Skeletal muscle regeneration and repair involve the recruitment and proliferation of resident satellite cells that exit the cell cycle during the process of myogenic differentiation to form myofibers. We demonstrate that the ubiquitin E3 ligase subunit FBXL2 is essential for skeletal myogenesis through its important effects on cell cycle progression and cell proliferative signaling. Further, we characterize a new mechanism whereby sustained stimulation by a major proinflammatory cytokine, TNF-α, regulates skeletal myogenesis by inhibiting the interaction of SP1 with the Fbxl2 core promoter in proliferating myoblasts. Our findings contribute to the understanding of skeletal muscle regeneration through the identification of Fbxl2 as both a critical regulator of myogenic proliferative processes and a susceptible gene target during inflammatory stimulation by TNF-α in skeletal muscle. Modulation of Fbxl2 activity may have relevance to disorders of muscle wasting associated with sustained proinflammatory signaling.
KEYWORDS: Fbxl2, SP1, TNF-α, myogenesis, proliferation, differentiation, cell cycle, myoblast, transcriptional regulation, ubiquitin
ABSTRACT
FBXL2 is an important ubiquitin E3 ligase component that modulates inflammatory signaling and cell cycle progression, but its molecular regulation is largely unknown. Here, we show that tumor necrosis factor alpha (TNF-α), a critical cytokine linked to the inflammatory response during skeletal muscle regeneration, suppressed Fbxl2 mRNA expression in C2C12 myoblasts and triggered significant alterations in cell cycle, metabolic, and protein translation processes. Gene silencing of Fbxl2 in skeletal myoblasts resulted in increased proliferative responses characterized by activation of mitogen-activated protein (MAP) kinases and nuclear factor kappa B and decreased myogenic differentiation, as reflected by reduced expression of myogenin and impaired myotube formation. TNF-α did not destabilize the Fbxl2 transcript (half-life [t1/2], ∼10 h) but inhibited SP1 transactivation of its core promoter, localized to bp −160 to +42 within the proximal 5′ flanking region of the Fbxl2 gene. Chromatin immunoprecipitation and gel shift studies indicated that SP1 interacted with the Fbxl2 promoter during cellular differentiation, an effect that was less pronounced during proliferation or after TNF-α exposure. TNF-α, via activation of JNK, mediated phosphorylation of SP1 that impaired its binding to the Fbxl2 promoter, resulting in reduced transcriptional activity. The results suggest that SP1 transcriptional activation of Fbxl2 is required for skeletal muscle differentiation, a process that is interrupted by a key proinflammatory myopathic cytokine.
IMPORTANCE Skeletal muscle regeneration and repair involve the recruitment and proliferation of resident satellite cells that exit the cell cycle during the process of myogenic differentiation to form myofibers. We demonstrate that the ubiquitin E3 ligase subunit FBXL2 is essential for skeletal myogenesis through its important effects on cell cycle progression and cell proliferative signaling. Further, we characterize a new mechanism whereby sustained stimulation by a major proinflammatory cytokine, TNF-α, regulates skeletal myogenesis by inhibiting the interaction of SP1 with the Fbxl2 core promoter in proliferating myoblasts. Our findings contribute to the understanding of skeletal muscle regeneration through the identification of Fbxl2 as both a critical regulator of myogenic proliferative processes and a susceptible gene target during inflammatory stimulation by TNF-α in skeletal muscle. Modulation of Fbxl2 activity may have relevance to disorders of muscle wasting associated with sustained proinflammatory signaling.
INTRODUCTION
Skeletal muscle development, regeneration, and repair require the expansion of resident satellite cells into a subpopulation of myoblasts that proliferate, migrate, and fuse to form myofibers during the process of myogenic differentiation (1). The stimulation of myogenic repair mechanisms by physiological proinflammatory cytokine secretion is an important phase during tissue repair (2, 3) and is regulated by a well-characterized set of myogenic regulatory factors (MRFs) (4–6). Tumor necrosis factor alpha (TNF-α), a central mediator of the inflammatory response in skeletal muscle, is released by injured and regenerating myofibers, as well as by infiltrating inflammatory cells, such as M1 macrophages and neutrophils (7). TNF-α stimulation of skeletal myoblasts results in specific biological effects characterized by biphasic activation of nuclear factor kappa B (NF-κB), which is dependent on the stage of skeletal myogenesis (8). Initially, TNF-α is both an important chemoattractant that recruits skeletal satellite cells to the site of injury (9, 10) and a mitogen for myoblast proliferation signaling (11). As NF-κB signaling is transiently required during the early phase of myoblast proliferation (12, 13), TNF-α is known to stimulate cell growth in skeletal muscle by promoting G1-to-S-phase transition through transcriptional upregulation of cyclin D1 (12, 14). Later, in the presence of sustained TNF-α-induced inflammatory stress, deleterious effects on skeletal myogenesis are characterized by intensified myoblast proliferation and inhibition of myogenic differentiation through downregulation of several essential MRFs (14–16). Sustained TNF-α stimulation thereby hinders cell cycle exit, which is a key event during the process of myogenic differentiation (6). Several of these proteins acting as checkpoints within the cell cycle progression apparatus are regulated at the level of protein stability (6).
The ubiquitin proteasome system controls cellular protein concentrations for pathways that regulate skeletal muscle mass, including degradation of sarcomeric proteins under atrophic conditions (17); degradation of MRFs, such as MyoD (18, 19); and modulation of proliferative signaling that is activated during muscle regeneration (20–22). TNF receptor-associated factor 6 (TRAF6) is a key adapter E3 ligase that is involved in MyD88-dependent and -independent signal transduction of the NF-κB pathway through the assembly of K63-linked polyubiquitin chains required for IκB kinase activation (21). TRAF6 can also directly effect K63-mediated ubiquitination of Akt and the mitogen-activated protein kinases (MAPKs) p38 and c-Jun N-terminal kinase (JNK), which are key regulators of cell survival, growth, and metabolism in muscle (23, 24). TRAF6 is activated by a number of proinflammatory cytokines, including TNF-α and interleukin 1β (IL-1β), corticosteroid treatment, and oxidative stress (25). As an important TNF-α signaling adaptor molecule, TRAF6 regulates NF-κB, p38 MAPK, and Akt signaling in skeletal muscle (13, 26–28) and is a key modulator of skeletal muscle wasting and repair in animal models of muscle injury (26, 28).
FBXL2 is a receptor component and member of the Skp-cullin-F box (SCF) ubiquitin E3 ligase superfamily that can moderate inflammation through the ubiquitination and proteasomal degradation of TRAF6 (29). FBXL2 mediates several other important molecular interactions that govern mitotic arrest and cell cycle exit through ubiquitin proteasomal degradation of cyclin D2, cyclin D3, and Aurora B (30–32). FBXL2 also targets the p85β regulatory subunit of the phosphoinositide 3-kinase (PI3K) pathway for proteasomal degradation (33) and regulates mitochondrial calcium influx by disposing of inositol 1,4,5-trisphosphate receptor 3 (IP3R3), located on the endoplasmic reticulum, through the proteasome to reduce sensitivity to calcium-dependent apoptotic stimuli (34). Together, these studies depict FBXL2 as a central adaptor E3 ligase subunit that facilitates inhibition of cellular proliferation and perhaps modulates cell survival. FBXL2 proteasomal degradation is controlled by another E3 ligase component, FBXO3; however, neither the transcriptional regulation of these E3 ligase components nor their role in skeletal myogenesis is known (29).
In this study, we demonstrate that induction of Fbxl2 is a marker of myogenic differentiation that is transcriptionally suppressed by TNF-α stimulation in skeletal myoblasts, which leads to increased cellular proliferation and a profound impairment of myotube formation. Hence, regulation of Fbxl2 production appears to be fundamental to the induction of a differentiated cellular phenotype in skeletal muscle by disposing of key effectors of cell cycle progression and proliferative signaling. Therefore, the findings of this study are relevant to our understanding of the inflammatory regulation of skeletal muscle development, regeneration, and repair in chronic disorders associated with muscle wasting.
RESULTS
FBXL2 is upregulated in myotubes during myogenic differentiation.
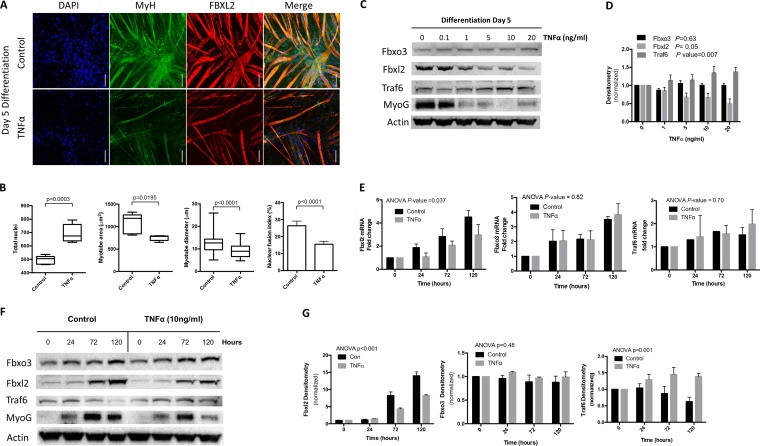
Serum starvation-mediated differentiation of C2C12 myoblasts was inhibited by TNF-α stimulation at day 5, as observed morphologically and characterized by increased myoblast proliferation (P < 0.0003), reduced myotube area (P < 0.02), decreased myotube diameter (P < 0.0001), and a lower nuclear fusion index (P < 0.0001) (Fig. 1A and B). After 5 days of culture, TNF-α blunted induction of FBXL2 and myogenin (MyoG) protein during differentiation and yet induced TRAF6 levels (Fig. 1C and D). We next measured total expression of Fbxl2, Fbxo3, and Traf6 mRNAs during myogenic differentiation at the indicated time points by quantitative PCR (qPCR). Fbxl2 mRNA levels increased 4-fold during differentiation but decreased by 34% in the presence of TNF-α (P = 0.037), yet Fbxo3 and Traf6 expression levels were not significantly different in the presence of TNF-α (Fig. 1E). We also examined protein expression of FBXL2, FBXO3, and TRAF6 and markers of myogenic differentiation during proliferation and differentiation. TNF-α also tended to reduce MyoG protein mass and FBXL2 with increased levels of TRAF6 (Fig. 1F and G).
