Abstract
Analysis of proteins and complexes under native mass spectrometric (MS) and solution conditions was typically performed using the time-of-flight analyzers, due to their routine high m/z transmission and detection capabilities. However, over recent years, the ability of Orbitrap-based mass spectrometers to transmit and detect a range of high molecular weight species is well documented. Herein, we describe how a 15 Tesla Fourier transform ion cyclotron resonance (15 Tesla FT-ICR) instrument is more than capable of analyzing a wide range of ions in the high m/z scale (>5000), in both positive and negative instrument polarities, ranging from the inorganic ceasium iodide salt clusters; a humanized IgG1k monoclonal antibody (mAb; 148.2 kDa); a IgG1-mertansine mAb drug conjugate (148.5 kDa, drug-to-antibody ratio, DAR 2.26); a siRNA-IgG1 mAb conjugate (159.1 kDa; ribonucleic acid to antibody ratio, RAR 1); the membrane protein aquaporin-Z (97.2 kDa) liberated from a C8E4 detergent micelle; the empty MSP1D1-nanodisc (142.5 kDa) and the tetradecameric chaperone protein complex GroEL (806.2 kDa; GroEL dimer at 1.6 MDa). We also investigate different regions of the FT-ICR which impact ion transmission and desolvation. Finally, we demonstrate how the transmission of these species and resultant spectra are highly consistent with those previously generated on both quadrupole time-of-flight (Q-ToF) and Orbitrap instrumentation. This report serves as an impactful example of how FT-ICR mass analyzers are competitive to Q-ToFs and Orbitraps for high mass detection at high m/z.
Keywords: Fourier transform ion cyclotron resonance, native-MS, monoclonal antibodies, cesium iodide, membrane proteins, antibody-drug-conjugates, nanodiscs, siRNA, GroEL
TOC
The TOC briefly summarizes the diverse samples that can be analyzed on a commercial 15 Tesla FT-ICR MS instrument
Introduction
Since the initial nano-electrospray ionization (nESI) mass spectrometric (MS) experiments demonstrating the preservation and transmission of biomolecular complexes from solution into the gas-phase, under native conditions over two decades ago [1–4], native-MS has become an increasingly important research tool in both industry [5–8] and academia [9–11]. As an analytical technique, native-MS lends its self particularly well to accurate protein complex molecular weight (MW) [10, 12] and subunit stoichiometry determination [13, 14]; protein-ligand stoichiometry and associated binding constant determinations [15]; gas-phase protein conformation measurements [14, 16] and gas phase stability assessment [17, 18]. Most recently, significant progress has been made characterizing membrane proteins, nanodiscs and associated complexes [19–22].
Incremental instrument improvements to MS performance, in the form of restricting ion source pumping [3, 23, 24]; source ion guide sleeves [25]; gas-bleed valves in to the source hexapole ion optics [24]; the use of more massive, mono and polyatomic collision gases [26–29]; low frequency quadrupoles [24] and lower frequency time-of-flight (ToF) pusher optics [24]. All of in the above modifications led to improved high m/z transmission, detection (> m/z 5000) and spectral quality, in terms of ion transmission and protein complex desolvation [30, 31] as well as preservation of multimeric protein stoichiometries [29, 30], some of which are in the MDa range [32–34].
Since the early 2000s, all of the aforementioned instrumental improvements were implemented exclusively on ToF [23] or quadrupole-ToF (Q-ToF [4, 24–27, 35]) instrumentation. During the subsequent decade of native-MS applications research and development, very little improvement to the ion source and ion optics were realized, specifically addressing charge state peak width, as a function of efficient ion desolvation; the charge states remained broadly adducted (much wider than the theoretical peak width; [30]). Previous native-MS experiments performed on the Q-ToF instrument, improved ion desolvation and cooling occurred in the ion source [3, 24, 27] and in the hexapole [25] or travelling wave [29] collision cell, using large monoatomic (xenon, Xe) or polyatomic (sulpurhexafluoride, SF6) collision gases. The ions were still far from fully desolvated, adding approximately 0.5% to 1.0% to the MW [30].
Only recently has the study of proteins and complexes under native-MS and solution conditions been transferred to trapping-based instruments such as the Orbitrap [8, 21, 31, 36–38] and the benefits been realized. In 2012 and 2013, Rose [36] and Kelleher [37] respectively, demonstrated a paradigm shift in native-MS protein spectral quality. Close to theoretical peak width distributions for GroEL [36] and pyruvate kinase [37] charge sates and base-line separation of sequential additions of nucleotide and sugar ligands, respectively, were achieved on the Orbitrap-EMR instrument. This level of spectral quality implied a superior level of ion desolvation, assumed to be occurring in the instrument source and HCD cell. Similar spectral quality was also achieved for monoclonal antibodies (mAbs) [36]. It must be noted however that Heck, in 2004, documented native-MS data acquisitions of vanillyl-alcohol oxidase on a modified linear ion trap [39] (LCQ-Deca) reporting well resolved charge states at m/z values 10,000 to 12,000. It is not absolutely clear which regions of the Orbitrap instrument are critical for the observed superior ion desolvation currently being described within the literature, as Rose [36] and Gault [31] have demonstrated that the HCD collision gas and HCD cell voltage, respectively, are important factors that should be considered for improved native protein desolvation and transmission. Further insights into the superior spectral quality can also be ascertained from Lange [40] where m/z data is displayed as combined absorption and magnitude mode spectrum from an apodised transient, therefore an improvement in spectral baseline noise levels and resolution are achieved by signal processing, however this is beyond the scope of this brief discussion and will be described in detail in a subsequent publication. A less frequently used instrument for native-MS analysis is the Fourier transform ion cyclotron (FT-ICR) and multiple groups have demonstrated its utility for mAbs, nanodiscs, membrane proteins and large multimeric complexes [7, 21, 41–44].
