Abstract
Tuberculosis, caused by the pathogenic bacterium Mycobacterium tuberculosis (Mtb), is the leading cause of death from an infectious disease, with a mortality rate of over a million people per year. This pathogen's remarkable resilience and infectivity is largely due to its unique waxy cell envelope, 40% of which comprises complex lipids. Therefore, an understanding of the structure and function of the cell wall lipids is of huge indirect clinical significance. This review provides a synopsis of the cell envelope and the major lipids contained within, including structure, biosynthesis and roles in pathogenesis.
Keywords: biosynthesis, cell wall, lipids, mycobacterium tuberculosis, mycolic acids
Introduction
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, was responsible for 1.5 million fatalities worldwide in 2018 [1]. Added to this are the growing concerns of a surge in strains resistant to front-line drugs (multidrug resistant; MDR) and those additionally resistant to second-line drugs (extremely drug resistant; XDR) and all known drugs (totally drug resistant; TDR) [2]. The distinctive cell envelope of Mtb is robust and waxy [3], thanks to the rich variety of lipids present, which bestow on this pathogen resilience [4], a hydrophobic barrier to antibiotic entry [5,6] and key virulence factors [7–10]. In view of these facts and since many of the existing antibiotics available for tuberculosis target the synthesis and transport of these complex lipids, an understanding of lipid structures and synthesis in Mtb is a major goal of current research, with promise of novel effective drug targets.
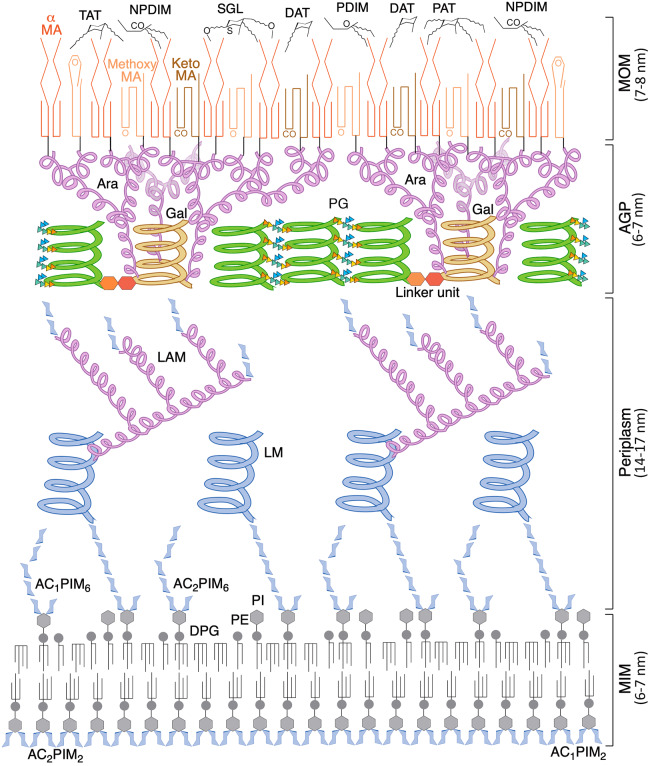
Neither fully Gram-negative nor Gram-positive, the envelope of mycobacteria (Figure 1) is proposed to adopt a remarkable dual membrane structure, with a specialised mycobacterial outer membrane or ‘MOM’, formed as an inner layer of mycolic acids (MAs) and an outer leaflet of free lipids [5,11–13]. While the free lipids are not attached and can be stripped from the envelope, the MA layer is covalently linked to the polysaccharide cell wall [5,11,14], a feature not seen in Gram-negative bacteria. This connection forms part of the mycoloylarabinogalactan–peptidoglycan complex (mAGP), consisting of branched arabinose domains esterified on the outer surface with MAs and linked to the peptidoglycan via the galactan helices of the arabinogalactan (AG) [15–18]. The periplasmic space of Mtb is sizeable, allowing for enzymatic reactions to take place and for the material to be processed and translocated to the MOM [13,19–21]. The inner membrane of Mtb was initially assumed to be conventional, though lipid studies have since revealed that the phospholipid composition is predominantly phosphatidyl-myo-inositol mannosides (PIMs). More than half is present as PIM2 (with two mannose sugars on the inositol moiety), which appears to be exclusively within the inner leaflet of the inner membrane; while PIM6 (six mannose residues) is thought to occupy the outer leaflet [22]. This unusual arrangement increases the content of fatty acid chains in the inner membrane as Ac1/Ac2PIM2 and Ac1/Ac2PIM6, each has three or four (Ac1 or Ac2) acyl chains. The subsequent tight packing of these chains could lead to a less fluid, more stable membrane with reduced permeability to drugs [22], coining a new term mycobacterial inner membrane or ‘MIM’ [23]. The lipoglycans, lipomannan (LM) and lipoarabinomannan (LAM) have lipid anchors based on PIMs and probably project into the periplasm from the MIM. Some models of the Mtb cell envelope depict a capsule as the outermost layer beyond the MOM, which consists mainly of polysaccharides and proteins, with only small quantities of lipids [24]. This has been observed using cryo-electron microscopy to have a depth of ∼30 nm, but only in cultures grown under optimal conditions [25].
Figure 1. Structure of the cell envelope of Mtb.
In the MIM (mycobacterial inner membrane), the inner leaflet mostly consists of AC1/2PIM2 (tri-/tetra-acylated phosphatidyl-myo-inositol-dimannoside). The outer leaflet also contains PIMs, in the form of AC1/2PIM6 (tri-/tetra-acylated phosphatidyl-myo-inositol-hexamannoside), as well as the more conventional phospholipids of the inner membrane, DPG (diphosphatidylglycerol), PE (phosphatidylethanolamine) and PI (phosphatidylinositol). In the periplasm, LM (lipomannan) and LAM (lipoarabinomannan) extend outwards, from lipid anchors in the MIM, though additional anchorage in the outer membrane has been demonstrated, which is contentious; the mannose core helix and individual sugar molecules of the PIMs and mannose caps are coloured blue and the branched arabinan domain is pink. The glycan backbone of the PG (peptidoglycan), green, is represented by helices positioned perpendicular to the membrane, according to the scaffold model [281, 282]. Peptide cross-links between the glycan helices are shown as coloured triangles (orange = l-alanine, yellow = d-isoglutamine, green = meso-diaminopimelate and blue = d-alanine). The PG is connected to the Gal (galactan) domain, yellow, of the arabinogalactan (AG) via a linker unit. The branched Ara (arabinan) domain of the AG, pink, is esterified with MA (mycolic acids), comprising the inner leaflet of the MOM (mycobacterial outer membrane); the typical conformation adopted by each class of MA is represented. The outer leaflet of the MOM is characterised by DAT, TAT and PAT (di-/tri- and pentaacyl trehalose); PDIM and NPDIM (dimycocerosates of phthiocerols and phthiodiolones); and SGL (sulphated trehalose glycolipids) [23,283]. The diagram is roughly to scale using dimensions obtained from cryo-electron microscopy [13].
Mycolic acids
MAs are an abundant feature of the cell envelope of mycobacteria, representing 30% of the dry weight [26]; they provide a hydrophobic permeability barrier to antibiotics [6,27,28] and are essential for viability [29,30] and pathogenesis [31–35]. Characterised as long-chain α-alkyl-β-hydroxy fatty acids (70–90 carbons), most are covalently bound to the AG, forming the inner leaflet of the MOM [5,36]. While two-thirds of the AG termini are esterified with MAs [5,36], some MAs are present as extractable lipids in the outer leaflet of the MOM, mainly linked to trehalose as trehalose dimycolates (TDM) and trehalose monomycolates (TMM), or as free mycolates (Figure 2b). While these are thought to be mostly intermediates in the attachment of the MA to the AG [37], there is evidence to suggest that TDM, the so-called ‘cord-factor’ and most highly expressed glycolipid in Mtb, also has dedicated roles in pathogenicity through interactions with the C-type lectin receptor Mincle on the macrophage, which prevents phagosome acidification and promotes granuloma formation [38–41]. Free mycolates are more abundant in latent phase cells and could also play a role in biofilm formation [42,43].
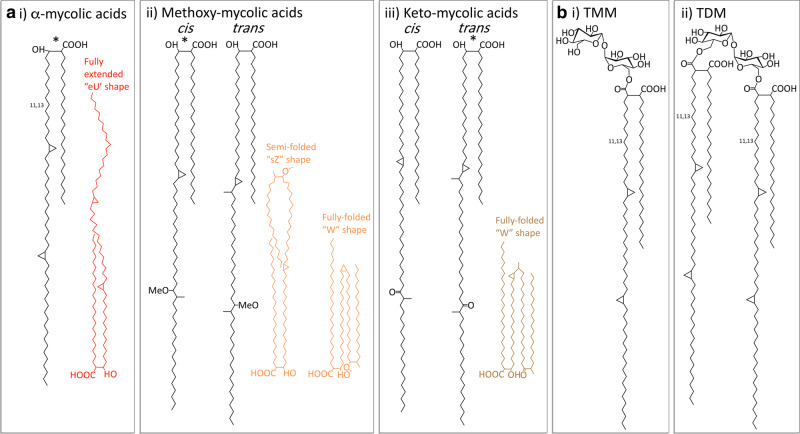
Figure 2. Structure of the major mycolic acid (MA) components from Mtb.
(a) MA structures and the typical conformations adopted, (i) α-MAs, (ii) Methoxy-MAs and (iii) Keto-MAs. The main components are denoted with a *. (b) (i) TMM (trehalose monomycolates) and (ii) TDM (trehalose dimycolates) [284].
MA structure can be separated into two parts: the meromycolate moiety consists of a long-chain meroaldehyde (up to 56 carbons) and usually has two functional groups, whereas the α-branch is a shorter, saturated alkyl chain (24–26 carbons) [3]. There are three classes of MAs in Mtb: α-mycolates are the most abundant and are present with only cis-cyclopropane rings, while oxygenated methoxymycolates and ketomycolates both have subclasses with predominantly either cis- or trans-cyclopropane rings and an adjacent methyl branch [5,6] (Figure 2a).
The MA chains are packed tightly into parallel arrays perpendicular to the surface of the cell envelope, which may be stabilised by hydrogen bonds between the β-OH groups [5,6,36,44]. Within these arrays, the MAs fold into energetically favourable conformations, modulated by the cis-double bonds and the cyclopropane rings, which introduce kinks into the long chains [6]; stabilising hydrophilic interactions are also formed through the oxygen functions [45]. This enables the keto-MAs to adopt a ‘W-form’ conformation, in which four chains are packed together in parallel [45,46]. Though the α-MAs and methoxy-MAs can also form a ‘W’ shape, they are more flexible, particularly the α-MAs, adopting more open forms at higher temperatures (Figure 2a) [47]. This tight packing provides the MA monolayer with appropriate rigidity, leading to a hydrophobic permeability barrier, which could be regulated by the MAs assuming altered conformations depending on their structure and the ambient temperature [45]. Each mycobacterial species has a specific MA profile, suggesting that the different structural elements have an important biological role, both physiologically and in pathogenicity [48].
Free lipids
Phthiocerol dimycocerosate (PDIMs) and phenolic glycolipids (PGLs) (Figure 3a,b) are large ( ∼90 carbons), complex, hydrophobic molecules found in the outer leaflet of the MOM [49,50]. The unusual lipid core consists of a long-chain β-diol (phthiocerol) esterified with two multimethyl branched long-chain fatty acids (mycocerosic acids) [49,51–53]. Whilst possessing a similar lipid core, PGLs differ through a phenolic residue that forms a link between this core and a chain of up to four saccharide units; the composition of the oligosaccharide is species-specific and confers antigenic properties [5]. The stereochemistry of the diol units of PDIMs and PGLs also vary by species; M. marinum and M. ulecerans both have erythro stereochemistry, while all other strains of mycobacteria have the threo configuration [54–56]. PDIMs are produced by all virulent strains of Mtb and have important roles in pathogenicity: forming an impermeable barrier on the outer surface of the cell [57,58], masking the cell from the host's immune system and lysing the phagosome in order to escape the macrophage and disseminate [59,60]. Interestingly, PDIMs give a negative advantage to growth in vitro, leading to their spontaneous loss, though they are never absent from clinical strains [61]. This could be partly due to the hydrophobic barrier that PDIMs form, which though useful in the hostile environment of the host, also prevents the diffusion of useful solutes. PE/PPE proteins, a family named for their N-terminal repeated regions of proline-(proline)-glutamate, have recently been shown to compensate for this, forming pores that selectively uptake the necessary nutrients [58].
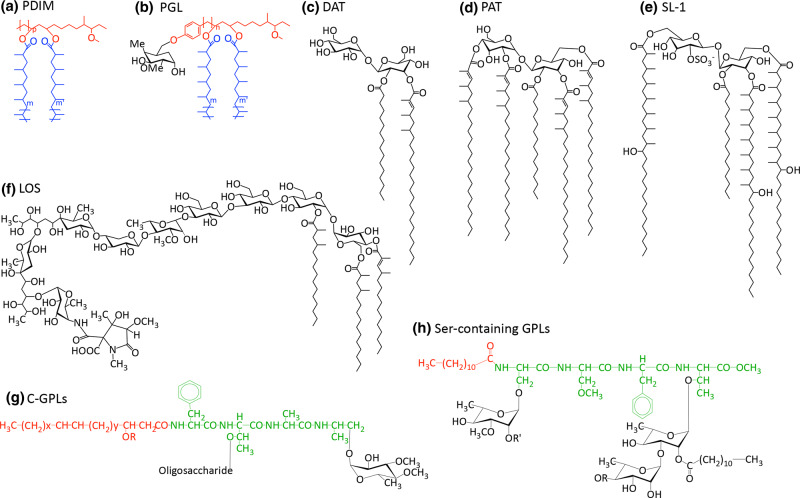
Figure 3. Free lipid structures.
(a) PDIM (Phthiocerol dimycocerosate) and (b) PGL (phenolic glycolipids, general structure): black = saccharide; red = phenolphthiocerol/phthiocerol; blue = multimethyl-branched fatty acids (mycocerosic acids) [285], (c) DAT (diacyl trehaloses) [286], (d) PAT (pentaacyl trehaloses) [286], (e) Sulfolipid-1, SL-1 (sulphated trehalose glycolipid, SGL) [286] (f) Lipooligosaccharides (LOS) from M. marinum [142], (g) C-GPLs (general structure) [103] and (h) Ser-containing GPL from M. xenopi: red = lipid; green = peptide; black = saccharide [106].
PGLs are less prevalent in Mtb isolates [62,63], though are present in the Mtb Canetti strain [63], M. leprae [64], M. marinum [65] and M. bovis [66], and could be responsible for the hyper-virulence and drug resistance of the W-Beijing family of Mtb [8,10]. PGLs affect virulence by suppressing the release of cytokines involved in the inflammatory response of the immune system [8]. In M. leprae, PGLs are also implicated in the Schwann cell invasion [67] and nerve demyelination [68]. The presence of PGLs is directed by a single fused polyketide synthase (PKS), Pks15/1, which is knocked out in strains devoid of PGLs by an insertion that splits the pks15/1 gene in two [62]. PKS proteins are required to condense, extend and modify nascent fatty acids; this particular PKS, Pks15/1, is proposed to extend from the phenol group, producing an intermediate in the synthesis of the phenolphthiocerol moiety, p-hydroxyphenylalkanoate (PHPA) [69]. This is extended by the PpsABCDE PKS proteins to phenolphthiocerol [70]. Mycocerosic acid synthase (MAS), a PKS located in the same cluster as the pps operon, produces the methyl-branched mycocerosic acids [71]. The latter two PKS systems are responsible for the synthesis of the lipid core common to PDIMs and PGLs.
Acyl trehaloses, trehalose glycolipids (SGLs) acylated with multimethyl branched fatty acids, are present in the MOM of all virulent strains of mycobacteria [49,72–74]. As such, they are important in virulence with a role in blocking phagosome maturation [75] and suppressing the host's immune system by inhibiting T cell proliferation and down-regulating cytokine secretion in activated monocytes [76–78]. The core structure consists of a central trehalose moiety that is esterified on the 2-position with a saturated fatty acid (C16 or C18); the trehalose is additionally esterified with as many as four multimethyl branched long-chain fatty acids. The simplest forms are the diacyl trehaloses (DATs) (Figure 3c), which are based on mycosanoic (C24) and mycolipanolic (C27) acids [79]. Pentaacyl trehaloses (PATs) (Figure 3d) contain mycolipenates (C27), whereas sulphated SGLs are made up of the longer fatty acids, phthioceranates (C37) and hydroxyphthioceranates (C40), and the trehalose moiety is sulphated at the 2′-position (Figure 3e) [80,81].
