Abstract
Purpose
Recently, the world has been dealing with a new type of coronavirus called COVID-19 that in terms of symptoms is similar to the SARS coronavirus. Unfortunately, researchers could not find a registered therapy to treat the infection related to the virus yet. Regarding the fact that drug repurposing is a good strategy for epidemic viral infection, we applied the drug repurposing strategy using virtual screening to identify therapeutic options for COVID-19. For this purpose, five proteins of COVID-19 (3-chymotrypsin-like protease (3CLpro), Papain-Like protease (PLpro), cleavage site, HR1 and RBD in Spike protein) were selected as target proteins for drug repositioning.
Methods
First, five proteins of COVID-19 were built by homology modeling. Then FDA-approved drugs (2471 drugs) were screened against cleavage site and RBD in Spike protein via virtual screening. One hundred and twenty-eight FDA-approved drugs with the most favorable free-binding energy were attached to the cleavage site and RBD in Spike protein. Of these 128 drugs, 18 drugs have either been used currently as antiviral or have been reported to possess antiviral effects. Virtual screening was then performed for the 18 selected drugs with ACE2, 3CLpro and PLpro and HR1 and TMPRSS2.
Results
According to the results, glecaprevir, paritaprevir, simeprevir, ledipasvir, glycyrrhizic acid, TMC-310911, and hesperidin showed highly favorably free binding energies with all tested target proteins.
Conclusion
The above-mentioned drugs can be regarded as candidates to treat COVID-19 infections, but further study on the efficiency of these drugs is also necessary.
Keywords: COVID-19, Drug repositioning, Virtual screening, FDA-approved drugs
Introduction
In 2019, Wuhan, China, was the host of a new type of coronavirus that caused SARS-like symptoms in human beings. In order to better know this new virus, phylogenetic analysis was conducted considering the complete viral genome (29,903 nucleotides). The results revealed that the new virus has 89.1% nucleotide similarity to genus Betacoronavirus -subgenus Sarbecovirus- that had previously caused an epidemic infectious disease known as SARS [1]. The new virus is called COVID-19 which contains major structural proteins (Spike (S) protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein.). The schematic representation of a coronavirus is shown in Fig. 1. Among them, S protein can promote fusion of the viral and cellular membranes. So, it facilitates the entry of coronavirus into the host cells [1].
Fig. 1.
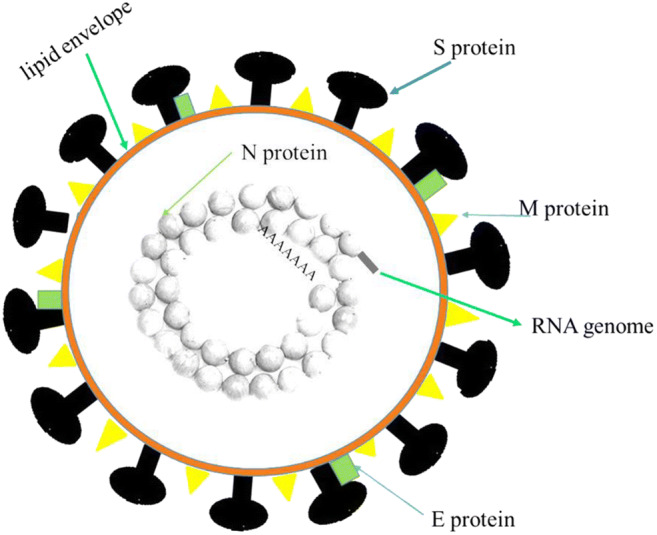
Schematic representation of a coronavirus. COVID-19 contains at least four structural proteins: Spike (S) protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein
S protein with 1288 amino acid consists of S1 and S2 subunits that mediates the fusion between the viral and host cell membranes. These subunits include the receptor-binding domain (RBD) and fusion peptide (FP), heptad repeat 1 (HR1), heptad repeat 2 (HR2), transmembrane domain (TM) and cytoplasmic domain fusion (CP). S1 and S2 have different roles in this entry; S1 plays a vital role in binding cellular receptor and S2 can facilitate viral fusion and entry [2]. A Conserved Receptor Binding Domain (RBD) connects to angiotensin-converting enzyme 2 (ACE2) that is known to interact with 14 aa in the S1 of SARS-CoV. Studies also show that ACE2 is a receptor of COVID-19. Moreover, sequence variations of the Spike protein cleavage site are