Abstract
Background
Coronavirus disease 2019 (COVID-19) has already affected 2883603 and killed 198842 people, as of April 27, 2020. Because there is no specific therapeutic drug, drug repurposing has been proposed. RNA-dependent RNA polymerase (RdRp) is a promising drug against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to control its replication, and several compounds have been suggested. The present study predicts relative efficacies of thirty known or repurposed compounds in inhibiting the RdRp.
Methods
The three-dimensional structure of the target enzyme was loaded into Molegro virtual docker software, followed by chemical structures of the test compounds. The docking was performed between the compounds and the active site of the enzyme to determine docking scores, and the energy liberated when the two dock. Thus, docking scores signify the affinity of ligand(s) with the active site of enzyme(s) and thus its inhibitory potential.
Results
Among known inhibitors, remdesivir was found to have the highest affinity for the active site of the RdRp. Among all compounds, chlorhexidine was predicted as the most potent inhibitor. Furthermore, the results predict the relative efficacy of different drugs as inhibitors of the drug target.
Conclusion
While the study identifies several compounds as inhibitors of RdRp of SARS-CoV-2, the prediction of their relative efficacies may be useful in future studies. While nucleoside analogs compete with the natural substrate of RdRp, thereby terminating RNA replication, other compounds would physically block entry of the natural substrates into the active site. Thus, based on the findings, we recommend in vitro and in vivo studies and clinical trials to determine their effectiveness against COVID-19.
Keywords: 2019-nCoV, Betacoronavirus, COVID-19, Molecular docking, Pneumonia
Introduction
A novel coronavirus emerging from Wuhan in China, in December 2019 has now spread to over 213 countries/territories across the globe (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). The virus, originally named 2019-nCoV, has now been renamed by taxonomists as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) owing to its genomic and structural similarities with the earlier severe acute respiratory syndrome coronavirus (SARS-CoV).1, 2, 3 The disease caused by this virus has been named by the World Health Organizationas coronavirus disease 2019 (COVID-19), and the outbreak was declared a pandemic on 11th March, 2020.
As of April 27 (2020), the virus has affected over 2883603 people and killed more than 198842, and the figures are increasing rapidly. Today, the COVID-19 pandemic has become the largest global health crisis, a burden on healthcare systems, and is affecting global trade and economy. So far, there exists no specific drug or vaccine for the treatment of COVID-19. Because de novo drug discovery is a time-consuming venture, researchers have suggested drug repurposing as a strategy to find a therapeutic against the virus. The approved drugs against other viruses, including the similar SARS-CoV, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), human immunodeficiency virus (HIV), and hepatitis C virus (HCV), have been suggested for further evaluation and clinical studies.1, 2, 3, 4, 5, 6, 7, 8 In addition, the knowledge gained through studies of these related viruses has been vital in designing therapeutics against SARS-CoV-2.
Structural biology approaches have deciphered the structures of different proteins/enzymes of the SARS-CoV-2, and at least three of them such as RNA-dependent RNA polymerase (RdRp), papain-like protease, and the main protease are important drug targets.5,6 RdRp is the key enzyme which replicates the viral RNA genome and is thus the most promising drug target. The RdRp of the SARS-CoV-2 shares 96% sequee identity with the SARS-CoV,9 and thus, the compounds or drugs effective against the RdRp of SARS-CoV are surmised to be effective against the novel CoV as well. This makes the RdRp the most important therapeutic target against SAR-CoV-2. Recent studies have suggested several known RdRp-inhibiting antivirals, other FDA-approved drugs, and phytochemicals for repurposing against SARS-CoV-2 using molecular docking studies.7 Some of the commonly repurposed drugs include ritonavir/lopinavir, remdesivir, hydroxychloroquine, ribavirin, and so on. In view of the previously mentioned fact, the present study investigates the relative efficacy of the known RdRp inhibitors, as well as other drugs/compounds which have been predicted to have RdRp-inhibiting potential, using computational modeling.
