Abstract
The Drug Design Data Resource (D3R) aims to identify best practice methods for computer aided drug design through blinded ligand pose prediction and affinity challenges. Herein, we report on the results of Grand Challenge 4 (GC4). GC4 focused on proteins beta secretase 1 and Cathepsin S, and was run in an analogous manner to prior challenges. In Stage 1, participant ability to predict the pose and affinity of BACE1 ligands were assessed. Following the completion of Stage 1, all BACE1 co-crystal structures were released, and Stage 2 tested affinity rankings with co-crystal structures. We provide an analysis of the results and discuss insights into determined best practice methods.
Keywords: D3R, docking, scoring, ligand ranking, free-energy, blinded prediction challenge
2. INTRODUCTION
Drug discovery remains a time consuming and costly venture. Among the central goals of computer aided drug design (CADD) technologies are amelioration of these issues through more rapid and cost effective in silico experiments. Given availability of three-dimensional (3D) atomic coordinates of a protein target of interest, two primary activities of CADD programs are to both predict (1) the bound conformation (pose) of candidate ligands, and (2) the binding affinity, or affinity ranking, of those ligands to the target macromolecule [1–3]. The CADD community has witnessed an explosion in methodologies and software, which seek to accomplish (1) and (2) [4–39]. However, these new technologies are rarely if ever compared on an equal footing, instead relying mainly on retrospective benchmark datasets that are subject to bias [40], and this remains a challenge for prospective application of these tools.
To address these challenges, the Drug Design Data Resource (D3R; www.drugdesigndata.org) was established. D3R, built upon the prior work of Community Structure Activity Resource (CSAR) [41–44], provides opportunities for blinded prospective methods benchmarking and comparison on hitherto privately-held data sets kindly provided by (primarily) industrial partners. To date, the D3R has conducted four major challenges [6, 30, 45]. The results of Grand Challenge 4 (GC4) reported herein. In total, GC4 saw our broadest participation levels to date, with 51 unique participants responsible for a total of 407 submissions. Herein, we outline the datasets employed, challenge assessment procedures, and prediction results, while seeking best practices methods for the field. A complementary set of articles from individual challenge participant laboratories accompanies this overview in the present special issue of the Journal of Computer-Aided Molecular Design.
3. METHODS
3.1. DATASETS AND SUBCHALLENGES
Grand Challenge 4 (GC4) is a blinded prediction challenge with components addressing pose prediction, affinity ranking, and free energy calculations. GC4 is based on two different protein targets, beta secretase 1 (BACE1) and Cathepsin S (CatS). These data sets were generously contributed by Novartis Institutes for Biomedical Research and Janssen Pharmaceuticals, Inc., respectively.
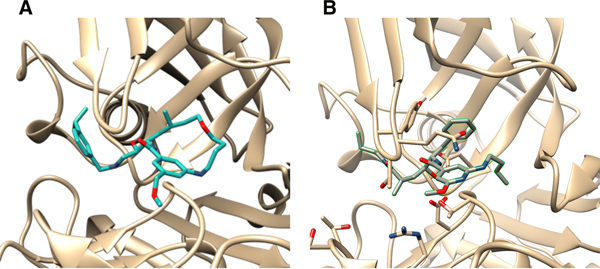
The BACE1 dataset encompasses 154 small molecule inhibitors with previously undisclosed crystallographic structures. Specifically, the BACE1 challenge included all three challenge components, and was based on 154 BACE1 inhibitors for affinity ranking, 20 for pose prediction, and 34 for free energy computation. Many of the ligands are large and flexible macrocycles. For pose prediction, the 20 ligands constituted a diverse set with many macrocycles. All the ligands bind the same region of the protein, with Figure 1 providing illustrative views of the binding modes for two of the BACE1 ligands. In total, 19 distinct Bemis-Murcko scaffolds are present in this set. Four of the BACE1 docking ligands are Lipinski rule-of-5 (Ro5) compliant [46], and 19 are Veber rule compliant [47]. We find that the physiochemical properties of BACE1 docking ligands span a diverse range of values: 1.5–7.5 ClogP, 350–670 Da, 3–16 rotatable bonds, for example. Supplementary Figure 1 presents histograms of the number of heavy atoms, ClogP, molecular weight, number of hydrogen bond acceptors/donors, number of rotatable bonds, number of rings, and topological surface area (TPSA). We next sought to identify how similar the BACE1 docking ligands were to ligands in publicly available BACE1 co-crystal structures. This was done by calculating Tanimoto coefficients to the nearest neighbor ligand (1nn) in the PDB. All Tanimoto coefficients were calculated using ECFP6 Morgan fingerprints (radius=3, nBits=1024) with RDKit [48]. Here, the nearest neighbor ligand is defined to be the ligand with the maximum similarity to the query ligand, and these maximum similarities span a broad range, with a maximum of 0.79, and a minimum of 0.28. Thus, the BACE1 docking ligands spanned a range of similarity to publicly known ligands in co-crystal structures.
Figure 1:
Binding pose from the BACE_10 (A) and BACE_20 (B) co-crystal structures used in GC4
For affinity prediction, the ligand affinities span a three order of magnitude (nM to μM) range of IC50s (Supplementary Figure 2), and the BACE1 free energy set involves a scaffold hopping challenge of cyclic (macrocycles) and non-cyclic (linear) compounds, making for a challenging free energy prediction component [49]. This is illustrated in Figure 1A and 1B. Detailed descriptions of the crystallization conditions for all 20 BACE1 co-crystal structures used in the challenge can be found in the supplementary material of the following literature [50]; and those of the assay conditions employed for GC4 BACE1 affinity data generation can be found in reference [51].
The CatS dataset constitutes a follow-on challenge to GC3, composed of non-peptidic, non-covalent, small molecule inhibitors with measured binding affinities ranging over three orders of magnitude range (nM to μM) of IC50s for CatS. A histogram of the pIC50 values is provided in Supplementary Figure 2. In all, the D3R data set provides 459 CatS inhibitors for affinity ranking, and 39 molecules for free energy prediction. Unlike BACE1, the CatS free energy data set focuses on a single chemical scaffold. A detailed description of the binding assay conditions was published in our previous GC3 publication and a Janssen publication [6, 30, 45, 52].
3.2. POSING THE CHALLENGE
GC4 constitutes the fourth D3R Grand Challenge to date. It followed a similar format to previous challenges [6, 30, 45], including pose prediction, affinity ranking, and free energy prediction components. GC4 followed a two-stage format. Since BACE1 is associated with new co-crystal structures, it included pose prediction component in Stage 1 and an affinity ranking and free energy prediction components in both Stages 1 and 2. CatS has no previously undisclosed co-crystal structures and was hence presented in only one stage that only included affinity ranking and free energy prediction. As in GC3, the pose prediction component in Stage 1 was further divided into two sub-stages, wherein structural information was released incrementally to evaluate different aspects of docking. Stage 1a constituted the crossdocking component, in which participants were asked to dock 20 BACE1 ligands whose co-crystal structures were withheld. Participants thus needed to select their own receptors for docking from the Protein Data Bank (PDB) archive (rcsb.org). Following Stage 1a, all 20 BACE1 co-crystal structures were unblinded, and participants were asked to redock each ligand to its associated crystal structure as part of a self-docking challenge in Stage 1b. In both Stage 1a and 1b, participants were allowed to submit up to 5 poses per ligand, with their “best guess” being designated as “Pose 1.” Participants were asked to align their poses to a designated structure to facilitate evaluation.
3.3. EVALUATION OF POSE AND AFFINITY PREDICTIONS
Prediction evaluations followed the same procedure as previous Grand Challenges, with all evaluation scripts available on Github (drugdesigndata.org/about/workflows-and-scripts). Pose predictions were evaluated in terms of the symmetry-corrected RMSD between predicted and crystallographic poses. These were calculated with the binding site alignment tool in the Maestro Prime Suite (align-binding-sites), where a secondary structure alignment of the full proteins is performed, followed by an alignment of the binding site Cα atoms within 5 Å of the ligand atoms [53]. The pose prediction evaluation results discussed herein are restricted to Pose 1 RMSDs, unless otherwise noted. Additional statistics, including lowest RMSD (“Closest Pose”) and mean pose (“All Poses”), are provided on the D3R website. Affinity predictions were evaluated in terms of the ranking statistics Kendall’s τ [54, 55], Spearman’s ρ [56], and the centered root-mean-square error (RMSEc) for the free energy sets, recomputed in 10,000 rounds of resampling with replacement to generate error bars based in experimental uncertainty following the same procedure seen in all previous challenges [6, 30, 45]. Experimental uncertainties were added to the free energy, ΔG, as a random offset δG drawn from a Gaussian distribution of mean zero and standard deviation RTln(Ierr). In this evaluation, the value of Ierr was set to 2.5, based on the estimated experimental uncertainty. As also noted in previous challenges, two null models were used as performance baselines for ranking ligand potencies [6, 30, 45]. The null models are “Mwt”, in which the affinities were ranked by decreasing molecular weight; and “clogP,” in which affinities were ranked based on increasing octanol–water partition coefficient estimated computationally by RDKit [48, 57].
Machine learning methods were also compared to another null model, a standard random forest regression model, to establish a baseline of performance for machine learning in the context of publicly available data. Using scikit-learn-0.14.1 [58–60], models were built for each target (CatS and BACE1) using publicly available IC50 data from ChEMBL25 [61] and a concatenated feature vector of Morgan fingerprints (radius=3, 4096 bits) and other molecular descriptors (molecular weight, the topological polar surface area, number of hydrogen donors, number of hydrogen acceptors, clogP, number of heavy atoms, number of rotatable bonds, and number of rings) built using RDKit [48].
4. Results
Grand Challenge 4 (GC4) garnered excellent community participation, with 51 unique participants contributing a total of 407 submissions. Details of the methods employed and of the performance statistics can be found in the supplementary materials. All information, including raw protocol files, identities of participants (for those who are not anonymous), and additional analysis statistics can be found on the D3R website (drugdesigndata.org). Finally, many of the submissions and methods are discussed by those responsible in this special issue.