FIG 1.
FBXL2 is a marker of myogenic differentiation. (A and B) C2C12 myoblasts were imaged by confocal microscopy in the presence or absence of TNF-α treatment (A), and the parameters of cellular proliferation and morphology were quantitated and shown graphically (B). Total nuclei, myotube morphology, and the nuclear fusion index were objectively quantified using the thresholding function in Fiji. A minimum of 5 random fields in each treatment group were acquired for analysis. Scale bars = 100 μm. (C and D) Cell lysates from C2C12 cells were obtained at day 5 of differentiation in the presence or absence of TNF-α stimulation and immunoblotted for protein expression (C), and band intensity was quantitated and graphed (D). (E) Changes in levels of Fbxl2, Fbxo3, and Traf6 mRNA expression were quantified by real-time qPCR during myogenic differentiation of C2C12 myoblasts under control conditions and with TNF-α treatment at 0, 24, 72, and 120 h. (F and G) Dose response effects of TNF-α stimulation on FBXO3, FBXL2, TRAF6, and MyoG protein levels (F), with band intensities quantitated by densitometry shown graphically (G). The data from each quantitated bar graph are representative of the results of at least three independent experiments. The P values shown represent the significance of trend analysis over time or concentrations as analyzed by ANOVA. Densitometry data are shown as mean and standard error of the mean (SEM). The box plot extends from the 25th to the 75th centile, and the whiskers extend from the minimum value to the maximum.
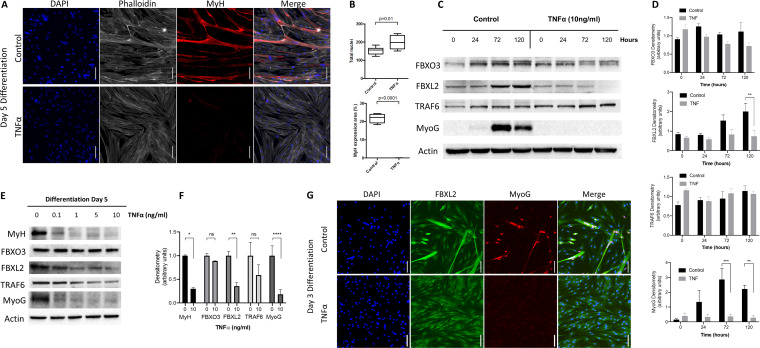
To increase the relevance of the findings, we confirmed our results in primary human myoblasts isolated from patient samples. As has been previously reported, we observed a profound inhibitory effect of TNF-α stimulation on myogenic differentiation with a concurrent increase in cellular proliferation (Fig. 2A and B) (15). In addition, expression of the early and late differentiation markers MyoG and myosin heavy chain (MyH) was significantly reduced with TNF-α at day 5 of differentiation (Fig. 2C to E). As observed in the murine C2C12 myoblasts, FBXL2 expression was blunted in the presence of TNF-α but FBXO3 levels were preserved (Fig. 2E and F). Evaluation of myotube formation revealed that FBXL2 expression was markedly upregulated during myogenic differentiation, colocalized to myogenin-expressing cells, and decreased significantly in the presence of TNF-α (P < 0.001) (Fig. 2G).
FIG 2.
Fbxl2 expression is upregulated during primary human myofibroblast differentiation. (A) Primary human myoblasts were isolated from the vastus lateralis muscles of heathy volunteers and differentiated for up to 5 days under control conditions or in the presence of TNF-α, and the morphological characteristics were evaluated by confocal microscopy (scale bars = 100 μm). (B) TNF-α stimulation (10 ng/ml) resulted in increased cellular proliferation as assessed by numbers of nuclei per field and complete abrogation of myogenic differentiation as evident from absence of MyH expression and myotube formation. The data were quantitated, and the results are shown graphically. (C and D) Cell lysates were obtained at the indicated time points and immunoblotted for FBXO3, FBXL2, TRAF6, and MyoG protein expression (C), with band intensities quantitated and graphed (D). (E and F) The dose response effect of TNF-α stimulation on MyH, FBXO3, FBXL2, TRAF6, and MyoG protein expression was evaluated (E), with band intensities quantitated and graphed (F). (G) Primary human myotubes differentiated for 3 days demonstrating upregulation of FBXL2 expression and colocalization restricted to MyoG-expressing cells (scale bars = 100 μm). The data from each quantitated bar graph are representative of the results of at least three independent experiments. ns, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by ANOVA. The data are shown as means and SEM. The box plot extends from the 25th to the 75th centile, and the whiskers extend from the minimum value to the maximum.
Fbxl2 and TNF-α uniquely alter transcriptomic profiles during differentiation.
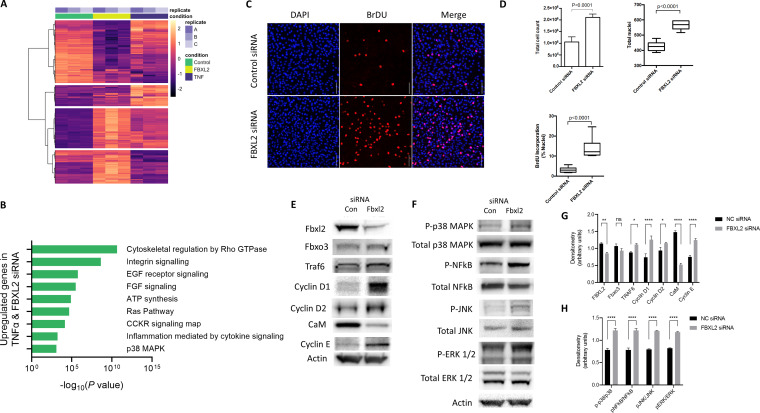
RNA sequencing was performed on C2C12 cells at 48 h postinduction of differentiation (control), after Fbxl2 gene silencing with small interfering RNA (siRNA) at 48 h, and after treatment with TNF-α for 48 h. Of 13,948 total transcripts identified, there were 569 differentially expressed genes (DEGs) with a log fold change of >1.5 and a P value of <0.05, of which 6,704 genes were differentially regulated after Fbxl2 gene silencing and TNF-α stimulation (see Table S1 in the supplemental material). Heat map visualization of the top 500 DEGs between the control, Fbxl2 knockdown, and TNF-α stimulation was performed with associated Panther (http://pantherdb.org) pathway analysis following gene set enrichment, demonstrating substantial overlap of differentially expressed genes involved in growth factor and inflammatory signaling observed between the Fbxl2 siRNA treatment and TNF-α groups (Fig. 3A and B). To determine the functional consequences of Fbxl2 depletion, we performed immunocytochemistry of C2C12 cells following a 2-h bromodeoxyuridine (BrdU) pulse (Fig. 3C). Total cell counts at 48 h were 2-fold higher after Fbxl2 knockdown than with control RNA (2.1 × 106 versus 1.1 × 106; P < 0.0001). The average number of nuclei per imaged field increased (563 versus 427; P < 0.0001) following Fbxl2 knockdown, and BrdU incorporation by cells significantly increased (14.3% versus 3.2%; P < 0.0001) (Fig. 3D). To assess the signaling events linked to Fbxl2 depletion, gene silencing of Fbxl2 demonstrated increased TRAF6, cyclin D1/cyclin D2, and cyclin E and reduced calmodulin protein levels in skeletal myoblasts (Fig. 3E and G) (29, 31). Not unexpectedly, known TRAF6 downstream targets, phosphorylated p38 MAPK and NF-κB, were increased by Fbxl2 depletion, in addition to MAPK pathways, JNK, and extracellular signal-regulated kinase (ERK) (Fig. 3F and H). Collectively, these results indicate a prostimulatory effect of Fbxl2 depletion on the proliferative behavior of C2C12 cells.
FIG 3.
Depletion of endogenous Fbxl2 promotes myoblast proliferation. C2C12 cells were seeded at a density of 5 × 104/ml and transfected at 60% confluence with 40 pM negative-control RNA or Fbxl2 siRNA in 6-well plates. The cells were induced to differentiate in control medium or treated with 10 μM TNF-α. Cellular lysates were prepared at confluence (proliferation) and at 48 h postinduction of differentiation (control). (A) Heat map visualization of the top 500 DEGs between control and Fbxl2 siRNA stimulation and TNF-α. (B) Panther pathway analysis following gene set enrichment demonstrating common enrichment of genes involved in growth factor and inflammatory signaling following Fbxl2 knockdown and TNF-α stimulation. Significantly differentially expressed genes were determined by filtering at a threshold defined by a minimum absolute change of 1.5-fold and a false-discovery rate P value of <0.05. (C) To assess the effect of Fbxl2 gene silencing on cellular proliferation, live cells were counted 48 h after differentiation using trypan blue exclusion staining or pulsed with BrdU for 2 h, fixed, and permeabilized after DNA hydrolysis. Images were captured on a confocal microscope after nuclear staining with BrdU antibody and counterstaining with DAPI (scale bar = 100 μm). (D) Total nuclei and BrdU-stained nuclei were objectively quantified using the thresholding function in Fiji from a minimum of 5 random fields. (E and G) Lysates were prepared from control RNA (Con) or Fbxl2 siRNA 48 h after differentiation and immunoblotted for FBXL2, FBXO3, TRAF6, cyclin D1, cyclin D2, calmodulin (CaM), and cyclin E protein expression (E), with band intensities quantitated and graphed (G). NC, nontargeting control RNA. (F and H) Fbxl2 gene silencing increased total and phosphorylated forms of p38-MAPK, NF-κB, JNK, and ERK1/2 protein expression (F), with band intensities quantitated and graphed (H). (G and H) Data from each quantitated bar graph are representative of the results of at least three independent experiments. ns, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by ANOVA. The data are shown as means and SEM. The box plot extends from the 25th to the 75th centile, and the whiskers extend from the minimum value to the maximum.
