Highlights
-
•
COVID-19 can deteriorate at the 2nd week of illness despite decreasing viral loads.
-
•
We analyzed PBMCs of severe or mild COVID-19 cases at the 1st and 3rd week of illness.
-
•
Cellular responses were not different between two groups at the 1st week of illness.
-
•
Higher T-cell proliferation, activation, and cytotoxicity was noted in severe cases at the 3rd week.
-
•
Aberrant hyperactivation of cytotoxic T-cell may contribute to severe COVID-19 pneumonia.
Keywords: COVID-19, Severity, Contraction, Cytotoxic T cell, Perforin, Granzyme B
Abstract
Objectives
We hypothesized that immune response may contribute to progression of coronavirus disease-19 (COVID-19) at the second week of illness. Therefore, we compared cell-mediated immune (CMI) responses between severe and mild COVID-19 cases.
Methods
We examined peripheral blood mononuclear cells of laboratory-confirmed COVID-19 patients from their first and third weeks of illness. Severe pneumonia was defined as an oxygen saturation ≤93% at room air. Expressions of molecules related to T-cell activation and functions were analyzed by flow cytometry.
Results
The population dynamics of T cells at the first week were not different between the two groups. However, total numbers of CD4+ and CD8+ T cells tended to be lower in the severe group at the third week of illness. Expressions of Ki-67, PD-1, perforin, and granzyme B in CD4+ or CD8+ T cells were significantly higher in the severe group than in the mild group at the third week. In contrast to the mild group, the levels of their expression did not decrease in the severe group.
Conclusions
Severe COVID-19 had a higher degree of proliferation, activation, and cytotoxicity of T-cells at the late phase of illness without cytotoxic T-cell contraction, which might contribute to the development of severe COVID-19.
Introduction
A pandemic of coronavirus disease-19 (COVID-19), which was first reported from Wuhan, China, in December 2019, is a global health threat (Zhu et al., 2020). As of 18 May, 2020, 4,618,821 laboratory-confirmed cases have occurred, and the number is still increasing, with new epicenters (World Health Organization, 2020).
The Chinese Center for Disease Control and Prevention reported that among 44,415 COVID-19 patients, more than 80% of the patients experienced mild disease. However, about 20% of the patients developed severe pneumonia or respiratory failure requiring mechanical ventilation (Wu and McGoogan, 2020). Although the recruitment of activated CD4+ and CD8+ T cells, decreased number of T cells, or increased expressions of cytotoxic granules have been reported (Qin et al., 2020, Thevarajan et al., 2020, Xu et al., 2020), cell-mediated immune (CMI) responses in COVID-19 are not well understood. Understanding immunopathogenesis of progression of COVID-19 pneumonia is urgently needed to accelerate the development of therapeutic strategies.
Recent viral shedding kinetics studies showed that virus titers usually reach peak during the first week of illness, and decline during the second week of illness (Liu et al., 2020, Zou et al., 2020). It is of note that patients with COVID-19 pneumonia may suddenly deteriorate 7‒10 days after illness onset, resulting in intensive care unit admission (Yang et al., 2020, Zhou et al., 2020). Therefore, we hypothesized that the immune response may play an important role for this temporally discrepant clinical deterioration in COVID-19 patients. In this study, we aimed to compare CMI responses between severe and mild COVID-19 patients at their first and third week of illness.
Methods
Patients and samples
We analyzed peripheral blood mononuclear cells (PBMCs) of laboratory-confirmed COVID-19 patients who had been treated at Seoul National University Hospital and Seoul National University Bundang Hospital. We collected peripheral blood samples from the patients at the first and third week of illness. A severe case was defined as having radiological pneumonia and having an oxygen saturation of 93% or less at room air during illness, while others were classified as a mild case (Wu and McGoogan, 2020). We also analyzed healthy volunteers’ PBMCs to accurately assess COVID-19-related CMI responses. Institutional Review Boards of Seoul National University Hospital and Seoul National University Bundang Hospital approved the study (IRB No. 2003-011-1105 and B-2004/607-401), and waived the need for written informed consent of COVID-19 patients on public health grounds.
Flow cytometry
PBMCs were purified from heparinized peripheral blood using a Ficoll–Histopaque gradient (1.077 g/mL; GE Healthcare Life Sciences, Piscataway, NJ, USA) (Kim et al., 2006). The PBMCs were then stored in liquid nitrogen after the addition of 50% fetal bovine serum (FBS), 10% dimethyl sulfoxide, and 40% RPMI-1640 (Life Technologies, Carlsbad, CA, USA) until assayed (Cho et al., 2012). Cryopreserved PBMCs were thawed and stained with following anti-human antibodies (Abs): V450–anti-HLA-DR, phycoerythrin (PE)–anti-CD25, fluorescein isothiocyanate (FITC)–anti-Ki-67, allophycocyanin–indotricarbocyanine (Cy7)–anti-CD8, FITC–anti-perforin, Alexa Fluor® 700–anti-granzyme B, Alexa Fluor® 647–anti-FoxP3, FITC–anti-IL-2, PE–anti-IL-4 from BD Biosciences (San Jose, CA, USA); eFluor® 450–anti-IL-7Rα, peridinin-chlorophyll proteins (PerCP)-cyanine 5.5 (Cy5.5)–anti-CD45RA, PE-Cy7–anti-interferon-γ (IFN-γ), eFluor® 660–anti-IL-17 from eBioscience (San Diego, CA, USA); Brilliant Violet (BV) 605–anti-CD4, Alexa Fluor® 700–anti-PD-1, BV605–anti-CD4, BV21–anti-CD8 from BioLegend (San Diego, CA, USA). FACS staining was performed as described previously (Cho et al., 2012, Kim et al., 2006). A fluorescence minus one (FMO) control, a tube of cells stained with all fluorochromes used in the experiment except one, was used to determine the cut-off point between the background fluorescence and the positive population in this experiment.
For staining the effector molecules and the proliferation marker, cells were fixed with 2% paraformaldehyde and permeabilized with BD Perm/Wash™ buffer (BD Biosciences). Next, the cells were stained with fluorescently labeled Abs for perforin, granzyme B, Ki-67, or surface molecules.
