Abstract
Runx2 (Runt-related transcription factor 2) is a key transcription factor which is associated with osteoblast differentiation and expressed in ER+ (estrogen receptor positive) human breast cancer cell lines. Runx2 also participates in mammary gland development. Deregulation of RNA Pol III genes (polymerase III-dependent genes) is tightly linked to tumor development, while Brf1 (TFIIB-related factor 1) specifically regulates these gene transcription. However, nothing is known about the effect of Runx2 on Brf1 expression and Pol III gene transcription. Expression of Runx2, Brf1 and Pol III genes from the samples of human breast cancer and cell culture model were determined by the assays of RT-qPCR, immunoblot, luciferase reporter activity, immunohistochemistry, chromatin immunoprecipitation and Immunofluorescence. High expression of Runx2 is observed in the cases of breast cancer. The patients of high Runx2 expression at early stages display longer survival period, whereas the cases of high Runx2 at advanced stages reveal faster recurrence. The identification of signaling pathway indicates that JNK1 and c-Jun mediate Runx2 transcription. Repression of Runx2 reduces Brf1 expression and Pol III gene transcription. Further analysis indicates that Runx2 is colocalized with Brf1 in nucleus of breast cancer tissue. Both Runx2 and Brf1 synergistically modulate Pol III gene transcription. These studies indicate that Brf1 overexpression is able to be used as an early diagnosis biomarker of breast cancer, while high Runx2 expression indicates long survival period and faster recurrence. Runx2 mediates the deregulation of Brf1 and Pol III genes and its abnormal expression predicts the worse prognosis of breast cancer.
Keywords: Runx2, Brf1, Pol III genes, alcohol, breast cancer
1. Introduction
Breast cancer is the first malignant disease in females. In the United States, over 200,000 cases of breast cancer are diagnosed each year. Approximately 80% breast cancer cases are ER+ (estrogen receptor positive) and 20% ER− (estrogen receptor negative) (1–2). Studies have indicated that alcohol consumption is consistently associated with increased risk for breast cancer in women (3–6). However, the mechanism for this observation remains to be established. Epidemiological studies have shown that the relative increase in risk ranges from 5–10% to 40% with alcohol drinking elevation (7–8). Alcohol administration promotes mammary tumor formation in mice (9–11). Alcohol has been classified as carcinogenic to humans by IARC (the International Agency for Research on Cancer) (12–14).
Runx2 (Runt-related transcription factor 2) is a key transcription factor which is associated with osteoblast differentiation (15) and has also been described as an oncogene (16). Emerging evidence shows that Runx2 is associated with mammary gland development and ER+ breast cancer (16). High level of Runx2 is found in breast cancer cell lines (17). Both E2 (17-β estradiol) and ERα (estrogen receptor α) upregulate Runx2 transcription (18). Our studies have shown that alcohol increases ERα activity to enhance Brf1 (TFIIB-related factor 1) expression and Pol III gene (RNA polymerase III-dependent gene) transcription (19). JNK1 (c-Jun N-terminal kinase 1) is a member of JNK subfamily, while c-Jun is a component of JNK downstream and a subunit of AP-1 (activated protein-1). Alcohol induces activation of JNK1 and increases c-Jun expression to elevate Brf1 expression and Pol III gene transcription (20), whereas both JNK1 and c-Jun upregulate Runx2 expression (21). Studies have revealed that Runx2 controls the expression of genes, which are associated with tumor cell growth and migration (22). It implies that alcohol is able to increase the cellular level of Runx2 which may be involved in the alcohol-induced the deregulation of Brf1 and Pol III genes to promote breast cancer development. However, nothing is known about the effects of Runx2 on Brf1 and Pol III genes.
Brf1 and products of Pol III genes, such as 5S rRNA and tRNAs, control the translational and growth capacity of cells (23–24). The deregulation of Pol III genes is tightly associated with cell transformation and tumor development (19–20, 23–29). Studies from our laboratory and others have demonstrated that oncogenic proteins, such as Ras, c-Jun, and c-Myc, stimulate Pol III gene transcription (25–28), whereas tumor suppressors, such as BRCA1, PTEN, pRb, p53 and Maf1, repress the transcription of this class of genes (26–30). The ability of these oncogenic and tumor suppressor proteins to regulate Pol III gene transcription result from their capacity to modulate the TFIIIB complex. TFIIIB complex consists of TBP (TATA box-binding protein) and its associated factors, Bdp1 and Brf1. TBP transcribes three polymerases-dependent genes, Pol I-, Pol II- and Pol III genes, whereas Brf1 and Bdp1 specifically regulate Pol III gene transcription. Our studies demonstrated that regulation of Bdp1, but not Brf1, occurred through JNK1-mediated alteration of TBP expression (24). Further analysis reveals that alcohol-induced ERα activity modulates Brf1 expression, but not TBP (19). Studies have indicated that specific tRNAs are upregulated in human breast cancer cells as promoters of breast cancer metastasis (31), while increased tRNAiMet within cancer cells drives cell migration and invasion to enhance the metastatic potential in cancer (32). Our study has demonstrated that Brf1 is overexpressed in ER+ cases of breast cancer (33). High expression of Brf1 in HCC (hepatocellular carcinoma) displays short survival period (34). Tam (Tamoxifen) is widely used as hormone therapy in post-menopausal ER+ women with breast cancer. Tam acts as an estrogen agonist, leading to certain adverse effects. Our early study has demonstrated that Tam represses Brf1 expression to decrease alcohol-induced Pol III gene transcription (35). Interestingly, high Brf1 expression in most of ER+ cases of breast cancer display longer survival times after Tam treatment (33). Given that ERα mediates Runx2 transcription (17–19), we explore whether Runx2 mediates Brf1 and Pol III genes and determine Runx2 expression in breast cancer cases.
Here, we report that Runx2 is overexpressed in breast cancer cases, high Runx2 expressions of these cases reveal long survival period and worse prognosis. Alcohol increases Runx2 expression and inhibitions of JNK1, c-Jun and ERα reduce Runx2 transcription, while repressing Runx2 decreases alcohol-induced Brf1 expression and Pol III gene transcription. Further analysis shows that Runx2 and Brf1 colocalize in nucleus of breast cancer cells, both Runx2 and Brf1 occupy the promoters of tRNALeu and 5S rRNA to synergistically upregulate these gene transcription and to promote breast cancer development.
2. Materials and methods
2.1. Cell lines, reagents and antibodies
ER+ human breast cancer cell lines (MCF-7) were from ATCC (University Boulevard, Manassas, VA, USA). Cell culture medium DMEM/F12, Lipofectamine 2000, TRIzol reagent and OPTI-MEM were from Life Technology (Van Allen Way, Carlsbad, CA, USA). Antibodies against Runx2 and b-actin and siRNA of Runx2 were obtained from Santa Cruz (Santa Cruz, CA, USA). Brf1 antibody was from Bethyl laboratories Inc (West FM, Montgomery, TX, USA). JNK inhibitor, SP600125 was from A.G. Scientific, Inc (Lusk Blvd, San Diego, CA, USA). The siRNAs of c-Jun and Runx2 were bought from Santa Cruz (Santa Cruz, CA, USA). The sequences of JNK1 and Brf1 siRNA (Table S1) and primers of Pol III genes (Table S2) were listed in Supplements (36–37). E2 (17b-estradiol) was from Sigma-Aldrich (St. Louis, MO, USA). Plasmid of Runx2 expression and Runx2-luciferase reporter construct were kindly provided by Dr. Baruch Frenkel (University of Southern California, USA) (38).
2.2. Patients and samples
Paraffin-embedded tumor tissue samples were obtained from 219 women diagnosed with breast carcinoma who underwent surgical resection between July 2001 and December 2007 in the Department of Breast and Thyroid Surgery and Department of Pathology at the First Affiliated Hospital of Sun Yat-sen University. We obtained prior patient’s consent and approval from the Medical Ethical Committee of the First Affiliated Hospital, Sun Yat-sen University, for use in these clinical materials in this study.