Lipooligosaccharides (LOS) are extremely polar lipids, which are associated with biofilm formation and motility [82]. They consist of a common trehalose-containing saccharide core, acylated on the trehalose moiety with multimethyl branched fatty acids (Figure 3f). The trehalose is further glycosylated with a mono-, or more commonly, an oligosaccharide of 2–6 sugars, which is species-specific [83,84]. LOS are lacking in many Mtb strains, though are present in Mtb Canetti strain [85], M. kansasii [86], M. gordonae, M. smegmatis [87], M. marinum [88,89] and a member of the M. fortuitum complex [90,91], later classified as M. houstonense. Those strains that do produce LOS are characterised by a smooth appearance, whereas those lacking these glycolipids are rough and have increased hydrophobicity over the outer membrane [92,93]. LOS synthesis requires the presence of two proximal pks5 gene homologues flanking an associated pap (PKS associated protein) gene. Strains lacking LOS expression appear to have undergone a recombination-deletion event between the two pks5 homologues, losing a pks5-pap segment [94]. Studies have demonstrated that LOS reduces virulence; smooth M. kanasii was unable to establish the chronic infection in the lungs of mice that their rough counterparts could [92]. This could be due to the observed interference by LOS of host-pathogen interactions, perhaps by covering up critical glycolipids, and it has been speculated that this has driven their loss in virulent strains [94]. The associated hydrophobicity detected in mycobacteria lacking LOS may also have the added benefit to the pathogen of enhancing aerosol transmission [93].
Glycopeptidolipids (GPLs), present in the outer leaflet of non-tuberculosis causing mycobacteria, consist of a lipopeptidyl core, glycosylated on the peptide unit. They are important in biofilm formation, aggregation, motility, cell wall integrity and interactions with the host's immune system [95–98]. Interestingly, strains producing smaller amounts of GPL are more invasive, denoting a role in pathogenicity [99–101]. C-type GPLs (Figure 3g), which form a sub-group that are alkaline-stabile, are found in M. smegmatis, M. avium, M. chelonae and M. abscessus [91,102]; their general structure consists of a core tripeptide-amino alcohol (d-Phe-d-allo-Thr-D-Ala-l-alaninol), esterified with a long fatty acyl chain (C28) [103]. The peptide is glycosylated with a 6-deoxytalose on the allo-threonine and a rhamnose unit linked to the alaninol. This basic core can be further acylated, methylated and glycosylated, depending on the strain, producing different serotypes [95,104,105]. The other group of GPLs, found in M. xenopi, are serine-containing and alkaline-labile, consisting of a tetrapeptide core with two serines (l-Ser-l-Ser-l-Phe-d-allo-Thr), esterified with a simpler fatty acyl chain (C12) [106,107] (Figure 3h). The N-terminal serine residue is methylated, acting as the primary glycosylation site for a 6-deoxytalose unit; a di/tetrasaccharide of rhamnose (terminating in a glucose for the tetrasaccharide) is attached to the allo-threonine [106,107].
Fatty acid and mycolic acid biosynthesis
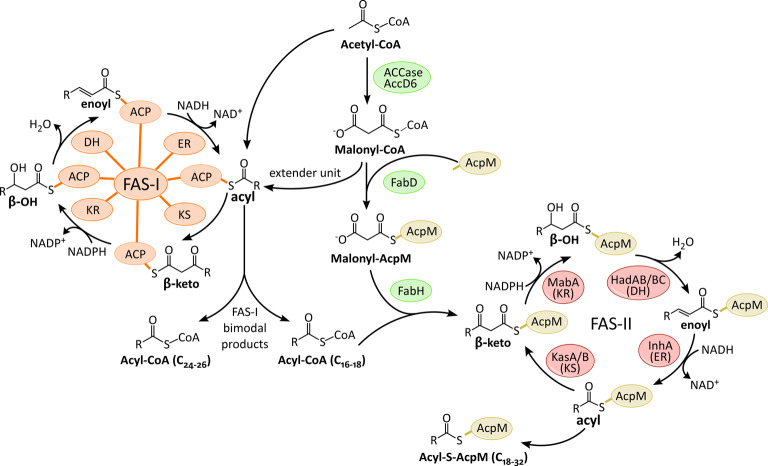
The biosynthesis of fatty acids and MAs is carried out by two fatty acid synthases, FAS-I and FAS-II (Figure 4). FAS-I is a multi-domain ‘eukaryotic-like’ fatty acid synthase, a single large protein comprising all the catalytic domains required for fatty acid synthesis. Whereas FAS-II is a ‘prokaryotic-like’ fatty acid synthase, a large complex of discrete enzymes. FAS-I is the only system capable of synthesising fatty acids de novo and uniquely to mycobacteria, the synthesis is bimodal, generating fatty acids with two different chain lengths: C24–C26 (corresponds to the α-branch in MA synthesis) and C16–C18 (extended by FAS-II to produce the meromycolate chain in MA synthesis) [108–110]. These fatty acids are either shuttled to FAS-II for MA production or to other PKS systems for the further extensions and modifications required to manufacture the various complex lipids.
Figure 4. Biosynthesis of Fatty acids.
For example in the production of mycolic acids (MAs). The FAS-I domains/FAS-II enzymes: KS (β-ketoacyl synthase), KR (β-keto reductase), DH (dehydratase), ER (enoyl reductase) and ACP (acyl carrier protein) [287,288].
Both FASs extend the nascent acyl chain through sequential additions of acetate (2 carbons) from the extender unit malonyl-Coenzyme A (CoA), though FAS-II requires the malonate to be presented on an acyl carrier protein (AcpM; Rv2244) [111]. The first step in fatty acid synthesis is the production of this unit through the carboxylation of acetyl-CoA, catalysed by the acyl-CoA carboxylase complex (ACCase) [112]. FabD (Rv2243), a transacylase, next generates the malonyl-AcpM extender unit required by FAS-II, transferring the malonyl group from the phosphopantetheine group of CoA to the same prosthetic group present on the active site serine of the AcpM [113].
The FAS-I gene present in Mtb (fas; Rv2524c) contains seven domains for fatty acid synthesis: acyl transferase (AT), enoyl reductase (ER), dehydratase (DH), malonyl/palmitoyl transferase (MPT), acyl carrier protein (ACP), β-keto reductase (KR) and β-ketoacyl synthase (KS) [114]. Synthesis is initiated with an acetyl group, which is transferred from the CoA to the ACP domain. During synthesis, the growing acyl chain remains bound to this domain.
In the FAS-II system, the discrete enzymes have corresponding functions to the domains of FAS-I. The first step of synthesis in this system is an important link between FAS-I and FAS-II. FabH (Rv0533c), a β-ketoacyl ACP synthase, plays a pivotal role in initiating FAS-II synthesis through the Claisen condensation of the shorter product from FAS-I (acyl-CoA, C16–18) with a malonyl-AcpM [115]. This forms a β-ketoacyl-AcpM, which can then be presented to the FAS-II enzymes, commencing a round of modifications: reduction in the keto group by the keto reductase (MabA; Rv1483), dehydration of the β-hydroxylacyl-AcpM intermediate by the dehydratase heterodimers (HadAB and HadBC; Rv0635–Rv0636 and Rv0636–Rv0637), and finally reduction in the enoyl-AcpM to acyl-AcpM by the enoyl reductase (InhA; Rv1484) [30,116–118]. After the first cycle, further rounds are initiated by KasA/B (Rv2245 and Rv2246), ketosynthases which then elongate the acyl chain by the addition of two carbons each cycle, forming a β-ketoacyl-AcpM intermediate [119,120]. The completed Acyl-AcpM undergoes further rounds of elongations, desaturations and also modifications, including cis-/trans-cyclopropanation and methoxy/keto group additions [31,34,121,122].
How the FAS systems regulate the length of the growing acyl chains is not clear. Though not essential, mutations in kasB result in MAs shorter by 2–4 carbons, suggesting that KasB is required for full-length extensions [120,123]. The HadBC heterodimer is also associated with later elongation cycles and is similarly expected to play a role in regulating the acyl chain length in FAS-II [117,124]. The mechanism by which FAS-I is able to synthesise two products with different chain lengths (C16–C18 and C24–C26) is likely to be a complex interplay between products and enzymes. Replacing the M. smegmatis FAS-I system with the Mtb homologue, did not alter the length of the longer fatty acid chains produced in M. smegmatis from the usual C24 to the C26 produced by Mtb; this indicates that regulation is not restricted to FAS-I and may involve interactions between FabH and FAS-II with FAS-I [125]. Endogenous polysaccharides also affect FAS-I product formation, favouring the synthesis of shorter chain fatty acids (C16–C18) as well as increasing the overall rate of synthesis; these polysaccharides, containing 3-O-methyl-d-mannose or 6-O-methyl-d-glucose, are proposed to enhance product release, a rate-limiting step in the synthesis, by forming a complex specifically with the shorter length product and thereby facilitating its release [126–128]. In the absence of these polysaccharides, the synthesis of the longer fatty acid chain is favoured (C24–C26), which as longer, shows an increased ability to self-interact, forming stable aggregates that also enable product release from FAS-I [126].
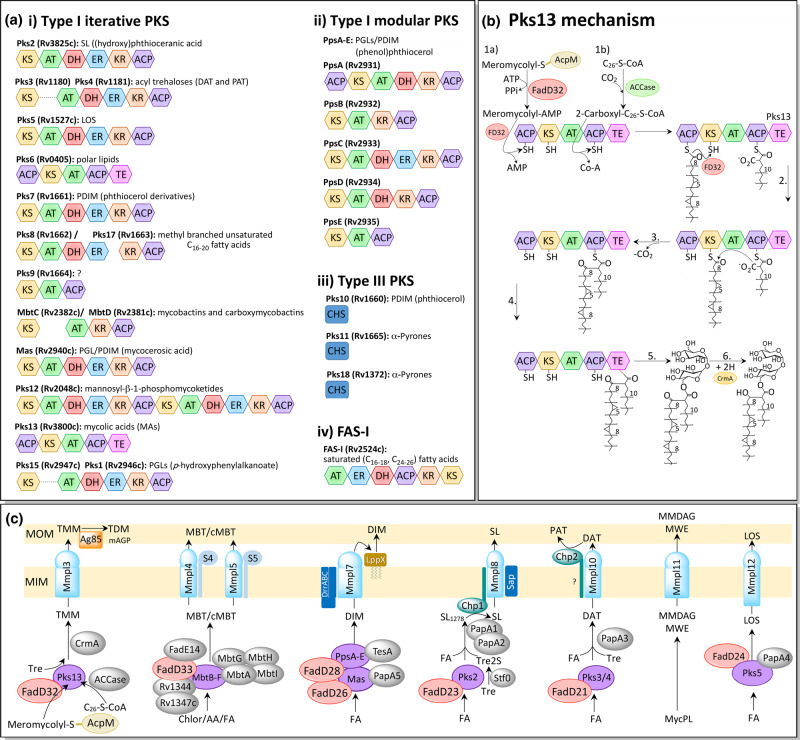
Polyketide synthases (PKS)
The mycobacterial genome boasts a large number of PKS, fundamental in the synthesis of the cell envelope's complex lipids, performing condensations, extensions and modifications of fatty acid chains (Figure 5a) [129]. There are three types of PKS systems, with distinct organisations and mechanisms. Multifunctional type I PKSs are large polypeptides with several catalytic domains. Pks12, for example, is the largest encoded polypeptide in Mtb, at 4151 amino acids [129]. There are two varieties of type I PKSs: iterative systems cycle the product several times through the same domains in a mechanism resembling FAS-I, whereas modular PKSs possess a single domain for each catalytic step of the entire synthesis. Type II PKSs are similar to FAS-II, comprising a large complex of enzymes, each responsible for a catalytic step. There are also three type III plant-like PKSs present in the genome of Mtb [129], condensing enzymes from the chalcone synthase (CHS) superfamily [130]. These enzymes are distinct from the other PKS classes, both in their structure, which is typically a homodimer of KS domains, and mechanism, acting on free acyl-CoA thioesters with no ACP involvement [131].
Figure 5. Polyketide synthase (PKS)/FAS-I domains and synthesis pathways.
(a) Domains. The PKS domains are: AT (acyl transferase; green), ER (enoyl reductase; blue), DH (dehydratase; red), ACP (acyl carrier protein; purple), KR (β-keto reductase; brown), KS (β-ketoacyl synthase; yellow), TE (thioesterase; pink) and CHS (chalcone synthase; dark blue). Other abbreviations: SL = sulfolipids, DAT = diacyl trehaloses, PAT = pentaacyl trehaloses, LOS = lipooligosaccharides, PGL = phenolic glycolipids, PDIM = phthiocerol dimycocerosate [289–291]. (b) Suggested mechanism of Claisen-type condensation by Pks13. 1.a FadD32 (FD32) activates the meromycolyl-S-AcpM with AMP and loads it on the N-terminal ACP (acyl carrier protein) domain of Pks13. 1.b The acyl-CoA (C26) produced by FAS-I is carboxylated by ACCase and loaded onto the C-terminal ACP domain of Pks13 by the AT (acyltransferase) domain. 2. The meromycolyl chain is transferred, in the presence of FD32, to the KS (ketosynthase) domain of PKS13. 3. The Claisen-type condensation reaction: the acidic α-carbon of the carboxyl-acyl group attacks the carbonyl group of the meromycolyl chain, producing 3-oxo-C78-α-mycolate, releasing CO2 and the meromycolyl chain from the KS domain. 4. The TE (thioesterase) domain cleaves the product from the ACP domain, possibly forming a transient covalent bond via an active site serine. 5. The TE domain transfers the product onto a trehalose unit. 6. The keto group is reduced by CmrA, producing the mature trehalose monomycolate (TMM) [138]. (c) MmpL transport of complex lipids across the cytoplasmic membrane in Mtb. PKS (polyketide synthase) proteins are purple, FadD are red and MmpL/S blue. S4 and S5 are MmpS4 and MmpS5, respectively; MmpL4 and 5 are involved in iron scavenging, exporting two siderophores, lipophilic mycobactin (MBT) and hydrophilic carboxymycobactin (cMBT). MmpL11 exports monomero-mycolyl diacylglycerol (MMDAG) and mycolate wax ester (MWE) from the intermediate mycolyl phospholipid (MycPL). Other abbreviations include FA = fatty acid, Tre = trehalose, mAGP = mycoloylarabinogalactan-peptidoglycan [292–294].
PKSs do not carry out de novo fatty acid synthesis, instead using fatty acid precursors from FAS-I. These are usually activated and loaded onto the ACP domain of the PKS by an associated FadD protein. The attached precursor is then modified by the catalytic domains of the PKS, cycling through successive extensions by the KS (β-ketoacyl synthase) domain, usually from a (methyl)malonyl-CoA extender unit via a Claisen-type condensation reaction; reduction in the keto group by the KR (β-keto reductase) domain; dehydration by the DH (dehydratase) domain and reduction in the enoyl group by the ER (enoyl reductase) domain. Some more complex lipids in mycobacteria require several PKSs to synthesise the different lipid components. For instance, in PGL synthesis, Pks15/1 synthesises the PHPA intermediate [62], PpsABCDE extends this to phenolphthiocerol [70] and mycocerosic acid synthase (MAS), produces the methyl-branched mycocerosic acids that are attached to the diols [71].
Pks13, an iterative type I PKS, is involved in the synthesis of MAs, joining together the acyl-CoA (C24–26) product of FAS-I with the meromycolyl-AcpM product from FAS-II, via a Claisen-type condensation reaction (Figure 5b). In this process, both substrates are first attached covalently as thioesters to the ACP domains of Pks13. The meromycolyl-AcpM is activated by FadD32 (Rv3801c) to an AMP derivative and loaded on to the N-terminal ACP of Pks13 [132,133]; the acyl-CoA fatty acid is carboxylated, by an ACCase [134,135], and loaded onto the C-terminal ACP of Pks13, via the AT domain [136]. The meromycolyl group is then transferred, in the presence of FadD32 [136], to the KS domain of Pks13 [132]. The carboxylation step is essential for the Claisen-type reaction that follows, activating the α-carbon and enabling a decarboxylative condensation reaction [137]. This proceeds via a nucleophilic attack by the acidic α-carbon of the carboxyl-acyl group towards the carbonyl group of the meromycolyl chain, releasing CO2 and the meromycolyl chain from the KS domain [138]. The thioesterase (TE) domain of Pks13 next cleaves the α-alkyl β-ketoacyl product from the ACP domain, possibly forming a transient covalent bond with the product via a serine in the active site; the TE domain then catalyses the transfer of the acyl chain onto a trehalose, forming α-alkyl β-ketoacyl trehalose monomycolate (TMMk) [139]. The final mature TMM is formed when the keto group is reduced by CmrA (Rv2509) [140,141].
Trehalose is a significant sugar in mycobacterial lipid production (for review see [142]); in addition to its role in MA synthesis, trehalose constitutes the core of the polyacyl-trehaloses, sulfolipids and LOS [138,143–147]. A disaccharide of α(1 → 1)-linked glucose, trehalose is well-known to protect against the stresses of heat, freezing, desiccation and γ-radiation [148–150]. During MA synthesis, the trehalose could be acting to shield the extreme hydrophobicity of the very long fatty acid chains, enabling export and attachment to the AG. For this reason, it would seem unlikely that the free mycolates found in the MOM, particularly during biofilm formation [42], are released from Pks13 as free mycolates, but perhaps produced and transported as TMM, and the trehalose removed by enzymatic hydrolysis in the outer membrane. After the MA is attached to the AG by the mycolyltransferases of the antigen 85 complex (Ag85) [151], the trehalose is recycled back into the cytoplasm by the ABC sugar transporter, LpqY-SugA-SugB-SugC (Rv1235–Rv1236–Rv1237–Rv1238); this transporter is essential for virulence and again demonstrates the importance of trehalose to the bacterium [152].
