Abstract
Bacterial ribosomal RNAs (rRNAs) are transcribed as precursors and require processing by Ribonucleases (RNases) to generate mature and functional rRNAs. Although the initial steps of rRNA processing in Escherichia coli (E. coli) were described several decades ago, the enzymes responsible for the final steps of 5S and 23S rRNA 5′-end maturation have remained unknown. Here, I show that RNase AM, a recently identified 5′ to 3′ exonuclease, performs the last step of 5S rRNA 5′-end maturation. RNase AM was also found to generate the mature 5′ end of 23S rRNA, subsequent to a newly identified prior processing step. Additionally, RNase AM was found to mature the 5′ end of 16S rRNA, a reaction previously attributed to RNase G. These findings indicate a major role for RNase AM in cellular RNA metabolism and establish a biological role for the first 5′ to 3′ RNA exonuclease identified in E. coli.
INTRODUCTION
The rRNAs represent the most abundant class of the RNAs, accounting for up to 90% of the total RNA content and nearly 20% of the dry weight of the cell (1). Therefore, the processing of these RNAs constitutes a major function of the cellular RNases. In E. coli, the three rRNAs are transcribed from seven operons with a 16S–23S–5S arrangement, in some cases, with tRNA genes interspersed between the rRNA genes (2,3). The initial separation of the rRNAs is performed by RNase III, a double-strand specific RNase, which cleaves within stem regions formed by the base-pairing of rRNA precursor sequences (4–6). The RNase III-cleaved products retain residual precursor sequences at both the 5′ and 3′ ends and require processing by several additional RNases before mature rRNAs can be produced.
Over several years, the enzymes that perform 3′-end maturation of the rRNAs have been identified, and they include RNase T for 5S and 23S rRNA, and multiple 3′ to 5′ exonucleases for 16S rRNA (7–11). The 5′-end maturation pathway, on the other hand, is less well defined. Specifically, the 5′ ends of 5S and 23S rRNA were shown to retain three and seven nucleotides (nts) of precursor sequences, respectively, after RNase III cleavage and further 5S rRNA processing by RNase E nearly 40 years ago (6,12). However, the mechanisms by which these precursor residues are subsequently removed have remained unknown.
During the course of studies to identify the basis for the 5′-end maturation of 5S rRNA, it was noticed that wild-type strains contained precursors with not only three unprocessed nts at the 5′ end, but also one and two nts, which suggested that maturation of this RNA might be proceeding via an exonucleolytic mechanism. Recently, the first 5′ to 3′ RNA exonuclease in E. coli, RNase AM, was identified, but no biological function has been attributed to this enzyme (13). Here, I show that RNase AM performs the 5′-end maturation of each of the 5S, 23S and 16S rRNAs.
MATERIALS AND METHODS
Strains
The parental strain used in this study is a derivative of MG1655, which contain a single engineered base-pair insertion that corrects a frame-shift in the rph gene, encoding RNase PH. Derivatives of this strain were made by P1 transduction of deletion alleles from the Keio strains collection (14).
RNase digests
Digestion of total RNA or ribosomal particles was performed on 10 ng of total RNA or 10–20 ng of 70S ribosomes, which were isolated by ultracentrifugation of cell extracts, as described (15). RNase AM, free of any detectable contaminants, was obtained from Dr. Frank Raushel (Texas A&M university). RNase AM was added to substrates in 25 mM HEPES (pH 7.5), 200 mM KCl, 0.5 mM MgCl2 and 50 μM MnCl2 in a final volume of 10 μl. The reactions were incubated at 37°C for 1 h and terminated by adding 5 μl of phenol. The reactions were centrifuged and the aqueous layer was supplemented with 50 μl water and 10 μg of yeast tRNA, extracted with one drop of chloroform, and ethanol precipitated. Digestions on pre-5S rRNA were performed similarly, except that the RNA was gel purified from a ΔyciV strain and labeled at the 5′ end with 32P prior to assay. Digestions with RNase G were performed as for RNase AM, except that the reaction buffer contained 25 mM Tris (pH 8.0), 150 mM KCl, 10 mM MgCl2 and 0.1% triton X-100. RNase G was purified using a tagged over-expression construct from the ASKA collection, as described (16,17).
RNA analysis
Primer extension was performed as described (18). The oligonucleotides used for primer extension analysis contained the sequences 5′ TTCTGAGTTCGGCATGGGG 3′ (for 5S rRNA), 5′ CGTTCAATCTGAGCATGATC 3′ (for 16S rRNA) and 5′ CCTTCATCGCCTCTGACTGCC 3′ or 5′ ACCGTGTACGCTTAGTC 3′ (for 23S rRNA). Sequencing ladders were prepared using a USB cycle-sequencing kit and 32P-labeled primers. 3′-end analysis of 23S rRNA was performed as described (19). All RNA analyses were performed a minimum of three times, with representative experiments depicted in figures.
