Abstract
Cardiomyopathy caused by A-type lamins gene (LMNA) mutations (LMNA cardiomyopathy) is associated with dysfunction of the heart, often leading to heart failure. LMNA cardiomyopathy is highly penetrant with bad prognosis with no specific therapy available. Searching for alternative ways to halt the progression of LMNA cardiomyopathy, we studied the role of calcium homeostasis in the evolution of this disease. We showed that sarcolipin, an inhibitor of the sarco/endoplasmic reticulum (SR) Ca2+ ATPase (SERCA) was abnormally elevated in the ventricular cardiomyocytes of mutated mice compared with wild type mice, leading to an alteration of calcium handling. This occurs early in the progression of the disease, when the left ventricular function was not altered. We further demonstrated that down regulation of sarcolipin using adeno-associated virus (AAV) 9-mediated RNA interference delays cardiac dysfunction in mouse model of LMNA cardiomyopathy. These results showed a novel role for sarcolipin on calcium homeostasis in heart and open perspectives for future therapeutic interventions to LMNA cardiomyopathy.
Keywords: Sarcolipin, Calcium handling, Nuclear envelope, LMNA, Dilated cardiomyopathy
Highlights
-
•
Sarcolipin, an inhibitor of the sarco/endoplasmic reticulum (SR) Ca2+ ATPase (SERCA) was abnormally elevated in the cardiac muscle of a mouse model of cardiomyopathy caused by LMNA mutations.
-
•
The elevation of sarcolipin expression leads to an alteration of calcium handling.
-
•
Down regulation of sarcolipin using adeno-associated virus (AAV) 9-mediated RNA interference delays cardiac dysfunction in mouse model of cardiomyopathy caused by LMNA mutations.
1. Introduction
Mutations in the gene encoding A-type nuclear lamins (LMNA) cause dilated cardiomyopathy associated with conduction defects (LMNA cardiomyopathy) [[1], [2], [3]]. Lamins are intermediate filament proteins that polymerized to form the nuclear lamina, a fibrous meshwork underlining the inner nuclear membrane of most eukaryotic cells [4,5]. Despite optimization of conventional therapeutic strategies, terminal heart failure and post transplantation complications represent by far the most frequent cause of death. There are currently no effective treatments for LMNA cardiomyopathy. Thus, it is necessary to clarify its pathological mechanisms and to search for potential compounds to protect against LMNA cardiomyopathy.
To study the pathophysiology of LMNA cardiomyopathy and to test possible therapeutics, murine models of the disease [6] have enabled insights into pathological mechanisms [[7], [8], [9], [10], [11]]. All these mechanisms have been targeted with small-molecules in mice and showed some beneficial impact. However, these approaches were not curative. Searching for alternative ways to slow down the cardiac disease progression, we here examined the involvement of calcium homeostasis in the LmnaH222P/H222P mouse model of the disease. Work from others converges to the conclusion that unhinged calcium (Ca2+) handling in the cardiomyocytes plays a central role in initiation and progression of dilated cardiomyopathy (12). The regulation of Ca2+ cycling is finely tuned by various proteins and regulatory processes, and is unhinged in cardiomyopathy. Therefore, the components of the Ca2+ cycling apparatus have been widely studied, aiming to restore the Ca2+ cycling process and ultimately improve the cardiac function. We here showed that sarcolipin (SLN), an inhibitor of the sarco/endoplasmic reticulum (SR) Ca2+ ATPase (SERCA), was abnormally elevated in LMNA cardiomyopathy, leading to an alteration of calcium cycling. These results establish a novel role for sarcolipin on calcium homeostasis in cardiac muscle and provide a rationale for future therapeutic interventions in LMNA cardiomyopathy.
2. Results
2.1. Activation of cardiac SLN expression in LmnaH222P/H222P mice
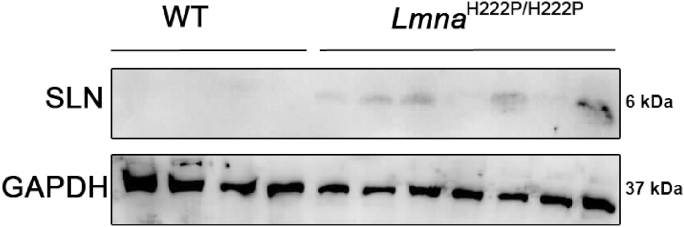
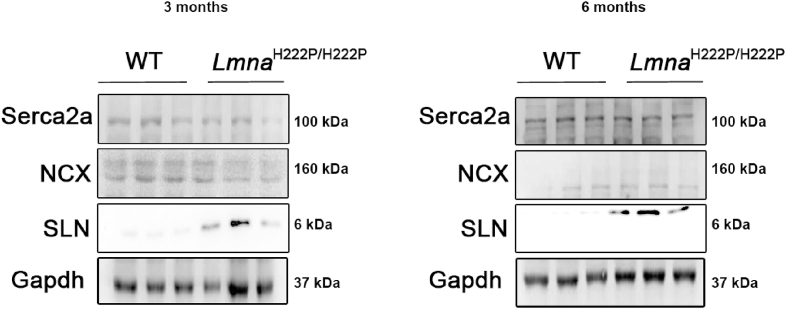
To explore the role of calcium signaling in the development of LMNA cardiomyopathy, we studied a mouse model that display a Lmna mutation, the LmnaH222P/H222P mouse model [13]. LmnaH222P/H222P mice develop a progressive left ventricular dysfunction that recapitulates human LMNA cardiomyopathy (Fig. S1) [13]. Lmna p.H222P mutation was found associated with dilated cardiomyopathy in human [14]. We previously studied expression of mRNAs isolated from hearts from young LmnaH222P/H222P mice, at 10 weeks of age [7]. This analysis highlighted activation of the expression of Sln gene, encoding sarcolipin (SLN), in LmnaH222P/H222P mice compared with wild type mice [7]. The up-regulation of SLN was also observed at 6 months of age in cardiac ventricles from LmnaH222P/H222P mice (Fig. 1), when the left ventricular function was altered. SLN activation was also observed at a younger age, when no left ventricular dysfunction was reported (Fig. 2).
Fig. 1.
Activation of SLN in LMNA cardiomyopathy
Representative immunoblot showing SLN expression in hearts from 6 months old male LmnaH222P/H222P and wild type (WT) mice. GAPDH was shown as loading control.
Fig. 2.
Expression of Serca2a and NCX in LMNA cardiomyopathy
Representative immunoblots showing SLN, Serca2a and NCX expression in ventricles from 3 and
6 months old male LmnaH222P/H222P and wild type (WT) mice. GAPDH was shown as loading control.