important in terms of cellular tropism and pathogenesis. The cleavage site in the S-protein sequence is involved in the pathogenesis of virus. The sequence of S-protein in COVID-19 has 12 extra nucleotides upstream to the single Arg¯ cleavage site 1 which results in PRRAR↓SV sequence that is solvent-exposed and resembles a cleavage site [3]. The reason for the well-organized spread of the virus among the population is this new cleavage site. Previous coronaviruses lacked this system. As mentioned earlier, the S2 subunit in COVID-19 is responsible for the facilitation of viral fusion and entry into the host cell. There exists a kind of interaction regarding cellular membranes between heptad repeats 1 (HR1) and heptad repeat 2 (HR2) which leads to fusion. The changes in the interaction pattern between HR1 and HR2 domains in COVID-19 S2 (in terms of fusion with cell membranes) are caused by mutations inside the HR1 core region [4]; 8 residues out of 21 indicate mutation (~38% difference). It is currently claimed that ACE2 could be the host receptor for COVID-19 as well. Perhaps, the susceptibility, symptoms, and outcome of COVID-19 infection are resulted by the expression level and pattern of ACE2 in tissues. According to the recent research, single-cell analysis of RNA-sequencing (RNA-seq) shows a higher expression of ACE2 in Asian males [5]. COVID-19 engages ACE2 as a receptor for entry into the host cell and employs the cellular transmembrane protease serine 2 (TMPRSS2) for S protein priming. TMPRSS2 activates the viral S protein which facilitates virus-cell membrane fusions [6]. Residues near lysine 31, and tyrosine 41, 82–84 and 353–357 in human ACE2 play an important role in the binding of S-protein coronavirus. The adipose tissue might be in a weak position regarding COVID-19 because ACE2 expression in adipose tissue was higher than that in lung tissue. Considering five different types of cancers, the researchers realized that the expression of ACE2 remarkably increases in tumor tissues, compared to other neighboring tissues. Hence, it is suggested that during COVID-19, obese individuals and patients who suffer from these five types of cancer should be prioritized [7]. In terms of gender and age, it should be mentioned that the expression of ACE2 is positively associated with age [8, 9]. Eight hundred kDa polypeptide is produced during transcription of the COVID-19 genome. This polypeptide is cleaved to produce various proteins. Meanwhile, papain-like protease (PLpro) and 3-chymotrypsin-like protease (3CLpro) facilitate proteolytic processing. Papain-like proteinase (PLpro) is responsible for the cleavages at the N-terminus of replicase polyprotein. 3CLpro plays an important role in coronaviruses and is strongly preserved in this family of viruses. Proteins necessary for viral replication are nonstructural and are produced by 3CLpro which activate the polyprotein at 11 distinct sites. This process would prepare the ground for virus replication; hence, to find anti-viral activities in compounds, researchers pay careful attention to 3CLpro [10]. Some options are suggested as a treatment for COVID-19, among which drug repositioning (repurposing) is considered as the only answer to the epidemic of sudden infectious diseases. Drug repurposing is an effective strategy to treat new diseases caused by infectious agents that spread rapidly. In this paper, the drug repurposing strategy is applied using virtual screening to identify therapeutic options for COVID-19.