To stain intracellular cytokines, cells were resuspended in RPMI 1640 medium containing 10% FBS and 1% penicillin/streptomycin (all reagents from Life Technologies), incubated for 30 min at 37 °C, and then stimulated for 4 h at 37 °C with phorbol 12-myristate 13-acetate (PMA; 50 ng/mL) and ionomycin (1 μg/mL) (both reagents from Sigma-Aldrich, St. Louis, MO, USA) in the presence of Golgistop® (BD Biosciences).
After being stained with the appropriate Abs, the cells were analyzed on a BD LSRFortessa™ (BD Biosciences) instrument with FACSDiva™ software, and data were analyzed using FlowJo® software (TreeStar, Ashland, OR, USA).
Statistical analysis
All laboratory data are expressed as the mean ± standard error of the mean (SEM). Data were compared using the two-tailed Mann–Whitney U test or Wilcoxon signed rank test. In all analyses, P < 0.05 was considered to indicate statistical significance. All analyses were performed using PASW for Windows (version 25.0; SPSS Inc., Chicago, IL, USA) and all graphs were plotted using GraphPad Prism 6.01 (GraphPad Software, La Jolla, CA, USA).
Results
Patients and samples
Four severe and eight mild cases were included in this study. Median (range) ages in severe and mild groups were 62 (35–82) and 55 (25–79), respectively (P = 0.552). Male proportions were 50.0% (2/4) and 25.0% (2/8) in these groups (P = 0.547). All of them had no prior signs of immunodeficiency. PBMCs of one severe patient were available only at his third week of illness. Clinical and laboratory characteristics when the first- and third-week samples were collected are summarized in Table 1 .
Table 1.
Clinical and laboratory characteristics of patients in mild and severe groups.
| Mild (n = 8) | Severe (n = 4) | P | |
|---|---|---|---|
| Male, n (%) | 2 (25.0) | 2 (50.0) | 0.547 |
| Age, median years (range) | 55 (25–79) | 62 (35–82) | 0.552 |
| White blood cells, cells/mm3, median (range) | |||
| At week 1 | 4125 (2780–6500) | 5210 (3160–8760) | 0.540 |
| At week 3 | 5080 (4220–7750) | 7535 (3900–14,180) | 0.396 |
| Hemoglobin, g/dL, median (range) | |||
| At week 1 | 13.5 (11.0–16.0) | 12.0 (11.0–13.1) | 0.179 |
| At week 3 | 12.0 (11.5–15.0) | 11.7 (9.0–13.0) | 0.218 |
| Platelets, 1000 cells/mm3, median (range) | |||
| At week 1 | 192 (159–396) | 263 (166–316) | 0.307 |
| At week 3 | 237 (185–386) | 317 (210–459) | 0.234 |
| C-reactive protein, mg/dL, median (range) | |||
| At week 1 | 0.7 (0.0–6.6) | 1.6 (1.6–14.9) | 0.041 |
| At week 3 | 0.1 (0.0–0.7) | 7.6 (4.5–28.5) | 0.006 |
| LDH, U/L, median (range) | |||
| At week 1 | 162 (131–350) | 439 (223–455) | 0.053 |
| At week 3 | 163 (129–411) | 393 (254–561) | 0.023 |
| Ct for E gene from NP swabs, median (range) | |||
| At week 1 | 24.78 (17.3–40.0) | 29.3 (26.8–32.1) | 0.221 |
| At week 3 | 39.3 (29.1–40.0) | 35.4 (29.7–40.0) | 0.725 |
| Oxygen therapy | – | ||
| Start date, day of illness, median (range) | – | 9 (4–14) | – |
| At week 3, median (range) | – | 6 L/min (4 L/min to MV) | – |
| Peak oxygen demand, median (range) | – | 6 L/min (6 L/min to MV) | – |
LDH, lactate dehydrogenase; Ct, cycle threshold; E gene, SARS-CoV-2 envelope gene; NP, nasopharyngeal; MV, mechanical ventilation.
Median [range] cycle threshold (Ct) values of quantitative real-time reverse transcription polymerase chain reaction for SARS-CoV-2 envelope (E) gene from nasopharyngeal swabs in severe patients were increased from 29.3 [26.8–32.1] at the first week to 35.4 [29.7–40.0] (negative results were converted to 40) at the third week, respectively. While all the patients in the severe group had minimal oxygen requirements at the first week, they all had worsened pneumonic infiltration and were receiving oxygen therapy when the third week samples were collected (three nasal prongs 4–6 L/min and one mechanical ventilation). All four severe patients and three of eight mild patients received lopinavir/ritonavir; however, corticosteroid was not administered in all patients during the course. No 30-day in-hospital mortality was noted among those patients.
PBMCs from five healthy volunteers were also analyzed. Their median (range) age was 35 (28–47), and three of them (60%) were male. Their results are described in Table 2 and shown in Figures as gray boxes to indicate their interquartile range.
Table 2.
Leukocyte populations and characterization of CD4+ and CD8+ T cells at the first week of illness according to the severity.