MmpL/S
MmpL (mycobacterial membrane proteins large) proteins belong to the RND (resistance, nodulation and cell division) superfamily of membrane proteins, which are typically characterised by 12-transmembrane (TM) helices and two large substrate-specific periplasmic loops between TM1–2 and TM7–8 [153,154]. The energy for their activity is derived from a proton gradient across the inner membrane [155] and they usually function as antiporters, coupling the energy from proton movement into the cytoplasm, with substrate efflux [156,157]. The Mtb genome encodes 13 MmpL proteins, many of which are found in the same loci as PKS proteins and other proteins involved in lipid synthesis [129,158]. Though MmpL5 and MmpL7 have been shown to have roles in drug resistance and efflux [159–161], the primary function of the MmpL proteins is to export the complex lipids synthesised in the cytoplasm across the inner membrane (Figure 5c). Some MmpL proteins (1, 2, 4 and 5) have smaller, transcriptionally coupled accessory proteins, MmpS (mycobacterial membrane proteins small) [129]. Other MmpL proteins also require additional membrane proteins; MmpL8 interacts with the membrane protein, Sap [162] and transport by MmpL7 involves the ABC transporter, DrrABC, and lipoprotein, LppX [57,163].
Knock-out studies have shown that MmpL3 (Rv0206c) is the only essential MmpL protein, though others are required for virulence [158]. MmpL3 is responsible for transporting the MA precursor, TMM, across the inner membrane. Inhibitors that lead to TMM accumulation in the cytoplasm and simultaneous loss of TDM in the periplasm also selected for spontaneous resistance mutations in mmpL3 [164,165], findings that have since been confirmed by a mmpL3 conditional mutant in M. smegmatis [164,166]. Several inhibitors of MmpL3 have recently been found though spontaneous resistance based screening [164,165,167], though some of these are broad-spectrum, targeting bacterial and fungal species lacking MAs, leading to speculation that the real target could be elsewhere. The crystal structure of the homologue from M. smegmatis, however, has demonstrated that many of these inhibitors, including SQ109, do bind directly to MmpL3 [168]. The resolved structure consists of a central proton translocating channel, formed between 2 TM helices (TM4 and TM10) that intertwine and are linked by hydrogen-bonding between two sets of Asp-Tyr residues. These interactions, which are essential for proton relay, were disrupted by binding of the inhibitors, all of which co-crystallised in the central pore [168]. While many RND transporters form homo-dimers or -trimers [168,169–171], and a homology model refined with electron microscopy (EM) suggested MmpL3 is a trimer [172], in the crystal structure MmpL3 was monomeric [168].
MmpL protein expression is controlled by transcriptional repressors, which are in turn regulated by the availability of fatty acids; MmpL3 is regulated by three TetR regulators: Rv3249c, Rv0302 and Rv1816, all of which dissociate from DNA upon binding palmitic acid, relieving transcriptional repression [173–175]. Phosphorylation of the large C-terminal domain possessed by MmpL3 has also been implicated as a post-translational regulation system [176].
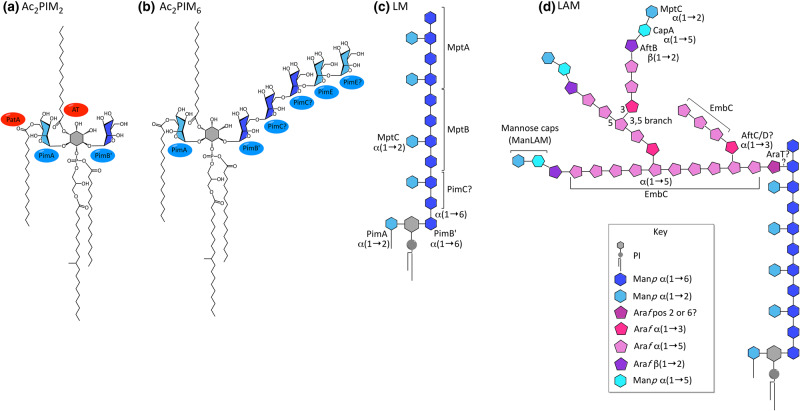
Phosphatidyl-myo-inositol mannosides (PIMs), lipomannan (LM) and lipoarabinomannan (LAM)
These lipoglycans (Figure 6) comprise a phosphatidyl-myo-inositol (PI) core decorated with mannose residues (Manp, pyranose ring form), with additional branches of arabinose (Araf, furanose ring) in LAM. Acyl chains on the inositol, glycerol and mannose moieties provide lipid anchors into the inner and possibly also the outer membranes [25,74,177], though the inner membrane is perhaps their principal location as LM and LAM possess the same lipid anchors as the PIMs, which share this location [22,23]. They are ubiquitous in mycobacteria, with important roles in stability, permeability and cell division [178–180]. LM and LAM are also important in virulence, modulating interactions with the immune system [7,9]; LAM, in particular, plays a key role in the infection life cycle of Mtb, blocking phagosome maturation and therefore avoiding lysis and antigen presentation, enabling Mtb to avoid the host's defences whilst providing a habitable environment in which to replicate [181]. Synthesis proceeds via PIM → LM → LAM [182], yet PIM2 and PIM6 are not only intermediates in the synthesis of LM and LAM, but end products in their own right, forming much of the cytoplasmic membrane, enhancing the stability and reducing drug permeability [22]. In fact, PIM6 is unlikely to be intermediary, possessing two α(1 → 2) linked mannose residues not present in LM and LAM; instead PIM4 is the likely branch point between the two pathways [183]. Structurally, in PIM2 (Figure 6a), the 2 and 6 positions of the myo-inositol ring each have a single linked mannose residue, whereas in PIM6 (Figure 6b), a chain of five mannose units resides at the 6-position in addition to the mannose at position 2 [184]. Both LM and LAM possess a core chain of 21–34 α(1 →6) linked mannose residues from position 6 of the myo-inositol ring, dotted with single α(1 → 2) linked mannose units (Figure 6c) [185]. LAM has an additional arabinan domain of highly branched arabinose residues, similar to AG (Figure 6d) [186]. The non-reducing termini of these arabinose branches can be capped mannose residues to form ManLAM [187].
Figure 6. Structure of phosphatidyl-myo-inositol mannosides, Lipomannan and Lipoarabinomannan.
(a) Ac2PIM2, (b) Ac2PIM6, (c) LM and (d) LAM (and ManLAM). See key for component sugars [23].
The biosynthesis of PIMs, LM and LAM is compartmentalised, which is achieved by the presence of both soluble, membrane-associated glycosyltransferases (GTs) in the cytoplasm (from the GT-A/B superfamily) and membrane-bound GTs (from the GT-C superfamily) in the periplasm [188,189]. This enables the initial glycosylation of the lower PIMs to occur within the cell; the lipid-anchored PI sits on the cytoplasmic face of the inner membrane, with the inositol moiety pointing inwards. Here, mannose is transferred from a GDP-Mannose sugar donor, initially with α(1 → 2) linkage to position 2 of the myo-inositol by PimA (Rv2610c), forming PIM1 [190,191]; and then with α(1 → 6) linkage to position 6 by PimB’ (Rv2188c), forming PIM2 (Figure 6a) [192,193]. An acyl chain (palmitate) is transferred by PatA (Rv2611c) to position 6 of the mannose added by PimA, forming Ac1PIM2, which has three acyl chains [194,195]; position 3 of the myo-inositol ring can also be acylated by an unknown acyl transferase to Ac2PIM2, a tetra-acylated form, though LM and LAM are predominantly triacylated [183]. PIM2 either remains in the inner leaflet of the cytoplasm where it forms the majority of the lipid content [22], or is further mannosylated on position 6 with α(1 → 6) linkages to PIM4. The GT(s) responsible for this has yet to be identified; PimC was found in the genome of Mtb CDC1551 as a possible candidate for the mannosylation to PIM3, though there is no homologue in some strains of mycobacteria (Mtb H37Rv and M. smegmatis) and a knock-out in M. bovis did not reduce the levels of PIMs, LM or LAM, indicating that another pathway must exist [196].
The biosynthesis of the higher PIMs, LM and LAM occurs outside of the cytoplasm, though it is not known whether the juncture for this is PIM2, PIM3 or PIM4 [197]. The PIM is flipped across the inner membrane so that the inositol and mannose moieties now point into the periplasm. A candidate for this transport is the putative ABC transporter, Rv1747, a knock-out of which had low levels of PIMs and showed reduced virulence [198]. However, this mutant still had WT levels of LM and LAM, suggesting the presence of an additional transport mechanism [199]. In the periplasm, membrane-bound GTs transfer further mannose units from the lipid-linked donor, polyprenyl-mannose [182]. The formation of PIM6 from PIM4 (Figure 6b) involves the addition of two α(1 →2) linked mannose residues, the first or both of which are transferred by PimE (Rv1159) [200]. In LM and LAM biosynthesis, the α(1 → 6) linked mannose core is extended from PIM4 by MptB (Rv1459c) [201], followed by MptA (Rv2174), an order determined by knock-outs. The deletion of MptA in C. glutamicum resulted in a truncated LM and halted LAM production, which was complemented by Rv2174 or the C. glutamicum homologue [202]. The deletion of MptB in C. glutamicum completely abolished all LM and LAM synthesis, which was also successfully complemented [201]. A similar knock-out of MptB in M. smegmatis, however, did not affect LM/LAM production, allowing for some redundancy in this pathway [201]. The α(1 → 2) linked mannose residues that decorate the mannose core, producing mature LM, are added by MptC (Rv2181) [203,204], a knock-out of which abolished all LM synthesis in M. smegmatis, though still produced a truncated version of LAM; the immature LM is speculated to be either degraded or processed into LAM [201]. The regulation of the branch point of PIM4 to either PIM6 or LM is controlled by LpqW (Rv1166). Disruption of the corresponding gene in M. smegmatis reduced LM/LAM synthesis, producing an unstable mutant that restored LM/LAM levels through a spontaneous mutation disrupting pimE, which simultaneously abolished the dependent PIM6 pathway [205,206]. A similar deletion in Corynebacterium glutamicum, which lacks a PimE homologue and higher PIMs, was stabilised by a point mutation in mptB. MptB, which is required for the initial mannosylation of the LM core, was subsequently shown to have reduced activity in the absence of LpqW and authors speculate that this mechanism could regulate MptB and subsequently LM synthesis [207]. GDP could also be a regulator, it has been shown to inhibit early PIM synthesis and reduce levels of the polyprenyl-mannose substrate, perhaps by reversing the synthesis of this substrate to GDP-Mannose [197].
The arabinan domain of LAM is similar to that of arabinoagalactan (AG), sharing the same linkages and branched core of 18–22 arabinose units [208], though LAM is more variable in structure [208–211] and is characterised by the presence of more linear chains at the reducing end. AG is terminated by a distinctive branched hexa-arabinoside unit (β-d-Araf(1 → 2)α-d-Araf]2-3,5-α-d-Araf-(1 → 5)-α-d-Araf) [212], whereas LAM also has the linear tetra-arabinoside motif (Araf4; a chain of four α(1 → 5) linked Araf) [186]. This is unsurprising since both domains are constructed by a similar set of membrane-bound arabinosyltransferases, using the lipid-linked donor, decaprenylphosphoryl-d-arabinose (DPA) [213]. During synthesis, the mannan core of the LM is first primed, though the arabinosyltransferase responsible and the location of this attachment are not known; while the O-2 of the mannose was originally implicated [214], more recent studies suggest that the linkage is to the O-6 position [215], which would restrict the attachment of the arabinan domain to the very end of the mannan core. Clarification requires further analysis with longer chain saccharides. After the initial priming, EmbC (Rv3793) adds an arabinan core of 12–15 arabinose residues with α(1 → 5) linkage [208,216,217]. AftC (Rv2673), which has α(1 → 3) activity, introduces 3,5-arabinose branches in both LAM [217] and AG synthesis [218]. AftD (Rv0236c) has been shown to have similar activity to AftC and may also introduce branch points [219], though α(1 → 5) activity in AG synthesis has also been demonstrated [220]. EmbC further extends these branch points by up to 8–9 arabinose units with α(1 → 5) linkage [208–216]. Finally, AftB (Rv3805c) adds the terminal arabinose residue, with β(1 → 2) linkage, to the non-reducing ends of the arabinan [221].
Another difference in the arabinan domains of LAM and AG is the nature of the reducing end. In AG, most are esterified with MAs [5,36], while LAM is capped with mannose, producing ManLAM. This mannose capping is species-specific: in Mtb and other slow-growing virulent strains, the LAM is capped with up to three linear α(1 → 2) linked mannose residues (Manp) on the 5-position of the terminal arabinose [187]; in faster-growing mycobacteria, such as M. smegmatis, much of the LAM is uncapped, while a portion is capped with phosphoinositide (PILAM) [222]; M. leprae has much reduced mannose capping and M. chelonae has none at all [222,223]. Capping is initiated by CapA (Rv1635c), which adds the first mannose residue with α(1 → 5) linkage [224,225]. Further residues are next attached by MptC, the mannosyltransferase responsible for adding the α(1 → 2) mannose to the mannan core [226].
Cell surface labelling, using either biotin followed by lipid extraction or antibodies and cryo-electron microscopy, has shown the presence of arabinan mannan (AM; non-acylated LAM), and also PIMs, LM and LAM, on the surface of the mycobacterial envelope; the PIMs and LM/LAM have lipid anchors and are thought to sit within the lipids of the MOM, projecting into the capsule, while the AM lacks this anchor and is free within the capsular layer [25,74,177]. However, this issue is debateable and not always reproducible; a study using atomic force microscopy with immunogold labelling, for example, was only able to demonstrate the presence of surface-exposed LAM after treatment with drugs that target the cell wall [227]. Also, if PIMs, LM and LAM do indeed locate to the MOM, a yet undiscovered transport system would be required to extract these lipoglycans from their inner membrane anchoring and then relocate them across the periplasmic space and through the lipids of the MOM, a system hypothesised to be similar to the one used by Escherichia coli in the transport of LPS [177,228]. The presence of PIMs, LM and LAM in the MOM would certainly be reassuring, since surface exposure is essential for their observed roles in infection and interactions with the host's immune system, especially since the PIMs, LM and LAM located in the inner membrane were proven not to be surface-exposed [177]. Studies with purified ManLAM and PILAM demonstrated that the mannose cap blocks the fusion of the phagosome and expression of pro-inflammatory cytokines [181,229–231], though these results were not replicated with live bacteria deleted of capA and lacking mannose caps [225]. Similar mutants did, however, demonstrate that ManLAM is recognised by Dectin-2, initiating the expression of pro-inflammatory cytokines [232]. LM has also been shown to activate the Toll-like receptor 2 (TLR2), triggering the innate immune system. This response is proportional to the chain length of the mannan core but is reduced with LAM, possibly because the mannan core is obscured by the arabinan domain [233–236]; truncations of the arabinan domain of LAM restored this activation [217,221]. While initial stimulation of the TLR2 activates the immune response, prolonged stimulation in macrophages has been shown to have the opposite effect, preventing antigen presentation by major histocompatibility complex class II molecules (MHC) [237,238]. This could be a crucial part of the mechanism employed by Mtb to evade the host's immune system and establish a latent phase infection in macrophages [239].
Mycobacterial antigens and CD1 presentation
For many years, it had been assumed that proteins are the sole antigens, activating T cells via the T cell receptor (TCR), and lipids were limited to stimulating the innate immune system. More recently, it has been recognised that many mycobacterial lipids, including mycolates [240], PIM [241–243], LAM [241] and phosphatidylglycerol (PG) [244], activate T cells through the CD1 (cluster of differentiation molecules) presentation pathway (Table 1). CD1 proteins are similar to MHC molecules [245], which present peptide antigens on antigen-presenting cells (APCs) to conventional T cells via the TCR, though CD1 presentation is limited to CD1-restricted T cells. There are five human CD1 proteins: CD1a, -b, -c and -d are membrane proteins involved in presenting lipids and are expressed on APCs, such as B cells, Langerhans cells and dendritic cells as well as cortical thymocytes [246]. CD1e is a soluble protein that processes the glycolipids in order to increase the versatility of the CD1 proteins. CD1e functions by binding directly to the glycolipids and aiding their enzymatic digestion, for example by α-mannosidase in PIM6 presentation by CD1b [243].
Table 1. Human CD1 and the mycobacterial lipids they present [249].
| Human CD1a | Human CD1b | Human CD1c | Human CD1d | Human CD1e |
|---|---|---|---|---|
| Dideoxy-mycobactin [259] | Mycolate [240] | Mannosyl mycoketide [260] | Phosphatidylglycerol [244] | PIM [243] |
| Glucose monomycolate [261,262] | Phosphomycoketide [263] | Cardiolipin [244,264] | ||
| Glycerol monomycolate [253] | Phosphatidylinositol [244,264] | |||
| Sulfoglycolipid [266] | Mannosyl-β-1-phosphoisoprenoids [265] | |||
| PIM [243] | PIM [242] | |||
| LAM [241] | ||||
| Phosphatidylglycerol [267] |
The expression of the CD1 proteins is regulated by the TLR, via interleukin-1β, a pathway that is triggered upon Mtb infection [247,248]. The lipid antigen binds to CD1 either directly at the cell surface or within the endosome, after the antigen or whole bacteria has been phagocytosed into the cell, a mechanism that enables additional processing by CD1e [249]. If binding occurs in the endosome, the CD1-lipid complex is then trafficked to the cell surface in order to present the antigen to the TCR of CD1-restricted T cells [249]. CD1b and CD1c are better adapted to binding to lipids within endosomal compartments [240,250], whereas CD1a is more suited to binding to antigens at the cell surface [251]. Studies have shown that there are higher levels of CD1-restricted T cells present in individuals exposed to Mtb [252,253] and their activation by CD1 presenting molecules is thought to function in the host defences. The T cell response to this activation is not known in humans, though in vitro effector mechanisms have been demonstrated to include the secretion of alpha and gamma interferon and granulysin production, which results in lysis of infected cells and macrophage activation [254–256]. CD1d is recognised by CD1d-restricted natural killer (NK) T cells, which activate both the innate and the adaptive immune system, with responses that include proliferation and cytokine secretion, including gamma interferon and interleukin-4. Invariant NKT cells are also important in granuloma formation and are recruited to the site by lipid antigens, though the evidence suggests that CD1d is not involved in their presentation [257,258].