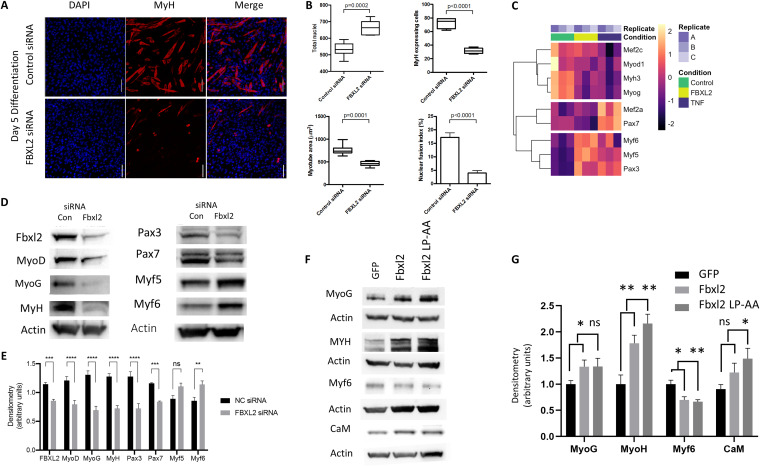
Marked impairment of myogenic differentiation was observed following Fbxl2 gene silencing, effectively inhibiting myotube formation (Fig. 4A). The characteristics of myotube formation were decreased by Fbxl2 knockdown as assessed morphologically by the number of myosin-expressing cells (P < 0.0001), average myotube area (P < 0.0001), and nuclear fusion index (P < 0.0001) (Fig. 4B). Transcriptomic profiling of a subset of myogenic regulator factors demonstrated similar profiles of gene expression in C2C12 cells subjected to Fbxl2 gene silencing and TNF-α stimulation (Fig. 4C). Gene silencing of Fbxl2 resulted in reduced protein expression of the transcriptional activators Pax3 and Pax7 and the myogenic regulatory factors MyoD, MyoG, and MyH. However, protein levels of Myf5 and Myf6 were increased at 48 h in the Fbxl2 knockdown cells (Fig. 4D and E). To examine the role of FBXL2 E3 ligase subunit activity in cellular differentiation, we overexpressed both murine Fbxl2 and an Fbxl2 mutant that contains a mutation in two residues in the F box domain demonstrated to inhibit Skp-1 binding and thereby prevents degradation by the ubiquitin machinery (35). Overexpression of both Fbxl2 and the ligase-inactive mutant Fbxl2 increased myogenic differentiation, suggesting that ubiquitin E3 ligase activity may not be required for this subunit to mediate effects on myoblast differentiation (Fig. 4F and G). These effects resemble the actions of the E3 ubiquitin ligase TRIP12, which also promotes cell cycle progression independently of its catalytic (ligase) activity (36).
FIG 4.
Fbxl2 expression is required for myogenic differentiation. (A) Gene silencing of Fbxl2 was performed in C2C12 cells, and the morphological characteristics of myocyte formation were evaluated at 120 h by immunocytochemistry using confocal microscopy. Scale bars = 100 μm. (B) Total nuclei, the number of myosin-expressing cells, and the average myotube area were objectively quantified using the thresholding function in Fiji; the nuclear fusion index was determined as the percentage of nuclei located within ≥50 myotubes. A minimum of 5 random fields in each treatment group were acquired for analysis. (C) Fbxl2 gene silencing and TNF-α stimulation demonstrating dysregulation of a transcriptomic subset of key myogenic regulator factors that regulate myogenesis in C2C12 cells. (D) Fbxl2 gene silencing decreased protein expression of the transcriptional activators Pax3 and Pax7 and the myogenic regulatory factors MyoD and MyoG. Protein levels of Myf5 and Myf6 were increased as measured at 48 h post-Fbxl2 siRNA transfection. (E) Protein expression with band intensities quantitated and graphed. NC, nontargeting control RNA. (F) Cells were ectopically expressed with wild-type Fbxl2 or an Fbxl2 double point mutant lacking the ability to mediate substrate ubiquitylation (Fbxl2 LP-AA), and effects on myocyte differentiation were assessed by immunoblotting. (G) Protein expression and band intensities were quantitated and graphed. (E and G) Data from each quantitated bar graph shown are representative of the results of at least three independent experiments. ns, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by ANOVA. The data are shown as means and SEM. The box plot extends from the 25th to the 75th centile, and the whiskers extend from the minimum value to the maximum.
Identification of a TNF-α-responsive cis-acting element in the Fbxl2 core promoter.
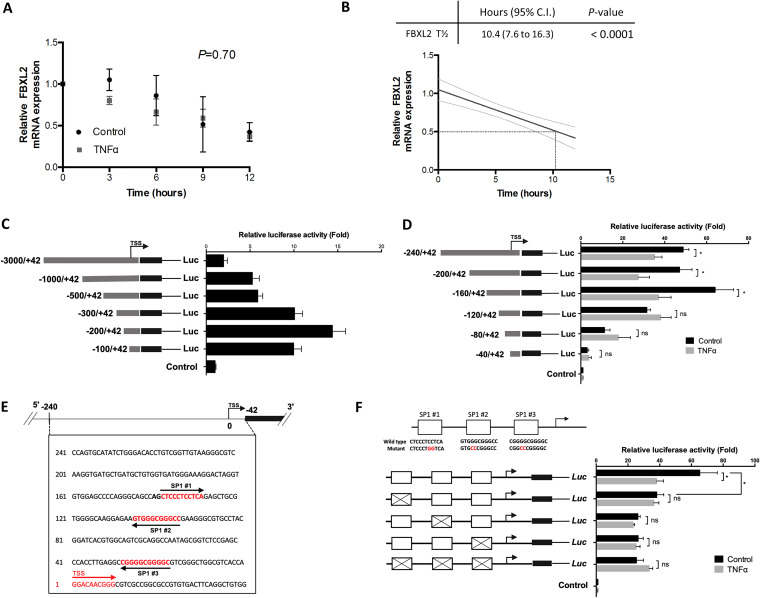
In earlier experiments, TNF-α decreased steady-state Fbxl2 mRNA levels, suggesting alterations in either its mRNA turnover or its transcription rate. To examine mRNA turnover, we measured the half-life of Fbxl2 mRNA by actinomycin D chase. The mRNA half-life of Fbxl2 was determined to be 10.4 h (95% confidence interval, 7.6 to 16.3 h) by linear regression analysis, and TNF-α stimulation did not alter Fbxl2 mRNA stability (P = 0.70) (Fig. 5A and B). Progressive deletional constructs of a 3,000-bp region of DNA corresponding to the proximal Fbxl2 5′ flanking sequence were cloned into the PGL3 basic firefly luciferase reporter plasmid. In transient-transfection experiments, we identified high-level promoter-reporter activity localized within 200 bp proximal to the transcription start site (TSS) (Fig. 5C). Progressive 5′ truncations of a −160 to +42 fragment demonstrated loss of peak reporter activity and reduction of TNF-α responsiveness after cellular expression of promoter-reporter plasmids (Fig. 5D). Sequence alignment of the proximal promoter region of the human and murine Fbxl2 gene demonstrated a high degree of sequence conservation. Three SP1 transcription factor binding motifs were detected in the proximal Fbxl2 promoter (Fig. 5E) (37, 38). We therefore evaluated the roles of individual SP1 binding sites in the Fbxl2 promoter by site-directed mutagenesis of individual putative binding elements. In C2C12 myoblasts, mutation at each SP1 binding site resulted in substantial loss of constitutive reporter activity, yet there was no additional reduction of promoter-reporter activity when all three SP1 sites were mutated (Fig. 5F). In the presence of TNF-α, reporter activities from expression plasmids harboring single or multiple SP1 mutations resulted in abrogation of TNF-α responsiveness (Fig. 5F). These results are highly suggestive that consensus SP1 cis-acting elements confer constitutive Fbxl2 promoter activity during differentiation and that the core promoter spans a region from bp −160 to +42 within the proximal 5′ flanking region of the gene. The results suggest the possibility that TNF-α impairs molecular interaction of SP1 with cis-acting elements in the Fbxl2 gene, though they do not exclude the ability of the cytokine to trigger Fbxl2 gene silencing by a repressor.
FIG 5.
Identification and characterization of the proximal Fbxl2 promoter. (A) There was no significant difference in Fbxl2 mRNA degradation measured by qPCR at 3, 6, 9, and 12 h under control and TNF-α treatment conditions in an actinomycin D assay. (B) The half-life of Fbxl2 mRNA was estimated as 10.4 h (95% confidence interval [gray lines], 7.6 to 16.3) by linear regression analysis. (C) A 3,000-nucleotide (nt) region was cloned into the PGL3 basic firefly luciferase (Luc) reporter plasmid and then cotransfected into C2C12 cells with a nanoluciferase reporter as a transfection control. The cells were allowed to differentiate for 24 h prior to harvesting and lysis, and luminescence was measured using the NanoLuc dual-reporter assay (Promega). Sequential deletion of the reporter plasmid identified peak activity in the Fbxl2 promoter at nt −200 to +42 proximal to the TSS. (D) Additional deletion analysis of the reporter plasmid in the Fbxl2 promoter at nt −200 to +42 proximal to the TSS. Loss of constitutive reporter activity was identified between nt +160 and +120, with further loss of activity occurring with progressive deletion of the core promoter. Loss of TNF-α responsiveness also occurred between nt +160 and +120 of the TSS. (E) Schematic of a nt +240 to −42 insert within the PGL3 basic reporter construct showing three SP1 motifs proximal to the TSS. (F) Site-directed mutagenesis was performed to evaluate the impact of individual SP1 mutations on Fbxl2 core promoter activity; mutation in each SP1 site (X) resulted in a similar 25-fold loss of reporter activity, yet there was no additional loss of activity when all three SP1 sites were mutated. In the presence of TNF-α, each SP1 mutant plasmid did not show any additional loss of reporter activity, suggesting that SP1 is a TNF-α-responsive cis-acting element within the Fbxl2 promoter. The data are representative of the results of three independent experiments. ns, not significant (P > 0.05); *, P < 0.05. The data are shown as means and SEM.
