Abstract
Knowledge about established breast carcinogens can support improved and modernized toxicological testing methods by identifying key mechanistic events. Ionizing radiation (IR) increases the risk of breast cancer, especially for women and for exposure at younger ages, and evidence overall supports a linear dose–response relationship. We used the Adverse Outcome Pathway (AOP) framework to outline and evaluate the evidence linking ionizing radiation with breast cancer from molecular initiating events to the adverse outcome through intermediate key events, creating a qualitative AOP. We identified key events based on review articles, searched PubMed for recent literature on key events and IR, and identified additional papers using references. We manually curated publications and evaluated data quality. Ionizing radiation directly and indirectly causes DNA damage and increases production of reactive oxygen and nitrogen species (RONS). RONS lead to DNA damage and epigenetic changes leading to mutations and genomic instability (GI). Proliferation amplifies the effects of DNA damage and mutations leading to the AO of breast cancer. Separately, RONS and DNA damage also increase inflammation. Inflammation contributes to direct and indirect effects (effects in cells not directly reached by IR) via positive feedback to RONS and DNA damage, and separately increases proliferation and breast cancer through pro-carcinogenic effects on cells and tissue. For example, gene expression changes alter inflammatory mediators, resulting in improved survival and growth of cancer cells and a more hospitable tissue environment. All of these events overlap at multiple points with events characteristic of “background” induction of breast carcinogenesis, including hormone-responsive proliferation, oxidative activity, and DNA damage. These overlaps make the breast particularly susceptible to ionizing radiation and reinforce that these biological activities are important characteristics of carcinogens. Agents that increase these biological processes should be considered potential breast carcinogens, and predictive methods are needed to identify chemicals that increase these processes. Techniques are available to measure RONS, DNA damage and mutation, cell proliferation, and some inflammatory proteins or processes. Improved assays are needed to measure GI and chronic inflammation, as well as the interaction with hormonally driven development and proliferation. Several methods measure diverse epigenetic changes, but it is not clear which changes are relevant to breast cancer. In addition, most toxicological assays are not conducted in mammary tissue, and so it is a priority to evaluate if results from other tissues are generalizable to breast, or to conduct assays in breast tissue. Developing and applying these assays to identify exposures of concern will facilitate efforts to reduce subsequent breast cancer risk.
Electronic supplementary material
The online version of this article (10.1007/s00204-020-02752-z) contains supplementary material, which is available to authorized users.
Keywords: Ionizing radiation, Breast carcinogenesis, DNA damage, Inflammation, Oxidative stress, Cell proliferation, Genomic instability, Epigenetics, Hormones, Characteristics of carcinogens, Adverse outcome pathway, Mechanisms, Key events, In vitro assays, In vivo assays
Introduction
Breast cancer imposes a significant burden on women worldwide and is an important focus for prevention. It is the most common invasive cancer in women with the highest rates found in North America and Europe (Ervik et al. 2016), and incidence is increasing globally (Forouzanfar et al. 2011). Around 60% of all cancers are related to non-heritable factors (Colditz and Wei 2012; Moller et al. 2016; Ronckers et al. 2005), so significant opportunities exist to reduce risk. Understanding breast carcinogenesis from an established breast carcinogen will help to identify early effect markers that can be used in screening chemicals and drugs to identify other breast carcinogens, prevent exposures, and reduce future breast cancers.
Ionizing radiation (IR) is a well-studied carcinogen that increases the risk of breast cancer in people (Bijwaard et al. 2010; Eidemuller et al. 2015; Henderson et al. 2010; Little and McElvenny 2017; Ma et al. 2008; Moskowitz et al. 2014; Neta et al. 2012) and mammary gland tumors in rodents (Imaoka et al. 2009; Rivina et al. 2016; Russo 2015; Wagner 2004), so we selected it for detailed analysis using the Adverse Outcome Pathway (AOP) framework. AOPs provide a structure and methodology for the systematic organization of mechanistic data into a series of discrete measurable events leading from the initial molecular perturbation by a stressor to an adverse outcome of regulatory relevance (OECD 2017; Villeneuve et al. 2014). AOPs are simplified representations of complex disease processes that identify intermediate events that are essential, biologically relevant, and testable.
This paper will identify and evaluate the evidence and dose–response for key events leading from IR exposure to breast cancer, and describe available assays and opportunities for measuring these events. Hormone exposure during development and into adulthood influences the risk of spontaneous breast cancer and breast cancer following IR, so this paper will review the role of hormones in IR carcinogenesis, and the intersection of hormonal effects with the key events identified here.
Methods
We used the approach outlined in Kushman and Kraft et al. (Kushman et al. 2013) to construct this AOP. We used review papers (Barcellos-Hoff and Kleinberg 2013; Barcellos-Hoff and Mao 2016; Barcellos-Hoff and Nguyen 2009; Cadet et al. 2017; Committee to Assess Health Risks from Exposure to Low Levels of Ionizing Radiation 2006; Hanahan and Weinberg 2011; Imaoka et al. 2009; Kadhim et al. 2013; Mukherjee et al. 2014; Nguyen et al. 2011a; Ravanat et al. 2014; Ruhm et al. 2016; Smith et al. 2016; Sridharan et al. 2015) to identify the mechanisms by which exposure to ionizing radiation may cause breast cancer. We then used a multipronged approach to identify literature relating to each mechanism. We conducted comprehensive literature searches of PubMed for each mechanism or mechanisms in combination with ionizing radiation focusing on papers since 2006, and used review and primary literature references as well as PubMed and Google Scholar queries to identify additional publications. The resulting publications were manually curated to identify human or animal in vivo or in vitro peer-reviewed primary publications relevant to ionizing radiation and to the events of interest. Papers were additionally evaluated for data quality and clarity before inclusion. Specifically, we looked for studies with three or more samples in each group, and excluded studies with inadequate controls or unclear methods. We ultimately extracted data from over 500 papers. Supplemental Table #1 summarizes the key studies for each topic.