| Severe cases (n = 3) | Mild cases (n = 8) | Healthy controls (n = 5) |
P |
|||
|---|---|---|---|---|---|---|
| Severe vs. mild | Severe vs. controls | Mild vs. controls | ||||
| White blood cells (/μL) | 5710.0 (± 1635.8) | 4541.3 (± 568.5) | 6422.5 (± 1260.0) | 0.540 | 0.724 | 0.174 |
| Neutrophil (/μL) | 4134.7 (± 1453.8) | 2803.9 (± 543.5) | 3581.0 (± 996.4) | 0.307 | 1.000 | 0.396 |
| Monocyte (/μL) | 429.3 (± 127.0) | 414.6 (± 51.5) | 486.5 (± 103.1) | 1.000 | 0.724 | 0.497 |
| Lymphocyte (/μL) | 1131.7 (± 112.6) | 1256.3 (± 130.4) | 2214.5 (± 380.6) | 0.683 | 0.034 | 0.011 |
| CD4+ T cells (/μL) | 306.2 (± 79.0) | 435.5 (± 57.5) | 768.3 (± 236.1) | 0.307 | 0.077 | 0.062 |
| Ki-67+ CD4+ (%) | 4.1 (± 1.3) | 4.0 (± 1.2) | 0.8 (± 0.2) | 0.683 | 0.025 | 0.008 |
| HLA-DR+CD4+ (%) | 2.2 (± 1.1) | 2.9 (± 0.9) | 0.6 (± 0.3) | 0.683 | 0.101 | 0.013 |
| PD-1+CD4+ (%) | 1.4 (± 0.4) | 0.8 (± 0.3) | 0.2 (± 0.1) | 0.153 | 0.025 | 0.057 |
| CD38+CD4+ (%) | 1.0 (± 1.0) | 0.3 (± 0.2) | 0.1 (± 0.0) | 0.540 | 0.881 | 0.079 |
| FoxP3+CD4+ (%) | 7.1 (± 1.2) | 6.6 (± 0.5) | 7.9 (± 0.8) | 0.838 | 0.655 | 0.242 |
| Perforin+CD4+ (%) | 7.0 (± 1.2) | 6.9 (± 3.1) | 6.1 (± 2.0) | 0.221 | 0.655 | 0.770 |
| Granzyme B+CD4+ (%) | 7.8 (± 1.8) | 11.6 (± 3.6) | 10.0 (± 1.8) | 0.683 | 0.297 | 0.464 |
| IFN-γ+CD4+ (%) | 4.7 (± 1.3) | 5.8 (± 1.2) | 8.8 (± 1.9) | 0.838 | 0.157 | 0.126 |
| IL-2+CD4+ (%) | 23.2 (± 10.5) | 20.2 (± 4.9) | 14.0 (± 3.4) | 0.683 | 0.456 | 0.558 |
| IL-4+CD4+ (%) | 3.4 (± 1.0) | 3.2 (± 0.6) | 2.1 (± 0.6) | 0.838 | 0.297 | 0.242 |
| IL-17+CD4+ (%) | 0.5 (± 0.2) | 0.4 (± 0.1) | 0.2 (± 0.1) | 0.838 | 0.297 | 0.188 |
| CD8+ T cells (/μL) | 143.2 (± 13.1) | 296.1 (± 60.9) | 660.5 (± 132.4) | 0.102 | 0.034 | 0.042 |
| Ki-67+CD8+ (%) | 6.4 (± 1.8) | 8.7 (± 3.6) | 0.8 (± 0.1) | 0.683 | 0.025 | 0.003 |
| HLA-DR+CD8+ (%) | 2.2 (± 1.1) | 6.0 (± 2.4) | 0.2 (± 0.1) | 0.540 | 0.025 | 0.013 |
| PD-1+CD8+ (%) | 1.0 (± 0.2) | 1.1 (± 0.4) | 0.2 (± 0.1) | 0.414 | 0.025 | 0.013 |
| CD38+CD8+ (%) | 5.9 (± 5.2) | 1.6 (± 0.4) | 0.4 (± 0.2) | 0.838 | 0.297 | 0.057 |
| Perforin+CD8+ (%) | 54.2 (± 7.1) | 40.0 (± 5.7) | 39.9 (± 7.7) | 0.102 | 0.180 | 0.884 |
| Granzyme B+CD8+ (%) | 65.2 (± 2.9) | 53.3 (± 7.0) | 46.2 (± 7.2) | 0.540 | 0.053 | 0.558 |
| IFN-γ+CD8+ (%) | 12.7 (± 7.8) | 13.2 (± 2.8) | 17.5 (± 1.5) | 0.540 | 0.480 | 0.174 |
| IL-2+CD8+ (%) | 11.0 (± 8.4) | 7.6 (± 2.4) | 5.5 (± 1.5) | 1.000 | 0.881 | 0.884 |
| IL-4+CD8+ (%) | 8.0 (± 6.5) | 4.4 (± 0.9) | 1.9 (± 0.8) | 0.838 | 0.297 | 0.079 |
| IL-17+CD8+ (%) | 0.2 (± 0.1) | 0.2 (± 0.1) | 0.1 (± 0.0) | 0.540 | 0.297 | 0.019 |
| Effector memory CD8+ (%) | 77.1 (± 4.1) | 75.5 (± 6.2) | 69.9 (± 6.8) | 0.838 | 0.456 | 0.420 |
| IL-7Rαhigh (%) | 20.6 (± 4.4) | 30.9 (± 3.9) | 32.1 (± 7.0) | 0.152 | 0.297 | 0.660 |
| IL-7Rαlow (%) | 79.0 (± 4.4) | 68.7 (± 3.9) | 65.6 (± 6.6) | 0.153 | 0.101 | 0.661 |
All data are presented as mean ± standard error of the mean.