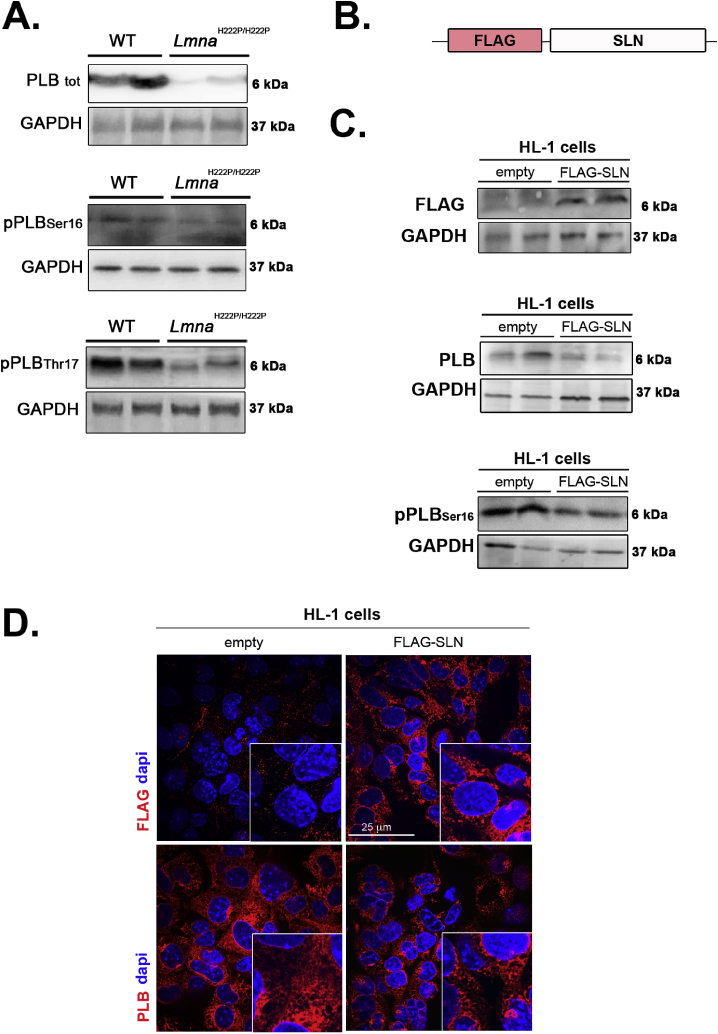
Given that several proteins tightly regulate defective [Ca2+]i cycling, we investigated whether the expression of calcium handling proteins was altered in LMNA cardiomyopathy. We found that expression of SERCA2a (Fig. 2, Fig. S2A) and NCX (Fig. 2, Fig. S2A) were not affected in hearts of 6-month old LmnaH222P/H222P mice. We also reported that expression of these proteins was not different from wild type mice at 3 months of age, when the left ventricular function was normal (Fig. 2). The expression of phospholamban (PLB), a regulator of SERCA2a, was decreased in the hearts from the LmnaH222P/H222P mice (Fig. 3A, Fig. S2B). This was ensuing an increased relative phosphorylation of PLB on residues Ser16 and Thr17 in the hearts from LmnaH222P/H222P mice (Fig. 3B, Fig. S2B). We then used a cellular model to assess the balance between SLN and PLB expression. We generated HL-1 cells stably expressing FLAG-tagged SLN (Fig. 3C). PLB expression was increased in these cells relative to HL-1 (Fig. 3D). These results demonstrated a correlation between enhanced SLN expression and decreased PLB expression in cardiac cells.
Fig. 3.
Expression of protein involved in calcium handling in LMNA cardiomyopathy.
(A) Representative immunoblots showing total PLB, phosphorylated PLB(Ser16) and phosphorylated PLB(Thr17) expressions in hearts from 6 months-old male LmnaH222P/H222P and wild type (WT) mice. GAPDH was shown as loading control.
(B) Schematic representation of the construct.
(C) Representative immunoblots showing total PLB and phosphorylated PLB(Ser16) expressions in HL-1 cells stably expressing FLAG epitope-tagged SLN (FLAG-SLN).
(D) Micrographs showing PLB labeling in HL-1 cells (empty) and HL-1 cells stably expressing FLAG epitope-tagged SLN (FLAG-SLN). Nuclei are counter-stained with 4′,6-diamidino-2-phenylindole (dapi).
2.2. Altered calcium homeostasis in LmnaH222P/H222P mice caused by SLN up-regulation
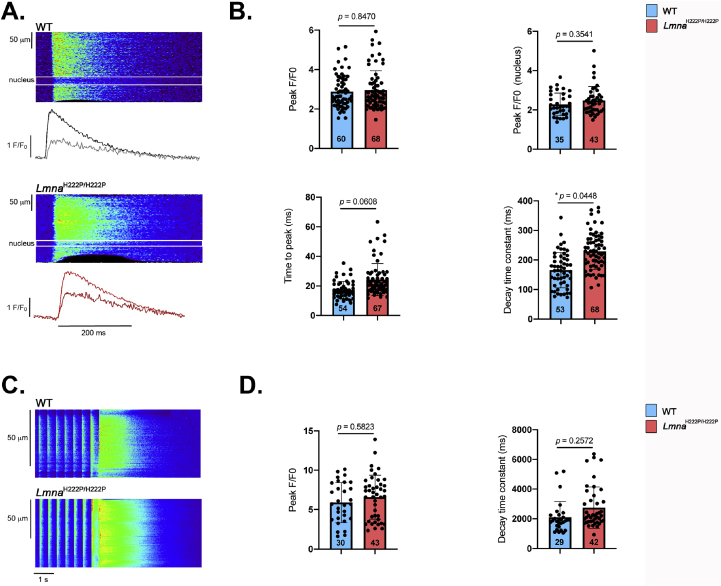
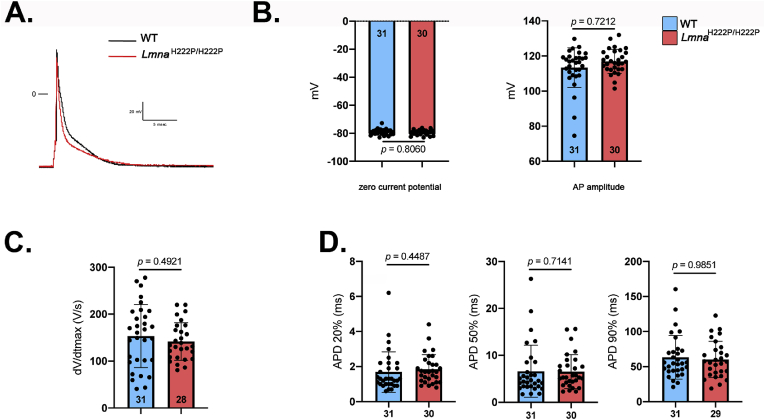
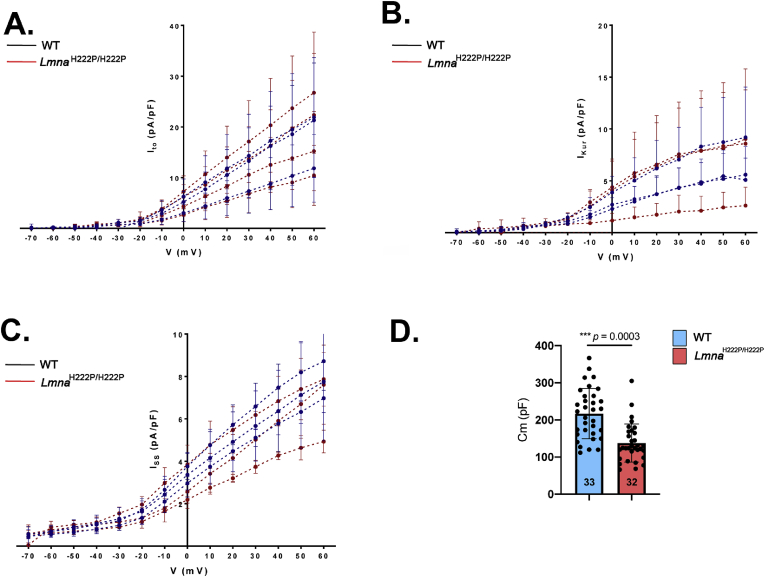
Given that SLN is a regulator of cardiac calcium handling through its inhibitory role on SERCA2a, we next hypothesized that over-expression of SLN in LmnaH222P/H222P mice could affect calcium homeostasis before detectable cardiac dysfunction. [Ca2+]i transients were elicited by electrical stimulation in ventricular cardiomyocytes from 3-month old LmnaH222P/H222P mice. Given that A-type lamins are important in nuclear pores organization [15], through which Ca2+ is diffused into the nucleus, and that nuclear Ca2+ is involved in gene transcription activation, we therefore set-up the scanning line through the nucleus to simultaneously record cytosolic [Ca2+]i and nuclear [Ca2+]n transients (Fig. 4A). The peak [Ca2+]i transient (F/F0) was not significantly different between ventricular cardiomyocytes from LmnaH222P/H222P mice and wild type mice (Fig. 4B), which is consistent with the normal heart contractile function reported in LmnaH222P/H222P mice at early stage (3 months) (Fig. S1). We showed that kinetics of the [Ca2+]i transients were slowed in ventricular cardiomyocytes from LmnaH222P/H222P mice compared with cardiomyocytes from wild type mice. The [Ca2+]i transient decay time was significantly prolonged in myocytes from LmnaH222P/H222P mice compared with those from wild type mice, suggesting slower Ca2+ uptake by the SERCA2a pump. However, the nuclear [Ca2+]n transient was not significantly different between the two types of cardiomyocytes (Fig. 4B). This suggests that sarcoplasmic reticulum (SR) calcium uptake was slowed in LmnaH222P/H222P mice, although the amplitude of the [Ca2+]i transient was maintained at this age. Because [Ca2+]i transients are triggered by the action potentials (AP), we next investigated