT G4 has type 1a looping (-ppp) and type VIIIa G-tetrads with four medium (MMMM) groves with widths of 15.4–15.8 Å (Figure 2). The glyosidic bond angles of the guanine residues in the G-tetrad were in the anti:anti:anti:anti conformation.
Figure 2.
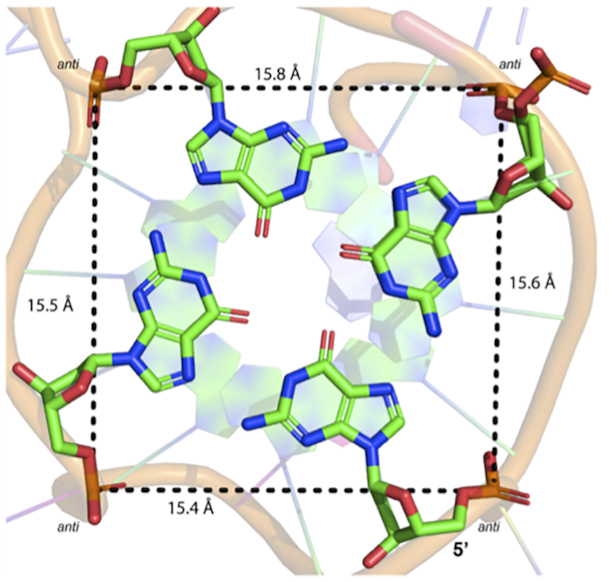
Measured distances of medium groove sizes (MMMM) of the first G-tetrad (G2•G6•G11•G18). Distances were measured from the 5′ phosphate groups. All guanine residues are in the anti-conformation.
An interesting observation of the G4 dimer structure is a lensing effect of the tetrad planes observed in each monomer, beginning as straight at the head (5′ end), and becoming progressively more concave into the tail (3′ end) of the monomer (Supplementary Figure S16). The straight tetrad plane beginning allows for a favourable head to head dimer interface. The more concave plane of the tails of each monomer would be less likely to accommodate any further oligomeric plane to plane interactions. This would suggest that under the geometric constraints seen in these crystal structures, that the G4 dimer represents some limit of oligomerization.
Poly-A stacking was observed in the 4nt propeller loop
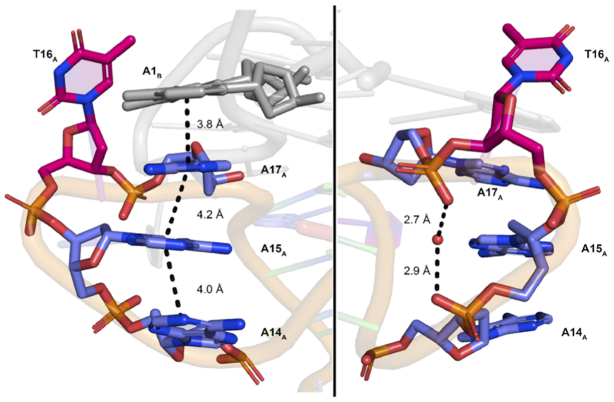
Like the NMR solution structure, the crystal structure of KRAS-22RT contains a bulge (T8) and three propeller loops: two single-residue loops (C5 and T10) and one four residue loop (A14, A15, T16, A17) (See Supplementary Figure S7 for schematic). A unique feature of the KRAS-22RT crystal structure is the extensive poly-A π-stacking interactions within the four-residue propeller loop (A14, A15 and A17) of one quadruplex and with A1 of the other quadruplex in the dimer, a feature which may add increased stability to the quadruplex (Figure 3). Upon simulation of the dimer, substantial variations in the orientation of the four-residue loop are observed, however two orientations remain predominant (Figure 4). Specifically, the geometry that is observed in the crystal structure is formed for 29% of the total simulation time. While for 65% of the total simulation time the propeller loop is bent such that A14 and A15 are stacking and T16, A17 and A1 are stacking (Figure 4). Between these two groups of stacking bases there are T-shaped interactions involving A15 and a hydrogen bond is formed between A15(N7) and A17(N6H) for 44% of the overall simulation time (Supplementary Table S3). In contrast, in the monomeric NMR solution structure residues A1 and A17 form a cap over the 5′ face of the quadruplex (29), which was absent in both the crystallographic and simulated dimer.
Figure 3.
(Left) The four-nucleotide loop forming π-π stacking with adenosine bases. A1B π–π stacking on A14A, A15A and A17A. Residue T16A is oriented away and does not appear to be involved in π-stacking. Distances from each ring displayed. (Right) The same four-nucleotide loop with water molecule shown to be pincered by the phosphate group.