Leukocyte populations in peripheral blood
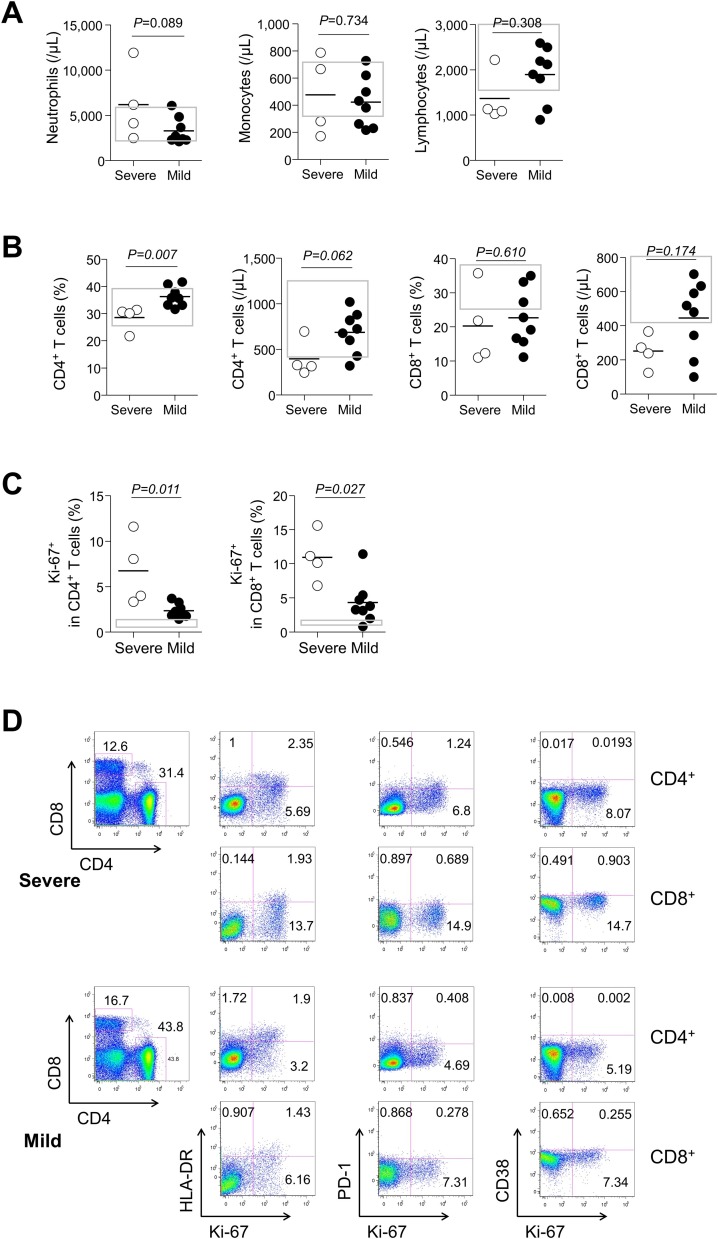
In general, neutropenia or lymphopenia can occur during the early stages of respiratory viral infection (Cheng et al., 2019, Colamussi et al., 1999). Thus, to confirm how they change in COVID-19 patients, we analyzed the changes in leukocyte subpopulations at the first week and third week of illness. The absolute numbers (cells/μL) of lymphocytes were comparable between the severe and mild groups in both time points, although they were lower than that from healthy controls (Table 2 and Figure 1 A). Although CD4+ and CD8+ T cell counts tended to be lower in the severe group than in the mild group at the third week of illness (mean ± standard error of the mean [SEM]; CD4+ T cell, 396.5 ± 101.9 in the severe group vs. 684.3 ± 82.0 in the mild group, P = 0.062; CD8+ T cell, 250.9 ± 50.1 in the severe group vs. 445.0 ± 76.1 in the mild group, P = 0.174, Figure 1B), there was no difference at the first week (Table 2). This raises the question of whether there are significant changes in immune-cell subsets according to different clinical courses.
Figure 1.
Leukocyte populations and CD4+ and CD8+ T cell counts and Ki-67 expressions at the third week of illness in 12 patients with COVID-19 according to severity. (A) The counts (cells/μL) of neutrophils, lymphocyte, and monocytes in severe (n = 4, open circle) or mild cases (n = 8, closed circle). (B) Frequencies and counts (cells/μL) of CD4+ and CD8+ T cells. (C) Ki-67 expressions in CD4+ and CD8+ T cells. (D) Representative dot plots show the identification of Ki-67+, HLA-DR+, PD-1+, or CD38+ cells in CD4+ (1st line of each panel) or CD8+ (2nd line of each panel) T cells in severe (upper panels) or mild (lower panels) cases. Bars denote mean values. Gray boxes represent interquartile ranges of healthy controls.
Expression of markers for proliferation and activation of T-cell subsets
To investigate dynamics of the antigen-specific T cells according to severity of COVID-19, we determined the frequency of proliferating or activated T cells in the two groups. Although we could not attempt staining with peptide-MHC tetramer (Altman et al., 1996) for SARS-CoV-2, it was thought that analysis of these markers would sufficiently trace antigen-specific T cells as previously known (Ndhlovu et al., 2015, Soares et al., 2010). Although frequencies (%) of Ki-67+ CD4+ and CD8+ T cells were higher than those from healthy controls in both groups, they were significantly higher in the severe group than in the mild group (mean ± SEM, the severe group vs. the mild group; Ki-67+ in CD4+ T cells, 6.7 ± 1.9 vs. 2.4 ± 0.3, P = 0.011; Ki-67+ in CD8+ T cells, 10.9 ± 1.8 vs. 4.3 ± 1.1, P = 0.027; Figure 1C and D), signifying a higher rate of T-cell turnover in the severe group.
