Abstract
BACKGROUND
Maternal obesity prior to or during pregnancy influences fetal growth, predisposing the offspring to increased risk for obesity across the life-course. Placental epigenetic mechanisms may underlie these associations. We conducted an epigenome-wide association study to identify placental DNA methylation changes associated with maternal pre-pregnancy body mass index (BMI) and rate of gestational weight gain at first (GWG1), second (GWG2) and third trimester (GWG3).
METHOD
Participants of the NICHD Fetal Growth Studies with genome-wide placental DNA methylation (n=301) and gene expression (n=75) data were included. Multivariable-adjusted regression models were used to test the associations of 1kg/m2 increase in pre-pregnancy BMI or 1kg/week increase in GWG with DNA methylation levels. Genes harboring top differentially methylated CpGs (FDR P<0.05) were evaluated for placental gene expression. We assessed whether DNA methylation sites known to be associated with BMI in child or adult tissues were also associated with maternal pre-pregnancy BMI in placenta.
RESULTS
Pre-pregnancy BMI was associated with DNA methylation at cg14568196[EGFL7], cg15339142[VETZ] and cg02301019[AC092377.1] (FDR P<0.05, P ranging from 1.4×10−10 to 1.7×10−9). GWG1 or GWG2 was associated with DNA methylation at cg17918270[MYT1L], cg20735365[DLX5] and cg17451688[SLC35F3] (FDR P<0.05, P ranging from 6.4×10−10 to 1.2×10−8). Both pre-pregnancy BMI and DNA methylation at cg1456819 [EGFL7] were negatively correlated with EGFL7 expression in placenta (P<0.05). Several CpGs previously implicated in obesity traits in children and adults were associated with pre-pregnancy BMI in placenta. Functional annotations revealed that EGFL7 is highly expressed in placenta and the differentially methylated CpG sites near EGFL7 and VEZT were cis-meQTL targets in blood.
CONCLUSIONS
We identified placental DNA methylation changes at novel loci associated with pre-pregnancy BMI and GWG. The overlap between CpGs associated with obesity traits in placenta and other tissues in children and adults suggests that epigenetic mechanisms in placenta may give insights to early origins of obesity.
Keywords: obesity, gestational weight gain, DNA methylation, EWAS, gene expression, placenta, DOHaD
INTRODUCTION
Approximately 40 million pregnant women were overweight and obese globally in 2014, of which 1.1 million pregnant women with obesity were in the United States.1 One in two pregnant women (~47%) were reported to gain more than the recommended amount of gestational weight gain (GWG).2 Higher maternal pre-pregnancy body mass index (BMI) and inadequate or excessive GWG are associated with aberrant fetal growth,3 and increased risk of obesity across the life course.4, 5 The observed associations of maternal adiposity-related factors with fetal growth and adiposity in later life can be mediated through changes in epigenetic regulation of specific genes.6, 7 Epigenetic regulation occurs through a number of processes including DNA methylation, the addition of a methyl group to cytosines in cytosine-guanine dinucleotide (CpG) sites.8 DNA methylation can regulate gene expression, playing crucial roles essential for cell fate, differentiation, and tissue integrity and embryonic development.6–8
The placenta, a transient organ at the maternal-fetal interface with endocrine and substrate transport functions,9 is sensitive to in-utero environmental influences. Maternal pre-pregnancy obesity and inadequate or excessive GWG lead to compromised placental function by increasing oxidative stress, vascular endothelium thickening, and maternal inflammatory lesions.3, 10, 11 Previous studies of maternal obesity and placental DNA methylation examined global DNA methylation levels or a few candidate genes. Global DNA methylation in human placentas was found to be more abundant in obese compared to lean pregnant women.12 Another study found increased epigenetic alterations of transcription regulators in placentas among women with inadequate GWG compared to women with normal GWG.13 A study that measured global methylation using long interspersed nuclear elements 1 (LINE-1) found no association between maternal pre-pregnancy BMI or GWG and methylation in either placenta or cord blood.14 To our knowledge, there is no published placental epigenome-wide association study (EWAS) of pre-pregnancy BMI or GWG.
In the present study, we performed EWAS to identify placental DNA methylation CpG sites associated with maternal pre-pregnancy BMI and trimester-specific rate of GWG. We also tested correlations between the DNA methylation levels of the top significant CpG sites and expression of the corresponding genes. Finally, we examined our data to determine 1) whether CpG sites in placenta, cord blood and adolescent blood, which were previously known to be associated with maternal adiposity traits, were also associated with maternal pre-pregnancy BMI and trimester specific rate of GWG in placenta, and 2) whether DNA methylation sites previously known to be associated with child or adult BMI were also associated with maternal pre-pregnancy BMI in placenta.
METHODS
The study protocol was reviewed and approved by institutional review boards at the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and each of the participating clinical sites. Written informed consent was obtained from all participants. The study was conducted in accordance with the principles of the Helsinki Declaration for human research.
Study population
The present analysis involved pregnant women from the NICHD Fetal Growth Studies-Singletons cohort who provided placenta samples at delivery (n=312). The NICHD Fetal Growth Studies – Singletons is a prospective longitudinal cohort of 2 802 pregnant woman from four self-reported race/ethnic groups recruited between July 2009 and January 2013 from 12 clinical sites in the US. Details of the cohort profile have been previously reported.15 Gestational age was determined using the date of the last menstrual period and confirmed by ultrasound between 8–13 weeks and 6 days of gestation. None of the 312 pregnant women smoked cigarettes and only two women reported alcohol consumption in the past six months prior to pregnancy.
Measures of maternal pre-pregnancy BMI and rate of gestational weight gain
Women’s pre-pregnancy BMI was calculated using self-reported pre-pregnancy weight in kilograms divided by measured height in meters squared (kg/m2). Women’s weight during pregnancy was recorded at each research visit and abstracted from prenatal care records. The rates of GWG in the first trimester (GWG1: 0 to 13 weeks and 6 days), second trimester (GWG2: 14 to 27 weeks and 6 days) and third trimester (GWG3: 28 weeks to delivery) were estimated in kg/week as previously described.16
Placenta sample collection, DNA methylation measurement and quality control
Placental parenchymal biopsies were collected from the fetal side approximately 5 cm from the umbilical cord insertion site within one hour of delivery. Samples were placed in RNALater and frozen at −80°C for molecular analysis. DNA was extracted from the placental biopsies and was assayed using Illumina’s Infinium Human Methylation450 Beadchip (Illumina Inc., San Diego, CA) array. SNP genotyping was done using HumanOmni2.5 Beadchips (Illumina Inc., San Diego, CA), followed by initial data processing using Illumina’s Genome Studio, as previously described.17 A total of 11 samples showing sex discrepancies between the phenotype and the genotype (n=4), that were outliers from the distribution of the samples’ genetic clusters based on multi-dimensional scaling plots (n=6), and with a mismatching sample identifier (n=1) were excluded. A total of 301 placental samples that passed the quality control filters were included in the EWAS.