All patients’ ages ranged from 24 to 79 (median = 50), including 12 cases of in situ carcinoma (DCIS), 197 cases of invasive ductal carcinoma (IDC), four cases of invasive lobular carcinoma (ILC), and six cases of metastatic breast cancer (MBC). The patients didn’t receive chemotherapy or radiotherapy before surgery. Clinicopathological information, such as age, tumor size, lymph node status, ER, PR, and HER2 status, was obtained by reviewing medical records and pathology reports.
Fresh tumor specimens were obtained from the patients who underwent resection of the primary breast cancer in the Department of Breast and Thyroid Surgery at the First Affiliated Hospital of Sun Yat-Sen University. Representative blocks from both the tumor foci and tumor foci adjacent noncancerous tissues from each specimen were stored in liquid nitrogen for RNA and protein extraction. Informed consent was obtained from each patient, and the study was approved by the Institute Research Ethics Committee of Sun Yat-Sen University. The patients hadn’t previously received chemotherapy or radiation therapy.
2.3. Immunohistochemistry
Immunohistochemical staining was carried out on formalin-fixed, paraffin-embedded sections (4 μm thick) which were deparaffinized in xylene and rehydrated in decreasing concentrations of ethanol and rinsed in phosphate buffered saline, and then retrieval antigen with microwave treatment in 10 mM citrate buffer (pH 6.0). Immunohistochemistry staining was carried out using the EnVision™ Kit (DAKO, Hamburg, Denmark) following the manufacturer’s instructions. The endogenous peroxidase activity was quenched by 3% hydrogen peroxide for 15 minutes. The sections were incubated with primary antibody Runx2 (1:400) mouse antibody or Brf1 (1:200) rabbit antibody over night at 4°C. Then the tissue sections were sequentially incubated with ready to use HRP- immunoglobulin (Evision™) for 30 min and were developed with 3,3’-diaminobenzidine (DAB) as a chromogen substrate. The nuclei were counterstained with Meyer’s hematoxylin.
The level of Runx2 immunostaining was evaluated independently by two pathologists blinded to the survival outcomes of the participants based on the proportion of positively stained tumor cells (stain area) and intensity of staining. Immunoreactivity of Runx2 was detected in the nuclei. Staining intensity was scored as: 0 (no staining), 1 (weak staining) for light yellow color, 2 (moderate staining) for yellow brown color, and 3 (strong staining) for brown color. The positive tumor cell proportion was scored as: 0 (no positive tumor cells), 1 (<20% positive tumor cells), 2 (20~50% positive tumor cells) and 3 (>50% positive tumor cells). A modified immunoreactivity score method to evaluate the immunostaining results was performed by multiplying stain intensity by stain area (staining index, SI) as previously described. The Runx2 expression level in breast carcinoma lesions were determined by the staining index (SI) which are 0, 1, 2, 3, 4, 6 or 9. An optimal cutoff value was identified as follows: the SI score of > 6 was used to define tumors as high Runx2 expression, and the SI score of ≤ 6 as low Runx2 expression (39).
2.4. RNA isolation and RT-qPCR
Total RNAs were isolated from breast cancer cell lines treated with ethanol using single step extraction method TRIzol reagent (Invitrogen). Total RNA samples were quantified and reverse-transcribed in a 20 μl reaction containing 1 × RT (reverse transcription) buffer. After first-strand cDNA synthesis, the cDNAs were diluted in DNase-free water and real time qPCR (RT-qPCR) were performed with specific primers as described previous (20,34,36) and PCR reagent kits (Bio-Rad Biotech) in the ABI prism 7700 Sequence Detection System. tRNALeu and 5S rRNA transcripts and mRNAs of Runx2 and Brf1 were measured by RT-qPCR as described previously (19–20).
2.5. Transfection and Runx2-luciferase reporter assays
For transient transfection assays, cells were transfected with plasmids and/or siRNAs as described previously (19–20). Serum-free medium was added to each dish with Lipofectin-DNA or Lipofectamine2000-siRNA complexes, and cells were further incubated for 4h. The medium was changed with 10% FBS DMEM/F12 (phenol red-free) (19) and cells were incubated for 48h before harvesting. Protein concentrations of the resultant lysates were measured by the Bradford method. For Runx2-luciferase reporter assays, cells were transfected with 0.2 μg of the Runx2-luciferase constructs or plus JNK1, c-Jun or ERα siRNA for 24 h. Cells were starved in DMEM/F12 for 4 h and treated with 25mM ethanol for another 60min. Cell pellets were resuspended in Promega reporter lysis buffer. The lysates were analyzed for luciferase activity using a luminometer and the Promega Luciferase Assay System as described (Promega). Resultant luciferase activities were normalized to the amount of protein in each lysate as described (25). The fold change in luciferase activity was calculated by determining the level of luciferase activity in the absence of alcohol, its value will be set at 1 for each independent experiment. Values are means ± SE of at least three independent experiments.
2.6. SDS-PAGE and immunoblot analysis.
Human breast cancer cells were incubated with 25mM ethanol for 60 min after starvation 4h. Cells were collected with lysis buffer and sonicated. The suspensions were centrifuged to save the supernatants. Protein concentrations were determined by the Bradford method using Fluostar Omega spectrometer (Cell Biology Core Laboratory of University of Southern California Research Center for Liver Diseases, P30DK DK48522). Lysates (50 μg of protein) were subjected to sodium dodecyl sulfatepolyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred from the SDS-PAGE gel to Hybond-P membrane and immunoblot analysis were performed with specific antibodies. Membranes were probed with either antibodies against Runx2, Brf1 and β-actin as described (19–20). Bound primary antibody was visualized using horseradish peroxidase-conjugated secondary antibody (Vector Laboratories) and enhancing chemiluminscence reagents (Thermo-Fisher).
2.7. Immunofluorescence
The collected tumors were fixed with 4% paraformaldehyde, embedded in paraffin, and cut into 4-μm-thick sections using a microtome. After removing paraffin wax with xylene, and then retrieval antigen with microwave treatment in 10 mM EDTA buffer (pH 9.0). The tumor sections were blocked with 5% BSA for 1h at room temperature, and then incubated with a rabbit monoclonal anti-human Brf1 antibody (BETHYL laboratories, Inc. USA), anti-human Runx2 polyclonal antibody (Santa Cruz, California, USA) over night at 4°C. and then incubated with anti-mouse IgG FITC and anti-rabbit IgG CY3 (Invitrogen Life Technologies Corporation, USA) as secondary antibody. Nuclear staining of cells was done using 4,6-diamidino-2-phenylindole (DAPI) (Beyotime Biotechnology, SHANGHAI, CHINA). The slides were mounted in antifade reagent (Invitrogen Life Technologies Corporation, USA). The photomicrographs were captured using Olympus BX63 fluorescent microscopy (Germany).
2.8. Cell Anchorage-independent growth
MCF-7 cells were transfected with mismatch RNA (siMM) or Runx2 siRNAs as described (19). The transfected MCF-7 cells (1 × 104 cells/well in 6-well plate) were suspended in 0.35% (w/v) agar in 10% FBS/DMEM/F12 with or without 25mM ethanol and the ethanol was over a bottom layer of media with 0.5% (w/v) agar. Cells were fed fresh complete media with ethanol twice weekly. Colonies were counted 2–3 weeks or longer after plating as previously described (36).
2.9. Statistical analysis
All statistical analyses were performed by using the SPSS 22.0 statistical software package. The Pearson χ2 and Fisher’s exact tests were used to analyze the relationship between Runx2 expression and clinicopathologic feature. Survival curves were plotted using Kaplan-Meier method and compared with the log-rank test. Univariate and multivariate Cox regression analyses were used to calculate the survival data. The forest plot was performed with R software. All tests were two sided. A P-value <0.05 in all cases was considered statistically significant.