The AOP was constructed following Organization for Economic Co-operation and Development (OECD) guidelines (OECD 2017, 2018b) entered into the OECD AOP Wiki as two separate AOPs for RONS or DNA damage, and submitted to the OECD AOP Development Programme for inclusion in their work plan. Readers may look there for additional details omitted here because of space constraints.
We evaluated the strength of evidence based on the essentiality of each key event to the pathway and to breast cancer using the modified Bradford Hill considerations as outlined in the OECD AOP handbook (Becker et al. 2015; OECD 2017, 2018b). In other words, do studies show that interfering with the key event reduces the occurrence of downstream events and increasing the key event likewise increases downstream events? We highlight the quality and amount of evidence, noting missing or conflicting evidence and identifying factors not addressed in this AOP. We compiled assays available for each of the key events and outcomes and considered their generalizability to mammary tissue.
The IR and breast cancer AOP
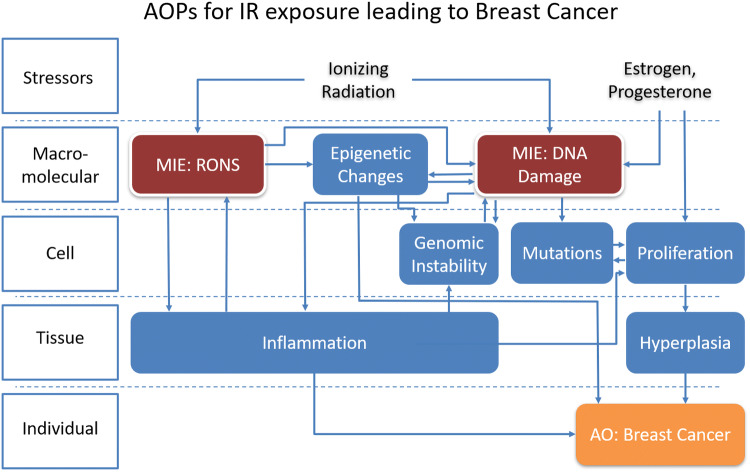
The sequence of key events by which ionizing radiation leads to breast cancer is shown in Fig. 1. IR induces two molecular initiating events via oxidation of DNA and proteins: an increase in DNA damage and an increase in reactive oxygen and nitrogen species (RONS). DNA damage leads to increased genomic instability (GI), mutations, proliferation (including clonal selection), hyperplasia, and risk of breast cancer. The increase in RONS also contributes to this pathway via increasing epigenetic changes, GI and DNA damage. Both RONS and DNA damage also initiate multiple inflammatory reactions in the tissue. Inflammation contributes to direct and indirect effects (effects in cells not directly reached by IR) via positive feedback to RONS, DNA damage, and GI and separately increases breast cancer through increased proliferation of cells and tumorigenesis and invasion.
Fig. 1.
Adverse outcome pathway showing the key events linking the stressor ionizing radiation to breast cancer. Arrows indicate directionality of events: e.g., an increase in the upstream event leads to an increase in the downstream event. The intersection of additional stressors (estrogen and progesterone) with this pathway is included on the right. MIE molecular initiating event, the original action caused by the stressor IR in tissue that leads to subsequent events. AO adverse outcome. While this pathway is focused on breast cancer as an adverse outcome, DNA damage and GI, mutations, and hyperplasia can be considered adverse outcomes in their own right
Ionizing radiation as stressor
Exposure to ionizing radiation (IR) comes from natural and industrial sources such as cosmic rays, radon, or radioactive fuels and wastes and from medical radiation methods such as X-ray imaging, mammography and CT scans, and radiation therapy for cancers. The pattern of energy transferred by IR to matter (linear energy transfer per unit length or LET) (1970) varies between sources. Lower or no LET IR such as mammographic X-rays and some radiation therapies sparsely deposit many individual excitations or small clusters of excitations of low energy (Goodhead 1988) deep into tissue. In contrast, high LET such as alpha particles from radioactive isotopes readily transfer their energy (Goodhead 1994) and, therefore, deposit dense clusters of energy closer to the tissue surface (Goodhead 1988). These different energy deposition patterns contribute to differences in radiation effects including the pattern of DNA damage.
Breast cancer as adverse outcome (AO)
Exposure to ionizing radiation is a well-established risk factor for many cancers including breast cancer (Ozasa et al. 2012; Preston et al. 2007). Women exposed to the atomic bomb in Japan (exposure is mostly low LET gamma IR with some inhaled high LET alpha and beta particles) (Little and McElvenny 2017), or to therapeutic radiation for benign disorders (Eidemuller et al. 2015), childhood cancer (Henderson et al. 2010; Moskowitz et al. 2014), or contralateral breast cancer (Neta et al. 2012) (often low LET X-rays but also higher LET beta radiation), or to frequent chest X-rays including TB fluoroscopy (Bijwaard et al. 2010; Ma et al. 2008) show a significant increase in breast cancer with radiation exposure. Ionizing radiation also increases mammary tumors in rodents, and sensitivity varies by species and strain (Bijwaard et al. 2010; Ma et al. 2008). See Supplemental Tables 2 and 3 for a summary of key studies examining ionizing radiation and breast and solid cancers in epidemiology and rodent studies, including interactions between IR and hormonal manipulation.
Why breast?
The key events described here are likely relevant to all tissues after exposure to IR, and are particularly relevant to the breast. While ionizing radiation causes many kinds of cancers including leukemia, lung, bladder, and thyroid cancers (Committee to Assess Health Risks from Exposure to Low Levels of Ionizing Radiation 2006; Preston et al. 2007), breast cancers are among the cancers most increased by exposure to ionizing radiation (Preston et al. 2007).