Methods and materials
Homology modeling was used to generate five proteins of COVID-19)3CLpro, PLpro, cleavage site, HR1 and RBD in Spike protein (and TMPRSS2. The homology models of these proteins were done in the SWISS-MODEL workspace [11, 12] (https://swissmodel.expasy.org/docs/references). Protein Data Bank (https://www.rcsb.org) was used to have ACE2 coordinates [13] (https://www.rcsb.org/structure/1r42). First, the water molecules were extracted from protein and hydrogen atoms were added to optimized hydrogen bonds. 3Drefine server and the package of GROMACS 5.1.4 were applied to minimize energy in the optimized model of the protein [14]. The section of small-molecules of the Drug Bank database [15] (https://www.drugbank.ca/about) was used to obtain all FDA-approved drugs. The Non-unique structures that were removed during the process are as follows: the compounds containing rare atoms (selenium, platinum, gold, silicon) and organometallic compounds. Finally, the screening database had 2471 compounds. Pyrx tool was used for energy minimizing and preparing ligands in PDBQT format, and AutoDock Vina was applied for virtual screening. It was used AutoDock Vina for drug discovery to screen libraries of compounds against targets [16, 17]. The box was centered on a specific area. This position using PockDrug server and it was determined by using data in uniprot about the main amino acids in the bond. According to previous evidence, the cleavage site in the S-protein and the conserved Receptor Binding Domain (RBD) is important role in the pathogenesis of virus. So at the first, these two targets were selected for virtual screening with all FDA-approved drugs. The drugs that had the desired binding energy at this stage and were reported to have antiviral properties were then selected for the next step. Therefore, virtual screening was performed for the selected drugs with ACE2, 3CLpro and PLpro and HR1 and TMPRSS2 in the next step. 2-D representations of protein/ligand complexes are shown using LIGPLOT program [18] (https://www.ebi.ac.uk/thornton-srv/software/LIGPLOT/).
Results and discussion
FDA-approved drugs were screened against cleavage site and RBD in Spike protein via virtual screening in this present study. It was observed that 128 FDA-approved drugs with the most suitable free-binding energy (out of 2471 drugs) were attached to the cleavage site and RBD in Spike protein. Of these 128 drugs, 18 drugs have either been used currently as antiviral or have been reported to possess antiviral effects. Virtual screening was then performed for the 18 selected drugs with ACE2 (major COVID_19 receptor), 3CLpro and PLpro (main enzymes in viral replication) and HR1 (main domain in virus fusion with the host cell membrane) and TMPRSS2 (main enzyme in activating the viral spike glycoproteins). According to the results, 7 drugs with all the target proteins achieved the most favorable free binding energy (Table 1). The protein complexes with these seven drugs are shown in Figs. 2, 3, 4, 5, 6, 7, and 8. Hydrophobic and hydrogen bonding interactions of docked molecules were visualized using LigPlot program (Table 2). As shown in Table 2, glycyrrhizic acid and hesperidin showed highest number of bonds with ACE2 and RBD. As shown in Table 1, paritaprevir and simeprevir have favorable binding energy to most target proteins and may be considered as therapeutic options for the coronavirus. These drugs were originally used in the treatment of chronic hepatitis C and can effectively bind to target proteins [10, 19, 20]. There are other drugs with good binding energy to target protein; one of them is ledipasvir. According to an in silico study, ledipasvir is an antiviral drug with the minimum side effects that can be applied in the treatment of COVID19 [21]. In the present study, medicinal compounds of plant origin were also attached with favorable binding energy to the target proteins. Glycyrrhizic acid which has plant origins is obtained from the roots of the licorice plant. Triterpene glycoside and glycyrrhetinic acid are used pharmacologically. The latter is applied as a hepatoprotective option to treat chronic hepatitis in Japan and China (https://pubchem.ncbi.nlm.nih.gov/compound/Glycyrrhizic-acid). In addition, glycyrrhizin, being non-toxic [22], is also used widely as a SARS treatment option. According to a study in 2003, glycyrrhizin has a preventive role in SARS [23]. Furthermore, based on studies, it plays an effective role in the hepatitis C virus (HCV) because it can influence the release phase associated with the infection of cells [24]. Using glycyrrhizin to treat hepatitis C is not a new method and it has been applied as a treatment for 20 years in Japan. Fortunately, the studies have shown its preventive role in the progression of cirrhosis and hepatocarcinoma and only few side effects [25]. The positive effects of glycyrrhizin have been proven on many viral diseases, such as herpes simplex type 1 (HSV-1), varicella-zoster virus (VZV), hepatitis A (HAV) and B Virus (HBV), human immunodeficiency virus (HIV), severe acute respiratory syndrome (SARS) and coronavirus, Epstein–Barr virus (EBV), human cytomegalovirus and influenza virus [24]. Glycyrrhizic acid’s preventive impact on ACE2 has been reported in some studies [26]. Aside from glycyrrhizic acid, hesperidin and tannic acid are also considered in medical circles; they have plant origins as well and can attach to proteins with good binding energy. Hesperidin is a flavonoid extracted from citrus fruit. It has multiple biological and antimicrobial activities against human viruses. Based on a study conducted in 2009, glucosyl hesperidin has an inhibiting role against the replication of influenza A virus [27]. In a study aimed at analyzing therapeutic targets for COVID-19 by computational methods, it was shown that hesperidin disrupts in the interaction of ACE2 with RBD [1]. As mentioned earlier, the 3CLpro can be an effective target for treatment of coronavirus. A study on investigation of the effect of active compounds on the inhibition of 3CLpro showed that hesperidin was the most potent inhibitor of 3CLpro among the compounds which were tested. Based on docking score, it was reported that hesperidin and glycyrrhizin are favorable in terms of binding to ACE2 and 3CLpro [28]. TMC-310911 is another FDA-approved drug that has good binding with target proteins. TMC-310911 is a new investigational protease inhibitor that is being investigated for use in HIV-1 infections. TMC-310911 is currently being investigated as a potential treatment for COVID-19 (https://www.drugbank.ca/).
