Highlights
-
•
Development and complete validation of a one-step RT-qPCR method for the detection and quantification of PEDV.
-
•
Broad range detection of S-INDEL and S-non-INDEL strains.
-
•
Optimization of Primer concentrations reduce primer dimer formation.
-
•
Addition of a proteinase K treatment allow good reproducibility.
Keywords: Porcine epidemic diarrhea virus, RT-qPCR, NF U47-600, Validation
Abstract
Since 2014, porcine epidemic diarrhea virus (PEDV) has reemerged in Europe. RT-PCR methods have been described for the detection of PEDV, but none have been validated according to a norm. In this study we described the development and validation of a SYBR™ Green one-step RT-qPCR according to the French norm NF U47-600, for the detection and quantification of PEDV viral RNA. The method was validated from sample preparation (feces or jejunum) through to nucleic acid extraction and RT-qPCR detection. Specificity and sensitivity, limit of detection (LoD), limit of quantification (LQ), linearity, intra and inter assay variability were evaluated using transcribed RNA and fecal and jejunum matrices spiked with virus. The analytical and diagnostic specificities and sensitivities of this RT-qPCR were 100% in this study. A LoD of 50 genome copies/5 μl of extract from fecal matrices spiked with virus or RNA transcript and 100 genome copies/5 μl of extract from jejunum matrices spiked with virus were obtained. The Lower LQ (LLQ) was 100 genome copies/5 μl and the Upper LQ (ULQ) 108 copies/5 μl. This method is the first, validated according a norm for PEDV and may serve as a global reference method to harmonize detection and quantification of PEDV viral RNA in both field and experimental settings.
1. Introduction
Porcine Epidemic Diarrhea (PED) was first described in Europe in 1971. It is characterized by watery diarrhea, vomiting, dehydration, and is most notable in young piglets. The etiologic agent, porcine epidemic diarrhea virus (PEDV) which was first identified by electron microscopy (EM) in 1977 (Chasey and Cartwright, 1978; Debouck and Pensaert, 1980) is now characterized as an enveloped virus with a single stranded positive sense RNA genome, member of the order Nidovirales, suborder Cornidovirinae, family Coronaviridae, subfamily Orthocoronavirinae, genus Alphacoronavirus, subgenus Pedacovirus (Walker et al., 2019).
In the 1980’s, PEDV was detected for the first time in Asia whilst in Europe it was endemic. During the 90’s only few sporadic cases were reported in Europe and most of these were reported in Italy were it remains endemic (Martelli et al., 2008). During the last two decades new PEDV strains have appeared in China and some of these strains have caused extremely severe outbreaks characterized by a morbidity of 100% and a mortality of 80–100% on suckling piglets (Sun et al., 2012). This has led to the naming of PEDV as either S-non-INDEL or S-INDEL genotypes. In general the more virulent viruses belong to the S-non-INDEL group. In the last decade both S-non-INDEL and S-INDEL viruses have emerged in the USA with serious consequences for the industry. Throughout Europe, the predominant types are now closely related to the viruses circulating in Asia and North and Central America (Boniotti et al., 2016). Furthermore, all viruses reported in Europe since 2014 belong to the S-INDEL group (Grasland et al., 2015; Stadler et al., 2015; Steinrigl et al., 2015; Theuns et al., 2015) except for one in the Ukraine (Dastjerdi et al., 2015). This data highlights the importance of PEDV diversity across several continents.
In France, since 2014, PED caused by S-non-INDEL is a notifiable disease. For territory monitoring purpose, all PEDV suspicions have to be notified to French Ministry of Agriculture and the PEDV genotype has to be confirmed by the national reference laboratory at the French agency for food, environmental and occupational health safety (Anses). Until today, no official method has been validated for the detection and quantification of the PEDV viral RNA. Since the 2000s, real-time PCR emerged as a tool of choice for the detection and quantification of viral RNA and has multiple benefits: i) these tests are highly specific ii) are easily standardized compared to “classical” virology procedures, iii) are much less time consuming, and iv) are highly reproducible. Several RT-PCRs have been described for the detection of PEDV RNA (Kim et al., 2007; Miller et al., 2016).
For a rapid, accurate and reliable diagnosis of PED in the veterinary laboratory, a method for the detection of PEDV viral RNA has been developed and more importantly validated according to the “Association Francaise de NORmalisation” (AFNOR) French NF U47-600 norm entitled “requirement and recommendation for the implementation, development and validation of PCR in animal health”(AFNOR, 2015a, b). This validated SYBR™ Green one-step RT-qPCR was based on a previously published TaqMan® probe real time RT-qPCR (Kim et al., 2007) and targeted the same zones of sequence in the conserved N open reading frame (ORF) as this had previously allowed for broad range detection and the capability to differentiate between the closely related virus Transmissible Gastro-Enteric virus (TGEV). The method developed in the current study under NF U47-600, unlike other molecular tests developed for PEDV, evaluates the whole process from sample preparation through to the detection and quantification by RT-qPCR. This method should help harmonize detection and quantification of viral RNA from PEDV belonging to both S-non-INDEL and S-INDEL strains in both field and experimental settings.