Herein, we present the use of a 15 Tesla FT-ICR instrument to acquire extended m/z range data of the humanized NIST IgG1k mAb; caesium iodide (CsI) clusters; the detergent encapsulated membrane protein aquaporin (AqpZ); an IgG1-siRNA conjugate, an IgG1-mertansine drug (DM1) conjugate; an empty MSP1D1-nanodisc (MSP1D1-Nd); and the tetradecameric chaperone protein complex GroEL, all under native-MS and solution conditions on an unmodified instrument. We have also investigated the regions within the FT-ICR instrument which may afford improved levels of mAb desolvation such as the source ion transfer capillary temperature, source skimmer voltage, collision cell voltage, collision cell and ICR-cell trapping times and monoatomic versus polyatomic collision gases. Finally, we highlight the spectral quality is highly comparable to that obtained on the Q-ToF and Orbitrap instruments, for similar protein complexes.
Material and Methods
Humanized IgG1k mAb was acquired from NIST (RM-8671[45]) at a stock concentration of 10 μg/μL. CsI was acquired from Sigma-Aldrich and a MS-working solution of 50 μg/μL in 50% (v/v) acetonitrile was used (no formic acid). AqpZ was purified and prepared using a previously described method [22]. The antibody drug conjugate (ADC) is an IgG1 covalently modified (via native lysine residues) with DM1 [46]. The siRNA-IgG1 SEFL2 [47] conjugate was prepared and purified as previously described [48]. Briefly, the IgG1 (SELF2 format) was conjugated with duplex siRNA (7.5 kDa) via an engineered cysteine. The empty MSP1D1-Nd was assembled and purified as described previously [21]. GroEL was purchased from Sigma-Aldrich and prepared as previously described [29]. All protein samples were chromatographed through an 5/150 GL Superdex column (GE Healthcare; 25 mM Tris, 100 mM NaCl) as a final purification step and re-concentrated to a low volume, high concentration stock. Immediately prior to native-MS, samples were buffer exchanged into 200mM ammonium acetate (0.5 % v/v C8E4, 2 x CMC, for AqpZ) using a BioRad P6 spin column. All working native-MS solutions were approximately 5 μM.
Mass Spectrometry
All data were acquired on a Bruker SolariX 15T FT-ICR MS system (Bruker Daltonics, Germany) operated in positive nESI mode over the m/z range 153 to 20,000 at 512 k (and negative nESI mode for the siRNA-IgG1 conjugate) equating to a resolution value of 4100 FWHM at m/z 6108.3 ([Cs23I23]Cs+). Argon (Ar) or Sulphurhexafluoride (SF6) were used as the collision gas. All samples were infused into the MS system using gold-coated glass capillaries (Waters narrow thin-walled M956232AD1-S) using the Bruker nESI source. Typical instrument voltage and pressures are noted in either the discussion or the figure legends. All data were acquired in magnitude mode and the free induction decay (FID) transients were processed using a symmetrical Hann (F = 0.5) function. Additionally, the effect of FID transient apodization is demonstrated in Figure S1 (Supporting Information). Detailed instrument parameters are documented in the Supporting Information.
Results and Discussion
High m/z and Mass Optimization on the 15 Tesla FT-ICR Instrument
There are a number of different instrument parameters which can influence the level of ion desolvation and transmission, such as the ion transfer glass capillary temperature, applied source skimmer and collision cell voltage, collision cell storage and ICR residence times (Figure 1a), all of which were systematically investigated using the NIST IgG1k mAb standard.
Figure 1.
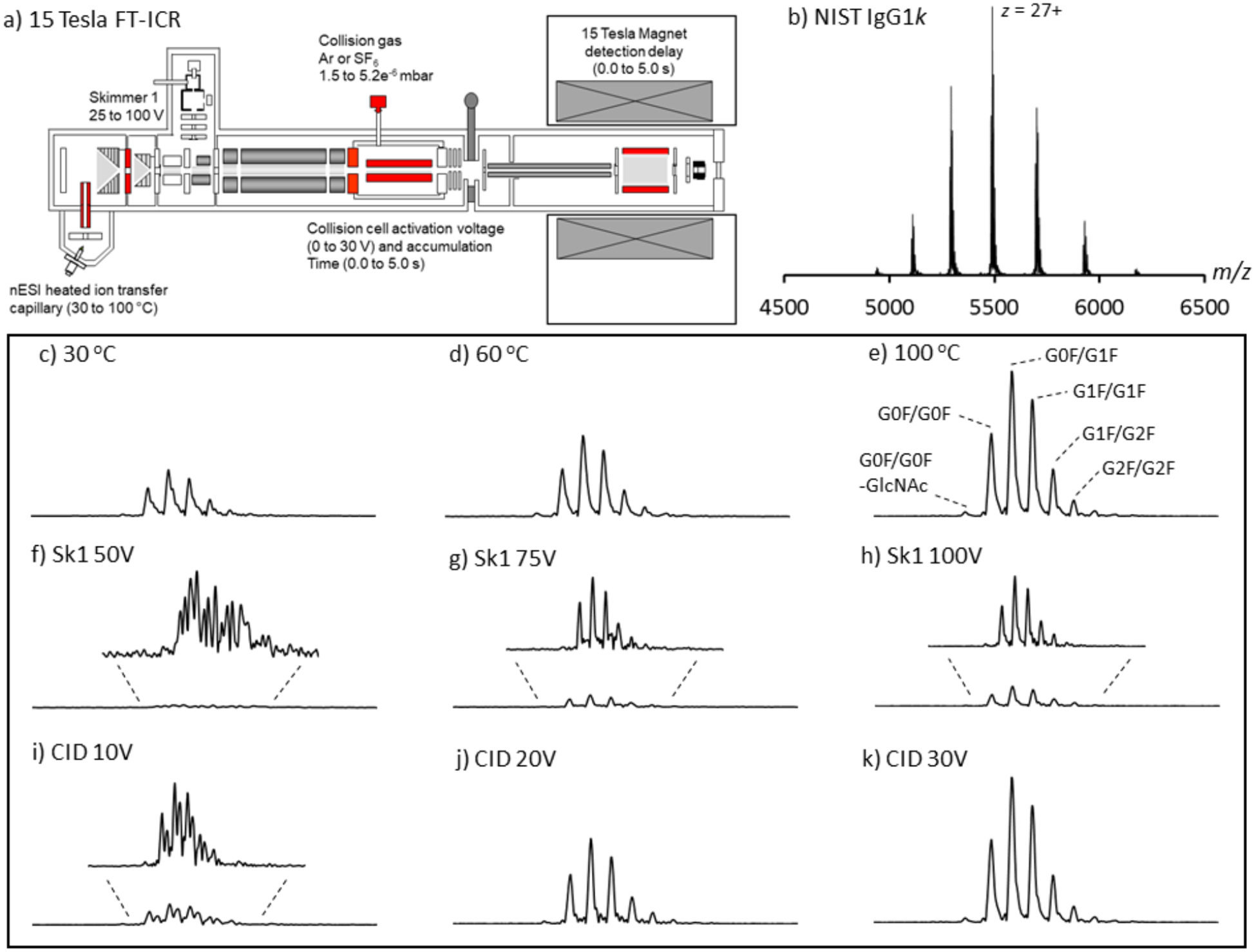
The 15 Tesla SolariX instrument schematics and measured effect of highlighted instrument regions on NIST mAb glycoform desolvation and transmission: a) The specific regions tested and optimized for high MW and high m/z transmission are highlighted in red. Heated ion transfer capillary (30 to 100 °C); source Skimmer 1 activation (25 to 100 V); collision cell activation (0 to 30 V); collision gas (Ar or SF6 indicated pressures 1.5 to 5.2e−6 mbar); collision cell accumulation time (0 to 5 s) and ICR detection delay time (0 to 5 s). Image courtesy of Bruker Daltonics, Billerica, MA, USA; b) native-MS spectrum of the NIST IgGk