RESULTS
RNase AM matures the 5′ end of 5S rRNA
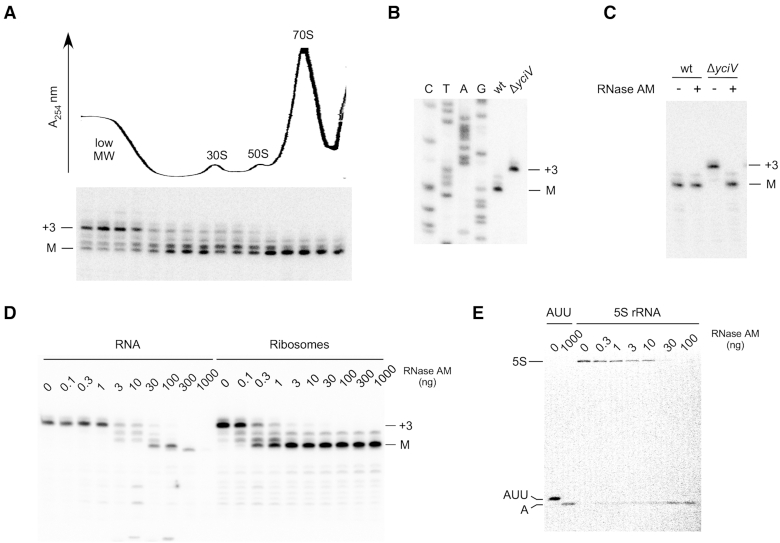
After RNase III cleaves the rRNA operon transcript, the pre-5S rRNA product is further cleaved by the endonuclease RNase E three nts upstream of the mature 5′ end and three nts downstream of the mature 3′ end, which is followed by removal of the 3′-end precursor residues by RNase T (8,12). To identify the mechanism for the removal of the 5′-end precursors, cell extracts from a wild-type strain were ultracentrifuged and primer extension was performed on RNA isolated from different fractions using a 5S rRNA-specific primer (Figure 1A). The low molecular weight fractions, which contain newly synthesized 5S rRNA, were found to harbor high levels of 5S precursors with three unprocessed 5′-end residues. In contrast, the 5S rRNA in ribosomal fractions contained predominantly mature 5′ ends, consistent with maturation occurring as the rRNA is incorporated into ribosomes. Significantly, precursors containing one and two nt extensions at the 5′ end could also be visualized in the intermediate fractions. These findings suggested that 5′-end maturation of 5S rRNA might be occurring through a 5′ to 3′ exonucleolytic mechanism.
Figure 1.
RNase AM matures the 5′ end of 5S rRNA. (A) Primer extension analysis of 5S rRNA in ribosomal fractions. Top, cell extracts derived from a wild-type strain were ultracentrifuged and the ribosomal profile was determined at 254 nm. The positions of the 70S ribosome, the 30S and 50S ribosomal subunits, and the low molecular weight fractions, are indicated. Bottom, RNA was extracted from 16 fractions and analyzed by primer extension using a 5S rRNA-specific primer. The positions of the mature 5′ end (M), and of a precursor containing three unprocessed 5′-end residues (+3), are indicated. The amounts of primer extension products loaded for gel analysis were adjusted to compensate for differences in 5S rRNA abundance in the different fractions. (B) Total RNA was extracted from wild-type or ΔyciV strains and analyzed by primer extension using a 5S rRNA-specific primer. A DNA sequence ladder generated using the same primer was run in parallel. (C) 70S ribosomes from wild-type or ΔyciV strains were treated with RNase AM or left untreated, followed by primer extension analysis. (D) Total cellular RNA or 70S ribosomes from a ΔyciV strain were treated with different amounts of RNase AM and analyzed by primer extension. (E) Precursor 5S rRNA was isolated from a ΔyciV strain and labeled with 32P at the 5′ end. The labeled product was treated with different amounts of RNase AM and fractionated by gel electrophoresis. A 5′ labeled trinucleotide (5′ AUU 3′) that corresponds to the hypothetical product of endonucleolytic 5′-end maturation and 5′-labeled adenosine monophosphate (A), generated by RNase AM digestion of the labeled trinucleotide, were run in parallel.
A recent report described the identification of the first 5′ to 3′ RNA exonuclease in E. coli, RNase AM (13). This enzyme is encoded by the E. coli yciV/trpH locus, which was previously thought to have a role in tryptophan metabolism. To determine whether RNase AM has a role in 5S rRNA maturation, total RNA was extracted from wild-type and ΔyciV strains, and analyzed by primer extension. Significantly, the 5S rRNA in the ΔyciV strain was found to retain three unprocessed nts at the 5′ end, suggesting that RNase AM is required to remove these precursor nts in vivo (Figure 1B). To determine directly whether RNase AM can remove these precursor residues from immature 5S rRNA, 70S ribosomal particles were isolated from wild-type and ΔyciV strains and treated with purified RNase AM. No effect of RNase AM was observed on ribosomes isolated from a wild-type strain, but the addition of this enzyme to ribosomes from a ΔyciV strain efficiently converted the unprocessed 5′ ends in 5S rRNA to a fully mature form (Figure 1C). Collectively, these results demonstrate that the enzyme that matures the 5′ end of 5S rRNA is RNase AM.