Figure 4.

MD representative structures of the quadruplex dimer. Chain A coloured (A = blue, G = green, T = pink, C = yellow and K+ = purple) and chain B in grey.
Bulges and caps compared to the solution structure
The thymine residue forming the bulge (T8) displayed a different orientation in the crystal structure when compared to the MD simulations and previously reported NMR solution structure. In the crystal structures T8 and BrU8 was observed to base-pair with A22 to form a 3′-cap (Supplementary Figure S8), whereas during MD simulations of the crystal structure and in the previously reported NMR solution structure T8 was projected out and away from the G-tetrad core (Figures 4 and 5) (29). Capping of the terminal G-tetrads has also been observed with previously reported crystal structures and is thought to contribute to the stability of the quadruplex (55–57).
Figure 5.
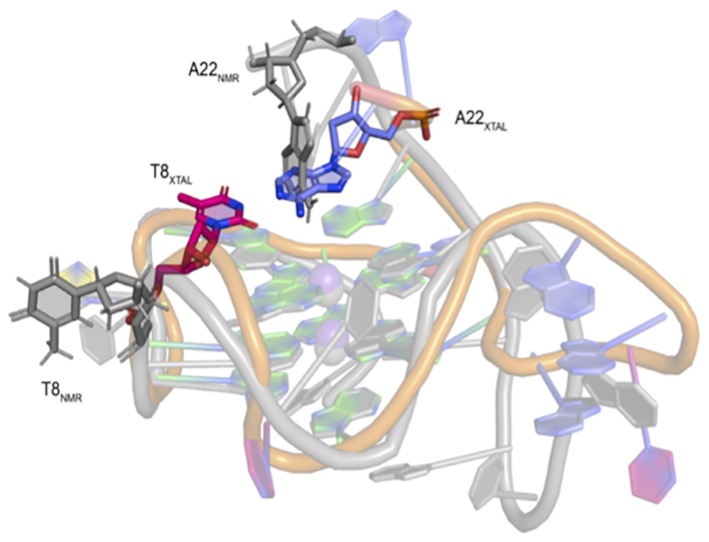
Crystal structure of KRAS-22RT G-quadruplex (A = blue, G = green, T = pink, C = yellow, K+ = purple) aligned with the NMR solution structure (PDB: 5I2V, grey). Residue T8 in the NMR structure is projected out and away of the G-tetrad core (T8NMR), while in the crystal structure forms a 3′-cap over the G-tetrad core (T8XTAL).
Conserved water molecules and stacking arrangements
Comparison between the crystal structure of 22RT and the 22RBrU showed that a majority of water molecules are conserved between the two G4 structures in the groove regions, suggesting water may be required to stabilize the structure (Supplementary Figure S9). We observed a single water molecule pincered by the phosphate back bone of A15 and A17 (∼2.7–2.9 Å), which was conserved in the crystal structure of 22RT across the dimer, which we speculate may be required to stabilize the stacking-loop (A14-A15-T16-A17) structure (Figure 3). Additionally, during the MD simulations a single water molecule, which exchanged during the simulation, is bridging a hydrogen bond between A15 and A17 for the entire simulation (Supplementary Table S3). During the simulation water hydrogen bonds with the phosphate backbone, as well as N1 and N6H of the nucleobases. Due to the conserved nature of the water molecule, we speculate that it is playing a role in stabilizing the stacking-loop (A14-A15-T16-A17) structure.
Comparison between other G-quadruplex structures
Interestingly, dimeric G-quadruplexes have been reported for the crystal structures of telomeric G4 (50,58) and promoter G4s such as c-KIT (47), c-MYC (49), BRAF (48) and the NMR solution structure of c-KIT2 (59), with the majority displaying head-to-head stacking, with c-KIT2 and BRAF being an exception. The first residues in the c-KIT and c-MYC crystal structures are also oriented away from the G-tetrad, to enable the head-to-head stacking interactions to occur.
It has been noted that in a biological setting, where genomic DNA is involved, the capping of the first G-tetrad may be unfavorable (49). In our crystal structures capping of the first G-tetrad (G2•G6•G11•G18) from A1, was not observed, which allowed the second G4 to stack on the surface. In a biological setting, only one copy of the KRAS near G4 sequence exists, therefore homodimerization is not possible. However, due the three G4 forming regions in the KRAS promoter (near, mid and far), we question if heterodimerization was possible with the other G4s and thus whether this plays a biological role.