whether AP characteristics were altered in cardiomyocytes from LmnaH222P/H222P mice. We showed that the zero current potential (Fig. 5A and B), amplitude (Fig. 5B), maximum dV/dt (Fig. 5C) and duration (Fig. 5D) were not significantly different between cardiomyocytes from LmnaH222P/H222P mice and wild type mice. The densities of outward potassium currents (transient outward Ito, ultrarapid IKur and sustained Iss) involved in AP repolarization phase were also similar between cardiomyocytes from LmnaH222P/H222P mice and wild type mice (Fig. 6A, B, 6C) despite the fact that the total membrane surface, measured as membrane capacitance, was smaller in cardiomyocytes isolated from LmnaH222P/H222P mice (Fig. 6D). Altogether, these data suggest that the alterations in [Ca2+]i handling were independent on an alteration of the electrophysiological characteristics of the cardiomyocyte from LmnaH222P/H222P mice.
Fig. 4.
Alteration of calcium cycling in LMNA cardiomyopathy.
(A) Line-scan images during field stimulation at 2 Hz of representative isolated cardiomyocytes from 3 months-old wild type (WT) and LmnaH222P/H222P mice. Below each image, there are represented the fluorescence traces in black for the cytosolic [Ca2+]i transient, and in gray for the ca transient inside the nucleus in the WT cell. The same for the LmnaH222P/H222P in red (cytosolic) and in dark red (nuclear).
(B) Graph showing values of peak calcium transients (F/F0), peak nuclear calcium transients (F/F0), time to peak and decay time constant of steady-state Ca2+ transients from isolated cardiomyocytes from 3 months-old wild type (WT) and LmnaH222P/H222P mice. The number of cardiomyocytes is indicated. (t-test).
(C) Line-scan images of caffeine-evoked intracellular Ca2+ transients after field stimulation at 2 Hz of representative isolated cardiomyocytes from 3 months-old wild type (WT) and LmnaH222P/H222P mice obtained by the application of 10 mmol/L caffeine.
(D) Graph showing values of maximum amplitude of caffeine-induced SR Ca2+ release, indicative of SR Ca2+ load (F/F0) and decay time constant of caffeine-induced SR Ca2+ release, indicative of cytosolic Ca2+ extrusion Ca2+ from isolated cardiomyocytes from 3 months-old wild type (WT) and LmnaH222P/H222P mice. The number of cardiomyocytes is indicated. (t-test).
Fig. 5.
Electrophysiological characteristics in cardiomyocytes isolated from LmnaH222P/H222Pmice
(A) Examples of action potentials (AP) recorded in wild type (WT) (black trace) and LmnaH222P/H222P (red trace) cells. Membrane resting potential measured as the Zero current potential (left) and AP amplitudes (right) (B), maximum AP upstroke velocity (measured as dV/dt) (C), and AP duration (APD) at 20, 50, and 90% of repolarization. (D). The number of cardiomyocytes is indicated. Data are represented as the mean ± standard deviation of individuals. (t-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
Potassium currents density in cardiomyocytes isolated from LmnaH222P/H222Pmice.
(A) Transient outward potassium current (Ito) recorded in 27 cells from wild type (WT) (black circles) and 22 LmnaH222P/H222P cells (red circles) from 3 mice for each genotype. Current amplitude (pA) was normalized by the membrane capacitance (pF), which is an indirect measurement of cell surface.
(B) Transient ultra-rapid potassium current (IKur) recorded in 18 wild type (WT) and 15 LmnaH222P/H222P cells from 3 mice for each genotype. Current amplitude (pA) was normalized by the membrane capacitance (pF), which is an indirect measurement of cell surface.
(C) Transient sustained potassium current (Iss) recorded in 20 wild type (WT) and 15 LmnaH222P/H222P cells from 3 mice for each genotype. Current amplitude (pA) was normalized by the membrane capacitance (pF), which is an indirect measurement of cell surface.