As mature 5S rRNA was predominantly found in ribosomal fractions (Figure 1A), it suggested that 5′-end maturation of 5S rRNA might be more efficient within the context of ribosomal particles as compared to free RNA. To test this hypothesis, total RNA or 70S ribosomes were isolated from a ΔyciV strain and treated with different amounts of RNase AM. When free RNA was used as a substrate, significant amounts of mature rRNA were produced with 30–100 ng of RNase AM, but increased levels of RNase AM reduced the amount of mature rRNA produced, suggesting RNA over-digestion (Figure 1D). In contrast, when ribosomal particles were used as a substrate, ten-fold lower levels of RNase AM were found to be sufficient for 5S rRNA maturation. Moreover, ribosomal 5S rRNA was also found to be resistant to over-digestion, likely because further RNase AM action is blocked by ribosomal proteins that bind near the 5′ end of this RNA or by rRNA structure in a ribosomal context. These findings indicate that 5S rRNA maturation occurs more efficiently and more accurately on ribosomal particles as compared to free RNA. Finally, to confirm that RNase AM digests the 5S rRNA precursor as an exonuclease, 5S rRNA was isolated from a ΔyciV strain, labeled at its 5′ end, and treated with different amounts of RNase AM. The sole labeled product observed was a mononucleotide with no evidence of a trinucleotide product, consistent with an exonucleolytic mode of 5S rRNA maturation (Figure 1E). These observations suggest that RNase AM acts on a biological substrate using the same mode of RNA digestion as has been described for this enzyme using synthetic oligonucleotide substrates in vitro (13).
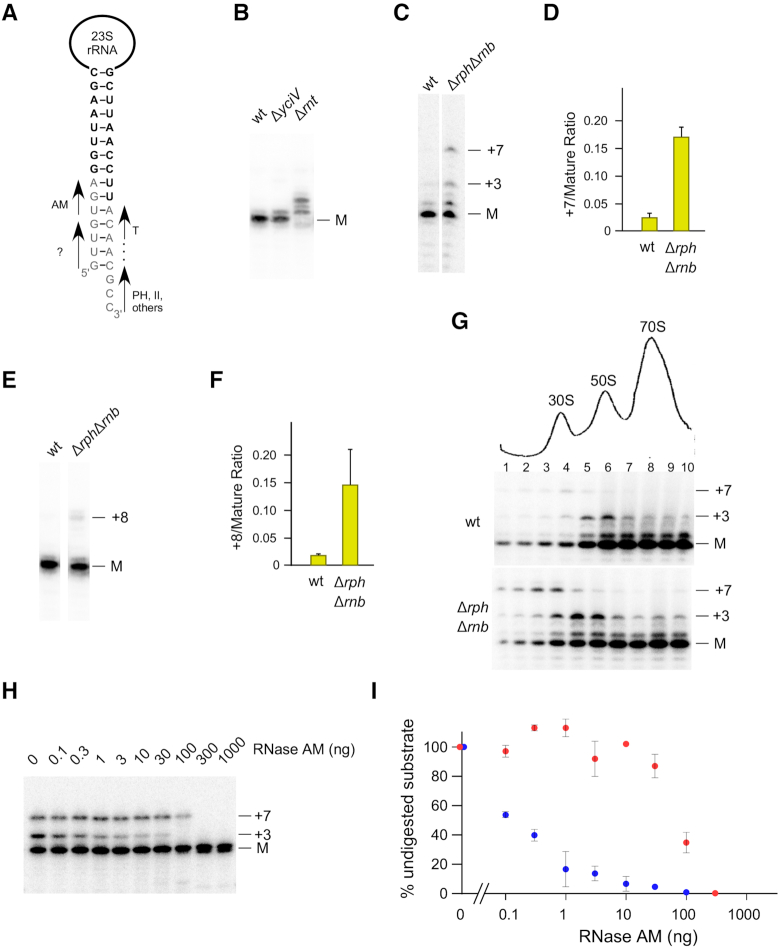
RNase AM matures the 5′ end of 23S rRNA
Many years ago it was shown that RNase III cleavage upstream of the 23S rRNA leaves seven unprocessed nts at the 5′ end, but the mechanism by which the RNA is further processed has eluded identification (6). To test whether RNase AM is involved in 23S rRNA maturation, primer extension was performed on total RNA isolated from wild-type and ΔyciV strains using a 23S rRNA-specific primer. No mature rRNA could be visualized in the ΔyciV strain, indicating that RNase AM is also necessary for processing the 5′ end of this RNA (Figure 2A). Surprisingly, the product accumulating in the mutant strain was found to be extended by three precursor nts, rather than seven, suggesting that RNase AM removes only the last three precursor nts from the 23S rRNA precursor. To confirm the activity of RNase AM on 23S rRNA precursors, 70S ribosomes from the wild-type and ΔyciV strains were treated with RNase AM. As was observed for 5S rRNA, the addition of RNase AM had no effect on mature 23S rRNA, but it effectively converted the +3 rRNA precursor to a mature form (Figure 2B). To compare the efficiency of RNase AM processing on free vs. ribosomal 23S rRNA substrates, total RNA or 70S particles were isolated from a ΔyciV strain and treated with different amounts of RNase AM (Figure 2C). When total RNA was used, a small amount of mature 23S rRNA was produced with 10–30 ng of RNase AM, whereas additional amounts of enzyme led to digestion into the body of the RNA without increasing the amount of mature product. In contrast, when ribosomal particles were used, nearly complete 5′-end maturation was observed using 1 ng of RNase AM. Significant over-digestion, the removal of one extra nt, was observed only when a thousand-fold higher amount of enzyme was used. These results indicate that 5′-end maturation of 23S rRNA by RNase AM occurs with much greater efficiency and accuracy within the context of ribosomal particles as compared to free RNA.