Structure of monomeric unit of G4
In order to determine whether the poly-A-π-stacking observed in the KRAS-22RT G4 dimer crystal structure could occur in a G4 monomer, simulations were performed on each of the monomeric units of the dimer, as well as the previously reported monomeric NMR solution structure. Regardless of the starting structure, the distances from each potassium ion is elongated in comparison to the dimer (on average ∼3.7 Å) and for guanines participating in G-tetrad formation the K+ to O6 guanine distances are similar to that observed in the dimer (on average 2.7 Å, Supplementary Table S2). Similar to the dimer there is substantial variation in the orientation of the four-residue loop. However, regardless of the starting structure, stacking interactions are typically observed between residues in the four-residue loop (Figure 6, Supplementary Figures S10 and S11). Similar solvation of the loop regions is observed to that of the dimer (Supplementary Tables S4–S6), which further supports the role of water in stabilizing this region of the quadruplex. Additionally, as seen for the simulation of the dimer T8 predominantly adopts an orientation that is projected out and away from the G-tetrad core. In contrast to the dimer simulation, T8 transiently forms hydrogen bonding interactions with A21 (Supplementary Tables S3–S5).
Figure 6.

MD representative structures of the quadruplex monomer simulation started from chain A (coloured as A = blue, G = green, T = pink, C = yellow and K+ = purple). See Supplementary Figures S10 and S11 for representative structures of the simulations started from chain B and the NMR solution structure.
Ligand binding to the near KRAS G4 dimer
The unique poly-A π-stacking interactions within the four-residue loop of the KRAS G4 may be of interest for drug design. Indeed, we have previously reported functionalized multicarbazoles (2b and 2f) that bind selectively to quadruplex DNA over duplex DNA and display a higher stabilization for the near KRAS G4 compared to telomeric and other promoter G-quadruplexes (30). Therefore, to probe the binding of these functionalized multicarbazoles to the near KRAS G4 dimer molecular docking followed by 1 μs molecular dynamics simulations were performed. Regardless of the ethyl or propyl functionalization, the ligand docked to one of the poly-A π-stacking loops (Figure 7). Upon simulation stacking is observed in the four-residue loop and this region is solvated as seen for the isolated dimer (Figure 8, Supplementary Figures S12 and S13 and Supplementary Tables S7 and S8). Nevertheless, there is substantial variation in the ligand binding location (Figure 8, Supplementary Figures S12 and S13). This indicates that the ligand is not interacting in a stable manner with the dimer. Indeed, upon calculation of the MMGBSA binding energy the ligand binds to the quadruplex with a stability of ∼–18 kcal/mol (Supplementary Table S9). Additionally, while there is no substantial change to the average distances between the potassium ions (3.6 or 3.7 Å), there is greater variation in these distances with a standard deviation up to 0.6 Å (Supplementary Table S2). For the guanines participating in G-tetrad formation the average K+ to O6 guanine distances is similar to that of the isolated dimer (2.7–2.8 Å), but the standard deviations are double that seen when the ligand is not interacting with the G4 (Supplementary Table S2). The high standard deviations indicate that the binding of the ligand to the quadruplex is increasing the flexibility of the overall quadruplex structure.
Figure 8.
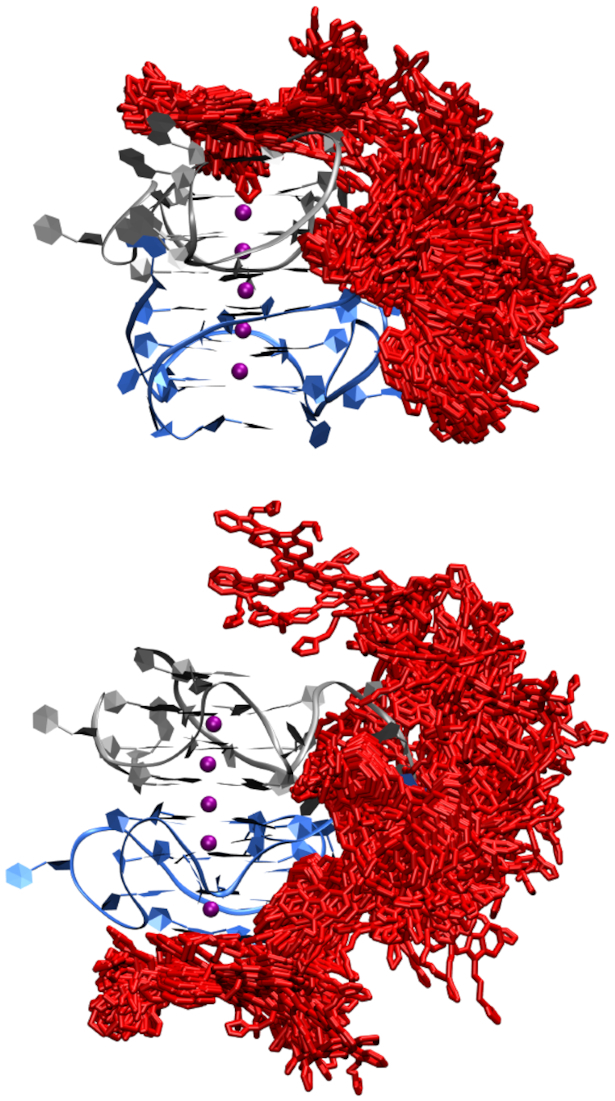
Dynamics in the binding location of the functionalized multicarbazoles 2b (top) or 2f (bottom) to the KRAS G4 dimer (chain A in blue, chain B in grey, K+ in purple and functionalized multicarbazoles in red).