4.1. POSE PREDICTIONS
4.1.1. Overview of pose prediction accuracy
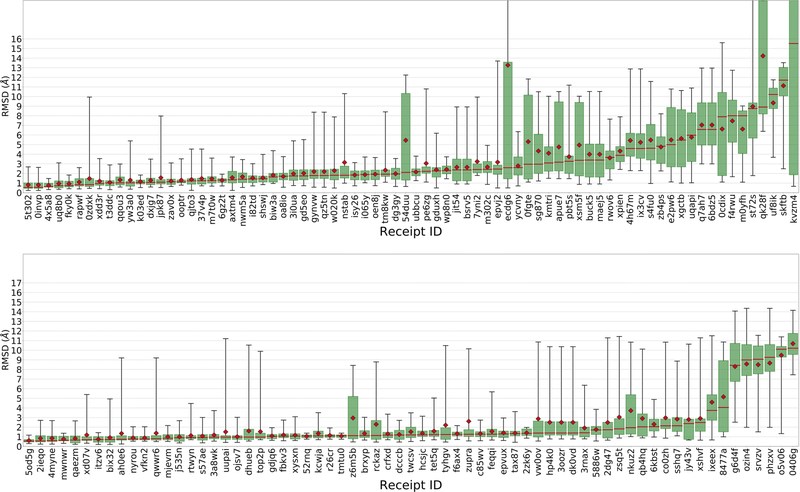
The RMSD statistics of the 20 BACE1 ligands demonstrated excellent pose prediction capabilities, despite the apparent complexity of the ligands (Figure 2). In total, 60% of all Stage 1a submissions achieved a median Pose 1 RMSD < 2.5 Å. Few submissions in Grand Challenge 3 (GC3)[30], achieved this level of cross docking accuracy. For self-docking, Stage 1b saw 59 out of 71 submissions (83%) obtain a median Pose 1 RMSD<2.5 Å. Interestingly, the top performing submission in Stage 1a performed as well as the top performing submission in Stage 1b, with a 0.5 median Pose 1 RMSD. Furthermore, the top 10 performing submissions in Stage 1a performed as well as those in Stage 1b.
Figure 2.
A. Box plots of Pose 1 RMSD statistics for all Stage 1a pose prediction submissions. B. Box plots of pose 1 RMSD statistics for all Stage 1b pose prediction submissions. X-axis labels are Receipt IDs, anonymized identifiers for the various submissions. All data are for BACE1.
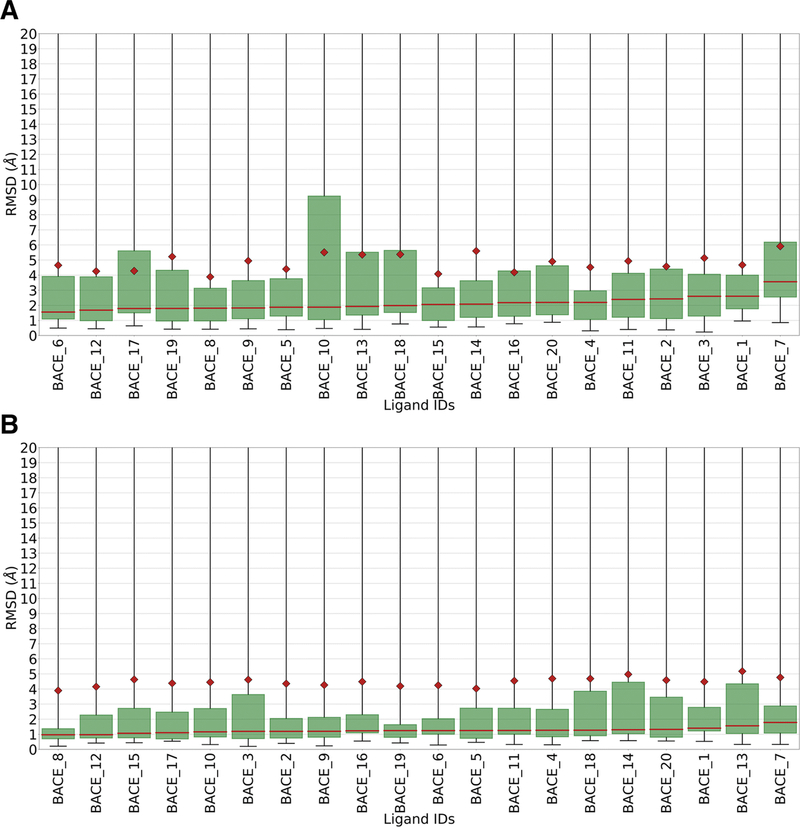
When viewed by ligand pose prediction performance as opposed to method performance, we observed no statistically significant variation in the ligand pose prediction performance RMSD metric. Instead, all ligands performed statistically equally well. Per ligand performance statistics are shown in figure 3.
Figure 3.
A. Box plots of RMSD statistics for each ligand in Stage 1a. B. Box plots of RMSD statistics for each ligand in Stage 1b.
BACE1 is an extensively studied target, due to its potential role in Alzheimer’s disease [62]. Indeed, over 300 BACE1 crystal structures were present in the PDB at the time of this challenge. In principal then, one possible explanation for excellent performance metrics could be that co-crystal structures were available for highly similar ligands, which could be used to guide the D3R docking exercise. However, we find a wide range of 1nn Tanimoto coefficients between the present ligands and the ligands available in the PDB; thus, if the availability of similar ligand structures were central to success, we should not have seen such similar performance across ligands. In fact, we find that the 1nn distribution is unable to distinguish difficult from easy docking challenges, such as CatS in GC3[30], as all ligands have essentially equivalent RMSD statistics (see above). We note that CatS, the target in GC3 [30], had a similar 1nn distribution, yet exhibited considerably poorer docking predictive accuracy. All 1nn distributions are shown in supplementary figure 3.
In summary, pose prediction results were of high accuracy across nearly all submissions. Performance was similar across all ligands, despite the wide range of maximum-similarity ligands available in BACE co-crystal structures in the PDB. Thus, the performance metrics discussed herein suggest that the docking methods themselves performed well.
4.1.2. Analysis by docking methodology
Our experience shows that multiple docking/cheminformatics software tools produced submissions of comparable accuracy. These software packages include docking software such as AutoDock Vina [63], Glide [64, 65], ICM [66], Corina [67], Gold [68], Cactvs[69], Rosetta [70], and EFindSite [71], and ligand preparation software such as Brikard [72], RDKit [48], Open Babel [73] and Maestro [74]. In addition, almost all submissions in Table 1 used more than one type of software package in their workflow, preferring rather to combine multiple docking software packages. New to the list of top performing methods is the application of deep learning from Guowei Wei’s group. However, insufficient detail regarding the deep learning methodology was provided in the submitted protocol file to permit elaboration on the method. When viewed by median Pose 1 RMSD, and accounting for the standard deviations in Table 1, it is apparent that all of the methods appear to be statistically similar.
Table 1.
Top-performing pose predictions for BACE1 Stage 1a (A) and Stage 1b (B), based on median Pose 1 RMSD (Å). The standard deviations (SD RMSD) of the Pose 1 RMSDs are also provided as measure of scatter. Software lists the software listed by the participants in their protocol files. Submitter/PI: names of submitter and principal investigator (PI) provided with submission. Organization: institution of PI provided with submission. Visual Inspection lists the participant’s response to the standard question “Did you use visual inspection to select, eliminate, and/or manually adjust your final predicted poses?” Similar Ligands lists the participant’s response to the standard question “Did you use publicly available co-crystal structures of this protein with similar ligands to guide your pose predictions?”
| (A) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Median RMSD | Mean RMSD | SD RMSD | Software | Submitter/PI | Organization | Visual Inspection | Similar Ligands | Submission ID |
| 0.55 | 0.81 | 0.58 | efindsite 1.3, openbabel 2.4.1, discover studio visualizer 4.5, maestro 10.2, shafts, gaussian 09, amber 16, homemade deep learning | K. Gao/G. Wei | Michigan State | yes | yes | 5t302 |
| 0.56 | 0.81 | 0.57 | efindsite 1.3, openbabel 2.4.1, discover studio visualizer 4.5, maestro 10.2, shafts, gaussian 09, amber 16, homemade deep learning | K. Gao/G. Wei | Michigan State | yes | yes | 0invp |
| 0.64 | 0.77 | 0.34 | brikard, libmol, rdkit, openbabel | D. Kozakov/D. Kozakov | Stony Brook | yes | yes | 4x5a8 |
| 0.72 | 0.94 | 0.65 | rdkit/torch/macromodel | Anonymous | Anonymous | yes | yes | uq8b0 |
| 0.74 | 0.93 | 0.45 | molsoft icm 3.8–7b | P. Lam/M. Totrov | Molsoft | no | yes | fky0k |
| 0.83 | 1.46 | 2.07 | cactvs chemoinformatics toolkit v3.433/schrodinger suite 2018–1/corina v3.60/ucsf chimera v1.10.2/gold v5.2 | I. Bogdan/I. Bogdan | Institut de Chimie des Substances Naturelles | no | yes | 0zdxk |
| 0.83 | 1.1 | 0.63 | cactvs chemoinformatics toolkit v3.433/schrodinger suite 2018–1/corina v3.60/ucsf chimera v1.10.2/gold v5.2 | I. Bogdan/I. Bogdan | Institut de Chimie des Substances Naturelles | no | yes | rapwf |
| 0.96 | 1.2 | 0.8 | Rosetta/corina classic, webserver version/openbabel-2.4.1/antechamber-17.3 | H. Park/ | University of Washington | no | yes | xdd3r |
| 1.02 | 1.33 | 0.71 | htmd1.13.8/acemd2/rdkit2018.03.4 | A. Rial/G. Fabritiis | Accelera | yes | yes | qqou3 |
| 1.02 | 1.06 | 0.57 | maestro/openeye/mgltools/autodock vina | X.Xu/X. Zou | University of Missouri-Columbia | yes | yes | t3ddc |
| (B) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Median RMSD | Mean RMSD | SD RMSD | Software | Submitter/PI | Organization | Visual Inspection |
Similar Ligands | Submission ID |
| 0.56 | 0.61 | 0.23 | molsoft icm 3.8–7b | L.Polo/M.Totrov | Molsoft | No | no | 5od5g |
| 0.57 | 0.84 | 0.58 | efindsite 1.3, openbabel 2.4.1, discover studio visualizer 4.5, maestro 10.2, shafts, guassian 09, amber 16, homemade deep learning | K. Gao/G.Wei | Michigan State | yes | yes | 2ieqo |
| 0.59 | 0.85 | 0.56 | efindsite 1.3, openbabel 2.4.1, discover studio visualizer 4.5, maestro 10.2, shafts, guassian 09, amber 16, homemade deep learning | K. Gao/G.Wei | Michigan State | yes | yes | 4myne |
| 0.64 | 0.76 | 0.32 | brikard, libmol, rdkit, openbabel, vina, libsampling | D.Kozakov/D.Kozakov | Stonybrook | yes | yes | mwnwr |
| 0.66 | 0.84 | 0.63 | rdkit/torch/macromodel | Anonymous | Anonymous | no | yes | qaezm |
| 0.69 | 1.18 | 1.27 | moe2016.08/autodock4/mgltools1.5.7rc1/amber16/ligprep/rdkit2018–3 | M.Carlos/M.Marti | yes | yes | xd07v | |
| 0.71 | 0.92 | 1.01 | schrodinger/in-house deep learning | D.Nguyen/G.Wei | Michigan State | no | yes | bix32 |
| 0.71 | 0.74 | 0.37 | schrodinger/in-house deep learning | D.Nguyen/G.Wei | Michigan State | no | yes | itzv6 |
| 0.74 | 1.34 | 1.92 | openbabel 2.3.2 maestro 2018–3 prime smina apr 2 2016 | B.Wingert/C.Camacho | university of pittsburgh | yes | yes | ah0e6 |
| 0.76 | 0.89 | 0.35 | autodock vina with in-house modifications (convex-pl as a scoring function), rdkit 2018, scipy, pymol 1.8.4, unreleased version of convex-pl scoring function | M.Kadukova/S.Grudinin | inria grenoble, mipt moscow | no | yes | nyrou |
We note that the docking of cyclic molecules presents a conformational sampling problem for docking [75]. Participants adopted diverse strategies to address this. For example, both Brikard [72] and Gold [68] were used to sample macrocyclic molecules during docking. Elsewhere, AutoDock4 [76] was used in a workflow that docked ligands in an open conformation and used a linear potential to restore broken bonds. Also, RDKit [48] and Open Babel [73] were each used for macrocycle conformer generation.
