Abstract
Genome-editing technologies hold tremendous potential for treating genetic diseases. However, the efficient and safe delivery of genome-editing elements to the location of interest, and the achievement of specific targeted gene correction without off-target side effect remains a big challenge. In this perspective, we highlight recent developments and discuss the challenges of non-viral nanoparticles for the delivery of genome-editing tools. Finally, we will propose promising strategies to improve the delivery efficacy and advance the clinical translation of gene-editing technology.
Keywords: CRISPR, gene editing, non-viral nanoparticles, lipid-like nanoparticles, nucleic acid delivery
Graphcial Abstract
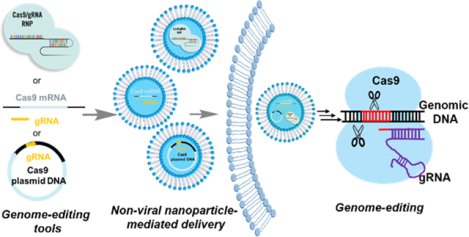
1. INTRODUCTION
The clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) system has emerged as one of the most revolutionary biological tools in recent years.1–3 The CRISPR/Cas9 system contains two components: a nuclease protein Cas9 that binds to DNA and initiates double-strand breaks (DSBs), and a very shot single guide RNA (sgRNA) that directs the Cas9 nuclease to the targeted genomic locus.4–6 Nuclease-induced DSBs can be naturally repaired by the cell through two different mechanisms:4 nonhomologous end-joining (NHEJ), which often causes gene coding sequence disruption by inducing indel mutations, and homology directed repair (HDR), which precisely correct gene mutations by using an exogenous DNA template (Figure 1). The CRISPR/Cas9 technology has widespread application in characterization of disease targets,7 creation of human cellular and animal models of disease,8, 9 and site-specific therapeutic genome editing to correct and treat a wide range of diseases.10 The first CRISPR/Cas9-based human clinical trial is already under way in China11 and the first CRISPR-for-cancer clinical trial in the US was recently authorized by the FDA (Clinicaltrials.gov Identifier: NCT03399448).
Figure 1.
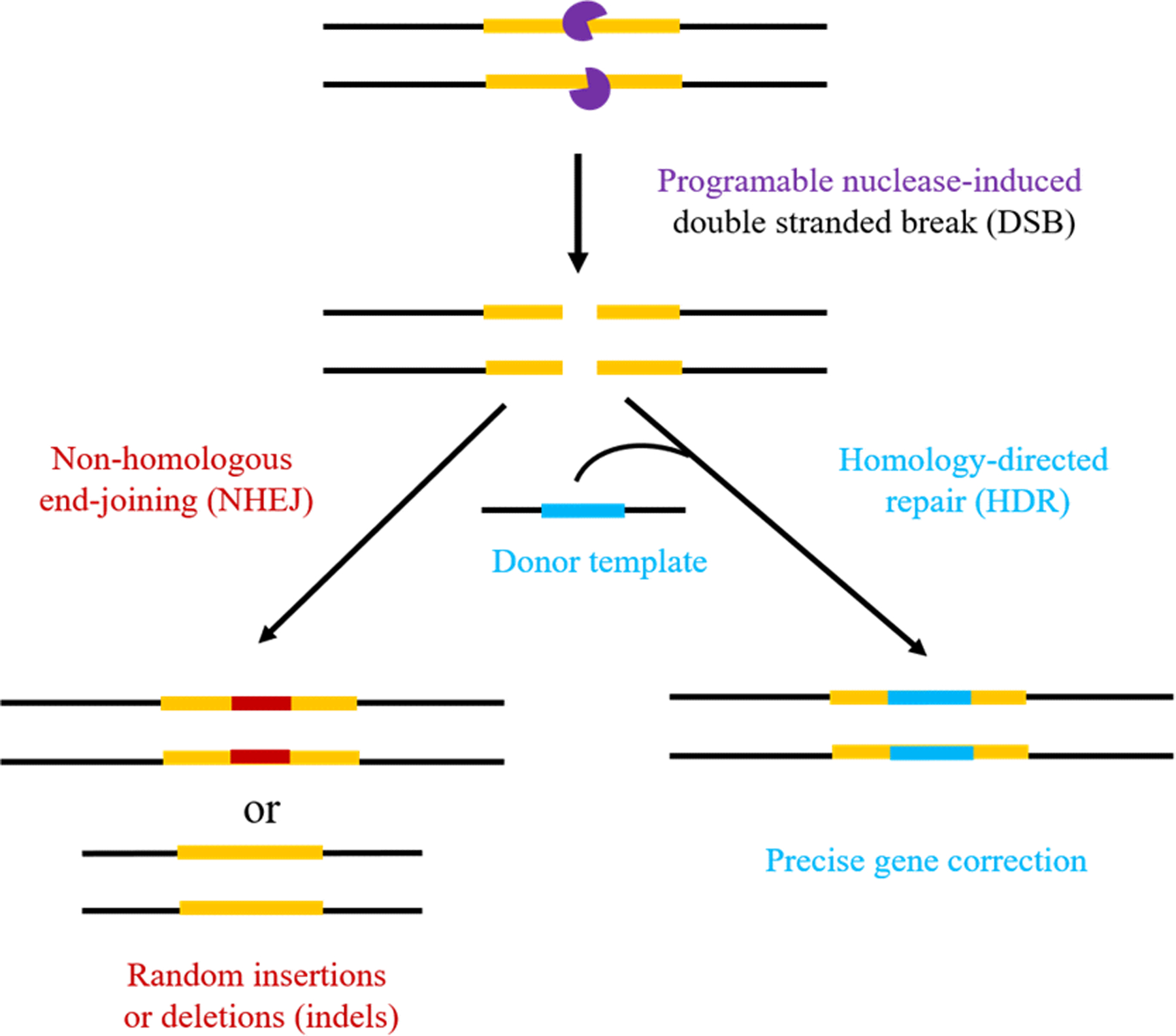
Schematic illustration of genome editing using double stranded break (DSB) induced by a programmable nuclease, and the two different pathways (NHEJ and HDR) for DSB repair.