Ligand binding to the KRAS near G4 monomeric unit
Since poly-A-π-stacking within the four-residue loop was also observed upon simulation of the monomer, and the binding of the ligand was shown to increase the flexibility of the quadruplex dimer, molecular docking and 1 μs molecular dynamics simulations were also performed. In contrast to the dimer, the functionalized multicarbazoles bind to the 5′-G-tetrad of the G4 while still maintaining interactions with the poly-A loop residues (A1 and A17 (Figures 9 and 10) and while the ligand shows some variability in orientation, it remains bound to the 5′-G-tetrad of the G4 upon simulation (Figure 10). The geometry about the potassium ions is similar to that of the geometry in the absence of the ligand (K+···K+ = ∼ 3.7 Å and K+···O6(G) = ∼2.7 Å, Supplementary Table S2). Stacking interactions still occur within the four-residue loop and the water mediated interactions are maintained, however the hydrogen-bonding between the loop residues is reduced (Supplementary Figures S14, S15 and Supplementary Tables S10 and S11). The ligand binding to the monomeric KRAS G4 is predominantly stabilized by the 5′ G-tetrad (G2, G6, G11 and G18) and A residues in the loops (A1 and A17; Figure 11 and Supplementary Table S12). Specifically, the multicarbazoles stack with the 5′ G-tetrad and A1, while the pyrrolidine forms stacking interactions with A17 (Supplementary Figures S14 and S15). 2f binds stronger to the G4 than 2b (–51.0 and –57.5 kcal/mol, respectively; Supplementary Table S12), and the stronger binding likely occurs due to the longer linker chains in 2f allowing for the pyrrolidine to interact with the loop regions without disrupting the stacking between the 5′ G-tetrad and the multicarbazoles. This higher binding of 2f correlates well with the previously reported higher stabilization of the near KRAS G4 by 2f versus 2b (30). Nevertheless, regardless of the functionalization, the binding of the ligand to the monomer is up to 40 kcal/mol stronger than the binding of the ligand to the dimer. Given G-tetrads are a conserved feature of all G4s, sequence and structural variability in the loop regions provide the opportunity for small-molecule targeting of specific G4s. Since the studied functionalized multicarbazoles show higher stabilization of the KRAS G4 over other quadruplexes (30), this could be due to the additional stacking interactions that occur with the poly-A loop residues.
Figure 9.