Standard Illumina protocols were followed for background correction, normalization to internal control probes, and quantile normalization. The resulting intensity files were processed with Illumina’s Genome Studio, which generated average beta-values for each CpG site (i.e., the fraction of methylated sites per sample by taking the ratio of methylated and unmethylated fluorescent signals) and detection P values that characterized the chance that the target sequence signal was distinguishable from the negative controls. The method was corrected for the probe design bias in the Illumina Infinium HumanMethylation450 BeadChip and achieved between-sample normalization.18 Normalization was performed using the modified Beta MIxture Quantile dilation (BMIQ) method to correct the probe design bias in the Illumina Infinium Human Methylation450 Beadchip and achieve between-sample normalization. After BMIQ normalization, missing CpGs were imputed by the k-nearest neighbors method setting k=10. Beta values with an associated detection P ≥ 0.05 were set to missing. In addition, probes with mean detection P ≥ 0.05 (n=36), cross-reactive (n=24 491), non-autosomal (n=14 589), and CpG sites located within 20 base pair from known single nucleotide polymorphisms (SNPs) (n=37 360) were removed.19 After these QC procedures, methylation data with 409 101 CpGs were available for analysis. As recommended by Du et al.,20 methylation beta values were logit transformed to M-value scale before analysis and the effect estimates were reported as the change in methylation beta value for a 1 kg/m2 increase in BMI or 1kg/week increase in GWG.
RNA extraction and quantification
RNA was extracted from 80 placentas using TRIZOL reagent (Invitrogen, MA), and sequenced using the Illumina HiSeq2000 system as previously described.17 Participants with both DNA methylation and RNA sequence data (n=75) were included to test correlation between DNA methylation and gene expression.
Statistical analysis of association between CpG sites and obesity traits
Epigenome-wide analyses were performed by fitting robust linear regression models for each CpG site as the response variable on the M-value scale and each of pre-pregnancy BMI, GWG1, GWG2, and GWG3 as a predictor as implemented in the R/Bioconductor package limma, version 3.24.15. So far, there is no reference for placental cell type composition; therefore, we used the ComBat function from the surrogate variable analysis (SVA) package to account for heterogeneity in cell-type composition.21 Placental genome-wide SNP data were used to estimate 10 genotype-based principal components (PCs) representing population structure. All analyses included maternal age (continuous), race/ethnicity (Hispanic, Black, White, Asian), offspring sex (male, female), methylation sample plate (n=5), ten genotype-based PCs, three methylation-based PCs, and components representing putative cell-mixture as adjustment factors in the models. The differentially methylated CpG sites were mapped to the nearby gene using R/Bioconductor package with a background reference comprising the whole set of genes present in the Illumina 450k platform. The analyses were controlled for false discovery rate (FDR) at 5% using the Benjamini-Hochberg method. To minimize genomic inflation (λ) and bias, a Bayesian method available as an R/Bioconductor package (BACON) was applied and used to correct the nominal P values.22 Quantile-Quantile (Q-Q) plots of P values and the corresponding λ were compared before and after BACON correction. All λs were close to 1 after BACON adjustments (Supplementary Figure S1, A–H). Associations with BACON-corrected FDR P<0.05 were considered statistically significant.
Correlation between DNA methylation and gene expression in placenta
The correlations between DNA methylation at the top differentially methylated CpG sites and expression of the corresponding genes in placenta were tested using the Pearson correlation test. We further evaluated associations between expression of genes mapping to the top differentially methylated CpGs and pre-pregnancy BMI or GWG using linear regression adjusted for maternal race/ethnicity, fetal sex and ten genotype-based PCs as implemented in DEseq2.23
Evaluation of CpG sites known to be associated with obesity traits
PubMed literature search was done to identify EWASs of obesity traits published between January 2013 and February 2019 (see details in Supplementary Table S1). A total of 19 publications met our inclusion criteria.6, 24–41 These included two EWAS of pre-pregnancy BMI in cord, child and adolescent blood 6, 24, two whole blood EWAS of childhood obesity,25, 26 13 whole blood EWAS of adult obesity,27–39 and two adipose tissue EWAS of adult obesity.40, 41 No studies included placental EWAS of maternal pre-pregnancy BMI or GWG.
Pathway enrichment analysis
To understand the biological functions of genes annotated to the top CpG sites associated with maternal pre-pregnancy BMI or GWG in our study (FDR P<0.01), we looked for enrichment of biological pathways using the Ingenuity Pathway Analysis (IPA) bioinformatics resource (IPA, Qiagen, Redwood City, CA, USA). Pathways that were significantly enriched at Benjamini-Hochberg corrected FDR P<0.05 were considered to be significant.
Functional annotation
The top-significant CpG sites (BACON-adjusted FDR P<0.05) were queried in the methylation quantitative loci (meQTL) database (http://www.mqtldb.org/) that documents meQTL at serial time points across the life-course, in order to identify single nucleotide polymorphisms (SNPs) that may influence DNA methylation at the CpG sites. The meQTL SNPs identified through the query were annotated using Haploreg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php), Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA, https://fuma.ctglab.nl/), and the Human Protein Atlas, version 18.1 (HPA, https://www.proteinatlas.org/) tools and databases to understand their functional importance, tissue-specific gene expression, and relevance in diseases based on published GWAS. All analysis was carried out using R/Bioconductor version 3.8.
RESULTS
Participants’ characteristics
The 301 pregnant women who participated in this study included 102 self-identified Hispanics, 77 non-Hispanic Whites, 72 non-Hispanic Blacks, and 50 Asians. The mean (s.d.) of women’s age was 27.7 (6.0) years, pre-pregnancy BMI was 25.1 (4.5) kg/m2, GWG1 was 0.2 (0.2) kg/week, GWG2 was 0.3 (0.2) kg/week, and GWG3 was 0.4 (0.2) kg/week. Further details of study participants are presented in Table 1.