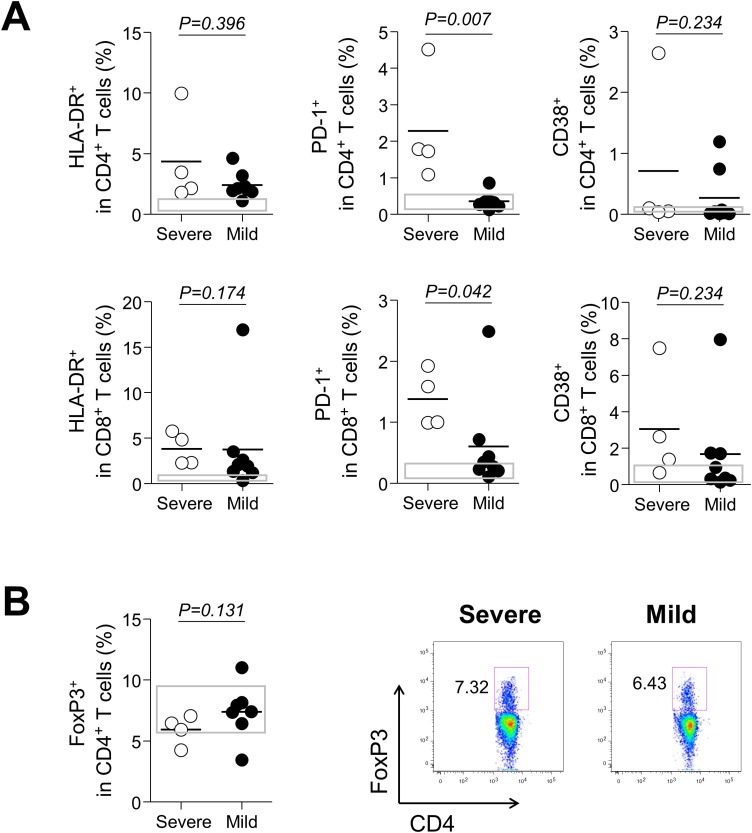
We also examined frequencies (%) of HLA-DR, PD-1, and CD38 as makers of activated T cells. Although they tended to be higher in both groups than in the healthy control group, there were no differences at the first week between the two groups (Table 2). However, activated T cells tended to be higher in the severe group than in the mild group at the third week, especially in terms of PD-1 expression (mean ± SEM, the severe group vs. the mild group; HLA-DR+ in CD4+ T cell, 4.3 ± 1.9 vs. 2.4 ± 0.4, P = 0.396; HLA-DR+ in CD8+ T cells, 3.8 ± 0.9 vs. 3.7 ± 1.9, P = 0.174; PD-1+ in CD4+ T cells, 2.3 ± 0.8 vs. 0.4 ± 0.1, P = 0.007; PD-1+ in CD8+ T cells, 1.4 ± 0.2 vs. 0.6 ± 0.3, P = 0.042; CD38+ in CD4+ T cells, 0.7 ± 0.6 vs. 0.3 ± 0.2, P = 0.234; CD38+ in CD8+ T cells, 3.0 ± 1.5 vs. 1.7 ± 0.9, P = 0.234; Figure 2 A). Taken together, the severe group showed a higher degree of proliferation and activation of T cells.
Figure 2.
Expressions of activation markers and FoxP3+ CD4+ T cells at the third week of illness according to severity. (A) HLA-DR, PD-1, and CD38 expressions in CD4+ and CD8+ T cells, in severe (n = 4, open circle) and mild cases (n = 8, closed circle). (B) Frequencies of FoxP3+CD4+ T cells and representative dot plots showing the identification of the population. Bars denote mean values. Gray boxes represent interquartile ranges of healthy controls.
Given the fact that regulatory T cells (Tregs), FoxP3+CD4+ T cells, suppress activation, proliferation and cytokine production of CD4+ and CD8+ T cells (Sakaguchi, 2004), the possibility was raised of whether excessive T-cell activation in the severe group could be due to a decrease in the number of Tregs or due to a decrease in inhibitory capacity. To address this, we measured frequency of FoxP3+CD4+ Tregs. As a result, although statistically not significant, frequencies (%) of FoxP3+CD4+ Tregs tended to be lower in severe patients at the third week of illness (mean ± SEM, 5.9 ± 0.6 in the severe group vs. 7.4 ± 0.9 in the mild group, P = 0.131, Figure 2B), suggesting that the overall inhibitory capacity for effector T cells would be reduced.
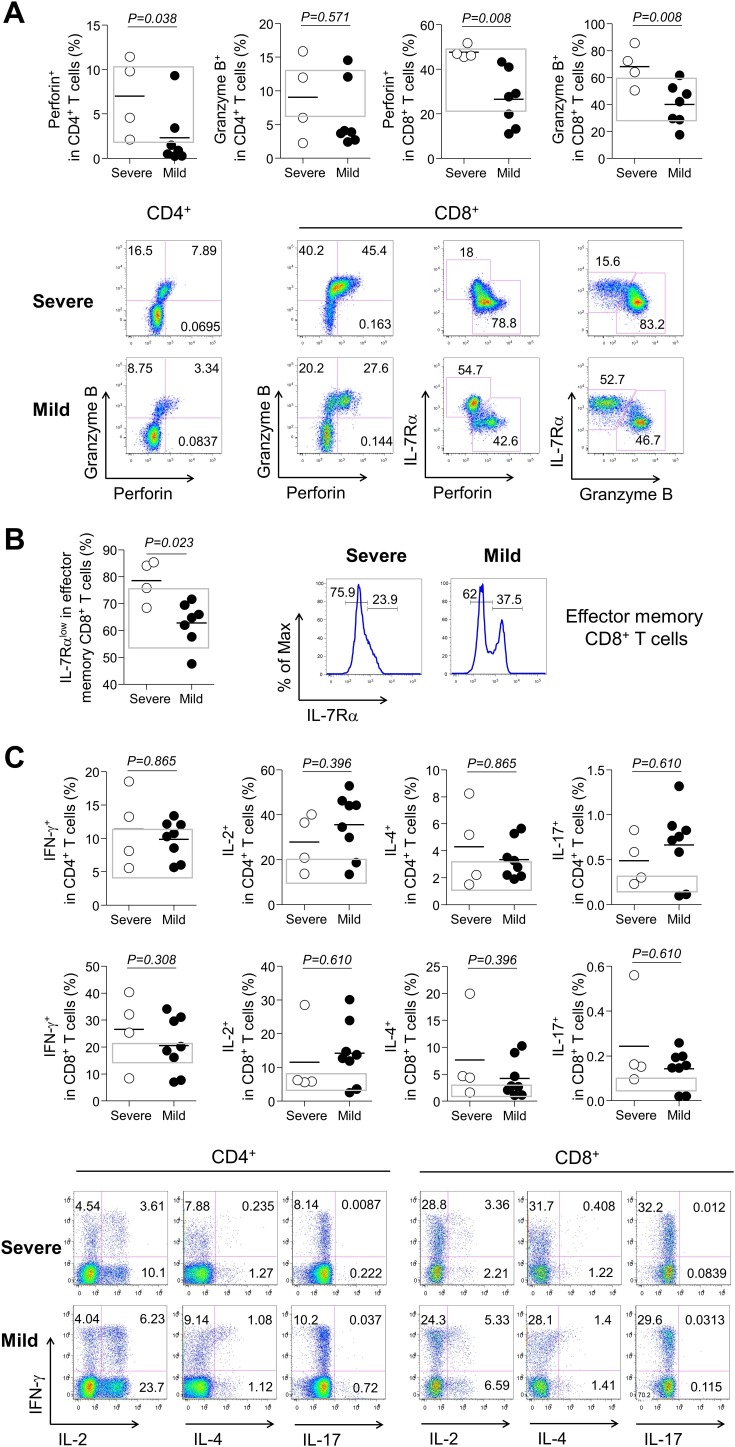
Expressions of effector granules and cytokines
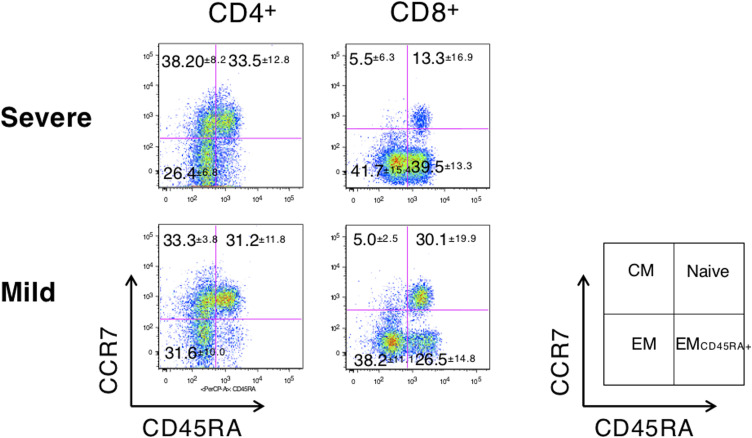