Despite the historic milestones which have already been made in the rapidly developing field of CRISPR/Cas9-based gene editing, the safe and effective delivery of the CRISPR/Cas9 gene editing cargoes, however, remains a big challenge. Until now, physical (electroporation, microinjection, and hydrodynamic injection) and viral (lentivirus, adenovirus, and adeno-associated virus (AAV)) approaches are the most widely used strategies for CRISPR/Cas9 delivery.12–15 Physical methods often afford high transfection efficiency but are most suited for ex vivo cell engineering, such as CAR-T engineering,16 and are not clinically-applicable techniques for systemic therapeutics delivery.14 Viral vectors show high efficacy on CRISPR/Cas9 delivery but suffer from limitations related to the restricted packaging ability, potent immunogenicity, risk of unwanted insertional mutagenesis, as well as the safety concerns such as the constant expression of the Cas9 protein in clinical translation.13, 17 Non-viral nanoparticles, like polymeric nanoparticles, liposomes, lipid nanoparticles (LNPs), and inorganic nanoparticles, have emerged as promising alternatives because they have the potential to be engineered to overcome many of these limitations.9, 18–20 Thus, in this perspective, we will highlight recent developments and discuss the challenges and limitations that non-viral nanoparticles faced in the delivery of genome-editing tools. Finally, promising strategies to improve the delivery efficacy and advance the clinical translation of gene-editing technology will be proposed.
2. APPROACHES DEVELOPED FOR NON-VIRAL DELIVERY OF DNA AND RNA
The past few decades have witnessed remarkable progress in the development of non-viral vectors for effective delivery of nucleic acid therapeutics such as plasmid DNA, mRNA, and siRNA.21–23 The ability of naked plasmid DNA and mRNA to induce sustainable protein expression in mice via intramuscular administration was first demonstrated in 1990.24 Since then, many delivery approaches have been developed for non-viral therapeutic nucleic acid delivery, including physical methods (such as electroporation, gene gun, hydrodynamic delivery, and others.),14, 25 and chemical approaches (such as lipids, liposomes, polymeric nanoparticles, and inorganic nanoparticles).26–30 Commercially available transfection reagents, such as Lipofectamine (which is sold in a variety of proprietary formulations) and JetPEI are the most commonly used non-viral vectors for in vitro intracellular nucleic acids delivery. In August 2018, FDA approved the first of a new class of RNA-based therapies, Patisiran™, for the treatment of polyneuropathy caused by hereditary transthyretin-mediated amyloidosis (hATTR) in adults.31 Patisiran™, which is based on LNPs, uses siRNA to interfere with the production of TTR protein via intravenous administration. In addition to Patisiran™, there are many nucleic acids-based therapies currently undergoing clinical trials at various stages. As an alternative to nanoparticles, another approach has been the direct conjugation of bioactive ligands to the RNAs, which endow them with the ability to enter the cells of interests. N-acetylgalactosamine (GalNAc) are the most clinically advanced RNA-conjugates, which are typically dosed subcutaneously and have the ability to target to hepatocytes in liver.32, 33
CAN WE DIRECTLY ADAPT WHAT HAS BEEN DEVELOPED FOR NUCLEIC ACID DELIVERY FOR CRISPR-BASED GENE EDITING?
Traditional nucleic acid-based therapy is a single component system. For example, plasmid DNA or mRNA can act alone express the desired protein once introduced into the cells and exert its biological function. Small RNAs and DNAs, such as siRNA, microRNA (miRNA), or anti-sense oligonucleotide (ASO), must complex with the RNA-induced silencing complex (RISC) or the Ribonuclease H in order to exert biological function, but these components are natively expressed in the target cells, so no additional therapeutic molecule needs be delivered. In contrast, CRISPR-based gene editing requires two non-native components, the endonuclease protein and the targeting sgRNA, which both need to be delivered to the cell in order to exert biological function. The Cas9 and sgRNA both need to be delivered to the target cell, and then they must form complex in order to have biological function, Hence, there are several unique factors to consider when designing a CRISPR delivery vector.
3.1. Using Cas9 encoding plasmid, mRNA or Cas9 protein directly?
CRISPR/Cas9 components can be delivered into cells in different strategies (Figure 2): (1) DNA gene-based delivery, in which plasmids or viral vectors that encode for the expression of both Cas9 and sgRNA are delivered; (2) RNA-based delivery, in which Cas9 mRNA with a sgRNA are delivered using traditional RNA delivery vectors, and (3) protein-based delivery, in which the Cas9 protein is complexed with a synthetic sgRNA (ribonucleoprotein complex, RNP) ex vivo, and the entire complex is delivered together.34, 35 Plasmid DNA delivery is an appealing delivery approach for gene editing due to the good stability and easy design and preparation of plasmids. However, to generate the final gene-editing protein complex, the plasmid DNA needs to be delivered into the nucleus, and the target cells native transcription mechanism must be recruited to transcribe the gene into mRNA, transport the mRNA into the cytoplasm where it will be translated into the protein, and then transport the protein back into the nucleus where the CRISPR mechanism can exert its effect on the cell’s genomic DNA. Due to this long biological process, the peak protein production from plasmid DNA delivery are usually observed 24–48 hours after plasmid delivery. Moreover, the plasmid delivery may induce more off-targeting editing than other delivery approaches, and may risk triggering an immunogenic response.36 As compared to plasmid DNA, the delivery of Cas9 mRNA results in quicker expression, with peak protein expression typically seen within 6 hours of deliver, due to the fact that the Cas9 mRNA bypasses the first steps of nuclear delivery and DNA transcription; translation of mRNA into protein occur immediately following cytosolic delivery.37 This approach mitigates the risk of off-target effects and reduced the probability of insertional mutagenesis. However, mRNA is less stable than plasmid DNA both during storage prior to delivery and also in vivo after the delivery, which is a big challenge for this type of delivery strategy. Another unique consideration for mRNA delivery is the timing to introduce the gRNA. The gRNA molecule cannot exert an effect until the mRNA has been translated into protein, and due to the instability of RNA, it may be optimal to first deliver the mRNA alone, allow some time for the mRNA to be translated, and only then deliver the gRNA. This may maximize the formation of functional Cas9/sgRNA complexes, but requires the sgRNA to be delivered into the cells at the right place as well as the right time. Delivery of Cas9 directly as a ribonucleoprotein complex is more straightforward. Unlike gene delivery, protein delivery is instantaneous and transient, and therefore avoids the concern of permanently integrating CRISPR genes into the host genome. Cas9 protein is usually preassembled with sgRNA to form RNP complex in vitro and delivered in a single unit. This avoids some of the timing and delivery issues of mRNA/gRNA delivery, and allows the therapeutic effect to be exerted nearly instantaneously.