SP1 binds to the Fbxl2 core promoter to regulate Fbxl2 gene expression.
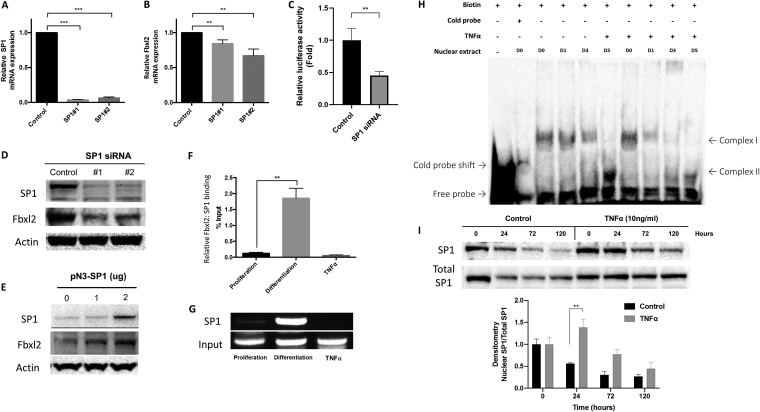
Gene silencing of Sp1 with two separate siRNA constructs resulted in a marked reduction of Sp1 mRNA (Fig. 6A). Sp1 gene silencing resulted in modest depletion of endogenous Fbxl2 mRNA at 48 h (Fig. 6B) and a significant reduction in the luciferase reporter activity of the Fbxl2 −160 to +42 fragment core promoter (Fig. 6C). Sp1 depletion also resulted in a significant decrease in both SP1 and FBXL2 proteins at 48 h postdifferentiation (Fig. 6D). Conversely, overexpression of Sp1 plasmid in C2C12 cells led to an increase in FBXL2 protein (Fig. 6E). To study the interaction between SP1 and a 371-bp region of the Fbxl2 promoter, we performed chromatin immunoprecipitation (ChIP) assays. C2C12 lysates were immunoprecipitated with SP1 antibody, and the precipitated DNA was amplified using primers specific to the proximal Fbxl2 promoter region and measured by qPCR. Quantification of the ChIP analysis showed that SP1 binds to the Fbxl2 promoter during differentiation (1.95%) but not during proliferation (0.12%) or in the presence of TNF-α stimulation (0.06%); the values shown are expressed as a percentage of DNA in the SP1 fractions compared to the input (Fig. 6F). The 371-bp region of the Fbxl2 promoter was amplified by PCR from SP1 input lysates and visualized on an agarose gel (Fig. 6G). An electromobility shift assay (EMSA) demonstrated DNA-protein complex formation after incubating nuclear extracts from proliferating and differentiating myoblasts, cultured in the presence or absence of TNF-α, with a 25-nucleotide biotin-labeled fragment that is conserved in the human Fbxl2 and mouse Fbxl2 core promoters (5′-CTTGGGGACGGGGCGGGGCGCCTGG-3′) and corresponds to the proximal putative SP1 binding element. The levels of complex I decreased as cells became terminally differentiated, while the more mobile complex II was elevated at 120 h following initiation of the differentiation process (Fig. 6H). TNF-α stimulation significantly decreased the formation of both complexes I and II during myogenic differentiation (Fig. 6H). Despite the decrease in Fbxl2 core promoter complex formation under TNF-α-stimulated conditions, we observed a significant increase in nuclear SP1 at 24 h (Fig. 6I). Hence, during the differentiation of myoblasts, SP1 binds and transactivates the Fbxl2 gene, but this functional association is impaired in the presence of TNF-α despite nuclear accumulation of the transcription factor.
FIG 6.
SP1 binds to the Fbxl2 promoter region to regulate its gene expression during myogenic differentiation. (A) Sp1 mRNA abundance following knockdown of SP1 in C2C12 cells. (B) Fbxl2 mRNA abundance by qPCR after SP1 silencing at 48 h postdifferentiation. (C) Relative luminescence after SP1 depletion in C2C12 cells overexpressing the +240 to −42 Fbxl2 promoter reporter construct. (D and E) SP1 and FBXL2 protein levels following depletion (D) and overexpression (E) of SP1 in C2C12 cells. The data are representative of the results of three independent experiments. (F) C2C12 lysates were immunoprecipitated with SP1 antibody. The precipitated DNA was amplified using specific primers to the proximal Fbxl2 promoter region and measured by qPCR; values are expressed as percentages of DNA in the SP1 fractions compared to input. Shown is a quantification graph for ChIP assays using qPCR data demonstrating significant SP1 binding to a region including the Fbxl2 promoter under differentiation conditions (**, P = 0.002) but not following TNF-α stimulation. The data are representative of the results of two independent experiments. (G) The 371-bp region of the Fbxl2 core promoter was amplified by PCR from SP1 and input lysates and visualized on an agarose gel. (H) Nuclear extracts were isolated from C2C12 myoblasts during proliferation and differentiation at the indicated time points in the presence or absence of TNF-α stimulation and then incubated with a biotin-labeled 25-nucleotide segment that corresponds to the proximal putative SP1 binding element of the human Fbxl2 and mouse Fbxl2 core promoters. An EMSA demonstrated the formation of two DNA-protein complexes during myogenic differentiation, which was decreased in the presence of TNF-α-stimulated cells. The data are representative of the results of two independent experiments. (I) Nuclear SP1 expression in C2C12 cells after differentiation with and without TNF-α. (Bottom) Protein levels and band intensities were quantitated and graphed as shown. (A to C, F, and I) **, P < 0.01; ***, P < 0.001 by ANOVA. The data are shown as means and SEM.
Phosphorylation of SP1 by JNK inhibits Fbxl2 promoter binding and transcriptional activity.
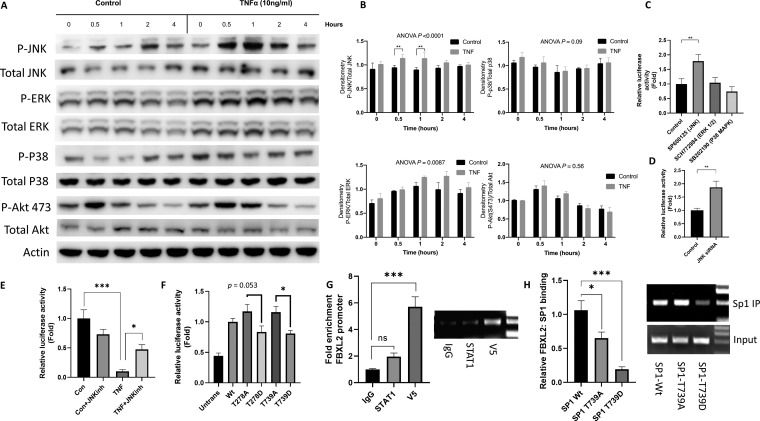
Because accumulation of nuclear SP1 after TNF-α stimulation was not sufficient to increase Fbxl2 promoter activity (Fig. 6I), the data suggested that SP1 might be posttranslationally modified by TNF-α. Phosphorylation is a well-described mechanism through which the association of SP1 with DNA is altered (39). TNF-α treatment of C2C12 increased JNK and ERK phosphorylation (Fig. 7A and B). Chemical inhibitors of JNK, but not of ERK1/2 or p38, increased Fbxl2 promoter-reporter activity (Fig. 7C). Similarly, gene silencing of JNK increased Fbxl2 promoter-reporter activity (Fig. 7D). In addition, pretreatment of C2C12 cells with the JNK inhibitor SP600125 partially restored TNF-α-mediated repression of Fbxl2 promoter-reporter activity (Fig. 7E). JNK phosphorylates SP1 at residues T278 and T739 (40), and SP1 phosphorylation at T739 inhibits its ability to bind DNA and enhance transcription (41, 42). Therefore, we examined if SP1 phosphorylation point mutant constructs, when expressed in C2C12 cells, might modulate Fbxl2 promoter activity. Overexpression of phosphorylation-deficient T739A, and to a lesser extent T278A, mutants resulted in increased promoter-reporter activity. The converse effect was observed with the overexpression of constitutive SP1 phosphorylation mimics (T278D and T739D) that exhibited reduced activity relative to the phosphorylation-deficient mutants (Fig. 7F). Lastly, in ChIP assays, ectopic expression of wild-type SP1 or the T739A or T739D mutant displayed differential association with the Fbxl2 promoter, with the SP1 phosphorylation mimic showing lower binding to the F box promoter region than wild-type SP1 or T739A (Fig. 7G and H). These observations collectively suggest that TNF-α-mediated repression of Fbxl2 gene transcription is mediated by JNK phosphorylation of SP1, which impairs its ability to bind the Fbxl2 promoter.
FIG 7.