Conclusions
Mtb commands a complex array of lipids, many of which are crucial to its pathogenicity and survival, with roles in stability, drug permeability, cell division, biofilm formation, infection and host interactions. Many of the lipids present, such as the MAs and the lipid anchors of the PIMs, are packed together tightly in parallel arrangements, a feature that greatly enhances structural stability, while providing a generous hydrophobic barrier that reduces permeability to drugs. Free lipids provide yet more hydrophobic coating, constituting the outer layer of the MOM, an unconventional second membrane. The distinctiveness of the Mtb cell envelope does not end here as the bulk of the complex lipids present form part of much larger complexes, mAGP and LM/LAM, structures essential for viability that are unique to the Corynebacterium genus.
While it might be easy to consider the cell envelope to be a rigid, unchanging entity, it is in fact dynamic. For instance, the conformation of the MAs is temperature dependent. Also the lipid content of the envelope varies according to the infectivity cycle (for review see [268]). PIMs are up-regulated during early infection, while LM and LAM are more likely to predominate during latent phase [43,268]; the high levels of LAM expressed in the granulomas may guard against the host's defences, masking the mannan core [233–235]. Prolonged stimulation of the TLR2 by LM could also assist in establishing and maintaining the granulomas, by inhibiting antigen presentation in the macrophages and thus evading the host's immune response [239]. MAs, as an essential component of the cell wall, are synthesised in line with growth, though their structural composition is thought to vary throughout [268,269]. The TDM and TMM levels, however, decrease in stationary phase, accompanied by an increase in the levels of free mycolates [43], which could be an initial step in biofilm formation [42]. In addition, PDIM is highly synthesised in actively growing cells, but not in stasis [268,269], correlating with the crucial role that it plays during active infection [270].
Many of the proteins involved in lipid synthesis and transport are essential and as such represent ideal drug targets. Indeed, many the current antibiotics already inhibit some of the enzymes involved, including isoniazid (INH) and ethionamide (ETH), both of which target InhA, abolishing MA biosynthesis [271–273]; ethambutol inhibits the arabinosyltransferases involved in assembling the branched arabinan domains of both LAM and AG, blocking MA attachment to the latter [274–278]; the recently approved second-line drugs delamanid and pretomanid also affect MA synthesis, though the exact targets are not known at present [279,280]; and not to mention the plethora of compounds in the pipe-line that look to be inhibiting MA transport by MmpL3 [164,165,167]. Future research into the area would benefit by studying the MmpL proteins, key lipid transporters present in the inner membrane, one of which is essential for survival (MmpL3) and many more are required for pathogenicity. Searching for the enzymes missing from some of the biosynthesis pathways could also prove fruitful, as would structural data and in vitro assay development for all appropriate targets. Today much of the current drug discovery works backwards from the drug to the target; though in the future, as our knowledge of this organism grows and enzyme assays are developed, screening could become increasingly tailored to the target. The cell envelope of Mtb is truly unique, contributing to its triumph as the most successful pathogen of our time. An understanding of its structure and biosynthesis, therefore, is crucial in the search for new drugs to tackle the ever-growing tide of resistance.
Acknowledgements
G.S.B. acknowledges support in the form of a Personal Research Chair from Mr James Bardrick, a Royal Society Wolfson Research Merit Award, The Wellcome Trust (081569/Z/06/Z) and funding from the MRC grants: MR/S000542/1 and MR/R001154/1.
Abbreviations
- ACP
acyl carrier protein
- AG
arabinogalactan
- AT
acyl transferase
- DH
dehydratase
- ER
enoyl reductase
- GPLs
glycopeptidolipids
- GTs
glycosyltransferases
- KR
keto reductase
- KS
ketoacyl synthase
- LAM
lipoarabinomannan
- LM
lipomannan
- LOS
lipooligosaccharides
- mAGP
mycoloylarabinogalactan–peptidoglycan complex
- MAS
mycocerosic acid synthase
- MPT
malonyl/palmitoyl transferase
- PDIMs
phthiocerol dimycocerosate
- PGLs
phenolic glycolipids
- PHPA
p-hydroxyphenylalkanoate
- PIMs
phosphatidyl-myo-inositol mannoside
- PKS
polyketide synthase
- RND
resistance, nodulation and cell division
- SGLs
sulphated trehalose glycolipids
- TCR
T cell receptor
- TDM
trehalose dimycolates
- TLR2
toll-like receptor 2
- TMM
trehalose monomycolates
- XDR
extremely drug resistant
Open Access
Open access for this article was enabled by the participation of University of Birmingham in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Global Tuberculosis Report (2019) World Health Organization, Geneva, Switzerland [Google Scholar]
- 2.Da Silva A., and Palomino P.E. and C J. (2011) Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J. Antimicrob. Chemother. 66, 1417–1430 10.1093/jac/dkr173 [DOI] [PubMed] [Google Scholar]
- 3.Asselineau J. and Lederer E. (1950) Structure of the mycolic acids of Mycobacteria. Nature 166, 782–783 10.1038/166782a0 [DOI] [PubMed] [Google Scholar]
- 4.Ratledge C. (1982) Lipids: cell composition, fatty acid biosynthesis In The Biology of the Mycobacteria (Ratledge C. and Stanford J., eds), pp. 53–94, Academic Press, London, U.K [Google Scholar]
- 5.Minnikin D.E. (1982) Lipids: complex lipids, their chemistry, biosynthesis and role In The Biology of Mycobacteria (Ratledge C. and Stanford J., eds), pp. 95–184, Academic Press, London, U.K [Google Scholar]
- 6.Brennan P.J. and Nikaido H. (1995) The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63 10.1146/annurev.bi.64.070195.000333 [DOI] [PubMed] [Google Scholar]
- 7.Maeda N., Nigou J., Herrmann J.-L., Jackson M., Amara A., Lagrange P.H. et al. (2003) The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J. Biol. Chem. 278, 5513–5516 10.1074/jbc.C200586200 [DOI] [PubMed] [Google Scholar]
- 8.Reed M. B., Domenech P., Manca C., Su H., Barczak A. K., Kreiswirth B. N. et al. (2004) A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431, 84 10.1038/nature02837 [DOI] [PubMed] [Google Scholar]
- 9.Schlesinger L.S., Hull S.R. and Kaufman T.M. (1994) Binding of the terminal mannosyl units of lipoarabinomannan from a virulent strain of Mycobacterium tuberculosis to human macrophages. J. Immunol. 152, 4070–4079 PMID: [PubMed] [Google Scholar]
- 10.Mohandas P., Budell W.C., Mueller E., Au A., Bythrow G.V. and Quadri L.E.N. (2016) Pleiotropic consequences of gene knockouts in the phthiocerol dimycocerosate and phenolic glycolipid biosynthetic gene cluster of the opportunistic human pathogen Mycobacterium marinum. FEMS Microbiol. Lett. 363, fnw016 10.1093/femsle/fnw016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen H., Garton N.J., Horobin R.W., Minnikin D.E. and Barer M.R. (1999) Lipid domains of mycobacteria studied with fluorescent molecular probes. Mol. Microbiol. 31, 1561–1572 10.1046/j.1365-2958.1999.01304.x [DOI] [PubMed] [Google Scholar]
- 12.Barksdale L. and Kim K.S. (1977) Mycobacterium. Bacteriol. Rev. 41, 217–372 10.1128/MMBR.41.1.217-372.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuber B., Chami M., Houssin C., Dubochet J., Griffiths G. and Daffe M. (2008) Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 190, 5672–5680 10.1128/JB.01919-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J., Rosenberg E.Y. and Nikaido H. (1995) Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc. Natl. Acad. Sci. U.S.A. 92, 11254–11258 10.1073/pnas.92.24.11254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barry C.E., Crick D.C. and McNeil M.R. (2007) Targeting the formation of the cell wall core of M. tuberculosis. Infect. Disord. Drug Targets 7, 182–202 10.2174/187152607781001808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crick D.C. and Brennan P.J. (2008) Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11, 107R–118R 10.1093/glycob/11.9.107R [DOI] [PubMed] [Google Scholar]
- 17.Crick D., Chatterjee D., Scherman M.S. and McNeil M.R. (2010) Structure and biosynthesis of the mycobacterial cell wall. Compr. Nat. Prod. II Chem. Biol 6, 381–406 10.1016/B978-008045382-8.00173-8 [DOI] [Google Scholar]
- 18.Daffé M. (2008) The global architecture of the mycobacterial cell envelope In The Mycobacterial Cell Envelope (Daffé M., Reyrat J. and Avenir G., eds), pp. 3–11, ASM Press, Washington, DC [Google Scholar]
- 19.Mishra A.K., Driessen N.N., Appelmelk B.J. and Besra G.S. (2011) Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol. Rev. 35, 1126–1157 10.1111/j.1574-6976.2011.00276.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur D., Guerin M.E., Skovierova H., Brennan P.J. and Jackson M. (2009) Chapter 2: biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv. Appl. Microbiol. 69, 23–78 10.1016/S0065-2164(09)69002-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jankute M., Grover S., Rana A.K. and Besra G.S. (2012) Arabinogalactan and lipoarabinomannan biosynthesis: structure, biogenesis and their potential as drug targets. Future Microbiol. 7, 129–147 10.2217/fmb.11.123 [DOI] [PubMed] [Google Scholar]
- 22.Bansal-Mutalik R. and Nikaido H. (2014) Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc. Natl. Acad. Sci. U.S.A. 111, 4958–4963 10.1073/pnas.1403078111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minnikin D.E., Lee O.Y.-C., Wu H.H.T., Nataraj V., Donoghue H., Ridell M. et al. (2015) Pathophysiological implications of cell envelope structure in Mycobacterium tuberculosis and related taxa In Tuberculosis - Expanding Knowledge (Ribon W., ed.), BoD [Google Scholar]
- 24.Ortalo-Magne A., Dupont M.A., Lemassu A., Andersen A.B., Gounon P. and Daffe M. (1995) Molecular composition of the outermost capsular material of the tubercle bacillus. Microbiology 141, 1609–1620 10.1099/13500872-141-7-1609 [DOI] [PubMed] [Google Scholar]
- 25.Sani M., Houben E.N.G., Geurtsen J., Pierson J., de Punder K., van Zon M., et al. (2010) Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 6, e1000794 10.1371/journal.ppat.1000794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouhi M. (1999) Tuberculosis: a tough adversary. Chem. Eng. News 77, 52–70 10.1021/cen-v077n020.p052 [DOI] [Google Scholar]
- 27.Jarlier V. and Nikaido H. (1994) Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123, 11–18 10.1111/j.1574-6968.1994.tb07194.x [DOI] [PubMed] [Google Scholar]
- 28.Liu J. and Nikaido H. (1999) A mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids accumulates meromycolates. Proc. Natl. Acad. Sci. U.S.A. 96, 4011 10.1073/pnas.96.7.4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhatt A., Kremer L., Dai A.Z., Sacchettini J.C. and Jacobs W.R. (2005) Conditional depletion of KasA, a key enzyme of mycolic acid biosynthesis, leads to mycobacterial cell lysis. J. Bacteriol. 187, 7596 10.1128/JB.187.22.7596-7606.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown A.K., Bhatt A., Singh A., Saparia E., Evans A.F. and Besra G.S. (2007) Identification of the dehydratase component of the mycobacterial mycolic acid-synthesizing fatty acid synthase-II complex. Microbiology 153, 4166–4173 10.1099/mic.0.2007/012419-0 [DOI] [PubMed] [Google Scholar]
- 31.Glickman M.S., Cox J.S. and Jacobs W.R.J. (2000) A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5, 717–727 10.1016/S1097-2765(00)80250-6 [DOI] [PubMed] [Google Scholar]
- 32.Rao V., Gao F., Chen B., Jacobs W.R.J. and Glickman M.S. (2006) Trans-cyclopropanation of mycolic acids on trehalose dimycolate suppresses Mycobacterium tuberculosis-induced inflammation and virulence. J. Clin. Invest. 116, 1660–1667 10.1172/JCI27335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan Y., Zhu Y., Crane D.D. and Barry C.E. III (1998) The effect of oxygenated mycolic acid composition on cell wall function and macrophage growth in Mycobacterium tuberculosis. Mol. Microbiol. 29, 1449–1458 10.1046/j.1365-2958.1998.01026.x [DOI] [PubMed] [Google Scholar]
- 34.Dubnau E., Chan J., Raynaud C., Mohan V.P., Laneelle M.A., Yu K. et al. (2000) Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 36, 630–637 10.1046/j.1365-2958.2000.01882.x [DOI] [PubMed] [Google Scholar]
- 35.Peyron P., Vaubourgeix J., Poquet Y., Levillain F., Botanch C., Bardou F., et al. (2008) Foamy macrophages from tuberculous patients’ granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 4, e1000204 10.1371/journal.ppat.1000204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNeil M., Daffe M. and Brennan P.J. (1991) Location of the mycolyl ester substituents in the cell walls of mycobacteria. J. Biol. Chem. 266, 13217–13223 PMID: [PubMed] [Google Scholar]
- 37.Kremer L. and Besra G.S (2005) Transport processes In Tuberculosis and the Tubercle Bacillus (Cole S.T., Eisenach K.D., McMurray D.N. and Jacobs W.R. Jr, eds), pp. 379–401, American Society for Microbiology, Washington, DC [Google Scholar]
- 38.Hunter R.L., Olsen M.R., Jagannath C. and Actor J.K. (2006) Multiple roles of cord factor in the pathogenesis of primary, secondary, and cavitary tuberculosis, including a revised description of the pathology of secondary disease. Ann. Clin. Lab. Sci. 36, 371–386 PMID: [PubMed] [Google Scholar]
- 39.Indrigo J., Hunter R.L. and Actor J.K. (2002) Influence of trehalose 6,6′-dimycolate (TDM) during mycobacterial infection of bone marrow macrophages. Microbiology 148, 1991–1998 10.1099/00221287-148-7-1991 [DOI] [PubMed] [Google Scholar]
- 40.Ishikawa E., Ishikawa T., Morita Y.S., Toyonaga K., Yamada H., Takeuchi O. et al. (2009) Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin mincle. J. Exp. Med. 206, 2879–2888 10.1084/jem.20091750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patin E.C., Geffken A.C., Willcocks S., Leschczyk C., Haas A., Nimmerjahn F. et al. (2017) Trehalose dimycolate interferes with FcγR-mediated phagosome maturation through Mincle, SHP-1 and FcγRIIB signalling. PLoS One 12, e0174973 10.1371/journal.pone.0174973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ojha A.K., Baughn A.D., Sambandan D., Hsu T., Trivelli X., Guerardel Y. et al. (2008) Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 69, 164–174 10.1111/j.1365-2958.2008.06274.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bacon J., Alderwick L.J., Allnutt J.A., Gabasova E., Watson R., Hatch K.A., et al. (2014) Non-replicating Mycobacterium tuberculosis elicits a reduced infectivity profile with corresponding modifications to the cell wall and extracellular matrix. PLoS One 9, e87329 10.1371/journal.pone.0087329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durand E., Welby M., Laneelle G. and Tocanne J.-F. (1979) Phase behaviour of cord factor and related bacterial glycolipid toxins. Eur. J. Biochem 93, 103–112 10.1111/j.1432-1033.1979.tb12799.x [DOI] [PubMed] [Google Scholar]
- 45.Villeneuve M., Kawai M., Kanashima H., Watanabe M., Minnikin D.E. and Nakahara H. (2005) Temperature dependence of the Langmuir monolayer packing of mycolic acids from Mycobacterium tuberculosis. Biochim. Biophys. Acta 1715, 71–80 10.1016/j.bbamem.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 46.Villeneuve M., Kawai M., Watanabe M., Aoyagi Y., Hitotsuyanagi Y., Takeya K. et al. (2007) Conformational behavior of oxygenated mycobacterial mycolic acids from Mycobacterium bovis BCG. Biochim. Biophys. Acta 1768, 1717–1726 10.1016/j.bbamem.2007.04.003 [DOI] [PubMed] [Google Scholar]