(D) Membrane capacitance. The number of cardiomyocytes is indicated. (t-test).
The amount of Ca2+ released in each twitch depends on the amount of Ca2+ stored in the sarcoplasmic reticulum (SR), which can be estimated by rapid caffeine application (Fig. 4C). Fig. 4D showed no significant difference in SR Ca2+ load (Peak [Ca2+]i transient (F/F0)), which is consistent with the normal [Ca2+]i transient amplitude. Moreover, the decay time constant during caffeine perfusion between cardiomyocytes isolated from LmnaH222P/H222P mice and wild type mice was also similar, showing no changes in Ca2+ extrusion though the NCX in myocytes from the mutated mice, and thus indicating that the slowing on the decay phase of the electrically evoked [Ca2+]i transient (Fig. 4B) is due to SERCA slowing.
2.3. SLN up-regulation induces cardiac dysfunction
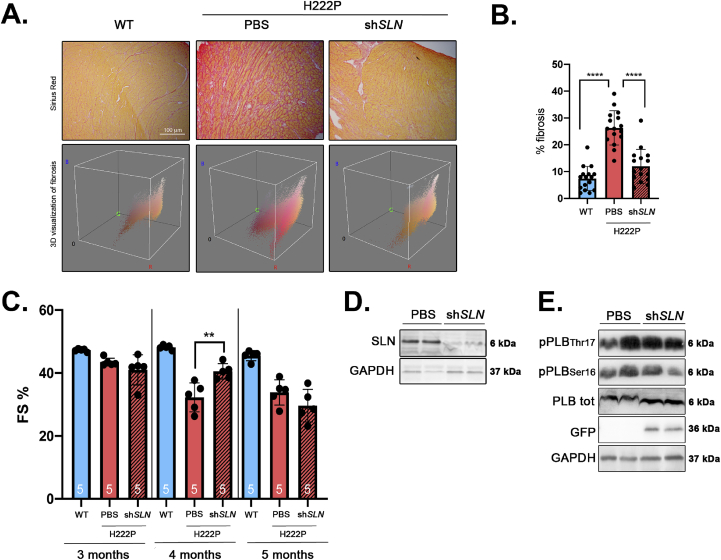
We next tested the hypothesis that SLN overexpression influences left ventricular function in vivo. We injected adeno-associated virus (AAV) vector expressing GFP-tagged shRNA against SLN (shSLN) in 1 month-old LmnaH222P/H222P mice. Following four months of treatment, the mice were analyzed by echocardiography and sacrificed for biochemical and histological studies. There was less alteration of myocardial fibrosis in hearts from LmnaH222P/H222P mice infected with GFP-tagged shSLN relative to mice injected with PBS (Fig. 7A and B). Compared with non-transduced LmnaH222P/H222P mice, for which the fractional shortening decreased overtime, fractional shortening in shSLN-transduced LmnaH222P/H222P mice did not change at 4 months of age (Fig. 7C, Table 1). This effect on the left ventricular function was transient as shSLN-transduced LmnaH222P/H222P mice showed an altered cardiac function no significantly different to non-transduced LmnaH222P/H222P mice at 5 months of age (Fig. 7C, Table 1). The inhibition of SLN expression in LmnaH222P/H222P mice (Fig. 7D) was followed by an increased cardiac expression of PLB (Fig. 7E, Fig. S3). This was followed by a decreased phosphorylation of PLB on residues Ser16 and Thr17 reported to total PLB expression in the hearts from shSLN-transduced LmnaH222P/H222P mice compared with non-transduced mice. (Fig. 7E, Fig. S3). These results showed that partially inhibiting SLN in vivo is efficient to delay the left-ventricular function in a mouse model of LMNA cardiomyopathy.
Fig. 7.
Inhibition of SLN expression delays cardiac dysfunction in a mouse model of LMNA cardiomyopathy.
(A) Representative Sirius Red staining and 3D visualization of fibrosis (Fiji software) of cross sections of hearts from PBS-treated LmnaH222P/H222P and shSLN-treated LmnaH222P/H222P mice. Cross sections of hearts from wild type (WT) mice were shown as controls.
(B) Graph showing fibrosis quantification from PBS-treated LmnaH222P/H222P and shSLN-treated LmnaH222P/H222P mice.
(C) Fractional shortening evolution from 3 months to 5 months from PBS-treated LmnaH222P/H222P (n = 5) and shSLN-treated LmnaH222P/H222P mice (n = 5). Data are represented as the mean ± standard deviation of individuals. **P ≤ 0.01 between PBS-injected LmnaH222P/H222P mice and wild type mice at each time point (ANOVA).
(D) Representative immunoblot SLN expression in heart from PBS-treated LmnaH222P/H222P and shSLN-treated LmnaH222P/H222P mice. GAPDH was shown as loading control.
(E) Representative immunoblot showing GFP, total PLB, phosphorylated PLB(Ser16) and phosphorylated PLB(Thr17)expressions in heart from PBS-treated LmnaH222P/H222P and shSLN-treated LmnaH222P/H222P mice. GAPDH was shown as loading control.
Table 1.
Echocardiographic parameters for male LmnaH222P/H222P mice at 3, 4 and 5 months of age after AAV9-shSLN injection.
| Genotype | Treatment | n | Heart rate (beats/min) | LVEDD (mm) | LVESD (mm) | FS (%) |
|---|---|---|---|---|---|---|
| 3 months | ||||||
| Lmna WT | none | 5 | 317.2 ± 24.3 | 3.6 ± 0.1 | 1.9 ± 0.1 | 47.3 ± 0.4 |
| Lmna H222P | PBS | 5 | 346.2 ± 36.3 | 3.6 ± 0.1 | 2.0 ± 0.1 # | 43.5 ± 1.3 ## |
| Lmna H222P | shSLN | 5 | 286.0 ± 31.8 | 4.0 ± 0.3* | 2.3 ± 0.3 | 41.0 ± 4.8 |
|
Genotype |
Treatment |
n |
Heart rate (beats/min) |
LVEDD (mm) |
LVESD (mm) |
FS (%) |
| 4 months | ||||||
| Lmna WT | none | 5 | 367.0 ± 26.2 | 3.4 ± 0.2 | 1.8 ± 0.1 | 48.2 ± 0.7 |
| Lmna H222P | PBS | 5 | 326.4 ± 34.3 # | 3.8 ± 0.2### | 2.6 ± 0.2 ### | 32.3 ± 4.6 ## |
| Lmna H222P | shSLN | 5 | 331.4 ± 9.9 | 3.8 ± 0.1 | 2.2 ± 0.1 * | 40.5 ± 2.5 ** |
|
Genotype |
Treatment |
n |
Heart rate (beats/min) |
LVEDD (mm) |
LVESD (mm) |
FS (%) |
| 5 months | ||||||
| Lmna WT | none | 5 | 352.0 ± 32.8 | 3.7 ± 0.1 | 2.0 ± 0.1 | 45.5 ± 1.6 |
| Lmna H222P | PBS | 5 | 348.2 ± 25.2 | 3.9 ± 0.2 | 2.6 ± 0.2 ## | 33.9 ± 4.0 ## |
| Lmna H222P | shSLN | 5 | 313.8 ± 45.8 | 4.2 ± 0.5 | 3.0 ± 0.5 | 29.6 ± 5.2 |
Mean ± standard deviation are presented for each parameter, timepoint, genotype and treatment.