Materials and methods
The drug target
The drug target for the present study is RdRp of SARS-CoV-2. The three-dimensional structure of the enzyme in complex with cofactors was obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (www.rcsb.org/pdb), bearing PDB id 6M71. The structure was determined using electron microscopy, at resolution of 2.90 Å, by Gao et al,10 and deposited to the database on March 16, 2020. The structure bears three non-structured proteins (NSPs) such as one NSP7 and two NSP8 as cofactors. The chain A is the NSP12, which is the RdRp, and it contains 851 amino acids. The structure was downloaded from the database in .pdb format.
The drugs
Thirty compounds were selected from the available literature as test drugs. The list includes known RdRp inhibitors, as well as other drugs/compounds which have been predicted to inhibit the drug target (Table 1). The three-dimensional conformers of the compounds were obtained from the National Center for Biotechnology Information (NCBI) PubChem compounds database (www.pubchem.ncbi.nlm.nih.gov/) and were downloaded in .sdf format.
Table 1.
Details of the compounds used in the present study and their docking scores at the active site of RdRp of SARS-CoV-2. The scores were obtained following molecular docking using MoleGro virtual docker software.
| Name of compound | PubChem ID | Rerank score | HBond | Remarks | Reference |
|---|---|---|---|---|---|
| Chlorhexidine | 9552079 | −132.846 | −7.996 | ZINC database | Wu et al, 20207 |
| Remdesivir | 121304016 | −114.469 | −5.644 | Anti-HIV | Elfiky 20205 |
| Novobiocin | 54675769 | −109.756 | −7.079 | ZINC database | Wu et al, 20207 |
| Ceftibuten | 5282242 | −103.087 | −6.154 | ZINC database | Wu et al, 20207 |
| Ribavirin | 37542 | −101.338 | −8.079 | Anti-HCV | Elfiky 20205 |
| Atovaquone | 74989 | −97.992 | −7.190 | ZINC database | Wu et al, 20207 |
| Valganciclovir | 135413535 | −97.215 | −8.392 | ZINC database | Wu et al, 20207 |
| Cromolyn | 2882 | −95.818 | −5.473 | ZINC database | Wu et al, 20207 |
| Bromocriptine | 31101 | −91.861 | −13.639 | ZINC database | Wu et al, 20207 |
| Silybin | 31553 | −88.239 | −8.832 | ZINC database | Wu et al, 20207 |
| Cefuroxime | 5479529 | −86.779 | −11.057 | ZINC database | Wu et al, 20207 |
| Fludarabine | 657237 | −86.756 | −12.101 | ZINC database | Wu et al, 20207 |
| Galidesivir | 10445549 | −85.484 | −9.768 | Anti-HCV | Elfiky 20205 |
| Sofosbuvir | 45375808 | −84.239 | −2.700 | Anti-HCV | Elfiky 20205 |
| Oseltamivir | 65028 | −82.248 | −5.074 | Known inhibitor | Lai et al, 202034 |
| Fenoterol | 3343 | −81.704 | −6.239 | ZINC database | Wu et al, 20207 |
| Nitazoxanide | 41684 | −77.894 | −12.989 | Anti-diarrhea | Wang et al, 202015 |
| Itraconazole | 3793 | −77.853 | −1.965 | ZINC database | Wu et al, 20207 |
| Benzylpenicilloyl G | 119212 | −76.869 | −6.351 | ZINC database | Wu et al, 20207 |
| Pancuronium bromide | 27350 | −74.816 | 2.339 | ZINC database | Wu et al, 20207 |
| Penciclovir | 135398748 | −74.741 | −9.713 | Known inhibitor | Wang et al, 202015 |
| 7-Deaza-2′-C-methyladenosine | 3011893 | −73.955 | −5.306 | West Nile virus | Eyer et al, 201935 |
| Idarubicin | 42890 | −73.555 | −4.887 | ZINC database | Wu et al, 20207 |
| Diphenoxylate | 13505 | −70.108 | 2.995 | ZINC database | Wu et al, 20207 |
| Ganciclovir | 135398740 | −68.529 | −12.727 | Known inhibitor | Lai et al, 202034 |
| Tenofovir | 464205 | −66.629 | −9.697 | Known inhibitor | Elfiky 20205 |
| Favipiravir | 492405 | −57.417 | −0.726 | Known inhibitor | Wang et al, 202015 |
| Tibolone | 444008 | −51.645 | −0.395 | ZINC database | Wu et al, 20207 |
| Chenodeoxycholic acid | 10133 | −20.482 | −7.573 | ZINC database | Wu et al, 20207 |
| Cortisone | 222786 | 79.677 | −1.491 | ZINC database | Wu et al, 20207 |
MW: molecular weight; HBD: number of hydrogen bond donor group; HBA: number of hydrogen bond acceptor group; TPSA: topological polar surface area; LogP: octanol/water partition coefficient; HBond: hydrogen bond energy; RdRp: RNA-dependent RNA polymerase; SARS-CoV-2: severe acute respiratory syndrome; HIV: human immunodeficiency virus; HCV, hepatitis C virus; ZINC: ZINC is Not Commercial.