Figure 2.
RNase AM matures the 5′ end of 23S rRNA. (A) Total RNA was extracted from wild-type or ΔyciV strains and analyzed by primer extension using a 23S rRNA-specific primer. The positions of the mature 5′ end and of precursors that contain three or seven unprocessed nts at the 5′ end (+3 and +7, respectively) are indicated. RNA from a control ΔsrmB strain (cont.), which accumulates both +3 and +7 precursors at low temperatures (32), was analyzed in parallel. (B) Ribosomes from wild-type or ΔyciV strains were treated with RNase AM or left untreated, followed by primer extension analysis. (C) Total cellular RNA or 70S ribosomes, isolated from a ΔyciV strain, were treated with varying amounts of RNase AM, followed by primer extension analysis.
Coordination of 5′-end and 3′-end processing of 23S rRNA
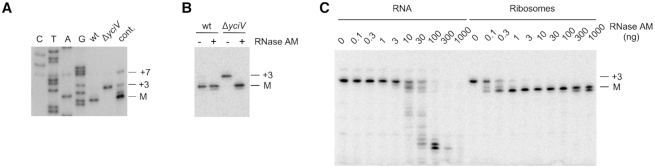
It has been established that precursor residues at the 5′ and 3′ ends of 23S rRNA base-pair with each other (6) (Figure 3A). Because most E. coli ribonucleases, with the exception of RNase III, recognize single-stranded regions more efficiently than double-stranded regions (20), we had suggested that such base-pairing might be inhibitory to rRNA processing (19). In that event, it would be expected that the eventual removal of precursors from one end of the RNA would be necessary to permit processing at the other end. With the identification of RNase AM as a 5′-end processing enzyme and prior knowledge of 3′-end processing enzymes, it became feasible to address the order with which processing reactions are performed at the two ends of 23S rRNA.
Figure 3.
Coordinated processing of the 3′- and 5′-ends of 23S rRNA. (A) Schematic description of the processing reactions that occur at the 5′ and 3′ ends of 23S rRNA. The sequence of the RNase III-cleaved 23S rRNA precursor is shown with the base-pairing between the 5′ and 3′ ends indicated. Mature rRNA sequences are shown in bold and precursor sequences in grey text. The RNases that remove the 5′ and 3′ precursor residues are indicated, whereas the unknown enzyme that removes the first four 5′ nts from the RNase III-generated product is denoted by a question mark. (B) 3′-end analysis of 23S rRNA. Total RNA from wild-type and ΔyciV strains were cleaved with RNase H using an oligonucleotide that binds ∼50 nts upstream of the 23S rRNA 3′ end. The resulting products were analyzed by northern blot using a labeled probe complementary to the 3′ end of 23S rRNA. RNA from a Δrnt strain, which accumulates 1–3 nt 3′-end precursors, was included as a control. (C) 5′-end analysis of 23S rRNA. Total RNA from wild-type or ΔrphΔrnb strains was analyzed by primer extension using a 23S rRNA-specific oligonucleotide. The positions of the mature 5′ end, and of the +3 and +7 5′-end precursors, are indicated. (D) Quantification of +7 precursor levels in (C). (E) 3′-end analysis of 23S rRNA from wild-type and ΔrphΔrnb strains. (F) Quantification of the +8 3′-end precursor levels in (E). (G) 23S rRNA precursor levels in ribosomal fractions. Ten ribosomal fractions, which included the 70S, 50S and 30S particles, were collected after ultracentrifugation of cell extracts from wild-type and ΔrphΔrnb strains. RNA was isolated from these fractions and analyzed by primer extension using a 23S rRNA-specific oligonucleotide. The positions of the mature 5′ end, and of the +3 and +7 precursors, are indicated. (H) Ribosomal particles from a ΔrphΔrnb strain were treated with different amounts of RNase AM and analyzed by primer extension. (I) Quantification of the extent of +3 and +7 substrate processing in (H) as a function of RNase AM concentration, based on three replicates. Blue data points, +3 substrates; red data points, +7 substrates. The amounts of the +3 and +7 substrates that were present prior to RNase AM treatment were each normalized to 100%. Error bars too small to be clearly depicted are not shown.