A limitation of many docking methods is their inability to account for the fact that different ligands lead to different binding-site conformations [77–79]. As noted for “Lessons Learned” in previous D3R GCs, as well as by others [80], one strategy to reduce this problem is ligand-guided similarity docking, where receptors co-crystallized with similar ligands to the one under question are selected as the docking receptor. Examination of the protocols provided by participants revealed that all of top-performing methods in Stage 1a used ligand-guided similarity docking, and 8/10 top-performing methods used this approach in Stage 1b. In total, 56/78 submissions in Stage 1a, and 48/71 submissions in Stage 1b, employed ligand-guided similarity docking. To determine impact on performance, we first omitted submissions with a median Pose 1 RMSD > 5 Å to exclude submissions that simply docked to the wrong pocket. We then recorded the mean of the median pose 1 RMSDs for submissions that used ligand-guided similarity docking, and those that did not use ligand-guided similarity docking. Submissions that employed ligand-guided similarity docking had a mean median Pose 1 RMSD of 2.1 +/− 1.2 Å. Submissions that did not employ ligand guided similarity docking had a mean median Pose 1 RMSD of 2.0 +/− 1.0 Å. The Mann-Whitney U statistic and p-value of these two submission subsets are 311.0 and 0.25 respectively, not allowing us to reject the null hypothesis that these statistics come from the same distribution. Moreover, although 10/10 of the top performing methods in Stage 1a used ligand guided similarity docking, so did 7/10 of the bottom performing methods. In aggregate, these results are inconclusive regarding whether or not ligand-guided similarity docking aided performance in GC4.
Visual inspection is frequently used to select final poses, with the impact of this having mixed results across previous Grand Challenges [6, 30, 45]. Six of the top 10 submissions employed visual inspection to select their final pose versus only two of the bottom ten performing submissions. This finding suggests that visual inspection augmented docking accuracy, albeit dependent on the quality of the scientist’s intuition and experience, as noted in GC3[30] and GC2[6].
In summary, multiple software packages achieve similar accuracy. The results herein are inconclusive as to whether ligand-guided similarity docking improved performance for the BACE1 ligands. Finally, visual inspection appeared to provide a slight performance benefit.
4.2. AFFINITY PREDICTIONS
In this section, we evaluate the accuracy of the predicted ligand-potency rankings and binding free energies for protein targets BACE1 and CatS. Because detailed binding free energy calculations are computationally demanding, these were limited to focused subsets of the ligands, termed Free Energy Sets. BACE1 was presented as a two-stage challenge, where new co-crystal structures involving 16 of the challenge ligands were not disclosed to participants until the opening of Stage 2. This allowed us to probe the performance of structure-based methods in ranking the ligands in the presence and absence of added structural data. For CatS, only one stage was presented, as no previously undisclosed co-crystal structures were available to D3R. However, the CatS data used for GC4 were drawn from a large dataset, provided by Janssen, which was also used in GC3. Thus, it was of interest to see whether the availability of more data and structures, as well as possible improved computational methodology, would lead to improved performance in GC4.
4.2.1. Overview of Potency Ranking
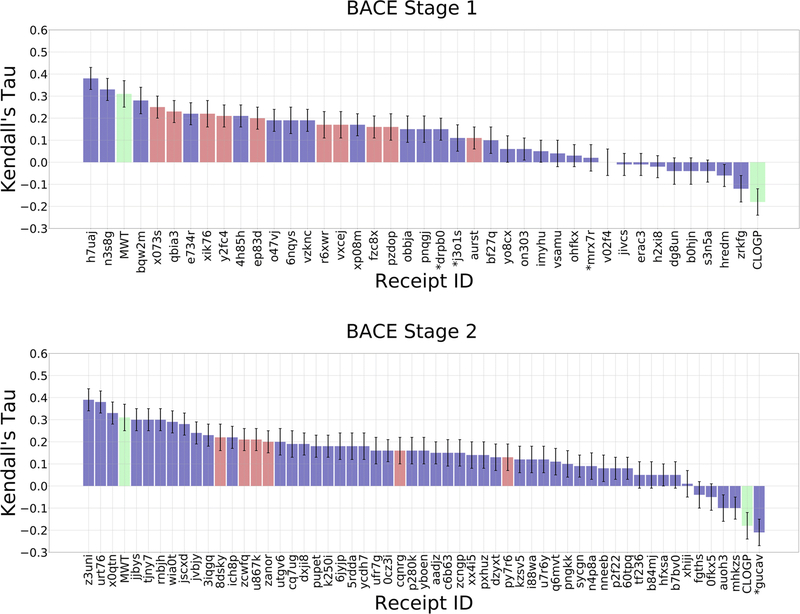
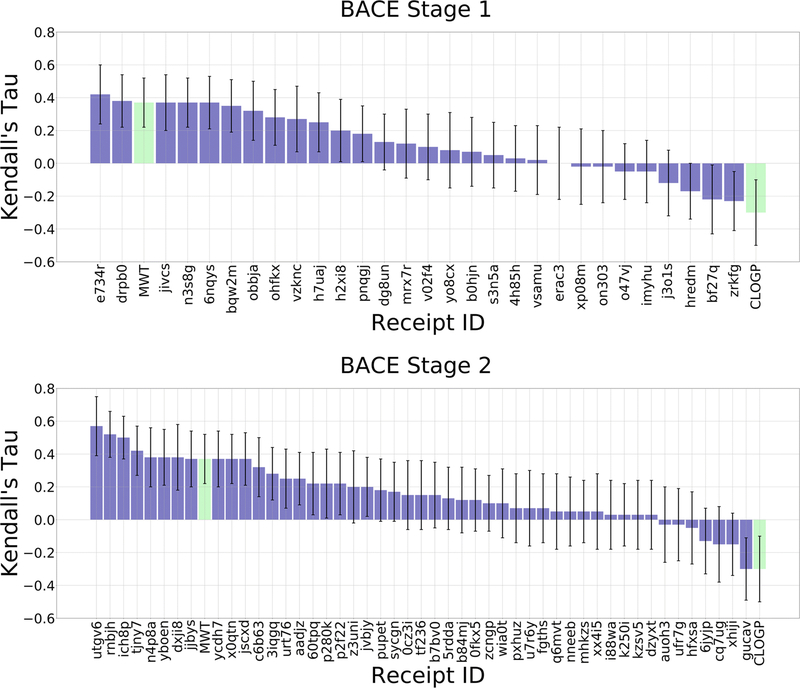
As in all previous D3R challenges, the majority of submissions give positive correlations with experimental data across all targets (Figures 4 and 5). Here, Kendall’s τ reaches values of 0.38 ± 0.05, 0.39 ± 0.05, and 0.54 ± 0.02 for BACE1 Stage 1, BACE1 Stage 2, and CatS, respectively (Table 2). The top methods clearly outperform the molecular weight and clogP null models, which achieve Kendall’s τ values of 0.26 ± 0.03 and −0.15 ± 0.03, respectively, for CatS; and 0.31 ± 0.06 and −0.18 ± 0.06, respectively, for BACE1. However, the molecular weight null model does outperform the mean Kendall’s τ values across all submissions, for all targets and Stages. The mean Kendall’s τs are 0.11, −0.14, and 0.20 for BACE1 Stage1, BACE1 Stage 2, and CatS, respectively. Thus, many methods still underperform the null model, where ligands were simply ranked based on molecular weight. However, we observed a difference in the number of methods outperforming the null models between the two targets. For BACE1, only two or three methods had greater Kendall’s τ values than the molecular weight null model, in Stages 1 and 2 respectively, while for CatS, 18 methods outperformed this null model. In this sense, the computational methods performed better for CatS than for BACE1.
Figure 4.
Kendall’s τ ranking correlation coefficients between predicted IC50 rankings and experimental IC50 rankings for the BACE1 dataset in Stages 1 and 2. Purple columns are for structure-based scoring methods, red bars are for ligand-based scoring, and green bars are for the null models where ligands are ranked based on molecular weight (MW) and the computed logarithm of the partition coefficient between n-octanol and water (CLOGP), as indicated in the axis labels. Receipt IDs labeled by an asterisk did not use the full set of challenge ligands. The error bars are 1σ confidence intervals based on 10,000 bootstrap samples
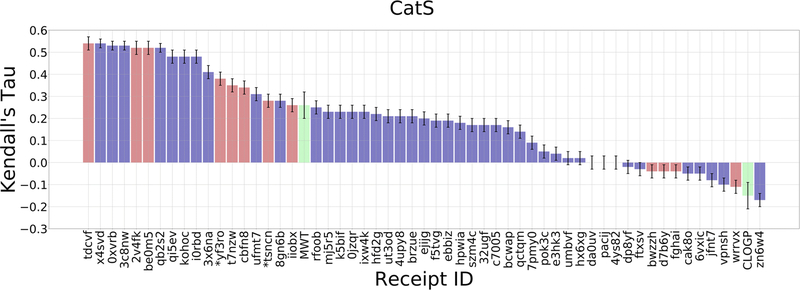
Figure 5.