Table 1.
Demographic Characteristics of Study participants (n=301)
| Maternal age, years, mean ± SD | 27.7 ± 5.3 |
| Maternal self-reported race/ethnicity, n (%) | |
| White | 77 (25.6) |
| Black | 72 (23.9) |
| Hispanic | 102 (33.9) |
| Asian/Pacific Islander | 50 (16.6) |
| Maternal BMI status, n (%) | |
| <25.0 | 188 (63.7) |
| 25.0–29.9 | 76 (25.8) |
| ≥30.0 | 31 (10.5) |
| Trimester specific rate of GWG, kg/wk, mean ± SD | |
| 1st Trimester | 0.22 ± 0.18 |
| 2nd Trimester | 0.30 ± 0.16 |
| 3rd Trimester | 0.37 ± 0.16 |
| Placental weight in grams, median (Q1-Q3) | 423.00 (360.0–488.0) |
| Placental length in cm, median (Q1-Q3) | 20.00 (18.5–22.0) |
| Placental width in cm, median (Q1-Q3) | 17.00 (15.0–18.5) |
| Birth weight in grams, mean ± SD | 3165.2 ± 450.5 |
| Gestational age at delivery, weeks, mean ± SD | 39.5 ± 1.1 |
| Deliver Mode, n (%) | |
| Vaginal | 205 (68.1) |
| Cesarean section | 96 (31.9) |
| Offspring sex, n (%) | |
| Male | 152 (50.5) |
| Female | 149 (49.5) |
| Alcohol | |
| Nondrinkers, n (%) | 298 (99.0) |
| Drinkers, n (%) | 2 (1.0) |
Associations of maternal pre-pregnancy BMI and GWG with DNA methylation in placenta
Each 1 kg/m2 increase in maternal pre-pregnancy BMI was associated with 0.09% (95% confidence interval (CI): 0.06, 0.12) higher methylation at cg14568196 [EGFL7]; 0.13% (0.07, 0.19) higher methylation at cg15339142 [VEZT], and 0.07 % decrease in methylation at cg02301019 [AC092377.1] (BACON-corrected FDR P<0.05; nominal P ranging from 3.4×10−10 to 1.7×10−9). Further, a 1kg/week increase in GWG1 was associated with 24.32% (14.15, 34.48) higher methylation at cg17918270 [MYT1L] and 1.01% (−1.47, −0.55) lower methylation at cg20735365 [DLX5] (BACON-corrected FDR P<0.05; nominal P ranging from 1.4×10−9 to 3.3×10−9). Each 1 kg/week increase in GWG2 was associated with 28.91% (18.08, 39.73) higher methylation at cg17918270 [MYT1L] and 13.01% (−17.52, −8.49) lower decrease in methylation at cg17451688 [SLC35F3] (BACON-corrected FDR P < 0.05; nominal P ranging from 6.4×10−10 to 1.2×10−8) (Table 2, Supplementary Figure S2).
Table 2.
Differentially methylated placental CpG sites associated with maternal pre-pregnancy BMI and trimester-specific gestational weight gain
| Traita | CpGa | Chra | Positionb | Relation to Island | Nearest Gene | Feature (or distance from gene)c | % Change in methylation (95% CI) | P | FDR P | BACON corrected FDR P |
|---|---|---|---|---|---|---|---|---|---|---|
| Pre-pregnancy BMI | cg14568196 | chr9 | 139552716 | N_Shore | EGFL7 | 0.6 kb | 0.09 (0.06, 0.12) | 1.41E-10 | 5.79E-05 | 0.01 |
| Pre-pregnancy BMI | cg15339142 | chr12 | 95651472 | OpenSea | VEZT | Body | 0.13 (0.07,0.19) | 3.47E-10 | 7.11E-05 | 0.01 |
| Pre-pregnancy BMI | cg02301019 | chr16 | 85465191 | Island | AC092377.1 | 78.6 kb | −0.07 (−0.09, −0.04) | 1.70E-09 | 0.0002 | 0.02 |
| GWG1 | cg17918270 | chr2 | 1983484 | OpenSea | MYT1L | Body | 24.32 (14.15, 34.48) | 3.30E-09 | 0.0006 | 0.01 |
| GWG1 | cg20735365 | chr7 | 96657023 | S_Shelf | DLX5 | 2.9 kb | −1.01 (−1.47, −0.55) | 1.41E-09 | 0.0005 | 0.01 |
| GWG2 | cg17451688 | chr1 | 234293071 | OpenSea | SLC35F3 | Body | −13.01 (−17.52, −8.49) | 1.20E-08 | 0.002 | 0.04 |
| GWG2 | cg17918270 | chr2 | 1983484 | OpenSea | MYT1L | Body | 28.91 (18.08, 39.73) | 6.36E-10 | 0.0002 | 0.01 |
Abbreviations: CpG, Cytosine phosphate Guanine; Chr, chromosome; Pre-pregnancy BMI, Pre-pregnancy body mass index; GWG1, Rate of gestational weight gain at 1st trimester; GWG2, Rate of gestational weight gain at 2nd trimester; kb, distance in kilo bases
position of genome build is GRCh37 (hg19)
Based on manifest feature annotation from Illumina
For each trait, model was adjusted for offspring sex, maternal age, maternal race/ethnicity, plates, first three principal components from methylation and ten principal components from genotypes.