The lengthy and hormone-dependent developmental trajectory of the breast is likely to be a major factor in its susceptibility to breast cancer in general and to breast cancer from ionizing radiation in particular. Although at first examination, breast cancer from ionizing radiation and hormones involve very different processes, in fact the hormone-dependent and ionizing radiation pathways of carcinogenesis intersect at multiple points along the trajectory of breast development and function, leaving the hormone-exposed breast especially vulnerable to radiation.
One major mechanism promoting breast cancer from ionizing radiation is the proliferation of breast stem cells. Most breast cancers form from epithelial cells, and the growth of these cells in the breast is limited until hormones rise during puberty, when stem cells proliferate to form ductal structures (Hinck and Silberstein 2005; Sternlicht et al. 2005). Stem cells are considered important to initiation because of their long life and capacity to pass on mutations to many progeny (Imaoka et al. 2009; Russo 2015). Breast tissue is responsive to estrogen and progesterone, reproductive hormones that rise at puberty and stimulate cellular proliferation with each reproductive cycle and in pregnancy. These hormonal proliferative cycles increase the risk of cancer in breast tissue (Brisken et al. 2015). IR increases the long-term proliferation of stem cells in pubertal but not adult mammary gland (Datta et al. 2012a; Nguyen et al. 2011b; Snijders et al. 2012; Suman et al. 2012; Tang et al. 2014). Replication of stem cells in the IR-exposed breast is, therefore, particularly elevated during puberty, likely contributing to the increased susceptibility to breast cancer from IR at this age.
Development continues to a lesser degree after puberty with each menstrual cycle until pregnancy, when another major hormone-dependent proliferation and subsequent differentiation and decline of stem cells is coupled with a decline in hormone-sensing epithelial cells (Anderson et al. 2007; Dall et al. 2017; Oakes et al. 2006b). This pregnancy-related decrease in hormone-sensing and stem cells does not occur in first pregnancies at older ages and may explain the lack of protection against spontaneous breast cancer afforded by the first parity in older women (Dall et al. 2017). This theory underlies the current efforts to prevent breast cancers by induction of terminal differentiation (mimicking pregnancy) in teenagers (Santucci-Pereira et al. 2013).
Another special vulnerability of the breast to IR is a byproduct of proliferation:mutations. Replication itself increases the likelihood of mutations, which add to mutations arising from IR and increase the likelihood of oncogenic transformation (Atashgaran et al. 2016). Furthermore, the high replication rate of mammary stem cells during puberty and pregnancy increases reliance on homologous recombination pathways (Kass et al. 2016) which shift to error-prone non-homologous end joining to respond to DNA damage from IR (Chang et al. 2015). Long-term disruption of these homologous repair (HR) processes by polymorphisms in genes like BRCA or by IR-induced mutation can also increase mutation rates and increase tumorigenesis (Mahdi et al. 2018). The consequence of mutations in stem cells is significant, since these cells can clonally expand to generate many mutated progeny. However, errors in stem cell division may not be the sole or primary factor driving cancer from radiation, since excess cancer risk for solid cancers at different sites from the atomic bomb is not clearly related to the number of stem cell divisions at that site (Tomasetti et al. 2017).
The elevated estrogen associated with normal development and the estrous cycle may also have direct effects that further complement the carcinogenic effects of IR. Estrogen directly increases oxidative stress in virgin (but not parous) mice (Yuan et al. 2016), interferes with DNA repair (Li et al. 2014; Pedram et al. 2009) increases DNA damage and mutations (Mailander et al. 2006), and increases TGF-β (Jerry et al. 2010). These effects are seen at physiological concentrations of estrogen, and each creates the potential for additive or synergistic interactions.
Inflammation from the estrous cycle may also contribute to tumorigenesis following IR. Cytokines and macrophages play an integral role in mammary gland development and ductal elaboration, with alternating inflammatory, immune surveillance, and phagocytic activity occurring over each estrous cycle (Atashgaran et al. 2016; Brady et al. 2016; Hodson et al. 2013). This inflammation could theoretically increase IR-induced DNA damage and mutations and promote tumorigenic and invasive characteristics.
The enhancement of IR-induced tumorigenesis by the estrous cycle may be replicated or further enhanced by exogenous endocrine disrupting chemicals. Indeed, evidence suggests that BPA (and presumably other estrogenic chemicals) exposure in utero can increase the mammary gland’s response to progesterone during puberty (Brisken et al. 2015). This enhancement would presumably also increase the risk of breast cancer from ionizing radiation, which rises with estrogen exposure and the number of menstrual cycles (De Bruin et al. 2009; Inskip et al. 2009; Moskowitz et al. 2014; Sigurdson et al. 2009; Travis et al. 2003).
Breast cancer risk factors
Younger age of exposure, exposure to estrogen and progesterone, pregnancy, and genetic susceptibility influence breast cancer risk from IR in people (Berrington de Gonzalez et al. 2010; Boice et al. 1991; Land et al. 2003; Ma et al. 2008; Miller et al. 1989; Preston et al. 2007; Stovall et al. 2008). In rodents, risk is highest for IR exposures during mammary gland development and puberty (Imaoka et al. 2011, 2013, 2017) and lower for embryonic, adult (Imaoka et al. 2011, 2013), and post-estrous rats (Bartstra et al. 1998).
While IR exposure during breast development increases risk, pregnancy or parity is protective against breast cancer from later radiation in people (Brooks et al. 2012a; Land et al. 1994), similar to spontaneous and chemical carcinogen-induced mammary tumors in rodents (Britt et al. 2007; Dall et al. 2017; Imaoka et al. 2009; Rajkumar et al. 2007; Russo 2015). In rodents, IR exposure during or shortly after pregnancy increases mammary cancer (Holtzman et al. 1982; Inano et al. 1991, 1996b; McDaniel et al. 2006; Suzuki et al. 1994), consistent with increased susceptibility due to development and proliferation in the mammary gland during pregnancy. There is also a “risk bump” of pregnancy-associated breast cancer observed in humans (Lambe et al. 1994).