mAb; c-e) the effect of the ion transfer capillary temperature (CID and Skimmer1 held constant at 30 V and 50 V, respectively); f-h) the effect of source Skimmer 1 voltage (CID and capillary temperature held constant at 0 V and 100 °C, respectively); i-k) and the effect of collision cell voltage (Skimmer 1 and capillary temperature held constant at 50 V and 100 °C, respectively) on desolvation, separation and transmission of the NIST IgG1k ion series m/z 5470–5520 z = 27+, glycoforms G0F/G0F, G0F/G1F, G1F/G1F, G1F/G2F and G2F/G2F. All data normalized to an ion count of 8.0e9. Zoom-ins are included for low intensity spectra. All data displayed in magnitude mode (512 kW) using the Hann windowing function of F = 0.5.
The effect of the Source Temperature, Source Skimmer and Collision Cell Voltage:
The NIST IgIk mAb [49] (Figure 1b) is represented by five well resolved glycoforms, G0F/G0F, G0F/G1F, G1F/G1F, G1F/G2F and G2F/G2F for each charge state (Figure 1e). Figures 1c to 1e display the effect of the ion source transfer capillary gas temperature (30 °C, 60 °C and 100 °C) on glycoform desolvation and transmission for charge state z = 27+. Increased ion transfer capillary temperature results in reduced levels of adducting. This reduction in charge state adducting and therefore desolvation can also be achieved by increasing Skimmer 1 (Figures 1f to 1h) and the collision cell voltage (Figure 1i to 1k). Improved ion transmission is also observed in all cases; the ion current is spread over less channels/adducts. Skimmer 1 voltage is less efficient at removing adducts. For example, 100V applied to the skimmer (CID set to 0 V) is required for efficient adduct removal and transmission (Figure 1h) as opposed to 30 V applied to the collision cell (Figure 1k; skimmer set to 50 V) where increased transmission is observed. These changes in levels of charge state adducting as a function of ion activation are consistent with those previously reported on different MS instruments [30, 31, 36] where higher activation voltages were required for improved transmission, desolvation and MW determination of large protein complex ions. Additionally, Figure S2 (Supporting Information) graphically demonstrates the improvement in ion desolvation (and peak width FWHM).
General Source Considerations:
Increasing the source backing pressure and subsequent improved high m/z ion transmission by increasing both radial and axial collisional cooling was previously documented [23–25]. Typically the source backing pressure of the Z-Spray ion source (Waters Corporation) is increased from the standard operating pressure of 1.5–2.0 mbar to 6.0 mbar, with dramatic effects [23, 30]. For efficient high m/z transmission on the FT-ICR described herein (Figures 1b–k and 2a–f) the source pressure was not adjusted (held constant at an indicated pressure of 2 mbar). This indicates that a significant amount of ion cooling occurs in the heated ion transfer capillary and the region of the source immediately prior to entry into the first ion funnel. It is also conceivable that the ion funnels are very efficient at focusing and transmitting ions of high m/z value. The pressure in the ion transfer capillary is not measured on the commercial 15 T SolariX instrument. However, it is assumed that the pressure is significantly higher than the ion funnel region which is held at an indicated 2.0 mbar.
Figure 2.

A selection of proteins and complexes displaying charge states over a wide m/z range, acquired under standard instrument voltages and pressures. All data displayed in magnitude mode using the Hann windowing function of F = 0.5. a) CsI, 50 μg/uL; b) the tetrameric AqpZ complex liberated from a C8E4-micelle, 10 μM (1RC2, downloaded from www.rcsb.org); c) a siRNA-IgG1 conjugate, 5 μM (displaying a single cysteamine modification); d) an IgG1-DM1 conjugate, 5 μM; e) the empty MSP1D1 nanodisc, 10 μM and e) the tetradecameric chaperone complex GroEL, 5 μM (4V43, downloaded from www.rcsb.org). All data acquired over the m/z range 153–20,000 (m/z 1000 to 25,000 for CsI). Transient times ranged from 0.04 s (128 kW) to 0.17 s (512 kW). All spectral zoom-ins and FIDs are displayed in Figures S10 and S11 (Supporting Information).