- 47.Groenewald W., Baird M.S., Verschoor J.A., Minnikin D.E. and Croft A.K. (2014) Differential spontaneous folding of mycolic acids from Mycobacterium tuberculosis. Chem. Phys. Lipids 180, 15–22 10.1016/j.chemphyslip.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 48.Minnikin D.E. and Goodfellow M. (1980) Lipid composition in the classification and identification of acid-fast bacteria. Soc. Appl. Bacteriol. Symp. Ser. 8, 189–256 PMID: [PubMed] [Google Scholar]
- 49.Daffe M. and Laneelle M.A. (1988) Distribution of phthiocerol diester, phenolic mycosides and related compounds in mycobacteria. J. Gen. Microbiol. 134, 2049–2055 10.1099/00221287-134-7-2049 [DOI] [PubMed] [Google Scholar]
- 50.Onwueme K.C., Vos C.J., Zurita J., Ferreras J.A. and Quadri L.E.N. (2005) The dimycocerosate ester polyketide virulence factors of mycobacteria. Prog. Lipid. Res. 44, 259–302 10.1016/j.plipres.2005.07.001 [DOI] [PubMed] [Google Scholar]
- 51.Minnikin D.E., Dobson G., Goodfellow M., Magnusson M. and Ridell M. (1985) Distribution of some mycobacterial waxes based on the phthiocerol family. J. Gen. Microbiol. 131, 1375–1381 10.1099/00221287-131-6-1375 [DOI] [PubMed] [Google Scholar]
- 52.Minnikin D.E., Dobson G., Parlett J.H., Goodfellow M. and Magnusson M. (1987) Analysis of dimycocerosates of glycosylphenolphthiocerols in the identification of some clinically significant mycobacteria. Eur. J. Clin. Microbiol. 6, 703–707 10.1007/BF02013082 [DOI] [PubMed] [Google Scholar]
- 53.Brennan P.J. (1988) Mycobacterium and other actinomycetes In Microbial Lipids (Ratledge C. and Wilkinson S.G., eds), pp. 203–298, Academic Press, London, U.K [Google Scholar]
- 54.Besra G.S., Mallet A.I., Minnikin D.E. and Ridell M. (1989) New members of the phthiocerol and phenolphthiocerol families from Mycobacterium marinum. J. Chem. Soc. Chem. Commun., 1451–1452 10.1039/c39890001451 [DOI] [Google Scholar]
- 55.Besra G.S., Minnikin D.E., Sharif A. and Stanford J.L. (1990) Characteristic new members of the phthiocerol and phenolphtiocerol families from Mycorbacterium ulcerans. FEMS Microbiol. Lett. 66, 11–13 10.1111/j.1574-6968.1990.tb03964.x [DOI] [PubMed] [Google Scholar]
- 56.Maskens K., Minnikin D.E. and Polgar N. (1966) Studies relating to phthiocerol. Part VI. Stereochemical studies. J. Chem. Soc. Chem. Commun., 2113–2115 10.1039/J39660002107 [DOI] [Google Scholar]
- 57.Camacho L.R., Constant P., Raynaud C., Laneelle M.A., Triccas J.A., Gicquel B. et al. (2001) Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276, 19845–19854 10.1074/jbc.M100662200 [DOI] [PubMed] [Google Scholar]
- 58.Wang Q., Boshoff H.I.M., Harrison J.R., Ray P.C., Green S.R., Wyatt P.G. et al. (2020) PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis. Science 367, 1147–1151 10.1126/science.aav5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murry J.P., Pandey A.K., Sassetti C.M. and Rubin E.J. (2009) Phthiocerol dimycocerosate transport is required for resisting interferon-γ-independent immunity. J. Infect. Dis. 200, 774–782 10.1086/605128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quigley J., Hughitt V.K., Velikovsky C.A., Mariuzza R.A., El-Sayed N.M. and Briken V. (2017) The cell wall lipid PDIM contributes to phagosomal escape and host cell exit of Mycobacterium tuberculosis. mBio 8, e00148-17 10.1128/mBio.00148-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Domenech P. and Reed M.B. (2009) Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: implications for virulence studies. Microbiology 155, 3532–3543 10.1099/mic.0.029199-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Constant P., Perez E., Malaga W., Laneelle M.-A., Saurel O., Daffe M. et al. (2002) Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. Evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J. Biol. Chem. 277, 38148–38158 10.1074/jbc.M206538200 [DOI] [PubMed] [Google Scholar]
- 63.Daffé M., Lacave C., Lanéelle M.-A. and Lanéelle G. (1987) Structure of the major triglycosyl phenol-phthiocerol of Mycobacterium tuberculosis (strain Canetti). Eur. J. Biochem. 167, 155–160 10.1111/j.1432-1033.1987.tb13317.x [DOI] [PubMed] [Google Scholar]
- 64.Hunter S.W. and Brennan P.J. (1981) A novel phenolic glycolipid from Mycobacterium leprae possibly involved in immunogenicity and pathogenicity. J. Bacteriol. 147, 728–735 10.1128/JB.147.3.728-735.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Villé C. and Gastambide-Odier M. (1970) Le 3-O-méthyl-l-rhamnose, Sucre isolé du mycoside G de Mycobacterium marinum. Carbohydr. Res. 12, 97–107 10.1016/S0008-6215(00)80229-3 [DOI] [Google Scholar]
- 66.Jarnagin J.L., Brennan P.J. and Harris S.K. (1983) Rapid identification of Mycobacterium bovis by a thin-layer chromatographic technique. Am. J. Vet. Res. 44, 1920–1922 PMID: [PubMed] [Google Scholar]
- 67.Ng V., Zanazzi G., Timpl R., Talts J.F., Salzer J.L., Brennan P.J. et al. (2000) Role of the cell wall phenolic glycolipid-1 in the peripheral nerve predilection of Mycobacterium leprae. Cell 103, 511–524 10.1016/S0092-8674(00)00142-2 [DOI] [PubMed] [Google Scholar]
- 68.Rambukkana A., Zanazzi G., Tapinos N. and Salzer J.L. (2002) Contact-dependent demyelination by Mycobacterium leprae in the absence of immune cells. Science 296, 927–931 10.1126/science.1067631 [DOI] [PubMed] [Google Scholar]
- 69.He W., Soll C.E., Chavadi S.S., Zhang G., Warren J.D. and Quadri L.E.N. (2009) Cooperation between a coenzyme A-independent stand-alone initiation module and an iterative type I polyketide synthase during synthesis of mycobacterial phenolic glycolipids. J. Am. Chem. Soc. 131, 16744–16750 10.1021/ja904792q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Azad A.K., Sirakova T.D., Fernandes N.D. and Kolattukudy P.E. (1997) Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J. Biol. Chem. 272, 16741–16745 10.1074/jbc.272.27.16741 [DOI] [PubMed] [Google Scholar]
- 71.Trivedi O.A., Arora P., Vats A., Ansari M.Z., Tickoo R., Sridharan V. et al. (2005) Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol. Cell 17, 631–643 10.1016/j.molcel.2005.02.009 [DOI] [PubMed] [Google Scholar]
- 72.Fujiwara N. (1997) [Distribution of antigenic glycolipids among Mycobacterium tuberculosis strains and their contribution to virulence]. Kekkaku 72, 193–205 PMID: [PubMed] [Google Scholar]
- 73.Munoz M., Laneelle M.A., Luquin M., Torrelles J., Julian E., Ausina V. et al. (1997) Occurrence of an antigenic triacyl trehalose in clinical isolates and reference strains of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 157, 251–259 10.1016/S0378-1097(97)00483-7 [DOI] [PubMed] [Google Scholar]
- 74.Ortalo-Magné A., Lemassu A., Lanéelle M.A., Bardou F., Silve G., Gounon P., Marchal G. and Daffé M. (1996) Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J. Bacteriol. 178, 456–461 10.1128/JB.178.2.456-461.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brodin P., Poquet Y., Levillain F., Peguillet I., Larrouy-Maumus G., Gilleron M., et al. (2010) High content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog. 6, e1001100 10.1371/journal.ppat.1001100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee K.-S., Dubey V.S., Kolattukudy P.E., Song C.-H., Shin A.-R., Jung S.-B., et al. (2007) Diacyltrehalose of Mycobacterium tuberculosis inhibits lipopolysaccharide- and mycobacteria-induced proinflammatory cytokine production in human monocytic cells. FEMS. Microbiol. Lett. 267, 121–128 10.1111/j.1574-6968.2006.00553.x [DOI] [PubMed] [Google Scholar]
- 77.Saavedra R., Segura E., Leyva R., Esparza L.A. and López-Marı´n L.M. (2001) Mycobacterial Di-O-Acyl-Trehalose inhibits mitogen- and antigen-induced proliferation of murine T cells in vitro. Clin. Diagn. Lab. Immunol. 8, 1081 10.1128/CDLI.8.6.1-91-1088.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saavedra R., Segura E., Tenorio E.P. and Lopez-Marin L.M. (2006) Mycobacterial trehalose-containing glycolipid with immunomodulatory activity on human CD4+ and CD8+ T-cells. Microbes Infect. 8, 533–540 10.1016/j.micinf.2005.08.005 [DOI] [PubMed] [Google Scholar]
- 79.Besra G.S., Bolton R.C., McNeil M.R., Ridell M., Simpson K.E., Glushka J. et al. (1992) Structural elucidation of a novel family of acyltrehaloses from Mycobacterium tuberculosis. Biochemistry 31, 9832–9837 10.1021/bi00155a040 [DOI] [PubMed] [Google Scholar]
- 80.Bertozzi C. and Schelle M (2008) Sulfated Metabolites from M. tuberculosis: sulfolipid-1 and beyond In The Mycobacterial Cell Envelope (Daffe M. and Reyrat J.-M., eds), pp. 291–304, American Society for Microbiology, Washington, DC [Google Scholar]
- 81.Goren M.B. and Brennan P.J. (1979) Mycobacterial lipids: chemistry and biologic activities In Tuberculosis (Youmans G.P., ed.), pp. 63–193, W.B. Saunders Company, Philadelphia, PA [Google Scholar]
- 82.Ren H., Dover L.G., Islam S.T., Alexander D.C., Chen J.M., Besra G.S. et al. (2007) Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum. Mol. Microbiol. 63, 1345–1359 10.1111/j.1365-2958.2007.05603.x [DOI] [PubMed] [Google Scholar]
- 83.Hunter S.W., Jardine I., Yanagihara D.L. and Brennan P.J. (1985) Trehalose-containing lipooligosaccharides from mycobacteria: structures of the oligosaccharide segments and recognition of a unique N-acylkanosamine-containing epitope. Biochemistry 24, 2798–2805 PMID: [DOI] [PubMed] [Google Scholar]
- 84.Gilleron M., Vercauteren J. and Puzo G. (1994) Lipo-oligosaccharidic antigen from Mycobacterium gastri. Complete structure of a novel C4-branched 3,6-dideoxy-.alpha.-xylo-hexopyranose. Biochemistry 33, 1930–1937 10.1021/bi00173a041 [DOI] [PubMed] [Google Scholar]
- 85.Daffe M., McNeil M. and Brennan P.J. (1991) Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry 30, 378–388 10.1021/bi00216a011 [DOI] [PubMed] [Google Scholar]
- 86.Hunter S.W., Murphy R.C., Clay K., Goren M.B. and Brennan P.J. (1983) Trehalose-containing lipooligosaccharides. A new class of species-specific antigens from Mycobacterium. J. Biol. Chem. 258, 10481–10487 [PubMed] [Google Scholar]
- 87.Saadat S. and Ballou C.E. (1983) Pyruvylated glycolipids from Mycobacterium smegmatis. Structures of two oligosaccharide components. J. Biol. Chem. 258, 1813–1818 PMID: [PubMed] [Google Scholar]
- 88.Minnikin D.E., Ridell M., Wallerstrom G., Besra G.S., Parlett J.H., Bolton R.C. et al. (1989) Comparative studies of antigenic glycolipids of mycobacteria related to the leprosy bacillus. Acta Leprol. 7, 51–54 PMID: [PubMed] [Google Scholar]
- 89.Burguière A., Hitchen P.G., Dover L.G., Kremer L., Ridell M., Alexander D.C. et al. (2005) Losa, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum. J. Biol. Chem. 280, 42124–42133 10.1074/jbc.M507500200 [DOI] [PubMed] [Google Scholar]
- 90.Besra G.S., McNeil M.R. and Brennan P.J. (1992) Characterization of the specific antigenicity of Mycobacterium fortuitum. Biochemistry 31, 6504–6509 10.1021/bi00143a021 [DOI] [PubMed] [Google Scholar]
- 91.Tsang A.Y., Barr V.L., McClatchy J.K., Goldberg M., Drupa I. and Brennan P.J. (1984) Antigenic relationships of the Mycobacterium fortuitum-Mycobacterium chelonae complex. Int. J. Syst. Bacteriol. 34, 35–44 10.1099/00207713-34-1-35 [DOI] [Google Scholar]
- 92.Belisle J.T. and Brennan P.J. (1989) Chemical basis of rough and smooth variation in mycobacteria. J. Bacteriol. 171, 3465–3470 10.1128/JB.171.6.3465-3470.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jankute M., Nataraj V., Lee O.Y.-C., Wu H.H.T., Ridell M., Garton N.J. et al. (2017) The role of hydrophobicity in tuberculosis evolution and pathogenicity. Sci. Rep. 7, 1315 10.1038/s41598-017-01501-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boritsch E.C., Frigui W., Cascioferro A., Malaga W., Etienne G., Laval F., et al. (2016) pks5-recombination-mediated surface remodelling in Mycobacterium tuberculosis emergence. Nat. Microbiol. 1, 15019 10.1038/nmicrobiol.2015.19 [DOI] [PubMed] [Google Scholar]
- 95.Villeneuve C., Etienne G., Abadie V., Montrozier H., Bordier C., Laval F. et al. (2003) Surface-exposed glycopeptidolipids of Mycobacterium smegmatis specifically inhibit the phagocytosis of mycobacteria by human macrophages. identification of a novel family of glycopeptidolipids. J. Biol. Chem. 278, 51291–51300 10.1074/jbc.M306554200 [DOI] [PubMed] [Google Scholar]
- 96.Etienne G., Villeneuve C., Billman-Jacobe H., Astarie-Dequeker C., Dupont M.-A. and Daffe M. (2002) The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology 148, 3089–3100 10.1099/00221287-148-10-3089 [DOI] [PubMed] [Google Scholar]
- 97.Recht J., Martinez A., Torello S. and Kolter R. (2000) Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 182, 4348–4351 10.1128/JB.182.15.4348-4351.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barrow W.W., Davis T.L., Wright E.L., Labrousse V., Bachelet M. and Rastogi N. (1995) Immunomodulatory spectrum of lipids associated with Mycobacterium avium serovar 8. Infect. Immun. 63, 126–133 10.1128/IAI.63.1.126-133.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Byrd T.F. and Lyons C.R. (1999) Preliminary characterization of a Mycobacterium abscessus mutant in human and murine models of infection. Infect. Immun. 67, 4700–4707 10.1128/IAI.67.9.4700-4707.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Howard S.T., Rhoades E., Recht J., Pang X., Alsup A., Kolter R. et al. (2006) Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 152, 1581–1590 10.1099/mic.0.28625-0 [DOI] [PubMed] [Google Scholar]
- 101.Catherinot E., Clarissou J., Etienne G., Ripoll F., Emile J.-F., Daffe M. et al. (2007) Hypervirulence of a rough variant of the Mycobacterium abscessus type strain. Infect. Immun. 75, 1055–1058 10.1128/IAI.00835-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bozic C.M., McNeil M., Chatterjee D., Jardine I. and Brennan P.J. (1988) Further novel amido sugars within the glycopeptidolipid antigens of Mycobacterium avium. J. Biol. Chem. 263, 14984–14991 PMID: [PubMed] [Google Scholar]
- 103.Laneelle G. and Asselineau J. (1968) Structure d'un glycoside de peptidolipide isolé d'une mycobactérie. Eur. J. Biochem. 5, 487–491 10.1111/j.1432-1033.1968.tb00396.x [DOI] [PubMed] [Google Scholar]
- 104.Patterson J.H., McConville M.J., Haites R.E., Coppel R.L. and Billman-Jacobe H. (2000) Identification of a methyltransferase from Mycobacterium smegmatis involved in glycopeptidolipid synthesis. J. Biol. Chem. 275, 24900–24906 10.1074/jbc.M000147200 [DOI] [PubMed] [Google Scholar]
- 105.Ripoll F., Deshayes C., Pasek S., Laval F., Beretti J.-L., Biet F., et al. (2007) Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. BMC Genom. 8, 114 10.1186/1471-2164-8-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Besra G.S., McNeil M.R., Rivoire B., Khoo K.H., Morris H.R., Dell A. et al. (1993) Further structural definition of a new family of glycopeptidolipids from Mycobacterium xenopi. Biochemistry 32, 347–355 10.1021/bi00052a043 [DOI] [PubMed] [Google Scholar]
- 107.Riviere M. and Puzo G. (1991) A new type of serine-containing glycopeptidolipid from Mycobacterium xenopi. J. Biol. Chem. 266, 9057–9063 PMID: [PubMed] [Google Scholar]
- 108.Peterson D.O. and Bloch K. (1977) Mycobacterium smegmatis fatty acid synthetase. Long chain transacylase chain length specificity. J. Biol. Chem. 252, 5735–5739 PMID: [PubMed] [Google Scholar]
- 109.Kikuchi S., Rainwater D.L. and Kolattukudy P.E. (1992) Purification and characterization of an unusually large fatty acid synthase from Mycobacterium tuberculosis var. bovis BCG. Arch. Biochem. Biophys. 295, 318–326 10.1016/0003-9861(92)90524-Z [DOI] [PubMed] [Google Scholar]
- 110.Bloch K. (1977) Control mechanisms for fatty acid synthesis in Mycobacterium smegmatis. Adv. Enzymol. Relat. Areas Mol. Biol. 45, 1–84 10.1002/9780470122907.ch1 [DOI] [PubMed] [Google Scholar]
- 111.Mdluli K., Slayden R.A., Zhu Y., Ramaswamy S., Pan X., Mead D. et al. (1998) Inhibition of a Mycobacterium tuberculosis β-Ketoacyl ACP synthase by isoniazid. Science 280, 1607 10.1126/science.280.5369.1607 [DOI] [PubMed] [Google Scholar]
- 112.Cronan J.E.J. and Waldrop G.L. (2002) Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 41, 407–435 10.1016/S0163-7827(02)00007-3 [DOI] [PubMed] [Google Scholar]