Furthermore, we examined the expression levels of effector molecules which could eradicate infected cells as well as cause tissue damage (Cho et al., 2012, Kim et al., 2012, Kim and Shin, 2019). Thus, we examined the expression of effector granules and cytokines to reveal the consequences of increased T-cell activation at the third week of illness. Frequencies (%) of perforin and granzyme B expression in both CD4+ and CD8+ T cells were higher in the severe group than in the mild group (mean ± SEM, the severe group vs. the mild group; perforin+ in CD4+ T cells, 7.0 ± 2.2 vs. 2.3 ± 1.2, P = 0.038; granzyme B+ in CD4+ T cells, 9.0 ± 3.0 vs. 6.2 ± 1.9, P = 0.571; perforin+ in CD8+ T cells, 47.6 ± 1.4 vs. 26.5 ± 4.8, P = 0.008; granzyme B+ in CD8+ T cells, 68.1 ± 7.4 vs. 40.0 ± 5.8, P = 0.008; Figure 3 A). Proportions (%) of IL-7Rαlow effector memory (CCR7−CD45RA+/−) CD8+ T cells, which are known for high perforin and granzyme B expression (Kim et al., 2012) (Figure 3A, lower panel), were significantly higher in the severe group than in the mild group (mean ± SEM, 78.5 ± 3.9 in the severe group vs. 62.8 ± 3.1 in the mild group, P = 0.023; Figure 3B), implying a higher cytotoxic potential in the severe groups at the third week of illness. Similarly, increased IL-7Rαlow effector memory CD8+ T cells might be secondary to the expansion of effector memory CD8+ T cells (Supplementary Figure S1), which is known to be produced by prolonged antigenic stimulation (Kim et al., 2006).
Figure 3.
Effector granules or cytokine expressions at the third week of illness according to severity. (A) Perforin or granzyme B expressions in severe (n = 4, open circle) and mild cases (n = 8, closed circle) and representative dot plots showing the identification of perforin+, granzyme B+, IL-7Rαhigh, or IL-7Rαlow cells in CD4+ or CD8+ T cells in severe (upper panels) or mild (lower panels) cases. (B) Frequencies of IL-7Rαlow effector memory (CCR7-CD45RA+/−) CD8+ T cells and representative histograms. (C) IFN-γ, IL-2, IL-4, and IL-17 expressions in CD4+ and CD8+ T cells and representative dot plots showing the identification of IFN-γ+, IL-2+, IL-4+, and IL-17+ cells in CD4+ or CD8+ T cells in severe (upper panels) or mild (lower panels) cases. Bars denote mean values. Gray boxes represent interquartile ranges of healthy controls.
Compared to the difference in cytotoxic molecules, expression levels of IFN-γ, IL-2, IL-4, and IL-17 were comparable between the two groups at both time points (Table 2 and Figure 3C). It is presumed that the effector cytokine expression alone did not significantly affect severity of COVID-19.