Monoatomic versus a polyatomic collision gases:
Historically, larger more massive, monoatomic collision gases (Ar, Kr, Xe) were used for small molecule ion trap CID experiments [50]. More recently, monoatomic and polyatomic collision gases (Xe, N2, SF6) were reported to improve collisional focusing, cooling and transmission of large monomeric and multimeric protein complexes on Q-ToF instruments, in both MS and tandem-MS modes of operation [25] [26, 28, 29, 51]. In 2012, Rose demonstrated that Xe could be used in the HCD cell of the Orbitrap EMR instrument [36]. The larger, polyatomic collision gas SF6 as opposed to Ar, was selected to test transmission gains on the FT-ICR described herein. The results demonstrated a modest improvement in ion transmission (~10%; Figures S3a–c, Supporting Information) for the NIST IgG1k mAb. However, the signal improvement for CsI cluster transmission was dramatic (Figures S3d–f; Supporting Information). One important point to note is that the SolariX quadrupole has an effective upper selection limit of m/z 6000, making it impossible to test the effects of lager polyatomic collision gases in protein complex tandem-MS experiments [28].
Collision Cell and ICR trapping times:
There were a number of gas-phase trapping experiments performed on proteins and their structural evolution was measured. The early work of Clemmer [52] demonstrates the gas-phase structural transition of ubiquitin was measured as a function of trapping time (10 ms to 30 s) prior to ion mobility. In addition the work of McLafferty [53] demonstrates trapping cytochrome-c in a 7 Tesla ICR from picoseconds to minutes and probing the structure by electron capture dissociation. There are two specific regions within the 15 Tesla SolariX where ions are trapped and accumulated prior to release or detection, the collision cell and the ICR cell. It was hypothesized that trapping times within different regions of the 15 Tesla FT-ICR may have an impact on the levels of ion desolvation. The increased residence time within the ICR cell was achieved by setting a delay prior to ion detection at 512 kW. Additionally, a low level of declustering potential was induced by raising the source skimmer to 75 V (CID was set to 0 V) only to allow for observable glycoform separation. Figures S4 (Supporting Information) demonstrate the effects of increasing trapping times and residence times within the trap hexapole and the ICR cell respectively. In all cases, increasing the times from 0.0 sec to 5.0 sec had no measurable effect on the level of adducting/declustering of the detected charge states. Due to lack of improvement over 0.0 to 5.0 sec, longer trapping times were not investigated.
High Mass Analysis
Figures 2a–f demonstrate the high mass and high m/z transmission capabilities of an unmodified commercial 15 Tesla SolariX FT-ICR MS instrument. CsI represents a sample which displays multiple ion clusters and charge states (z = 1+ to 5+ [29]) over a wide m/z range 500 to 20,000. The peak series labelled [Cs29I29]Cs+, [Cs33I33]Cs+, [Cs49I49]Cs+, [Cs62I62]Cs+ and [Cs74I74]Cs+ represent the stable cubic clusters (magic-numbers [54, 55]). They also display signal enhancement due to constructive overlap [29, 55], with corresponding m/z values 7667.5, 8706.8, 12,863.9, 16,241.8 and 19,359.8. This spectrum is highly consistent with those previously reported on Q-ToF [24, 29, 55] and Orbitrap [36] systems and historically, on a 3 Tesla ICR instrument [56]. However, ions above m/z 20,000, as previously reported by Lebrilla [57] are not observed. Since the instrument described herein (15 T SolariX FT-ICR) is commercial, it is possible enhancements may be required to improve ultra-high m/z transmission, such as increased source pressure for improved collisional cooling and ion transmission of the larger CsI clusters. It is also interesting to note that the level of CsI cluster detection between m/z values 13,000 to 20,000 is sparse compared to data previously described [24, 29, 55]. Only the larger stable clusters are detected (m/z 16,241.8 and 19.359.8, as less stable clusters may not survive the transmission and subsequent detection within this instrument.
Over the past fifteen years the Q-ToF instrument was the instrument of choice for native membrane protein analysis [58, 59]. Only since 2015, was the Orbitrap used for membrane protein and nanodisc analysis [31, 60]. However, the FT-ICR is more than capable of analyzing membrane proteins liberated from a detergent micelle [21, 61]. Figure 2b shows the tetrameric water channel AqpZ liberated from the detergent C8E4, as a function of skimmer and collision cell voltage (50 V and 30 V respectively). Between m/z 5500 to 7000, a series of well resolved charge states (z = 12+ to 18+) correspond to a tetrameric MW of 97.2 kDa, consistent with previously reported data in C8E4 [62]. Each charge state consists of an additional superposition of peaks corresponding to multiple N-terminal formylation modifications [61] (Figure S5, Supporting Information).
Two therapeutically relevant mAb conjugates were chosen: a siRNA conjugate [48] and a DM1 [46] conjugate, analysed in negative and positive nESI modes, respectively. The position of the charge states in the m/z scale differ from negative to positive modes of operation: m/z 7,000–9,000 (charge states z = 24- to 19-) for the siRNA-IgG1 conjugate and m/z range 5000–7000 (charge states z = 28+ to 22+) for the mAb-DM1 conjugate. The IgG1-siRNA conjugate is in the ribonucleic acid-to-antibody ratio of 1 (RAR1). Since the siRNA duplex is highly negatively charged, it was decided to measure and display this molecule in negative nESI mode. This siRNA-IgG1 conjugate was demonstrated to ionize equally well in positive nESI mode [63]. The IgG1-DM1 conjugate displays up to 8 DM1 covalent additions (Figure 2d), all of which are baseline separated. The calculated DAR for this native nESI FT-ICR data is 2.26, compared to a previously calculated DAR value 3.1 to 3.3 [46]. The differences between native-MS and denaturing-MS derived DAR values have previously been addressed [7, 64]. In both cases, mild voltages are used to induce efficient ion desolvation (Skimmer 1 50V; collision cell 30V).
Figure 1e represents the empty MSP1D1 nanodisc (MW 142.5 kDa) and was described elsewhere, using different data processing parameters [21, 42]. Briefly, a highly polydisperse spectrum is displayed over the m/z range 6000–8000, representing charge states in the range of z = 21+ to 24+ (Figure S6, Supporting Information). The use of a symmetric Hann apodization function (F = 0.5) improves the spectral quality over that previously described [21, 42].