Kendall’s τ ranking correlation coefficients between predicted IC50 rankings and experimental IC50 rankings for the CatS dataset. See Figure 4 for details.
Table 2.
Top 10 submissions, based on Kendall’s τ, for each affinity ranking challenge. See Table 1 for details.
| Kendall’s τ | Software | Submitter/PI | Organization | Submission ID |
|---|---|---|---|---|
| BACE1 Stage 1 | ||||
| 0.38 | cactvs chemoinformatics toolkit v3.433/schrodinger suite 2018–1/corina v3.60/ucsf chimera v1.10.2/gold v5.2 | B. Iorga/B. Iorga | Institut De Chimie Des Substances Naturelles, Cnrs, Gif-Sur-Yvette, France | h7uaj |
| 0.33 | cactvs chemoinformatics toolkit v3.433/schrodinger suite 2018–1/corina v3.60/ucsf chimera v1.10.2/gold v5.2 | B. Iorga/B. Iorga | Institut De Chimie Des Substances Naturelles, Cnrs, Gif-Sur-Yvette, France | n3s8g |
| 0.28 | openbabel, multiconf-dock and pl-patchsurfer2 | W. Shin/D. Kihara | Purdue University, Department of Biological Science | bqw2m |
| 0.25 | python2.7, scikit-learn, rdkit | J. Shamsara/J. Shamsara | Mashhad Unicversity Of Medical Sciences, Mashhad, Iran | x073s |
| 0.23 | rdkit (2018.03.4), keras (2.2.2) | J. Yin/A. Shabani | Qulab Inc. | qbia3 |
| 0.22 | bcl v3.5, corina v4.1.0 | B. Brown/J. Meiler | Vanderbilt University | xik76 |
| 0.22 | lead finder 1808 | O. Stroganov/Biomoltech inc. | Biomoltech Inc. | e734r |
| 0.21 | bcl v3.5, corina v4.1.0 | B. Brown/J. Meiler | Vanderbilt University | y2fc4 |
| 0.21 | molsoft icm 3.8–7b | P. lam/M. Totrov | Molsoft, San Diego | 4h85h |
| 0.2 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | ep83d |
| BACE1 Stage 2 | ||||
| 0.39 | htmd1.13.8/rdkit2018.03.4/kdeep | A. Varela Rial/G. De Fabritiis | Acellera | z3uni |
| 0.38 | cactvs chemoinformatics toolkit v3.433/schrodinger suite 2018–1/corina v3.60/ucsf chimera v1.10.2/gold v5.2 | B. Iorga/B. Iorga | Institut De Chimie Des Substances Naturelles, Cnrs, Gif-Sur-Yvette, France | urt76 |
| 0.33 | cactvs chemoinformatics toolkit v3.433/schrodinger suite 2018–1/corina v3.60/ucsf chimera v1.10.2/gold v5.2 | B. Iorga/B. Iorga | Institut De Chimie Des Substances Naturelles, Cnrs, Gif-Sur-Yvette, France | x0qtn |
| 0.3 | ag/dg/tdl-bp/schrodinger | K. Gao; D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | tjny7 |
| 0.3 | smina/data-warrior4.7.2/pycharmce4.5/sklearn | B. Wang/H. NG | Kansas State University | rnbjh |
| 0.3 | rdkit/gnina/smina | P. Francoeur/D. Koes | University of Pittsburgh | jjbys |
| 0.29 | rosetta (pre-release version, git hash fc616bc278565f41a234093f1dee53b196432524)/corina classic, webserver version/openbabel-2.4.1/antechamber-17.3 | H. Park/Institute for Protein Design | University of Washington | wia0t |
| 0.28 | openbabel, multiconf-dock, and pl-patchsurfer2 | W. Shin/D. Kihara | Purdue University, Department of Biological Science | jscxd |
| 0.24 | ag/dg/tdl-bp/schrodinger | M. Wang; D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | jvbjy |
| 0.23 | ag/dg/tdl-bp/schrodinger | M. Wang; D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | 3iqgq |
| CatS | ||||
| 0.54 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | tdcvf |
| 0.54 | molsoft icm 3.8–7b | P. lam/M. Totrov | Molsoft, San Diego | x4svd |
| 0.53 | ag/dg/tdl-bp/schrodinger | D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | 0xvrb |
| 0.53 | ag/dg/tdl-bp/schrodinger | K. Gao; D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | 3c8nw |
| 0.52 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | 2v4fk |
| 0.52 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | be0m5 |
| 0.52 | ag/dg/tdl-bp/schrodinger | K. Gao; D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | qb2s2 |
| 0.48 | ag/dg/tdl-bp/schrodinger | D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | qi5ev |
| 0.48 | ag/dg/tdl-bp/schrodinger | M. Wang; D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | i0rbd |
| 0.48 | ag/dg/tdl-bp/schrodinger | D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | kohoc |
As CatS was previously provided as a target for Grand Challenge 3 (GC3), we compared the accuracy of predictions herein to those in GC3. The best performing methods in GC4 have a Kendall’s τ of 0.54 ± 0.02, compared to 0.45 ± 0.05 in GC3, and the mean Kendall’s τ for the top 20% of methods in GC4 is 0.50, compared to 0.38 in GC3 (Figure 4). This improvement might result from prior participant experience with GC3, and the availability of the GC3 data to help guide the GC4 calculations. (N.B.: Few participants explicitly mentioned any use of the GC3 data.)
4.2.2. Analysis by affinity prediction methodology
We now review the top-performing method for each challenge target and Stage (Figures 4 and 5, Table 2). In BACE1 Stage 1, the top performing method clearly outperforms all other methods with a Kendall’s τ of 0.38 ± 0.05 (Figure 4, Table 2). This submission from the Iorga lab (D3R Receipt ID: h7uaj) used the Gold docking software and Goldscore scoring function [81]. For BACE1 Stage 2, the top submissions include two methods that perform similarly, with Kendall’s τ of 0.39 ± 0.05 (D3R Receipt ID: z3uni) and 0.38 ± 0.05 (D3R Receipt ID: urt76). One, from the Iorga lab, again uses the Gold docking software and Goldscore scoring function; the other, from Accelera, uses docking and affinity tools called SkeleDock and Kdeep, respectively [82]. For CatS, we received 10 submissions with similar performance around a mean Kendall’s τ of 0.51 ± 0.02 (Figure 5, Table 2). These methods were submitted from three different groups that presented slight variations of their methods. One submission uses a custom ICM-docking procedure and iterative 3D atomic property field quantitative structure–activity relationships (QSAR) model from Molsoft LLC (D3R Receipt ID: x4svd) [19, 32, 83–85]. Three submissions from the Evangelidis lab (D3R Receipt IDs: tdcvf, 2v4fk, be0m5) used variations of their DeepScaffOpt method, where an ensemble of deep neural networks was trained on CatS data from ChEMBL. Lastly, six of these submissions are from the Wei lab (D3R Receipt IDs: 0xvrb, 3c8nw, qb2s2, qi5ev, i0rbd, kohoc) and used variations of their topology-based deep learning methods where features were generated by algebraic graphs, differential geometry, and algebraic topology scores [35, 86–90].
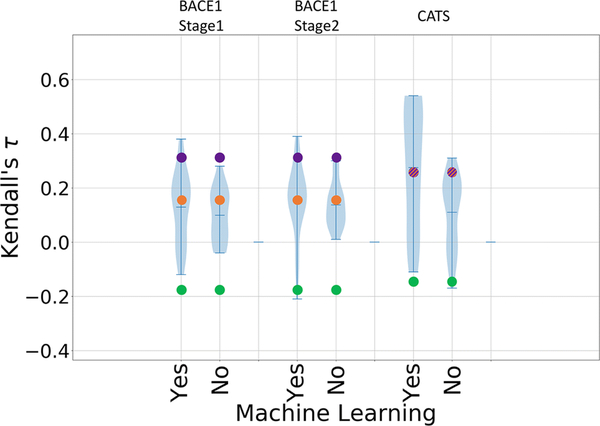
Although fewer than ten submissions used machine learning in GC2, D3R has seen a surge in the development and application of such methods in subsequent challenges. As in GC3, a number of submissions used machine learning in GC4, with 56% of all submissions mentioning use of such methods. Figure 6 shows violin plots of Kendall’s τ values for methods that do and do not use machine learning in each of challenge component. Although it is not clear that there is much difference between the two sets of submissions for BACE1, submissions that used machine learning tended to perform better for CatS, with the top performing methods in the “Yes” category outperforming those in the “No” category. It is also important to note that the Kendall’s τ values of the machine learning methods (marked Yes in Figure 6) have a broader range than those of the other methods (marked No in Figure 6). Interestingly, our own random forest regression null model, constructed with ChEMBL data to establish a baseline of performance for machine learning methods, is outperformed by many of the machine learning methods for BACE1 and even for CatS, where one might have expected the large quantity of available data to support a relatively accurate regression model.
Figure 6.
Violin plots of Kendall’s τ between predicted and experimental IC50 rankings for submissions that use machine learning and those that do not in each target dataset: BACE1 Stages 1 and 2, and CATS. Mean, minimum, and maximum Kendall’s τ for each dataset are shown by whiskers. Null models based on clogP, molecular weight, and a random forest regression model are shown in green, purple, and orange, respectively. Note that the molecular weight and regression model data points are overlapping for CatS.
4.2.3. Relationship between affinity ranking accuracy and pose prediction
A recurring question in our GCs has been whether knowledge of crystallographic poses would improve affinity rankings. We compared the ranking evaluations using only the 16 BACE1 ligands for which crystallographic poses were released between the two stages (BACE 1 and BACE 4 to 20, excluding BACE 17 and 18) (Figure 7). We observe an increase in Kendall’s τs for the top methods from 0.42 ± 0.18 in Stage 1 to 0.57 ±0.18 in Stage 2; and a significant increase in the number of methods with Kendall’s τ greater than or equal to the molecular weight null model of 0.37 ± 0.15 Kendall’s τ totaling 2 methods in Stage 1 versus 8 methods in Stage 2. This indicates a general improvement of the accuracy of affinity rankings coinciding with the knowledge of the crystallographic poses of the ligands released in Stage 2. It should be noted that the methods outperforming the molecular weight null model in the whole set for Stage 2 (z3uni, urt76, x0qtn) did not outperform that null model in this subset of 16 ligands, indicating that the benefit of knowing the crystallographic poses is method dependent.