As with spontaneous breast cancer, the reproductive hormones estrogen and progesterone increase the risk of breast cancer following ionizing radiation. Breast cancer rates following IR are lower in women who undergo premature menopause (De Bruin et al. 2009; Inskip et al. 2009; Moskowitz et al. 2014; Travis et al. 2003). Genetic variation in estrogen signaling also affects risk (Sigurdson et al. 2009), Similarly, exposure to estrogen or the synthetic estrogen diethylstilbestrol (DES) is associated with more tumors (particularly adenocarcinomas) in rats following IR (Broerse et al. 1987; Holtzman et al. 1979, 1981; Inano et al. 1991; Segaloff and Maxfield 1971; Shellabarger et al. 1976; Solleveld et al. 1986) including male (Inano et al. 1996a) and ovariectomized rats (Inano et al. 1995; Yamanouchi et al. 1995). Conversely, ovariectomy (Clifton et al. 1985; Cronkite et al. 1960; Solleveld et al. 1986) and the anti-estrogen tamoxifen (Lemon et al. 1989; Peterson et al. 2005; Welsch et al. 1981) reduce tumors from IR.
Breast cancer risk from IR in postmenopausal atomic bomb survivors increases with and may be partially mediated by increased serum estrogen (Grant et al. 2018), which may in turn be mediated by inflammatory factors (Frasor et al. 2008; Salama et al. 2009). In addition, one study reports that IR can increase circulating estrogen in rodents (Suman et al. 2012). To our knowledge, no publications have attempted to replicate these findings. Progesterone also promotes mammary carcinogenesis after IR in rodents, but its effect depends on the developmental state of the mammary gland: moderate doses promote carcinogenesis in the mature but not immature mammary gland, but high doses can be protective, possibly by differentiating the gland in a manner akin to pregnancy (Inano et al. 1995; Takabatake et al. 2018; Yamanouchi et al. 1995).
IR may increase the risk of ER- breast cancer in particular (Alkner et al. 2015; Castiglioni et al. 2007; Dores et al. 2010; Horst et al. 2014; Huang et al. 2000; Neta et al. 2012; VoPham et al. 2017), although some studies report no difference between risk of ER+ and ER− cancers (Ma et al. 2008; Miura et al. 2008). This effect depends on the presence of estrogen or developmental state: in animals, tumors formed after IR in the absence of estrogen (ovariectomized animals) are often ER−, while those formed in the presence of estrogen or DES are often ER + (Inano et al. 1995) and those formed in the presence of estrogen and progesterone are almost always ER + (Inano et al. 1995; Yamanouchi et al. 1995). Breast tumors can change from ER+ to ER− as the tumor advances or in response to ER antagonists (Lindstrom et al. 2012; Szostakowska et al. 2019).
An early study of women exposed to the atomic bomb suggested that a surge in early onset cancers arose from genetically susceptible populations (Land et al. 1993). Studies of genetic polymorphisms in people and strain variation in rodents support a contribution of genetic background to breast cancer risk from IR (Andrieu et al. 2006; Bernstein et al. 2010; Bernstein et al. 2013; Broeks et al. 2007; Brooks et al. 2012b; Imaoka et al. 2007; Millikan et al. 2005; Pijpe et al. 2012; Rivina et al. 2016; Shellabarger 1972; Sigurdson et al. 2009; Vogel and Turner 1982).
RONS (MIE)
Evidence of activation by IR
Reactive oxygen and nitrogen species (RONS) are oxygen- and nitrogen-based molecules that often contain or generate free radicals, including superoxide ([O2]•−), hydrogen peroxide (H2O2), hydroxyl radical ([OH]•), lipid peroxide (ROOH), nitric oxide ([NO]•, and peroxynitrite ([ONOO–]) (Dickinson and Chang 2011; Egea et al. 2017). RONS are generated in the course of cellular respiration, metabolism, cell signaling, and inflammation (Dickinson and Chang 2011; Egea et al. 2017).
We include both reactive oxygen (ROS) and nitrogen (RNS) species in this key event for several reasons. First, reactive species of both types interact and produce overlapping effects (Calcerrada et al. 2011; Mikkelsen and Wardman 2003). Second, because the most common methods used to detect their presence are fairly non-selective (Kalyanaraman et al. 2012; Mikkelsen and Wardman 2003) and thus the majority of evidence for their activation by IR does not distinguish between them. Third, evidence suggests that both ROS and RNS are produced after IR and are involved in the generation of downstream events (addressed below).