The chaperone protein GroEL (Figure 2f) represents the largest molecule analyzed within this study (806.2 kDa) with charge states ranging from z = 79+ to 70+ detected between m/z 10,000 and 12,000. The GroEL dimer is also observed in the m/z range 13,500 to 15,000 (Figure S7, Supporting Information). The individual tetradecamer charge states are fully baseline resolved. However, the resolution, or more precisely, the peak width, previously reported on the Orbitrap-EMR [36, 37], is not observed here. There may be several reasons for this. First, the GroEL sample analysed here displayed multiple N-terminal methionine oxidations (Figures S8a–d, Supporting Information), which leads to sample micro-heterogeneity, therefore broader individual charge state peak width. It is also possible that the Orbitrap source and HCD cell are more efficient at ion desolvation. Furthermore, the GroEL construct and purification procedure were most likely different. Figure S9 (Supporting Information) displays GroEL analysed at increasing levels of instrument resolution (32 kW to 4 MW). In the context of the aforementioned results, it is important to note that the Orbitrap data are displayed as a function of mixed absorption and magnitude modes [40] resulting in increased spectral resolution (approximately 1.7 times [65, 66]). Figures S10 and S11 (Supporting Information) display zoom-in m/z regions and FID transients for all spectra discussed above.
Conclusions
The 15 Tesla SolariX FT-ICR MS is more than capable of transmitting and detecting high m/z ions and high MW species efficiently, and importantly, on an unmodified commercial instrument. The data presented herein are highly consistent with historical data acquired on Q-ToF and more recently Orbitrap-EMR and UHMR instruments. Additioally, there is an increased level of therapeutic biomolecule MS characterization required from regulatory agencies [67] [68] [69] [70] therefore, the data presented herein further demonstrates that the 15 Tesla SolariX is a bona fide platform for membrane protein, protein complex and pharmaceutically relevant protein (under native-MS and solution conditions) and high MW analysis.
The highest detected and assigned ion was CsI, m/z 19,359.8, [Cs74I74]Cs+. The highest MW detected was the GroEL dimer measured at 1.61 MDa, consistent with calculated trapping limit mcritical = 1.20607×107zB2a2/Vtrapα [44, 71]. Additionally, Li recently demonstrated electron transfer dissociation charge reduced generated ions of m/z values 40,000 detected on a 15 Tesla magnet [44]. However, one must note that GroEL can also be detected on a smaller 7 Tesla magnet (Figure S12). The NIST IgG1k mAb and the IgG1-DM1 conjugate demonstrated baseline resolved glycoforms and DM1 covalent modifications, respectively, at m/z values of 5000–6500. The empty MSP1D1-Nd spectrum is highly polydisperse and consistent with previously published data. The membrane protein AqpZ liberated from the C8E4-micelle can be efficiently transmitted and detected as an intact tetrameric complex. All the aforementioned species were measured under nESI positive instrument polarity. The siRNA-mAb conjugate was measured effectively under negative instrument polarity. Data quality, in terms of charge state peak widths are significantly improved when the FID transients are apodized using a symmetric Hann function (F = 0.5).
To achieve efficient protein complex and high charge state transmission, specifically desolvation for reduced levels of charge state adducting, instrument parameter optimizations were minimal. The most effective optimizations were increasing the source Skimmer 1 voltage (up to 100 V) in combination with mild collisional activation (collision cell; up to 30 V). Operating the source ion transfer capillary to 100 °C resulted in maximum charge state desolvation. Using SF6, as opposed to Ar, led to (species dependent) improvements in high m/z and high MW transmission. Increasing ion residence times within the hexapole trap and the ICR cell, made no measurable difference to levels of native charge state desolvation.
Finally, we wish to clarify that care should be taken when referring to native-MS data as high resolution. High resolution implies obtaining isotopic resolution and observing isotopic fine structure. To date, most, if not all Orbitrap-EMR (and UHMR) native protein data lack evidence of isotopic fine structure. These are typically acquired at a spectral resolution ranging from 3200 FWHM (12 Hz) to 25,000 FWHM (1.5 Hz) at m/z 6000 [36], where many of the charge states are detected; this is far from high resolution data. We therefore suggest using the terms “highly desolvated” or “superior spectral quality” to describe the native-MS protein data currently published and acquired on both Orbitrap and FT-ICR instruments.
Supplementary Material
Acknowledgements
The authors would like to thank Pascal Egea (UCLA) for the kind donation of AqpZ. Jennifer L. Lippens (Amgen) is thanked for her useful discussions regarding aquaporin-Z detergent exchange. Tawnya Flick (Amgen) is thanked for her useful discussions and guidance regarding ADC characterization by MS. We also wish to thank Steve Van Orden (Bruker Daltonics) and David Kilgour (Nottingham Trent University) for their many discussions regarding FT-ICR operation, performance and data processing. Peter Tieleman (University of Calgary) is thanked for the coarse-grain empty nanodisc image, which is used in Figure 2e. Finally, the authors would like to thank Philip Tagari (Amgen) and Peter Grandsard (Amgen) for their leadership and continued support for novel MS applications, external collaborations and technology development effort within Amgen Research. Support from the US National Institutes of Health (R01GM103479 and S10RR028893 to J.A.L.), the US Department of Energy for the UCLA/DOE Institute for Genomics and Proteomics (DE-FC02-02ER63421 to J.A.L.) are gratefully acknowledged.
Footnotes
Supporting Information
Detailed mass spectrometer, liquid chromatographic parameters and sample handling conditions, effects of Ar and SF6 on mAb and CsI transmission, effect of hexapole ion storage and ICR residence time on NIST mAb, AqpZ acquired at different instrument resolution values, UniDec deconvolved empty nanodisc MSP1D1, 1.61 MDa GroEL dimer, GroEL monomer oxidation characterization, GroEL acquired at different instrument resolution values, zoom-ins and FIDs for all samples analysed, GroEL analysis on a 7 Tesla FT-ICR instrument.