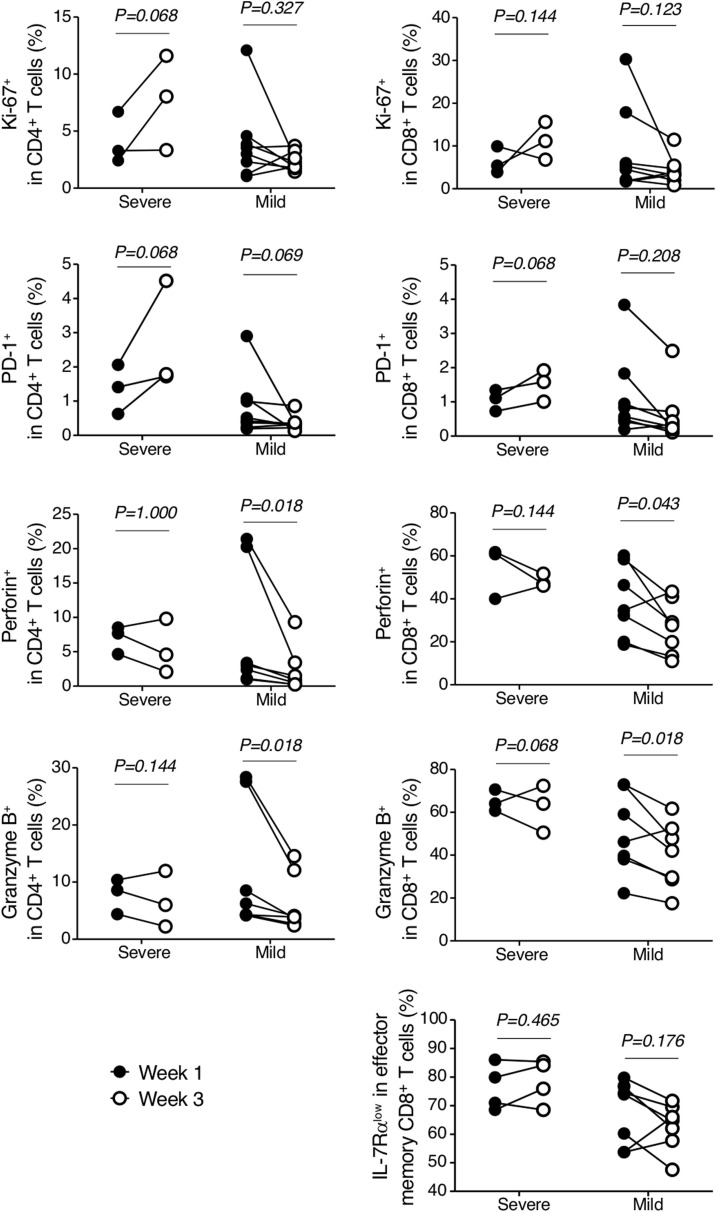
Lack of T-cell contraction in severe COVID-19 groups
Interestingly, when we compared the above immune responses between the first and third weeks of illness in each group, the mild group tended to show substantial reduction of Ki-67, PD-1, perforin, and granzyme B expressions compared to those in the severe group (Figure 4 ). In contrast, the expression levels of those molecules in the severe group did not decrease or even tended to increase at the third week. The proportion of IL-7Rαlow effector memory CD8+ T cells was not reduced in the severe group. Since cells expressing such markers are believed to represent cells that have recently been stimulated with antigen (Kim et al., 2007b, Ndhlovu et al., 2015, Soares et al., 2010), these findings imply a lack of cytotoxic T-cell contraction or delayed hyperactivation of T cells in the severe group.
Figure 4.
Temporal changes of cell-mediated immune responses in 11 patients with COVID-19 according to severity. Frequencies of Ki-67+, PD-1+, perforin+, granzyme B+ CD8+ T cells, and IL-7Rαlow effector memory CD8+ T cells in severe (n = 3) or mild (n = 8) cases from the first week (closed circle) to the third week (open circle).
Discussion
Therapeutic strategies for severe COVID-19 should be developed in a timely manner, and must be preceded by an understanding of its immunopathogenesis. In this study, we observed higher turnover and activation of T cells and higher expression of cytotoxic effectors such as perforin and granzyme B in the severe group than in the mild group at the third week of illness. This might be due to the lack of cytotoxic T-cell contraction, and it is possible that such an immune response contributed to clinical deterioration at the second or third week of illness in severe COVID-19 patients.
Median days from symptom onset to dyspnea, development of acute respiratory distress syndrome, and ICU admission in COVID-19 patients were 7, 12, and 12 days, respectively (Yang et al., 2020, Zhou et al., 2020). Interestingly, viral load of SARS-coronavirus-2 (SARS-CoV-2) peaks during the first 3–5 days of illness and declines thereafter (Liu et al., 2020, Zou et al., 2020). Although initial viral loads are higher in severe cases, they also decline after the early peak (Liu et al., 2020). This viral shedding kinetics of COVID-19 is markedly different from that of MERS, which peaks during the second week of illness (Oh et al., 2016). Such a time lag between the peak of viral load and the clinical deterioration implies the importance of immune response in the pathogenesis of severe COVID-19. And persistent or delayed hyperactivation of cytotoxicity could be explained in CD8+ T cells as well as CD4+ T cells (Juno et al., 2017). Although there is no difference in the expression of cytokines between the severe and the mild groups, if IFN-γ-producing CD8+ T (Tc1) cells express highly cytotoxic molecules such as perforin and granzyme B, they may exhibit high cytotoxicity in the severe group.
There were a few studies that explored CMI response in COVID-19 (Qin et al., 2020, Thevarajan et al., 2020, Xu et al., 2020). Although they reported activation of CD4+ and CD8+ T cells, decreased numbers of T cells, or high concentrations of cytotoxic granules, and some are consistent with the results of the present study, data on severe COVID-19 patients or temporal changes of CMI response were lacking (Qin et al., 2020, Thevarajan et al., 2020, Xu et al., 2020). We believe that our results, focusing on immune responses characterizing severe COVID-19 cases with a temporal consideration, may elucidate the pathogenesis of severe COVID-19.
In addition, unknown factors that may affect the immune response could be related to severe COVID-19. In this regard, Cornberg et al. suggested an important possibility that the degree of antigen exposure can regulate the balance between protective immunity and immunopathology by presenting the medium-dose virus-induced immunopathology and T-cell exhaustion by massive virus exposure (Cornberg et al., 2013). It seems consistent with the development of severe COVID-19, which clinically deteriorates when viral loads are declining. In addition, activated bystander CD8+ T cells induced by IL-15 can participate in protective immunity (Kim et al., 2007a, Sim et al., 2017), but they also can cause excessive tissue damage because of its lack of specificity for the pathogen (Kim and Shin, 2019). In this regard, an increase in IL-7Rαlow effector memory CD8+ T cells, which are known to express activating receptors of natural killer cells and have cytotoxic ability through this receptor (Cho et al., 2012), with high expression of cytotoxic molecules in severe groups suggests that there is a possibility of CMI response-mediated tissue damage. Nonetheless, further studies are needed to explore whether these factors contribute to pathogenesis of severe COVID-19.
There are several therapeutic agents for the treatment of COVID-19. When considering the viral shedding kinetics (Liu et al., 2020, Zou et al., 2020), antiviral agents such as lopinavir/ritonavir, remdesivir, or hydroxychloroquine (Cao et al., 2020, Wang et al., 2020, Yao et al., 2020) may be effective during the first week of illness when the virus is actively replicating. In contrast, our study suggests that immune-modulating agents may be effective during the second week of illness.