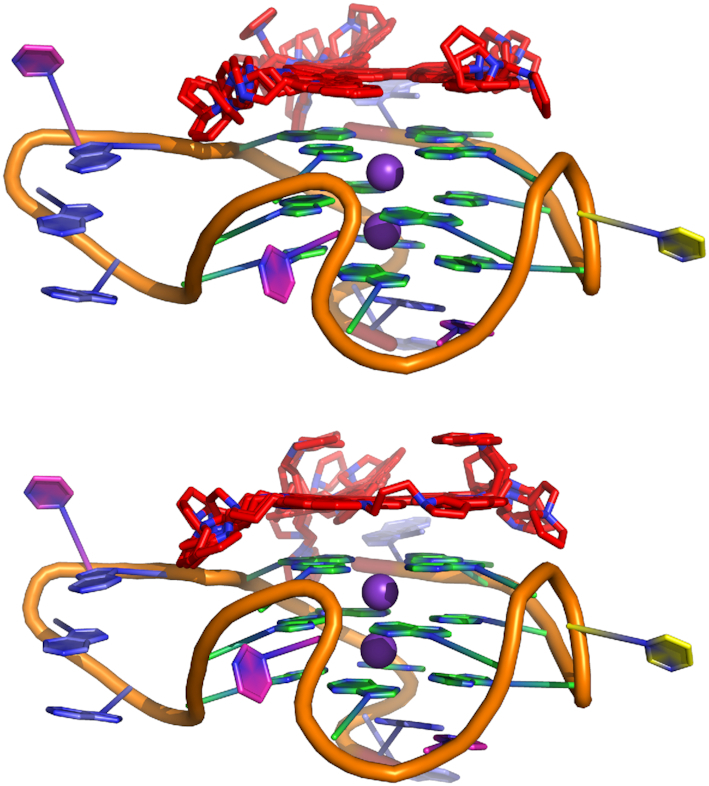
KRAS G4 monomer with 2b (top) or 2f (bottom) docked. Coloured as A = blue, G = green, T = pink, C = yellow, K+ = purple, and the functionalized multicarbazoles = red.
Figure 10.

Dynamics in the binding location of the functionalized multicarbazoles 2b (top) or 2f (bottom) to the KRAS G4 monomer (KRAS in blue, K+ in purple and functionalized multicarbazoles in red).
Figure 11.
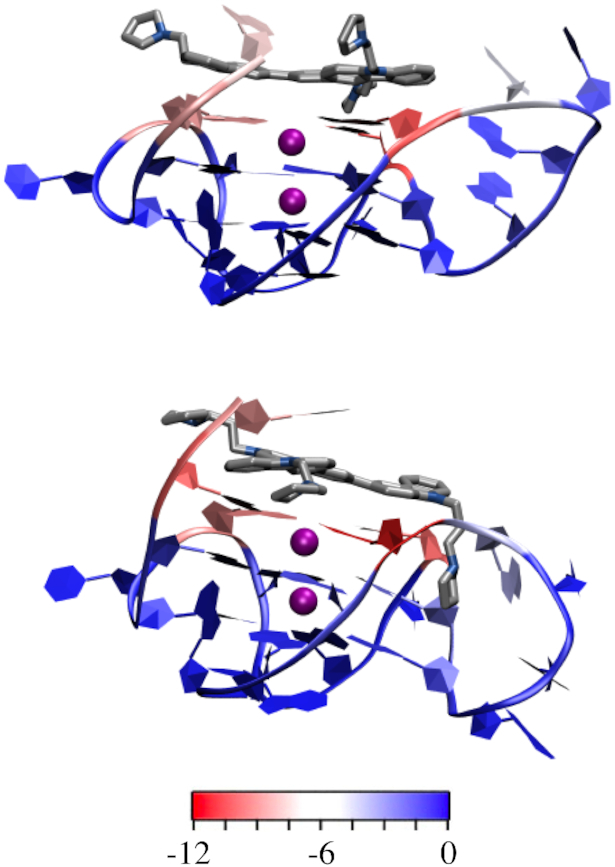
MD representative structures showing the energetic contribution (kcal/mol) of each nucleoside to binding of 2b (top) or 2f (bottom) to the KRAS G4 monomer. See Supplementary Table S12 for exact binding energies.
Ligand binding to the KRAS A17T mutant
To further clarify the role of the poly-A-π-stacking within the four-residue loop on the structure of the KRAS quadruplex and the role of this loop in ligand binding, simulations were performed on the A17T mutant. For both the mutant dimer and monomer, the potassium ions are positioned similarly to the KRAS_22RT (K+ to K+ = 3.7 Å) and the guanines participating in G-tetrad maintain their interactions with the ions (K+ to O6 = 2.7 Å, Supplementary Table S2). There is less variation in the orientation of the four-residue loop of the A17T quadruplexes compared to the native quadruplex monomer or dimer. In the A17T quadruplex dimer or monomer, A15 and A14 form a stacking interaction and T17 and T16 are not interacting with the stacked adenine residues (Supplementary Figures S17 and S18). This loss of poly-A-π-stacking within the four-residue loop has the potential to reduce the binding propensity of the ligands to the quadruplex.