2. Materials and methods
All commercial methods were performed according to the manufacturers’ recommendations unless otherwise stated.
2.1. Primer design
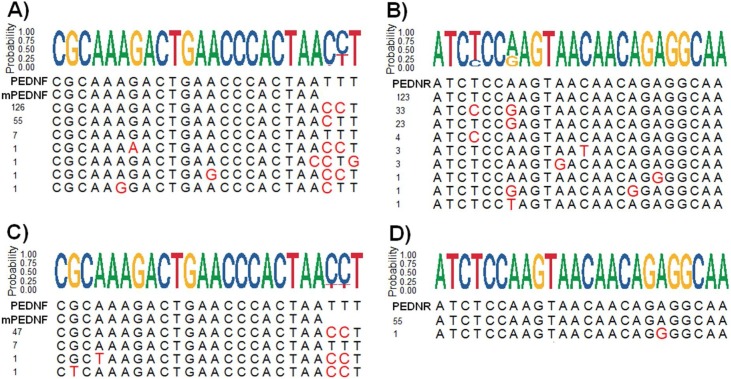
An alignment of 192 PEDV N ORF sequences that were available on the data base at the time of the study (2014) was made using MAFFT (Katoh and Standley, 2013) and the probabilities of the nucleotides at the priming zones defined by Kim et al. (2007) (PEDNF : 5’-CGCAAAGACTGAACCCACTAATTT-3’, and PEDNR : 5’-TTGCCTCTGTTGTTACTT-GGAGAT-3’) were calculated using R (Wagih, 2017) (Fig. 1 ). Based on these probabilities forward primer mPEDNF (5’-CGCAAAGACTGAACCCACTAA-3’) and reverse primer PEDNR were chosen (Fig. 1). These primers were subsequently checked against N ORFs of the S-INDEL and S-non-INDEL PEDV strains circulating in Europe (Dastjerdi et al., 2015; Grasland et al., 2015; Hanke et al., 2017; Martelli et al., 2008; Stadler et al., 2015; Steinrigl et al., 2015; Theuns et al., 2015).
Fig. 1.
Alignment of primer hybridization sequences within 192 N ORFs available in May 2014 (A and B). Alignment of primer hybridization sequences within 56 N ORFs of the S-INDEL and S-non-INDEL PEDV strains circulating in Europe (C and D). Nucleotide probabilities at each position are shown as coloured text above the alignments. Red text in the alignment sequences represent a mismatch. Sequences of primers are shown above the alignment (PEDNF, mPEDNF or PEDNR). PEDNR is shown as reverse complement. Each line represents a hybridization sequence, the number of strains presenting this sequence is indicated to the left of the sequence (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
2.2. Viruses
Original CV777, the PEDV reference strain isolated in 1977, was collected from perfused jejunum performed in 1981 and kept at −80 °C. This stock was named wtCV777. wtCV777 was propagated in cell culture as previously described (Hofmann and Wyler, 1988) and was named ccCV777. A stock of ccCV777 was produced as follows: 20 × 175 mm2 confluent monolayer of Vero cells (ATCC® CCL-81) were infected each with 500 μl of 6.8 × 104 TCID50 of ccCV777 in infection media; EMEM (ThermoFisher Scientific, France) supplemented with 0.3% tryptone phosphate broth, 0.02% yeast extract, 1% Penicillin/Streptomycin and 10 μg/ml trypsin. After 24 h of infection, cells were subjected to three freeze thaw cycles and the culture medium was clarified by centrifugation at 10,000 g for 10 min. A total volume of 1 l of supernatant was then centrifuged for four hours at 20,000 g to pellet the virus. The pellet was then re-suspended in 100 ml of PBS. The infectious viral titer of ccCV777 was determined by immune-peroxidase monolayer assay according Kärber’s method (Kärber, 1931). The virus stock solution was titrated by immuno-peroxidase monolayer assay to 1.2 × 107 TCID50/ml.
Four other PEDV strains were used: three French field strains (PEDV/FR/001/2014 Genbank accession number (GB acc) KR011756, PEDV/FR/001/2017 and PEDV/FR/001/2019 GB acc MN056942), and one American strain (PEDV/USA/2014/IOWA GB acc MF373643, kindly provided by Dr P.GAUGER from IOWA State University). Nine other ‘non-PEDV’ RNA viruses were also used: one pig alpha-coronavirus (Porcine Respiratory Coronavirus, PRCV), and two gamma-coronaviruses (infectious bronchitis virus (IBV) GB acc FJ904713), turkey coronavirus (TCoV) GB acc KR822424) as well as other pig viruses: a pig artevirus (Porcine Reproductive and Respiratory Syndrome Virus (PRRSV), GB acc KY366411), a pestivirus (classical swine fever virus (CSFV)), three pig ortomyxoviruses (swine influenza viruses H1NI, H1N2, H3N2), and two Swine DNA virus, one circovirus (porcine circovirus type 2 (PCV2) GB acc AF201311), and an asfavirus (African swine fever virus (ASFV) BankIt1774827 ANSES-MADA68322).