#P < 0.05, ##P < 0.01, ###P < 0.001 between PBS-injected LmnaH222P/H222P mice and wild type mice.
*P < 0.05, **P < 0.01 between PBS and shSLN-injected LmnaH222P/H222P mice. (ANOVA).
3. Discussion
We showed in this study that expression of cardiac SLN, an inhibitor of the sarco/endoplasmic reticulum (SR) Ca2+ ATPase (SERCA), was up regulated in ventricles from LmnaH222P/H222P mice and [Ca2+]i transients were slowed down early before the development of cardiomyopathy symptoms. We therefore hypothesized that chronic SERCA2a inhibition contributes to cardiac muscle pathogenesis in LMNA cardiomyopathy. We tested our hypothesis using shRNA approach in LmnaH222P/H222P mice, and found that reduction in SLN expression was sufficient to delay left ventricular dysfunction. These results suggest a novel mechanism in the regulation of pathophysiology in LMNA cardiomyopathy.
We have reported for the first time an alteration in the Ca2+ handling in cardiomyocytes from LmnaH222P/H222P mice. The prolongation of the [Ca2+]i transient duration can be explained either by impairment in Ca2+ recapture by the SR through SERCA2a or by a decrease in Ca2+ extrusion though NCX1 [16,17]. Our data suggest that only the recapture by the SR mechanisms was altered in cardiomyocytes from LmnaH222P/H222P mice. A decreased SERCA2a function is commonly observed in models of heart failure, with both reduced and preserved ejection fraction (HFpEF) [18,19]. While in overt heart failure SR Ca2+ content and [Ca2+]i transient amplitudes are depressed, accounting for reduced contractile function, in HFpEF those parameters are normal as in LmnaH222P/H222P mice. The preservation in SR Ca2+ load in LmnaH222P/H222P mice suggests that SERCA2a can keep pumping enough Ca2+ to maintain cell contraction, notwithstanding an alteration of its function. Thus the decreased Ca2+ re-uptake in LmnaH222P/H222P mice is still not enough to decrease SR Ca2+ at normal heart rate, so contractile function can be sustained for a while, but the left ventricular function will ultimately decline overtime (Fig. S1).
SERCA2a function in cardiac muscle is regulated by SR membrane proteins, PLB and SLN, both inhibiting SERCA function [20,21]. Here, we showed that the abnormal activation of SLN in cardiac cells and in the heart of a murine model of LMNA cardiomyopathy was followed by a decreased PLB expression. Our results are divergent from work from others [22], as we showed that SLN overexpression induced changes in the expression of PLB. Because a number of conditions may affect PLB expression including sample preparation, disease state, and model, these findings may require additional experimentation. Our findings suggest that the stoichiometry between PLB and SLN is important for the regulation SERCA2a in cardiac pathophysiology, according to work from others [23]. It has been previously described that overexpression of SLN is slowing calcium reuptake by the SR, demonstrating that SLN inhibits SERCA2a, which leads to decreased rates of calcium uptake and decreased contractility [21,24]. Nevertheless, our data show that the decrease in PLB expression was not sufficient to maintain the physiological Ca2+ re-uptake in cardiomyocytes from LmnaH222P/H222P mice, suggesting that the slowed SERCA2a function was related to the activation of SLN expression.
Ca2+ handling alteration may be involved in cardiac dysfunction that appears progressively in LmnaH222P/H222P mice. [Ca2+]i plays an important role in the progression of cardiac disease by activating the so-called excitation-transcription coupling [25]. This mechanism is dependent on calcineurin activation by Ca2+ in the cytosol and/or Ca2+/calmodulin-dependent protein kinase II (CaMKII) in the nucleus [26,27]. The impeded [Ca2+]i transient duration could activate calcineurin signalling, leading to LMNA cardiomyopathy. Supporting this hypothesis, it has been shown that SLN overexpression increases calcineurin activity [28] while SLN deletion tends to inhibit calcineurin activity [29,30].
Regulation of SERCA2a by targeting SLN expression in LMNA cardiomyopathy could be an interesting approach to normalize Ca2+ homeostasis. Consistent with our data, recent work in Duchenne muscular dystrophy (DMD), a severe form of muscular dystrophy with cardiomyopathy [31], reported a cardiac SLN overexpression [32]. In this study, Voit et al. described a decreased SERCA2a function activity in the heart of a DMD mouse model, the mdx mice [32]. Furthermore, these authors and others showed that the reduction in SLN expression in mdx mice was sufficient to improve the SERCA function in heart [32] and in skeletal muscle [33]. These data, together with the present study, suggest that SLN could be a target to improve cardiac function in inherited cardiomyopathies. However, the impact of reduction in SLN expression on SERCA2a function remains to be tested in LMNA cardiomyopathy. The partial effect on cardiac function by reduction in SLN expression could reflect the fact that other molecular and cellular defects reported in LMNA cardiomyopathy [[7], [8], [9], [10], [11],34,35] were not targeted by our approach and could still hamper the cardiac function. Therefore, our work encourages further approaches to mechanistically assess the role played by SLN on calcium handling.
Our findings suggest that drugs that could correct unhinged calcium handling would ameliorate LMNA cardiomyopathy. In this regard, we previously showed that the use of pyridazinone derivative Ca2+ sensitizing agent SCH00013 was beneficial on cardiac function in LmnaH222P/H222P mice, inducing increased life expectancy and decreased fibrosis [36]. We therefore brought further evidence that the role of calcium handling is of importance in the development of LMNA cardiomyopathy. Moreover, SLN overexpression has been detected in humans with different cardiac pathologies contributing to the contractile dysfunction [37,38]. Therefore, we could speculate that the regulation of SLN may not be specific to LMNA cardiomyopathy but occurs in several cardiomyopathies.
In conclusion, we showed for the first time that the calcium handling is disturbed in LMNA cardiomyopathy before overt signs of cardiac dysfunction, which can be, at least partially due to SLN overexpression. This work opens novel perspectives for further therapeutic interventions. How mutated A-type lamins regulate SLN expression remains an unanswered question. A-type lamins is known to have regulatory roles on gene transcription [[39], [40], [41]], through interaction with gene promoters [42]. LMNA mutants could therefore lead to alteration of A-type lamins–chromatin associations and transcriptional defects [43], which could have downstream effects on cardiac function [44]. Whether mutated A-type lamins alter SLN promoter is unclear and would requires further investigation.