To determine the effect of the loss of poly-A-π-stacking within the four-residue loop, the functionalized multicarbazoles (2b and 2f) were docked to the mutant dimer and monomer and 1 μs simulations were performed. 2b displays weaker binding to the A17T quadruplex dimer and monomer by 3–4 kcal/mol (Supplementary Tables S13 and S14). The weaker binding is predominantly due to loss of stabilizing interactions with A17 due to the change in orientation of the loop relative to the bound ligand (Figure 12 and Supplementary Figure 19). The same drastic change is not observed for binding of 2f to the A17T quadruplex dimer or monomer compared to KRAS_22RT, although in both cases there are no interactions between the 2f ligand and the four residue loop, hence binding to the loop is completely disrupted (Supplementary Figure S20). The simulations on ligand binding to the A17T KRAS quadruplex indicates a key role of the poly-A sequence of the four-residue loop in ligand binding.
Figure 12.

MD representative structures showing the stacking between the A residues of the loops and the functionalized multicarbazole 2b (red) in the native KRAS quadruplex (top) and A17T mutant KRAS quadruplex (bottom).
Native DNA PAGE of the KRAS near G4
Native DNA PAGE of the KRAS-22RT G4 indicated that at both low and high oligo concentrations two G4 species were observed in solution, a lower band which displayed a faster mobility and corresponds to the monomeric G4 and an upper band which displayed a much slower mobility and corresponded to the higher order G4 (Supplementary Figure S21). Both structures were observed at oligo concentrations as low as 100 μM, confirming that the higher order G4 structure is present in solution even at low concentrations. In contrast, the unfolded KRAS-22RT mutant (AGAGCAGTGTGAGAATAAGGAA) displays no higher order structure. The presence of compound 2f results in retardation of the higher order structure, where increasing the ligand:oligo ratio results in a greater band retardation (Supplementary Figure 21). The lower band which corresponds to the monomeric G4 appears to be unaffected by the presence of compound 2f.
CONCLUSION
We reported two crystal structures of the near KRAS G4 (22RT and 22RBrU) found in the promoter region at 1.6 and 1.8 Å resolution, respectively. The structure was found to be a parallel G4 head-to-head dimer, in contrast to the previously-reported monomeric NMR solution structure. The crystal structure displayed unique elements which were not observed in the solution structure: the four-nucleotide propeller loop of one quadruplex contained extensive π-stacking interactions between adenines (A14A, A15A and A17A) that extended onto the second quadruplex by π-stacking with the first residue (A1B) of the second G4. Furthermore, the crystal structure was capped at the 3′-ends by T8 and A22 through Watson Crick base-pairing, whereas this was not observed in the previously reported NMR solution structure. We demonstrated using Native DNA PAGE, that in solution the KRAS 22RT G4 contains two main G4 species, a monomeric G4 and a higher order G4. Using molecular dynamics simulation with two multicarbazole ligands that display high stabilization of the near KRAS G4 we demonstrated that the poly-A loop residues are important sites for ligand binding to the G4 dimer and monomer in addition to the 5′-G-tetrad. We envisage these stacking interactions with the poly-A loop residues are important contributors to the observed high stabilization of the near KRAS G4 with these ligands.
DATA AVAILABILITY
Atomic coordinates and structure factors were deposited in the RCPB PDB with accession codes of 6N65 and 6WCK.
Supplementary Material
ACKNOWLEDGEMENTS
This research was undertaken in part using the MX1 and MX2 beamlines at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector; and with the assistance of resources and services from the National Computational Infrastructure (NCI), which is supported by the Australian Government. The authors would like to thank Prof. Tracy Bryan (Children's Medical Research Institute) for help with analyzing the native DNA PAGE gel.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council; Australian Research Council; National Breast Cancer Foundation; A.O. would like to thank the Australian Government and UWA for scholarships and financial support. Funding for open access charge: University of Western Australia.
Conflict of interest statement. None declared.