Figure 2.
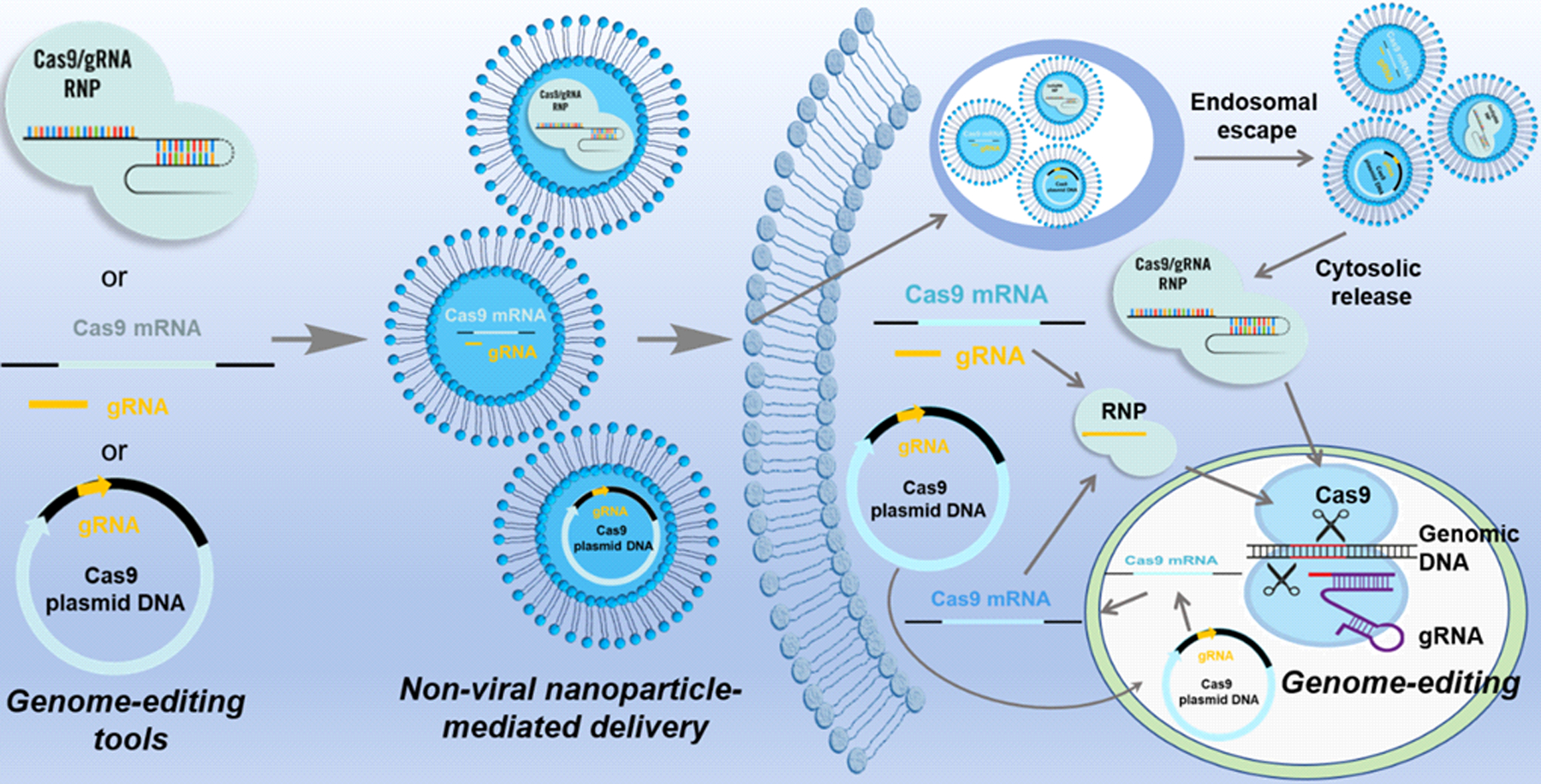
CRISPR/Cas9 delivery as plasmid DNA, mRNA, or protein. CRISPR/Cas9 components can be delivered into cells in either DNA, mRNA, or protein format to achieve a specific gene editing function. For non-viral nanoparticle-mediated delivery, each format should be encapsulated in nanocarriers for efficient intracellular uptake. After endocytosis into the cell, nanoparticles need to escape from endo-/lysosome. For plasmid DNA delivery, DNA needs to be delivered into the nucleus, and the target cells native transcription mechanism must be recruited to transcribe the gene into mRNA, transport the mRNA into the cytoplasm where it will be translated into the protein, and then transport the protein back into the nucleus where the CRISPR mechanism can exert its effect on the cell’s genomic DNA. For mRNA delivery, cargo should be released in the cytosol to enable mRNA translation to protein. Protein delivery is instantaneous and transient, results in the most immediate onset of gene editing, and therefore avoids the concern of permanently integrating CRISPR genes into the host genome.