Table 1.
Docking results of 18 FDA-approved drugs with ACE2 (major COVID_19 receptor), 3CLpro and PLpro (main enzymes in viral replication) and HR1(main domain in virus fusion with the host cell membrane) and TMPRSS2 (main enzyme in activation the viral spike glycoproteins) by using PyRx (PyRx 0.8) software
| Proposed drugs | Complexes of SPRRARSVAS (Docking score) |
Complexes of ACE2_31_41 (Docking score) |
Complexes of ACE2_82_84 (Docking score) |
Complexes of 3CLpro (Docking score) |
Complexes of RBD (Docking score) |
Complexes of HR1 (Docking score) | Complexes of PLpro (Docking score) |
Complexes of TMPRRS2 (Docking score) |
|---|---|---|---|---|---|---|---|---|
| Glecaprevir | −10.6 | −10.4 | −7.6 | −8.2 | −8.9 | −8.1 | −10.2 | −7.9 |
| Simeprevir | −9.5 | −9 | −8.7 | −8.6 | −8.4 | −8.6 | −12 | −6.8 |
| Ledipasvir | −9.5 | −9.6 | −8.6 | −8.4 | −7.9 | −9.2 | −11.8 | −5.3 |
| Paritaprevir | −10.1 | −11.6 | −8.6 | −8.7 | −8.5 | −8.5 | −11.1 | −7.3 |
| Glycyrrhizic acid | −9.4 | −8.8 | −7.8 | −8.8 | −8.6 | −7.4 | −10.7 | −6.6 |
| Daclatasvir | −8.8 | −8.7 | −7.6 | −7.3 | −7.5 | −8.1 | −10.3 | −5.2 |
| Ombitasvir | −9.1 | −8.3 | −7.5 | −7.1 | −7 | −6.7 | −10.2 | −6.2 |
| TMC-310911 | −9.1 | −9.4 | −9.2 | −8.2 | −6.7 | −8 | −10.1 | −6.1 |
| Maraviroc | −8.5 | −9.6 | −7.8 | −7.6 | −7.6 | −7.9 | −10 | −6.5 |
| Dasabuvir | −8.1 | −8.9 | −7.4 | −7.3 | −7.1 | −8.1 | −10 | −6.3 |
| Baloxavir marboxil | −8.1 | −8.3 | −7 | −7.2 | −7.3 | −6.9 | −9.9 | −5.4 |
| Hesperidin | −8.5 | −11 | −8 | −8 | −7.4 | −8.1 | −9.4 | −6.1 |
| Cobicistat | −7.7 | −7.4 | −8.2 | −6.2 | −6.4 | −7.3 | −9.3 | −5.4 |
| Pibrentasvir | −10.1 | −10.8 | −6.4 | −8 | −6.7 | −7.5 | −9.1 | −6.2 |
| Ritonavir | −7.7 | −7.9 | −7.2 | −7.8 | −6.4 | −5.7 | −9.1 | −5.6 |
| Lopinavir | −7.5 | −7.7 | −9.5 | −8 | −6.8 | −6.6 | −9 | −5.6 |
| Tannic acid | −7.6 | −10.3 | −8.3 | −7.5 | −6.4 | −6 | −8.4 | −4.6 |
| Rifabutin | −9.5 | −10.4 | −7.1 | −7.2 | −7.4 | −7.7 | −7.9 | −5.8 |
* Bold numbers indicate the favorable binding energy of drugs with Target
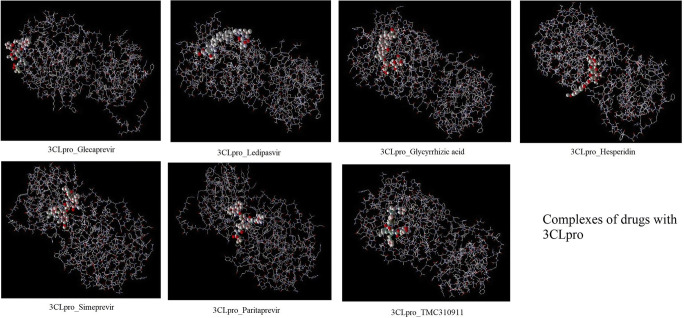
Fig. 2.