The molecular docking
Molecular docking is a powerful computational modeling tool which determines the affinity of binding of any compound with the active site of a receptor/enzyme. The molecular docking also determines the orientation of binding of the compound at the active site of the enzyme. The amount of energy thus liberated, which is a negative value, determines the stability of the ligand–receptor complex and is referred to as a docking score. A compound with a higher (i.e. more negative) docking score forms a more stable ligand–protein complex and is surmised to be a better inhibitor.11,12 Furthermore, when a compound shows higher affinity for the active site of an enzyme compared with the natural substrate, it interferes with the activity of the enzyme and may bring about competitive inhibition. In the present study, the molecular docking was performed using Molegro virtual docker software 6.0 (MVD), following standard procedures.11,12 MVD is one of the most widely used and powerful tools in determining ligand–protein interactions and has higher accuracy (87%) than many other similar tools.13
Briefly, the structure of the enzyme was loaded into the software with all the cofactors. This was followed by addition of the structures of the test compounds into the workspace. Docking was performed between the ligands and the active site of the receptor/enzyme, with Grid resolution of 0.30 Å, MolDock scoring function, and including amino acids within a radius of 20 Å around X: 122.74, Y: 106.69, Z: 155.25 as the active site residues. Once the docking scores were obtained, the best pose in terms of the Rerank score for each compound was retained, and the binding geometry or conformation was assessed.
Results
Ligand–receptor interaction
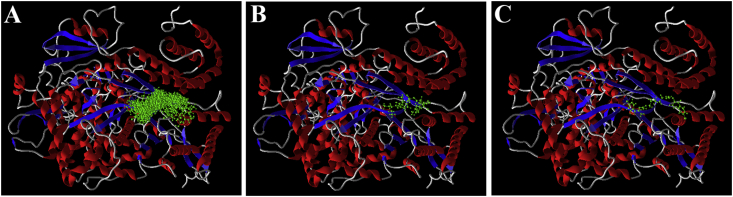
While performing docking, incorporation of remdesivir as a test ligand for the present study has two implications. First, it was included to see its relative affinity compared with the other compounds for the active site of the enzyme and thereby predict its relative efficacy as an inhibitor of RdRp of SARS-CoV-2. Second, the drug, being a nucleoside analog, interacts at the active site of the enzyme and thus serves as a reference ligand. In the present study, all the test compounds (except cortisone) were found to interact at the same active site or binding pocket (Fig. 1A) as that of the remdesivir (Fig. 1B). This demonstrates that all the compounds have the potential to interact at the same active site of the enzyme and may interfere with the activity of the enzyme.
Fig. 1.