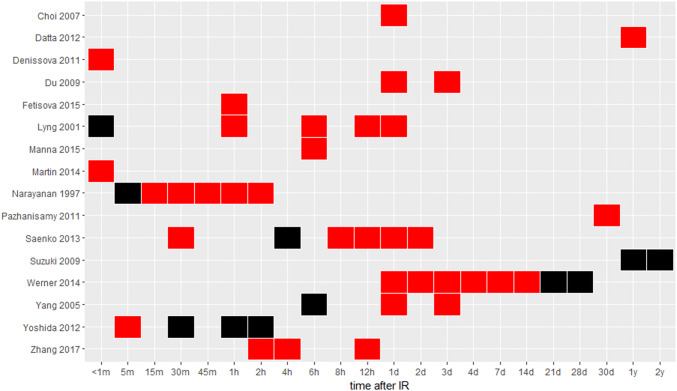
RONS levels increase at multiple time points in vitro and in vivo following IR indicating a persistent effect (Fig. 2) (Ameziane-El-Hassani et al. 2015; Choi et al. 2007; Das et al. 2014; Datta et al. 2012b; Denissova et al. 2012; Du et al. 2009; Lyng et al. 2001; Manna et al. 2015; Martin et al. 2014; Narayanan et al. 1997; Pazhanisamy et al. 2011; Saenko et al. 2013; Werner et al. 2014; Yang et al. 2005; Yoshida et al. 2012; Zhang et al. 2017). Ionizing radiation initially generates RONS by direct hydrolysis of water molecules. An early (15 min) and later (days to weeks) elevation in RONS is associated with increased NADPH-oxidase production of superoxide and H2O2 (Ameziane-El-Hassani et al. 2015; Narayanan et al. 1997). Intermediate (hours to days) and chronic RONS elevation has been associated with mitochondrial respiration (Datta et al. 2012b; Dayal et al. 2009; Saenko et al. 2013) driven by nitric oxide signals and cell division (Suzuki et al. 2009). IR-exposed cells show elevated oxidative activity up to a year after IR exposure of the animal (Datta et al. 2012b; Pazhanisamy et al. 2011). For RNS in particular, mitochondrial and NOS1-dependent NOS increase NO and peroxynitrate at 1–30 min after IR, peaking around 5 min (Kostyuk et al. 2012; Leach et al. 2002). Mitochondrial and NF-κB dependent NO concentrations increase again around 8–24 h (Dong et al. 2015; Ha et al. 2010; Saenko et al. 2013; Zhou et al. 2008). RONS (Buonanno et al. 2011; Lyng et al. 2001; Narayanan et al. 1997; Yang et al. 2005) including RNS (Shao et al. 2008; Zhou et al. 2008) is also increased in neighboring or bystander cells—cells not directly exposed to IR—via H2O2, NO, TNF-α, and TGF-β (Narayanan et al. 1997; Shao et al. 2008; Zhou et al. 2008) as well as COX2 and NF-κB activation and decreased SOD2 (Chai et al. 2013b; Tian et al. 2015; Zhou et al. 2008). Measures of RONS after IR in breast or breast cancer cells are rare, but generally support increased RONS minutes or days after IR (Bensimon et al. 2016; Jones et al. 2007; Leach et al. 2001).
Fig. 2.
Intracellular RONS at varying times after IR. RONS are measured with DCFH (dichlorodihydrofluorescein diacetate), a common but non-specific fluorescent indicator of intracellular oxidants. Red indicates elevated RONS; black indicates that RONS was measured but not elevated. Each reference is listed on the y-axis. Cell types are mostly embryonic or immortalized fibroblasts, keratinocytes, epithelial cells, and immune cells—none are mammary/breast (color figure online)
A few studies have measured RONS at multiple or lower doses (< 1 Gy) of ionizing radiation to establish a clear dose–response relationship, and those that have tested multiple doses performed their measures at a range of time points after IR, making it difficult to establish a clear narrative. Immediately after IR, one study at doses above 1 Gy finds that IR increases the number of cells with elevated RONS rather than the degree of RONS in each cell (Leach 2001) and another at doses below 1 Gy reports non-linear dose-dependence of RONS species with a maximum response around 0.4 Gy (Narayanan et al. 1997). RONS increases between 0.5 and 12 Gy IR at 1–2 days (Jones et al. 2007; Saenko et al. 2013) but plateaus at 1 Gy (Werner et al. 2014) at 7 days. The only study explicitly measuring reactive nitrogen species (RNS) at different doses of IR (in endothelial cells) also reports non-linear dose-dependence at 3 h, with a nadir around 0.1 Gy (Kostyuk et al. 2012). RONS increases dose-dependently in extracellular media (Driessens et al. 2009) and in bystander cells not directly exposed to IR (Buonanno et al. 2011; Narayanan et al. 1997).
Essentiality of RONS
Essentiality is high. RONS are sufficient to trigger subsequent key events in this AOP. Extracellularly applied or intracellularly generated ROS (which also facilitates the formation of RNS) are capable of creating DNA damage in vitro including base damage, single- and double-strand breaks, and chromosomal damage (Berdelle et al. 2011; Dahm-Daphi et al. 2000; Driessens et al. 2009; Gradzka and Iwanenko 2005; Ismail et al. 2005; Lorat et al. 2015; Nakamura et al. 2003; Oya et al. 1986; Stanicka et al. 2015) and mutations (Ameziane-El-Hassani et al. 2010; Sandhu and Birnboim 1997; Seager et al. 2012; Sharma et al. 2016). Similarly, decreased antioxidant activity and higher RONS are observed in cells with GI (Bensimon et al. 2016; Buonanno et al. 2011; Datta et al. 2012b; Dayal et al. 2008; Kim et al. 2006; Limoli and Giedzinski 2003; Limoli et al. 2003; Natarajan et al. 2007; Owens et al. 2012; Pazhanisamy et al. 2011; Thomas et al. 2012). RONS are also involved in the activation of inflammation (Nakao et al. 2008; Zhang et al. 2017) and epigenetic effects (Bernal et al. 2013; Galligan et al. 2014; Kloypan et al. 2015; Simone et al. 2009; Yang et al. 2014).
The strongest evidence for the essentiality of RONS in breast cancer from IR comes from studies, showing that reducing RONS with antioxidants including catalase or SOD or NOX/DUOX inhibitors also reduces downstream key events including DNA damage and GI in multiple types of irradiated cells and bystander cells in vivo and in vitro after IR (Ameziane-El-Hassani et al. 2010, 2015; Azzam et al. 2002; Bensimon et al. 2016; Choi et al. 2007; Dayal et al. 2008, 2009; Douki et al. 2006; Jones et al. 2007; Manna et al. 2015; Ozyurt et al. 2014; Pazhanisamy et al. 2011; Tartier et al. 2007; Tian et al. 2015; Weyemi et al. 2015; Winyard et al. 1992; Yang et al. 2007b, 2005), while inhibiting NO in particular inhibits DNA damage and mutations in bystander cells (Han et al. 2010; Tartier et al. 2007; Yang et al. 2007b; Zhou et al. 2008). Antioxidant activity also reduces the inflammatory response to IR in animals and cultured skin cells (Berruyer et al. 2004; Das et al. 2014; Ezz et al. 2018; Haddadi et al. 2017; Khan et al. 2015; Ozyurt et al. 2014; Sinha et al. 2011, 2012; Zetner et al. 2016; Zhang et al. 2017) and the epigenetic effects of IR in vitro (Bernal et al. 2013; Kloypan et al. 2015; Simone et al. 2009). RONS also drives proliferation directly (Han et al. 2010) and via cell death and compensatory proliferation following damage to DNA and important cellular macromolecules (Nishida et al. 2013).