4. CURRENT NON-VIRAL NANOPARTICLES DELIVERY APPROACHES FOR GENOME EDITING
4.1. Lipid nanoparticles
So far, some well-developed non-viral platforms for nucleic acids delivery have been adapted for genome-editing applications. Commercialized cationic lipid nucleic acid transfection reagents, like Lipofectamine 2000™, Lipofectamine 3000™, RNAiMAX ™ etc., can potently intracellularly deliver protein or nucleic acids to cells for genome-editing. Liu and coworkers reported the use of Lipofectamine 2000™ and RNAiMAX ™ for efficient delivery of engineered Cre-recombinase, TALENs, and Cas9/sgRNA RNPS into mammalian cells; they demonstrated that delivery of Cas9/sgRNA into the mouse inner ear resulted in 20% modification efficiency in mouse cochlea hair cells.38 Although the commercially available transfection reagents show good potential in genome-editing application, they are limited by their high toxicity and inflammatory side effects. Cationic lipids synthesized in a combinatorial method have the potential to overcome these drawbacks.39 We recently reported the development of a combinatorial library of cationic bioreducible lipids to deliver a supernegatively charged genome-editing protein. The library is generated through the Michael addition reaction between an amine head group and an acrylate tail group bearing a bioreducible disulfide bond.40 The incorporated disulfide bond in lipids can be cleaved under the high GSH condition in the cells, which facilities the endosomal escape of genome-editing tools encapsulated nanoparticles into cytosol and nucleus for genome-editing. The simple chemical reaction allows a variety of amine and acrylate groups to be reacted in a combinatorial fashion to generate a large library of lipid-like materials which can be used as delivery vectors. We used the library screening strategy to evaluate the ability of these lipids for Cas9/sgRNA delivery for genome editing. The 8-O14B lipid, one of the leading candidates screened out from 12 bioreducible lipids, showed excellent ability to deliver negatively supercharged Cre ((−27)GFP-Cre)), achieving ~80% GFP-Cre-mediated recombination efficiency in vitro, which was higher than that of Lipofectamine 2000™. Additionally, the in vivo local delivery of (−27)GFP-Cre) into mouse brain demonstrated functional modification in vivo. This demonstrates that the lipids have the potential to be a promising platform for protein-mediated genome-editing therapy for brain diseases. Moreover, we also identified lipids 3-O14B, 4-O14B, and 6-O14B which delivered the Cas9/sgRNA complex at a comparable genome editing efficiency to that of Lipofectamine 2000™.
In addition to the delivery of genome-editing proteins, lipid materials have also been used to deliver Cas9 mRNA and sgRNAs both in vitro and in vivo. For example, zwitterionic amino lipids (ZALs) were employed for the co-delivery of Cas9 mRNA and sgRNAs for genome-editing.41 The intravenous co-delivery of Cas9 mRNA and sgLoxP achieved detectable expression of floxed tdTomato in the liver, lung, and kidneys of genetically engineered mice. In another study, Cas9 mRNA and sgRNA were delivered by lipid-like NPs to the liver to treat HBV infection and hypercholesterolemia systemically in adult animals.37
4.2. Polymeric nanoparticles
For the delivery of genome-editing tools, cationic polymers have been commonly used to improve cellular uptake, facilitate endosomal escape, and enhance the overall transfection efficiency. In one study, Wei and colleagues reported the preparation of a multifunctional nucleus-targeting “core-shell” artificial virus (RRPHC) for the delivery of CRISPR-Cas9 system.42 RRPHC contained a fluorinated polymer (PF33) core to bind with the CRISPR-Cas9 plasmid and a multifunctional shell RGD-R8-PEG-HA (RRPH). Cas9-hMTH1 system loaded RRPHC showed effective MTH1 gene disruption and significantly inhibited the tumor growth in vivo. Murthy and colleagues reported a CRISPR delivery system in which Cas9/sgRNA RNPs were adsorbed onto DNA-conjugated gold nanoparticles, and then coated with the cationic endosomal disruptive polymer poly(N-(N-(2-aminotehyl)-2aminoethyl) aspartamide) (PAsp(DET)). This system, which they named CRISPR-Gold, was used to directly deliver Cas9 RNP and donor DNA in vivo via local injection in a mouse model of Duchenne Muscular Dystrophy (DMD) and induced 5.4% of dystrophin gene correction in mdx mice.43 Substantial progress has been made in using this system to deliver CRISPR-Cas9 and Cpf1 (another RNA guided endonucleases) to edit genes in the brains of adult mice via a local intracranial administration.44 Recently, Leong and Cheng demonstrated the use of a PEGylated nanoparticles (P-HNPs) based on a water-soluble, α-helical cationic polypeptide poly(γ−4-(2-(piperidin-1-yl)ethyl)aminomethyl)benzyl-L-glutamate) (PPABLG) to deliver Cas9 and synthesized sgRNA plasmids for genome-editing.45
4.3. Inorganic nanoparticles
Gold nanoparticles are a good nanocarrier for RNP delivery. In one study, gold nanoparticles were co-assembled with a glutamate peptide tag (E-tag) engineered Cas9 protein and a sgRNA into nano-assemblies for highly efficient direct cytoplasmic/nuclear RNP delivery.46 In recent years, metal-organic frameworks (MOFs) have gained significant attention as nanocarriers in biomedical applications for their tunable pore size and rigid structure, which have been demonstrated to be able to not only effectively encapsulate but also protect nucleic acids and proteins from degradation.47, 48 Recently, these have also been explored to deliver CRISPR/Cas9 RNP for genome editing.49
5. CHALLENGES FOR NON-VIRAL NANOPARTICLE-MEDIATED DELIVERY OF GENOME-EDITING SYSTEMS
Even though huge progress has been made in non-viral nanoparticle design for intracellular protein and gene delivery, the efficient and safe delivery of genome-editing tools remains a large challenge. Effective packing of the editing tools into a single vector is the first major challenge. As mentioned above, at least two critical elements should be encapsulated into the nanoparticles. For protein-based delivery, the commonly used Cas9 is positively charged (net charge: +20) with a molecular weight of ~160 KDa, which was larger than that of the most proteins, sgRNA is negatively charged with a molecular weight of ~31 KDa.46, 50 Thus, it is difficult to encapsulate these components by using the developed non-viral gene or protein delivery systems directly. Moreover, the incorporation of additional DNA template for HDR applications will further challenge the nanocarrier design. As with the protein form, Cas9 mRNA and plasmids also have large sizes, which hinders effective packing in most vectors. To address this challenge, new strategies have been developed. For example, Daniel and coworkers reported the combination of non-viral lipid nanoparticles-mediated Cas9 mRNA delivery with AAV (viral) encoded sgRNA and HDR repair template to cure Fahmut/mut mice via systemic administration.51
Systemic delivery is the second challenge due to the intrinsic fragile properties of proteins, RNAs, and even DNA plasmids. Editing tools usually have low plasma stability, so they will be rapidly degraded after exposure to blood. This can be due to both active degradation by nucleases and proteases in the blood, as well as passive adsorption of blood proteins blocking the function of these cargoes. Recent research has also identified the endogenous presence of anti-Cas9 antibodies in some humans.52 Thus, how to encapsulate the editing tools into the interior of nanoparticle to protect them from degradation, recognition, and clearance is challenging. However, after systemic administration the nanocomplexes themselves are also prone to absorb circulating proteins onto their surface, which will induce the recognition and clearance of these particles by the reticuloendothelial system (RES). The most commonly-used strategy to reduce RES-mediated clearance is PEGylation, in which the antifouling hydrophilic molecule PEG is coated onto the surface of the nanoparticles.53 Although PEGylation could increase the circulation time as well as the accumulation of nanoparticle in the targeted sites, the accompanied decrease of cellular uptake should also be noted.