Correlation between DNA methylation and gene expression in placenta
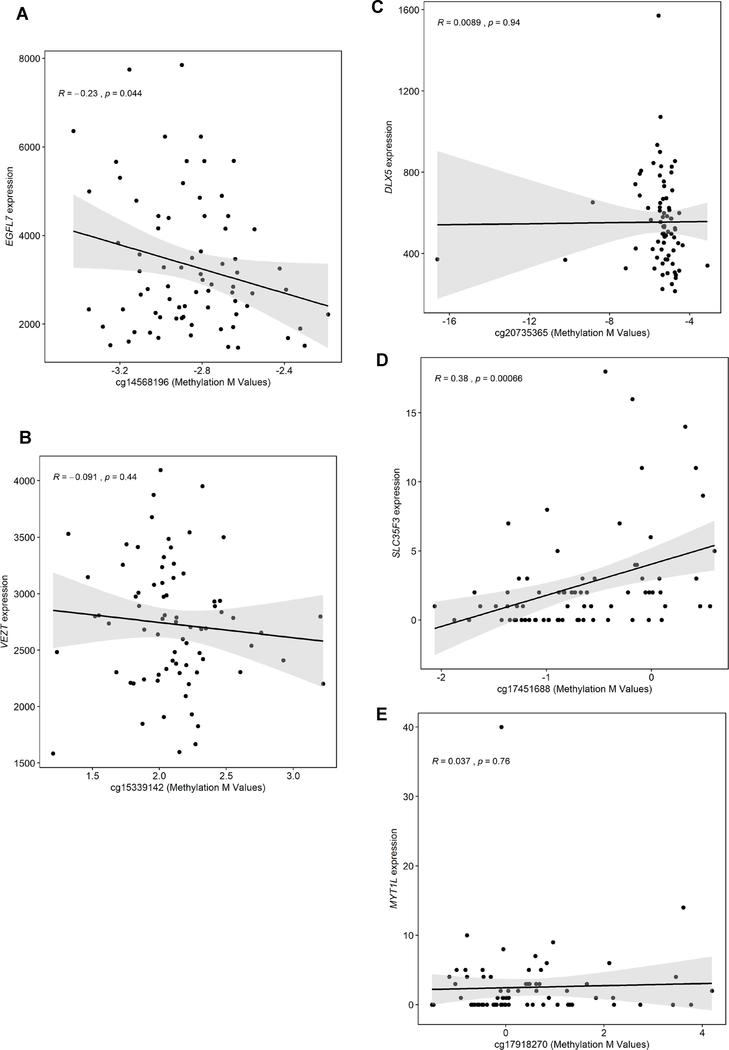
We tested the correlations between DNA methylation at each of the six CpG sites we identified to be associated with pre-pregnancy BMI or GWG and expression of the corresponding annotated genes. DNA methylation at cg1456819 (which was hypermethylated with higher pre-pregnancy BMI) was negatively correlated with EGFL7 expression in placenta (r=−0.23; P=0.04). DNA methylation at cg17451688 (which was hypomethylated with higher GWG2) was positively correlated with SLC35F3 expression in placenta (r=0.38, P=0.0007) (Figure 1). Gene expression analysis found that higher pre-pregnancy BMI was associated with reduced expression of EGFL7 in placenta (log2FC=−0.03, P=0.02) (Table 3).
Figure 1.
Correlation between DNA methylation at top significant CpG sites and expression of the corresponding gene. A. EFGL7 B. VETZ C. DLX5 D. SLC35F3 E. MYT1L
Table 3.
Associations of maternal pre-pregnancy BMI and gestational weight gain with placental gene expression
| Traitsa | Gene symbol | Log2FCb | SE | P |
|---|---|---|---|---|
| Pre-pregnancy BMI | EGFL7 | −0.03 | 0.01 | 0.02 |
| VEZT | 0.01 | 0.01 | 0.16 | |
| GWG1 | MYT1L | −2.46 | 1.90 | 0.19 |
| DLX5 | −0.35 | 0.45 | 0.43 | |
| GWG2 | MYT1L | −1.81 | 1.99 | 0.35 |
| SLC35F3 | 1.76 | 1.48 | 0.24 |
Abbreviations: pre-pregnancy BMI, Pre-pregnancy body Mass Index; GWG1, Rate of gestational weight gain at 1st trimester; GWG2, Rate of gestational weight gain at 2nd trimester; SE, standard error
Log2FC, Log2 fold change
Offspring CpG sites previously associated with maternal obesity traits
196 out of 1834 CpG sites in cord blood and 17 out of 143 CpG sites in adolescent blood, associated with maternal pre-pregnancy BMI in previous EWASs,6, 24 were also associated with maternal pre-pregnancy BMI in placenta in our study (P<0.05). The associations in placenta at 66 out of the 196 CpG sites (36.3%) and at 3 out of the 17 CpG sites (15.1%) were directionally consistent with the published findings (Supplementary Table S2).
CpG sites previously associated with child and adult obesity traits
15 out of 120 CpG sites in whole blood associated with childhood obesity,25, 26 152 out of 941 CpG sites in whole blood associated with adult obesity,27–39 and 641 out of 4606 CpG sites in adipose tissue 41 associated with adult obesity in previous EWASs were also associated with maternal pre-pregnancy BMI in placenta in our study (P < 0.05). The associations in placenta at 8 out of the 15 CpG sites in children blood, 84 out of the 152 CpG sites in adult blood and 414 out of the 641 CpG sites in adipose tissue were directionally consistent with the published findings (Supplementary Table S2).
Enrichment of pathways
Pathway enrichment analyses included 64 genes for pre-pregnancy BMI, 9 genes for GWG1, 8 genes for GWG2, and 5 genes for GWG3 (Supplementary Tables S3). The top canonical IPA networks included disease and function networks related to connective tissue disorders, skeletal and muscular disorders, cell cycle, cellular assembly and organization, DNA replication, lipid metabolism and metabolic diseases (Table 4). The 3-phosphoinositide degradation IPA canonical pathway was significantly enriched (FDR P<0.05) with pre-pregnancy BMI-associated genes in placenta (Supplementary Table S4).
Table 4.