None of the studies addressing RONS and inflammation are in mammary gland/breast, but a limited number of studies in mammary and breast cells do support the essentiality of RONS to DNA damage or cancer (Bensimon et al. 2016; Dutta et al. 2014) including after IR (Inano et al. 1999; Jones et al. 2007).
RONS act in multiple ways to promote downstream key events, not only directly damaging DNA and proteins but also modifying signal transduction pathways and epigenetic mechanisms (Gao and Schottker 2017; Mikkelsen and Wardman 2003). Examples include inflammatory pathways like TNF-α and IL-6, transcription factors like NF-κB, and methylation of histones (Eskiocak et al. 2010; Galligan et al. 2014; Kloypan et al. 2015; Yang et al. 2014; Zhou et al. 2008).
Data gaps and uncertainties
Although many studies support the link between IR, RONS, and DNA damage, a few RONS studies use mammary tissue, multiple doses, doses below 1 Gy, or explicitly measure RNS presenting an addressable data gap.
Despite the effectiveness of antioxidants in decreasing all forms of DNA damage, externally applied RONS is less effective than IR at generating double-strand breaks and mutations (Dahm-Daphi et al. 2000; Gradzka and Iwanenko 2005; Ismail et al. 2005; Sandhu and Birnboim 1997). One possible explanation for this discrepancy is that IR may elicit a higher concentration of localized RONS than can be achieved with external application of H2O2. IR deposits energy and oxidizes molecules within a relatively small area over a rapid timescale potentially permitting a very high local concentration which could precede or overwhelm local buffering capacity. In contrast, extracellularly applied H2O2 would interact with many antioxidants and other molecules on its way to the nucleus, where the concentration would slowly reach a lower steady state.
To our knowledge, no experiments have tested whether elevating intracellular RONS alone in one group of cells can cause bystander effects in another.
DNA damage, GI, and mutation (MIE & KEs)
Evidence of activation by IR and RONS
IR and RONS cause immediate and delayed DNA damage and GI. Both of these forms of DNA damage contribute to changes in DNA sequence, i.e., mutations. DNA changes include to the base of the nucleotide (with guanine particularly vulnerable because of its low redox potential), a break in the sugar (deoxyribose)-phosphate backbone creating a single-strand break (David et al. 2007), or simultaneous proximal breaks in both strands of DNA to form double-strand breaks (Ameziane-El-Hassani et al. 2015; Berdelle et al. 2011; Dahm-Daphi et al. 2000; Driessens et al. 2009; Haegele et al. 1998; Han et al. 2010; Lorat et al. 2015; Nakamura et al. 2003; Oya et al. 1986; Pouget et al. 2002; Roots et al. 1990; Rothkamm and Lobrich 2003; Sandhu and Birnboim 1997; Seager et al. 2012; Sharma et al. 2016; Stanicka et al. 2015; Ward 1988; Werner et al. 2014; Yang et al. 2013). Double-strand breaks are considered more destructive and mutagenic than lesions or single-strand breaks, and higher densities of double-strand breaks generate chromosomal abnormalities including changes in chromosomal number, breaks and gaps, translocations, inversions, and deletions (Behjati et al. 2016; Haag et al. 1996; Morishita et al. 2016; Ponnaiya et al. 1997b; Unger et al. 2010; Yang et al. 1992, 1997).
The type and amount of DNA damage depends on both the quality and dose of radiation. The pattern of energy transferred by IR to matter (linear energy transfer per unit length or LET) (1970) varies between sources. Lower or no LET IR such as mammographic X-rays and some radiation therapies sparsely deposit many individual excitations or small clusters of excitations of low energy (Goodhead 1988) deep into tissue. In contrast, high LET such as alpha particles from radon readily transfer their energy (Goodhead 1994) and, therefore, deposit dense clusters of energy closer to the tissue surface (Goodhead 1988).
These different energy deposition patterns contribute to differences in radiation effects including the pattern of DNA damage. Higher LET radiation such as alpha particles generates more complex clusters of damage including more frequent double-strand breaks (Nikitaki et al. 2016; Ottolenghi et al. 1997; Rydberg et al. 2002; Watanabe et al. 2015) and other chromosomal abnormalities (Anderson et al. 2002; Yang et al. 1997), while lower LET radiation (gamma rays, X-rays) and RONS generate more oxidized base damage and single-strand breaks (Douki et al. 2006). Although complex damage is more likely to occur from high LET IR, low LET IR and RONS can also generate complex damage, even from a single particle or photon (Ravanat et al. 2014; Sharma et al. 2016; Sutherland et al. 2002). Complex damage is more difficult to repair, making it more detrimental to the cell and the organism than a single point of damage (Kuhne et al. 2000; Pinto et al. 2005; Rydberg et al. 2005; Stenerlow et al. 2000).
DNA damage is also observed in in cells not directly in the path of ionizing radiation and at a delay following exposure. These indirect or bystander effects are mediated by multiple factors including RONS (Yang et al. 2005), TGF-β (Dickey et al. 2009), and other cytokines (Havaki et al. 2015).