JNK-mediated phosphorylation of SP1 inhibits Fbxl2 transcriptional activity in response to TNF-α. (A) Expression of total and phosphorylated JNK, ERK1/2, p38, and AKT with and without TNF-α treatment in C2C12 cells. (B) Protein densitometry was quantitated and graphed. The P values shown represent significance as indicated by the brackets between bars or that of trend analysis over time as analyzed by ANOVA. (C) Fbxl2 promoter-reporter activity in C2C12 cells treated with chemical inhibitors of JNK, ERK1/2, and p38 (1 to 10 μM) for 24 h. (D) Fbxl2 promoter-reporter activity in C2C12 cells after knockdown of JNK with siRNA for 48 h. (E) Fbxl2 promoter-reporter activity in C2C12 cells pretreated with the JNK inhibitor SP600125 (1 μM) 30 min prior to TNF-α treatment. Cells were harvested at 48 h posttreatment. (F) Fbxl2 promoter-reporter activity in HEK cells transfected with SP1 plasmids encoding phosphorylation-deficient mutants (T278A and T739A) or phosphorylation mimics (T278D and T739D) for 48 h. (G) C2C12 cells were transfected with plasmids expressing V5-tagged SP1. The cells were fixed and sonicated, and DNA was immunoprecipitated with isotype control (IgG), V5, or STAT-1 antibodies. (Left) Relative enrichment of the Fbxl2 promoter was quantified by qPCR. (Right) Agarose gel of a 193-bp region of the Fbxl2 core promoter amplified by PCR. (H) C2C12 cells were transfected with plasmids expressing V5-tagged SP1 wild type (Wt), SP1-T739A, and SP1-T739D for 48 h. The cells were fixed and sonicated, and DNA was immunoprecipitated (IP) with V5 antibody. (Left) Relative enrichment of the Fbxl2 promoter was quantified by qPCR. (Right) Agarose gel of the 193-bp region of the Fbxl2 core promoter amplified by PCR. ns, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001 by ANOVA. The data are shown as means and SEM.
DISCUSSION
This is the first study that demonstrates the molecular regulation of the ubiquitin E3 ligase component FBXL2 and uncovers its role in the modulation of skeletal muscle maturation during inflammatory stress. The new contributions of this study are (i) finding that FBXL2 has an essential role in governing the process of myogenic differentiation, (ii) finding that TNF-α and Fbxl2 cellular depletion produces marked similarities in the gene expression profile of MRFs required for myoblast proliferation and myogenic differentiation, (iii) identifying and characterizing the molecular behavior of Fbxl2 expression through its gene transcription rate and defining its core promoter regulation by critical binding elements, (iv) finding that SP1 is indispensable for constitutive activity of the Fbxl2 gene, and (v) finding that TNF-α stimulation disrupts SP1 interaction with the Fbxl2 promoter, resulting in decreased Fbxl2 expression through JNK-mediated phosphorylation of SP1, likely at residues Thr739 and Thr278. The results collectively underscore the importance of Fbxl2 mRNA synthesis as a fundamental mechanism that is needed to permit maturation of skeletal muscle and that this process may be severely disrupted under sustained proinflammatory conditions.
The findings here show that FBXL2 protein mass increased gradually in both primary human myoblasts and C2C12 myoblasts with differentiation but that induction of these levels was blunted with TNF-α exposure. One consequence of depleted cellular FBXL2 levels was alterations in TRAF6 levels, because this TNF adaptor molecule is a known substrate of the SCFFBXL2 E3 ligase (29). TRAF6 levels tended to decrease in myoblasts with differentiation but were stabilized after TNF-α exposure. The reduced FBXL2 protein levels correlated with augmented cellular proliferative responses and impaired myogenic differentiation in primary human myoblasts and in C2C12 cells, raising the possibility that SCFFBXL2 E3 ligase acts to eliminate an inhibitor of myofilament generation in concert with other crucial myogenic transcription factors, such as MYF5, MyoD, myogenin, and MRF4 (43). Further, the observations here that Fbxl2 gene silencing potentiated NF-κB and p38 MAPK activities is consistent with consequences of TRAF6 stabilization, as they are downstream effectors of this adaptor molecule in cellular proliferative signaling (12, 23, 27, 44).
Analysis of the human and mouse Fbxl2 gene promoter regions revealed a high degree of conservation, particularly within 200 bp of the transcription start site. We identified three SP1 transcription factor motifs in both species within this region, as well as two CRE binding motifs. Dual SP1 cis-acting elements have been described in the proximal cyclin D1 promoter of vascular endothelial cells, resulting in constitutive CRE-mediated activation with further transcriptional activation through G1-inducible SP1 activation via a Ras-dependent pathway (45, 46). Cooperation between SP1 and CRE in the promoter regions of several other genes has also been demonstrated, including fibronectin (47), neurofibromatosis 1 (48), and hepatic growth factor receptor (49) genes and the TNF-α gene itself (50). Mirroring the findings in this study, the cis-acting response element to various stimuli, including nitric oxide and TNF-α exposure, was identified as the SP1 binding site and not the CRE site.
SP1 is a ubiquitously and constitutively expressed zinc finger transcription factor that binds GC-rich DNA motifs to regulate thousands of genes that are involved in critical cellular functions, such as cell growth, differentiation, and apoptosis (39). Posttranslational modifications, such as phosphorylation, acetylation, ubiquitination, and glycosylation, influence the binding of SP1 to DNA and thus determine its activator or repressor transcriptional activity (51). As shown here, SP1 phosphorylation at specific molecular sites by JNK likely impaired its ability to associate with the Fbxl2 gene in response to TNF-α. SP1 regulation of several components of the cell cycle has been established, including downregulation of cyclin D2 and upregulation of cyclin G2 and cdkn2c/p18 (52). Our observation that SP1 regulates transcription of the Fbxl2 gene reveals a cooperative interaction between gene transcription events linking with the ubiquitin proteasome system to affect the cell cycle. Our data, together with prior studies of the molecular behavior of FBXL2, suggest a key role for SP1 regulation of cell cycle components to promote mitotic arrest through transcriptional events and initiation of cyclin D2 and D3 disposal via FBXL2-mediated ubiquitin proteasomal degradation (30, 31).
An important observation in our work is that mutation of any one of the three SP1 sites in the Fbxl2 promoter resulted in similar reductions in reporter activity. This suggests that for complete transcriptional activation of Fbxl2 by SP1, the formation of multimeric SP1 complexes is required through the documented process of SP1 synergism and superactivation whereby multimeric SP1 complexes form via interaction across different domains within the protein (53). Synergistic interactions between SP1 and a variety of other transcription factors have been described, including cAMP response-element binding protein (CREB) (54), MEF2 (55), and NF-κB (56), which promote gene transcription in a sequence- and cell-specific manner. Notably, TNF-α can regulate SP1 binding to the growth hormone receptor promoter (49) and also counteracts the stimulatory effects of SP1 on adiponectin gene transcription during adipocyte differentiation (57). As with FBXL2, the mechanism of TNF-α responsiveness is mediated through altered transcriptional SP1 binding affinity to these gene promoter regions, which our study suggests is regulated by the phosphorylation by JNK of SP1 threonine residues at positions 739 and 278 in the presence of TNF-α (58). Our EMSA data support the premise of altered SP1 binding affinity, as we noted a shift in complex formation from proliferation through terminal differentiation of C2C12 myoblasts utilizing a sequence that corresponds to the proximal Fbxl2 core promoter. Further, we demonstrate diminished protein-DNA complex formation in the presence of TNF-α stimulation.
Our work supports the role of FBXL2 in the arrest of satellite cell proliferation and the modulation of known ubiquitin proteasome signaling pathways that regulate cell growth and differentiation through the complex process of muscle regeneration. This is clinically relevant, as systemic elevation of TNF-α in blood has been associated with a number of disease states related to muscle wasting, including septic shock (59), chronic obstructive pulmonary disease (COPD) (60), and cancer (61), leading to its initial characterization as a “cachectin” (62). Though our findings are of particular importance to models of skeletal muscle regeneration, repair, and differentiation, it is notable that the function of FBXL2 may have relevance to other tissues and disease states, given its broad tissue expression profile in humans (63). Previous studies have reported a role for FBXL2 in the prevention of cancer progression through its varied functions that affect cell cycle progression, as well as proliferative and cell survival signaling (31, 33, 34). Since cancer development and progression are strongly linked to inflammation (64), it is significant that, as a promising drug target, FBXL2 protein levels are rendered especially susceptible to inflammatory modulation by both transcriptional suppression and FBXO3-mediated ubiquitin proteasomal degradation (65).
Conclusions.
Ubiquitin proteasome signaling has a critical role in the control of cell cycle exit, proliferative signaling, and the promotion of cell growth and survival during skeletal muscle regeneration. In this study, we described the previously uncharacterized yet essential role of FBXL2 in the regulation of skeletal myoblast proliferation and myogenic differentiation. We found that transcriptional activation of cis-regulatory elements in the Fbxl2 proximal promoter during differentiation is affected by SP1. Further, we demonstrated a novel mechanism whereby TNF-α abolished myogenic differentiation through disruption of SP1 transactivator binding to the Fbxl2 promoter through JNK-triggered site-specific phosphorylation of SP1. This study contributes to our understanding of the regenerative potential of skeletal muscle in health and disease by uncovering a highly relevant mechanism for the transcriptional suppression of differentiation in skeletal muscle by TNF-α, with consequent sustained proliferative signaling and cell cycle activation.
MATERIALS AND METHODS
Antibodies and reagents.
FBXL2 antibody was obtained from GeneTex (Irvine, CA; catalog no. GTX17018). Antibodies against FBXO3 (catalog no. sc-514625), MyoD (catalog no. sc-71629), Myh3 (catalog no. sc-376157), myogenin (catalog no. sc-52903), and Myf6 (catalog no. sc-514379) were purchased from Santa Cruz Biotechnologies (Dallas, TX). Myf5 antibody (catalog no. 04766) was obtained from Avivasysbio. TRAF6 antibody (catalog no. 06-1110) and SP1 antibody with isotype control antibody (catalog no. 17-601) were purchased from Millipore (Cambridge, MA). p38-MAPK (catalog no. 9212) and phosphorylated p38-MAPK (p-p38-MAPK) (catalog no. 4511), Akt (catalog no. 2920S), p-Akt-Ser473 (catalog no. 4060S), p-Akt-Thr308 (catalog no. 13038S), ERK1/2 (catalog no. 9101S), and p-ERK1/2 (catalog no. 4696S) were obtained from Cell Signaling (Beverly, MA). NF-κB (catalog no. ab16502) and phospho-NF-κB p65 (catalog no. ab28856), and horseradish peroxidase (HRP)-tagged β-actin (catalog no. ab49900) antibodies were obtained from Abcam (Cambridge, MA). HRP-conjugated anti-mouse IgG, anti-rabbit IgG, and anti-goat IgG were obtained from Bio-Rad (Hercules, CA). MF20, Pax3, and Pax7 antibodies were obtained from the Developmental Studies Hybridoma Bank (DSHB) (Iowa City, IA). Anti-BrdU antibody (catalog no. 555627) and the BrdU flow cytometry assay were purchased from BD Pharmingen (San Diego, CA). Miniprep kits were purchased from Invitrogen (Grand Island, NY). The MAPK inhibitors SB202190, SCH772984, and SP600125 were purchased from Sigma (Sigma-Aldrich, St. Louis, MO).