3. Results
3.1. The expression of Runx2 in breast invasive duct carcinoma lesions.
The expressions of Runx2 in breast invasive duct carcinoma were examined by immunohistochemistry. In 219 paraffin-embedded breast carcinoma tissue samples, Runx2 immunoreactivity was detected in the nuclei of breast carcinoma cells (Fig 1). 71 cases (32.4%) have strong staining in lesion tissues with SI>6, which was classified as high expression level group. The other 148 cases (67.6 %) of breast carcinoma have moderate, weak or negative staining in lesion tissues with SI≤6, which was classified as low expression level group. In addition, we have found fresh IDC (invasive duct carcinoma) samples with strong Runx2 staining in the carcinoma tissues (Fig. 1A). In contrast, weak staining is observed in para-cancerous breast duct (Fig. 1B).
Fig. 1. Runx2 immunohistochemistry (IHC) staining and Kaplan-Meier survival curve and log-rank test analysis of the association between Runx2 expression and breast cancer patient survival.
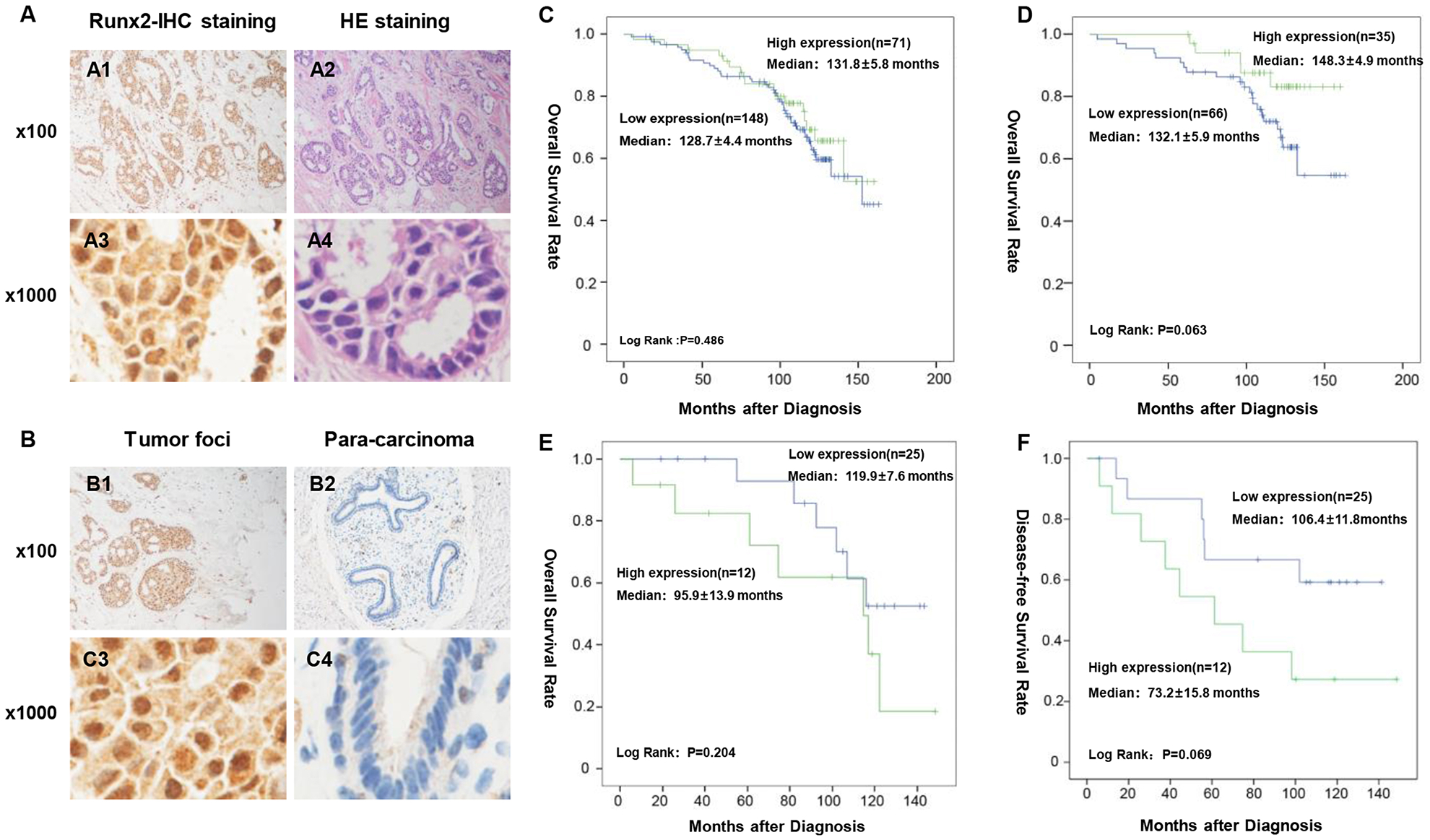
(A). Runx2 staining. (A1 and A3) IHC staining of Brf1 of HBC tumor tissues; (A2 and A4) H&E staining of HBC tumor tissues. (A1 & A2) × 100 magnification; (A3 & A4) × 1000 magnification. A representative Runx2 staining of human breast cancer samples. (B). Comparison of Runx2 staining in tumor foci and para-can tissue of breast cancer. (B1 & B3): strong staining signals of Runx2 expression are seen in tumor foci of breast cancer; (B2 & B4): week signals of Runx2 staining are detected in para-carcinoma tissue of breast cancer. (B1 & B3) × 100 magnification; (B2 & B4) × 1000 magnification. (C). Runx2 expression was determined in 219 cases of breast cancer. All cases of breast cancer were determined by pathological analysis and immunohistochemistry staining. (D), The cases of breast cancer are at clinic stage 2b. (E and F): The cases at stage III of this disease. n = number of patients in the subgroup, M = median survival in months of the subgroup. P-values were calculated by log-rank test. The group of high Runx2 expression display shorter survival period and faster recurrence.
3.2. Relationship of Runx2 up-regulation with the Clinic pathologic features of breast cancer
Next, we have determined that relationship of Runx2 up-regulation with Clinic pathologic features of 219 cases, the information is summarized in Table S4 in Supplements. The correlation between Runx2 protein expression and clinicopathologic features were analyzed by Chi-square (χ2) test except the distant metastasis using Fisher’s exact test. As the results shown in Table S4, the levels of Runx2 expression correlated with tumor sizes (P = 0.057).
3.3. Association between Runx2 expression with patient survival
Reports indicate that Runx2 expression is increased in human breast cancer cell lines (16–17). However, the relationship of levels of Runx2 expression and survival period in the human cases of breast cancer is unclear. We have performed the Kaplan-Meier survival curves and the log-rank test survive analysis for the 219 cases (Fig. 1C–1F). The results showed that survival of patients in clinical stage II-b with high Runx2 levels was longer than patients with low Runx2 levels (P =0.063) (Fig. 1D). High expression of Runx2 predicted a faster recurrence in the groups in clinical advanced stage. (P =0.069) (Fig. 1F).
3.4. Alcohol induces Runx2 expression in ER+ breast cancer cell line
Studies have demonstrated that alcohol consumption is associated with the risk of breast cancer. We found that alcohol increases Brf1 expression and Pol III gene transcription to promote cell transformation and high expression of Brf1 is observed in most of ER+ cases of breast cancer (19,33). To determine whether alcohol affects Runx2 expression, we treated the cells of ER+ breast cancer line, MCF-7 with alcohol. The results indicate that alcohol increases Runx2 promoter activity (Fig. 2A). Further analysis reveals that alcohol enhances the cellular levels of Runx2 mRNA (Fig. 2B) and protein in MCF-7 cells (Fig. 2C). These results show that alcohol is able to increase Runx2 transcription.
Fig. 2. Alcohol mediates Runx2 expression and signaling events in ER+ breast cancer cells.