Most evidence suggests that nucleotide damage, single-strand breaks, clustered damage, and double-strand breaks increase linearly with IR dose over a wide range (0.001–80 Gy) (Asaithamby and Chen 2009; Beels et al. 2010; Ojima et al. 2008; Roots et al. 1990; Rothkamm and Lobrich 2003; Rydberg et al. 2002; Sutherland et al. 2002; Yang et al. 2005). The low LET dose–response of chromosomal aberrations and other mis-rejoining of DNA is linear or supralinear (Iwasaki et al. 2011; Ryu et al. 2016; Tanaka et al. 2009; Yang et al. 1997) at lower doses (0.01 Gy) (Iwasaki et al. 2011; Schiestl et al. 1994) and dose–response upwardly curving at high-dose rates (0.89 Gy/day) and higher doses (10–80 Gy) (Olipitz et al. 2012; Rydberg et al. 2005; Tanaka et al. 2009), while the response to high LET radiation is linear (Jones et al. 2007; Yang et al. 1997). DNA damage measured in bystander cells 1 h to 3 days after exposure is also dose-dependent at very low doses, but may approach a maximum around 0.1 Gy (Ojima et al. 2008; Yang et al. 2007b).
GI is the recurrent appearance of DNA and chromosomal damage and mutations. DNA damage following ionizing radiation in directly and indirectly damaged cells is repaired over the first few hours or days (Nikitaki et al. 2016), but GI can appear weeks, months, or even years after the initial exposure and persist in subsequent generations of cells in vivo (Datta et al. 2012b; Mukherjee et al. 2012; Pazhanisamy et al. 2011; Snijders et al. 2012) and in vitro (Bensimon et al. 2016; Buonanno et al. 2011; Moore et al. 2005; Natarajan et al. 2007). DNA damage response and dysfunctional telomeres are implicated in the formation of GI (Sishc et al. 2015; Williams et al. 2009; Yu et al. 2001), but the delayed appearance and high frequency of GI and its transmission through media to bystander cells strongly suggest the involvement of non-mutational (epigenetic) mechanisms (Al-Mayah et al. 2012; Rugo et al. 2011; Ullrich and Davis 1999). RONS is also involved in the production of GI (Bensimon et al. 2016; Buonanno et al. 2011; Datta et al. 2012b; Dayal et al. 2008; Kim et al. 2006; Limoli and Giedzinski 2003; Limoli et al. 2003; Natarajan et al. 2007; Owens et al. 2012; Pazhanisamy et al. 2011; Thomas et al. 2012).
Mutations occur when DNA damage is converted to permanent changes in nucleotide sequence. Nucleotide lesions contribute to point mutations when a different nucleotide is mistakenly inserted in place of a damaged nucleotide, sometimes altering the resulting amino acid. Nucleotide lesions also contribute to single-strand breaks, and both lesions and single-strand breaks contribute to double-strand breaks if not repaired before replication. Double-strand breaks contribute to deletions, inversions, translocations, and duplications, and can lead to major chromosomal damage. Complex DNA damage (multiple damaged sites in close proximity) is more difficult and time-consuming to repair, so complex damage is more likely to persist until replication and be converted into mutations.
Because DNA damage and mutation increase the risk of all types of cancers, these events are the subject of established guideline assays (Table 1) and are considered adverse outcomes in their own right.
Table 1.
Guideline assays and other methods measuring key events in the pathway to breast cancer
| Event | Guideline test | Methods | Tissue |
|---|---|---|---|
| Breast cancer | Yes | OECD Test No. 451 and 453, 2-year bioassay for carcinogenicity and combined toxicity and carcinogenicity (OECD 2009a, b) | Mammary |
| US National Toxicology Program (NTP), FDA, EPA guidelines for 2-year cancer bioassay and risk assessments (EPA 2005; FDA 2007; NTP 2011b) | Mammary | ||
| RONS | No | Fluorescent protein-based probes (Dickinson and Chang 2011; Griendling et al. 2016; Kalyanaraman et al. 2012; Wang et al. 2013) | Any* |
| Boronate-based small molecule probes (Dickinson and Chang 2011; Griendling et al. 2016; Kalyanaraman et al. 2012; Wang et al. 2013) | Any* | ||
| EPA ToxCast assay for mitochondrial membrane potential and intracellular superoxide (DHE) (Giuliano et al. 2010) | Primary rat hepatocyte* | ||
| DNA damage | Yes | OECD DNA synthesis Test No. 486 for nucleotide excision repair (OECD 1997b) | Mammalian liver cells—only informs primary damage to liver (EFSA Scientific Committee et al. 2017) |
| OECD Alkaline comet assay Test No. 489 for single- and double-strand breaks and nucleotide damage (OECD 2016f) | Any*,** | ||
| OECD Chromosomal damage and micronuclei Test No. 473, 475, 483, and 487 (OECD 2016a; OECD 2016b; OECD 2016d; OECD 2016e) | 473, 487: Stable cell line, esp lymphocyte (no validated mammary)* | ||
| 475: Mammalian bone marrow*,** | |||
| 483: Rodent sperm cells*,** | |||
| No | Electrophoretic methods for finding strand breaks or specific DNA lesions: high-throughput comet assay (Ge et al. 2014; Sykora et al. 2018), pulsed field gel electrophoresis (PFGE) (Nikitaki et al. 2015; Sutherland et al. 2003) | Any* | |
| Direct measurement of DNA lesions via HPLC–MS/MS (Ravanat 2012) | Any** | ||
| Immunostaining using antibodies to label DNA damage repair proteins (H2AX, XRCC2, OGG1, etc.) coupled with microscopy or flow cytometry (Lobrich et al. 2005; Nikitaki et al. 2015, 2016; Ojima et al. 2008; Rothkamm and Lobrich 2003). Can detect clustered lesions (Nikitaki et al. 2016) | Any*,** | ||