4. Methods
4.1. Animals
All animal experiments were approved by the French Ministry of Health at the Center for Research in Myology for the Care and Use of Experimental Animals (approval number #5556). Experiments were performed according to guidelines from directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes. LmnaH222P/H222P mice were fed standard chow and housed in a disease-free barrier facility with 12h/12h light/dark cycles. Mice were sacrificed with cervical dislocation.
4.2. Isolation of mouse cardiomyocytes
Wild type and LmnaH222P/H222P mice were anesthetized with intraperitoneal injection of sodium pentobarbital (50 mg/kg). Ventricular cardiomyocytes were isolated as described earlier [45].
4.3. Confocal microscopy and electrophysiological measurements
Isolated ventricular cardiomyocytes were loaded with the fluorescence Ca2+ dye Fluo-3 AM and viewed with a Leica microscopy in the line scan mode as earlier described45. [Ca2+]i transients were elicited by field stimulation through two Pt electrodes at 2 Hz. Ca2+ sparks were recorded in quiescent myocytes and SR Ca2+ load estimated by rapid caffeine (10 mM) application right after electrical stimulation at 2 Hz. Images were analyzed by home-made routines in IDL (Harrys Aerospatiale). The patch-clamp method in the configuration of whole cell was used to record action potentials in the current clamp configuration and potassium currents in the voltage-clamp configuration. Protocols and solutions were previously described [45].
4.4. Protein extraction and western blot analysis
Total proteins were isolated from heart tissue or cultured cell in extraction buffer (Cell Signaling). The heart samples were extracted with Lysis-D tube using Fast-Prep (3 pulses of 45 s), the cells samples were sonicated (5 pulses of 10 s at 30% amplitude) to allow dissociation of protein from chromatin and solubilization. Sample protein content was determined by the BiCinchoninic Acid Assay protein assay (Thermo Fisher Scientific). Extracts were analyzed by SDS-PAGE using Tris-glycine gels (Life Technologies) and transferred onto nitrocellulose 0.2 μm membrane (novex, Life Technologies). Membranes were blocked 1 h with 5% bovine serum albumin (BSA) in phosphate buffer saline containing 1% Tween 20 (PBS-T). Subsequently to blocking, the membranes were incubated with primary antibodies overnight at 4 °C. After TBS-T washes membranes were incubated with anti-chicken, anti-mouse or anti-rabbit antibodies for 1h at room temperature and washed again with TBS-T. The signal was then revealed using Immobilon Western Chemiluminescent HorseRadish Peroxidase (HRP) Substrate (Millipore) on a G-Box system with GeneSnap software (Ozyme).
4.5. Cell culture and reagents
HL-1 (Merck Millipore) were maintained at 37 °C with 5% CO2. HL-1 cells were culture in Claycomb Medium (Sigma-Aldrich) supplemented with 10% FBS (Invitrogen), 1% penicillin/streptomycin (Invitrogen), norepinephrine 0.1 mM (Sigma-Aldrich) in 30 mM ascorbic acid (Sigma-Aldrich) and l-glutamine 2 mM with the required gelatin/fibronectin coating. HL-1-SLN cells were created using lentiviral vector EF1.GOI (Sarcolipin Tag Flag) (Vectalys). MOI: 100, cells were incubated with the lentiviral vector for 24h followed by a 2-day incubation with normal medium and a 3–5 days puromicin selection.
4.6. Antibodies
Primary antibodies used were: SLN for Western blot (Merck Millipore, #ABT13), SERCA2a (Abcam, #ab2861), PLB (Cell Signaling, #14562), phospho-PLB(Ser16) (Santa Cruz, SC-12963-R), phospho-PLB(Thr17) (Santa Cruz, SC-17024-R), FLAG (Sigma, F3165), GAPDH (Santa Cruz, sc-47724). Secondary antibodies for immunoblotting were HRP-conjugated donkey anti-chicken (Invitrogen), rabbit anti-mouse or goat anti-rabbit IgG (Jackson ImmunoResearch). Secondary antibodies for immunofluorescence were Alexa Fluor 488 conjugated goat anti-rabbit IgG, Alexa Fluor 568-conjugated goat anti-mouse IgG, Alexa Fluor 488-conjugated goat anti-chicken IgG (Life Technologies).
4.7. Immunofluorescence microscopy
HL-1 cells were grown in coated (gelatin/fibronectin) coverslips. Cells were fixed (10 min, 4% paraformaldehyde in PBS at room temperature), permeabilized (8 min, 0,5% Triton X-100 in PBS), blocked (1h, in PBS with 0,3% triton X-100 and 5% BSA) and incubated with primary antibodies (1h30 at RT, in PBS with 1% triton X-100 and 1% BSA). The section were then washed in PBS (3 times 5 min) and incubated with secondary antibodies (1 h at RT, in PBS with 1% triton X-100 and 1% BSA) and washed (3 times 5 min).
4.8. Histology
Hearts were fixed in 4% formaldehyde for 48h, embedded in paraffin, sectioned at 5 mm and stained with Sirius red and hematoxylin eosin. Representative stained sections were photographed using a Microphot SA (Nikon) light microscope attached to a Spot RT Slide camera (Diagnostic Instruments). Images were processed using Adobe Photoshop CS (Adobe Systems) [35].
4.9. Echocardiography
Mice were anesthetized with 0.75% isoflurane in O2 and placed in a heating pad. Transthoracic echocardiography was performed using a Vivid 7 Dimension/Vivid7 PRO ultrasound with an 11 MHz transducer applied to the chest wall [34,35].
4.10. Construction and injection of AAV enconding sh-SLN
AAV9-GFP-U6-mSLN-shRNA (VectorBiolabs) recombinant AAV viruses were produced through co-transfecting HEK293 cells with (i) the AAV plasmid encoding sh-SLN, (ii) the adenovirus helper plasmid and (iii) the AAV packaging plasmid encoding the rep2 and cap. Two days after transfections, cells were harvested, and AAV vectors were purified through CsCl-gradient ultra-centrifugation. Vector titers (GC/ml - genome copies/ml) were determined by real-time PCR. One month-old mice were injected with AAV9-GFP-U6-mSLN-shRNA into the retro-orbital vein (5 × 1013 viral genomes/kg in 100 μl).