Figure 7.
Kendall’s τ ranking correlation coefficients between predicted IC50 rankings and experimental IC50 rankings for BACE1 ligands with co-crystal structures released at the end of Stage 1: ligands 1 and 4 to 20, excluding ligands 17 and 18. See Figure 4 for details.
Another assessment of the importance of structural data can be done by looking at the differences in performance between structure-based and ligand-based approaches for BACE1 and CatS affinity rankings. In BACE1, the only methods outperforming the molecular weight null-model in Stages 1 and 2 are structure-based methods (Figure 4). In BACE1 Stage 2, the top-performing methods result in Kendall’s τ of 0.39 ± 0.05 and 0.22 ± 0.06 for structure-based and ligand-based methods, respectively. In CatS, both structure-based and ligand-based approaches perform similarly with top-performing Kendall’s τs of ~0.54 ± 0.02 (Figure 5). Thus, the impact of knowing the crystallographic pose on affinity ranking accuracy could be positive for certain targets and with certain methods.
4.2.4. Binding free energy predictions
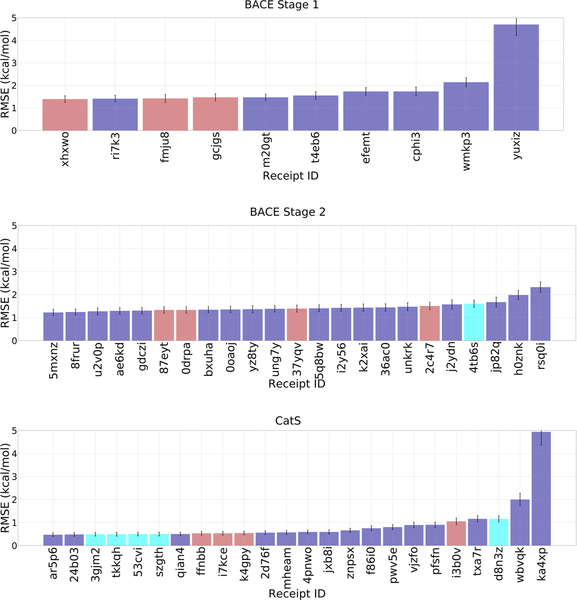
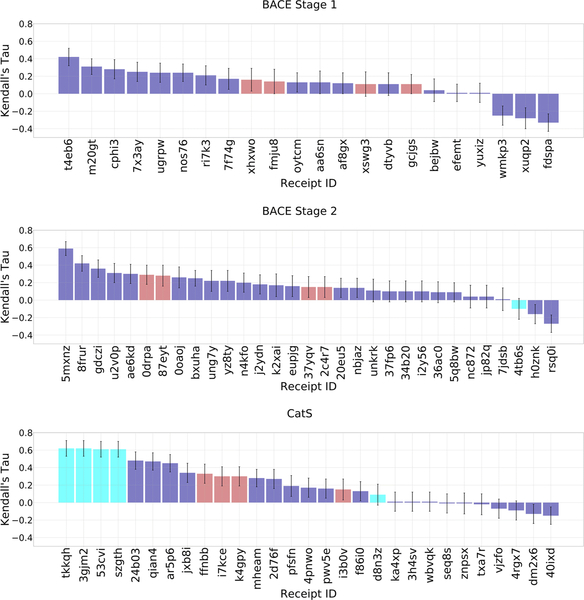
In GC4, we evaluated computationally demanding alchemical methods of predicting relative binding free energy [91–96] within sets of chemically similar ligands binding two targets: BACE1 (34 ligands) and CatS (39 ligands), respectively. The BACE1 free energy set involves scaffold hopping, while the CatS free energy set includes only one chemical scaffold. Although the challenge was designed to test explicit solvent alchemical free energy methods, as in prior GCs, only one and five methods were of this type, for BACE1 Stage 2 and CatS, respectively, while the rest of the submissions were structure-based and ligand-based scoring methods that provided relative binding free energies between pairs of ligands. When compared with experiment, most of the predictions yield RMSEc values of less than 2 kcal/mol and positive Kendall’s tau values (Figures 8 and 9, and Tables 3 and 4). The methods with RMSEc< 2 kcal/mol have values of Kendall’s τ from −0.07 to 0.62, but it is not clear how much this variation reflects random noise versus meaningful differences among the methods, given the small numerical range. For CatS, the top-performing methods include four submissions from two participants in the Simmerling lab using explicit solvent alchemical free energy methods (D3R Receipt IDs: 3gjm2, tkkqh, 53cvi, szgth), where all four methods have a Kendall’s τ of ~0.62 ± 0.09. Indeed, when assessed with Kendall’s τ, these predictions outperformed all other methods, as the next highest value of Kendall’s τ is 0.48 ± 0.1. These four predictions also performed well in terms of RMSEc, yielding an average error of 0.5 kcal/mol, which is within statistical error of the top performing method based on this metric (D3R Receipt ID: ar5p6; RMSEc = 0.47 ± 0.08). Unfortunately, the Simmerling lab did not compete in BACE1. For BACE1, the one alchemical free energy method submitted resulted in a less impressive Kendall’s τ of −0.1 ± 0.12 and RMSEc of 1.6 ± 0.16.
Figure 8.
RMSEc values for the compounds in the free energy prediction sets. Purple bars are for structurebased scoring with free energy estimates, red bars are for ligand-based scoring with free energy estimates, and cyan bars are for methods using explicit solvent alchemical free energy simulations. Receipt IDs that resulted in an RMSEc greater than 5 Å have been omitted for clarity. 12, 8, and 5 submissions were omitted in BACE1 Stage 1, BACE1 Stage2, and CatS, respectively. Receipt IDs labeled with an asterisk did not use the full set of FE ligands. The error bars are 1σ confidence intervals based on 10,000 bootstrap samples
Figure 9.
Kendall’s τ ranking correlation coefficients between predicted IC50 rankings and experimental IC50 rankings for the free energy prediction set ligands. See Figure 8 for details.
Table 3.
Top 10 submissions, based on Kendall’s τ, for each free energy prediction challenge. See Table 3 for details.
| Kendall’s τ | Software | Submitter/PI | Organization | Submission ID |
|---|---|---|---|---|
| BACE1 Stage 1 | ||||
| 0.42 | amber16, ambertools17, as-ie (developed by our own laboratory, doi10.1021/acs.jctc.7b01295) protein forcefield ff14sb ligand forcefield gaff water model tip3p | J. Bao/J. Zhang | East China Normal University | t4eb6 |
| 0.31 | rdkit / mgltools/ smina (modified) | R. Quiroga/M. Villarreal | Universidad Nacional De Cordoba Argentina | m20gt |
| 0.28 | gromacs, g_mmpbsa, acpype protein forcefield amber ligand forcefield gaff amber14 water model tip3p | A. Stander/A. Stander | University of Pretoria | cphi3 |
| 0.25 | homoligalign, sqm/cosmo protein forcefield amber14sb ligand forcefield gaff2 water model tip3p | T. evangelidis/P. Hobza | Uochb & Ceitec | 7x3ay |
| 0.24 | homoligalign, sqm/cosmo protein forcefield amber14sb ligand forcefield gaff2 water model tip3p | T. evangelidis/P. Hobza | Uochb & Ceitec | nos76 |
| 0.24 | omega 3/datawarrior 4.7.3/smina version apr. 29, 2017 with vinardo scoring function/rf-score vs v2 | B. Wang/H. NG | Kansas State University | ugrpw |
| 0.21 | itscore | X. Xu/X. Zou | University of Missouri-Columbia | ri7k3 |
| 0.17 | amber18 for md, amber16 for mmpbsa.py protein forcefield amberff19sb ligand forcefield gaff2 water model tip3p | S. Sasmal; L. El Khoury/D. Mobley | University of California, Irvine | 7f74g |
| 0.16 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | xhxwo |
| 0.14 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | fmju8 |
| BACE1 Stage 2 | ||||
| 0.59 | htmd1.13.8/rdkit2018.03.4/kdeep | A. Varela Rial/G. De Fabritiis | Acellera | 5mxnz |
| 0.42 | ag/dg/tdl-bp/schrodinger | K. Gao;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | 8frur |
| 0.36 | rdkit/gnina/smina | P. Francoeur/D. Koes | University of Pittsburgh | gdczi |
| 0.31 | rdkit/gnina/smina | P. Francoeur/D. Koes | University of Pittsburgh | u2v0p |
| 0.3 | ag/dg/tdl-bp/schrodinger | M. Wang;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | ae6kd |
| 0.29 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | 0drpa |
| 0.28 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | 87eyt |
| 0.26 | ag/dg/tdl-bp/schrodinger | D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | 0oaoj |
| 0.25 | ag/dg/tdl-bp/schrodinger | M. Wang;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | bxuha |
| 0.22 | rdkit/gnina/smina | P. Francoeur/D. Koes | University of Pittsburgh | yz8ty |
| CatS | ||||
| 0.62 | maestro/schrodinger,pmemd.gti,antechamber protein forcefield amber ff14sb ligand forcefield gaff2 water model tip3p | J. Zou/C. Simmerling | Stony Brook University | |
| 0.62 | maestro/schrodinger,pmemd.gti,antechamber protein forcefield amber ff14sb ligand forcefield gaff2 water model tip3p | T. Chuan/C. Simmerling | Stony Brook University | 3gjm2 |
| 0.61 | maestro/schrodinger,pmemd.gti,antechamber protein forcefield amber ff14sb ligand forcefield gaff2 water model tip3p | J. Zou/C. Simmerling | Stony Brook University | tkkqh |
| 0.61 | maestro/schrodinger,pmemd.gti,antechamber protein forcefield amber ff14sb ligand forcefield gaff2 water model tip3p | T. Chuan/C. Simmerling | Stony Brook University | 53cvi |
| 0.48 | ag/dg/tdl-bp/schrodinger | D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | szgth |
| 0.47 | ag/dg/tdl-bp/schrodinger | K. Gao;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | 24b03 |
| 0.45 | ag/dg/tdl-bp/schrodinger | K. Gao;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | qian4 |
| 0.34 | ag/dg/tdl-bp/schrodinger | M. Wang;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | ar5p6 |
| 0.33 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | jxb8i |
| 0.3 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | ffnbb |
Table 4.