Therefore, efficient cellular uptake is the next challenge that should be taken into consideration while designing the delivery vehicles. Like most other proteins and nucleic acids, the naked genome-editing tools are cell membrane impermeable. To improve the cellular uptake of gene-editing elements, active targeting strategies are often employed. Many types of ligands, including peptides, antibodies, aptamers, transferrin, and mono-/poly-saccharide can be introduced onto the surface of non-viral nanoparticles to enhance the cellular uptake via receptor-mediated endocytosis.54, 55 To address the need for targeted RNP delivery, Dounna and coworkers developed an engineered CRISPR-Cas9 RNP harboring a ligand for the asialoglycoprotein receptor ligands (ASGPrL) (Cas9-ASGPrL RNP) for hepatocyte-specific RNP delivery.56 The Cas9-ASGPrL RNP resulted in receptor-facilitated, cell -type specific gene editing in HEPG2 cells, an ASGPr positive liver-derived cell line.
In addition to cellular uptake, nanoparticle entrapment and degradation within endosomes or lysosomes is also a critical barrier for efficient delivery of genome-editing systems. Vectors with proton buffer capacity, such as cationic lipids, polyethyleneimine (PEI), or poly(2-(dimethylamino) ethyl metharylate) (PDMAEMA), have been widely used to disrupt the endosomal or lysosomal membrane, facilitating the endosomal/lysosomal escape of the cargo. Membrane fusion peptides (such as GALA peptide) and cell penetrating peptides (such as TAT and R8 peptide) have also been extensively exploited to facilitate endo-/lysosomal escape.57–59
Immunogenicity is another major challenge since the most widely used orthologs of Cas9 proteins are derived from Staphylococcus aureus and Streptococcus pyogenes. These two bacterial species are common naturally and have high infection frequencies in mammals, including humans. When humans are exposed to these pathogens over the course of daily life, they are typically expected to elicit an immunogenic response.60–62 This immune response may result in the generation of antibodies targeting S. aureus and S. pyogenes proteins, which will then recognize and disable the therapeutically beneficial Cas9 proteins. This phenomenon may hinder the safe and efficacious use of CRISPR/Cas9 in human patients, and may cause significant side effects. Thus, further work may be needed to minimize this immune response in human patients.
Off-target editing effects have always been the major concern regarding the in vivo application or clinical translation of CRISPR technology. Off-target editing concerns can be broken into two broad categories: biological off-target editing (in which the CRISPR complex results in a DSB at an off-target locus in the genome), and off-target delivery effects (in which the CRISPR complex is delivered to a target organ where it is not necessary to exert its therapeutic effect). From the biological point of view, scientists have developed several strategies, including using the Cpf1 enzyme (also known as Cas12a) which has higher specificity than that of spCas9 variant and other advantages,63, engineering of the Cas9 protein itself to improve specificity, and using paired Cas9 nickase enzymes,64 to reduce the off-target genome editing. From the delivery point of view, as discussed above, targeted delivery of CRISPR machinery to the specific organs or cell types using non-viral nanoparticles could be helpful to mitigate the off-target effect. Engineering the Cas9 protein to be inactive unless specifically in the presence of certain activating conditions, for example light or exogenous small molecules, may also improve the delivery targeting of therapeutic CRISPR65, 66 Meanwhile, the direct delivery of RNP complexes, which offers transient Cas9 activity than virus or plasmid transfection, can results in reduced off-target events.67
6. CONCLUSIONS AND FUTURE PERSPECTIVE
In conclusion, genome-editing technology is now revolutionizing the area of gene therapy. The final clinical translation of this technology, however, is largely reliant on the development of highly safe and efficient delivery systems. Non-viral nanoplatforms could expand the scope of genome-editing therapies into the next level. However, as discussed in this perspective, delivery of genome editing tools is much more difficult than traditional nucleic acid delivery. While the existing delivery systems for RNA or DNA delivery can be adapted for genome editing applications, the adaptation may not be straightforward, and a number of changes or developments may be necessary. Although many genome-editing delivery platforms have been developed so far, and a number of them have showed promising in vivo efficacy, but they all limited to local administration, the effective systemic delivery of genome-editing components in vivo into specifically targeted cells is still a large challenge.