References
- 1.Katta V, Chait BT: Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun Mass Spectrom. 5, 214–217 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Loo JA: Observation of large subunit protein complexes by electrospray ionisation mass spectrometry. Journal of Mass Spectrometry. 30, 180–183 (1995). [Google Scholar]
- 3.Rostom AA, Robinson CV: Detection of the Intact GroEL Chaperonin Assembly by Mass Spectrometry. Journal of the American Chemical Society. 121, 4718–4719 (1999). [Google Scholar]
- 4.van Berkel WJ, van den Heuvel RH, Versluis C, Heck AJ: Detection of intact megaDalton protein assemblies of vanillyl-alcohol oxidase by mass spectrometry. Protein Sci. 9, 435–439 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagal D, Valliere-Douglass JF, Balland A, Schnier PD: Resolving disulfide structural isoforms of IgG2 monoclonal antibodies by ion mobility mass spectrometry. Anal Chem. 82, 6751–6755 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Haberger M, Leiss M, Heidenreich AK, Pester O, Hafenmair G, Hook M, Bonnington L, Wegele H, Haindl M, Reusch D, Bulau P: Rapid characterization of biotherapeutic proteins by size-exclusion chromatography coupled to native mass spectrometry. MAbs. 8, 331–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campuzano IDG, Netirojjanakul C, Nshanian M, Lippens JL, Kilgour DPA, Van Orden S, Loo JA: Native-MS Analysis of Monoclonal Antibody Conjugates by Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal Chem. 90, 745–751 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Bailey AO, Han G, Phung W, Gazis P, Sutton J, Josephs JL, Sandoval W: Charge variant native mass spectrometry benefits mass precision and dynamic range of monoclonal antibody intact mass analysis. MAbs. 10, 1214–1225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson CV: From molecular chaperones to membrane motors: through the lens of a mass spectrometrist. Biochem Soc Trans. 45, 251–260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leney AC, Heck AJ: Native Mass Spectrometry: What is in the Name? J Am Soc Mass Spectrom. 28, 5–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcoux J, Champion T, Colas O, Wagner-Rousset E, Corvaia N, Van Dorsselaer A, Beck A, Cianferani S: Native mass spectrometry and ion mobility characterization of trastuzumab emtansine, a lysine-linked antibody drug conjugate. Protein Sci. 24, 1210–1223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehmood S, Allison TM, Robinson CV: Mass spectrometry of protein complexes: from origins to applications. Annu Rev Phys Chem. 66, 453–474 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, Doudna JA, Robinson CV: Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci U S A. 105, 18139–18144 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benesch JL: Collisional activation of protein complexes: picking up the pieces. J Am Soc Mass Spectrom. 20, 341–348 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Kitova EN, El-Hawiet A, Schnier PD, Klassen JS: Reliable determinations of protein-ligand interactions by direct ESI-MS measurements. Are we there yet? J Am Soc Mass Spectrom. 23, 431–441 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Heck AJ: Native mass spectrometry: a bridge between interactomics and structural biology. Nat Methods. 5, 927–933 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Ruotolo BT, Hyung SJ, Robinson PM, Giles K, Bateman RH, Robinson CV: Ion mobility-mass spectrometry reveals long-lived, unfolded intermediates in the dissociation of protein complexes. Angew Chem Int Ed Engl. 46, 8001–8004 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Hopper JT, Oldham NJ: Collision induced unfolding of protein ions in the gas phase studied by ion mobility-mass spectrometry: the effect of ligand binding on conformational stability. J Am Soc Mass Spectrom. 20, 1851–1858 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Bolla JR, Agasid MT, Mehmood S, Robinson CV: Membrane Protein-Lipid Interactions Probed Using Mass Spectrometry. Annu Rev Biochem. 88, 85–111 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Hoi KK, Robinson CV, Marty MT: Unraveling the Composition and Behavior of Heterogeneous Lipid Nanodiscs by Mass Spectrometry. Anal Chem. 88, 6199–6204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campuzano ID, Li H, Bagal D, Lippens JL, Svitel J, Kurzeja RJ, Xu H, Schnier PD, Loo JA: Native MS Analysis of Bacteriorhodopsin and an Empty Nanodisc by Orthogonal Acceleration Time-of-Flight, Orbitrap and Ion Cyclotron Resonance. Anal Chem. 88, 12427–12436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lippens JL, Nshanian M, Spahr C, Egea PF, Loo JA, Campuzano IDG: Fourier Transform-Ion Cyclotron Resonance Mass Spectrometry as a Platform for Characterizing Multimeric Membrane Protein Complexes. J Am Soc Mass Spectrom. 29, 183–193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tahallah N, Pinkse M, Maier CS, Heck AJ: The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun Mass Spectrom. 15, 596–601 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV: A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 74, 1402–1407 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Chernushevich IV, Thomson BA: Collisional cooling of large ions in electrospray mass spectrometry. Anal Chem. 76, 1754–1760 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Tito MA, Robinson CV: Improvements in or relating to microfluidic sample preparation and mass spectrometry. WO 2000-GB2625 (2000). [Google Scholar]
- 27.Sobott F, Robinson CV: Characterising electrosprayed biomolecules using tandem-MS-the noncovalent GroEl chaperonin assembly. Internation journal of mass spectrometry. 236, 25–32 (2004). [Google Scholar]
- 28.Lorenzen K, Versluis C, Van Duijn E, Van den Heuvel R, Heck AJR: Optimizing macromolecular tandem mass spectrometry of large non-covalent complexes using heavy collision gases. Internation journal of mass spectrometry. 268, 198–206 (2007). [Google Scholar]