Complexes of drugs with 3CLpro
Fig. 3.
Complexes of drugs with ACE2
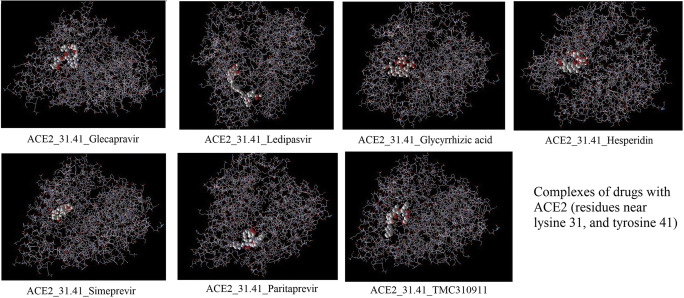
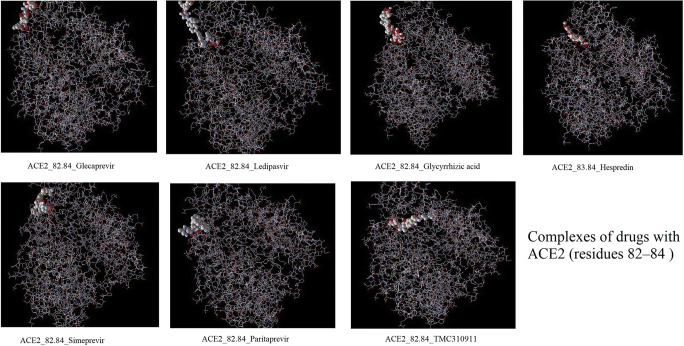
Fig. 4.
Complexes of drugs with ACE2
Fig. 5.
Complexes of drugs with HR1
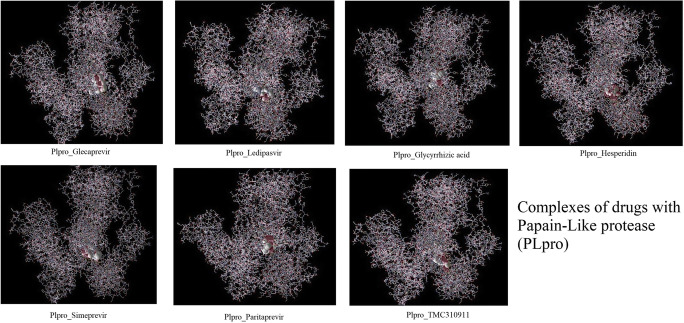
Fig. 6.
Complexes of drugs with PLpro
Fig. 7.
Complexes of drugs with RBD
Fig. 8.
Complexes of drugs with cleavage site in Spike protein
Table 2.