2.3. Matrices
Jejunum and fecal samples were collected from both specific pathogen free (SPF) pigs confirmed negative for coronavirus RNA by deep sequencing and from PEDV infected pigs positive for PEDV RNA. The PEDV positive samples had been collected during previous experimental studies (Gallien et al., 2018a, b; Gallien et al., 2019). SPF samples were used as negative controls or were spiked with PEDV produced in vitro as described in section 2.2. Spiked SPF samples were used for the validation of the method and are later referred to as ‘infectious reference materials’.
For each jejunum sample, 200 mg were homogenized in 1 ml of Phosphate Buffered Saline (PBS) (Merck, France) with 4 mm stainless steel beads in a TissueLyserII (Qiagen, France). Samples were then clarified by centrifugation at 10,000 g for 10 min. For each fecal sample, 1 ml was diluted in 9 ml of PBS and vortexed for 5 min before clarification by centrifugation as describe above.
2.4. Method description
2.4.1. Production of standards
To determine the limit of quantification (LQ) of the PCR and produce standard for quantification, a RNA transcript was produced by in vitro transcription of the PEDV wtCV777 N ORF sequence.
wtCV777 RNA was extracted using Trizol (ThermoFisher Scientific, France). Viral RNA extract was subjected to reverse transcription using hexanucleotide primers and superscript III reverse transcriptase (ThermoFisher Scientific, France). Reverse transcription was performed at 55°C for 1 h followed by enzyme inactivation at 70°C for 15 min. To amplify the N ORF, 5μl of RT were subjected to PCR amplification in 50μl reaction containing 400nM of primers oGVB160−f (GTCGGATCCACTTTATGGCTTCT) and oGVB160−r (GTCCTCGAGATT GTTTAATTTCCT), 2.5 units of Platinum Taq HiFi (Invitrogen, France), 5 μl of 10x High Fidelity PCR Buffer, and MgSO4 at a final concentration of 2 mM. The PCR was performed as follows: 95 °C for 2 min for initial denaturation, 5 cycles of 95 °C for 15 s, 30 s at 55 °C decreasing by 2.5 °C per cycle and then 68 °C for 2 min, follow by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 68 °C for 2 min. Amplified PEDV N cDNA was separated on 2% agarose gel and extracted using Montage gel extraction kit (Millipore, France). 100 ng of extracted product were cloned in pCR4-TOPO vector (Invitrogen, France). Plasmid DNA was prepared using NucleoSpin® plasmid kit (Macherey Nagel, France). In vitro transcription was performed with MAXIscript™ T7 transcription kit (ThermoFisher Scientific, France) using 1 μg of precipitated SpeI linearized N ORFs plasmid. RNA was purified with Agencourt® RNAclean XP kit (BeckmanCoulter, France), and quantified using Qubit® fluorometer (Life Technology, France, Saint Aubin). Stock of in vitro transcribed RNA was stored at −80 °C. Number of molecular copies was calculated according the following formula:
RNA transcript was diluted to 109 molecules/5 μl, aliquoted in 100 μl, supplemented with 20 μl of RNAstable® (M, France) and dried in SpeedVac® vacuum concentrator (ThermoElectron, France). The standard transcript was resuspended in 1 ml in deionized nuclease-free water and then Log10 serially diluted from 108 to 102 copies/5 μl and stored at −80 °C.
2.4.2. RNA extraction
All RNA extractions were performed using RNeasy® Mini kit (Qiagen, France) with the following modifications. 120 μl of sample mixture containing 100 μl of sample, 10 μl of an External Exogenous Control (EEC) and 10 μl of proteinase K were used as opposed to 100 μl of sample alone as recommended by the kit. RNA was eluted with 50 μl of nuclease-free water and stored at −80 °C until use. EEC used in this study was viral RNA genome (Mengovirus).
2.4.3. Conditions of the one-step PEDV RT-qPCR
Reactions were carried out in an Applied Biosystems 7500 Real-Time PCR system, with Power SYBR™ Green RNA-to-Ct™ 1-Step Kit (Applied Biosystems, Saint Aubin, France). The final PCR mix volume was composed of 12.5 μl of master mix (2x), 0.2 μl of enzyme mix, 5 μl of RNA template, primers mPEDNF and PEDNR at 300 nM or 600 nM, H2O to final volume of 25 μl. RT-PCR cycles were as follows: reverse transcription at 48 °C for 30 min, followed by 95 °C for 10 min, then 40 cycles of 95 °C for 15 s, 60 °C for 1 min, and a final melting curve analysis step as defined by the applied 7500 software V2.3. All sample amplifications with a melting temperature corresponding to the standard with a viral RNA concentration equal to, or above to the limit of detection (LoD) were considered positive.
2.5. Method validation
All of the following tests were performed using primers at 300 nM.