- 29.Campuzano I, Giles K Nanospray Ion Mobility Mass Spectrometry of Selected High Mass Species Nanoproteomics: Methods and Protocols, Methods in Molecular Biology, (Eds: Toms SA, Weil R), Humana Press, a part of Springer Science+Business Media, LLC, New York: 790, 57–70 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Hernandez H, Robinson CV: Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2, 715–726 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Gault J, Donlan JA, Liko I, Hopper JT, Gupta K, Housden NG, Struwe WB, Marty MT, Mize T, Bechara C, Zhu Y, Wu B, Kleanthous C, Belov M, Damoc E, Makarov A, Robinson CV: High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat Methods. 13, 333–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uetrecht C, Versluis C, Watts NR, Wingfield PT, Steven AC, Heck AJ: Stability and shape of hepatitis B virus capsids in vacuo. Angew Chem Int Ed Engl. 47, 6247–6251 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoemaker GK, van Duijn E, Crawford SE, Uetrecht C, Baclayon M, Roos WH, Wuite GJ, Estes MK, Prasad BV, Heck AJ: Norwalk virus assembly and stability monitored by mass spectrometry. Mol Cell Proteomics. 9, 1742–1751 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uetrecht C, Watts NR, Stahl SJ, Wingfield PT, Steven AC, Heck AJ: Subunit exchange rates in Hepatitis B virus capsids are geometry- and temperature-dependent. Phys Chem Chem Phys. 12, 13368–13371 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris HR, Paxton T, Dell A, Langhorne J, Berg M, Bordoli RS, Hoyes J, Bateman RH: High sensitivity collisionally-activated decomposition tandem mass spectrometry on a novel quadrupole/orthogonal-acceleration time-of-flight mass spectrometer. Rapid Commun Mass Spectrom. 10, 889–896 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJ: High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods. 9, 1084–1086 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Belov ME, Damoc E, Denisov E, Compton PD, Horning S, Makarov AA, Kelleher NL: From protein complexes to subunit backbone fragments: a multi-stage approach to native mass spectrometry. Anal Chem. 85, 11163–11173 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Debaene F, Boeuf A, Wagner-Rousset E, Colas O, Ayoub D, Corvaia N, Van Dorsselaer A, Beck A, Cianferani S: Innovative native MS methodologies for antibody drug conjugate characterization: High resolution native MS and IM-MS for average DAR and DAR distribution assessment. Anal Chem. 86, 10674–10683 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Heck AJR, van den Heuvel R: Investigation of intact protein complexes by mass spectrometry. Mass Spectrometry Reviews. 23, 368–389 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Lange O, Damoc E, Wieghaus A, Makarov A: Enhanced Fourier transform for Orbitrap mass spectrometry. International Journal of Mass Spectrometry. 369, 16–22 (2014). [Google Scholar]
- 41.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML: Native electrospray and electron-capture dissociation in FTICR mass spectrometry provide top-down sequencing of a protein component in an intact protein assembly. J Am Soc Mass Spectrom. 21, 1966–1968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marty MT, Zhang H, Cui W, Blankenship RE, Gross ML, Sligar SG: Native mass spectrometry characterization of intact nanodisc lipoprotein complexes. Anal Chem. 84, 8957–8960 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Wongkongkathep P, Van Orden SL, Ogorzalek Loo RR, Loo JA: Revealing ligand binding sites and quantifying subunit variants of noncovalent protein complexes in a single native top-down FTICR MS experiment. J Am Soc Mass Spectrom. 25, 2060–2068 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Nguyen HH, Ogorzalek Loo RR, Campuzano IDG, Loo JA: An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes. Nat Chem. 10, 139–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campuzano IDG, Larriba C, Bagal D, Schnier PD: Ion Mobility and Mass Spectrometry Measurements of the Humanized IgGk NIST Monoclonal Antibody. ACS Symposium Series. 1202, 75–112 (2015). [Google Scholar]
- 46.Luo Q, Chung HH, Borths C, Janson M, Wen J, Joubert MK, Wypych J: Structural Characterization of a Monoclonal Antibody-Maytansinoid Immunoconjugate. Anal Chem. 88, 695–702 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Jacobsen FW, Stevenson R, Li C, Salimi-Moosavi H, Liu L, Wen J, Luo Q, Daris K, Buck L, Miller S, Ho SY, Wang W, Chen Q, Walker K, Wypych J, Narhi L, Gunasekaran K: Engineering an IgG Scaffold Lacking Effector Function with Optimized Developability. J Biol Chem. 292, 1865–1875 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Humphreys SC, Thayer MB, Campuzano IDG, Netirojjanakul C, Rock BM: Quantification of siRNA-Antibody Conjugates in Biological Matrices by Triplex-Forming Oligonucleotide ELISA. Nucleic Acid Ther. 29, 161–166 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Formolo T, Ly M, Levy M, Kilpatrick L, Lute S, Phinney K, Marzilli L, Brorson K, Boyne M, Davis D, Schiel J: Determination of the NISTmAb Primary Structure. State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 2 ACS Symposium Series; American Chemical Society: Washington, DC, doi: 10.1021/bk-2015-1201.ch001. (2015). [DOI] [Google Scholar]
- 50.Schey KL, Kenttamaa HI, Wysocki VH, Cooks RG: LOW-ENERGY COLLISIONAL ACTIVATION OF POLYATOMIC IONS WITH DIFFERENT TARGET GASES International Journal of Mass Spectrometry and Ion Processes. 90, 71–83 (1989). [Google Scholar]