Docking poses of the ligands at the active site of RdRp of SARS-CoV-2. (A): Poses of all the ligands; (B) docking pose of remdesivir; and (C) docking pose of chlorhexidine. The poses were obtained following molecular docking using MoleGro virtual docker software. RdRp, RNA-dependent RNA polymerase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Inhibition of enzyme activity
Binding or interaction of the ligands at the active site of an enzyme implicates that the ligands would interfere with the activity of the enzyme, thereby bringing about inhibition of the enzyme. The free energy of binding or the docking score implicates the efficacy, with which the inhibition is affected. Chlorhexidine was found to have the highest affinity among all the studied compounds for the active site of RdRp, followed by remdesivir, novobiocin, ceftibuten, and ribavirin (Table 1). The docking score of chlorhexidine (−132.85) was found to be 1.16-fold higher than that of remdesivir (−114.47), while the hydrogen bond score of chlorhexidine (−7.996) was found to be 1.42-fold higher than that of remdesivir (−5.644). Furthermore, the docking score of remdesivir was found to be 1.04-fold, 1.11-fold and 1.13-fold higher than that of novobiocin, ceftibuten, and ribavirin, respectively (Table 1). Among all the compounds, cortisone was found to have a positive Rerank score. This indicates that the compound may not be able to interact with and inhibit the RdRp.
Discussion
Using molecular docking studies, researchers have identified several known and novel compounds as inhibitors of different drug targets of the SARS-CoV-2. For the viruses using RdRp for replication of their genetic material (RNA), including CoVs and HIV, inhibition of RdRp is one of the most crucial therapeutic strategies, which is primarily affected by nucleoside analogs. The RdRp of SARS-CoV-2 and SARS-CoV have 97% sequence identity, while the active site of the RdRp of most CoVs and RNA viruses is highly conserved.5 This has been the basis for drug repurposing against SARS-CoV-2, using those compounds which are known to inhibit the RdRp of related viruses.
The present study reveals that remdesivir has the highest potential in binding and therefore competitively inhibiting RdRp of SARS-CoV-2, among all known RdRp inhibitors (Table 1). The inhibitory potential of the same on the RdRp has also been reported from other molecular modeling studies.7,14 Remdesivir is an adenosine analog, and its incorporation into the growing viral RNA leads to termination of the RNA replication. High affinity of the compound for the active site of RdRp of the SARS-CoV-2 indicates that the compound would compete with the natural substrates (ribonucleotides) of the RdRp and get incorporated into the growing RNA chain. Remdesivir has been suggested by several researchers for use in patients with COVID-19 to reduce viral load.1, 2, 3, 4, 5, 6, 7 The compound has already been reported to inhibit replication of the virus in vitro in Vero E6 cells.15 Remdesivir has earlier been reported to inhibit a spectrum of human RNA viruses, including SARS-CoV, MERS-CoV, Marburg virus, Hendra virus, Nipah virus, Ebola virus, and HIV.2,16, 17, 18, 19, 20, 21 Remdesivir was used for successful treatment of the first COVID-19 case of the US.22 A large number of clinical trials are going on in different countries to evaluate the effectiveness of this compound in patients with COVID-19.
In the present study, chlorhexidine has been found to have highest affinity for the active site of the RdRp of SARS-CoV-2 (Table 1; Fig. 1C). The compound was found to bind to the same active site of the RdRp and is therefore predicted to be a promising inhibitor. Nevertheless, the mode of inhibition of this compound would be different from that of nucleoside analogs (such as remdesivir). This is because nucleoside analogs mimic the natural substrates of RdRp and act as chain terminators, unlike chlorhexidine. Chlorhexidine can not mimic natural substrates of RdRp and will not be incorporated into the viral RNA. Rather, it may block the active site of the enzyme and thereby inhibit the entry of the substrates (i.e. nucleotides) at the site. This will prevent activity of RdRp, thereby preventing replication of the viral genome, rather than terminating it. This compound has also been predicted by Wu et al7 as an inhibitor of RdRp, using molecular docking approaches. Chlorhexidine is one of the most widely used drugs against dental plaques, gingivitis, pharyngitis, candidiasis, and tonsillitis.23 It is a bactericidal, fungicidal, and virucidal agent and is known to alter membrane potential24 and inhibit ATPase.25 At concentrations ranging from 0.001% to 2%, the drug is known to inhibit a wide range of viruses, and for coronaviruses, it shows effectiveness at a concentration of 0.01%.23 However, the concentration at which the drug can inhibit RdRp of SARS-CoV-2 remains to be determined. While this may be predicted using quantitative structure–activity relationship (QSAR) modeling studies, in vitro, in vivo, and clinical studies would be required to ascertain the same.