Cell culture and transfection.
Primary human myoblasts were isolated from the vastus lateralis muscles of healthy donors as previously described (66). Institutional review board approval was granted by the University of Pittsburgh for the study of human subjects (no. PRO17010337). In brief, muscle biopsy samples were digested with collagenase, filtered through a 30-μm mesh, plated, and maintained in Dulbecco’s modified Eagle’s medium (DMEM)–F-12 from Gibco (Life Technologies, Grand Island, NY) with 20% fetal bovine serum (FBS) from Gemini (Sacramento, CA) and 1% penicillin-streptomycin. Nuclear MyoD staining assessment confirmed >95% primary myoblast cell purity using this technique. Murine C2C12 cells were obtained from the ATCC (Manassas, VA). During myoblast proliferation, cells were supplemented daily with growth medium (GM) (DMEM with 10% FBS and 1% penicillin-streptomycin). Cells were counted using trypan blue exclusion staining and seeded at a density of 5 × 105 in 6- or 12-well plates. Myogenic differentiation was induced when myoblasts reached 90 to 100% confluence by replacing GM with differentiation medium (DM) (DMEM with 2% horse serum; Sigma-Aldrich, St. Louis, MO) and 1% penicillin-streptomycin. The myoblasts were treated with TNF-α (catalog no. 210-TA-020; R&D Systems) at the concentrations indicated during myoblast differentiation. For gene silencing, negative-control siRNA (catalog no. 51-01-14-04), Fbxl2 siRNA (mm.Ri.Fbxl2.13.3), SP1 (mm.Ri.Sp1.13.2; mm.Ri.Sp1.13.3), and JNK (mm.Ri.Mapk8.13.1) were purchased from Integrated DNA Technologies (Coralville, IA), and 20 to 40 nM was transfected into cells using GenMute (SignaGen, Rockville, MD) according to the manufacturer’s protocol. Lysates were prepared during proliferation and at various time points during differentiation (24 h, 48 h, and 120 h posttransfection). HEK293A (ATCC) cells were maintained in DMEM (Gibco) containing 20% FBS with 1% penicillin-streptomycin. The cells were cultured at 37°C under a humidified 5% CO2 atmosphere. Wild-type Fbxl2 and a ligase-deficient mutant of Fbxl2 were generated by cloning a gene fragment encoding murine Fbxl2 (Genewiz, South Plainfield, NJ) into a pcDNA3.1 vector containing a eukaryotic translation initiation factor 1 α subunit (eIF1α) promoter. LP-AA (residues 15 and 16 within the conserved F box domain) were generated by site-directed mutagenesis. Mutant C2C12 cells were transfected with XtremeGene transfection reagent (Roche Applied Science) according to the manufacturers’ protocols. pN3-Sp1FL was a gift from Guntram Suske (Addgene plasmid no. 24543). SP1 point mutants T739A and T739D were generated by site-directed mutagenesis on a pN3-Sp1FL plasmid. V5-tagged SP1 was generated by cloning SP1 into a pcDNA3.1 vector with restriction enzymes KpnI and BstBI (New England Biolabs).
Immunoblotting.
Cell lysates were prepared at various time points during proliferation (0 h) and differentiation (24 h, 48 h, 60 h, and 120 h posttransfection) in 200 μl of radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 10 μg/ml protease inhibitors, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin), sonicated on ice for 12 s, and centrifuged at 10,000 × g for 10 min at 4°C in a microcentrifuge. For gene silencing, lysates were prepared 48 h posttransfection. For immunoblotting, equal amounts of supernatant (25 μg) were loaded onto SDS-10% PAGE gels, transferred to nitrocellulose membranes, blocked with 2% (wt/vol) bovine serum albumin (BSA) in TBST (25 mM Tris-HCl [pH 7.4], 137 mM NaCl, and 0.1% Tween 20) for 1 h, and incubated with primary antibodies in TBST overnight. The membranes were washed three times with TBST at 10-min intervals, followed by 1 h of incubation with mouse, rabbit, or goat horseradish peroxidase-conjugated secondary antibody (1:2,000). The membranes were developed with an enhanced chemiluminescent substrate using the Chemidoc imaging system (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions.
Immunostaining.
Cells (5 × 105) were plated and grown to confluence on glass bottom culture slides. The cells were washed with phosphate-buffered saline (PBS), fixed with 2% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100, blocked with 2% BSA for 1 h, and then exposed to 1:250 primary antibodies overnight at 4°C, followed by incubation with Alexa Fluor-conjugated secondary antibodies (Thermo Fisher; 1:1,000 dilutions) and then briefly stained with DAPI (4′,6-diamidino-2-phenylindole) at room temperature. Immunofluorescent cell imaging was performed on a Nikon A1 confocal microscope (Nikon, Melville, NY) using a 405-nm, 458-nm, 488-nm, 514-nm, or 647-nm wavelength. Images were captured at 1,024- by 1,024-pixel density using Nikon Imaging Software Elements AR 3.2 with a 20× differential interference contrast objective lens. For BrdU proliferation quantification, an additional DNA hydrolysis step was performed with 2 M HCl for 15 min after permeabilization prior to anti-BrdU immunostaining. All images were analyzed using Fiji image software (68).
Quantitative RT-PCR.
Total cellular RNA was collected from C2C12 cells using an RNeasy minikit (Qiagen). RNA was processed for RNA sequencing analysis or used to create cDNA using a high-capacity cDNA reverse transcription (RT) kit (Applied Biosystems) according to the manufacturer’s protocol. qPCR was performed using SYBR select master mix (Applied Biosystems) according to the manufacturer’s protocol with 20 ng cDNA as a template and a primer concentration of 200 nM in a real-time DNA thermal cycler (CFX96; 10-μl reaction volume; Bio-Rad, Munich, Germany). Each biological replicate was performed in technical triplicate; data were analyzed using the ΔΔCq method. All the oligonucleotide primer sequences are available in Table S2 in the supplemental material. To assess for Fbxl2 mRNA degradation, 72-h-differentiated control and TNF-α-treated C2C12 cells were treated with 5 μg/ml actinomycin D to arrest the transcription machinery. Cells were lysed at five different time points after addition of actinomycin D (0, 3, 6, 9, and 12 h) for mRNA measurement.
RNA sequencing.
RNA integrity was determined with an Agilent 2100 Bioanalyzer (Carlsbad, CA). Samples with RNA integrity numbers (RINs) ranging from 9.7 to 10.0 were processed with an mRNA sequencing sample preparation kit (Illumina, Inc., San Diego, CA). Sequencing libraries were prepared from 1 μg of RNA using a Nextera XT DNA library preparation kit (Illumina), normalized at 2 nM using Tris-HCl (10 mM; pH 8.5) with 0.1% Tween 20, and diluted and denatured to a final concentration of 1.8 nM using the Illumina denaturing and diluting libraries for the NextSeq 500 protocol revision D (Illumina). Cluster generation and 1-by-75-bp forward-end read sequencing were performed on an Illumina NextSeq 500 system at the University of Pittsburgh Health Sciences Sequencing Core.
RNA sequence analysis.
Raw transcript data were imported into CLC Genomics Workbench 11, and quality control and trimming of files were performed prior to alignment of reads to the mouse reference genome (mm10). Genes with a mean raw count value of <10 were removed from further processing. DEGs between the experimental groups were determined with Deseq2, using filters to select genes with an absolute fold change (FC) of >1.5 and a false-discovery rate (FDR) P value of ≤0.05. Principal-component analysis, volcano plots, and heat maps were generated using R Studio. Hierarchical clustering of DEGs was performed using Ward’s method with Euclidean distance.
Pathway analysis.
Significantly differentially expressed genes were imported into Panther (http://pantherdb.org) for gene set enrichment analysis of the effects of Fbxl2 gene silencing and TNF-α treatment compared to a 48-h differentiation control. Fisher’s exact test with Benjamini-Hochberg correction for multiple testing using an FDR with a P value cutoff of <0.05 was used to test for pathway significance.
Fbxl2 promoter analysis.
The proximal promoter regions of the human and mouse Fbxl2 genes were aligned using T-Coffee (67). Potential transcription binding sites within this target region were evaluated using the TFBS tools in R Studio from the JASPAR 2018 database; a score of >7.0 was considered significant (37, 38).
Fbxl2 mutants and deletional promoter plasmids.
A 4-kb DNA sequence spanning 3 kb upstream and 1 kb downstream from the transcriptional start site of the Fbxl2 gene was amplified from mouse C57BL/6J genomic DNA (Zyagen, San Diego, CA) by a PCR-based approach using Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA). The primer pairs utilized are listed in Table S2 in the supplemental material. The amplified cDNA fragments were digested with the restriction enzymes MluI-HF and HindIII-HF (New England Biolabs) in Cutsmart buffer (New England Biolabs) according to the manufacturer’s instructions and cloned into the cloning site of the pGL3-basic luciferase reporter vector (Promega, Madison, WI). To control for transfection efficiency, cells were cotransfected with the NanoLuc reporter plasmid (Promega) using XtremeGene transfection reagent (Roche Applied Science) according to the manufacturers’ protocols. At 24 h posttransfection, a luciferase assay was performed using the Nano-Glo Dual-Luciferase assay system (N1110; Promega) on a Biotek Synergy 2 microplate luminometer.
Cell proliferation assay.