Top networks associated with A) pre-pregnancy BMI and B) trimester specific gestational weight gain
| Networks | Molecules in Network | Score | Molecules | Top Diseases and Functions |
|---|---|---|---|---|
| Pre-pregnancy BMI | ||||
| 1 | ANTXR1, Akt, CD3, Ck2, CollagentypeI, CollagentypeIV, DICER1, DNMT3L, EPAS1, ERK, ERK1/2, GPAM, GSN, HCAR1, Histoneh3, INPP4B, ITGB1BP1, Insulin,MAFK, MMP15, MYO16, NRF1, PI3K(complex), PTPRJ, RAB27B, RASGRF1, RIMS2, SCNN1B, SEC14L1, SLITRK1, TBX1, TNFRSF8,cytokine, estrogen receptor, mir-26 | 55 | 23 | Connective Tissue Disorders, Organismal Injury and Abnormalities, Skeletal and Muscular Disorders |
| 2 | AFF3, AFF4, ANKFY1, Atp5e, CHPF, CTDSPL, Cyb5r3, DENND3, EIF3B, FAM13C, FANCD2, KIAA1324, LRRC45, Lrrfip2, MED12L, MED19, MED30, MED4, MOB2, MPP4, NFkB (complex), NMRAL1, NUMBL, PDX1, PLEKHM2, POLN, SASH1, SDCBP2, ST18, ST3GAL1, Slc25a1, TMX2, UNC5CL, WRN, ZC3H13 | 32 | 15 | Cell Cycle, Cellular Assembly and Organization, DNA Replication, Recombination, and Repair |
| 3 | ADPGK, AGAP3, AGBL5, AGO2, ANKRD11, C10orf71, CAMTA1, CFAP206, CNNM3, CTC1, DYRK1A, EGFL7, FAM219A, FBXL18, GPAT3, GSE1, HAO2, HM13, KAZN, KLHL29, LZTS2, PI4K2B, PIGQ, QPCTL, RASAL2, SH3BP5L, SHPK, SLC25A13,SMG6, TMCO3, TRIM25, TROAP, ZMAT2, miR-589-5p (and other miRNAs w/seed GAGAACC), mir-589 | 29 | 14 | Developmental Disorder, Hereditary Disorder, Metabolic Disease |
| 4 | ALDH4A1, ALLC, ARRB1, ATP5F1D, Atp5k, C1orf116, CAPS, CDH1, CMTM8, COG7, DBH-AS1, DPP10, ESR1, Erk1/2 dimer, FAM160B2, FGF1, FGF22, IFNB1, LINC-ROR, LRRC17, MAPK3, MRPL1, Mta, Ndocosahexaenoylethanolamine, NAA16, PANK1, PXYLP1, RBBP9, SMAD2, TP53, TSKU, VEZT, goralatide,lewis Y, miR-515-5p (and other miRNAs w/seed UCUCCAA) | 21 | 11 | Cancer, Hereditary Disorder, Organismal Injury and Abnormalities |
| GWG1 | ||||
| 1 | ALLC, CD163L1, CGREF1, DLX5, Dlx6os1, Eda, GPR19, GPR85, HENMT1, HOOK1, IL17D, IL4, Ighg3, KIF3A, Ltbp, MFAP4, MIR155HG, MN1, MPV17L, MYT1L, N-arachidonoyl-dopamine, NFkB (complex), PPP4R3A, SLC2A6, SMARCA4, SMG7, Srrm2, TGFB1, TP53, Tnfrsf22/Tnfrsf23, ZNF385A, miR-515-3p (and other miRNAs w/seed AGUGCCU), mir-584, neopterin,sesamol | 17 | 6 | Cell Cycle, Cellular Development, Cellular Growth and Proliferation |
| 2 | DHDDS, FOXP4, HNF4A, LOX, MT-ND1, NPC2, NUS1, ditrans, polycis-polyprenyl diphosphate synthase [(2E,6E)-farnesyl diphosphate specific], dolichol monophosphate | 3 | 1 | Lipid Metabolism, Small Molecule Biochemistry, Vitamin and Mineral Metabolism |
| GWG2 | ||||
| 1 | ADAMTS8, AP3B1, C16dihydroceramide, CCDC88B, CD163L1, CEACAM3, CHI3L2, Ccl27a, FCGR1B, FRMD4A, H60a,IFNG, IL17D, IL1F10, LILRA4, MTHFD2L, MYT1L, Myhs, NR4A1, RFX1, Raet1b, SCUBE1, SCUBE2, SLC28A1, SLC28A2, TNF, TRDV2, TREM3, TRIL, Trbv13-1,d18:0/24:0 dihydroceramide, neopterin, palmitoylsphingomyelin, ubiquinone 9, vanillic acid | 13 | 5 | Drug Metabolism, Molecular Transport, Nucleic Acid Metabolism |
| 2 | IL7R, SLC35F3 | 3 | 1 | Cell Cycle, Cell Death and Survival, Cellular Function and Maintenance |
| 3 | DEPDC1B, EBPL, GLP1R, KMT2D, NLGN3, NPM1 | 3 | 1 | Cell Morphology, Cell-To-Cell Signaling and Interaction, Cellular Assembly and Organization |
| GWG3 | ||||
| 1 | ADAM19, ARHGEF17, BPGM, BUD23, DNAJC13, EBPL, EGR2, EMG1, FAU, FRMD4A, Foxo, GFRA2, GZMM, HSPA12A, HSPA13, Hnrnpa1, IL17D, IL17RD, IL1RL2, IL7R, ITGAE, Lymphotoxin, MR1, NPM1, NPM3, NRTN, OGN, POU3F2, RPL29, RPL7L1, SH3TC2, SLAMF6, SLC35F3, TNF, ZNF750 | 15 | 5 | Cell Morphology, Cellular Assembly and Organization, Cellular Development |
Abbreviations: Pre-pregnancy BMI, Body Mass Index, GWG1, Rate of Gestational weight gain at 1st trimester; GWG2, Rate of Gestational weight gain at 2nd trimester; GWG3, Rate of Gestational weight gain at 3rd trimester
Functional annotation results
In the meQTL database, 26 SNPs in the EGFL7 and AGPAT2 genes were cis-meQTL with the cg14568196 [EGFL7] locus that was associated with pre-pregnancy BMI in our study. In addition, 130 SNPs in the VEZT, NDUFA12, NR2C1, and FGD6 genes were cis-meQTL with the cg15339142 [VEZT] locus that was associated with pre-pregnancy BMI in our study. The EGFL7 gene had the highest expression in placenta (Supplementary Figure S3–S4, Supplementary Table S5).
DISCUSSION
To our knowledge, this is the first placental EWAS of maternal pre-pregnancy BMI and trimester-specific rate of GWG. We identified novel differentially methylated CpG sites associated with increased maternal pre-pregnancy BMI and rate of GWG at first and second trimester. Pre-pregnancy BMI-associated genes in placenta were enriched in 3-phosphoinositide degradation canonical pathway. The 3-phosphoinositides are particularly important in signaling cascades that influence a wide variety of cellular functions. Disruption of these signaling pathways plays a key role in pathophysiological conditions of metabolic diseases.42 Functional annotations revealed that the differentially methylated CpG sites near EGFL7 and VEZT genes that were found to be associated with maternal pre-pregnancy BMI were cis-meQTL targets in blood across the life course, suggesting genetic regulation on DNA methylation of the genes across the life course. Moreover, we found that several CpG sites previously implicated in obesity traits in children and adults were significantly associated with maternal pre-pregnancy BMI in placenta,6, 24–39, 41 providing clues for specific epigenetic markers of obesity in early life.