(A): Alcohol increase Runx2 promoter activity. MCF-7 cells were transfected with Runx2-Luc reporter and treated with ethanol to determine Luc activity; (B-C): MCF-7 cells were treated with ethanol (25mM) to extract total RNA and cell lysates. mRNA levels of Runx2 were measured by RT-PCR. The fold changes are calculated by normalizing to the amount of GAPDH mRNA (B). Western blots were performed to determine the levels of Runx2 and β-actin, which is represent Western blot (C). These results indicate that alcohol mediates Runx2 expression. On other hands, MCF-7 cells were transfected with Runx2-Luc-reporter plus mismatch RNA (siMM), JNK1 siRNA, c-Jun siRNA, ERα siRNA as indicated, respectively. The cells were treated with 25mM ethanol to determine Runx2 promoter activity. (E) MCF-7 cells were pretreated with JNK inhibitor, SP 600125 (5μM) for 1 hour and then treated with alcohol as described above. (F) JNK1 siRNA (siJNK1); (G) c-Jun siRNA (si-c-Jun) and (H) ERα siRNA (siERα), compared to siMM. Luciferase activities were normalized to total protein levels. The change in Runx2 promoter activities were calculated relative to that with no ethanol treatment of siMM-transfected cells. (H) Western blot to determine the levels of Runx2 protein. These results indicate that reductions of JNK1, c-Jun and ERα by their siRNA inhibit Runx2 transcription. The bars represent Mean ± SE at least three independent determinations. *: p<0.05; **: p<0.01.
3.5. Signaling events of affecting Runx2 transcription
Our studies have demonstrated that alcohol induces activation of JNK1 and increases c-Jun expression to upregulate Brf1 and Pol III gene transcription (19–20). Therefore, we have further determined whether this pathway mediates Runx2 activity. The results indicate that JNK chemical inhibitor SP600125 decreases alcohol-induced Runx2 promoter activity (Fig. 2D). Early studies have demonstrated that JNK1 positively, but JNK2 negatively, modulates Pol III gene transcription (25,37). Given that chemical inhibitors reveal non-specific inhibition, we have utilized a specific inhibitor, JNK1 siRNA (36). The result indicates that JNK1 siRNA significantly decreases Runx2 promoter activity, compared to mismatch RNA (siMM) as a control (Fig. 2E). Therefore, we further determined whether c-Jun mediates alcohol-induced Runx2 transcription. Fig. 2F shows that decreasing c-Jun expression dramatically inhibits Runx2 promoter activity. In addition, we also tested the role of ERα in Runx2 expression. The result indicates that repressing ERα by its siRNA (siERα) indeed decreases Runx2 transcription (Fig. 2G). Furthermore, immunoblot analysis shows that JNK1 and c-Jun siRNAs (siJNK1 and si-cJun) reduce the cellular levels of Runx2 protein, compared to mismatch RNA (Fig. 2H). These studies indicate that alcohol affects Runx2 expression through JNK1, c-Jun and ERα pathway.
3.6. Runx2 modulates Pol III gene transcription and Brf1 expression
Deregulation of Pol III genes is tightly associated with cell transformation and tumor development. Our studies have demonstrated that alcohol increases Brf1 expression and Pol III gene transcription (19–20). Given that alcohol enhances cellular levels of Runx2 mRNA and Protein (Fig. 2). We have further determined whether change in Runx2 expression affects Pol III gene activity. The results indicate that alcohol induces Pol III gene, either tRNALeu (Fig. 3A) or 5S rRNA (Fig. 3C), transcription. While enhancing Runx2 results in additional elevation of Pol III gene transcription (Fig. 3A, 3C). In contrast, repressing Runx2 by its siRNA (siRunx2) significantly decreases the induction of these genes caused by alcohol (Fig. 3B and 3D). Brf1 is a key transcription factor which specifically regulates Pol III genes. Thus, we further observed whether alteration of Runx2 affects Brf1 expression. When ER+ breast cancer cells were transfected with a Runx2 express construct to increase its cellular level, resulting in elevation of Brf1 transcription (Fig. 4B), whereas repressing Runx2 expression by its siRNA decreases the cellular levels of Brf1 mRNA and protein (Fig.4A, 4C). Together, these studies have demonstrated that Runx2 modulates Pol III gene transcription and Brf1 expression under alcohol treatment.
Fig. 3. Runx2 modulates Pol III gene transcription.

(A and C) ER+ MCF-7 breast cancer cells with Runx2 expression construct were treated with ethanol as described at reference (40). (B and D) MCF-7 cells were transfected with mismatch (mm) RNA, Runx2 siRNA for 48 h, respectively. The cells were treated as described above. The amounts of tRNALeu (A and C) and 5S rRNA (B and D) were measured by RT-qPCR. Repression of Runx2 (B and D) by its siRNAs decreases Pol III gene transcription. While increasing Runx2 expression elevates transcription of Pol III genes (A and C). The fold changes are calculated by normalizing to the amount of GAPDH mRNA. The bars represent Mean ± SE of at least three independent determinations. *: p<0.05; **: p < 0.01.
Fig. 4. Alteration of cellular level of Runx2 affects Brf1 expression.
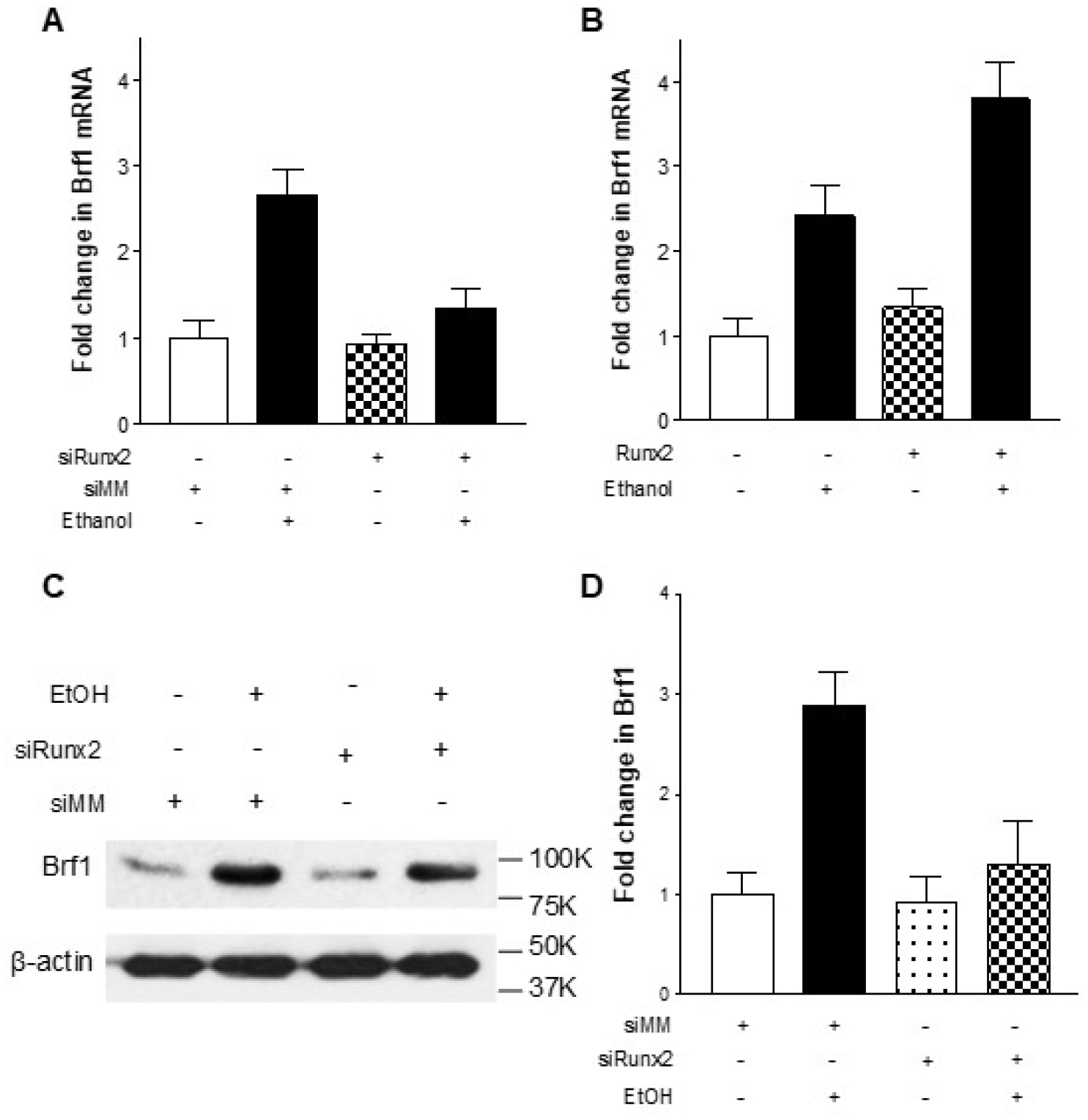
(A) MCF-7 cells were transfected with mismatch (mm) RNA, Runx2 siRNA for 48 h, and then treated with ethanol as described above. (B) MCF-7 cells with Runx2 expression construct were treated with ethanol as described above. The amounts of Brf1 mRNA (A and B) were measured by RT-qPCR. The fold changes are calculated by normalizing to the amount of GAPDH mRNA. The bars represent Mean ± SE of at least three independent determinations. *: p<0.05; **: p<0.01. (C) MCF-7 cells were transfected with siMM or Runx2 siRNA, respectively. The change in Brf1 protein levels were determined by Western blot. A representative Western blot is shown in (C). (D) Quantitation of the Western blot results of (C).