- 113.Kremer L., Nampoothiri K.M., Lesjean S., Dover L.G., Graham S., Betts J. et al. (2001) Biochemical characterization of acyl carrier protein (AcpM) and malonyl-CoA:AcpM transacylase (mtFabD), two major components of Mycobacterium tuberculosis fatty acid synthase II. J. Biol. Chem. 276, 27967–27974 10.1074/jbc.M103687200 [DOI] [PubMed] [Google Scholar]
- 114.Fernandes N.D. and Kolattukudy P.E. (1996) Cloning, sequencing and characterization of a fatty acid synthase-encoding gene from Mycobacterium tuberculosis var. bovis BCG. Gene 170, 95–99 10.1016/0378-1119(95)00842-X [DOI] [PubMed] [Google Scholar]
- 115.Choi K.H., Kremer L., Besra G.S. and Rock C.O. (2000) Identification and substrate specificity of beta -ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis. J. Biol. Chem. 275, 28201–28207 10.1074/jbc.M003241200 [DOI] [PubMed] [Google Scholar]
- 116.Marrakchi H., Ducasse S., Labesse G., Montrozier H., Margeat E., Emorine L. et al. (2002) Maba (FabG1), a Mycobacterium tuberculosis protein involved in the long-chain fatty acid elongation system FAS-II. Microbiology 148, 951–960 10.1099/00221287-148-4-951 [DOI] [PubMed] [Google Scholar]
- 117.Sacco E., Covarrubias A.S., O'Hare H.M., Carroll P., Eynard N., Jones T.A. et al. (2007) The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 104, 14628–14633 10.1073/pnas.0704132104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Banerjee A., Dubnau E., Quemard A., Balasubramanian V., Um K.S., Wilson T. et al. (1994) Inha, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263, 227–230 10.1126/science.8284673 [DOI] [PubMed] [Google Scholar]
- 119.Schaeffer M.L., Agnihotri G., Volker C., Kallender H., Brennan P.J. and Lonsdale J.T. (2001) Purification and biochemical characterization of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthases KasA and KasB. J. Biol. Chem. 276, 47029–47037 10.1074/jbc.M108903200 [DOI] [PubMed] [Google Scholar]
- 120.Kremer L., Dover L.G., Carrere S., Nampoothiri K.M., Lesjean S., Brown A.K. et al. (2002) Mycolic acid biosynthesis and enzymic characterization of the beta-ketoacyl-ACP synthase A-condensing enzyme from Mycobacterium tuberculosis. Biochem. J. 364, 423–430 10.1042/bj20011628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Glickman M.S. (2003) The mmaA2 gene of Mycobacterium tuberculosis encodes the distal cyclopropane synthase of the alpha-mycolic acid. J. Biol. Chem. 278, 7844–7849 10.1074/jbc.M212458200 [DOI] [PubMed] [Google Scholar]
- 122.Barkan D., Rao V., Sukenick G.D. and Glickman M.S. (2010) Redundant function of cmaA2 and mmaA2 in mycobacterium tuberculosis cis cyclopropanation of oxygenated mycolates. J. Bacteriol. 192, 3661–3668 10.1128/JB.00312-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Slayden R.A. and Barry C.E. III (2002) The role of KasA and KasB in the biosynthesis of meromycolic acids and isoniazid resistance in Mycobacterium tuberculosis. Tuberc. Edinb. Scotl. 82, 149–160 10.1054/tube.2002.0333 [DOI] [PubMed] [Google Scholar]
- 124.Marrakchi H., Bardou F. and Daffé M (2008) A comprehensive overview of mycolic acid structure and biosynthesis In The Mycobacterial Cell Envelop (Daffe M. and Reyrat J.-M., eds), pp. 41–62, American Society for Microbiology, Washington, DC [Google Scholar]
- 125.Zimhony O., Vilcheze C. and Jacobs W.R.J. (2004) Characterization of Mycobacterium smegmatis expressing the Mycobacterium tuberculosis fatty acid synthase I (fas1) gene. J. Bacteriol. 186, 4051–4055 10.1128/JB.186.13.4051-4055.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Papaioannou N., Cheon H.-S., Lian Y. and Kishi Y. (2007) Product-regulation mechanisms for fatty acid biosynthesis catalyzed by Mycobacterium smegmatis FAS I. Chembiochem 8, 1775–1780 10.1002/cbic.200700380 [DOI] [PubMed] [Google Scholar]
- 127.Banis R.J., Peterson D.O. and Bloch K. (1977) Mycobacterium smegmatis fatty acid synthetase. Polysaccharide stimulation of the rate-limiting step. J. Biol. Chem. 252, 5740–5744 PMID: [PubMed] [Google Scholar]
- 128.Wood W.I., Peterson D.O. and Bloch K. (1977) Mycobacterium smegmatis fatty acid synthetase. A mechanism based on steady state rates and product distributions. J. Biol. Chem. 252, 5745–5749 [PubMed] [Google Scholar]
- 129.Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 130.Moore B. and Hopke J. (2001) Discovery of a new bacterial polyketide biosynthetic pathway. Chembiochem 2, 35–38 [DOI] [PubMed] [Google Scholar]
- 131.Schröder J. (1997) A family of plant-specific polyketide synthases: facts and predictions. Trends Plant Sci. 2, 373–378 10.1016/S1360-1385(97)87121-X [DOI] [Google Scholar]
- 132.Trivedi O.A., Arora P., Sridharan V., Tickoo R., Mohanty D. and Gokhale R.S. (2004) Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature 428, 441–445 10.1038/nature02384 [DOI] [PubMed] [Google Scholar]
- 133.Léger M., Gavalda S., Guillet V., van der Rest B., Slama N., Montrozier H. et al. (2009) The dual function of the Mycobacterium tuberculosis FadD32 required for mycolic acid biosynthesis. Chem. Biol. 16, 510–519 10.1016/j.chembiol.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 134.Gande R., Gibson K.J.C., Brown A.K., Krumbach K., Dover L.G., Sahm H. et al. (2004) Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 279, 44847–44857 10.1074/jbc.M408648200 [DOI] [PubMed] [Google Scholar]
- 135.Portevin D., de Sousa-D'Auria C., Montrozier H., Houssin C., Stella A., Lanéelle M.-A. et al. (2005) The Acyl-AMP ligase FadD32 and AccD4-containing Acyl-CoA carboxylase are required for the synthesis of mycolic acids and essential for mycobacterial growth: identification of the carboxylation product and determination of the Acyl-CoA carboxylase components. J. Biol. Chem. 280, 8862–8874 10.1074/jbc.M408578200 [DOI] [PubMed] [Google Scholar]
- 136.Gavalda S., Leger M., van der Rest B., Stella A., Bardou F., Montrozier H., et al. (2009) The Pks13/FadD32 crosstalk for the biosynthesis of mycolic acids in Mycobacterium tuberculosis. J. Biol. Chem. 284, 19255–19264 10.1074/jbc.M109.006940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gande R., Dover L.G., Krumbach K., Besra G.S., Sahm H., Oikawa T. et al. (2007) The two carboxylases of Corynebacterium glutamicum essential for fatty acid and mycolic acid synthesis. J. Bacteriol. 189, 5257 10.1128/JB.00254-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Takayama K., Wang C. and Besra G.S. (2005) Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 18, 81 10.1128/CMR.18.1.81-101.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gavalda S., Bardou F., Laval F., Bon C., Malaga W., Chalut C. et al. (2014) The polyketide synthase Pks13 catalyzes a novel mechanism of lipid transfer in Mycobacteria. Chem. Biol. 21, 1660–1669 10.1016/j.chembiol.2014.10.011 [DOI] [PubMed] [Google Scholar]
- 140.Bhatt A., Brown A.K., Singh A., Minnikin D.E. and Besra G.S. (2008) Loss of a mycobacterial gene encoding a reductase leads to an altered cell wall containing beta-oxo-mycolic acid analogs and accumulation of ketones. Chem. Biol. 15, 930–939 10.1016/j.chembiol.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lea-Smith D.J., Pyke J.S., Tull D., McConville M.J., Coppel R.L. and Crellin P.K. (2007) The reductase that catalyzes mycolic motif synthesis is required for efficient attachment of mycolic acids to arabinogalactan. J. Biol. Chem. 282, 11000–11008 10.1074/jbc.M608686200 [DOI] [PubMed] [Google Scholar]
- 142.Nobre A., Alarico S., Maranha A., Mendes V. and Empadinhas N. (2014) The molecular biology of Mycobacterial trehalose in the quest for advanced tuberculosis therapies. Microbiology 160, 1547–1570 10.1099/mic.0.075895-0 [DOI] [PubMed] [Google Scholar]
- 143.Hatzios S.K., Schelle M.W., Holsclaw C.M., Behrens C.R., Botyanszki Z., Lin F. et al. (2009) Papa3 is an acyltransferase required for polyacyltrehalose biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 284, 12745–12751 10.1074/jbc.M809088200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kumar P., Schelle M.W., Jain M., Lin F.L., Petzold C.J., Leavell M.D. et al. (2007) Papa1 and PapA2 are acyltransferases essential for the biosynthesis of the Mycobacterium tuberculosis virulence factor sulfolipid-1. Proc. Natl. Acad. Sci. U.S.A. 104, 11221 10.1073/pnas.0611649104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jackson M., Stadthagen G. and Gicquel B. (2007) Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis: biosynthesis, transport, regulation and biological activities. Tuberc. Edinb. Scotl. 87, 78–86 10.1016/j.tube.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 146.Bhatt K., Gurcha S.S., Bhatt A., Besra G.S. and Jacobs W.R.J. (2007) Two polyketide-synthase-associated acyltransferases are required for sulfolipid biosynthesis in Mycobacterium tuberculosis. Microbiology 153, 513–520 10.1099/mic.0.2006/003103-0 [DOI] [PubMed] [Google Scholar]
- 147.Besra G.S., Khoo K.H., Belisle J.T., McNeil M.R., Morris H.R., Dell A. et al. (1994) New pyruvylated, glycosylated acyltrehaloses from Mycobacterium smegmatis strains, and their implications for phage resistance in mycobacteria. Carbohydr. Res. 251, 99–114 10.1016/0008-6215(94)84279-5 [DOI] [PubMed] [Google Scholar]
- 148.Lee J., Lin E.-W., Lau U.Y., Hedrick J.L., Bat E. and Maynard H.D. (2013) Trehalose glycopolymers as excipients for protein stabilization. Biomacromolecules 14, 2561–2569 10.1021/bm4003046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Iturriaga G., Suarez R. and Nova-Franco B. (2009) Trehalose metabolism: from osmoprotection to signaling. Int. J. Mol. Sci. 10, 3793–3810 10.3390/ijms10093793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Webb K.M. and DiRuggiero J. (2012) Role of Mn2+ and compatible solutes in the radiation resistance of thermophilic bacteria and archaea. Archaea 2012, 845756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Belisle J.T., Vissa V.D., Sievert T., Takayama K., Brennan P.J. and Besra G.S. (1997) Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420 10.1126/science.276.5317.1420 [DOI] [PubMed] [Google Scholar]
- 152.Kalscheuer R., Weinrick B., Veeraraghavan U., Besra G.S. and Jacobs W.R.J. (2010) Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 107, 21761–21766 10.1073/pnas.1014642108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Saier M.H.J., Tam R., Reizer A. and Reizer J. (1994) Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol. Microbiol. 11, 841–847 10.1111/j.1365-2958.1994.tb00362.x [DOI] [PubMed] [Google Scholar]
- 154.Chim N., Torres R., Liu Y., Capri J., Batot G., Whitelegge J.P. et al. (2015) The structure and interactions of periplasmic domains of crucial MmpL membrane proteins from Mycobacterium tuberculosis. Chem. Biol. 22, 1098–1107 10.1016/j.chembiol.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ma D., Cook D.N., Alberti M., Pon N.G., Nikaido H. and Hearst J.E. (1993) Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol 175, 6299–6313 10.1128/JB.175.19.6299-6313.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Seeger M.A., Schiefner A., Eicher T., Verrey F., Diederichs K. and Pos K.M. (2006) Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313, 1295–1298 10.1126/science.1131542 [DOI] [PubMed] [Google Scholar]
- 157.Murakami S., Nakashima R., Yamashita E., Matsumoto T. and Yamaguchi A. (2006) Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179 10.1038/nature05076 [DOI] [PubMed] [Google Scholar]
- 158.Domenech P., Reed M.B. and Barry C.E. III (2005) Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73, 3492–3501 10.1128/IAI.73.6.3492-3501.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Milano A., Pasca M.R., Provvedi R., Lucarelli A.P., Manina G., Luisa de Jesus Lopes Ribeiro A. et al. (2009) Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5–MmpL5 efflux system. Tuberculosis 89, 84–90 10.1016/j.tube.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 160.Halloum I., Viljoen A., Khanna V., Craig D., Bouchier C., Brosch R. et al. (2017) Resistance to thiacetazone derivatives active against Mycobacterium abscessus involves mutations in the MmpL5 transcriptional repressor MAB_4384. Antimicrob. Agents Chemother. 61, e02509-02516 10.1128/AAC.02509-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hartkoorn R.C., Uplekar S. and Cole S.T. (2014) Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 2979–2981 10.1128/AAC.00037-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Seeliger J.C., Holsclaw C.M., Schelle M.W., Botyanszki Z., Gilmore S.A., Tully S.E. et al. (2012) Elucidation and chemical modulation of sulfolipid-1 biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 287, 7990–8000 10.1074/jbc.M111.315473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sulzenbacher G., Canaan S., Bordat Y., Neyrolles O., Stadthagen G., Roig-Zamboni V., et al. (2006) Lppx is a lipoprotein required for the translocation of phthiocerol dimycocerosates to the surface of Mycobacterium tuberculosis. EMBO J. 25, 1436–1444 10.1038/sj.emboj.7601048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Grzegorzewicz A.E., Pham H., Gundi V.A.K.B., Scherman M.S., North E.J., Hess T., et al. (2012) Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 8, 334–341 10.1038/nchembio.794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tahlan K., Wilson R., Kastrinsky D.B., Arora K., Nair V., Fischer E. et al. (2012) SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56, 1797 10.1128/AAC.05708-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Varela C., Rittmann D., Singh A., Krumbach K., Bhatt K., Eggeling L. et al. (2012) Mmpl genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chem. Biol. 19, 498–506 10.1016/j.chembiol.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.La Rosa V., Poce G., Canseco J.O., Buroni S., Pasca M.R., Biava M. et al. (2012) Mmpl3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob. Agents Chemother. 56, 324 10.1128/AAC.05270-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang B., Li J., Yang X., Wu L., Zhang J., Yang Y., et al. (2019) Crystal structures of membrane transporter MmpL3, an anti-TB drug target. Cell 176, 636–648.e13 10.1016/j.cell.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 169.Sennhauser G., Bukowska M.A., Briand C. and Grutter M.G. (2009) Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 389, 134–145 10.1016/j.jmb.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 170.Murakami S., Nakashima R., Yamashita E. and Yamaguchi A. (2002) Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419, 587–593 10.1038/nature01050 [DOI] [PubMed] [Google Scholar]