Notably, we found that higher pre-pregnancy BMI was associated with increased methylation at cg14568196 [EGFL7] and with reduced expression of the EGFL7 gene in placenta. Moreover, higher DNA methylation at the CpG site was significantly correlated with decreased EGFL7 expression. These findings suggest that down-regulation of EGFL7 gene expression attributed to maternal adiposity may be mediated via epigenetic alteration at cg14568196. EGFL7 has the highest gene expression in placenta and encodes a protein that regulates vasculogenesis and promotes endothelial cell adhesion to the extracellular matrix in humans.43 In the homozygous Egfl7-knockout mice, vascular development is delayed in many organs despite normal endothelium cells proliferation, and 50% of the knockout embryos die in utero. Along with poor feto-placental perfusion, Egfl7 knockout resulted in reduced placental weight, 50% reduction in fetal blood space and fetal growth restriction.44 In the BPH/5 mouse model with preeclampsia, downregulation of Egfl7 in compromised placentas occurs prior to the onset of characteristic maternal signs of preeclampsia.43 An in vivo study suggested that there is a correlation between reduced expression of EGFL7 and inadequate trophoblast invasion observed in placentopathies.45 Another study showed that EGFL7 gene expression is regulated by hypoxia in trophoblast and EGFL7 expression was increased in maternal blood in women with early onset preeclampsia.46 However, the role of EGFL7 in placental development largely remains unknown. Future studies focused on this gene may give clues on mechanisms linking maternal obesity with placental vascular functional changes and altered fetal growth and development.
The relationship between DNA methylation and obesity traits is complicated by DNA sequence variation, which may contribute to both traits.47 Many obesity-associated SNPs were previously reported to be associated with proximal gene regulation.48 We found genetic loci in EGFL7 and AGPAT2 genes and loci in the VEZT, NDUFA12, NR2C1, and FGD6 genes that were cis-meQTL with two CpG sites associated with pre-pregnancy BMI in our study. The genes annotated to the cis-meQTL SNPs have functional relevance to obesity and related comorbidities. For example, AGPAT2 plays a role in regulation of growth and development of adipocytes, 49 NDUFA12 is down-regulated in obesity, 50 NR2C1 is highly expressed in adipose tissue and is associated with infant obesity, insulin resistance and inflammation, 51 and VEZT is highly expressed in the germ cell line and contributes to morphogenesis of preimplantation in the mouse embryo.52
DNA methylation changes in placenta may be relevant to understanding the developmental programming of human body weight regulation. Our study found overlapping CpG sites associated with pre-pregnancy BMI in cord and adolescent blood and with child and adult BMI in blood and adipose tissues,6, 24–41 consistent with a recent meta-analysis finding that reported consistent associations between higher maternal pre-pregnancy BMI and risk of obesity in early- mid- and late-childhood.53 Several CpG sites in genes such as FASN (adipose), HIF3A (adipose and blood), PHGDH (adipose and blood) were found to be significantly associated with maternal pre-pregnancy BMI in our data.54 For example, the hypoxia inducible factor 3 subunit alpha (HIF3A) gene expression is linked with vascular response to changes in oxygen tension, energy expenditure, metabolism and obesity 39 and adipose tissue dysfunction.55 CpG methylation annotated to HIF3A has been associated with birth weight and BMI at birth.56 Therefore, maternal adiposity-induced placental dysfunction may lead to hypoxia and oxidative stress in placenta 57 via epigenetic dysregulation of HIF3A expression, and the resulting fetal developmental response may be relevant for early programming obesity risk in later life.
Our study did not identify loci associated with GWG3. This may be due to the relatively smaller variation in GWG3 in our data (i.e., while mean GWG3 was larger than GWG1 and GWG2, the variation in GWG3 was not proportionally larger than that of GWG1 and GWG2). Fetal organogenesis occurs in early gestation during which the fetal epigenome is susceptible to environmental stimuli.58 Therefore, our findings may also be related to the possibility that maternal adiposity status in pre-pregnancy and early gestational weeks is more predictive of DNA methylation changes in placenta.53
An important limitation of our study is that because of absence of reference data for cell proportion in placenta, the findings may be confounded by unmeasured cellular heterogeneity. To counter this limitation, we implemented a validated reference-free adjustment for cell type proportion variation implemented in SVA.27 Furthermore, a Bayesian method was applied to correct the P-values for false positive rate and inflation.22 We were able to adjust for genotype-based ancestry estimates that have been found to be more robust in minimizing spurious associations due to population stratification.59 Another limitation was that the findings were not replicated in an independent population cohort because similarly designed studies are not common. Despite this limitation, we were reassured by the consistent corroboration of our EWAS findings with the results of the gene expression and in-silico annotations. The use of self-reported weight to calculate the pre-pregnancy BMI of the pregnant women may introduce inaccuracies. Finally, although large methylation changes associated with the maternal obesity traits could be identified by our EWAS, we acknowledge that studies with larger sample sizes are needed to detect CpGs of small effects associated with maternal obesity traits.
In conclusion, this is the first EWAS that identified placental DNA methylation loci associated with maternal pre-pregnancy BMI and GWG. Notably, the findings suggest that DNA methylation changes at the EGFL7 locus are associated with pre-pregnancy BMI may mediate down-regulation of the expression of placental EGFL7 gene. Placental epigenetic changes due to increase in maternal BMI may explain in-utero mechanisms in the developmental origins of obesity in the offspring and may be useful targets for early intervention.
Supplementary Material
Acknowledgements
The authors acknowledge the research teams at all participating clinical centers for the NICHD Fetal Growth Studies, including Christina Care Health Systems, Columbia University, Fountain Valley Hospital, California, Long Beach Memorial Medical Center, New York Hospital, Queens, Northwestern University, University of Alabama at Birmingham, University of California, Irvine, Medical University of South Carolina, Saint Peters University Hospital, Tufts University, and Women and Infants Hospital of Rhode Island. The authors also acknowledge C-TASC and The EMMES Corporations in providing data and imaging support. Genotyping was performed in the Department of Laboratory Medicine and Pathology, University of Minnesota. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
FUNDING: This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (contracts HHSN275200800013C; HHSN275200800002I; HHSN27500006; HHSN275200800003IC; HHSN275200800014C; HHSN275200800012C; HHSN275200800028C; HHSN275201000009C; HHSN27500008). Additional support was obtained from the National Institute on Minority Health and Health Disparities, and the National Institute of Diabetes and Digestive and Kidney Diseases. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Footnotes
COMPETING INTEREST STATEMENT: The authors declared no conflict of interest.