2.5.1. Analytical sensitivity and specificity
The analytical sensitivity and specificity were determined as described in the NF U47-600 norm. All nucleic acid extractions from viruses listed in 2.2 were tested. Five strains of PEDV were tested for inclusivity, and eleven other virus for exclusivity, among which, four coronaviruses, five other RNA viruses, and two DNA virus, all known as pathogens in pigs.
2.5.2. Diagnostic sensitivity and specificity
The diagnostic sensitivity and specificity were determined as described in the NF U47-600 norm as the true positive rate [number of true positive / (number of true positive + number of false negative)] ×100, and true negative rate [number of true negative / (number of true negative + number of false positive)] ×100. Thirty-six infected pigs from five different experimental studies were used as true positive samples. The true negative samples were the twenty-five SPF negative pigs of the same experiments. The experiments were carried out with two field PEDV strains, one French (PEDV/FR/001/2014, GB acc KR011756) and one American (PEDV/USA/2014/IOWA, GB acc MF373643). All pigs were sampled each day during the first week and thereafter at fourteen days post infection (dpi).
2.5.3. Limit of detection (LoD)
According to NF U47-600, LoD is the last dilution of reference material that allows a detection of the target with a confidence level of 95%. N RNA transcript dilutions were tested for the LoD of the PCR. Six points of a two-fold dilution series ranging from 400 to 12.5 genome copies/5 μl were analyzed in eight replicates. Three independent assays were performed for RNA transcripts (LoDPCR). To determine the LoD of the method, SPF jejunum and fecal samples spiked with ccCV777 from 106 to 10−2 TCID50/ml, were tested in two independent assays on a hundredfold serial dilution ranging from 108 to 102 and 50 N transcripts equivalent/5 μl, as infectious reference materials (LoDjejunum or LoDfeces). LoD’s were determined by Probit calculation (Finney and Stevens, 1948).
2.5.4. Limit of quantification (LQ)
According to NF U47-600, LQ is defined as the lowest (Lower LQ, LLQ) and highest level (Upper LQ, ULQ) between which, for each dilution, the statistical bias is under or equal to 0.25log10. The bias is the difference between the measured value and the theoretical value calculated by linear regression on all dilutions. Uncertainty is calculated as the variance of calculated point plus the medium bias value. The statistical bias is defined as the medium of uncertainty. For the LQ, seven points of a ten-fold serial dilution of N RNA transcript were tested (108 to102). Ten independent assays were performed on four independent serial dilutions. The LQ for organic matrices were calculated on results obtained for the LoD assessment (hundredfold dilution from 108 to 102).
2.5.5. PCR parameters
PCR efficiency was evaluated by plotting the Ct against an expected RNA copy number in respect to the TCID50/ml (data not shown) for infectious reference material or by Qubit quantification for RNA transcript. In agreement with the NF U47-600 norm, an efficiency of 75–125% was accepted.
3. Results
3.1. Alignments of 192 PEDV N ORFs
The forward primer of Kim et al. (Kim et al., 2007) (PEDNF) had perfect base pairing with 7 of the 192 (3.6%) N ORFs sequences. The forward primer designed in the current study (mPEDNF) which did not contain the last three bases of Kim et al. (2007) had perfect base pairing with 188 of 192 (979%) and of those that did not match at 100% only one had a mismatch at the last 3’ position (Fig. 1 A). Sequence of the reverse primer (PEDNR) had perfect base pairing with 123 of 192 sequence (64.1%) and those that did not match at 100% did not have any mismatches in the last three nucleotides of the 3’ end (Fig. 1 B).
Concerning the alignments with the European strains available after May 2014, PEDNF had perfect base pairing with 7 of 56 N ORFs sequences (Fig. 1 C). mPEDNF had perfect base pairing with 54 of 56 sequences, those sequences that did not match at 100% only contained one mismatch and these were localized close to the 5’ end (Fig. 1 C). PEDNR had perfect base pairing with 55 of 56 sequences and only one single miss-match with the remaining sequence at the 5’ end.
3.2. Analytical specificity and sensitivity
Amongst the different viruses strains listed in 2.2, only the PEDV strains (CV777, American field strain, and three French field strains) were positive. wtCV777 (Ct = 20), ccCV777 (Ct = 12), all with a Tm of 79.5 ± 0.5 °C which is the expected Tm for the PEDV sequence amplicon according to the in vitro transcription control. All the other viruses were negative. The analytical specificity and sensibility were both 100%.