Details of docking results using LIGPLOT program
| Ligands | 3CLpro | ACE2 | PLpro | RBD | ||||
|---|---|---|---|---|---|---|---|---|
| H-bond Donor/acceptor | H-bond Interactions (Interacting residues) | H-bond Donor/acceptor | H-bond Interactions (Interacting residues) | H-bond Donor/acceptor | H-bond Interactions (Interacting residues) | H-bond Donor/acceptor | H-bond Interactions (Interacting residues) | |
| Glecaprevir | 2 | His 80 | 2 | Thr 78 | 3 | Asn 1552 | 5 | Arg 457 |
| Ser 81 | His 818 | Arg 454 | ||||||
| Ile468 | ||||||||
| Simeprevir | – | – | 2 | Thr 196 | 1 | Glu 819 | 3 | Arg 454 |
| Gln 98 | Glu 471 | |||||||
| Ledipasvir | 2 | Thr 26 | 6 | Gln 98 | 3 | Ser 20 | 1 | Lys 458 |
| Ser 46 | Asn 194 | Gln 168 | ||||||
| Tyr 19 | Glu 815 | |||||||
| Ser 105 | ||||||||
| Tyr 202 | ||||||||
| Asp 350 | ||||||||
| Paritaprevir | 1 | Lys 5 | 1 | Lys 94 | 2 |
Lys 945 Asn 1552 |
1 | Phe 490 |
| Glycyrrhizic acid | 7 | Gln 189 | 14 | Thr 196 | 7 | Glu 487 | 11 | Arg 457 |
| His 164 | Glu 208 | Ser 925 | Asp 467 | |||||
| Glu 166 | Glu 87 | Ser 1560 | Ile 488 | |||||
| His 163 | Asn 194 | Asp 924 | Cys 480 | |||||
| Ser 144 | Gln 101 | Arg 1546 | Gly 482 | |||||
| Asn 142 | Glu 87 | Thr 1456 | Ser 469 | |||||
| Asn 119 | Asn 397 | Glu 411 | ||||||
| Glu 402 | Arg 454 | |||||||
| His 401 | ||||||||
| Asp 382 | ||||||||
| Ala 348 | ||||||||
| Ser 47 | ||||||||
| Ser 43 | ||||||||
| Gly 395 | ||||||||
| Hesperidin | 6 | Gln 110 | 19 | Gln 208 | 6 | Gln 168 | 9 | Phe 456 |
| Thr 111 | Gly 205 | Glu 177 | Ile 472 | |||||
| Arg 105 | Gln 98 | Gln 180 | Arg 454 | |||||
| Asp 295 | ||||||||
| Asn 194 | Asn 79 | Arg 457 | ||||||
| Asn 103 | Asp 467 | |||||||
| Gln 101 | Ile 468 | |||||||
| His 195 | Gln 474 | |||||||
| Thr 196 | Phe 456 | |||||||
| Asn 394 | Ile 472 | |||||||
| Glu 402 | ||||||||
| His 401 | ||||||||
| Asp 382 | ||||||||
| Ala 348 | ||||||||
| Ser 47 | ||||||||
| Ser 43 | ||||||||
| Gly 395 | ||||||||
| TMC-310911 | 2 | Glu 166 | 8 | Gly 205 | 1 | Gln 168 | 4 | Lys 458 |
| Thr 26 | Lys 562 | Gln 474 | ||||||
| Gln 98 | Arg 457 | |||||||
| Asp 382 | Asp 467 | |||||||
| His 401 | ||||||||
| Asn 394 | ||||||||
| Gly 395 | ||||||||
| Tyr 385 | ||||||||
| Phe 390 | ||||||||
Conclusion
Drug repurposing is an effective strategy to treat new diseases caused by infectious agents that spread rapidly. In this paper, the drug repurposing strategy was applied using virtual screening to identify therapeutic options for COVID-19. FDA-approved drugs were screened against cleavage site and RBD in Spike protein via virtual screening. It was observed that 128 FDA-approved drugs (out of 2471 drugs) with the most suitable free-binding energy were attached to the cleavage site and RBD in Spike protein. Out of these 128 drugs, 18 drugs were antiviral. Virtual screening was then performed for the 18 selected drugs with ACE2 (major COVID_19 receptor), 3CLpro and PLpro (main enzymes in viral replication), HR1 (main domain in virus fusion with the host cell membrane) and TMPRSS2 (main enzyme in activating the viral spike glycoproteins). According to the data of this study as well as the related literature, paritaprevir, simeprevir, ledipasvir, glycyrrhizic acid, TMC-310911, and hesperidin can be suitable candidates for further investigation on the treatment of COVID-19 infection. It seems that investigations of RNA polymerase inhibitors may be effective for better selection of drugs in the treatment of COVID-19.
Compliance with ethical standards
Conflict of interest
The authors corroborate that this study has no conflict of interest.