4.11. Statistics
Graphics were performed using GraphPad Prism 8 software. For both in vivo pharmacology studies and each parameter, statistical analyses were performed to evaluate the pathology effect by comparing LmnaH222P/H222P mice versus wild type mice, then for the study from 3 to 5 months to evaluate the effect of treatment on LmnaH222P/H222P mice by comparing the AAV9-shSLN treatment group to the PBS treatment group. For each objective an analysis of variance (ANOVA) with interaction was performed on factors “Time” (repeated) and “Group” or “Treatment” depending on the case. The variances heterogeneity was taken into account if necessary. Appropriate post-hoc analysis was then performed. The significance level is taken to 5%, except for the interaction test for which the significance level is taken to 10%. For in vitro pharmacology studies and each parameter, statistical analyses were performed to evaluate the pathology effect by comparing LmnaH222P/H222P mice versus wild type mice. A student t-test for mRNA expression and Cm parameter, a mixed effect model for calcium cycling, action potentials and sparks characteristics were carried out. The variances heterogeneity was taken into account if necessary. A log-transformation was performed for parameters peak calcium transients (peak F/F0), peak nuclear calcium transients (peak F/F0 (nucleus)), time to peak, decay time constant of caffeine-induced SR Ca2+ release, APD20%, APD50% and APD90%, sparks frequency, sparks peak F/F0 and D50. A rank-transformation was performed for parameters zero current potential and amplitude. Due to convergence issue of the mixed effect model on the raw data for Cm parameter, the mean of Cm by animal was analyzed. The significance level was taken to 5%. All statistical analyses were performed using SAS 9.4 (SAS Institute Inc., Cary, NC, USA) on Windows 7 PC.
Funding
This work was supported by the Association Française contre les Myopathies, the Institut National de la Santé et de la Recherche Médicale, Sorbonne Université and ANR (ANR-13-BSV1-0023-01 to A-M.G. and ANR-15-CE14-005 to JPB). B.M.R. was funded by Sanofi through a CIFRE PhD grant.
Author contributions
Conceptualization, A.M.; Investigation, B.M.R., A.D-R., J-P.B., T.M., N.M., A-M.G. and A.M.; Sample preparation, B.M.R. and F.L.; Writing – Original Draft, A.M. and A-M.G.; Writing – Review & Editing, B.M.R., V.B., G.B., A-M.G. and A.M.; Funding Acquisition, A-M.G. and A.M.; Supervision, P.B., G.B., V.B. and A.M.
Declaration of competing interest
B.M.R., V.B. and P.B. are employed by Sanofi, a global biopharmaceutical company focused on human health.
A.D-R., J-P.B., F.L., T.M., N.M., G.B., A-M.G. and A.M. do not have any conflict of interest.
Acknowledgements
We thank Dorothée Tamarelle for help with statistical analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2020.100767.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bonne G., Di Barletta M.R., Varnous S. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 2.Fatkin D., MacRae C., Sasaki T. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 3.Kumar S., Baldinger S.H., Gandjbakhch E. Long-term arrhythmic and nonarrhythmic outcomes of lamin A/C mutation carriers. J. Am. Coll. Cardiol. 2016;68:2299–2307. doi: 10.1016/j.jacc.2016.08.058. [DOI] [PubMed] [Google Scholar]
- 4.Gerace L., Blum A., Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J. Cell Biol. 1978;79:546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aebi U., Cohn J., Buhle L., Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- 6.Chatzifrangkeskou M., Bonne G., Muchir A. Nuclear envelope and striated muscle diseases. Curr. Opin. Cell Biol. 2015;32:1–6. doi: 10.1016/j.ceb.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Muchir A., Pavlidis P., Decostre V., Herron A.J., Arimura T. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J. Clin. Invest. 2007;117:1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muchir A., Wu W., Choi J.C., Iwata S., Morrow J.P. Abnormal p38α mitogen-activated protein kinase signaling in dilated cardiomyopathy caused by lamin A/C gene mutation. Hum. Mol. Genet. 2012;21:4325–4333. doi: 10.1093/hmg/dds265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi J.C., Muchir A., Wu W., Iwata S., Homma S. Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci. Transl. Med. 2012;4:144ra102. doi: 10.1126/scitranslmed.3003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatzifrangkeskou M., Le Dour C., Wu W., Morrow J.P., Joseph L.C. ERK1/2 directly acts on CTGF/CCN2 expression to mediate myocardial fibrosis in cardiomyopathy caused by mutations in the lamin A/C gene. Hum. Mol. Genet. 2016;25:2220–2233. doi: 10.1093/hmg/ddw090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Dour C., Macquart C., Sera F., Homma S., Bonne G. Decreased WNT/β-catenin signalling contributes to the pathogenesis of dilated cardiomyopathy caused by mutations in the lamin A/C gene. Hum. Mol. Genet. 2017;26:333–343. doi: 10.1093/hmg/ddw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marks A.R. Calcium cycling proteins and heart failure: mechanisms and therapeutics. J. Clin. Invest. 2013;123:46–52. doi: 10.1172/JCI62834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arimura T., Helbling-Leclerc A., Massart C., Varnous S., Niel F. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet. 2005;14:155–169. doi: 10.1093/hmg/ddi017. [DOI] [PubMed] [Google Scholar]
- 14.Bonne G., Mercuri E., Muchir A., Urtizberea A., Bécane H.M. Clinical and molecular genetic spectrum of autosomal dominant Emery-Dreifuss muscular dystrophy due to mutations of the lamin A/C gene. Ann. Neurol. 2000;48:170–180. [PubMed] [Google Scholar]
- 15.Xie W., Chojnowski A., Boudier T., Lim J.S., Ahmed S. A-type lamins form distinct filamentous networks with differential nuclear pore complex associations. Curr. Biol. 2016;26:2651–2658. doi: 10.1016/j.cub.2016.07.049. [DOI] [PubMed] [Google Scholar]
- 16.Periasamy M., Kalyanasundaram A. SERCA pump isoforms: their role in calcium transport and disease. Muscle Nerve. 2007;35:430–442. doi: 10.1002/mus.20745. [DOI] [PubMed] [Google Scholar]
- 17.Periasamy M., Bhupathy P., Babu G.J. Regulation of sarcoplasmic reticulum Ca2+ ATPase pump expression and its relevance to cardiac muscle physiology and pathology. Cardiovasc. Res. 2008;77:265–273. doi: 10.1093/cvr/cvm056. [DOI] [PubMed] [Google Scholar]