3.3. PCR efficiency and effects of PCR conditions
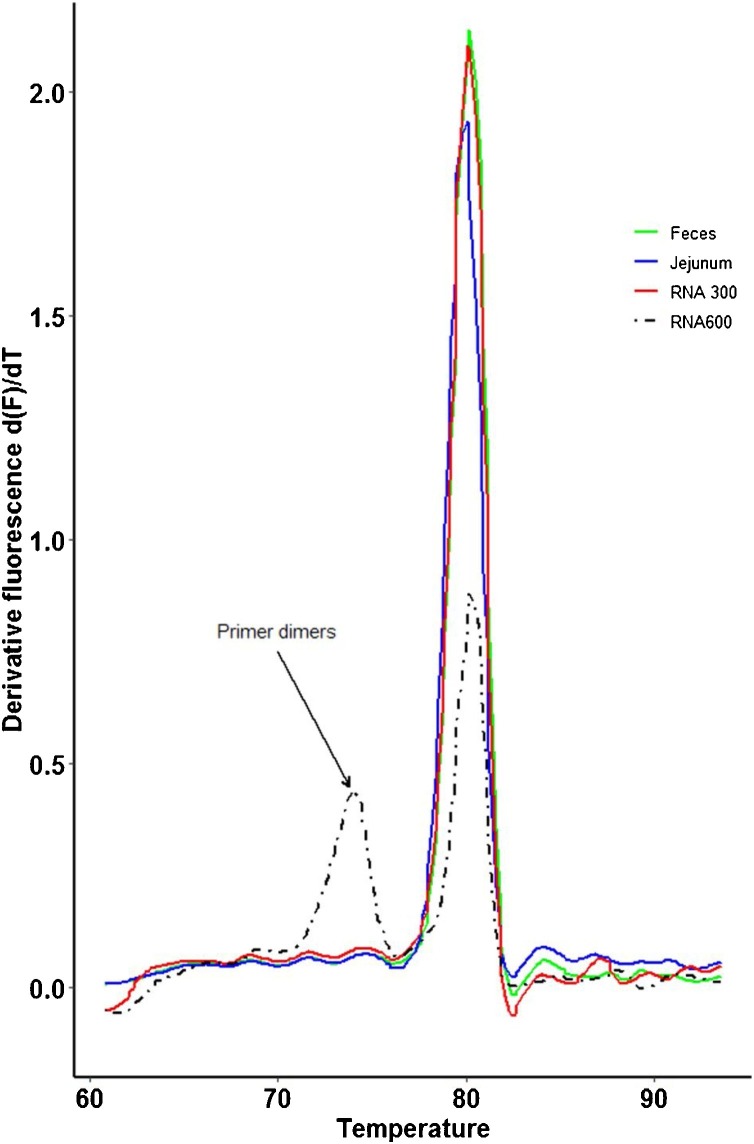
Efficiency of the method, calculated by linear regression, was 91.04% ± 1.31(0.01) for RNA transcripts, 93.51% ± 3.97(0.04) for spiked jejunum and 99.36% ± 5.12(0.05) for spiked feces. Different concentrations of primers had no effect on the efficiency of the method (data not shown), however melting curve analysis showed the presence of primer dimers at 600 nM and not at 300 nM (Fig. 2 ).
Fig. 2.
Measured Tm for in vitro transcribed RNA (Red), viral RNA in spiked feces (Green) and viral RNA in spiked jejunum (Blue). Two primer concentrations were tested with transcribed RNA, 300 nM (solid line), and 600 nM (dashed line). Primer dimers were detected at concentrations of 600 nM (first peak, dashed line). No primer dimers were observed at 300 nM (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
3.4. Limit of detection (LoD) and quantification (LQ)
The LoD was determined at 50 copies/5 μl for the RNA transcript, 50 copies/5 μl (0.5 × 100.01 TCID50/ml for the spiked feces and 100 copies/5 μl (100.01 TCID50/ml) for spiked jejunum (Table 1 ). For every selected RNA dilution tested, from 108 to 102 copies/5 μl, bias enlarged of uncertainty were included in the norm limits (-0.5 to 0.5) and statistical bias (mean of uncertainty) were < 0.25 log10 (Table 2 ). The ULQs and LLQ were 108 and 102 copies/5 μl respectively for all matrices.
Table 1.
Limit of detection for RNA transcripts, spiked jejunum and fecal samples, intra and inter-assay mean Ct, standard deviation (SD), and coefficient of variation (CV) for each point is calculated. In cases where the confidence level did not reach 95%, the number of positives from the number tested are shown.