3.7. Runx2 and Brf1 synergistically regulate Pol III gene transcription to increase the rate of colony formation
Above studies have shown that Runx2 expression is increased in the cases of breast cancer (Fig. 1) and change in Runx2 level affects Pol III gene transcription (Fig.3). As Brf1 is overexpressed in tumor foci of ER+ breast cancer cases (Fig.5A1, A3), compared to para-cancerous tissues (Fig.5A2, A4) (33). Thus, we further investigate whether there is synergistic role between Runx2 and Brf1 in the regulation of Pol III genes. We have performed immunofluorescence colocalization analysis of human breast cancer samples. The results indicate that both Brf1 (Fig.5B1) and Runx2 (Fig.5B2) localize in nucleus. The signals of Brf1 and Runx2 display clear colocalization in part of cells (Fig.5B4). Next, we have carried out chromatin immunoprecipitation with Brf1 and Runx2 antibodies and primers of Pol III genes (Table S3), respectively, to put down their binding chromatin. These results show that both Brf1 and Runx2 occupy the promoters of tRNALeu (Fig. 6A and 6B left panel) and 5S rRNA genes (Fig. 6A and 6B right panel). However, either repressing Brf1 or Runx2 reduces the occupancy of the promoters (Fig. 6A to 6D).
Fig. 5. Colocalization of Brf1 and Runx2 in human biopsy of breast cancer.

(A) Brf1 staining. IHC staining of Brf1 of tumor foci (A1 and A3) and para-carcinoma tissues (A2 and A4) H&E staining of breast cancer (A1 & A2) × 100 magnification; (A3 & A4) × 1000 magnification. A representative Brf1 staining of human breast cancer samples. (B) Colocalization: Brf1 (red) and Runx2 (green) of the human breast adenocarcinoma tumor tissues were determined by immunofluorescence staining. The white arrows indicate that both Brf1 and Runx2 are localized in nucleus of the cancer cells. Merging picture clearly shows that Brf1 and Runx2 reveal co-localization in nucleus of human breast cancer biopsy (B4).
Fig. 6. Runx2 occupy the promoters of Pol III genes to modulates their transcription.

(A-D): MCF-7 cells were transfected with siMM, Brf1 siRNA (A and B) or Runx2 siRNA (C and D) for 48 h, respectively. The cells were treated as described above. The chromatin was extracted from these cells and ChIP assays were performed with Brf1 or Runx2 antibodies, respectively to determine the occupancy of Brf1 or Runx2 at the promoters of pre-tRNALeu (A and C) and 5S rRNA (B and D) by RT-qPCR. These studies indicate that both Brf1 and Runx2 occupy the promoters to synergistically modulate transcription of tRNALeu (A and C) and 5S rRNA (B and D) and alcohol enhance the occupancy of Brf1 and Runx2. (E) Colony formation: MCF-7 cells were transfected with siMM or Runx2 siRNA for 48 h, respectively. The cells were seeded in 6x well plates and treated with ethanol (25 mM) previously described (26). The cells were analyzed for colony formation in soft agar. Colonies were counted at 2–3 weeks after plating. The bars represent Mean ± SE of at least three independent determinations. *: p<0.05; **: p<0.01.
As increasing Brf1 expression results in enhancement of Pol III gene transcription and is sufficient for cell transformation (19–20, 23). Induction of Brf1 expression allowed anchorage-independent colonies to form and promoted tumor formation in mouse (23). We have demonstrated that repressing ERα and/or Brf1 reduce the rates of cell transformation (19–20). To further assess potential of Runx2 in cellular phenotype, we have investigated whether Runx2 affects alcohol-induced colony formation. MCF-7 cells were transfected with siMM or siRunx2 and treated with alcohol. The results indicate that alcohol increases the rate of colony formation of MCF-7 cells transfected with siMM as a control (Fig. 6E), while repressing Runx2 expression by its siRNA dramatically decreases the rate of colony formation, compared to the cells transfected with siMM (Fig. 6E). Given that inhibiting ERα expression decreased cellular levels of Brf1 protein and mRNA (19), as well as Pol III gene transcription (19,33), reduction of Runx2 expression by its siRNA significantly decreased alcohol-induced Brf1 expression and Pol III gene transcription (Fig. 3,4), eventually resulting in reduction of colony formation of MCF-7 cells (Fig. 6E). These results demonstrate that alcohol increases Runx2 activity, which modulates Brf1 expression to enhance tRNA and 5S rRNA transcription, thereby promoting alcohol-induced colony formation of MCF-7 cells.
4. Discussion
In the present study, we have investigated the expression of Runx2 in 219 cases of breast cancer and its role in the deregulation of Brf1 and Pol III genes. We have performed the mechanism analysis characterizing how Runx2 mediates the endogenous Pol III gene, tRNA and 5S rRNA, transcription. The results indicate that Runx2 is overexpressed in the cases of breast cancer. High expression of Runx2 displays longer survival time of the cases in stage I and II, while high Runx2 expression in advanced stages (III and IV) reveals faster recurrence. Alcohol increases Runx2 expression through JNK1, c-Jun and ERα pathway (Fig. S1). Repression of Runx2 decreases the induction of Brf1 and Pol III genes caused by alcohol. Runx2 and Brf1 are colocalized in nucleus of human breast cancer cells, they synergistically modulate Pol III gene transcription. As the deregulation of Brf1 and Pol III genes is tightly linked to cell transformation and tumor formation (19–20,23–29), these findings here support that idea that the abnormal expression of Runx2 and its modulation of alcohol-induced deregulation of Brf1 and Pol III genes may play a key role in breast cancer development and metastasis, particularly in advanced cases of this disease.
Runx2 is a member of Runx family and important for bone development with tissue-specific functionality (40–42). Study indicates that Runx2 is associated with mammary gland development and ER+ breast cancer (16). Runx2 involves breast cancer metastasis by regulating the genes of metastasis and invasion, such as MMPs (43). Although there are reports on Runx2 associated with breast cancer and other carcinomas (44–47), the significance of Runx2 expression and its roles in human cases of breast cancer remain to be addressed. Here, we report that Runx2 is overexpressed in the human sample of breast cancer (Fig. 1). High expression of Runx2 at early stage betokens longer survival and fast recurrence of this disease at advanced stage (Fig. 1). These studies indicate that alteration of Runx2 level is able to be as a novel marker of breast cancer and its high expression in breast cancer cases implies worse prognosis.