C2C12 cells were transfected with negative control or Fbxl2 siRNA at 60 to 70% confluence. After 24 h, the cells were cultured in low-serum differentiation medium for 48 h. The cells were pulsed with 10 mM BrdU for 1 h and then fixed, permeabilized, and hydrolyzed with 2 M HCl for 30 min prior to staining with anti-BrdU antibody (Sigma-Aldrich, St. Louis, MO; catalog no. B8434).
Nuclear extract preparation.
Nuclear extracts were harvested from C2C12 cells at confluence and 24 h, 72 h, and 120 h of differentiation by centrifugation after collection in ice-cold PBS. The cells were lysed in 500 μl hypotonic buffer solution (20 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2) with 0.05% NP-40 and protease inhibitors. Nuclear pellets were separated from the cytosol by centrifugation for 10 min at 3,000 rpm at 4°C and then resuspended in nuclear extraction buffer containing 10 mM Tris (pH 7.4), 100 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10% glycerol, 0.1% SDS, and 0.5% deoxycholate. The extracts were aliquoted and stored at −80°C. The protein concentration was measured using the Lowry protocol.
Chromatin immunoprecipitation.
Chromatin precipitation was performed using an EpiTect ChIP OneDay kit (Qiagen) in accordance with the manufacturer’s instructions. In brief, C2C12 cells were grown in 10-cm dishes, fixed in 1% paraformaldehyde, and then lysed at confluence (proliferation) and 48 h after serum withdrawal (differentiation) or after 48 h of stimulation with TNF-α. Chromatin was sheared by sonication on ice to obtain 300- to 500-bp chromatin lengths, which were confirmed by 1.2% agarose gel separation. DNA was immunoprecipitated using SP1 antibody from Millipore (catalog no. 17-601) or isotype control and purified per protocol. Quantification of SP1-bound DNA was determined by qPCR using primers directed against the Fbxl2 promoter region (see Table S2). Analysis of the ChIP-qPCR data was performed using SP1 relative to the input fraction to normalize for background levels and input chromatin. Visualization of the PCR-amplified Fbxl2 promoter region from ChIP fractions was performed using 0.8% agarose gels. In separate experiments, C2C12 cells were transfected with plasmids expressing V5-tagged SP1 wild type, SP1-T739A, and SP1-T739D for 48 h. The cells were fixed and sonicated as described above. DNA was immunoprecipitated using isotype control, V5 (Thermo Fisher; R960-25), or STAT-1 (Cell Signaling; 9172P) antibody according to protocol and quantified via qPCR as described above. The Fbxl2 promoter region expression via qPCR was normalized to nonspecific genomic regions spanning the promoter of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene.
Electrophoretic mobility shift assays.
Ten micrograms of nuclear extracts was incubated with 2 μg poly(dI-dC) in a 20-μl volume of binding buffer containing 50 mmol/liter HEPES (pH 7.9), 6 mmol/liter MgCl2, 50 mmol/liter KCl, 5 mmol/liter dithiothreitol (DTT), 100 μg/ml BSA, and 0.01% NP-40. Biotinylated and unlabeled synthetic oligonucleotides were obtained from IDT, annealed into double-stranded DNA (dsDNA), added to the reaction mixture, and incubated for 20 min at room temperature. In competition experiments, nuclear extracts were preincubated for 10 min with a 10-fold molar excess of unlabeled competitor oligonucleotide. The DNA-protein complexes were resolved on a 6% polyacrylamide gel at 100 V using Tris-buffered EDTA (TBE) electrophoresis buffer. The DNA complexes were transferred to nylon membranes at 30 V for 90 min, blocked with blocking solution (730 mg NaCl, 240 mg Na2HPO4, 100 mg NaH2PO4, 5 g SDS), incubated with streptavidin for 1 h, and imaged after the addition of a chemiluminescent substrate.
Statistical analysis.
Descriptive statistics are reported with means and standard errors of the mean (SEM); a P value of <0.05 was indicative of significance. All analyses were performed using one-way analysis of variance (ANOVA) or an unpaired t test to compare means with Prism version 6.0 for Mac OS (GraphPad Software, La Jolla, CA).
Accession number(s).
All transcriptome sequencing (RNA-seq) data have been uploaded to NCBI’s Gene Expression Omnibus (GEO), series number GSE120600.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Donna Stoltz at the University of Pittsburgh Center for Biological Imaging. MF20 was deposited at the DSHB by D. A. Fischman (DSHB hybridoma product MF20); PAX7 was deposited at the DSHB by A. Kawakami (DSHB hybridoma product PAX7); Pax3 was deposited at the DSHB by C. P. Ordahl (DSHB hybridoma product Pax3). We are grateful for careful editorial suggestions from Anuradha Ray, Prabir Ray, and Janet S. Lee.
The work was supported by a Merit Review Award from the U.S. Department of Veterans Affairs and National Institutes of Health R01 grants HL096376, HL097376, HL098174, HL081784, 1UH2HL123502, and P01 HL114453 (to R.K.M.) and a CHEST Foundation grant in COPD (to M.O.B).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. 2004. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol 166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin H, Price F, Rudnicki MA. 2013. Satellite cells and the muscle stem cell niche. Physiol Rev 93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, Gherardi RK, Chazaud B. 2007. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med 204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lassar AB, Skapek SX, Novitch B. 1994. Regulatory mechanisms that coordinate skeletal muscle differentiation and cell cycle withdrawal. Curr Opin Cell Biol 6:788–794. doi: 10.1016/0955-0674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 5.Skapek SX, Rhee J, Spicer DB, Lassar AB. 1995. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science 267:1022–1024. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- 6.Walsh K, Perlman H. 1997. Cell cycle exit upon myogenic differentiation. Curr Opin Genet Dev 7:597–602. doi: 10.1016/s0959-437x(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 7.Tidball JG. 2005. Inflammatory processes in muscle injury and repair. Am J Physiol Regul Integr Comp Physiol 288:R345–R353. doi: 10.1152/ajpregu.00454.2004. [DOI] [PubMed] [Google Scholar]
- 8.Ladner KJ, Caligiuri MA, Guttridge DC. 2003. Tumor necrosis factor-regulated biphasic activation of NF-kappa B is required for cytokine-induced loss of skeletal muscle gene products. J Biol Chem 278:2294–2303. doi: 10.1074/jbc.M207129200. [DOI] [PubMed] [Google Scholar]
- 9.Torrente Y, El Fahime E, Caron NJ, Del Bo R, Belicchi M, Pisati F, Tremblay JP, Bresolin N. 2003. Tumor necrosis factor-alpha (TNF-alpha) stimulates chemotactic response in mouse myogenic cells. Cell Transplant 12:91–100. doi: 10.3727/000000003783985115. [DOI] [PubMed] [Google Scholar]
- 10.Lolmede K, Campana L, Vezzoli M, Bosurgi L, Tonlorenzi R, Clementi E, Bianchi ME, Cossu G, Manfredi AA, Brunelli S, Rovere-Querini P. 2009. Inflammatory and alternatively activated human macrophages attract vessel-associated stem cells, relying on separate HMGB1- and MMP-9-dependent pathways. J Leukoc Biol 85:779–787. doi: 10.1189/jlb.0908579. [DOI] [PubMed] [Google Scholar]
- 11.Li Y-P. 2003. TNF-alpha is a mitogen in skeletal muscle. Am J Physiol Cell Physiol 285:C370–C376. doi: 10.1152/ajpcell.00453.2002. [DOI] [PubMed] [Google Scholar]
- 12.Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS. 1999. NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol 19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueck T, Berger F, Buechsler I, Valchanova RS, Landuzzi L, Lollini P-L, Klingel K, Munz B. 2011. TRAF6 regulates proliferation and differentiation of skeletal myoblasts. Differentiation 81:99–106. doi: 10.1016/j.diff.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS. 2000. NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science 289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- 15.Miller SC, Ito H, Blau HM, Torti FM. 1988. Tumor necrosis factor inhibits human myogenesis in vitro. Mol Cell Biol 8:2295–2301. doi: 10.1128/mcb.8.6.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szalay K, Rázga Z, Duda E. 1997. TNF inhibits myogenesis and downregulates the expression of myogenic regulatory factors myoD and myogenin. Eur J Cell Biol 74:391–398. [PubMed] [Google Scholar]
- 17.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 18.Tintignac LA, Lagirand J, Batonnet S, Sirri V, Leibovitch MP, Leibovitch SA. 2005. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J Biol Chem 280:2847–2856. doi: 10.1074/jbc.M411346200. [DOI] [PubMed] [Google Scholar]
- 19.Csibi A, Leibovitch MP, Cornille K, Tintignac LA, Leibovitch SA. 2009. MAFbx/Atrogin-1 controls the activity of the initiation factor eIF3-f in skeletal muscle atrophy by targeting multiple C-terminal lysines. J Biol Chem 284:4413–4421. doi: 10.1074/jbc.M807641200. [DOI] [PubMed] [Google Scholar]