First, based on the accumulation of 23S 5′-end precursors in the ΔyciV strain (Figure 2A), I tested whether a failure to process the 5′ end has any effect on processing at the 3′ end, which was done by comparing the levels of precursors at the 3′ end of 23S rRNA in wild-type and ΔyciV strains. Apart from a small increase in a precursor that contains one unprocessed nt, no increase in the levels of any other precursors was noted in the latter strain (Figure 3B), suggesting that 3′-end maturation is not significantly impaired by a lack of 5′-end processing.
Next, I tested strains lacking the 3′ to 5′ exonucleases RNase PH and RNase II, as an absence of these two enzymes has been shown to retard the further 3′-end processing of 23S rRNA molecules after they have been cleaved by RNase III, eight nts downstream of the mature 3′ end (11). Apart from RNase PH and RNase II, other enzymes also contribute to the removal of these precursors, but testing strains additionally lacking these enzymes was not feasible because the processing enzymes are collectively essential for viability. Significantly, when total RNA was isolated from wild-type and ΔrnbΔrph strains and analyzed by primer extension, increased levels of the +7 5′-end precursor were observed in the ΔrnbΔrph strain, suggesting that inefficient processing at the 3′ end retards 5′-end processing (Figure 3C and D). To determine the relationship between the levels of precursors at each end, the 3′-end precursor/mature ratio was also quantified in the wild-type and the mutant strains (Figure 3E and F). Significantly, the relative levels of the 3′-end +8 precursors, both in the wild-type and mutant strains, were found to be almost identical to the respective levels of the 5′-end +7 precursors, suggesting that 5′-end processing is completely dependent upon prior 3′-end processing (Figure 3D and F). These observations can be explained in terms of the structure of the 23S rRNA precursor (Figure 3A), because a failure to remove the 3′-end precursors is expected to render the 5′-end precursor residues completely double-stranded, and presumably, refractory to processing by single-strand specific RNases. Additionally, initial processing at the 3′ end is consistent with the ability of one of the processing enzymes, RNase PH, to digest duplex regions of moderate stability (21). On the basis of these observations, it is suggested that 23S rRNA maturation initiates at the 3′ end, with the removal of 3′-end precursors necessary to generate a single-stranded 5′ end, which represents a substrate for the 5′-end processing enzymes.
Based on the observation that precursors with both three and seven unprocessed nts at the 5′ end were observed in the ΔrnbΔrph strain, and to a lower degree, in the wild-type strain (Figure 3C), these findings were used to define the stages at which the two different 5′-end processing reactions take place on 23S rRNA during ribosome assembly. For these purposes, cell extracts from wild-type and ΔrnbΔrph strains were fractionated by ultracentrifugation, and 23S rRNA, prepared from the individual fractions, was analyzed by primer extension (Figure 3G). Significant amounts of the +7 precursor, more clearly observed with the ΔrnbΔrph strain, were found in fractions 3 and 4, but were present at reduced levels in the later fractions, suggesting that processing of the +7 precursor occurs as incompletely formed subunits are converted to the mature 50S large subunit (LSU). In contrast, +3 precursors were predominantly identified in fractions 5 and 6, which correspond to the LSU, with reduced amounts observed in the 70S ribosomal fractions, suggesting that final maturation of the 23S rRNA 5′ end takes place as ribosomal particles transition from the LSU to 70S ribosomes. Thus, the two processing reactions appear to occur at distinct steps, indicating that the enzymes responsible for each reaction act on 23S rRNA at different stages of ribosome assembly.
The significant accumulation of +3 and +7 23S rRNA 5'-end precursors in the ΔrnbΔrph strain was also used to investigate whether RNase AM is capable of digesting the in vivo products of RNase III cleavage, which contain seven unprocessed nts. Thus, ribosomal particles, derived from 30S to 40S fractions of the ΔrnbΔrph strain, were treated with different amounts of RNase AM. As little as 0.1 ng of RNase AM was found to be sufficient to digest ∼50% of substrates containing a +3 end, whereas digestion of the +7 substrate to a similar extent required nearly 1000-fold higher amounts of RNase AM (Figure 3H and I). These observations indicate that 23S rRNA containing +7 ends represent a poor substrate for RNase AM, suggesting a need for another enzyme to perform this process in vivo.
RNase AM matures the 5′ end of 16S rRNA
Several years ago, the then newly discovered endonuclease, RNase G, was implicated in 5′-end maturation of 16S rRNA by two groups, based on the evidence that 16S rRNA is extended by 66 nts at the 5′ end when this enzyme is absent and the observed cleavage of those precursors by RNase G in vitro (22,23). However, the products of the in vitro reactions were not examined at nucleotide resolution to confirm that RNase G cleavage yields a precisely mature 5′ end. Based on the results obtained with 5S and 23 S rRNA, I investigated whether RNase AM could also have a role in 16S rRNA maturation by performing primer extension analysis on wild-type and ΔyciV strains using 16S rRNA-specific primers. Significantly, no mature 16S rRNA 5′ ends could be observed in the ΔyciV strain (Figure 4A). Instead, yet again, rRNA precursors containing three unprocessed nts were found to accumulate. These observations suggested that RNase AM is involved in 16S rRNA maturation as well.
Figure 4.