REFERENCES
- 1. Haigis K.M. KRAS alleles: the devil is in the detail. Trends Cancer. 2017; 3:686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rommel C., Vanhaesebroeck B., Vogt P.K.. Phosphoinositide 3-Kinase in Health and Disease. 2010; NY: Springer Verlag. [Google Scholar]
- 3. Hanahan D., Weinberg R.A.. The hallmarks of cancer. Cell. 2000; 100:57–70. [DOI] [PubMed] [Google Scholar]
- 4. Román M., Baraibar I., López I., Nadal E., Rolfo C., Vicent S., Gil-Bazo I.. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol. Cancer. 2018; 17:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Westcott P.M.K., To M.D.. The genetics and biology of KRAS in lung cancer. Chin. J. Cancer. 2013; 32:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gandhi J., Afridi A., Vatsia S., Joshi G., Joshi G., Kaplan S.A., Smith N.L., Khan S.A.. The molecular biology of prostate cancer: current understanding and clinical implications. Prostate Cancer Prostatic Dis. 2018; 21:22–36. [DOI] [PubMed] [Google Scholar]
- 7. Kondo Y., Hayashi K., Kawakami K., Miwa Y., Hayashi H., Yamamoto M.. KRAS mutation analysis of single circulating tumor cells from patients with metastatic colorectal cancer. BMC Cancer. 2017; 17:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barault L., Veyrie N., Jooste V., Lecorre D., Chapusot C., Ferraz J.-M., Lièvre A., Cortet M., Bouvier A.-M., Rat P. et al.. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int. J. Cancer. 2008; 122:2255–2259. [DOI] [PubMed] [Google Scholar]
- 9. Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C.D., Biankin A.V., Neale R.E., Tempero M., Tuveson D.A., Hruban R.H. et al.. Pancreatic cancer. Nat. Rev. Dis. Primers. 2016; 2:16022. [DOI] [PubMed] [Google Scholar]
- 10. Toss A., Cristofanilli M.. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res. 2015; 17:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y., Kaiser C.E., Frett B., Li H.. Targeting mutant KRAS for anticancer therapeutics: a review of novel small molecule modulators. J. Med. Chem. 2013; 56:5219–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kessler D., Gmachi M., Mantoulidis A., Martin L.J., Zoephel A., Mayer M., Gollner A., Covini D., Fischer S., Gerstberger T. et al.. Drugging an undruggable pocket on KRAS. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:15823–15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tran T.H., Alexander P., Dharmaiah S., Agamasu C., Nissley D.V., McCormick F., Esposito D., Simanshu D.K., Stephen A.G., Balius T.E.. The small molecule BI-2852 induces a nonfunctional dimer of KRAS. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:3363–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mattox T.E., Chen Xi., Maxuitenko Y.Y., Keeton A.B., Piazza G.A.. Exploiting RAS nucleotide cycling as a strategy for drugging RAS-Driven cancers. Int. J. Mol. Sci. 2019; 21:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J.. Drugging the undruggable RAS: mission possible. Nat. Rev. Drug Discov. 2014; 13:828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baines A.T., Xu D., Der C.J.. Inhibition of Ras for cancer treatment: the search continues. Future Med. Chem. 2011; 3:1787–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hänsel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279–284. [DOI] [PubMed] [Google Scholar]
- 19. Brown R.V., Danford F.L., Gokhale V., Hurley L.H., Brooks T.A.. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J. Biol. Chem. 2011; 286:41018–41027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cogoi S., Xodo L.E.. G4 DNA in ras genes and its potential in cancer therapy. Biochim. Biophys. Acta. 2016; 1859:663–674. [DOI] [PubMed] [Google Scholar]
- 22. Morgan R.K., Batra H., Gaerig V.C., Hockings J., Brooks T.A.. Identification and characterization of a new G-quadruplex forming region within the kRAS promoter as a transcriptional regulator. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 2016; 1859:235–245. [DOI] [PubMed] [Google Scholar]
- 23. Cogoi S., Xodo L.E.. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006; 34:2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cogoi S., Paramasivam M., Spolaore B., Xodo L.E.. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008; 36:3765–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cogoi S., Paramasivam M., Membrino A., Yokoyama K.K., Xodo L.E.. The KRAS promoter responds to Myc-associated zinc finger and Poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA -element. J. Biol. Chem. 2010; 285:22003–22016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cogoi S., Ferino A., Miglietta G., Pedersen E.B., Xodo L.E.. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic Acids Res. 2018; 46:661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fleming A.M., Ding Y., Burrows C.J.. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amato J., Madanayake T.W., Laccarino N., Novellino E., Randazzo A., Hurley L.H., Pagano B.. HMGB1 binds to the KRAS promoter G-quadruplex: a new player in oncogene transcriptional regulation. Chem. Commun. 2018; 54:9442–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kerkour A., Marquevielle J., Ivashchenko S., Yatsunyk L.A., Mergny J.-L., Salgado G.F.. High-resolution three-dimensional NMR structure of the KRAS proto-oncogene promoter reveals key features of a G-quadruplex involved in transcriptional regulation. J. Biol. Chem. 2017; 292:8082–8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ou A., Guédin A., Skelton B.W., Amrane S., Evans C.W., Norret M., Iyer K.S., Mergny J.-L., Smith N.M.. Multicarbazole scaffolds for selective G-quadruplex binding. Chem. Commun. 2018; 54:9647–9650. [DOI] [PubMed] [Google Scholar]
- 31. McPhillips T.M., McPhillips S.E., Chiu H.-J., Cohen A.E., Deacon A.M., Ellis P.J., Garman E., Gonzalez A., Sauter N.K., Phizackerley R.P. et al.. Blu-Ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 2002; 9:401–406. [DOI] [PubMed] [Google Scholar]
- 32. Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006; 62:72–82. [DOI] [PubMed] [Google Scholar]
- 34. Evans P.R., Murshudov G.N.. How good are my data and what is the resolution. Acta Crystallogr. D Biol. Crystallogr. 2013; 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A. et al.. Overview of the CCP 4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC 5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. The PyMOL Molecular Graphics System Schrödinger.