- 20.Cai D, Frantz JD, Tawa NE, Melendez PA, Oh B-C, Lidov HGW, Hasselgren P-O, Frontera WR, Lee J, Glass DJ, Shoelson SE. 2004. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell 119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 21.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. 2000. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351–361. doi: 10.1016/S0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 22.Sandri M. 2008. Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. 2008. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell 31:918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang W-L, Wang J, Chan C-H, Lee S-W, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, Lin H-K. 2009. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, Shevde NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M, Du K, Singh S, Pike JW, Darnay BG, Choi Y, Wu H. 2002. Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 26.Paul PK, Gupta SK, Bhatnagar S, Panguluri SK, Darnay BG, Choi Y, Kumar A. 2010. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol 191:1395–1411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao F, Wang H, Fu X, Li Y, Wu Z. 2012. TRAF6 promotes myogenic differentiation via the TAK1/p38 mitogen-activated protein kinase and Akt pathways. PLoS One 7:e34081. doi: 10.1371/journal.pone.0034081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hindi SM, Kumar A. 2016. TRAF6 regulates satellite stem cell self-renewal and function during regenerative myogenesis. J Clin Invest 126:151–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen BB, Coon TA, Glasser JR, McVerry BJ, Zhao J, Zhao Y, Zou C, Ellis B, Sciurba FC, Zhang Y, Mallampalli RK. 2013. A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat Immunol 14:470–479. doi: 10.1038/ni.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen BB, Glasser JR, Coon TA, Mallampalli RK. 2011. FBXL2 is a ubiquitin E3 ligase subunit that triggers mitotic arrest. Cell Cycle 10:3487–3494. doi: 10.4161/cc.10.20.17742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen BB, Glasser JR, Coon TA, Mallampalli RK. 2012. F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest. Oncogene 31:2566–2579. doi: 10.1038/onc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen BB, Glasser JR, Coon TA, Mallampalli RK. 2013. Skp-cullin-F box E3 ligase component FBXL2 ubiquitinates Aurora B to inhibit tumorigenesis. Cell Death Dis 4:e759. doi: 10.1038/cddis.2013.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuchay S, Duan S, Schenkein E, Peschiaroli A, Saraf A, Florens L, Washburn MP, Pagano M. 2013. FBXL2- and PTPL1-mediated degradation of p110-free p85β regulatory subunit controls the PI(3)K signalling cascade. Nat Cell Biol 15:472–480. doi: 10.1038/ncb2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchay S, Giorgi C, Simoneschi D, Pagan J, Missiroli S, Saraf A, Florens L, Washburn MP, Collazo-Lorduy A, Castillo-Martin M, Cordon-Cardo C, Sebti SM, Pinton P, Pagano M. 2017. PTEN counteracts FBXL2 to promote IP3R3- and Ca2+-mediated apoptosis limiting tumour growth. Nature 546:554–558. doi: 10.1038/nature22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell ID, Grancell AS, Sorger PK. 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J Cell Biol 145:933–950. doi: 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larrieu D, Brunet M, Vargas C, Hanoun N, Ligat L, Dagnon L, Lulka H, Pommier RM, Selves J, Jády BE, Bartholin L, Cordelier P, Dufresne M, Torrisani J. 2020. The E3 ubiquitin ligase TRIP12 participates in cell cycle progression and chromosome stability. Sci Rep 10:789. doi: 10.1038/s41598-020-57762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan G, Lenhard B. 2016. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 32:1555–1556. doi: 10.1093/bioinformatics/btw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, Baranasic D, Arenillas DJ, Sandelin A, Vandepoele K, Lenhard B, Ballester B, Wasserman WW, Parcy F, Mathelier A. 2018. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan NY, Khachigian LM. 2009. Sp1 phosphorylation and its regulation of gene transcription. Mol Cell Biol 29:2483–2488. doi: 10.1128/MCB.01828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuang J-Y, Wang Y-T, Yeh S-H, Liu Y-W, Chang W-C, Hung J-J. 2008. Phosphorylation by c-Jun NH2-terminal kinase 1 regulates the stability of transcription factor Sp1 during mitosis. Mol Biol Cell 19:1139–1151. doi: 10.1091/mbc.e07-09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuang J-Y, Wang S-A, Yang W-B, Yang H-C, Hung C-Y, Su T-P, Chang W-C, Hung J-J. 2012. Sp1 phosphorylation by cyclin-dependent kinase 1/cyclin B1 represses its DNA-binding activity during mitosis in cancer cells. Oncogene 31:4946–4959. doi: 10.1038/onc.2011.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, Ye W, Wu J, Liu L, Yang L, Gao L, Chen B, Zhang F, Yang H, Li Y. 2015. Sp1-CD147 positive feedback loop promotes the invasion ability of ovarian cancer. Oncol Rep 34:67–76. doi: 10.3892/or.2015.3999. [DOI] [PubMed] [Google Scholar]
- 43.Zammit PS. 2017. Function of the myogenic regulatory factors Myf5, MyoD, myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol 72:19–32. doi: 10.1016/j.semcdb.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Li Y-P, Chen Y, John J, Moylan J, Jin B, Mann DL, Reid MB. 2005. TNF-alpha acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. FASEB J 19:362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagata D, Suzuki E, Nishimatsu H, Satonaka H, Goto A, Omata M, Hirata Y. 2001. Transcriptional activation of the cyclin D1 gene is mediated by multiple cis-elements, including SP1 sites and a cAMP-responsive element in vascular endothelial cells. J Biol Chem 276:662–669. doi: 10.1074/jbc.M005522200. [DOI] [PubMed] [Google Scholar]
- 46.Lin H-M, Pestell RG, Raz A, Kim H-R. 2002. Galectin-3 enhances cyclin D(1) promoter activity through SP1 and a cAMP-responsive element in human breast epithelial cells. Oncogene 21:8001–8010. doi: 10.1038/sj.onc.1205820. [DOI] [PubMed] [Google Scholar]
- 47.Han S, Ritzenthaler JD, Rivera HN, Roman J. 2005. Peroxisome proliferator-activated receptor-gamma ligands suppress fibronectin gene expression in human lung carcinoma cells: involvement of both CRE and Sp1. Am J Physiol Lung Cell Mol Physiol 289:L419–L428. doi: 10.1152/ajplung.00002.2005. [DOI] [PubMed] [Google Scholar]
- 48.Mancini DN, Singh SM, Archer TK, Rodenhiser DI. 1999. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene 18:4108–4119. doi: 10.1038/sj.onc.1202764. [DOI] [PubMed] [Google Scholar]
- 49.Denson LA, Menon RK, Shaufl A, Bajwa HS, Williams CR, Karpen SJ. 2001. TNF-alpha downregulates murine hepatic growth hormone receptor expression by inhibiting Sp1 and Sp3 binding. J Clin Invest 107:1451–1458. doi: 10.1172/JCI10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, Wang W, Wesley RA, Danner RL. 1999. A Sp1 binding site of the tumor necrosis factor alpha promoter functions as a nitric oxide response element. J Biol Chem 274:33190–33193. doi: 10.1074/jbc.274.47.33190. [DOI] [PubMed] [Google Scholar]
- 51.Bouwman P, Philipsen S. 2002. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol 195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 52.Deniaud E, Baguet J, Chalard R, Blanquier B, Brinza L, Meunier J, Michallet M-C, Laugraud A, Ah-Soon C, Wierinckx A, Castellazzi M, Lachuer J, Gautier C, Marvel J, Leverrier Y. 2009. Overexpression of transcription factor Sp1 leads to gene expression perturbations and cell cycle inhibition. PLoS One 4:e7035. doi: 10.1371/journal.pone.0007035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pascal E, Tjian R. 1991. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev 5:1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- 54.Hong JS, Kim S-W, Koo JS. 2008. Sp1 upregulates cAMP response element-binding protein expression during retinoic acid-induced mucous differentiation of normal human bronchial epithelial cells. Biochem J 410:49–61. doi: 10.1042/BJ20070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grayson J, Bassel-Duby R, Williams RS. 1998. Collaborative interactions between MEF-2 and Sp1 in muscle-specific gene regulation. J Cell Biochem 70:366–375. doi:. [DOI] [PubMed] [Google Scholar]
- 56.Perkins ND, Agranoff AB, Pascal E, Nabel GJ. 1994. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol 14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barth N, Langmann T, Schölmerich J, Schmitz G, Schäffler A. 2002. Identification of regulatory elements in the human adipose most abundant gene transcript-1 (apM-1) promoter: role of SP1/SP3 and TNF-alpha as regulatory pathways. Diabetologia 45:1425–1433. doi: 10.1007/s00125-002-0895-5. [DOI] [PubMed] [Google Scholar]
- 58.Das A, Acharya S, Gottipati KR, McKnight JB, Chandru H, Alcorn JL, Boggaram V. 2011. Thyroid transcription factor-1 (TTF-1) gene: identification of ZBP-89, Sp1, and TTF-1 sites in the promoter and regulation by TNF-α in lung epithelial cells. Lung Cell Mol Physiol 301:L427–L440. doi: 10.1152/ajplung.00090.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Files DC, D'Alessio FR, Johnston LF, Kesari P, Aggarwal NR, Garibaldi BT, Mock JR, Simmers JL, DeGorordo A, Murdoch J, Willis MS, Patterson C, Tankersley CG, Messi ML, Liu C, Delbono O, Furlow JD, Bodine SC, Cohn RD, King LS, Crow MT. 2012. A critical role for muscle ring finger-1 in acute lung injury-associated skeletal muscle wasting. Am J Respir Crit Care Med 185:825–834. doi: 10.1164/rccm.201106-1150OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Remels AHV, Gosker HR, Langen RCJ, Schols A. 2013. The mechanisms of cachexia underlying muscle dysfunction in COPD. J Appl Physiol 114:1253–1262. doi: 10.1152/japplphysiol.00790.2012. [DOI] [PubMed] [Google Scholar]
- 61.Tisdale MJ. 2002. Cachexia in cancer patients. Nat Rev Cancer 2:862–871. doi: 10.1038/nrc927. [DOI] [PubMed] [Google Scholar]
- 62.Beutler B, Cerami A. 1986. Cachectin and tumour necrosis factor as two sides of the same biological coin. Nature 320:584–588. doi: 10.1038/320584a0. [DOI] [PubMed] [Google Scholar]
- 63.GTEx Consortium, Laboratory, Data Analysis andCoordinating Center, Battle A, Brown CD, Engelhardt BE, Montgomery SB. 2017. Genetic effects on gene expression across human tissues. Nature 550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coussens LM, Werb Z. 2002. Inflammation and cancer. Nature 420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mallampalli RK, Coon TA, Glasser JR, Wang C, Dunn SR, Weathington NM, Zhao J, Zou C, Zhao Y, Chen BB. 2013. Targeting F box protein Fbxo3 to control cytokine-driven inflammation. J Immunol 191:5247–5255. doi: 10.4049/jimmunol.1300456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Danoviz ME, Yablonka-Reuveni Z. 2012. Skeletal muscle satellite cells: background and methods for isolation and analysis in a primary culture system. Methods Mol Biol 798:21–52. doi: 10.1007/978-1-61779-343-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 68.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.