In addition to remdesivir, other known nucleoside analogs including sofosbuvir, ribavirin, penciclovir, ganciclovir, and favipiravir were found to interact with the active site of RdRp of SARS-CoV-2 (Fig. 1A) with high affinities (Table 1). Our findings are in compliance with the study of Elfiky,5 who reported ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir as effective inhibitors of SARS-CoV-2 RdRp, using molecular modeling. In the present study, sofosbuvir was also found to be a potent inhibitor of the enzyme. Ribavirin, an FDA-approved synthetic guanosine nucleoside, is a wide-spectrum inhibitor of RNA viruses, including SARS-CoV, MERS-CoV, and HCV.26, 27, 28 Penciclovir is also a synthetic nucleoside analog – acyclic guanine derivative –which has been reported to be effective against herpes simplex virus, and varicella-zoster virus.29 Ganciclovir is also an approved nucleoside analog and effective against HIV.30 Favipiravir is an investigational nucleoside analog and inhibitor of RdRp and has been reported to be effective against Ebola and influenza A (H1N1) viruses.31, 32, 33 However, the major limitation of the present study is that we have not predicted the inhibitory concentrations of the compounds, and thus, it is suggested that QSAR modeling studies are performed to determine the same. Furthermore, we recommend that in vitro and in vivo studies are performed to first ascertain the effectiveness of the compounds on SARS-CoV-2 before proceeding to clinical studies.
Conclusion
The present study reports relative efficacy of 30 compounds in interacting with the active site of RdRp of SARS-CoV-2. In vitro studies and molecular modeling approaches have indicated that some of the broad-spectrum antivirals, including remdesivir, can be effective therapeutics against the virus. While we report chlorhexidine to be the best inhibitor, remdesivir was found to be the most potent among known antivirals. Nucleoside analogs (such as remdesivir) mimic the natural substrates of the RdRp and consequently bring about chain termination. However, other compounds including chlorhexidine are predicted to physically block the active site of the enzyme and thereby affect its activity. While clinical trials with remdesivir are ongoing, it is recommended that clinical trials be initiated with chlorhexidine and other antivirals to determine their effectiveness against COVID-19. Thus, the present study is of immense significance in the therapeutic intervention against COVID-19.
Disclosure of competing interest
The authors have none to declare.
References
- 1.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo D. Old weapon for new enemy: drug repurposing for treatmentof newly emerging viral diseases. Virol Sin. 2020 doi: 10.1007/s12250-020-00204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko W.C., Rolain J.M., Lee N.Y. Arguments in favour of remdesivir for treating SARS-CoV-2 infections. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham A.C., Goh H.P., Koh D. Treatment of COVID-19: old tricks for new challenges. Crit Care. 2020;24:91. doi: 10.1186/s13054-020-2818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 7.Wu C., Liu Y., Yang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhury S., Mazumder M.K. Suggesting Ritonavir against COVID-19/SARS-CoV-2. Med Hypotheses. 2020;140:109764. doi: 10.1016/j.mehy.2020.109764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W., Morse J.S., Lalonde T., Xu S. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y., Yan L., Huang Y. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target. BioRxiv. 2020 doi: 10.1101/2020.03.16.993386. [DOI] [Google Scholar]
- 11.Mazumder M.K., Paul R., Borah A. β-Phenethylamine - a phenylalanine derivate in brain – contributes to oxidative stress by inhibiting mitochondrial complexes and DT-diaphorase: an in silico study. CNS Neurosci Ther. 2013;19:596–602. doi: 10.1111/cns.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazumder M.K., Choudhury S. Tea polyphenols as multi-target therapeutics for Alzheimer's disease: an in silico study. Med Hypotheses. 2019;125:94–99. doi: 10.1016/j.mehy.2019.02.035. [DOI] [PubMed] [Google Scholar]