| Matrix | Viral RNA | Assay 1 | Assay 2 | Assay 3 | Interassay |
|---|---|---|---|---|---|
| Copies / 5 μl | mean Ct ± SD(CV) | mean Ct ± SD(CV) | mean Ct ± SD(CV) | mean Ct ± SD(CV) | |
| RNA transcript | 400 | 32.45 ± 0.13 (0.004) | 31.14 ± 0.13 (0.004) | 30.69 ± 0.11 (0.004) | 31.44 ± 0.7 (0.022) |
| 200 | 33.93 ± 0.69 (0.02) | 31.84 ± 0.22 (0.007) | 31.75 ± 0.2 (0.006) | 32.51 ± 1.38 (0.042) | |
| 100 | 35.61 ± 0.46 (0.013) | 33.42 ± 1.08 (0.032) | 32.65 ± 0.27 (0.008) | 33.89 ± 2.18 (0.064) | |
| 50 | 36.24 ± 0.58 (0.016) | 34.3 ± 0.59 (0.017) | 34.11 ± 0.45 (0.013) | 34.82 ± 1.43 (0.041) | |
| 25 | 7+/8 | 6+/8 | 8+/8 | / | |
| 12.5 | 3+/8 | 3+/8 | 3+/8 | / | |
| Jejunum | 108 | 14.23 ± 0.04 (0.003) | 13.69 ± 0.04 (0.003) | / | 13.96 ± 0.11 (0.008) |
| 106 | 21.52 ± 0.06 (0.003) | 20.77 ± 0.16 (0.008) | / | 21.15 ± 0.25 (0.012) | |
| 104 | 28.55 ± 0.04 (0.002) | 27.57 ± 0.07 (0.002) | / | 28.06 ± 0.29 (0.010) | |
| 102 | 35.17 ± 0.82 (0.023) | 34.75 ± 1.21 (0.035) | / | 34.96 ± 1.05 (0.030) | |
| 50 | 3+/4 | 4+/4 | / | / | |
| Feces | 108 | 12.41 ± 0.22 (0.018) | 13.25 ± 0.20 (0.015) | / | 12.83 ± 0.39 (0.031) |
| 106 | 18.56 ± 0.03 (0.001) | 19.23 ± 0.01 (0.0004) | / | 18.90 ± 0.13 (0.007) | |
| 104 | 25.80 ± 0.14 (0.006) | 26.22 ± 0.14 (0.005) | / | 26.01 ± 0.19 (0.007) | |
| 102 | 32.88 ± 0.45 (0.014) | 32.67 ± 0.52 (0.016) | / | 32.78 ± 0.50 (0.015) | |
| 50 | 34.276 ± 1.07 (0.31) | 33.22 ± 1.11 (0.33) | / | 33.75 ± 1.15 (0.034) |
LoDs determined for each matrix are shown in bold text.
Table 2.
Bias, uncertainty and statistical bias for the linearity range selected.
| Matrix | Viral RNA | bias | Statistical Bias |
|---|---|---|---|
| copies/5 μl | mean ± uncertainty | mean of uncertainty | |
| RNA transcript | 108 | 0.06 ± 0.16 | 0.14 |
| 107 | −0.01 ± 0.07 | ||
| 106 | −0.04 ± 0.11 | ||
| 105 | −0.03 ± 0.17 | ||
| 104 | −0.03 ± 0.16 | ||
| 103 | 0.02 ± 0.11 | ||
| 102 | 0.04 ± 0.15 | ||
| Jejunum | 108 | 0.03 ± 0.13 | 0.22 |
| 106 | −0.03 ± 0.11 | ||
| 104 | −0.01 ± 0.30 | ||
| 102 | 0.02 ± 0.21 | ||
| Feces | 108 | −0.07 ± 0.12 | 0.21 |
| 106 | 0.11 ± 0.36 | ||
| 104 | −0.01 ± 0.21 | ||
| 102 | −0.03 ± 0.18 |
3.5. Repeatability and reproducibility
Calculations were done when a minimum of 23 out of 24 results were positive for the LoD and for all replicates for LQ. All coefficients of variation (CV) were below the 0.1 limit given by the norm NF U47-600 with 0.004 – 0.032, 0.002 – 0.035, 0.0004 – 0.018, for RNA transcript, jejunum and feces intra-assay CVs respectively and 0.022 – 0.064, 0.008 – 0.031, 0.007 – 0.031 for RNA transcript, jejunum and feces inter-assay CVs respectively (Table 1).
3.6. Diagnostic specificity and sensitivity
The diagnostic sensitivity was 100% at two and fourteen dpi, PEDV viral RNA were detected in all true positive pigs. The diagnostic specificity was 100% as all non PEDV infected pigs were found negative all along all experiments.
4. Discussion
PEDV is of global importance to the pig industry with many different strains and genotypes existing in different continents. After 2013 and the introduction of both S-INDEL and S-non-INDEL strains to North America and the resulting huge economic losses, the French ministry for agriculture classified PED caused by the S-non-INDEL virulent strains as a notifiable disease. Thus there was a need for a reliable method for rapid, accurate and specific detection and quantification of a broad range of PEDV strains and one that was completely validated according to French norm NF U47-600.
Many methods have been developed and used for PEDV detection and quantification as previously reviewed (Diel et al., 2016) such as direct viral isolation, but it is laborious, time consuming, and requires a reliable model for all possible strains. Furthermore, many PEDV strains cannot be isolated in vitro. Many immuno-assay tests have been developed to detect viral proteins (IFA, Blotting, ELISA) but all these methods are time consuming, have a low sensitivity and reaction, and are subject to cross reactivity decreasing the specificity. For these reasons the current study focused on developing and validating a specific and rapid diagnostic test for the detection of PEDV viral RNA. Basing this test on a TaqMan® multiplex RT-qPCR, published by Kim et al. (2007), we developed and validated a SYBR™ Green one-step RT-qPCR method.