Emerging studies indicate that alcohol consumption is consistently associated with the risk of breast cancer (4–6). Alcohol has been classed as a carcinogen to humans by IARC (12–14). However, the mechanism of alcohol-associated cancers still remains to be established. Alcohol is a good reagent to explore the underlying mechanism of breast cancer (19,33). Studies indicate that alcohol feeding promotes mammary tumor formation through MCP-1 and CRR2 (9–10). Our studies have demonstrated that alcohol activates JNK1 pathway and enhances c-Jun expression to increase Pol III gene transcription, resulting in the development of hepatocellular carcinoma of mice (20). Alcohol dramatically increases Pol III gene transcription in ER+ breast cancer cell lines, but not in ER− normal breast cell lines and ER− breast cancer cell lines (19,48–49). Further study shows that alcohol activates JNK1 to enhance ERα activity and increases Brf1 expression and Pol III gene transcription to facilitate cell transformation and tumor formation (19,33). These studies demonstrate that alcohol is indeed associated with tumor development. However, nothing is known about the role of Runx2 in alcohol-induced deregulation of Brf1 and Pol III genes. Here, we report that alcohol induces Runx2 transcription through JNK1, c-Jun and ERα, resulting in elevation of Brf1 expression and Pol III gene transcription. These novel findings further enhance our understanding in the mechanism of alcohol-associated breast cancer.
Our early studies have demonstrated that increases in TFIIIB subunit (Brf1, Bdp1 and TBP) expression to enhance Pol III gene transcription (19,25). Bdp1 regulation, but not Brf1, occurred through JNK1-mediated alteration in TBP expression (36), suggesting that Bdp1 and Brf1 may be regulated independently. Our recent study demonstrates that alcohol induces Pol III gene transcription in vivo and in vitro (19–20). The induction of Pol III genes promoted liver tumor development in HCV NS5A transgenic mouse (20). Further analysis indicates that ERα modulates alcohol-induced deregulation of Pol III gene transcription (19). Interestingly, alcohol-enhanced ERα activity increases Brf1 expression, but not TBP in ER+ breast cancer cells (19). This indicates that ERα activity does not directly affect TBP expression (19). In contrast, change in cellular level of ERα caused an alteration of Brf1 expression and ERα directly occupies the Brf1 promoter to modulate its expression (19). This finding is consistent with studies of human breast cancer biopsies, in which Brf1 is overexpressed in ER+ breast cancer cases (33,50). Given that Runx2 is mediated by ERα, the inhibition of ERα decreases Runx2 transcription in ER+ breast cancer cells (Fig. 2G). As approximately 80% cases of breast cancer are ER+, our recent study has demonstrated that ER+ cases with high Brf1 expression display longer survival times after hormone therapy with Tam (Tamoxifen) (33,35). This is because Tam is able to repress Brf1 expression and Pol III gene transcription (35). More interestingly, most of the cases with high Runx2 expression at early stages, reveal longer survival period, while the cases mainly are at ER+ status (Data not shown). In other hand, the cases with high Runx2 expression at advanced stages show faster recurrence, which matches the function of Runx2 speeding the metastasis of breast cancer (22). In addition, high Runx2 at advanced stages may produce the feedback role in ERα express to further enhance Brf1 and tRNA transcription, results in metastasis (32). These results are consistent with the idea which ERα positively modulates Runx2 transcription (18). It suggests that Brf1 is a target which is modulated by ERα and Runx2. ERα-mediated Runx2 transcription may play an important role in cell transformation and alcohol-associated breast cancer.
Studies have reported that tRNAs are upregulated in human breast cancer cells as promoters of breast cancer metastasis and increased tRNAs facilitates cancer cell migration and invasion to enhance metastatic potential in cancer (30–31). At the present study, our findings indicate that Runx2 positively modulates Brf1 expression and Pol III gene transcription (Fig. 2–4), whereas high expression of Runx2 in advanced stages of breast cancer displays faster recurrence of the disease (Fig. 1). It suggests that Runx2-mediated deregulation of Pol III genes may be critical important in tumor metastasis of this disease.
5. Conclusions
The present study indicates that Runx2 is overexpressed in breast cancer. High expression of Runx2 displays longer survival period and faster recurrence. Alcohol increases cellular levels of Runx2, the inhibitions of JNK1, c-Jun and ERα decrease Runx2 transcription (Fig. S1). The alteration of Runx2 level affects Brf1 expression and Pol III gene transcription. Runx2 and Brf1 are colocalized in nucleus of tumor tissue of breast cancer and synergistically modulate Pol III gene transcription. Repression of Runx2 reduces the rate of colony formation. These studies, for the first time, provide the evidence that Runx2 mediates alcohol-induced deregulation of Brf1 and Pol III genes, which are tightly linked to tumor development and cancer metastasis. These novel findings also suggest the possibility that inhibition of Runx2 and Brf1 expression may be potential approaches for therapy of breast cancer. These outcomes from this study will benefit scientific community and the patients of breast cancer.
Supplementary Material
Highlight:
Runx2 was overexpressed in human cases of breast cancer. High Runx2 expression of breast cancer cases display worse prognosis;
Alcohol increased Runx2 expression to enhance Brf1 and Pol III gene transcription. Inhibiting Runx2 repressed Brf1 expression and Pol III gene transcription;
Rubx2 and Brf1 colocalize in nucleus of tumor tissue of breast cancer to synergistically modulate Pol III gene transcription;
Decreasing Runx2 expression caused phenotypic changes in breast cancer cells.
Acknowledgements
We would like to thank Dr. B. Frenkel (USC), who provide Plasmids of Runx2 expression and Runx2-luciferase reporter construct. This work was supported by National Institutes of Alcohol Abuse and Alcoholism grants AA023247 and AA024169 to S. Zhong and by NSFC grants 81370368 and 81672417 to W. Li.
Abbreviations
- Runx2
Runt-related transcription factor 2
- ER+
estrogen receptor positive
- ER−
estrogen receptor negative
- E2
17-β estradiol
- Pol III genes
RNA polymerase III-dependent genes
- Brf1
TFIIB-related factor 1
- TBP
TATA box-binding protein
- Tam
Tamoxifen
- IARC
the International Agency for Research on Cancer
- IHC
immunohistochemistry
- IDC
invasive ductal carcinoma
- ILC
invasive lobular carcinoma
- MBC
metastatic breast cancer
- DAB
3,3’-diaminobenzidine
- SI
staining index
- DAPI
4,6-diamidino-2-phenylindole
- siMM
mismatch RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. Supplementary material
Supplementary data are available at Chemico-Biological Interaction online.
Declaration: The authors declare no competing financial interests.