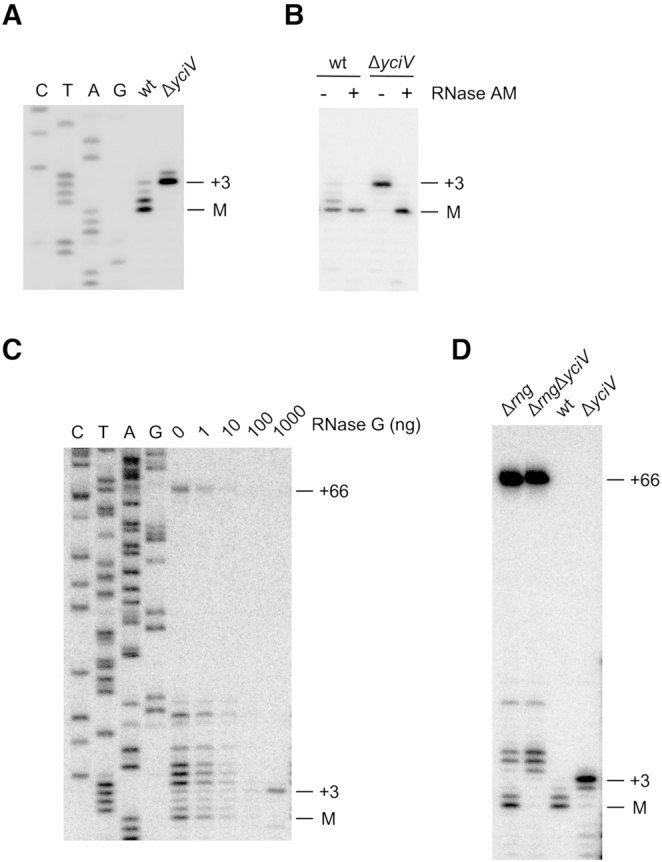
RNase AM matures the 5′ end of 16S rRNA. (A) Total RNA was extracted from wild-type or ΔyciV strains and analyzed by primer extension using a 16S rRNA-specific primer. A sequence ladder was run in parallel. (B) 70S ribosomes from wild-type or ΔyciV strains were treated with purified RNase AM or left untreated, followed by primer extension analysis. (C) 70S ribosomes from a Δrng strain were treated with different amounts of purified RNase G, followed by primer extension analysis using a 16S rRNA-specific primer. A sequence ladder was also run in parallel. The apparent increase of signal with 1000 ng of RNase G, observed in independent experiments, likely represents the conversion of multiple processing intermediates to the +3 product. (D) Total RNA was isolated from Δrng and ΔrngΔyciV strains and analyzed by primer extension. As controls, RNA isolated from wild-type and ΔyciV strains were also analyzed. The higher levels of the +66 5'-end product, as compared to in (C), likely reflect degradation of the extended precursor residues in the latter during the isolation of 70S ribosomes.
To verify that RNase AM digests the 16S rRNA precursor, 70S ribosomes from wild-type and ΔyciV strains were treated with purified RNase AM. As was observed with the 5S and 23S rRNAs, the addition of RNase AM had no effect on mature 16S rRNA but converted the unprocessed rRNA in the ΔyciV strain to a mature form (Figure 4B).
Because RNase G has been previously implicated in 16S rRNA maturation, the products generated by RNase G cleavage were re-evaluated by treating 70S particles isolated from a Δrng strain with different amounts of purified RNase G, followed by primer extension analysis. In the absence of RNase G, several products, including the +66 precursor could be visualized (Figure 4C). Additional shorter products were also present in the samples, which might have been generated by digestion of the 16S rRNA precursor by other enzymes in vivo (22,23), or in vitro during the isolation of the 70S particles. The addition of increasing amounts of RNase G resulted in the disappearance of these products and ultimately yielded a product that retains three precursor nts at the 5′ end. These observations suggest that RNase G cleaves 16S rRNA precursors three nucleotides upstream of the mature 5′ end, rather than at the mature terminus. Of note, a reduction in the amount of products following RNase G addition suggests that RNase G might also be cleaving ribosomal RNA non-specifically downstream of the mature rRNA in vitro.
Noting that small amounts of mature rRNA were observed in ribosomal preparations from the Δrng strain, I speculated that these might have been produced by RNase AM action on 16S rRNA molecules that have undergone partial processing by other enzymes in the absence of RNase G. To test that hypothesis, total RNA was isolated from Δrng and ΔrngΔyciV strains and analyzed by primer extension (Figure 4D). Apart from the +66 precursor, small amounts of mature rRNA were apparent in RNA isolated from the Δrng strain but not from the ΔrngΔyciV strain, indicating that RNase AM is responsible for their formation. In contrast, an increased accumulation of precursors containing 4–6 unprocessed residues was observed, suggesting that these might represent processing products that are generated through a default pathway when RNase G is absent, which are then digested by RNase AM to generate mature RNA. Collectively, the findings shown in Figure 4 indicate that RNase G cleaves a 66 nt 5′-end precursor three nts upstream of the mature end and that RNase AM removes these unprocessed nts subsequently to generate a mature 16S rRNA end.