- 13.Thomsen R., Christensen M.H. MolDock: A new technique for high-accuracy molecular docking. J Med Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L., Zhou R. Binding mechanism of remdesivir to SARS-CoV-2 RNA dependent RNA polymerase. Preprints. 2020 doi: 10.20944/preprints202003.0267.v1. [DOI] [PubMed] [Google Scholar]
- 15.Wang M., Cao R., Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown A.J., Won J.J., Graham R.L. 2019. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir Res. 2019;169:104541. doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Momattin H., Al-Ali A.Y., Al-Tawfiq J.A. A systematic review of therapeutic agents for the treatment of the Middle East respiratory syndrome coronavirus (MERS-CoV) Trav Med Infect Dis. 2019;30:9–18. doi: 10.1016/j.tmaid.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warren T.K., Jordan R., Lo M.K. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheahan T.P., Sims A.C., Graham R.L. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo M.K., Jordan R., Arvey A. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardon C.J., Tchesnokov E.P., Feng J.Y. The antiviral compound remdesivir potentially inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020 doi: 10.1074/jbc.AC120.013056. DOI: 10.10.74/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holshue M.L., DeBolt C., Lindquist S. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim K.S., Kam P.C. Chlorhexidine – pharmacology and clinical applications. Anaesth Intensive Care. 2008;36:502–512. doi: 10.1177/0310057X0803600404. [DOI] [PubMed] [Google Scholar]
- 24.Harold F.M., Baarda J.R., Baron C., Abrams A. Dio 9 and chlorhexidine. Inhibition of membrane bound ATPase and of cation transport in Streptococcus faecalis. Biochim Biophys Acta. 1969;183:129–136. doi: 10.1016/0005-2736(69)90136-9. [DOI] [PubMed] [Google Scholar]
- 25.Lambert P.A., Hammond S.M. Potassium fluxes. First indications of membrane damage in microorganisms. Biochem Biophys Res Commun. 1973;54:796–799. doi: 10.1016/0006-291x(73)91494-0. [DOI] [PubMed] [Google Scholar]
- 26.AASLD-IDSA HCV Guidance Panel Hepatitis C guidance 2018 update: AASLD-IDSA recommendations for testing, managing, and treating hepatitis C virus infection. Clin Infect Dis. 2018;67:1477–1492. doi: 10.1093/cid/ciy585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsang K., Zhong N.S. SARS: pharmacotherapy. Respirology. 2003;8(suppl 1):S25–S30. doi: 10.1046/j.1440-1843.2003.00525.x. [DOI] [PubMed] [Google Scholar]
- 28.Arabi Y.M., Shalhoub S., Mandourah Y. Ribavirin and interferon therapy for critically ill patients with middle east respiratory syndrome: a multicenter observational study. Clin Infect Dis. 2019 doi: 10.1093/cid/ciz544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiraki K. Antiviral drugs against alphaherpesvirus. Adv Exp Med Biol. 2018;1045:103–122. doi: 10.1007/978-981-10-7230-7_6. [DOI] [PubMed] [Google Scholar]
- 30.Al-Badr A.A., Ajarim T.D.S. Ganciclovir. Profiles of Drug Substances, Excipients, and Related Methodology. 2018;43:1–208. doi: 10.1016/bs.podrm.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Furuta Y., Gowen B.B., Takahashi K. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldhill D.H., Te Velthuis A.J.W., Fletcher R.A. The mechanism of resistance to favipiravir in influenza. Proc Natl Acad Sci U S A. 2018;115:11613–11618. doi: 10.1073/pnas.1811345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardile A.P., Warren T.K., Martins K.A. Will there be a cure for Ebola? Annu Rev Pharmacol Toxicol. 2017;57:329–348. doi: 10.1146/annurev-pharmtox-010716-105055. [DOI] [PubMed] [Google Scholar]
- 34.Lai C.C., Shi T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eyer L., Fojtíková M., Nencka R., Rudolf I., Hubálek Z., Ruzek D. Viral RNA-Dependent RNA Polymerase Inhibitor 7-Deaza-2'-C-Methyladenosine Prevents Death in a Mouse Model of West Nile Virus Infection. Antimicrob Agents Chemother. 2019;63(3):e02093–e02118. doi: 10.1128/AAC.02093-18. [DOI] [PMC free article] [PubMed] [Google Scholar]