Conflict of Interest: All authors read and approved the final manuscript. The authors declare no conflict of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Reference
- [1].Singletary K, Nelshoppen J, Wallig M, Enhancement by chronic ethanol intake of N-methyl-N-nitrosourea-induced rat mammary tumorigenesis, Carcinogenesis, 16 (1995) 959–964 [DOI] [PubMed] [Google Scholar]
- [2].Watabiki T, Okii Y, Tokiyasu T, Yoshimura S, Yoshida M, Akane A, Shikata N, Tsubura A, Long-term ethanol consumption in ICR mice causes mammary tumor in females and liver fibrosis in males, Alcohol Clin Exp Res, 24 (2000) 117S–122S [PubMed] [Google Scholar]
- [3].Hamajima N, Hirose K, Tajima K, Rohan T, Calle EE, Heath CW Jr., Coates RJ, Liff JM, Talamini R, Chantarakul N, Koetsawang S, Rachawat D, Morabia A, Schuman L, Stewart W, Szklo M, Bain C, Schofield F, Siskind V, Band P, Coldman AJ, Gallagher RP, Hislop TG, Yang P, Kolonel LM, Nomura AM, Hu J, Johnson KC, Mao Y, De Sanjose S, Lee N, Marchbanks P, Ory HW, Peterson HB, Wilson HG, Wingo PA, Ebeling K, Kunde D, Nishan P, Hopper JL, Colditz G, Gajalanski V, Martin N, Pardthaisong T, Silpisornkosol S, Theetranont C, Boosiri B, Chutivongse S, Jimakorn P, Virutamasen P, Wongsrichanalai C, Ewertz M, Adami HO, Bergkvist L, Magnusson C, Persson I, Chang-Claude J, Paul C, Skegg DC, Spears GF, Boyle P, Evstifeeva T, Daling JR, Hutchinson WB, Malone K, Noonan EA, Stanford JL, Thomas DB, Weiss NS, White E, Andrieu N, Bremond A, Clavel F, Gairard B, Lansac J, Piana L, Renaud R, Izquierdo A, Viladiu P, Cuevas HR, Ontiveros P, Palet A, Salazar SB, Aristizabel N, Cuadros A, Tryggvadottir L, Tulinius H, Bachelot A, Le MG, Peto J, Franceschi S, Lubin F, Modan B, Ron E, Wax Y, Friedman GD, Hiatt RA, Levi F, Bishop T, Kosmelj K, Primic-Zakelj M, Ravnihar B, Stare J, Beeson WL, Fraser G, Bullbrook RD, Cuzick J, Duffy SW, Fentiman IS, Hayward JL, Wang DY, McMichael AJ, McPherson K, Hanson RL, Leske MC, Mahoney MC, Nasca PC, Varma AO, Weinstein AL, Moller TR, Olsson H, Ranstam J, Goldbohm RA, van den Brandt PA, Apelo RA, Baens J, de la Cruz JR, Javier B, Lacaya LB, Ngelangel CA, La Vecchia C, Negri E, Marubini E, Ferraroni M, Gerber M, Richardson S, Segala C, Gatei D, Kenya P, Kungu A, Mati JG, Brinton LA, Hoover R, Schairer C, Spirtas R, Lee HP, Rookus MA, van Leeuwen FE, Schoenberg JA, McCredie M, Gammon MD, Clarke EA, Jones L, Neil A, Vessey M, Yeates D, Appleby P, Banks E, Beral V, Bull D, Crossley B, Goodill A, Green J, Hermon C, Key T, Langston N, Lewis C, Reeves G, Collins R, Doll R, Peto R, Mabuchi K, Preston D, Hannaford P, Kay C, Rosero-Bixby L, Gao YT, Jin F, Yuan JM, Wei HY, Yun T, Zhiheng C, Berry G, Cooper Booth J, Jelihovsky T, MacLennan R, Shearman R, Wang QS, Baines CJ, Miller AB, Wall C, Lund E, Stalsberg H, Shu XO, Zheng W, Katsouyanni K, Trichopoulou A, Trichopoulos D, Dabancens A, Martinez L, Molina R, Salas O, Alexander FE, Anderson K, Folsom AR, Hulka BS, Bernstein L, Enger S, Haile RW, Paganini-Hill A, Pike MC, Ross RK, Ursin G, Yu MC, Longnecker MP, Newcomb P, Bergkvist L, Kalache A, Farley TM, Holck S, Meirik O, Collaborative C Group on Hormonal Factors in Breast, Alcohol, tobacco and breast cancer--collaborative reanalysis of individual data from 53 epidemiological studies, including 58,515 women with breast cancer and 95,067 women without the disease, Br J Cancer, 87 (2002) 1234–1245. 10.1038/sj.bjc.6600596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].MacMahon B, Epidemiology and the causes of breast cancer, Int J Cancer, 118 (2006) 2373–2378. 10.1002/ijc.21404 [DOI] [PubMed] [Google Scholar]
- [5].Petri AL, Tjonneland A, Gamborg M, Johansen D, Hoidrup S, Sorensen TI, Gronbaek M, Alcohol intake, type of beverage, and risk of breast cancer in pre- and postmenopausal women, Alcohol Clin Exp Res, 28 (2004) 1084–1090 [DOI] [PubMed] [Google Scholar]
- [6].Singletary KW, Gapstur SM, Alcohol and breast cancer: review of epidemiologic and experimental evidence and potential mechanisms, JAMA, 286 (2001) 2143–2151 [DOI] [PubMed] [Google Scholar]
- [7].Seitz HK, Pelucchi C, Bagnardi V, La Vecchia C, Epidemiology and pathophysiology of alcohol and breast cancer: Update 2012, Alcohol Alcohol, 47 (2012) 204–212. 10.1093/alcalc/ags011 [DOI] [PubMed] [Google Scholar]
- [8].Demark-Wahnefried W, Goodwin PJ, To your health: how does the latest research on alcohol and breast cancer inform clinical practice?, J Clin Oncol, 31 (2013) 1917–1919. 10.1200/JCO.2013.49.0466 [DOI] [PubMed] [Google Scholar]
- [9].Wang S, Xu M, Li F, Wang X, Bower KA, Frank JA, Lu Y, Chen G, Zhang Z, Ke Z, Shi X, Luo J, Ethanol promotes mammary tumor growth and angiogenesis: the involvement of chemoattractant factor MCP-1, Breast Cancer Res Treat, 133 (2012) 1037–1048. 10.1007/s10549-011-1902-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wong AW, Dunlap SM, Holcomb VB, Nunez NP, Alcohol promotes mammary tumor development via the estrogen pathway in estrogen receptor alpha-negative HER2/neu mice, Alcohol Clin Exp Res, 36 (2012) 577–587. 10.1111/j.1530-0277.2011.01654.x [DOI] [PubMed] [Google Scholar]
- [11].Polanco TA, Crismale-Gann C, Reuhl KR, Sarkar DK, Cohick WS, Fetal alcohol exposure increases mammary tumor susceptibility and alters tumor phenotype in rats, Alcohol Clin Exp Res, 34 (2010) 1879–1887. 10.1111/j.1530-0277.2010.01276.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cogliano VJ, Baan R, Straif K, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Wild CP, Preventable exposures associated with human cancers, J Natl Cancer Inst, 103 (2011) 1827–1839. 10.1093/jnci/djr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].I.W.G.o.t.E.o.C.R.t. Humans, IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol 100, A Review of Human Carcinogens, IARC Monogr Eval Carcinog Risks Hum, 100 (2011) v–vii, 1–412 [Google Scholar]
- [14].Shi G, Zhong S, Alcohol-associated cancer and deregulation of Pol III genes, Gene, 612 (2017) 25–28. 10.1016/j.gene.2016.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T, Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts, Cell, 89 (1997) 755–764 [DOI] [PubMed] [Google Scholar]
- [16].Ferrari N, McDonald L, Morris JS, Cameron ER, Blyth K, RUNX2 in mammary gland development and breast cancer, J Cell Physiol, 228 (2013) 1137–1142. 10.1002/jcp.24285 [DOI] [PubMed] [Google Scholar]
- [17].Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, Stein JL, Lian JB, Stein GS, Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo, Proc Natl Acad Sci U S A, 102 (2005) 1454–1459. 10.1073/pnas.0409121102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kammerer M, Gutzwiller S, Stauffer D, Delhon I, Seltenmeyer Y, Fournier B, Estrogen Receptor alpha (ERalpha) and Estrogen Related Receptor alpha (ERRalpha) are both transcriptional regulators of the Runx2-I isoform, Mol Cell Endocrinol, 369 (2013) 150–160. 10.1016/j.mce.2013.01.024 [DOI] [PubMed] [Google Scholar]
- [19].Zhang Q, Jin J, Zhong Q, Yu X, Levy D, Zhong S, ERalpha mediates alcohol-induced deregulation of Pol III genes in breast cancer cells, Carcinogenesis, 34 (2013) 28–37. 10.1093/carcin/bgs316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhong S, Machida K, Tsukamoto H, Johnson DL, Alcohol induces RNA polymerase III-dependent transcription through c-Jun by co-regulating TATA-binding protein (TBP) and Brf1 expression, J Biol Chem, 286 (2011) 2393–2401. 10.1074/jbc.M110.192955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang YF, Lin JJ, Lin CH, Su Y, Hung SC, c-Jun N-terminal kinase 1 negatively regulates osteoblastic differentiation induced by BMP2 via phosphorylation of Runx2 at Ser104, J Bone Miner Res, 27 (2012) 1093–1105. 10.1002/jbmr.1548 [DOI] [PubMed] [Google Scholar]