Top 10 submissions, based on RMSEc, for each free energy prediction challenge. See Table 3 for details.
| RMSEc | Software | Submitter/PI | Organization | Submission ID |
|---|---|---|---|---|
| BACE1 Stage 1 | ||||
| 1.39 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | xhxwo |
| 1.41 | itscore | X. Xu/X. Zou | University of Missouri-Columbia | ri7k3 |
| 1.42 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | fmju8 |
| 1.47 | rdkit / mgltools/ smina (modified) | R. Quiroga/M. Villarreal | Universidad Nacional De Cordoba Argentina | m20gt |
| 1.47 | htmd1.13.8/acemd3/rdkit2018.03.4/mmenergy/deltadelta protein forcefield amber ligand forcefield amber water model tip3 | A. Varela Rial/G. De Fabritiis | Acellera | gcjgs |
| 1.55 | amber16, ambertools17, as-ie (developed by our own laboratory, doi10.1021/acs.jctc.7b01295) protein forcefield ff14sb ligand forcefield gaff water model tip3p | J. Bao/J. Zhang | East China Normal University | t4eb6 |
| 1.73 | gromacs, g_mmpbsa, acpype protein forcefield amber ligand forcefield gaff amber14 water model tip3p | A. Stander/A. Stander | University of Pretoria | cphi3 |
| 1.73 | schrodinger (2018–2) protein forcefield opls_2005 ligand forcefield opls_2005 water model no water model is used in this method | P. Chen/Department of High Performance Computing Application, National Supercomputer Center in Guangzhou (NSCC-GZ) | Sun Yet-Sen University | efemt |
| 2.14 | tensorflow | X. Xu/X. Zou | University of Missouri-Columbia | wmkp3 |
| 4.7 | autodock vina 1.1.2/rdkit 2018.03.1/openbabel/pymol/fkcombu/acpype/gromacs 5.1.5/g_mmpbsa protein forcefield amber99sb-ildn ligand forcefield gaff water model tip3p | T. wang | None | yuxiz |
| BACE1 Stage 2 | ||||
| 1.22 | htmd1.13.8/rdkit2018.03.4/kdeep | A. Varela Rial/G. De Fabritiis | Acellera | 5mxnz |
| 1.24 | ag/dg/tdl-bp/schrodinger | K. Gao;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | 8frur |
| 1.27 | rdkit/gnina/smina | P. Francoeur/D. Koes | University of Pittsburgh | u2v0p |
| 1.29 | ag/dg/tdl-bp/schrodinger | M. Wang;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | ae6kd |
| 1.3 | rdkit/gnina/smina | P. Francoeur/D. Koes | University of Pittsburgh | gdczi |
| 1.33 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | 0drpa |
| 1.33 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | 87eyt |
| 1.34 | ag/dg/tdl-bp/schrodinger | M. Wang;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | bxuha |
| 1.35 | ag/dg/tdl-bp/schrodinger | D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | 0oaoj |
| 1.36 | rdkit/gnina/smina | P. Francoeur/D. Koes | University of Pittsburgh | yz8ty |
| CatS | ||||
| 0.47 | ag/dg/tdl-bp/schrodinger | K. Gao;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | ar5p6 |
| 0.48 | ag/dg/tdl-bp/schrodinger | D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | 24b03 |
| 0.49 | maestro/schrodinger,pmemd.gti,antechamber protein forcefield amber ff14sb ligand forcefield gaff2 water model tip3p | J. Zou/C. Simmerling | Stony Brook University | 3gjm2 |
| 0.49 | maestro/schrodinger,pmemd.gti,antechamber protein forcefield amber ff14sb ligand forcefield gaff2 water model tip3p | J. Zou/C. Simmerling | Stony Brook University | tkkqh |
| 0.5 | maestro/schrodinger,pmemd.gti,antechamber protein forcefield amber ff14sb ligand forcefield gaff2 water model tip3p | J. Zou/C. Simmerling | Stony Brook University | 53cvi |
| 0.5 | maestro/schrodinger,pmemd.gti,antechamber protein forcefield amber ff14sb ligand forcefield gaff2 water model tip3p | J. Zou/C. Simmerling | Stony Brook University | szgth |
| 0.5 | ag/dg/tdl-bp/schrodinger | K. Gao;D. Nguyen/W. Guo-Wei | Michigan State University, East Lansing, Mi | qian4 |
| 0.53 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | ffnbb |
| 0.53 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | i7kce |
| 0.54 | deepscaffopt | T. Evangelidis/T. Evangelidis | Uochb & Ceitec | k4gpy |
5. DISCUSSION
D3R aims to provide community-wide prospective studies for rigorous analysis of pose and affinity prediction protocols. To this end, D3R provided GC4 as a venue for participants to evaluate computational methods of their choosing. GC4 attracted robust community participation, with 55 participants submitting over 380 prediction sets. Novel to GC4 was the inclusion of CatS data drawn from a large dataset, provided by Janssen, which was also used in GC3. Thus, it was of interest to see whether the availability of more data and structures, as well as possible improved computational methodology, would lead to improved performance in GC4.
The pose prediction portion of GC4 consisted of 20 BACE1 ligands of high molecular weight and rotatable bond count. Notwithstanding the complexity of these ligands, docking performance in GC4 was particularly good. Notably, all top ten performing methods achieved a median pose 1 RMSD<1 Å in both Stages 1a and 1b. Despite having various 1nn Tanimoto coefficients, all BACE1 ligand poses were predicted nearly equally well across both stages. In this case, we observe no link between the predicted pose accuracy and the 1nn Tanimoto coefficient: even with low values for 1nn, one can get good predictive accuracy. Assessing the 1nn distribution at the start of a new project should therefore be considered informative (only) and should not deter from taking on the docking challenge. As seen in prior challenges, submission protocols for top-performing methods used a variety of software. Conversely, we did not find a statistically significant impact of using ligand guided similarity docking as for previous GCs [6, 30]. Finally, we do note a small performance benefit from computational docking followed by visual inspection of docked poses, with the result being heavily dependent on the individual operator. With respect to the matter of best practices, the docking statistics reported herein indicate that various approaches and software tools can yield high pose RMSD accuracy.
Unlike much of what we have seen in prior GCs, the availability of new structural data used in affinity rankings does appear to improve the potency ranking accuracy for BACE1. Many of the submissions evaluated in terms of affinity rank ordering of ligands for which crystallographic poses were provided in Stage 2 show higher Kendall’s τs compared to submissions in Stage 1. As CatS was presented as a target in two consecutive challenges, we have also observed an improvement in participant performance for CatS between GC3 and GC4. However, this improvement could not be attributed to either the use of GC3 data, or improvements in methodology for ranking ligand affinity. It is also important to note the continued impact of machine learning methods. Although similar performance was observed for methods utilizing machine learning or not for the BACE1 target, CatS methods that use machine learning tended to perform better than those that did not. The accuracy of affinity ranking methodology appears to be very target-dependent and dependent on specific practitioner and methodology. We cannot at this time propose a general best practices approach for this type of work with respect to incorporation of machinelearning or choosing between structure-based or ligand-based strong methods and alchemical free energy methods. Both machine-learning and alchemical free energy methods show performance in CatS, but not in BACE1.
6. CONCLUSIONS
BACE1 macrocycle inhibitor ligand docking accuracy was of high quality, with all top ten performing methods in both Stages 1a and 1b obtaining median pose 1 RMSD<2.0 Å.
Multiple methods and software packages (mostly open source) achieved high docking accuracy.
Computational methods in affinity ranking prediction performed better for CatS than for BACE1.
We see a performance improvement in CatS potency ranking for GC4 versus GC3.
Methods that use machine learning tended to perform better for CatS than alternative approaches.
Unlike much of what we have seen in prior GCs, the availability of structural data used in structure-based affinity rankings can improve the potency ranking accuracy for BACE1.
For CatS, alchemical free energy methods produced greater ranking accuracy than faster, less detailed scoring methods.