DISCUSSION
Ribosomal RNAs are the main components of the translational machinery and are synthesized at high levels to meet the cellular demands for protein synthesis, especially under conditions of rapid growth (24). However, a description of the pathways that lead to the generation of mature rRNAs in E. coli has remained incomplete even though the initial steps of rRNA processing were defined nearly 50 years ago. Based on the evidence suggesting that 5S rRNA undergoes 5′-end maturation using a 5′ to 3′ exonucleolytic mechanism, I examined a strain that lacks RNase AM, a 5′ to 3′ exonuclease, and found that 5′-end maturation of 5S rRNA is performed by this enzyme both in vivo and in vitro (Figure 1). I also found that RNase AM is responsible for the 5′-end maturation of 23S rRNA, but this enzyme removes only the final three nts of the seven precursor residues, indicating the presence of another enzyme that removes the preceding four nts (Figure 2). Finally, I found that RNase AM generates the mature 5′ end of 16S rRNA (Figure 4). This process was attributed to RNase G, but here it is shown that the RNase G cleavage product retains three unprocessed nts, which are then removed by RNase AM. Why RNase AM should remove precisely three unprocessed nts from each of the rRNAs remains unclear. Nonetheless, the identification of the enzyme that matures the 5' end of these rRNAs, in conjunction with the previously identified enzymes responsible for 3′-end maturation, now provides a full description of the enzymes that generate mature ends for each of the three rRNAs in E. coli.
Due to the high levels of ribosomes that are required for growth, a significant proportion of transcription, and therefore, of the energy generated by a cell, is used for rRNA synthesis. A question of interest is how rRNA degradation, which would contribute to inefficiencies in energy utilization, is minimized given a cellular environment that contains multiple RNases. It has been shown that once complete ribosomes are formed, rRNA becomes extraordinarily stable in the assembled form (25), but a small proportion of rRNA does get degraded during the assembly process (26,27). One factor that limits more extensive rRNA degradation during the assembly process could be through a restriction of the activity of rRNA processing enzymes to late stages of rRNA assembly, where over-digestion can be curbed by the presence of ribosomal proteins or rRNA structures that form during the assembly process. Supporting this view, I found that RNase AM maturation of 5S and 23S rRNA occurs not only more efficiently, but also with greater accuracy on ribosomal particles as compared to purified rRNA (Figure 1D and 2C). In the case of 23S rRNA, RNase AM processing was observed to occur at a late stage of assembly when 50S particles join with 30S particles to form the complete ribosome (Figure 3G). Similarly, it has been shown that processing of the 5S and 23S rRNAs at their 3′ ends by RNase T is also more efficient on ribosomal particles, as compared to free RNA (8,9). Identifying the mechanisms through which the activity of the processing RNases is restricted on free rRNA and/or channeled to ribosomal particles will be a topic of interest for the future.
We had previously provided experimental evidence that the 5′ and 3′ ends of 23S rRNA undergo maturation at similar rates under different growth conditions, and based on those observations, we suggested that the processing of the two ends might be coupled (19). This hypothesis was tested by using ΔyciV and ΔrnbΔrph strains to inhibit processing at the 5′ or 3′ ends, respectively. These analyses revealed that in a ΔrnbΔrph strain, 5′-end precursors accumulate to the same extent as 3′-end precursors, suggesting that 3′-end maturation occurs first and is necessary for processing at the 5′ end (Figure 3). Similarly, work on 16S rRNA has suggested that processing at one end facilitates maturation at the other end (10,28). Thus, the coupled processing of 3′ and 5′ ends may be a common feature of bacterial rRNAs, with potential relevance for increasing the efficiency of rRNA maturation at both ends.
Paralogs of RNase AM are found in a wide variety of organisms, including many other Gram-negative and positive bacteria, archaea and lower eukaryotes. However, the model Gram-positive bacterium, Bacillus subtilis lacks an RNase AM homolog. In this case, 5′-end maturation of the three rRNAs each proceeds through a different set of mechanisms, involving the enzymes Mini-III for 23S rRNA, RNase J1 for 16S rRNA and RNase M5 for 5S rRNA maturation (29–31). Interestingly, in some members of the delta-proteobacteria, RNase AM paralogs are found in fusion with RNase III, which suggests that multiple steps of rRNA processing might be performed by the same enzyme in these organisms. Future studies on this enzyme will be needed to clarify the rRNA maturation role of RNase AM in all domains of life. In summation, the findings described here provide answers to decades-old questions regarding the mechanism of 5′-end maturation of the E. coli rRNAs, complete the roster of the RNases required to yield mature rRNAs and describe the first biological function for a 5′ to 3′ RNA exonuclease in this organism.
ACKNOWLEDGEMENTS
I thank Dr. Frank Raushel for providing purified RNase AM, and Drs. Marc Dreyfus and Murray Deutscher for their comments on the manuscript. This work is dedicated to my mother, Asha Jain.