CLINICAL TRIAL REGISTRATION: ClinicalTrials.gov, NCT00912132.
Supplementary Data
Supplementary information is available at International Journal of Obesity’s website
Data Availability
The DNA methylation, genotype, and gene expression data are available through dbGaP with accession number phs001717.v1.p1.
REFERENCES
- 1.Chen C, Xu X, Yan Y. Estimated global overweight and obesity burden in pregnant women based on panel data model. PLoS One 2018; 13(8): e0202183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deputy NP, Sharma AJ, Kim SY. Gestational Weight Gain - United States, 2012 and 2013. Mmwr-Morbid Mortal W 2015; 64(43): 1215–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maffeis C, Morandi A. Effect of Maternal Obesity on Foetal Growth and Metabolic Health of the Offspring. Obes Facts 2017; 10(2): 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mamun AA, Mannan M, Doi SA. Gestational weight gain in relation to offspring obesity over the life course: a systematic review and bias-adjusted meta-analysis. Obes Rev 2014; 15(4): 338–47. [DOI] [PubMed] [Google Scholar]
- 5.Godfrey KM, Reynolds RM, Prescott SL, Nyirenda M, Jaddoe VW, Eriksson JG et al. Influence of maternal obesity on the long-term health of offspring. The lancet. Diabetes & endocrinology 2017; 5(1): 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharp GC, Lawlor DA, Richmond RC, Fraser A, Simpkin A, Suderman M et al. Maternal pre-pregnancy BMI and gestational weight gain, offspring DNA methylation and later offspring adiposity: findings from the Avon Longitudinal Study of Parents and Children. Int J Epidemiol 2015; 44(4): 1288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thakali KM, Faske JB, Ishwar A, Alfaro MP, Cleves MA, Badger TM et al. Maternal obesity and gestational weight gain are modestly associated with umbilical cord DNA methylation. Placenta 2017; 57: 194–203. [DOI] [PubMed] [Google Scholar]
- 8.Desai M, Jellyman JK, Ross MG. Epigenomics, gestational programming and risk of metabolic syndrome. Int J Obes (Lond) 2015; 39(4): 633–41. [DOI] [PubMed] [Google Scholar]
- 9.Thornburg KL, O’Tierney PF, Louey S. The Placenta is a Programming Agent for Cardiovascular Disease. Placenta 2010; 31: S54–S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desoye G The Human Placenta in Diabetes and Obesity: Friend or Foe? The 2017 Norbert Freinkel Award Lecture. Diabetes Care 2018; 41(7): 1362–1369. [DOI] [PubMed] [Google Scholar]
- 11.van Dijk SJ, Molloy PL, Varinli H, Morrison JL, Muhlhausler BS, Members of Epi S. Epigenetics and human obesity. Int J Obes (Lond) 2015; 39(1): 85–97. [DOI] [PubMed] [Google Scholar]
- 12.Mitsuya K, Parker AN, Liu L, Ruan J, Vissers MCM, Myatt L. Alterations in the placental methylome with maternal obesity and evidence for metabolic regulation. PLoS One 2017; 12(10): e0186115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai T, Yamada T, Abe K, Okamura K, Kamura H, Akaishi R et al. Increased epigenetic alterations at the promoters of transcriptional regulators following inadequate maternal gestational weight gain. Sci Rep 2015; 5: 14224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michels KB, Harris HR, Barault L. Birthweight, maternal weight trajectories and global DNA methylation of LINE-1 repetitive elements. PLoS One 2011; 6(9): e25254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grewal J, Grantz KL, Zhang C, Sciscione A, Wing DA, Grobman WA et al. Cohort Profile: NICHD Fetal Growth Studies-Singletons and Twins. Int J Epidemiol 2018; 47(1): 25–25l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinkle SN, Johns AM, Albert PS, Kim S, Grantz KL. Longitudinal changes in gestational weight gain and the association with intrauterine fetal growth. Eur J Obstet Gynecol Reprod Biol 2015; 190: 41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delahaye F, Do C, Kong Y, Ashkar R, Salas M, Tycko B et al. Genetic variants influence on the placenta regulatory landscape. Plos Genet 2018; 14(11): e1007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2012; 29(2): 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tekola-Ayele F, Workalemahu T, Gorfu G, Shrestha D, Tycko B, Wapner R et al. Sex differences in the associations of placental epigenetic aging with fetal growth. Aging 2019; 11(15): 5412–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010; 11: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012; 28(6): 882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Iterson M, van Zwet EW, Heijmans BT. Controlling bias and inflation in epigenome-and transcriptome-wide association studies using the empirical null distribution. Genome biology 2017; 18(1): 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology 2010; 11(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharp GC, Salas LA, Monnereau C, Allard C, Yousefi P, Everson TM et al. Maternal BMI at the start of pregnancy and offspring epigenome-wide DNA methylation: findings from the pregnancy and childhood epigenetics (PACE) consortium. Hum Mol Genet 2017; 26(20): 4067–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fradin D, Boelle PY, Belot MP, Lachaux F, Tost J, Besse C et al. Genome-Wide Methylation Analysis Identifies Specific Epigenetic Marks In Severely Obese Children. Sci Rep 2017; 7: 46311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang RC, Garratt ES, Pan H, Wu Y, Davis EA, Barton SJ et al. Genome-wide methylation analysis identifies differentially methylated CpG loci associated with severe obesity in childhood. Epigenetics 2015; 10(11): 995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhana K, Braun KVE, Nano J, Voortman T, Demerath EW, Guan W et al. An Epigenome-Wide Association Study of Obesity-Related Traits. Am J Epidemiol 2018; 187(8): 1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 2017; 541(7635): 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Pan Y, Zhu H, Hao G, Huang Y, Barnes V et al. An epigenome-wide study of obesity in African American youth and young adults: novel findings, replication in neutrophils, and relationship with gene expression. Clinical epigenetics 2018; 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geurts YM, Dugue PA, Joo JE, Makalic E, Jung CH, Guan W et al. Novel associations between blood DNA methylation and body mass index in middle-aged and older adults. Int J Obes (Lond) 2018; 42(4): 887–896. [DOI] [PubMed] [Google Scholar]