To date, the most prominent non-viral strategy to delivery nucleic acids or CRISPR machineries is the use of cationic materials. However, these particles face potential systemic administration challenges, such as unfavorable protein absorption, rapid clearance by RES, and severe systemic cytotoxicity due to their cationic nature. These issues may be addressed by developing non-cationic materials which possess intrinsic affinity to the CRISPR cargoes though supramolecular interactions. Our recently developed nitrilotriacetic acid-containing lipidoid nanoparticles exhibit high genome editing efficiencies and low cytotoxicity in mammalian cells.68 In addition, unlike traditional protein or siRNA delivery, CRISPR elements must be delivered into the cell nucleus where genome editing happens, so the size and the nuclear localization or permeation properties of the nanoparticles should be well controlled. Moreover, off-target concerns should always be taken into consideration when designing a new delivery platform. Thus, increasingly precise, specific, and capable non-viral nanosystems should be developed to further overcome these hurdles and to advance the application of genome-editing tools for basic biology research, biotechnology, and therapeutic development.
ACKNOWLEDGMENTS
We acknowledge the financial support from National Institutes of Health (NIH) Grants R01 EB027170-01 and UG3 TR002636-01.
Funding Sources
National Institutes of Health (NIH) Grants R01 EB027170-01 and UG3 TR002636-01.
ABBREVIATIONS
- CRISPR/Cas9
clustered regularly interspaced short palindromic repeats-associated protein 9
- RNP
ribonucleoprotein complex
- DSBs
double-strand breaks
- sgRNA
single guide RNA
- NHEJ
nonhomologous end-joining
- HDR
homology directed repair
- FDA
Food and Drug Administration
- AAV
adeno-associated virus
- LNPs
lipid nanoparticles
- hATTR
hereditary transthyretin-mediated amyloidosis
- GalNAc
N-acetylgalactosamine
- miRNA
microRNA
- ASO
anti-sense oligonucleotide
- RISC
RNA-induced silencing complex
- ZALs
zwitterionic amino lipids
- DMD
Duchenne Muscular Dystrophy
- MOFs
metal-organic frameworks
- E-tag
glutamate peptide tag
- RES
reticuloendothelial system
- ASGPrL
asialoglycoprotein receptor ligands
- PEI
polyethyleneimine
- PDMAEMA
poly(2-(dimethylamino) ethyl metharylate)
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Hsu PD; Lander ES; Zhang F, Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157 (6), 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Doudna JA; Charpentier E, The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346 (6213), 1258096. [DOI] [PubMed] [Google Scholar]
- (3).Sternberg SH; Doudna JA, Expanding the biologist’s toolkit with CRISPR-Cas9. Mol. Cell 2015, 58 (4), 568–574. [DOI] [PubMed] [Google Scholar]
- (4).Jiang FG; Doudna JA, CRISPR-Cas9 structures and mechanisms In Annu. Rev. Biophys, Dill KA, Ed. 2017; Vol. 46, pp 505–529. [DOI] [PubMed] [Google Scholar]
- (5).Gasiunas G; Barrangou R; Horvath P; Siksnys V, Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (39), E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mali P; Yang LH; Esvelt KM; Aach J; Guell M; DiCarlo JE; Norville JE; Church GM, RNA-guided human genome engineering via Cas9. Science 2013, 339 (6121), 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Fellmann C; Cowen BC; Lin PC; Doudna JA; Corn JE, Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov 2017, 16 (2), 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Xue W; Chen SD; Yin H; Tammela T; Papagiannakopoulos T; Joshi NS; Cai WX; Yang GL; Bronson R; Crowley DG; Zhang F; Anderson DG; Sharp PA; Jacks T, CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 2014, 514 (7522), 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wang HX; Li M; Lee CM; Chakraborty S; Kim HW; Bao G; Leong KW, CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem. Rev 2017, 117 (15), 9874–9906. [DOI] [PubMed] [Google Scholar]
- (10).Long CZ; Li H; Tiburcy M; Rodriguez-Caycedo C; Kyrychenko V; Zhou HY; Zhang Y; Min YL; Shelton JM; Mammen PPA; Liaw NY; Zimmermann WH; Bassel-Duby R; Schneider JW; Olson EN, Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci. Adv 2018, 4 (1), eaap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Cyranoski D, Chinese scientists to pioneer first human CRISPR trial. Nature 2016, 535 (7613), 476. [DOI] [PubMed] [Google Scholar]
- (12).Kay MA, State-of-the-art gene-based therapies: the road ahead. Nat. Rev. Genet 2011, 12 (5), 316–328. [DOI] [PubMed] [Google Scholar]
- (13).Yin H; Kanasty RL; Eltoukhy AA; Vegas AJ; Dorkin JR; Anderson DG, Non-viral vectors for gene-based therapy. Nat. Rev. Genet 2014, 15 (8), 541–555. [DOI] [PubMed] [Google Scholar]
- (14).Wells DJ, Gene therapy progress and prospects: Electroporation and other physical methods. Gene Ther. 2004, 11 (18), 1363–1369. [DOI] [PubMed] [Google Scholar]
- (15).Chen X; Goncalves M, Engineered viruses as genome editing devices. Mol. Ther 2016, 24 (3), 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yin H; Kauffman KJ; Anderson DG, Delivery technologies for genome editing. Nat. Rev. Drug Discov 2017, 16 (6), 387–399. [DOI] [PubMed] [Google Scholar]
- (17).Wu ZJ; Yang HY; Colosi P, Effect of genome size on AAV vector packaging. Mol. Ther 2010, 18 (1), 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Rui Y; Wilson DR; Green JJ, Non-viral delivery to enable genome editing. Trends Biotechnol. 2019, 37 (3), 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Li L; Hu S; Chen XY, Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 2018, 171, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wang M; Glass ZA; Xu Q, Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 2017, 24 (3), 144–150. [DOI] [PubMed] [Google Scholar]