FUNDING
National Institutes of Health [GM114540]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ehrenberg M., Bremer H., Dennis P.P.. Medium-dependent control of the bacterial growth rate. Biochimie. 2013; 95:643–658. [DOI] [PubMed] [Google Scholar]
- 2. Kiss A., Sain B., Venetianer P.. The number of rRNA genes in Escherichia coli. FEBS Lett. 1977; 79:77–79. [DOI] [PubMed] [Google Scholar]
- 3. Nomura M., Gourse R., Baughman G.. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 1984; 53:75–117. [DOI] [PubMed] [Google Scholar]
- 4. Dunn J.J., Studier F.W.. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. PNAS. 1973; 70:3296–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Young R.A., Steitz J.A.. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. PNAS. 1978; 75:3593–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bram R.J., Young R.A., Steitz J.A.. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980; 19:393–401. [DOI] [PubMed] [Google Scholar]
- 7. Apirion D., Lassar A.B.. A conditional lethal mutant of Escherichia coli which affects the processing of ribosomal RNA. J. Biol. Chem. 1978; 253:1738–1742. [PubMed] [Google Scholar]
- 8. Li Z., Deutscher M.P.. The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. PNAS. 1995; 92:6883–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Z., Pandit S., Deutscher M.P.. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA. 1999; 5:139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sulthana S., Deutscher M.P.. Multiple exoribonucleases catalyze maturation of the 3′ terminus of 16S ribosomal RNA (rRNA). J. Biol. Chem. 2013; 288:12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gutgsell N.S., Jain C.. Role of precursor sequences in the ordered maturation of E. coli 23S ribosomal RNA. RNA. 2012; 18:345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roy M.K., Singh B., Ray B.K., Apirion D.. Maturation of 5-S rRNA: ribonuclease E cleavages and their dependence on precursor sequences. Eur. J. Biochem. 1983; 131:119–127. [DOI] [PubMed] [Google Scholar]
- 13. Ghodge S.V., Raushel F.M.. Discovery of a previously unrecognized ribonuclease from escherichia coli that Hydrolyzes 5′-Phosphorylated fragments of RNA. Biochemistry. 2015; 54:2911–2918. [DOI] [PubMed] [Google Scholar]
- 14. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2:2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jagessar K.L., Jain C.. Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. RNA. 2010; 16:1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H.. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): unique resources for biological research. DNA Res. 2005; 12:291–299. [DOI] [PubMed] [Google Scholar]
- 17. Jain C. Overexpression and purification of tagged Escherichia coli proteins using a chromosomal knock-in strategy. Protein Expr. Purif. 2006; 46:294–298. [DOI] [PubMed] [Google Scholar]
- 18. Diwa A., Bricker A.L., Jain C., Belasco J.G.. An evolutionarily conserved RNA stem-loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes Dev. 2000; 14:1249–1260. [PMC free article] [PubMed] [Google Scholar]
- 19. Gutgsell N.S., Jain C.. Coordinated regulation of 23S rRNA maturation in Escherichia coli. J. Bacteriol. 2010; 192:1405–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bechhofer D.H., Deutscher M.P.. Bacterial ribonucleases and their roles in RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 2019; 54:242–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jain C. Novel role for RNase PH in the degradation of structured RNA. J. Bacteriol. 2012; 194:3883–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z., Pandit S., Deutscher M.P.. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 1999; 18:2878–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wachi M., Umitsuki G., Shimizu M., Takada A., Nagai K.. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem. Biophys. Res. Commun. 1999; 259:483–488. [DOI] [PubMed] [Google Scholar]
- 24. Bremer H., Dennis P.P.. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus. 2008; 3:doi:10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- 25. Piir K., Paier A., Liiv A., Tenson T., Maivali U.. Ribosome degradation in growing bacteria. EMBO Rep. 2011; 12:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gausing K. Regulation of ribosome production in Escherichia coli: synthesis and stability of ribosomal RNA and of ribosomal protein messenger RNA at different growth rates. J. Mol. Biol. 1977; 115:335–354. [DOI] [PubMed] [Google Scholar]
- 27. Jain C. Role of ribosome assembly in Escherichia coli ribosomal RNA degradation. Nucleic Acids Res. 2018; 46:11048–11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith B.A., Gupta N., Denny K., Culver G.M.. Characterization of 16S rRNA processing with Pre-30S subunit assembly intermediates from E. coli. J. Mol. Biol. 2018; 430:1745–1759. [DOI] [PubMed] [Google Scholar]
- 29. Condon C., Rourera J., Brechemier-Baey D., Putzer H.. Ribonuclease M5 has few, if any, mRNA substrates in Bacillus subtilis. J. Bacteriol. 2002; 184:2845–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Redko Y., Bechhofer D.H., Condon C.. Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol. Microbiol. 2008; 68:1096–1106. [DOI] [PubMed] [Google Scholar]
- 31. Britton R.A., Wen T., Schaefer L., Pellegrini O., Uicker W.C., Mathy N., Tobin C., Daou R., Szyk J., Condon C.. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 2007; 63:127–138. [DOI] [PubMed] [Google Scholar]
- 32. Charollais J., Pflieger D., Vinh J., Dreyfus M., Iost I.. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2003; 48:1253–1265. [DOI] [PubMed] [Google Scholar]