- [22].Reufsteck C, Lifshitz-Shovali R, Zepp M, Bauerle T, Kubler D, Golomb G, Berger MR, Silencing of skeletal metastasis-associated genes impairs migration of breast cancer cells and reduces osteolytic bone lesions, Clin Exp Metastasis, 29 (2012) 441–456. 10.1007/s10585-012-9462-8 [DOI] [PubMed] [Google Scholar]
- [23].Johnson SA, Dubeau L, Johnson DL, Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation, J Biol Chem, 283 (2008) 19184–19191. 10.1074/jbc.M802872200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhong S, Fromm J, Johnson DL, TBP is differentially regulated by c-Jun N-terminal kinase 1 (JNK1) and JNK2 through Elk-1, controlling c-Jun expression and cell proliferation, Mol Cell Biol, 27 (2007) 54–64. 10.1128/MCB.01365-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhong S, Zhang C, Johnson DL, Epidermal growth factor enhances cellular TATA binding protein levels and induces RNA polymerase I- and III-dependent gene activity, Mol Cell Biol, 24 (2004) 5119–5129. 10.1128/MCB.24.12.5119-5129.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Goodfellow SJ, Innes F, Derblay LE, MacLellan WR, Scott PH, White RJ, Regulation of RNA polymerase III transcription during hypertrophic growth, The EMBO journal, 25 (2006) 1522–1533. 10.1038/sj.emboj.7601040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].White RJ, RNA polymerase III transcription and cancer, Oncogene, 23 (2004) 3208–3216. 10.1038/sj.onc.1207547 [DOI] [PubMed] [Google Scholar]
- [28].Johnson DL, Johnson SA, Cell biology. RNA metabolism and oncogenesis, Science, 320 (2008) 461–462. 10.1126/science.1158680 [DOI] [PubMed] [Google Scholar]
- [29].Woiwode A, Johnson SA, Zhong S, Zhang C, Roeder RG, Teichmann M, Johnson DL, PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex, Mol Cell Biol, 28 (2008) 4204–4214. 10.1128/MCB.01912-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhong Q, Shi G, Zhang Y, Lu L, Levy D, Zhong S, Alteration of BRCA1 expression affects alcohol-induced transcription of RNA Pol III-dependent genes, Gene, 556 (2015) 74–79. 10.1016/j.gene.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF, Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression, Cell, 165 (2016) 1416–1427. 10.1016/j.cell.2016.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Birch J, Clarke CJ, Campbell AD, Campbell K, Mitchell L, Liko D, Kalna G, Strathdee D, Sansom OJ, Neilson M, Blyth K, Norman JC, The initiator methionine tRNA drives cell migration and invasion leading to increased metastatic potential in melanoma, Biol Open, 5 (2016) 1371–1379. 10.1242/bio.019075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fang Z, Yi Y, Shi G, Li S, Chen S, Lin Y, Li Z, He Z, Li W, Zhong S, Role of Brf1 interaction with ERalpha, and significance of its overexpression, in human breast cancer, Mol Oncol, 11 (2017) 1752–1767. 10.1002/1878-0261.12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhong Q, Xi S, Liang J, Shi G, Huang Y, Zhang Y, Levy D, Zhong S, The significance of Brf1 overexpression in human hepatocellular carcinoma, Oncotarget, 7 (2016) 6243–6254. 10.18632/oncotarget.6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhong Q, Shi G, Zhang Q, Lu L, Levy D, Zhong S, Tamoxifen represses alcohol-induced transcription of RNA polymerase III-dependent genes in breast cancer cells, Oncotarget, 5 (2014) 12410–12417. 10.18632/oncotarget.2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhong S, Johnson DL, The JNKs differentially regulate RNA polymerase III transcription by coordinately modulating the expression of all TFIIIB subunits, Proc Natl Acad Sci U S A, 106 (2009) 12682–12687. 10.1073/pnas.0904843106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yi Y, Huang C, Zhang Y, Tian S, Lei J, Chen S, Shi G, Wu Z, Xia N, Zhong S, Exploring a common mechanism of alcohol-induced deregulation of RNA Pol III genes in liver and breast cells, Gene, 626 (2017) 309–318. 10.1016/j.gene.2017.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chimge NO, Baniwal SK, Little GH, Chen YB, Kahn M, Tripathy D, Borok Z, Frenkel B, Regulation of breast cancer metastasis by Runx2 and estrogen signaling: the role of SNAI2, Breast Cancer Res, 13 (2011) R127 10.1186/bcr3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li W, Yu CP, Xia JT, Zhang L, Weng GX, Zheng HQ, Kong QL, Hu LJ, Zeng MS, Zeng YX, Li M, Li J, Song LB, Sphingosine kinase 1 is associated with gastric cancer progression and poor survival of patients, Clin Cancer Res, 15 (2009) 1393–1399. 10.1158/1078-0432.CCR-08-1158 [DOI] [PubMed] [Google Scholar]
- [40].Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ, Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development, Cell, 89 (1997) 765–771 [DOI] [PubMed] [Google Scholar]
- [41].Komori T, Regulation of skeletal development by the Runx family of transcription factors, J Cell Biochem, 95 (2005) 445–453. 10.1002/jcb.20420 [DOI] [PubMed] [Google Scholar]
- [42].Selvamurugan N, Shimizu E, Lee M, Liu T, Li H, Partridge NC, Identification and characterization of Runx2 phosphorylation sites involved in matrix metalloproteinase-13 promoter activation, FEBS Lett, 583 (2009) 1141–1146. 10.1016/j.febslet.2009.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS, Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone, Cancer Metastasis Rev, 25 (2006) 589–600. 10.1007/s10555-006-9032-0 [DOI] [PubMed] [Google Scholar]
- [44].Sancisi V, Borettini G, Maramotti S, Ragazzi M, Tamagnini I, Nicoli D, Piana S, Ciarrocchi A, Runx2 isoform I controls a panel of proinvasive genes driving aggressiveness of papillary thyroid carcinomas, J Clin Endocrinol Metab, 97 (2012) E2006–2015. 10.1210/jc.2012-1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Akech J, Wixted JJ, Bedard K, van der Deen M, Hussain S, Guise TA, van Wijnen AJ, Stein JL, Languino LR, Altieri DC, Pratap J, Keller E, Stein GS, Lian JB, Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions, Oncogene, 29 (2010) 811–821. 10.1038/onc.2009.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li H, Zhou RJ, Zhang GQ, Xu JP, Clinical significance of RUNX2 expression in patients with nonsmall cell lung cancer: a 5-year follow-up study, Tumour Biol, 34 (2013) 1807–1812. 10.1007/s13277-013-0720-4 [DOI] [PubMed] [Google Scholar]
- [47].Sase T, Suzuki T, Miura K, Shiiba K, Sato I, Nakamura Y, Takagi K, Onodera Y, Miki Y, Watanabe M, Ishida K, Ohnuma S, Sasaki H, Sato R, Karasawa H, Shibata C, Unno M, Sasaki I, Sasano H, Runt-related transcription factor 2 in human colon carcinoma: a potent prognostic factor associated with estrogen receptor, Int J Cancer, 131 (2012) 2284–2293. 10.1002/ijc.27525 [DOI] [PubMed] [Google Scholar]
- [48].Yi Y, Lei J, Shi G, Chen S, Zhang Y, Hong Z, The effects of liquor spirits on RNA Pol III genes and cell growth of human cancer lines., Food and Nutrition Sciences 9(2018) 208–220 [Google Scholar]
- [49].Chen S, Yi Y, Xia T, Hong Z, Zhang Y, Shi G, He Z, Zhong S, The influences of red wine in phenotypes of human cancer cells, Gene, (2018). 10.1016/j.gene.2018.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Julka PK, Chacko RT, Nag S, Parshad R, Nair A, Oh DS, Hu Z, Koppiker CB, Nair S, Dawar R, Dhindsa N, Miller ID, Ma D, Lin B, Awasthy B, Perou CM, A phase II study of sequential neoadjuvant gemcitabine plus doxorubicin followed by gemcitabine plus cisplatin in patients with operable breast cancer: prediction of response using molecular profiling, Br J Cancer, 98 (2008) 1327–1335. 10.1038/sj.bjc.6604322 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