- 171.Kumar N., Su C.-C., Chou T.-H., Radhakrishnan A., Delmar J.A., Rajashankar K.R. et al. (2017) Crystal structures of the Burkholderia multivorans hopanoid transporter HpnN. Proc. Natl. Acad. Sci. U.S.A. 114, 6557–6562 10.1073/pnas.1619660114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Belardinelli J.M., Yazidi A., Yang L., Fabre L., Li W., Jacques B., et al. (2016) Structure-function profile of MmpL3, the essential mycolic acid transporter from Mycobacterium tuberculosis. ACS Infect. Dis. 2, 702–713 10.1021/acsinfecdis.6b00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Delmar J.A., Chou T.-H., Wright C.C., Licon M.H., Doh J.K., Radhakrishnan A., et al. (2015) Structural basis for the regulation of the MmpL transporters of Mycobacterium tuberculosis. J. Biol. Chem. 290, 28559–28574 10.1074/jbc.M115.683797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chou T.-H., Delmar J.A., Wright C.C., Kumar N., Radhakrishnan A., Doh J.K., et al. (2015) Crystal structure of the Mycobacterium tuberculosis transcriptional regulator Rv0302. Protein Sci. 24, 1942–1955 10.1002/pro.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Galagan J.E., Minch K., Peterson M., Lyubetskaya A., Azizi E., Sweet L., et al. (2013) The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183 10.1038/nature12337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Prisic S., Dankwa S., Schwartz D., Chou M.F., Locasale J.W., Kang C.-M. et al. (2010) Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. U.S.A. 107, 7521–7526 10.1073/pnas.0913482107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pitarque S., Larrouy-Maumus G., Payre B., Jackson M., Puzo G. and Nigou J. (2008) The immunomodulatory lipoglycans, lipoarabinomannan and lipomannan, are exposed at the mycobacterial cell surface. Tuberculosis (Edinb) 88, 560–565 10.1016/j.tube.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Parish T., Liu J., Nikaido H. and Stoker N.G. (1997) A Mycobacterium smegmatis mutant with a defective inositol monophosphate phosphatase gene homolog has altered cell envelope permeability. J. Bacteriol. 179, 7827 10.1128/JB.179.24.7827-7833.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Patterson J.H., Waller R.F., Jeevarajah D., Billman-Jacobe H. and McConville M.J. (2003) Mannose metabolism is required for mycobacterial growth. Biochem. J. 372, 77–86 10.1042/bj20021700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fukuda T., Matsumura T., Ato M., Hamasaki M., Nishiuchi Y., Murakami Y., et al. (2013) Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis. MBio 4, e00472–e00412 10.1128/mBio.00472-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fratti R.A., Chua J., Vergne I. and Deretic V. (2003) Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc. Natl. Acad. Sci. U.S.A. 100, 5437–5442 10.1073/pnas.0737613100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Besra G.S., Morehouse C.B., Rittner C.M., Waechter C.J. and Brennan P.J. (1997) Biosynthesis of mycobacterial lipoarabinomannan. J. Biol. Chem. 272, 18460–18466 10.1074/jbc.272.29.18460 [DOI] [PubMed] [Google Scholar]
- 183.Khoo K.H., Dell A., Morris H.R., Brennan P.J. and Chatterjee D. (1995) Structural definition of acylated phosphatidylinositol mannosides from Mycobacterium tuberculosis: definition of a common anchor for lipomannan and lipoarabinomannan. Glycobiology 5, 117–127 10.1093/glycob/5.1.117 [DOI] [PubMed] [Google Scholar]
- 184.Severn W.B., Furneaux R.H., Falshaw R. and Atkinson P.H. (1998) Chemical and spectroscopic characterisation of the phosphatidylinositol manno-oligosaccharides from Mycobacterium bovis AN5 and WAg201 and Mycobacterium smegmatis mc2 155. Carbohydr. Res. 308, 397–408 10.1016/S0008-6215(98)00108-6 [DOI] [PubMed] [Google Scholar]
- 185.Hunter S.W. and Brennan P.J. (1990) Evidence for the presence of a phosphatidylinositol anchor on the lipoarabinomannan and lipomannan of Mycobacterium tuberculosis. J. Biol. Chem. 265, 9272–9279 PMID: [PubMed] [Google Scholar]
- 186.Chatterjee D., Bozic C.M., McNeil M. and Brennan P.J. (1991) Structural features of the arabinan component of the lipoarabinomannan of Mycobacterium tuberculosis. J. Biol. Chem. 266, 9652–9660 PMID: [PubMed] [Google Scholar]
- 187.Chatterjee D., Lowell K., Rivoire B., McNeil M.R. and Brennan P.J. (1992) Lipoarabinomannan of Mycobacterium tuberculosis. Capping with mannosyl residues in some strains. J. Biol. Chem. 267, 6234–6239 PMID: [PubMed] [Google Scholar]
- 188.Liu J. and Mushegian A. (2003) Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 12, 1418–1431 10.1110/ps.0302103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Morita Y.S., Velasquez R., Taig E., Waller R.F., Patterson J.H., Tull D. et al. (2005) Compartmentalization of lipid biosynthesis in Mycobacteria. J. Biol. Chem. 280, 21645–21652 10.1074/jbc.M414181200 [DOI] [PubMed] [Google Scholar]
- 190.Hill D.L. and Ballou C.E. (1966) Biosynthesis of mannophospholipids by Mycobacterium phlei. J. Biol. Chem. 241, 895–902 PMID: [PubMed] [Google Scholar]
- 191.Guerin M.E., Kaur D., Somashekar B.S., Gibbs S., Gest P., Chatterjee D. et al. (2009) New insights into the early steps of phosphatidylinositol mannoside biosynthesis in mycobacteria: PimB’ is an essential enzyme of Mycobacterium smegmatis. J. Biol. Chem. 284, 25687–25696 10.1074/jbc.M109.030593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kordulakova J., Gilleron M., Mikusova K., Puzo G., Brennan P.J., Gicquel B. et al. (2002) Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis. PimA is essential for growth of mycobacteria. J. Biol. Chem. 277, 31335–31344 10.1074/jbc.M204060200 [DOI] [PubMed] [Google Scholar]
- 193.Lea-Smith D.J., Martin K.L., Pyke J.S., Tull D., McConville M.J., Coppel R.L. et al. (2008) Analysis of a new mannosyltransferase required for the synthesis of phosphatidylinositol mannosides and lipoarbinomannan reveals two lipomannan pools in corynebacterineae. J. Biol. Chem. 283, 6773–6782 10.1074/jbc.M707139200 [DOI] [PubMed] [Google Scholar]
- 194.Korduláková J., Gilleron M., Puzo G., Brennan P.J., Gicquel B., Mikušová K. et al. (2003) Identification of the required acyltransferase step in the biosynthesis of the phosphatidylinositol mannosides of Mycobacterium species. J. Biol. Chem. 278, 36285–36295 10.1074/jbc.M303639200 [DOI] [PubMed] [Google Scholar]
- 195.Albesa-Jove D., Svetlikova Z., Tersa M., Sancho-Vaello E., Carreras-Gonzalez A., Bonnet P., et al. (2016) Structural basis for selective recognition of acyl chains by the membrane-associated acyltransferase PatA. Nat. Commun. 7, 10906 10.1038/ncomms10906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kremer L., Gurcha S.S., Bifani P., Hitchen P.G., Baulard A., Morris H.R. et al. (2002) Characterization of a putative alpha-mannosyltransferase involved in phosphatidylinositol trimannoside biosynthesis in Mycobacterium tuberculosis. Biochem. J. 363, 437–447 10.1042/bj3630437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Morita Y.S., Patterson J.H., Billman-Jacobe H. and McConville M.J. (2004) Biosynthesis of mycobacterial phosphatidylinositol mannosides. Biochem. J. 378, 589–597 10.1042/bj20031372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Glass L.N., Swapna G., Chavadi S.S., Tufariello J.M., Mi K., Drumm J.E., et al. (2017) Mycobacterium tuberculosis universal stress protein Rv2623 interacts with the putative ATP binding cassette (ABC) transporter Rv1747 to regulate mycobacterial growth. PLoS Pathog. 13, e1006515–e1006515 10.1371/journal.ppat.1006515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Spivey V.L., Whalan R.H., Hirst E.M.A., Smerdon S.J. and Buxton R.S. (2013) An attenuated mutant of the Rv1747 ATP-binding cassette transporter of Mycobacterium tuberculosis and a mutant of its cognate kinase, PknF, show increased expression of the efflux pump-related iniBAC operon. FEMS Microbiol. Lett. 347, 107–115 10.1111/1574-6968.12230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Morita Y.S., Sena C.B.C., Waller R.F., Kurokawa K., Sernee M.F., Nakatani F. et al. (2006) Pime is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J. Biol. Chem. 281, 25143–25155 10.1074/jbc.M604214200 [DOI] [PubMed] [Google Scholar]
- 201.Mishra A.K., Alderwick L.J., Rittmann D., Wang C., Bhatt A., Jacobs Jr W.R. et al. (2008) Identification of a novel α(1→6) mannopyranosyltransferase MptB from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan- and lipoarabinomannan-deficient mutant. Mol. Microbiol. 68, 1595–1613 10.1111/j.1365-2958.2008.06265.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mishra A.K., Alderwick L.J., Rittmann D., Tatituri R.V.V., Nigou J., Gilleron M. et al. (2007) Identification of an α(1→6) mannopyranosyltransferase (MptA), involved in Corynebacterium glutamicum lipomanann biosynthesis, and identification of its orthologue in Mycobacterium tuberculosis. Mol. Microbiol. 65, 1503–1517 10.1111/j.1365-2958.2007.05884.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mishra A.K., Krumbach K., Rittmann D., Appelmelk B., Pathak V., Pathak A.K. et al. (2011) Lipoarabinomannan biosynthesis in Corynebacterineae: the interplay of two α(1→2)-mannopyranosyltransferases MptC and MptD in mannan branching. Mol. Microbiol. 80, 1241–1259 10.1111/j.1365-2958.2011.07640.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kaur D., Berg S., Dinadayala P., Gicquel B., Chatterjee D., McNeil M.R. et al. (2006) Biosynthesis of mycobacterial lipoarabinomannan: Role of a branching mannosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 103, 13664 10.1073/pnas.0603049103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kovacevic S., Anderson D., Morita Y.S., Patterson J., Haites R., McMillan B.N.I. et al. (2006) Identification of a novel protein with a role in lipoarabinomannan biosynthesis in mycobacteria. J. Biol. Chem. 281, 9011–9017 10.1074/jbc.M511709200 [DOI] [PubMed] [Google Scholar]
- 206.Crellin P.K., Kovacevic S., Martin K.L., Brammananth R., Morita Y.S., Billman-Jacobe H. et al. (2008) Mutations in pimE restore lipoarabinomannan synthesis and growth in a Mycobacterium smegmatis lpqW mutant. J. Bacteriol. 190, 3690–3699 10.1128/JB.00200-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rainczuk A.K., Yamaryo-Botte Y., Brammananth R., Stinear T.P., Seemann T., Coppel R.L. et al. (2012) The lipoprotein LpqW is essential for the mannosylation of periplasmic glycolipids in Corynebacteria. J. Biol. Chem. 287, 42726–42738 10.1074/jbc.M112.373415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shi L., Berg S., Lee A., Spencer J.S., Zhang J., Vissa V. et al. (2006) The carboxy terminus of embC from Mycobacterium smegmatis mediates chain length extension of the arabinan in lipoarabinomannan. J. Biol. Chem. 281, 19512–19526 10.1074/jbc.M513846200 [DOI] [PubMed] [Google Scholar]
- 209.Treumann A., Xidong F., McDonnell L., Derrick P.J., Ashcroft A.E., Chatterjee D. et al. (2002) 5-methylthiopentose: a new substituent on lipoarabinomannan in Mycobacterium tuberculosis. J. Mol. Biol. 316, 89–100 10.1006/jmbi.2001.5317 [DOI] [PubMed] [Google Scholar]
- 210.Torrelles J.B., Khoo K.-H., Sieling P.A., Modlin R.L., Zhang N., Marques A.M. et al. (2004) Truncated structural variants of lipoarabinomannan in Mycobacterium leprae and an ethambutol-resistant strain of Mycobacterium tuberculosis. J. Biol. Chem. 279, 41227–41239 10.1074/jbc.M405180200 [DOI] [PubMed] [Google Scholar]
- 211.Turnbull W.B., Shimizu K.H., Chatterjee D., Homans S.W. and Treumann A. (2004) Identification of the 5-methylthiopentosyl substituent in Mycobacterium tuberculosis lipoarabinomannan. Angew. Chem. Int. Ed. 43, 3918–3922 10.1002/anie.200454119 [DOI] [PubMed] [Google Scholar]
- 212.McNeil M.R., Robuck K.G., Harter M. and Brennan P.J. (1994) Enzymatic evidence for the presence of a critical terminal hexa-arabinoside in the cell walls of Mycobacterium tuberculosis. Glycobiology 4, 165–174 10.1093/glycob/4.2.165 [DOI] [PubMed] [Google Scholar]
- 213.Wolucka B.A., McNeil M.R., de Hoffmann E., Chojnacki T. and Brennan P.J. (1994) Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J. Biol. Chem. 269, 23328–23335 PMID: [PubMed] [Google Scholar]
- 214.Chatterjee D., Khoo K.H., McNeil M.R., Dell A., Morris H.R. and Brennan P.J. (1993) Structural definition of the non-reducing termini of mannose-capped LAM from Mycobacterium tuberculosis through selective enzymatic degradation and fast atom bombardment-mass spectrometry. Glycobiology 3, 497–506 10.1093/glycob/3.5.497 [DOI] [PubMed] [Google Scholar]
- 215.Angala S.K., McNeil M.R., Zou L., Liav A., Zhang J., Lowary T.L. et al. (2016) Identification of a novel mycobacterial arabinosyltransferase activity which adds an arabinosyl residue to α-d-mannosyl residues. ACS Chem. Biol. 11, 1518–1524 10.1021/acschembio.6b00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhang N., Torrelles J.B., McNeil M.R., Escuyer V.E., Khoo K.-H., Brennan P.J. et al. (2003) The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol. Microbiol. 50, 69–76 10.1046/j.1365-2958.2003.03681.x [DOI] [PubMed] [Google Scholar]
- 217.Birch H.L., Alderwick L.J., Appelmelk B.J., Maaskant J., Bhatt A., Singh A. et al. (2010) A truncated lipoglycan from mycobacteria with altered immunological properties. Proc. Natl. Acad. Sci. U.S.A. 107, 2634–2639 10.1073/pnas.0915082107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Birch H.L., Alderwick L.J., Bhatt A., Rittmann D., Krumbach K., Singh A. et al. (2008) Biosynthesis of mycobacterial arabinogalactan: identification of a novel alpha(1 → 3) arabinofuranosyltransferase. Mol. Microbiol. 69, 1191–1206 10.1111/j.1365-2958.2008.06354.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Skovierova H., Larrouy-Maumus G., Zhang J., Kaur D., Barilone N., Kordulakova J., et al. (2009) Aftd, a novel essential arabinofuranosyltransferase from mycobacteria. Glycobiology 19, 1235–1247 10.1093/glycob/cwp116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Alderwick L.J., Birch H.L., Krumbach K., Bott M., Eggeling L. and Besra G.S. (2018) Aftd functions as an α1 → 5 arabinofuranosyltransferase involved in the biosynthesis of the mycobacterial cell wall core. Cell Surf. Amst. Neth. 1, 2–14 10.1016/j.tcsw.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Jankute M., Alderwick L.J., Noack S., Veerapen N., Nigou J. and Besra G.S. (2017) Disruption of Mycobacterial AftB results in complete loss of terminal β(1 → 2) arabinofuranose residues of lipoarabinomannan. ACS Chem. Biol. 12, 183–190 10.1021/acschembio.6b00898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Khoo K.-H., Dell A., Morris H.R., Brennan P.J. and Chatterjee D. (1995) Inositol phosphate capping of the nonreducing termini of lipoarabinomannan from rapidly growing strains of Mycobacterium. J. Biol. Chem. 270, 12380–12389 10.1074/jbc.270.21.12380 [DOI] [PubMed] [Google Scholar]
- 223.Guérardel Y., Maes E., Elass E., Leroy Y., Timmerman P., Besra G.S. et al. (2002) Structural study of lipomannan and lipoarabinomannan from Mycobacterium chelonae : presence of unusual components with α1,3-mannopyranose side chainS. J. Biol. Chem. 277, 30635–30648 10.1074/jbc.M204398200 [DOI] [PubMed] [Google Scholar]
- 224.Dinadayala P., Kaur D., Berg S., Amin A.G., Vissa V.D., Chatterjee D. et al. (2006) Genetic basis for the synthesis of the immunomodulatory mannose caps of lipoarabinomannan in Mycobacterium tuberculosis. J. Biol. Chem. 281, 20027–20035 10.1074/jbc.M603395200 [DOI] [PubMed] [Google Scholar]
- 225.Appelmelk B.J., den Dunnen J., Driessen N.N., Ummels R., Pak M., Nigou J. et al. (2008) The mannose cap of mycobacterial lipoarabinomannan does not dominate the Mycobacterium-host interaction. Cell. Microbiol. 10, 930–944 10.1111/j.1462-5822.2007.01097.x [DOI] [PubMed] [Google Scholar]
- 226.Kaur D., Obregon-Henao A., Pham H., Chatterjee D., Brennan P.J. and Jackson M. (2008) Lipoarabinomannan of Mycobacterium: mannose capping by a multifunctional terminal mannosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 105, 17973–17977 10.1073/pnas.0807761105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Alsteens D., Verbelen C., Dague E., Raze D., Baulard A.R. and Dufrene Y.F. (2008) Organization of the mycobacterial cell wall: a nanoscale view. Pflugers Arch. 456, 117–125 10.1007/s00424-007-0386-0 [DOI] [PubMed] [Google Scholar]
- 228.Dhiman R.K., Dinadayala P., Ryan G.J., Lenaerts A.J., Schenkel A.R. and Crick D.C. (2011) Lipoarabinomannan localization and abundance during growth of Mycobacterium smegmatis. J. Bacteriol. 193, 5802–5809 10.1128/JB.05299-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Vergne I., Chua J. and Deretic V. (2003) Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3 K hVPS34 cascade. J. Exp. Med. 198, 653–659 10.1084/jem.20030527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Knutson K.L., Hmama Z., Herrera-Velit P., Rochford R. and Reiner N.E. (1998) Lipoarabinomannan of Mycobacterium tuberculosis promotes protein tyrosine dephosphorylation and inhibition of mitogen-activated protein kinase in human mononuclear phagocytes. Role of the Src homology 2 containing tyrosine phosphatase. J. Biol. Chem. 273, 645–652 10.1074/jbc.273.1.645 [DOI] [PubMed] [Google Scholar]