- 51.Ruotolo BT, Benesch JL, Sandercock AM, Hyung SJ, Robinson CV: Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc. 3, 1139–1152 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Badman ER, Myung S, Clemmer DE: Gas-phase separations of protein and peptide ion fragments generated by collision-induced dissociation in an ion trap. Anal Chem. 74, 4889–4894 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Breuker K, McLafferty FW: Stepwise evolution of protein native structure with electrospray into the gas phase, 10(−12) to 10(2) s. Proc Natl Acad Sci U S A. 105, 18145–18152 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahmadi TS, El-Sayed MA: Effect of Lattice Energy Mismatch on the Relative Mass Peak Intensities of Mixed AlkaliHalide Nanocrystals. J phys Chem A. 101, 690–693 (1997). [Google Scholar]
- 55.Galhena AS, Jones CM, Wysocki VH: Influence of cluster size and ion activation method on the dissociation of cesium iodide clusters. Internation journal of mass spectrometry. 287, 105–113 (2009). [Google Scholar]
- 56.Amster JI, McLafferty FW, Castro ME, Russell DH, Cody RB, Ghaderi S: Detection of Mass 16 241 Ions by Fourier-Transform Mass Spectrometry. Analytical Chemistry. 58, 483–485 (1986). [DOI] [PubMed] [Google Scholar]
- 57.Lebrilla CB, Wang DT, Hunter RL, McIver RT Jr.: Detection of mass 31,830 ions with an external ion source Fourier transform mass spectrometer. Anal Chem. 62, 878–880 (1990). [DOI] [PubMed] [Google Scholar]
- 58.Barrera NP, Isaacson SC, Zhou M, Bavro VN, Welch A, Schaedler TA, Seeger MA, Miguel RN, Korkhov VM, van Veen HW, Venter H, Walmsley AR, Tate CG, Robinson CV: Mass spectrometry of membrane transporters reveals subunit stoichiometry and interactions. Nat Methods. 6, 585–587 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laganowsky A, Reading E, Hopper JT, Robinson CV: Mass spectrometry of intact membrane protein complexes. Nat Protoc. 8, 639–651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marty MT, Hoi KK, Gault J, Robinson CV: Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs. Angew Chem Int Ed Engl. 55, 550–554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lippens JL, Nshanian M, Spahr C, Egea PF, Loo JA, Campuzano IDG: Fourier Transform-Ion Cyclotron Resonance Mass Spectrometry as a Platform for Characterizing Multimeric Membrane Protein Complexes. J Am Soc Mass Spectrom. (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV: Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 510, 172–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Campuzano IDG, Lantz C, Netirojjanakul C, Humphreys SC, Thayer MB, Loo JA: Duplex and Triplex siRNA-mAb Conjugate Product Confirmation for Pharma: Positive or Negative Native nESI MS. Proc. 67th ASMS Conf. Mass Spectrometry and Applied Topics, Atlanta, 2019. TOH. [Google Scholar]
- 64.Chen J, Yin S, Wu Y, Ouyang J: Development of a native nanoelectrospray mass spectrometry method for determination of the drug-to-antibody ratio of antibody-drug conjugates. Anal Chem. 85, 1699–1704 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Vining BA, Bossio RE, Marshall AG: Phase correction for collision model analysis and enhanced resolving power of fourier transform ion cyclotron resonance mass spectra. Anal Chem. 71, 460–467 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Qi Y, Thompson CJ, Van Orden SL, O’Connor PB: Phase correction of Fourier transform ion cyclotron resonance mass spectra using MatLab. J Am Soc Mass Spectrom. 22, 138–147 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Rogstad S, Faustino A, Ruth A, Keire D, Boyne M, Park J: A Retrospective Evaluation of the Use of Mass Spectrometry in FDA Biologics License Applications. J Am Soc Mass Spectrom. 28, 786–794 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Rathore D, Faustino A, Schiel J, Pang E, Boyne M, Rogstad S: The role of mass spectrometry in the characterization of biologic protein products. Expert Rev Proteomics. 15, 431–449 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Rogstad S, Yan H, Wang X, Powers D, Brorson K, Damdinsuren B, Lee S: Multi-Attribute Method for Quality Control of Therapeutic Proteins. Anal Chem. 91, 14170–14177 (2019). [DOI] [PubMed] [Google Scholar]
- 70.De Leoz MLA, Duewer DL, Fung A, Liu L, Yau HK, Potter O, Staples GO, Furuki K, Frenkel R, Hu Y, Sosic Z, Zhang P, Altmann F, Gru Nwald-Grube C, Shao C, Zaia J, Evers W, Pengelley S, Suckau D, Wiechmann A, Resemann A, Jabs W, Beck A, Froehlich JW, Huang C, Li Y, Liu Y, Sun S, Wang Y, Seo Y, An HJ, Reichardt NC, Ruiz JE, Archer-Hartmann S, Azadi P, Bell L, Lakos Z, An Y, Cipollo JF, Pucic-Bakovic M, Stambuk J, Lauc G, Li X, Wang PG, Bock A, Hennig R, Rapp E, Creskey M, Cyr TD, Nakano M, Sugiyama T, Leung PA, Link-Lenczowski P, Jaworek J, Yang S, Zhang H, Kelly T, Klapoetke S, Cao R, Kim JY, Lee HK, Lee JY, Yoo JS, Kim SR, Suh SK, de Haan N, Falck D, Lageveen-Kammeijer GSM, Wuhrer M, Emery RJ, Kozak RP, Liew LP, Royle L, Urbanowicz PA, Packer NH, Song X, Everest-Dass A, Lattova E, Cajic S, Alagesan K, Kolarich D, Kasali T, Lindo V, Chen Y, Goswami K, Gau B, Amunugama R, Jones R, Stroop CJM, Kato K, Yagi H, Kondo S, Yuen CT, Harazono A, Shi X, Magnelli PE, Kasper BT, Mahal L, Harvey DJ, O’Flaherty R, Rudd PM, Saldova R, Hecht ES, Muddiman DC, Kang J, Bhoskar P, Menard D, Saati A, Merle C, Mast S, Tep S, Truong J, Nishikaze T, Sekiya S, Shafer A, Funaoka S, Toyoda M, de Vreugd P, Caron C, Pradhan P, Tan NC, Mechref Y, Patil S, Rohrer JS, Chakrabarti R, Dadke D, Lahori M, Zou C, Cairo C, Reiz B, Whittal RM, Lebrilla CB, Wu L, Guttman A, Szigeti M, Kremkow BG, Lee KH, Sihlbom C, Adamczyk B, Jin C, Karlsson NG, Ornros J, Larson G, Nilsson J, Meyer B, Wiegandt A, Komatsu E, Perreault H, Bodnar ED, Said N, Francois YN, Leize-Wagner E, Maier S, Zeck A, Heck AJR, Yang Y, Haselberg R, Yu YQ, Alley W, Leone JW, Yuan H, Stein SE: NIST Interlaboratory Study on Glycosylation Analysis of Monoclonal Antibodies: Comparison of Results from Diverse Analytical Methods. Mol Cell Proteomics. 19, 11–30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marshall AG, Hendrickson CL, Jackson GS: Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom Rev. 17, 1–35 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