- 31.Meeks KAC, Henneman P, Venema A, Burr T, Galbete C, Danquah I et al. An epigenome-wide association study in whole blood of measures of adiposity among Ghanaians: the RODAM study. Clinical epigenetics 2017; 9: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sayols-Baixeras S, Subirana I, Fernandez-Sanles A, Senti M, Lluis-Ganella C, Marrugat J et al. DNA methylation and obesity traits: An epigenome-wide association study. The REGICOR study. Epigenetics 2017; 12(10): 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansego ML, Milagro FI, Zulet MA, Moreno-Aliaga MJ, Martinez JA. Differential DNA Methylation in Relation to Age and Health Risks of Obesity. International journal of molecular sciences 2015; 16(8): 16816–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson LE, Harlid S, Xu Z, Sandler DP, Taylor JA. An epigenome-wide study of body mass index and DNA methylation in blood using participants from the Sister Study cohort. Int J Obes (Lond) 2017; 41(1): 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al Muftah WA, Al-Shafai M, Zaghlool SB, Visconti A, Tsai PC, Kumar P et al. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clinical epigenetics 2016; 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almen MS, Nilsson EK, Jacobsson JA, Kalnina I, Klovins J, Fredriksson R et al. Genome-wide analysis reveals DNA methylation markers that vary with both age and obesity. Gene 2014; 548(1): 61–7. [DOI] [PubMed] [Google Scholar]
- 37.Aslibekyan S, Demerath EW, Mendelson M, Zhi D, Guan W, Liang L et al. Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity (Silver Spring) 2015; 23(7): 1493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet 2015; 24(15): 4464–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet (London, England) 2014; 383(9933): 1990–8. [DOI] [PubMed] [Google Scholar]
- 40.Agha G, Houseman EA, Kelsey KT, Eaton CB, Buka SL, Loucks EB. Adiposity is associated with DNA methylation profile in adipose tissue. Int J Epidemiol 2015; 44(4): 1277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ronn T, Volkov P, Gillberg L, Kokosar M, Perfilyev A, Jacobsen AL et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet 2015; 24(13): 3792–813. [DOI] [PubMed] [Google Scholar]
- 42.Bridges D, Saltiel AR. Phosphoinositides: Key modulators of energy metabolism. Biochim Biophys Acta 2015; 1851(6): 857–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lacko LA, Massimiani M, Sones JL, Hurtado R, Salvi S, Ferrazzani S et al. Novel expression of EGFL7 in placental trophoblast and endothelial cells and its implication in preeclampsia. Mech Dev 2014; 133: 163–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lacko LA, Hurtado R, Hinds S, Poulos MG, Butler JM, Stuhlmann H. Altered feto-placental vascularization, feto-placental malperfusion and fetal growth restriction in mice with Egfl7 loss of function. Development 2017; 144(13): 2469–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Massimiani M, Vecchione L, Piccirilli D, Spitalieri P, Amati F, Salvi S et al. Epidermal growth factor-like domain 7 promotes migration and invasion of human trophoblast cells through activation of MAPK, PI3K and NOTCH signaling pathways. Mol Hum Reprod 2015; 21(5): 435–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitehead CL, Kaitu’u-Lino TJ, Binder NK, Beard S, De Alwis N, Brownfoot F et al. EGFL7 gene expression is regulated by hypoxia in trophoblast and altered in the plasma of patients with early preeclampsia. Pregnancy Hypertens 2018; 14: 115–120. [DOI] [PubMed] [Google Scholar]
- 47.Hernandez DG, Singleton AB. Using DNA methylation to understand biological consequences of genetic variability. Neurodegener Dis 2012; 9(2): 53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voisin S, Almen MS, Zheleznyakova GY, Lundberg L, Zarei S, Castillo S et al. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome medicine 2015; 7: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cortes VA, Curtis DE, Sukumaran S, Shao X, Parameswara V, Rashid S et al. Molecular mechanisms of hepatic steatosis and insulin resistance in the AGPAT2-deficient mouse model of congenital generalized lipodystrophy. Cell metabolism 2009; 9(2): 165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinonen S, Buzkova J, Muniandy M, Kaksonen R, Ollikainen M, Ismail K et al. Impaired Mitochondrial Biogenesis in Adipose Tissue in Acquired Obesity. Diabetes 2015; 64(9): 3135–45. [DOI] [PubMed] [Google Scholar]
- 51.Kang HS, Okamoto K, Kim YS, Takeda Y, Bortner CD, Dang H et al. Nuclear orphan receptor TAK1/TR4-deficient mice are protected against obesity-linked inflammation, hepatic steatosis, and insulin resistance. Diabetes 2011; 60(1): 177–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hyenne V, Harf JC, Latz M, Maro B, Wolfrum U, Simmler MC. Vezatin, a ubiquitous protein of adherens cell-cell junctions, is exclusively expressed in germ cells in mouse testis. Reproduction (Cambridge, England) 2007; 133(3): 563–74. [DOI] [PubMed] [Google Scholar]
- 53.Voerman E, Santos S, Patro Golab B, Amiano P, Ballester F, Barros H et al. Maternal body mass index, gestational weight gain, and the risk of overweight and obesity across childhood: An individual participant data meta-analysis. PLoS Med 2019; 16(2): e1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohde K, Keller M, la Cour Poulsen L, Bluher M, Kovacs P, Bottcher Y. Genetics and epigenetics in obesity. Metabolism: clinical and experimental 2019; 92: 37–50. [DOI] [PubMed] [Google Scholar]
- 55.Pfeiffer S, Kruger J, Maierhofer A, Bottcher Y, Kloting N, El Hajj N et al. Hypoxia-inducible factor 3A gene expression and methylation in adipose tissue is related to adipose tissue dysfunction. Sci Rep 2016; 6: 27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan H, Lin X, Wu Y, Chen L, Teh AL, Soh SE et al. HIF3A association with adiposity: the story begins before birth. Epigenomics 2015; 7(6): 937–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pereira RD, De Long NE, Wang RC, Yazdi FT, Holloway AC, Raha S. Angiogenesis in the placenta: the role of reactive oxygen species signaling. Biomed Res Int 2015; 2015: 814543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grindler NM, Vanderlinden L, Karthikraj R, Kannan K, Teal S, Polotsky AJ et al. Exposure to Phthalate, an Endocrine Disrupting Chemical, Alters the First Trimester Placental Methylome and Transcriptome in Women. Sci Rep 2018; 8(1): 6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barfield RT, Almli LM, Kilaru V, Smith AK, Mercer KB, Duncan R et al. Accounting for population stratification in DNA methylation studies. Genetic epidemiology 2014; 38(3): 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.