- 231.Nigou J., Zelle-Rieser C., Gilleron M., Thurnher M. and Puzo G. (2001) Mannosylated lipoarabinomannans inhibit IL-12 production by human dendritic cells: evidence for a negative signal delivered through the mannose receptor. J. Immunol. 166, 7477–7485 10.4049/jimmunol.166.12.7477 [DOI] [PubMed] [Google Scholar]
- 232.Decout A., Silva-Gomes S., Drocourt D., Blattes E., Rivière M., Prandi J. et al. (2018) Deciphering the molecular basis of mycobacteria and lipoglycan recognition by the C-type lectin Dectin-2. Sci. Rep. 8, 16840 10.1038/s41598-018-35393-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gibson K.J.C., Gilleron M., Constant P., Sichi B., Puzo G., Besra G.S. et al. (2005) A lipomannan variant with strong TLR-2-dependent pro-inflammatory activity in Saccharothrix aerocolonigenes. J. Biol. Chem. 280, 28347–28356 10.1074/jbc.M505498200 [DOI] [PubMed] [Google Scholar]
- 234.Vignal C., Guérardel Y., Kremer L., Masson M., Legrand D., Mazurier J. et al. (2003) Lipomannans, but not lipoarabinomannans, purified from Mycobacterium chelonae and Mycobacterium kansasii induce TNF-α and IL-8 secretion by a CD14-toll-like receptor 2-dependent mechanism. J. Immunol. 171, 2014–2023 10.4049/jimmunol.171.4.2014 [DOI] [PubMed] [Google Scholar]
- 235.Underhill D.M., Ozinsky A., Smith K.D. and Aderem A. (1999) Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc. Natl. Acad. Sci. U.S.A. 96, 14459 10.1073/pnas.96.25.14459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Nigou J., Vasselon T., Ray A., Constant P., Gilleron M., Besra G.S. et al. (2008) Mannan chain length controls lipoglycans signaling via and binding to TLR2. J. Immunol. 180, 6696–6702 10.4049/jimmunol.180.10.6696 [DOI] [PubMed] [Google Scholar]
- 237.Noss E.H., Pai R.K., Sellati T.J., Radolf J.D., Belisle J., Golenbock D.T. et al. (2001) Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19-kDa lipoprotein of Mycobacterium tuberculosis. J. Immunol. 167, 910 10.4049/jimmunol.167.2.910 [DOI] [PubMed] [Google Scholar]
- 238.Pai R.K., Convery M., Hamilton T.A., Boom W.H. and Harding C.V. (2003) Inhibition of IFN-gamma-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J. Immunol. 171, 175–184 10.4049/jimmunol.171.1.175 [DOI] [PubMed] [Google Scholar]
- 239.Harding C.V. and Boom W.H. (2010) Regulation of antigen presentation by mycobacterium tuberculosis: a role for Toll-like receptors. Nat. Rev. Microbiol. 8, 296–307 10.1038/nrmicro2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Beckman E.M., Porcelli S.A., Morita C.T., Behar S.M., Furlong S.T. and Brenner M.B. (1994) Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature 372, 691–694 10.1038/372691a0 [DOI] [PubMed] [Google Scholar]
- 241.Sieling P.A., Chatterjee D., Porcelli S.A., Prigozy T.I., Mazzaccaro R.J., Soriano T. et al. (1995) CD1-restricted t cell recognition of microbial lipoglycan antigens. Science 269, 227–230 10.1126/science.7542404 [DOI] [PubMed] [Google Scholar]
- 242.Fischer K., Scotet E., Niemeyer M., Koebernick H., Zerrahn J., Maillet S., et al. (2004) Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proc. Natl. Acad. Sci. U.S.A. 101, 10685–10690 10.1073/pnas.0403787101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.de la Salle H., Mariotti S., Angenieux C., Gilleron M., Garcia-Alles L.-F., Malm D., et al. (2005) Assistance of microbial glycolipid antigen processing by CD1e. Science 310, 1321 10.1126/science.1115301 [DOI] [PubMed] [Google Scholar]
- 244.Tatituri R.V.V., Watts G.F.M., Bhowruth V., Barton N., Rothchild A., Hsu F.-F., et al. (2013) Recognition of microbial and mammalian phospholipid antigens by NKT cells with diverse TCRs. Proc. Natl. Acad. Sci. U.S.A. 110, 1827–1832 10.1073/pnas.1220601110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Calabi F. and Milstein C. (1986) A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature 323, 540–543 10.1038/323540a0 [DOI] [PubMed] [Google Scholar]
- 246.Porcelli S.A. (1995) The CD1 family: a third lineage of antigen-presenting molecules. Adv. Immunol. 59, 1–98 10.1016/S0065-2776(08)60629-X [DOI] [PubMed] [Google Scholar]
- 247.Roura-Mir C., Wang L., Cheng T.-Y., Matsunaga I., Dascher C.C., Peng S.L. et al. (2005) Mycobacterium tuberculosis regulates CD1 antigen presentation pathways through. J. Immunol. 175, 1758–1766 10.4049/jimmunol.175.3.1758 [DOI] [PubMed] [Google Scholar]
- 248.Brightbill H.D., Libraty D.H., Krutzik S.R., Yang R.B., Belisle J.T., Bleharski J.R. et al. (1999) Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science 285, 732–736 10.1126/science.285.5428.732 [DOI] [PubMed] [Google Scholar]
- 249.Van Rhijn I. and Moody D.B. (2015) CD1 and mycobacterial lipids activate human T cells. Immunol. Rev. 264, 138–153 10.1111/imr.12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Jackman R.M., Stenger S., Lee A., Moody D.B., Rogers R.A., Niazi K.R. et al. (1998) The tyrosine-containing cytoplasmic tail of CD1b is essential for its efficient presentation of bacterial lipid antigens. Immunity 8, 341–351 10.1016/S1074-7613(00)80539-7 [DOI] [PubMed] [Google Scholar]
- 251.Manolova V., Kistowska M., Paoletti S., Baltariu G.M., Bausinger H., Hanau D. et al. (2006) Functional CD1a is stabilized by exogenous lipids. Eur. J. Immunol. 36, 1083–1092 10.1002/eji.200535544 [DOI] [PubMed] [Google Scholar]
- 252.Ulrichs T., Moody D.B., Grant E., Kaufmann S.H.E. and Porcelli S.A. (2003) T-cell responses to CD1-presented lipid antigens in humans with Mycobacterium tuberculosis infection. Infect. Immun. 71, 3076–3087 10.1128/IAI.71.6.3076-3087.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Layre E., Collmann A., Bastian M., Mariotti S., Czaplicki J., Prandi J., et al. (2009) Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating. Chem. Biol. 16, 82–92 10.1016/j.chembiol.2008.11.008 [DOI] [PubMed] [Google Scholar]
- 254.Rosat J.P., Grant E.P., Beckman E.M., Dascher C.C., Sieling P.A., Frederique D. et al. (1999) CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ alpha beta T cell pool. J. Immunol. 162, 366–371 PMID: [PubMed] [Google Scholar]
- 255.Stenger S., Mazzaccaro R.J., Uyemura K., Cho S., Barnes P.F., Rosat J.P. et al. (1997) Differential effects of cytolytic T cell subsets on intracellular infection. Science 276, 1684–1687 10.1126/science.276.5319.1684 [DOI] [PubMed] [Google Scholar]
- 256.Stenger S., Hanson D.A., Teitelbaum R., Dewan P., Niazi K.R., Froelich C.J. et al. (1998) An antimicrobial activity of cytolytic T cells mediated by granulysin. Science 282, 121–125 10.1126/science.282.5386.121 [DOI] [PubMed] [Google Scholar]
- 257.Gilleron M., Ronet C., Mempel M., Monsarrat B., Gachelin G. and Puzo G. (2001) Acylation state of the phosphatidylinositol mannosides from Mycobacterium bovis bacillus calmette guerin and ability to induce granuloma and recruit natural killer T cells. J. Biol. Chem. 276, 34896–34904 10.1074/jbc.M103908200 [DOI] [PubMed] [Google Scholar]
- 258.Apostolou I., Takahama Y., Belmant C., Kawano T., Huerre M., Marchal G., et al. (1999) Murine natural killer T(NKT) cells [correction of natural killer cells] contribute to the granulomatous reaction caused by mycobacterial cell walls. Proc. Natl. Acad. Sci. U.S.A. 96, 5141–5146 10.1073/pnas.96.9.5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Moody D.B., Young D.C., Cheng T.-Y., Rosat J.-P., Roura-Mir C., O'Connor P.B., et al. (2004) T cell activation by lipopeptide antigens. Science 303, 527–531 10.1126/science.1089353 [DOI] [PubMed] [Google Scholar]
- 260.Matsunaga I. and Sugita M. (2012) Mycoketide: a CD1c-presented antigen with important implications in mycobacterial infection. Clin. Dev. Immunol. 2012, 981821–981821 10.1155/2012/981821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Moody D.B., Reinhold B.B., Guy M.R., Beckman E.M., Frederique D.E., Furlong S.T., et al. (1997) Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science 278, 283–286 10.1126/science.278.5336.283 [DOI] [PubMed] [Google Scholar]
- 262.Moody D.B., Guy M.R., Grant E., Cheng T.Y., Brenner M.B., Besra G.S. et al. (2000) CD1b-mediated t cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J. Exp. Med. 192, 965–976 10.1084/jem.192.7.965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ly D., Kasmar A. G., Cheng T.-Y., de Jong A., Huang S., Roy S. Jr et al. (2013) CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J. Exp. Med. 210, 729–741. 10.1084/jem.20120624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Cox D., Fox L., Tian R., Bardet W., Skaley M., Mojsilovic D. et al. (2009) Determination of cellular lipids bound to human CD1d molecules. PLoS One 4, e5325–e5325 10.1371/journal.pone.0005325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Moody D.B., Ulrichs T., Muhlecker W., Young D.C., Gurcha S.S., Grant E., et al. (2000) CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature 404, 884–888 10.1038/35009119 [DOI] [PubMed] [Google Scholar]
- 266.Gilleron M., Stenger S., Mazorra Z., Wittke F., Mariotti S., Bohmer G. et al. (2004) Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating. J. Exp. Med. 199, 649–659 10.1084/jem.20031097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Van Rhijn I., van Berlo T., Hilmenyuk T., Cheng T.-Y., Wolf B.J., Tatituri R.V.V., et al. (2016) Human autoreactive T cells recognize CD1b and phospholipids. Proc. Natl. Acad. Sci. U.S.A. 113, 380–385 10.1073/pnas.1520947112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Dulberger C.L., Rubin E.J. and Boutte C.C. (2020) The mycobacterial cell envelope — a moving target. Nat. Rev. Microbiol. 18, 47–59 10.1038/s41579-019-0273-7 [DOI] [PubMed] [Google Scholar]
- 269.Betts J.C., Lukey P.T., Robb L.C., McAdam R.A. and Duncan K. (2002) Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731 10.1046/j.1365-2958.2002.02779.x [DOI] [PubMed] [Google Scholar]
- 270.Cambier C.J., Takaki K.K., Larson R.P., Hernandez R.E., Tobin D.M., Urdahl K.B. et al. (2014) Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505, 218–222 10.1038/nature12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Grumbach F., N RIST., Libermann D., Moyeux M., Cals S. and Clavel S. (1956) [Experimental antituberculous activity of certain isonicotinic thioamides substituted on the nucleus]. C R Hebd. Seances Acad. Sci. 242, 2187–2189 PMID: [PubMed] [Google Scholar]
- 272.Fox H.H. (1952) The chemical approach to the control of tuberculosis. Science 116, 129–134 10.1126/science.116.3006.129 [DOI] [PubMed] [Google Scholar]
- 273.Bernstein J., Lott W.A., Steinberg B.A. and Yale H.L. (1952) Chemotherapy of experimental tuberculosis. V. Isonicotinic acid hydrazide (nydrazid) and related compounds. Am. Rev. Tuberc. 65, 357–364 10.1164/art.1952.65.4.357 [DOI] [PubMed] [Google Scholar]
- 274.Mikusova K., Slayden R.A., Besra G.S. and Brennan P.J. (1995) Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob. Agents Chemother. 39, 2484–2489 10.1128/AAC.39.11.2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lee R.E., Mikusova K., Brennan P.J. and Besra G.S. (1995) Synthesis of the Arabinose Donor .beta.-D-Arabinofuranosyl-1-monophosphoryldecaprenol, development of a basic arabinosyl-transferase assay, and identification of ethambutol as an arabinosyl transferase inhibitor. J. Am. Chem. Soc. 117, 11829–11832 10.1021/ja00153a002 [DOI] [Google Scholar]
- 276.Belanger A.E., Besra G.S., Ford M.E., Mikusová K., Belisle J.T., Brennan P.J. et al. (1996) The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. U.S.A. 93, 11919–11924 10.1073/pnas.93.21.11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Telenti A., Philipp W.J., Sreevatsan S., Bernasconi C., Stockbauer K.E., Wieles B. et al. (1997) The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat. Med. 3, 567–570 10.1038/nm0597-567 [DOI] [PubMed] [Google Scholar]
- 278.Ramaswamy S.V., Amin A.G., Göksel S., Stager C.E., Dou S.-J., El Sahly H. et al. (2000) Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44, 326 10.1128/AAC.44.2.326-336.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Matsumoto M., Hashizume H., Tomishige T., Kawasaki M., Tsubouchi H., Sasaki H. et al. (2006) OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 3, e466 10.1371/journal.pmed.0030466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Stover C.K., Warrener P., VanDevanter D.R., Sherman D.R., Arain T.M., Langhorne M.H., et al. (2000) A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405, 962–966 10.1038/35016103 [DOI] [PubMed] [Google Scholar]
- 281.Dmitriev B., Toukach F. and Ehlers S. (2005) Towards a comprehensive view of the bacterial cell wall. Trends Microbiol. 13, 569–574 10.1016/j.tim.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 282.Dmitriev B.A., Ehlers S., Rietschel E.T. and Brennan P.J. (2000) Molecular mechanics of the mycobacterial cell wall: from horizontal layers to vertical scaffolds. Int. J. Med. Microbiol. 290, 251–258 10.1016/S1438-4221(00)80122-8 [DOI] [PubMed] [Google Scholar]
- 283.Jankute M., Cox J.A.G., Harrison J. and Besra G.S. (2015) Assembly of the mycobacterial cell wall. Annu. Rev. Microbiol. 69, 405–423 10.1146/annurev-micro-091014-104121 [DOI] [PubMed] [Google Scholar]
- 284.Minnikin D.E., Lee O.Y.-C., Wu H.H.T., Besra G.S., Bhatt A., Nataraj V. et al. (2015) Ancient mycobacterial lipids: key reference biomarkers in charting the evolution of tuberculosis. Tuberc. Edinb. Scotl. 95, S133–S139 10.1016/j.tube.2015.02.009 [DOI] [PubMed] [Google Scholar]
- 285.Vergnolle O., Chavadi S.S., Edupuganti U.R., Mohandas P., Chan C., Zeng J. et al. (2015) Biosynthesis of cell envelope-associated phenolic glycolipids in Mycobacterium marinum. J. Bacteriol. 197, 1040 10.1128/JB.02546-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Minnikin D.E., Kremer L., Dover L.G. and Besra G.S. (2002) The methyl-branched fortifications of Mycobacterium tuberculosis. Chem. Biol. 9, 545–553 10.1016/S1074-5521(02)00142-4 [DOI] [PubMed] [Google Scholar]
- 287.Marrakchi H., Lanéelle M.-A. and Daffé M. (2014) Mycolic acids: structures, biosynthesis, and beyond. Chem. Biol. 21, 67–85 10.1016/j.chembiol.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 288.Gago G., Diacovich L., Arabolaza A., Tsai S.-C. and Gramajo H. (2011) Fatty acid biosynthesis in actinomycetes. FEMS Microbiol. Rev. 35, 475–497 10.1111/j.1574-6976.2010.00259.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Quadri L.E.N. (2014) Biosynthesis of mycobacterial lipids by polyketide synthases and beyond. Crit. Rev. Biochem. Mol. Biol. 49, 179–211 10.3109/10409238.2014.896859 [DOI] [PubMed] [Google Scholar]
- 290.Rousseau C., Sirakova T.D., Dubey V.S., Bordat Y., Kolattukudy P.E., Gicquel B. et al. (2003) Virulence attenuation of two Mas-like polyketide synthase mutants of Mycobacterium tuberculosis. Microbiology 149, 1837–1847 10.1099/mic.0.26278-0 [DOI] [PubMed] [Google Scholar]
- 291.Dubey V.S., Sirakova T.D., Cynamon M.H. and Kolattukudy P.E. (2003) Biochemical function of msl5 (pks8 plus pks17) in Mycobacterium tuberculosis H37Rv: biosynthesis of monomethyl branched unsaturated fatty acids. J. Bacteriol. 185, 4620–4625 10.1128/JB.185.15.4620-4625.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Wells R.M., Jones C.M., Xi Z., Speer A., Danilchanka O., Doornbos K.S. et al. (2013) Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog. 9, e1003120 10.1371/journal.ppat.1003120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Chalut C. (2016) Mmpl transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberc. Edinb. Scotl. 100, 32–45 10.1016/j.tube.2016.06.004 [DOI] [PubMed] [Google Scholar]
- 294.Ma S., Huang Y., Xie F., Gong Z., Zhang Y., Stojkoska A. et al. (2020) Transport mechanism of Mycobacterium tuberculosis MmpL/S family proteins and implications in pharmaceutical targeting. Biol. Chem. 401, 331–334 10.1515/hsz-2019-0326 [DOI] [PubMed] [Google Scholar]