Supplementary Material
7. ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health (NIH) grant 1U01GM111528 for the Drug Design Data Resource (D3R). We also thank OpenEye Scientific Software for generously donating the use of their software. We thank Prof. William Jorgensen (Yale) for providing valuable insight into the selected free energy sets. The RCSB PDB is jointly funded by the National Science Foundation (DBI-1832184), the National Institutes of Health (R01GM133198), and the United States Department of Energy (DESC0019749).The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the federal funding agencies. MKG has an equity interest in, and is a co-founder and scientific advisor of, VeraChem LLC; REA has equity interest in and is a co-founder and scientific advisor of Actavalon, Inc.; and PW has an equity interest in Relay Pharmaceuticals, Inc. We also thank the reviewers for their helpful suggestions.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
8 REFERENCES
- 1.Macalino SJY, Gosu V, Hong S, Choi S (2015) Role of computer-aided drug design in modern drug discovery. Arch Pharm Res 38:1686–1701. 10.1007/s12272-015-0640-5 [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen WL (2004) The Many Roles of Computation in Drug Discovery. Science 303:1813–1818. 10.1126/science.1096361 [DOI] [PubMed] [Google Scholar]
- 3.Sliwoski G, Kothiwale S, Meiler J, Lowe EW (2013) Computational Methods in Drug Discovery. Pharmacological Reviews 66:334–395. 10.1124/pr.112.007336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irwin JJ, Shoichet BK (2016) Docking Screens for Novel Ligands Conferring New Biology. J Med Chem 59:4103–4120. 10.1021/acs.jmedchem.5b02008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao C, Sitzmann M, Pugliese A, Nicklaus MC (2011) Software and resources for computational medicinal chemistry. Future Medicinal Chemistry 3:1057–1085. 10.4155/fmc.11.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaieb Z, Liu S, Gathiaka S, et al. (2018) D3R Grand Challenge 2: blind prediction of protein–ligand poses, affinity rankings, and relative binding free energies. Journal of Computer-Aided Molecular Design 32:1–20. 10.1007/s10822-017-0088-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Athanasiou C, Vasilakaki S, Dellis D, Cournia Z (2018) Using physics-based pose predictions and free energy perturbation calculations to predict binding poses and relative binding affinities for FXR ligands in the D3R Grand Challenge 2. J Comput Aided Mol Des 32:21–44. 10.1007/s10822-017-0075-9 [DOI] [PubMed] [Google Scholar]
- 8.Baumgartner MP, Evans DA (2018) Lessons learned in induced fit docking and metadynamics in the Drug Design Data Resource Grand Challenge 2. J Comput Aided Mol Des 32:45–58. 10.1007/s10822-017-0081-y [DOI] [PubMed] [Google Scholar]
- 9.Bhakat S, Åberg E, Söderhjelm P (2018) Prediction of binding poses to FXR using multitargeted docking combined with molecular dynamics and enhanced sampling. J Comput Aided Mol Des 32:59–73. 10.1007/s10822-017-0074-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva Figueiredo Celestino Gomes P, Da Silva F, Bret G, Rognan D (2018) Ranking docking poses by graph matching of protein–ligand interactions: lessons learned from the D3R Grand Challenge 2. J Comput Aided Mol Des 32:75–87. 10.1007/s10822-017-0046-1 [DOI] [PubMed] [Google Scholar]
- 11.Ding X, Hayes RL, Vilseck JZ, et al. (2018) CDOCKER and $ $\lambda$ $λ-dynamics for prospective prediction in D3R Grand Challenge 2. J Comput Aided Mol Des 32:89–102. 10.1007/s10822-017-0050-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan R, Xu X, Zou X (2018) Lessons learned from participating in D3R 2016 Grand Challenge 2: compounds targeting the farnesoid X receptor. J Comput Aided Mol Des 32:103–111. 10.1007/s10822-017-0082-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fradera X, Verras A, Hu Y, et al. (2018) Performance of multiple docking and refinement methods in the pose prediction D3R prospective Grand Challenge 2016. J Comput Aided Mol Des 32:113–127. 10.1007/s10822-017-0053-2 [DOI] [PubMed] [Google Scholar]
- 14.Gao Y-D, Hu Y, Crespo A, et al. (2018) Workflows and performances in the ranking prediction of 2016 D3R Grand Challenge 2: lessons learned from a collaborative effort. J Comput Aided Mol Des 32:129–142. 10.1007/s10822-017-0072-z [DOI] [PubMed] [Google Scholar]
- 15.Hogues H, Sulea T, Gaudreault F, et al. (2018) Binding pose and affinity prediction in the 2016 D3R Grand Challenge 2 using the Wilma-SIE method. J Comput Aided Mol Des 32:143–150. 10.1007/s10822-017-0071-0 [DOI] [PubMed] [Google Scholar]
- 16.Kadukova M, Grudinin S (2018) Docking of small molecules to farnesoid X receptors using AutoDock Vina with the Convex-PL potential: lessons learned from D3R Grand Challenge 2. J Comput Aided Mol Des 32:151–162. 10.1007/s10822-017-0062-1 [DOI] [PubMed] [Google Scholar]
- 17.Kumar A, Zhang KYJ (2018) A cross docking pipeline for improving pose prediction and virtual screening performance. J Comput Aided Mol Des 32:163–173. 10.1007/s10822-017-0048-z [DOI] [PubMed] [Google Scholar]
- 18.Kurkcuoglu Z, Koukos PI, Citro N, et al. (2018) Performance of HADDOCK and a simple contactbased protein–ligand binding affinity predictor in the D3R Grand Challenge 2. J Comput Aided Mol Des 32:175–185. 10.1007/s10822-017-0049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam PC-H, Abagyan R, Totrov M (2018) Ligand-biased ensemble receptor docking (LigBEnD): a hybrid ligand/receptor structure-based approach. J Comput Aided Mol Des 32:187–198. 10.1007/s10822-017-0058-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mey ASJS, Jiménez JJ, Michel J (2018) Impact of domain knowledge on blinded predictions of binding energies by alchemical free energy calculations. J Comput Aided Mol Des 32:199–210. 10.1007/s10822-017-0083-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsson MA, García-Sosa AT, Ryde U (2018) Binding affinities of the farnesoid X receptor in the D3R Grand Challenge 2 estimated by free-energy perturbation and docking. J Comput Aided Mol Des 32:211–224. 10.1007/s10822-017-0056-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padhorny D, Hall DR, Mirzaei H, et al. (2018) Protein–ligand docking using FFT based sampling: D3R case study. J Comput Aided Mol Des 32:225–230. 10.1007/s10822-017-0069-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Réau M, Langenfeld F, Zagury J-F, Montes M (2018) Predicting the affinity of Farnesoid X Receptor ligands through a hierarchical ranking protocol: a D3R Grand Challenge 2 case study. J Comput Aided Mol Des 32:231–238. 10.1007/s10822-017-0063-0 [DOI] [PubMed] [Google Scholar]
- 24.Rifai EA, van Dijk M, Vermeulen NPE, Geerke DP (2018) Binding free energy predictions of farnesoid X receptor (FXR) agonists using a linear interaction energy (LIE) approach with reliability estimation: application to the D3R Grand Challenge 2. J Comput Aided Mol Des 32:239–249. 10.1007/s10822-017-0055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salmaso V, Sturlese M, Cuzzolin A, Moro S (2018) Combining self-and cross-docking as benchmark tools: the performance of DockBench in the D3R Grand Challenge 2. J Comput Aided Mol Des 32:251–264. 10.1007/s10822-017-0051-4 [DOI] [PubMed] [Google Scholar]
- 26.Schindler C, Rippmann F, Kuhn D (2018) Relative binding affinity prediction of farnesoid X receptor in the D3R Grand Challenge 2 using FEP+. J Comput Aided Mol Des 32:265–272. 10.1007/s10822-017-0064-z [DOI] [PubMed] [Google Scholar]
- 27.Selwa E, Elisée E, Zavala A, Iorga BI (2018) Blinded evaluation of farnesoid X receptor (FXR) ligands binding using molecular docking and free energy calculations. J Comput Aided Mol Des 32:273–286. 10.1007/s10822-017-0054-1 [DOI] [PubMed] [Google Scholar]
- 28.Wingert BM, Oerlemans R, Camacho CJ (2018) Optimal affinity ranking for automated virtual screening validated in prospective D3R grand challenges. J Comput Aided Mol Des 32:287–297. 10.1007/s10822-017-0065-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yakovenko O, Jones SJM (2017) Modern drug design: the implication of using artificial neuronal networks and multiple molecular dynamic simulations. J Comput Aided Mol Des 1–13. 10.1007/s10822-017-0085-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaieb Z, Parks CD, Chiu M, et al. (2019) D3R Grand Challenge 3: blind prediction of protein–ligand poses and affinity rankings. Journal of Computer-Aided Molecular Design. 10.1007/s10822-018-0180-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunseri J, King JE, Francoeur PG, Koes DR (2019) Convolutional neural network scoring and minimization in the D3R 2017 community challenge. J Comput Aided Mol Des 33:19–34. 10.1007/s10822-018-0133-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam PC-H, Abagyan R, Totrov M (2019) Hybrid receptor structure/ligand-based docking and activity prediction in ICM: development and evaluation in D3R Grand Challenge 3. J Comput Aided Mol Des 33:35–46. 10.1007/s10822-018-0139-5 [DOI] [PubMed] [Google Scholar]
- 33.Kumar A, Zhang KYJ (2019) Shape similarity guided pose prediction: lessons from D3R Grand Challenge 3. J Comput Aided Mol Des 33:47–59. 10.1007/s10822-018-0142-x [DOI] [PubMed] [Google Scholar]
- 34.Xie B, Minh DDL (2019) Alchemical Grid Dock (AlGDock) calculations in the D3R Grand Challenge 3. J Comput Aided Mol Des 33:61–69. 10.1007/s10822-018-0143-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen DD, Cang Z, Wu K, et al. (2019) Mathematical deep learning for pose and binding affinity prediction and ranking in D3R Grand Challenges. J Comput Aided Mol Des 33:71–82. 10.1007/s10822-018-0146-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koukos PI, Xue LC, Bonvin AMJJ (2019) Protein–ligand pose and affinity prediction: Lessons from D3R Grand Challenge 3. J Comput Aided Mol Des 33:83–91. 10.1007/s10822-018-0148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaput L, Selwa E, Elisée E, Iorga BI (2019) Blinded evaluation of cathepsin S inhibitors from the D3RGC3 dataset using molecular docking and free energy calculations. J Comput Aided Mol Des 33:93–103. 10.1007/s10822-018-0161-7 [DOI] [PubMed] [Google Scholar]
- 38.He X, Man VH, Ji B, et al. (2019) Calculate protein–ligand binding affinities with the extended linear interaction energy method: application on the Cathepsin S set in the D3R Grand Challenge 3. J Comput Aided Mol Des 33:105–117. 10.1007/s10822-0180162-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ignatov M, Liu C, Alekseenko A, et al. (2019) Monte Carlo on the manifold and MD refinement for binding pose prediction of protein–ligand complexes: 2017 D3R Grand Challenge. J Comput Aided Mol Des 33:119–127. 10.1007/s10822-018-0176-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallach I, Heifets A (2018) Most Ligand-Based Classification Benchmarks Reward Memorization Rather than Generalization. J Chem Inf Model 58:916–932. 10.1021/acs.jcim.7b00403 [DOI] [PubMed] [Google Scholar]