This study has some limitations. First, the number of patients analyzed in this study was small. Although we could find important differences of CMI responses in severe and mild COVID-19 cases which were consistent with clinical course, it would be better for this to be verified in a larger cohort. Second, it would be desirable to have more time points to be examined to elucidate the exact temporal changes of such responses or when the persistent cytotoxic T-cell activity returns to normal in severe cases. Finally, the potential immunomodulatory effects of lopinavir/ritonavir could not be adjusted because the drug was prescribed to all severe patients in this study.
Conclusion
We observed a higher degree of proliferation, activation, and cytotoxicity of T-cells accompanying the lack of cytotoxic T-cell contraction in severe COVID-19 cases than in mild cases, which may contribute to the progression of pneumonia during the second week of illness.
Ethics approval and consent to participate
Institutional Review Boards of Seoul National University Hospital and Seoul National University Bundang Hospital approved the study (IRB No. 2003-011-1105 and B-2004/607-401).
Funding
This work was supported in part by the Creative-Pioneering Researchers Program through Seoul National University (to H.-R. Kim), the Bio & Medical Technology Development Program of the National Research Foundation (NRF) & funded by the Korean government (MSIT) (RF-2018M3A9H4055197, H.-R. Kim), and Seoul National University Hospital Research Fund (04-2020-0030, M.-d. Oh).
Disclaimer
The funding agencies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Conflict of interest
The authors declare no competing interests.
Authors’ contributions
Chang Kyung Kang, Hang-Rae Kim, and Myoung-don Oh conceived the study, Chang Kyung Kang, Gi-Chan Han, Minji Kim, Gwanghun Kim, Hyun Mu Shin, Hang-Rae Kim, and Myoung-don Oh carried out the analysis, Chang Kyung Kang, Hang-Rae Kim, and Myoung-don Oh drafted the first manuscript, all authors discussed the results, critically read and revised the manuscript, and gave final approval for publication.
Acknowledgment
None.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2020.05.106.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Altman J.D., Moss P.A., Goulder P.J., Barouch D.H., McHeyzer-Williams M.G., Bell J.I. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Zhao H., Song P., Zhang Z., Chen J., Zhou Y.H. Dynamic changes of lymphocyte counts in adult patients with severe pandemic H1N1 influenza A. J Infect Public Health. 2019;12(6):878–883. doi: 10.1016/j.jiph.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B.A., Ko Y., Kim Y.S., Kim S., Choi M.S., Kim I.S. Phenotypic characterization of peripheral T cells and their dynamics in scrub typhus patients. PLoS Negl Trop Dis. 2012;6(8) doi: 10.1371/journal.pntd.0001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colamussi M.L., White M.R., Crouch E., Hartshorn K.L. Influenza A virus accelerates neutrophil apoptosis and markedly potentiates apoptotic effects of bacteria. Blood. 1999;93(7):2395–2403. [PubMed] [Google Scholar]
- Cornberg M., Kenney L.L., Chen A.T., Waggoner S.N., Kim S.K., Dienes H.P. Clonal exhaustion as a mechanism to protect against severe immunopathology and death from an overwhelming CD8 T cell response. Front Immunol. 2013;4:475. doi: 10.3389/fimmu.2013.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juno J.A., van Bockel D., Kent S.J., Kelleher A.D., Zaunders J.J., Munier C.M. Cytotoxic CD4 T cells-friend or foe during viral infection? Front Immunol. 2017;8:19. doi: 10.3389/fimmu.2017.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.R., Hong M.S., Dan J.M., Kang I. Altered IL-7Ralpha expression with aging and the potential implications of IL-7 therapy on CD8+ T-cell immune responses. Blood. 2006;107(7):2855–2862. doi: 10.1182/blood-2005-09-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.R., Hwang K.A., Kang I. Dual roles of IL-15 in maintaining IL-7RalphalowCCR7- memory CD8+ T cells in humans via recovering the phosphatidylinositol 3-kinase/AKT pathway. J Immunol. 2007;179(10):6734–6740. doi: 10.4049/jimmunol.179.10.6734. [DOI] [PubMed] [Google Scholar]
- Kim H.R., Hwang K.A., Kim K.C., Kang I. Down-regulation of IL-7Ralpha expression in human T cells via DNA methylation. J Immunol. 2007;178(9):5473–5479. doi: 10.4049/jimmunol.178.9.5473. [DOI] [PubMed] [Google Scholar]
- Kim J.S., Cho B.A., Sim J.H., Shah K., Woo C.M., Lee E.B. IL-7Ralphalow memory CD8+ T cells are significantly elevated in patients with systemic lupus erythematosus. Rheumatology (Oxford) 2012;51(9):1587–1594. doi: 10.1093/rheumatology/kes100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Shin E.C. The activation of bystander CD8(+) T cells and their roles in viral infection. Exp Mol Med. 2019;51(12):1–9. doi: 10.1038/s12276-019-0316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yan L.M., Wan L., Xiang T.X., Le A., Liu J.M. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20(6):656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndhlovu Z.M., Kamya P., Mewalal N., Kloverpris H.N., Nkosi T., Pretorius K. Magnitude and kinetics of CD8+ T cell activation during hyperacute HIV infection impact viral set point. Immunity. 2015;43(3):591–604. doi: 10.1016/j.immuni.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.D., Park W.B., Choe P.G., Choi S.J., Kim J.I., Chae J. Viral load kinetics of MERS coronavirus infection. N Engl J Med. 2016;375(13):1303–1305. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- Sim J.H., Kim K.S., Park H., Kim K.J., Lin H., Kim T.J. Differentially expressed potassium channels are associated with function of human effector memory CD8(+) T cells. Front Immunol. 2017;8:859. doi: 10.3389/fimmu.2017.00859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares A., Govender L., Hughes J., Mavakla W., de Kock M., Barnard C. Novel application of Ki67 to quantify antigen-specific in vitro lymphoproliferation. J Immunol Methods. 2010;362(1–2):43–50. doi: 10.1016/j.jim.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020 doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Novel coronavirus (2019-nCoV) situation reports. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ [Accessed at 19 May 2020] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. 2020;50(5):e13233. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.