Footnotes
In the original publication of the article, the affiliation of corresponding author Mahboobeh Zarrabi was published incorrectly. Now it has been corrected here.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/10/2023
A Correction to this paper has been published: 10.1007/s40200-023-01248-8
References
- 1.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Zheng M. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xia S, Xu W, Wang Q, Wang C, Hua C, Li W, Jiang S. Peptide-based membrane fusion inhibitors targeting HCoV-229E spike protein HR1 and HR2 domains. Int J Mol Sci. 2018;19(2):487. doi: 10.3390/ijms19020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020. [DOI] [PMC free article] [PubMed]
- 4.Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020:1–3. [DOI] [PMC free article] [PubMed]
- 5.Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discovery. 2020;6(1):1–4. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020. [DOI] [PMC free article] [PubMed]
- 7.Jia X, Yin C, Lu S, Chen Y, Liu Q, Bai J, Lu Y. Two things about COVID-19 might need attention. 2020. [Google Scholar]
- 8.Chen Y, Shan K, Qian W. Asians and other races express similar levels of and share the same genetic polymorphisms of the SARS-CoV-2 cell-entry receptor. 2020. [Google Scholar]
- 9.Guy JL, Jackson RM, Jensen HA, Hooper NM, Turner AJ. Identification of critical active-site residues in angiotensin-converting enzyme-2 (ACE2) by site-directed mutagenesis. FEBS J. 2005;272(14):3512–3520. doi: 10.1111/j.1742-4658.2005.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alamri, M. A., ul Qamar, M. T., & Alqahtani, S. M. (2020). Pharmacoinformatics and molecular dynamic simulation studies reveal potential inhibitors of SARS-CoV-2 main protease 3CLpro [DOI] [PMC free article] [PubMed]
- 11.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Lepore R. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, Schwede T. The SWISS-MODEL repository—New features and functionality. Nucleic Acids Res. 2017;45(D1):D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towler P, Staker B, Prasad SG, Menon S, Tang J, Parsons T, Patane MA. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem. 2004;279(17):17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharya D, Nowotny J, Cao R, Cheng J. 3Drefine: An interactive web server for efficient protein structure refinement. Nucleic Acids Res. 2016;44(W1):W406–W409. doi: 10.1093/nar/gkw336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., & Assempour, N. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res, 46(D1), D1074-D1082 [DOI] [PMC free article] [PubMed]
- 16.Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Chemical Biology: Humana press; 2015. [DOI] [PubMed] [Google Scholar]
- 17.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laskowski RA, Swindells MB. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. 2011. [DOI] [PubMed] [Google Scholar]
- 19.Hosseini FS, Amanlou M. Simeprevir, potential candidate to repurpose for coronavirus infection: Virtual screening and molecular docking study. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan RJ, Jha RK, Amera GM, Jain M, Singh E, Pathak A, Pathak A. Targeting novel coronavirus 2019: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2'-O-ribose Methyltransferase. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, Y. W., Yiu, C. P. B., & Wong, K. Y. (2020). Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: Virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research, 9(129), 129 [DOI] [PMC free article] [PubMed]
- 22.Pilcher H. Liquorice may tackle SARS. Nature. 2003.
- 23.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361(9374):2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto, Y., Matsuura, T., Aoyagi, H., Matsuda, M., Hmwe, S. S., Date, T., ... & Wake, K. (2013). Antiviral activity of glycyrrhizin against hepatitis C virus in vitro. PLoS One, 8(7) [DOI] [PMC free article] [PubMed]
- 25.Rossum TV, Man RD. Glycyrrhizin as a potential treatment for chronic hepatitis C. Aliment Pharmacol Ther. 1998;12(3):199–205. doi: 10.1046/j.1365-2036.1998.00309.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Du Q. Potential natural compounds for preventing 2019-nCoV infection. 2020. [Google Scholar]
- 27.Saha RK, Takahashi T, Suzuki T. Glucosyl hesperidin prevents influenza a virus replication in vitro by inhibition of viral sialidase. Biol Pharm Bull. 2009;32(7):1188–1192. doi: 10.1248/bpb.32.1188. [DOI] [PubMed] [Google Scholar]
- 28.Yan, Y. M., Shen, X., Cao, Y. K., Zhang, J. J., Wang, Y., & Cheng, Y. X. (2020). Discovery of Anti-2019-nCoV agents from Chinese patent drugs via docking screening