- (21).Chang J; Chen XH; Glass Z; Gao F; Mao LQ; Wang M; Xu QB, Integrating combinatorial lipid nanoparticle and chemically modified protein for intracellular delivery and genome editing. Acc. Chem. Res 2019, 52 (3), 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wang M; Sun S; Neufeld CI; Perez-Ramirez B; Xu QB, Reactive oxygen species-responsive protein modification and its intracellular delivery for targeted cancer therapy. Angew. Chem. Int. Ed 2014, 53 (49), 13444–13448. [DOI] [PubMed] [Google Scholar]
- (23).Wang M; Alberti K; Sun S; Arellano CL; Xu QB, Combinatorially Designed Lipid-like Nanoparticles for Intracellular Delivery of Cytotoxic Protein for Cancer Therapy. Angew. Chem. Int. Ed 2014, 53 (11), 2893–2898. [DOI] [PubMed] [Google Scholar]
- (24).Uchida S; Kataoka K, Design concepts of polyplex micelles for in vivo therapeutic delivery of plasmid DNA and messenger RNA. J. Biomed. Mater. Res. Part A 2019, 107 (5), 978–990. [DOI] [PubMed] [Google Scholar]
- (25).Mehier-Humbert S; Guy RH, Physical methods for gene transfer: Improving the kinetics of gene delivery into cells. Adv. Drug Deliv. Rev 2005, 57 (5), 733–753. [DOI] [PubMed] [Google Scholar]
- (26).Kamaly N; Yameen B; Wu J; Farokhzad OC, Degradable controlled-release polymers and polymeric nanoparticles: mechanisms of controlling drug release. Chem. Rev 2016, 116 (4), 2602–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Patel AK; Kaczmarek JC; Bose S; Kauffman KJ; Mir F; Heartlein MW; DeRosa F; Langer R; Anderson DG, Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv. Mater 2019, 31 (8), 1805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kanasty R; Dorkin JR; Vegas A; Anderson D, Delivery materials for siRNA therapeutics. Nat. Mater 2013, 12 (11), 967–977. [DOI] [PubMed] [Google Scholar]
- (29).Cheng L; Yang L; Meng FH; Zhong ZY, Protein nanotherapeutics as an emerging modality for cancer therapy. Adv. Health. Mater 2018, 7 (20), 1800685. [DOI] [PubMed] [Google Scholar]
- (30).Qiu M; Zhang ZQ; Wei YH; Sun HL; Meng FH; Deng C; Zhong ZY, Small-sized and robust chimaeric lipopepsomes: A simple and functional platform with high protein loading for targeted intracellular delivery of protein toxin in vivo. Chem. Mater 2018, 30 (19), 6831–6838. [Google Scholar]
- (31).Hoy SM, Patisiran: First global approval. Drugs 2018, 78 (15), 1625–1631. [DOI] [PubMed] [Google Scholar]
- (32).Yu RZ; Graham MJ; Post N; Riney S; Zanardi T; Hall S; Burkey J; Shemesh CS; Prakash TP; Seth PP; Swayze EE; Geary RS; Wang YF; Henry S, Disposition and pharmacology of a GalNAc(3)-conjugated ASO targeting human lipoprotein (a) in mice. Mol. Ther.-Nucl. Acids 2016, 5, e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Springer AD; Dowdy SF, GalNAc-siRNA conjugates: Leading the way for delivery of RNAi therapeutics. Nucl. Acid Ther 2018, 28 (3), 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Komor AC; Badran AH; Liu DR, CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 2017, 168 (1–2), 20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Glass Z; Lee M; Li YM; Xu QB, Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018, 36 (2), 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hughes TS; Langer SJ; Virtanen SI; Chavez RA; Watkins LR; Milligan ED; Leinwand LA, Immunogenicity of intrathecal plasmid gene delivery: cytokine release and effects on transgene expression. J. Gene. Med 2009, 11 (9), 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Jiang C; Mei M; Li B; Zhu XR; Zu WH; Tian YJ; Wang QN; Guo Y; Dong YZ; Tan X, A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017, 27 (3), 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Zuris JA; Thompson DB; Shu Y; Guilinger JP; Bessen JL; Hu JH; Maeder ML; Joung JK; Chen ZY; Liu DR, Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol 2015, 33 (1), 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Altinoglu S; Wang M; Xu QB, Combinatorial library strategies for synthesis of cationic lipid-like nanoparticles and their potential medical applications. Nanomedicine 2015, 10 (4), 643–657. [DOI] [PubMed] [Google Scholar]
- (40).Wang M; Zuris JA; Meng FT; Rees H; Sun S; Deng P; Han Y; Gao X; Pouli D; Wu Q; Georgakoudi I; Liu DR; Xu QB, Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (11), 2868–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Miller JB; Zhang SY; Kos P; Xiong H; Zhou KJ; Perelman SS; Zhu H; Siegwart DJ, Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed 2017, 56 (4), 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Li L; Song LJ; Liu XW; Yang X; Li X; He T; Wang N; Yang SLX; Yu C; Yin T; Wen YZ; He ZY; Wei XW; Su WJ; Wu QJ; Yao SH; Gong CY; Wei YQ, Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano 2017, 11 (1), 95–111. [DOI] [PubMed] [Google Scholar]
- (43).Lee K; Conboy M; Park HM; Jiang FG; Kim HJ; Dewitt MA; Mackley VA; Chang K; Rao A; Skinner C; Shobha T; Mehdipour M; Liu H; Huang WC; Lan F; Bray NL; Li S; Corn JE; Kataoka K; Doudna JA; Conboy I; Murthy N, Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng 2017, 1 (11), 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Lee B; Lee K; Panda S; Gonzales-Rojas R; Chong A; Bugay V; Park HM; Brenner R; Murthy N; Lee HY, Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomd. Eng 2018, 2 (7), 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Wang HX; Song ZY; Lao YH; Xu X; Gong J; Cheng D; Chakraborty S; Park JS; Li MQ; Huang DT; Yin LC; Cheng JJ; Leong KW, Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (19), 4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Mout R; Ray M; Tonga GY; Lee YW; Tay T; Sasaki K; Rotello VM, Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 2017, 11 (3), 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wang ZJ; Fu Y; Kang ZZ; Liu XG; Chen N; Wang Q; Tu YQ; Wang LH; Song SP; Ling DS; Song HY; Kong XQ; Fan CH, Organelle-specific triggered release of immunostimulatory oligonucleotides from intrinsically coordinated DNA-metal-organic frameworks with soluble exoskeleton. J. Am. Chem. Soc 2017, 139 (44), 15784–15791. [DOI] [PubMed] [Google Scholar]