The development and validation of the complete method, including the steps of sample preparation, RNA extraction, and RT-qPCR, were done according to the French standard NF U47-600. This norm is an adaptation to the French context of the Manual of diagnostic tests and vaccines for terrestrial animals (International Office of Epizootics, 2018) and respects the criteria stated by the World Organization for Animal Health (OIE). These standards describe the validation criteria for a PCR method in animal health and allows the characteristics not only of RT-qPCR to be determined, but also of the complete method, including sample preparation and extraction. For this, fecal and jejunum samples were used as this material has previously been described as the best matrices for detection of PEDV RNA in animals (Gallien et al., 2018a). Validating the complete method in this way means that the method is applicable for both experimental and diagnostic purposes.
In the current study the primers used by Kim et al. in 2007 were refined by in silico analysis. N ORF alignments of the priming site showed that the PEDNF forward primer of Kim et al. (2007) had mismatches with several different PEDV N ORFs and that the last three nucleotides at the 3’ end only matched with 3.6% of the sequences. Removing these three nucleotides in primer mPEDNF allowed a 100% match with 97.9% of international sequences and with 96.4% of European strains. The method using the new coupled primers demonstrated sufficient sensitivity to detect all tested PEDV strains (historical, S-INDEL and S-non-INDEL strains). Although SYBR™ Green PCRs are characteristically less specific than probe based PCRs, the specificity of the method was 100% against all viral types tested. Primer dimer formation, which are problematic for fluorescent dye based methods as they interfere dramatically with quantification, were eliminated by optimizing the primer concentration to 300 nM.
During validation, the sample preparation and RNA extraction step were optimized by the addition of a proteinase K treatment step which allowed the statistical bias to be maintained in acceptable limits (<0.25log10). The statistical bias obtain with the proteinase K treatment confirms a correct reproducibility at all quantification points, and guarantees a near or equivalent LoD (50 and 100 copies/5 μl for feces and jejunum) for the different matrices than for the transcribed RNA (50 copies/5 μl). In addition, the detection limit determined in this study (100.01 TCID50/ml) is very similar to other RT-qPCRs (100.03 TCID50/ml) (Miller et al., 2016).
In conclusion, many PCRs have been developed to detect and monitor the presence of PEDV, but, as yet to the authors’ knowledge none have been developed with a complete validation according to a norm such as the French NF U47-600. This fully validated method is the first of its kind for PEDV and should help harmonize detection and quantification of PEDV viral RNA in both field and experimental settings.
Authorship contributions
Category 1
Conception and design of study: Lionel Bigault, Béatrice Grasland
Acquisition of data: Lionel Bigault, Cécilia Bernard
Analysis and/or interpretation of data: Lionel Bigault, Béatrice Grasland
Category 2
Drafting the manuscript: Lionel Bigault
Revising the manuscript critically for important intellectual content: Paul Brown, Yannick Blanchard, Béatrice Grasland
Category 3
Approval of the version of the manuscript to be published (the names of all authors must be listed): Lionel Bigault, Paul Brown, Cécilia Bernard, Yannick Blanchard, Béatrice Grasland.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors wish to thanks Ms. Cherbonnel-Pansart for her help with AFNOR validation methodology, PhD. Le Guyader for the furniture of the MengoVirus and PhD P.Gauger for the S-INDEL strain furniture. This work was partially funded by “Direction générale de l’alimentation” of the French ministry of agriculture (project n°2014-145).
References
- AFNOR . AFNOR NF U47-600-601; 2015. AFNOR NF U47-600-1 ; Méthodes d’analyse en santé animale - PCR (réaction de polymérisation en chaîne) - Partie 1 : Exigences et recommandations pour la mise en oeuvre de la PCR en santé animale. [Google Scholar]
- AFNOR . AFNOR NF U47-600-602; 2015. AFNOR NF U47-600-2 ; Méthodes d’analyse en santé animale - PCR (réaction de polymérisation en chaîne) - Partie 2 : Exigences et recommandations pour le développement et la validation de la PCr en santé animale. [Google Scholar]
- Boniotti M.B., Papetti A., Lavazza A., Alborali G., Sozzi E., Chiapponi C., Faccini S., Bonilauri P., Cordioli P., Marthaler D. Porcine Epidemic Diarrhea Virus and Discovery of a Recombinant Swine Enteric Coronavirus, Italy. Emerg Infect Dis. 2016;22:83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasey D., Cartwright S.F. Virus-like particles associated with porcine epidemic diarrhoea. Res. Vet. Sci. 1978;25:255–256. doi: 10.1016/S0034-5288(18)32994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastjerdi A., Carr J., Ellis R.J., Steinbach F., Williamson S. Porcine Epidemic Diarrhea Virus among Farmed Pigs, Ukraine. Emerg Infect Dis. 2015;21:2235–2237. doi: 10.3201/eid2112.150272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouck P., Pensaert M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am. J. Vet. Res. 1980;41:219–223. [PubMed] [Google Scholar]