- 41. Trott O., Olson A.J.. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009; 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Case D.A., Betz R.M., Cerutti D.S., Cheatham T.E., Darden T., Duke R.E.. 2016; AMBER16 San Fransico.
- 43. Galindo-Murillo R., Robertson J.C., Zgarbová M., Šponer J., Otyepka M., Jurečka P., Cheatham T.E.. Assessing the current state of Amber force field modifications for DNA. J. Chem. Theory Comput. 2016; 12:4114–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang J., Wang W., Kollman P.A., Case D.A.. Antechamber: an accessory software package for molecular mechanical calculations. J. Am. Chem. Soc. 2001; 222:U403. [Google Scholar]
- 45. Humphrey W., Dalke A., Schulten K.. VMD: visual molecular dynamics. J. Mol. Graph. 1996; 14:33–38. [DOI] [PubMed] [Google Scholar]
- 46. DeLano W.L. PyMOL: an open-source molecular graphics tool. CCP4 Newslett. Protein Crystallogr. 2002; 40:82–92. [Google Scholar]
- 47. Wei D., Parkinson G.N., Reszka A.P., Neidle S.. Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation. Nucleic Acids Res. 2012; 40:4691–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wei D., Todd A.K., Zloh M., Gunaratnam M., Parkinson G.N., Neidle S.. Crystal structure of a promoter sequence in the B-raf gene reveals an intertwined dimer quadruplex. J. Am. Chem. Soc. 2013; 135:19319–19329. [DOI] [PubMed] [Google Scholar]
- 49. Stump S., Mou T.-C., Sprang S.R., Natale N.R., Beall H.D.. Crystal structure of the major quadruplex formed in the promoter region of the human c-MYC oncogene. PLoS One. 2018; 13:e0205584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Parkinson G.N., Lee M.P.H., Neidle S.. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002; 417:876–880. [DOI] [PubMed] [Google Scholar]
- 51. Chung W.J., Heddi B., Schmitt E., Lim K.W., Mechulam Y., Phan A.T.. Structure of a left-handed DNA G-quadruplex. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:2729–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guédin A., Lin L.Y., Armane S., Lacroix L., Mergny J.-L., Thore S., Yatsunyk L.A.. Quadruplexes in ‘Dicty’: crystal structure of a four-quartet G-quadruplex formed by G-rich motif found in the Dictyostelium discoideum genome. Nucleic Acids Res. 2018; 46:5297–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Haider S., Parkinson G.N., Neidle S.. Crystal structure of the potassium form of an oxytricha nova G-quadruplex. J. Mol. Biol. 2002; 320:189–200. [DOI] [PubMed] [Google Scholar]
- 54. Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chem. Eur. J. 2007; 13:9738–9745. [DOI] [PubMed] [Google Scholar]
- 55. Lim K.W., Amrane S., Bouaziz S., Xu W., Mu Y., Patel D.J., Luu K.N., Phan A.T.. Structure of the human telomere in K + solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009; 131:4301–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Phan A.T., Kuryavyi V., Luu K.N., Patel D.J.. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K + solution †. Nucleic Acids Res. 2007; 35:6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang Y., Patel D.J.. Solution structure of a parallel-stranded G-quadruplex DNA. J. Mol. Biol. 1993; 234:1171–1183. [DOI] [PubMed] [Google Scholar]
- 58. Nicoludis J.M., Miller S.T., Jeffrey P.D., Barrett S.P., Rablen P.R., Lawton T.J., Yatsunyk L.A.. Optimized end-stacking provides specificity of N-methyl mesoporphyrin IX for human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2012; 134:20446–20456. [DOI] [PubMed] [Google Scholar]
- 59. Kuryavyi V., Phan A.T., Patel D.J.. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010; 38:6757–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors were deposited in the RCPB PDB with accession codes of 6N65 and 6WCK.