- 41.Carlson HA (2016) Lessons Learned over Four Benchmark Exercises from the Community Structure–Activity Resource. Journal of Chemical Information and Modeling 56:951–954. 10.1021/acs.jcim.6b00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlson HA, Smith RD, Damm-Ganamet KL, et al. (2016) CSAR 2014: A Benchmark Exercise Using Unpublished Data from Pharma. Journal of Chemical Information and Modeling 56:1063–1077. 10.1021/acs.jcim.5b00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Damm-Ganamet KL, Smith RD, Dunbar JB, et al. (2013) CSAR Benchmark Exercise 2011–2012: Evaluation of Results from Docking and Relative Ranking of Blinded Congeneric Series. Journal of Chemical Information and Modeling 53:1853–1870. 10.1021/ci400025f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith RD, Damm-Ganamet KL, Dunbar JB, et al. (2016) CSAR Benchmark Exercise 2013: Evaluation of Results from a Combined Computational Protein Design, Docking, and Scoring/Ranking Challenge. Journal of Chemical Information and Modeling 56:1022–1031. 10.1021/acs.jcim.5b00387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gathiaka S, Liu S, Chiu M, et al. (2016) D3R grand challenge 2015: Evaluation of protein–ligand pose and affinity predictions. Journal of Computer-Aided Molecular Design 30:651–668. 10.1007/s10822-016-9946-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews 23:3–25. 10.1016/S0169409X(96)00423-1 [DOI] [PubMed] [Google Scholar]
- 47.Veber DF, Johnson SR, Cheng H-Y, et al. (2002) Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J Med Chem 45:2615–2623. 10.1021/jm020017n [DOI] [PubMed] [Google Scholar]
- 48.RDKit. https://www.rdkit.org/. Accessed 13 Aug 2019
- 49.Yu HS, Deng Y, Wu Y, et al. (2017) Accurate and Reliable Prediction of the Binding Affinities of Macrocycles to Their Protein Targets. J Chem Theory Comput 13:6290–6300. 10.1021/acs.jctc.7b00885 [DOI] [PubMed] [Google Scholar]
- 50.Machauer R, Laumen K, Veenstra S, et al. (2009) Macrocyclic peptidomimetic β-secretase (BACE-1) inhibitors with activity in vivo. Bioorganic & Medicinal Chemistry Letters 19:1366–1370. 10.1016/j.bmcl.2009.01.055 [DOI] [PubMed] [Google Scholar]
- 51.Hanessian S, Yang G, Rondeau J-M, et al. (2006) Structure-Based Design and Synthesis of Macroheterocyclic Peptidomimetic Inhibitors of the Aspartic Protease β-Site Amyloid Precursor Protein Cleaving Enzyme (BACE). J Med Chem 49:4544–4567. 10.1021/jm060154a [DOI] [PubMed] [Google Scholar]
- 52.Thurmond RL, Sun S, Sehon CA, et al. (2004) Identification of a Potent and Selective Noncovalent Cathepsin S Inhibitor. J Pharmacol Exp Ther 308:268–276. 10.1124/jpet.103.056879 [DOI] [PubMed] [Google Scholar]
- 53.A hierarchical approach to all‐atom protein loop prediction-Jacobson-2004-Proteins: Structure, Function, and Bioinformatics-Wiley Online Library; https://onlinelibrary.wiley.com/doi/full/10.1002/prot.10613. Accessed 9 Jun 2019 [DOI] [PubMed] [Google Scholar]
- 54.Kendall MG (1938) A New Measure of Rank Correlation. Biometrika 30:81–93. 10.2307/2332226 [DOI] [Google Scholar]
- 55.Kendall MG (1945) The Treatment of Ties in Ranking Problems. Biometrika 33:239–251. 10.2307/2332303 [DOI] [PubMed] [Google Scholar]
- 56.Zwillinger D, Kokoska S (1999) CRC Standard Probability and Statistics Tables and Formulae. CRC Press [Google Scholar]
- 57.Wildman SA, Crippen GM (1999) Prediction of Physicochemical Parameters by Atomic Contributions. J Chem Inf Comput Sci 39:868–873. 10.1021/ci990307l [DOI] [Google Scholar]
- 58.Breiman L (2001) Random Forests. Machine Learning 45:5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 59.Pedregosa F, Varoquaux G, Gramfort A, et al. (2011) Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research 12:2825–2830 [Google Scholar]
- 60.Buitinck L, Louppe G, Blondel M, et al. (2013) API design for machine learning software: experiences from the scikit-learn project. arXiv:13090238 [cs] [Google Scholar]
- 61.Gaulton A, Hersey A, Nowotka M, et al. (2017) The ChEMBL database in 2017. Nucleic Acids Res 45:D945–D954. 10.1093/nar/gkw1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das B, Yan R (2017) Role of BACE1 in Alzheimer’s synaptic function. Transl Neurodegener 6:. 10.1186/s40035-017-0093-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trott O, Olson AJ (2010) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry 31:455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friesner RA, Banks JL, Murphy RB, et al. (2004) Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. Journal of Medicinal Chemistry 47:1739–1749. 10.1021/jm0306430 [DOI] [PubMed] [Google Scholar]
- 65.Halgren TA, Murphy RB, Friesner RA, et al. (2004) Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. Journal of Medicinal Chemistry 47:1750–1759. 10.1021/jm030644s [DOI] [PubMed] [Google Scholar]
- 66.Abagyan R, Totrov M, Kuznetsov D (1994) ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. Journal of Computational Chemistry 15:488–506. 10.1002/jcc.540150503 [DOI] [Google Scholar]
- 67.Tetko IV, Gasteiger J, Todeschini R, et al. (2005) Virtual Computational Chemistry Laboratory – Design and Description. J Comput Aided Mol Des 19:453–463. 10.1007/s10822-005-8694-y [DOI] [PubMed] [Google Scholar]
- 68.Verdonk ML, Cole JC, Hartshorn MJ, et al. (2003) Improved protein–ligand docking using GOLD. Proteins: Structure, Function, and Bioinformatics 52:609–623. 10.1002/prot.10465 [DOI] [PubMed] [Google Scholar]
- 69.Computation and management of chemical properties in CACTVS: An extensible networked approach toward modularity and compatibility | Journal of Chemical Information and Modeling. https://pubs.acs.org/doi/pdf/10.1021/ci00017a013. Accessed 11 Jun 2019 [Google Scholar]
- 70.Chaudhury S, Gray JJ (2008) Conformer selection and induced fit in flexible backbone proteinprotein docking using computational and NMR ensembles. J Mol Biol 381:1068–1087. 10.1016/j.jmb.2008.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feinstein WP, Brylinski M (2014) e FindSite: Enhanced Fingerprint-Based Virtual Screening Against Predicted Ligand Binding Sites in Protein Models. Molecular Informatics 33:135–150. 10.1002/minf.201300143 [DOI] [PubMed] [Google Scholar]
- 72.Coutsias EA, Lexa KW, Wester MJ, et al. (2016) Exhaustive Conformational Sampling of Complex Fused Ring Macrocycles Using Inverse Kinematics. J Chem Theory Comput 12:4674–4687. 10.1021/acs.jctc.6b00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O’Boyle NM, Banck M, James CA, et al. (2011) Open Babel: An open chemical toolbox. Journal of Cheminformatics 3:33 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maestro, Schrödinger Release 2019–4. Schrödinger, New York, New York [Google Scholar]
- 75.Bonnet P, Agrafiotis DK, Zhu F, Martin E (2009) Conformational Analysis of Macrocycles: Finding What Common Search Methods Miss. J Chem Inf Model 49:2242–2259. 10.1021/ci900238a [DOI] [PubMed] [Google Scholar]
- 76.Morris GM, Huey R, Lindstrom W, et al. (2009) AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J Comput Chem 30:2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amaro RE, Baudry J, Chodera J, et al. (2018) Ensemble Docking in Drug Discovery. Biophysical Journal 114:2271–2278. 10.1016/j.bpj.2018.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amaro RE, Baron R, McCammon JA (2008) An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. Journal of Computer-Aided Molecular Design 22:693–705. 10.1007/s10822-007-9159-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Korb O, Olsson TSG, Bowden SJ, et al. (2012) Potential and Limitations of Ensemble Docking. J Chem Inf Model 52:1262–1274. 10.1021/ci2005934 [DOI] [PubMed] [Google Scholar]
- 80.Tuccinardi T, Botta M, Giordano A, Martinelli A (2010) Protein Kinases: Docking and Homology Modeling Reliability. J Chem Inf Model 50:1432–1441. 10.1021/ci100161z [DOI] [PubMed] [Google Scholar]
- 81.Jones G, Willett P, Glen RC, et al. (1997) Development and validation of a genetic algorithm for flexible docking11Edited by F. E. Cohen. Journal of Molecular Biology 267:727–748. 10.1006/jmbi.1996.0897 [DOI] [PubMed] [Google Scholar]
- 82.Jiménez J, Škalič M, Martínez-Rosell G, De Fabritiis G (2018) KDEEP: Protein–Ligand Absolute Binding Affinity Prediction via 3D-Convolutional Neural Networks. J Chem Inf Model 58:287–296. 10.1021/acs.jcim.7b00650 [DOI] [PubMed] [Google Scholar]
- 83.Abagyan R, Totrov M (1994) Biased Probability Monte Carlo Conformational Searches and Electrostatic Calculations for Peptides and Proteins. Journal of Molecular Biology 235:983–1002. 10.1006/jmbi.1994.1052 [DOI] [PubMed] [Google Scholar]
- 84.Totrov M, Abagyan R (1999) Derivation of sensitive discrimination potential for virtual ligand screening. In: Proceedings of the third annual international conference on Computational molecular biology-RECOMB ‘99 ACM Press, Lyon, France, pp 312–320 [Google Scholar]
- 85.Abagyan R, Totrov M, Kuznetsov D (1994) ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. Journal of Computational Chemistry 15:488–506. 10.1002/jcc.540150503 [DOI] [Google Scholar]
- 86.Bramer D, Wei G-W (2018) Multiscale weighted colored graphs for protein flexibility and rigidity analysis. J Chem Phys 148:054103. 10.1063/1.5016562 [DOI] [PubMed] [Google Scholar]
- 87.Nguyen DD, Xiao T, Wang M, Wei G-W (2017) Rigidity Strengthening: A Mechanism for Protein–Ligand Binding. J Chem Inf Model 57:1715–1721. 10.1021/acs.jcim.7b00226 [DOI] [PubMed] [Google Scholar]
- 88.Cang Z, Wei G-W (2018) Integration of element specific persistent homology and machine learning for protein-ligand binding affinity prediction. International Journal for Numerical Methods in Biomedical Engineering 34:e2914. 10.1002/cnm.2914 [DOI] [PubMed] [Google Scholar]
- 89.Cang Z, Wei G-W (2017) TopologyNet: Topology based deep convolutional and multi-task neural networks for biomolecular property predictions. PLOS Computational Biology 13:e1005690. 10.1371/journal.pcbi.1005690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cang Z, Mu L, Wei G-W (2018) Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLOS Computational Biology 14:e1005929. 10.1371/journal.pcbi.1005929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jorgensen WL, Thomas LL (2008) Perspective on Free-Energy Perturbation Calculations for Chemical Equilibria. J Chem Theory Comput 4:869–876. 10.1021/ct800011m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mobley DL, Klimovich PV (2012) Perspective: Alchemical free energy calculations for drug discovery. J Chem Phys 137:230901. 10.1063/1.4769292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chodera JD, Mobley DL, Shirts MR, et al. (2011) Alchemical free energy methods for drug discovery: progress and challenges. Current Opinion in Structural Biology 21:150–160. 10.1016/j.sbi.2011.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cournia Z, Allen B, Sherman W (2017) Relative Binding Free Energy Calculations in Drug Discovery: Recent Advances and Practical Considerations. J Chem Inf Model 57:2911–2937. 10.1021/acs.jcim.7b00564 [DOI] [PubMed] [Google Scholar]
- 95.Christ CD, Fox T (2014) Accuracy Assessment and Automation of Free Energy Calculations for Drug Design. J Chem Inf Model 54:108–120. 10.1021/ci4004199 [DOI] [PubMed] [Google Scholar]
- 96.Mobley DL, Gilson MK (2017) Predicting Binding Free Energies: Frontiers and Benchmarks. Annual Review of Biophysics 46:531–558. 10.1146/annurev-biophys070816-033654 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.