- Diel D.G., Lawson S., Okda F., Singrey A., Clement T., Fernandes M.H.V., Christopher-Hennings J., Nelson E.A. Porcine epidemic diarrhea virus: an overview of current virological and serological diagnostic methods. Virus Res. 2016;226:60–70. doi: 10.1016/j.virusres.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney D.J., Stevens W.L. A table for the calculation of working probits and weights in probit analysis. Biometrika. 1948;35:191–201. [PubMed] [Google Scholar]
- Gallien S., Andraud M., Moro A., Lediguerher G., Morin N., Gauger P.C., Bigault L., Paboeuf F., Berri M., Rose N., Grasland B. Better horizontal transmission of a US non-InDel strain compared with a French InDel strain of porcine epidemic diarrhoea virus. Transbound. Emerg. Dis. 2018 doi: 10.1111/tbed.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S., Moro A., Lediguerher G., Catinot V., Paboeuf F., Bigault L., Berri M., Gauger P.C., Pozzi N., Authie E., Rose N., Grasland B. Evidence of porcine epidemic diarrhea virus (PEDV) shedding in semen from infected specific pathogen-free boars. Vet. Res. 2018;49:7. doi: 10.1186/s13567-018-0505-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S., Moro A., Lediguerher G., Catinot V., Paboeuf F., Bigault L., Gauger P.C., Pozzi N., Berri M., Authie E., Rose N., Grasland B. Limited shedding of an S-InDel strain of porcine epidemic diarrhea virus (PEDV) in semen and questions regarding the infectivity of the detected virus. Vet. Microbiol. 2019;228:20–25. doi: 10.1016/j.vetmic.2018.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasland B., Bigault L., Bernard C., Quenault H., Toulouse O., Fablet C., Rose N., Touzain F., Blanchard Y. Complete genome sequence of a porcine epidemic diarrhea s gene indel strain isolated in france in december 2014. Genome Announc. 2015:3. doi: 10.1128/genomeA.00535-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke D., Pohlmann A., Sauter-Louis C., Hoper D., Stadler J., Ritzmann M., Steinrigl A., Schwarz B.A., Akimkin V., Fux R., Blome S., Beer M. Porcine epidemic diarrhea in Europe: in-detail analyses of disease dynamics and molecular epidemiology. Viruses. 2017:9. doi: 10.3390/v9070177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26:2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Office of Epizootics, B.S, Commission . 2018. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals: (mammals, Birds and Bees) [PubMed] [Google Scholar]
- Kärber G. Beitrag zur kollektiven Behandlung pharmakologisher Reihenversuche. Archiv f. experiment. Pathol. u. Pharmakol. 1931;162:480. [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Kim I.J., Pyo H.M., Tark D.S., Song J.Y., Hyun B.H. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol. Methods. 2007;146:172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli P., Lavazza A., Nigrelli A.D., Merialdi G., Alborali L.G., Pensaert M.B. Epidemic of diarrhoea caused by porcine epidemic diarrhoea virus in Italy. Vet. Rec. 2008;162:307–310. doi: 10.1136/vr.162.10.307. [DOI] [PubMed] [Google Scholar]
- Miller L.C., Crawford K.K., Lager K.M., Kellner S.G., Brockmeier S.L. Evaluation of two real-time polymerase chain reaction assays for Porcine epidemic diarrhea virus (PEDV) to assess PEDV transmission in growing pigs. J. Vet. Diagn. Invest. 2016;28:20–29. doi: 10.1177/1040638715621949. [DOI] [PubMed] [Google Scholar]
- Stadler J., Zoels S., Fux R., Hanke D., Pohlmann A., Blome S., Weissenbock H., Weissenbacher-Lang C., Ritzmann M., Ladinig A. Emergence of porcine epidemic diarrhea virus in southern Germany. BMC Vet. Res. 2015;11:142. doi: 10.1186/s12917-015-0454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinrigl A., Fernandez S.R., Stoiber F., Pikalo J., Sattler T., Schmoll F. First detection, clinical presentation and phylogenetic characterization of Porcine epidemic diarrhea virus in Austria. BMC Vet. Res. 2015;11:310. doi: 10.1186/s12917-015-0624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuns S., Conceicao-Neto N., Christiaens I., Zeller M., Desmarets L.M., Roukaerts I.D., Acar D.D., Heylen E., Matthijnssens J., Nauwynck H.J. Complete genome sequence of a porcine epidemic diarrhea virus from a novel outbreak in Belgium, january 2015. Genome Announc. 2015:3. doi: 10.1128/genomeA.00506-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagih O. Ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics. 2017;33:3645–3647. doi: 10.1093/bioinformatics/btx469. [DOI] [PubMed] [Google Scholar]
- Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Dempsey D.M., Dutilh B.E., Harrach B., Harrison R.L., Hendrickson R.C., Junglen S., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Nibert M., Rubino L., Sabanadzovic S., Simmonds P., Varsani A., Zerbini F.M., Davison A.J. Changes to virus taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2019) Arch. Virol. 2019;164:2417–2429. doi: 10.1007/s00705-019-04306-w. [DOI] [PubMed] [Google Scholar]