- 18.Zima A.V., Bovo E., Mazurek S.R., Rochira J.A., Terentyev D. Ca handling during excitation-contraction coupling in heart failure. Pflügers Archiv. 2014;466:1129–1137. doi: 10.1007/s00424-014-1469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peana D., Domeier T.L. Cardiomyocyte Ca2+ homeostasis as a therapeutic target in heart failure with reduced and preserved ejection fraction. Curr. Opin. Pharmacol. 2017;33:17–26. doi: 10.1016/j.coph.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babu G.J., Bhupathy P., Carnes C.A., Billman G.E., Periasamy M. Differential expression of sarcolipin protein during muscle development and cardiac pathophysiology. J. Mol. Cell. Cardiol. 2007;43:215–222. doi: 10.1016/j.yjmcc.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babu G.J., Zheng Z., Natarajan P., Wheeler D., Janssen P.M. Overexpression of sarcolipin decreases myocyte contractility and calcium transient. Cardiovasc. Res. 2005;65:177–186. doi: 10.1016/j.cardiores.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Asahi M., Kurzydlowski K., Tada M., MacLennan D.H. Sarcolipin inhibits polymerization of phospholamban to induce superinhibition of sarco(endo)plasmic reticulum Ca2+-ATPases (SERCAs) J. Biol. Chem. 2002;277:26725–26728. doi: 10.1074/jbc.C200269200. [DOI] [PubMed] [Google Scholar]
- 23.Asahi M., Sugita Y., Kurzydlowski K., De Leon S., Tada M. Sarcolipin regulates sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) by binding to transmembrane helices alone or in association with phospholamban. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5040–5043. doi: 10.1073/pnas.0330962100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asahi M., Otsu K., Nayakama H., Hikoso S., Takeda T. Cardiac-specific overexpression of sarcolipin inhibits sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA2a) activity and impairs function in mice. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9199–9204. doi: 10.1073/pnas.0402596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson M.E. Connections count: excitation-contraction meets excitation-transcription coupling. Circ. Res. 2000;86:717–719. doi: 10.1161/01.res.86.7.717. [DOI] [PubMed] [Google Scholar]
- 26.Molkentin J.D., Lu J.R., Antos C.L., Markham B., Richardson J. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X., Zhang T., Bossuyt J., Li X., McKinsey T.A. Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J. Clin. Invest. 2006;116:675–682. doi: 10.1172/JCI27374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurya S.K., Bal N.C., Sopariwala D.H., Pant M., Rowland L.A. Sarcolipin is a key determinant of the basal metabolic rate, and its overexpression enhances energy expenditure and resistance against diet-induced obesity. J. Biol. Chem. 2015;290:10840–10849. doi: 10.1074/jbc.M115.636878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fajardo V.A., Rietze B.A., Chambers P.J., Bellissimo C., Bombardier E. Effects of sarcolipin deletion on skeletal muscle adaptive responses to functional overload and unload. Am. J. Physiol. Cell Physiol. 2017;313:C154–C161. doi: 10.1152/ajpcell.00291.2016. [DOI] [PubMed] [Google Scholar]
- 30.Fajardo V.A., Chambers P.J., Juracic E.S., Rietze B.A., Gamu D. Sarcolipin deletion in mdx mice impairs calcineurin signalling and worsens dystrophic pathology. Hum. Mol. Genet. 2018;27:4094–4102. doi: 10.1093/hmg/ddy302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towbin J.A., Hejtmancik J.F., Brink P., Gelb B., Zhu X.M. X-linked dilated cardiomyopathy. Molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation. 1993;87:1854–1865. doi: 10.1161/01.cir.87.6.1854. [DOI] [PubMed] [Google Scholar]
- 32.Voit A., Patel V., Pachon R., Shah V., Bakhutma M. Reducing sarcolipin expression mitigates Duchenne muscular dystrophy and associated cardiomyopathy in mice. Nat. Commun. 2017;8:1068. doi: 10.1038/s41467-017-01146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanihata J., Nagata T., Ito N., Saito T., Nakamura A. Truncated dystrophin ameliorates the dystrophic phenotype of mdx mice by reducing sarcolipin-mediated SERCA inhibition. Biochem. Biophys. Res. Commun. 2018;505:51–59. doi: 10.1016/j.bbrc.2018.09.039. [DOI] [PubMed] [Google Scholar]
- 34.Vignier N., Chatzifrangkeskou M., Morales Rodriguez B., Mericskay M., Mougenot N. Rescue of biosynthesis of nicotinamide adenine dinucleotide (NAD+) protects the heart in cardiomyopathy caused by lamin A/C gene mutation. Hum. Mol. Genet. 2018;27:3870–3880. doi: 10.1093/hmg/ddy278. [DOI] [PubMed] [Google Scholar]
- 35.Chatzifrangkeskou M., Yadin D., Marais T., Chardonnet S., Cohen-Tannoudji M. Cofilin-1 phosphorylation catalyzed by ERK1/2 alters cardiac actin dynamics in dilated cardiomyopathy caused by lamin A/C gene mutation. Hum. Mol. Genet. 2018;27:3060–3078. doi: 10.1093/hmg/ddy215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arimura T., Sato R., Machida N., Bando H., Zhan D.-Y. Improvement of left ventricular dysfunction and of survival prognosis of dilated cardiomyopathy by administration of calcium sensitizer SCH00013 in a mouse model. J. Am. Coll. Cardiol. 2010;55:1503–1505. doi: 10.1016/j.jacc.2009.10.065. [DOI] [PubMed] [Google Scholar]
- 37.Nef H.M., Möllmann H., Troidl C., Kostin S., Voss S. Abnormalities in intracellular Ca2+ regulation contribute to the pathomechanism of Tako-Tsubo cardiomyopathy. Eur. Heart J. 2009;30:2155–2164. doi: 10.1093/eurheartj/ehp240. [DOI] [PubMed] [Google Scholar]
- 38.Zheng J., Yancey D.M., Ahmed M.I., Wei C.-C., Powell P.C. Increased sarcolipin expression and adrenergic drive in humans with preserved left ventricular ejection fraction and chronic isolated mitral regurgitation. Circ. Heart Fail. 2014;7:194–202. doi: 10.1161/CIRCHEARTFAILURE.113.000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harr J.C., Luperchio T.R., Wong X., Cohen E., Wheelan S.J., Reddy K.L. Directed targeting of chromatin of the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol. 2015;208:33–52. doi: 10.1083/jcb.201405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddy K.L., Zullo J.M., Bertolino E., Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 41.Andres V., Gonzalez J.M. Role of A-type lamins in signaling, transcription, and chromatin organization. J. Cell Biol. 2009;187:945–957. doi: 10.1083/jcb.200904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lund E., Oldenburg A.R., Delbarre E., Freberg C.T., Duband-Goulet I. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res. 2013;23:1580–1589. doi: 10.1101/gr.159400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perovanovic J., Dell'Orso S., Gnochi V.F., Jaiswal J.K., Sartorelli V. Laminopatghies disrupt epigenomic developmental programs and cell fate. Sci. Transl. Med. 2016;8:335ra38. doi: 10.1126/scitranslmed.aad4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salvarani N., Crasto S., Miragoli M., Bertero A., Paulis M. The K219T-Lamin mutation induces conduction defects through epigenetic inhibition of SCN5A in human cardiac laminopathy. Nat. Commun. 2019;10:2267. doi: 10.1038/s41467-019-09929-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz-Hurtado G., Li L., Fernández-Velasco M., Rueda A., Lefebvre F. Reconciling depressed Ca2+ sparks occurrence with enhanced RyR2 activity in failing mice cardiomyocytes. J. Gen. Physiol. 2015;146:295–306. doi: 10.1085/jgp.201511366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.