- (48).Peng S; Bie BL; Sun YZS; Liu M; Cong HJ; Zhou WT; Xia YC; Tang H; Deng HX; Zhou X, Metal-organic frameworks for precise inclusion of single-stranded DNA and transfection in immune cells. Nat. Commun 2018, 9, 1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Yang XT; Tang Q; Jiang Y; Zhang MN; Wang M; Mao LQ, Nanoscale ATP-responsive zeolitic imidazole framework-90 as a general platform for cytosolic protein delivery and genome editing. J. Am. Chem. Soc 2019, 141 (9), 3782–3786. [DOI] [PubMed] [Google Scholar]
- (50).Mout R; Ray M; Lee YW; Scaletti F; Rotello VM, In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: progress and challenges. Bioconjug. Chem 2017, 28 (4), 880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Yin H; Song CQ; Dorkin JR; Zhu LHJ; Li YX; Wu QQ; Park A; Yang J; Suresh S; Bizhanova A; Gupta A; Bolukbasi MF; Walsh S; Bogorad RL; Gao GP; Weng ZP; Dong YZ; Koteliansky V; Wolfe SA; Langer R; Xue W; Anderson DG, Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol 2016, 34 (3), 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Charlesworth CT; Deshpande PS; Dever DP; Camarena J; Lemgart VT; Cromer MK; Vakulskas CA; Collingwood MA; Zhang LY; Bode NM; Behlke MA; Dejene B; Cieniewicz B; Romano R; Lesch BJ; Gomez-Ospina N; Mantri S; Pavel-Dinu M; Weinberg KI; Porteus MH, Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med 2019, 25 (2), 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Suk JS; Xu QG; Kim N; Hanes J; Ensign LM, PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliver. Rev 2016, 99, 28–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Zhong YA; Meng FH; Deng C; Zhong ZY, Ligand-directed active tumor-targeting polymeric nanoparticles for cancer chemotherapy. Biomacromolecules 2014, 15 (6), 1955–1969. [DOI] [PubMed] [Google Scholar]
- (55).Sun HL; Dong YY; Jan FJ; Zhong ZY, Peptide-decorated polymeric nanomedicines for precision cancer therapy. J. Control. Release 2018, 290, 11–27. [DOI] [PubMed] [Google Scholar]
- (56).Rouet R; Thuma BA; Roy MD; Lintner NG; Rubitski DM; Finley JE; Wisniewska HM; Mendonsa R; Hirsh A; de Onate L; Barron JC; McLellan TJ; Bellenger J; Feng XD; Varghese A; Chrunyk BA; Borzilleri K; Hesp KD; Zhou KH; Ma NN; Tu MH; Dullea R; McClure KF; Wilson RC; Liras S; Mascitti V; Doudna JA, Receptor-mediated delivery of CRISPR-Cas9 endonuclease for cell-type-specific gene editing. J. Am. Chem. Soc 2018, 140 (21), 6596–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Yao PL; Zhang YF; Meng H; Sun HL; Zhong ZY, Smart polymersomes dually functionalized with cRGD and fusogenic GALA peptides enable specific and high-efficiency cytosolic delivery of apoptotic proteins. Biomacromolecules 2019, 20 (1), 184–191. [DOI] [PubMed] [Google Scholar]
- (58).Zhu YQ; Jian Z; Meng FH; Chao D; Ru C; Feijen J; Zhong ZY, cRGD/TAT dual-ligand reversibly cross-linked micelles loaded with docetaxel penetrate deeply into tumor tissue and show high antitumor efficacy in vivo. ACS Appl. Mater. Interfaces 2017, 9 (41), 35651–35663. [DOI] [PubMed] [Google Scholar]
- (59).Akishiba M; Takeuchi T; Kawaguchi Y; Sakamoto K; Yu HH; Nakase I; Takatani-Nakase T; Madani F; Graslund A; Futaki S, Cytosolic antibody delivery by lipid-sensitive endosomolytic peptide. Nat. Chem 2017, 9 (8), 751–761. [DOI] [PubMed] [Google Scholar]
- (60).Colque-Navarro P; Jacobsson G; Andersson R; Flock JI; Mollby R, Levels of antibody against 11 staphylococcus aureus antigens in a healthy population. Clin. Vaccine Immunol 2010, 17 (7), 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Kolata JB; Kuhbandner I; Link C; Normann N; Vu CH; Steil L; Weidenmaier C; Broker BM, The fall of a dogma? Unexpected high T-cell memory response to staphylococcus aureus in humans. J. Infect. Dis 2015, 212 (5), 830–838. [DOI] [PubMed] [Google Scholar]
- (62).Chew WL; Tabebordbar M; Cheng JKW; Mali P; Wu EY; Ng AHM; Zhu KX; Wagers AJ; Church GM, A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 2016, 13 (10), 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Kim D; Kim J; Hur JK; Been KW; Yoon SH; Kim JS, Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol 2016, 34 (8), 863–868. [DOI] [PubMed] [Google Scholar]
- (64).Mali P; Aach J; Stranges PB; Esvelt KM; Moosburner M; Kosuri S; Yang LH; Church GM, CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol 2013, 31 (9), 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Hemphill J; Borchardt EK; Brown K; Asokan A; Deiters A, Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc 2015, 137 (17), 5642–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Dow LE; Fisher J; O’Rourke KP; Muley A; Kastenhuber ER; Livshits G; Tschaharganeh DF; Socci ND; Lowe SW, Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol 2015, 33 (4), 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Liang XQ; Potter J; Kumar S; Zou YF; Quintanilla R; Sridharan M; Carte J; Chen W; Roark N; Ranganathan S; Ravinder N; Chesnut JD, Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol 2015, 208, 44–53. [DOI] [PubMed] [Google Scholar]
- (68).Li YM; Li A; Xu QB, Intracellular delivery of His-tagged genome-editing proteins enabled by nitrilotriacetic acid-containing lipidoid nanoparticles. Adv. Healthc. Mater 2019, 8 (6), 1800996. [DOI] [PMC free article] [PubMed] [Google Scholar]