| EPA ToxCast anti-p53 assay (Giuliano et al. 2010) | HEPG2 cells* | ||
| Mutation | Yes | OECD Ames Test No. 471 (OECD 1997a) | Bacteria* |
| OECD Hprt or Xprt Test No. 476 (OECD 2016c) | Selected lines, not mammary* | ||
| OECD Transgenic Rodent Somatic and Germ Cell Gene Mutation Assays Test No. 488 (OECD 2013) | Any (performed in mammary gland (Jakubczak et al. 1996)** | ||
| OECD Thymidine kinase Test No. 490 (OECD 2016g) | Lymphoma or lymphoblastoid cell lines* | ||
| No | Transgenic rodent in vivo/in vitro: RaDR-GFP for errors in homologous recombination (sister chromatid exchange) (Sukup-Jackson et al. 2014) | Any (performed in mammary gland (Sukup-Jackson et al. 2014))** | |
| Transgenic rodent in vitro: many tests, some metabolically characterized (White et al. 2019) | Any (some in mammary) (White et al. 2019) | ||
| Array CGH detects copy number variations in tumors or clonal cells (Bonnet et al. 2012; Gaudet et al. 2003) | Any* | ||
| Genomic Instability | No | Many of the above methods for detecting DNA and chromosomal damage and mutation are applied at a range of time points to detect GI (Datta et al. 2012b; Gaudet et al. 2003; Lee et al. 2015; Owens et al. 2012; Rugo et al. 2011; Sishc et al. 2015; Snijders et al. 2012). Multiple time points should be used to find the rate of ongoing damage, and multiple methods used to capture the range of possible GI outcomes (Limoli et al. 1997) | Any*,** |
| Proliferation, Hyperplasia | No | Change in cell numbers (DNA synthesis markers (in vivo or in vitro), impedance, time lapse imaging, colony formation assay (in vitro)) (Menyhart et al. 2016; Morgan et al. 2020) | Any* |
| Immunohistochemistry (Ki67 labeling) (Romar et al. 2016) | Any*,** | ||
| Histology (proliferation, hyperplasia) in in vivo toxicity studies such as 90-day subchronic, 28 day, or 14-day can include longitudinal sectioning of male and female mammary glands and BrDU injection and staining to better detect proliferation (Collins 2018; Makris 2011; Rudel et al. 2011) | Any*,** | ||
| EPA ToxCast BRDU assay (Giuliano et al. 2010) | HepG2 carcinoma line* | ||
| Inflammation | No | Detection of inflammatory proteins like IL6, TNF-α: ELISA (El-Saghire et al. 2013; Partridge et al. 2010; Siva et al. 2014), Western Blot (Chai et al. 2013b; Ha et al. 2010), electrophoretic mobility shift assay (Haase et al. 2003), immunostaining (Chai et al. 2013b; Wang et al. 2015), PCR based changes in expression of inflammation-related signals (Azimzadeh et al. 2011; Bouchet et al. 2013; Moravan et al. 2011; Snijders et al. 2012; Wang et al. 2014b) | Any (ideally both target tissue and leukocytes)*,** |
| Histology (leukocyte infiltration) (Ebrahimian et al. 2018; Monceau et al. 2013; Moravan et al. 2011) | Any*,** | ||
| EPA ToxCast BioMap assays (Houck et al. 2009) | Various, not including mammary* | ||
| Epigenetic changes | No | Global methylation: [3H]dCTP extension assay (Koturbash et al. 2016), HPLC for 5mdC (Wang et al. 2014a), quantitative immunoassay for 5-mc (Nzabarushimana et al. 2014) | Any*,** |
| Gene-specific methylation: MeDIP-on-chip (Hsu et al. 2015), methylation-sensitive/dependent restriction enzymes and qPCR (Nzabarushimana et al. 2014; Oakes et al. 2006a), bisulfite sequencing (Wang et al. 2014a) | Any*,** | ||
| miRNA expression (Mestdagh et al. 2014): qRT-PCR (Stankevicins et al. 2013; Szatmari et al. 2017), hybridization (e.g., microarray, etc.) (Aypar et al. 2011; Jacob et al. 2013), sequencing (Mestdagh et al. 2014) | Any*,** | ||
| Histone methylation: ChIP (Prior et al. 2016) and immunohistochemistry (Kutanzi and Kovalchuk 2013) | Any*,** | ||
| Gene expression: western blot (Wang et al. 2014a), qRT-PCR (Prior et al. 2016), whole transcriptomics e.g. RNA microarray, RNA-seq (Hrdlickova et al. 2017; Manzoni et al. 2018) | Any*,** |
*This test/measurement is not typically conducted in mammary tissue, so validation that results are generalizable is needed
**Verify that test substance reaches the mammary gland
Essentiality of DNA damage, GI, and mutation
Essentiality is high. Although not commonly measured in the same experiments, IR-induced DNA damage, GI, and mutation (Denissova et al. 2012; Fibach and Rachmilewitz 2015; Jones et al. 2007; Padula et al. 2016; Sandhu and Birnboim 1997; Ullrich and Davis 1999; Wazer et al. 1994) precede cell transformation (Ullrich et al. 1996; Unger et al. 2010; Wang et al. 2011; Wazer et al. 1994; Yang et al. 1992, 1997), and tumorigenesis (Adams et al. 1987; Andrieu et al. 2006; Bernstein et al. 2010; Bernstein et al. 2013; Broeks et al. 2007; Brooks et al. 2012b; de Ostrovich et al. 2008; Francis et al. 2009; Francis et al. 2011; Gustin et al. 2009; Little 2009; Millikan et al. 2005; Pijpe et al. 2012; Podsypanina et al. 2008; Poirier and Beland 1994; Radice et al. 1997; Tao et al. 2017; Umesako et al. 2005). Further support for including DNA damage, GI, and mutation in the mechanistic pathway linking ionizing radiation with breast cancer comes from the observation that variants in DNA repair genes increase the risk of mammary tumors in animals after IR (Umesako et al. 2005; Yu et al. 2001) and increase breast cancer after IR (Andrieu et al. 2006; Bernstein et al. 2010, 2013; Broeks et al. 2007; Brooks et al. 2012b; Millikan et al. 2005; Pijpe et al. 2012). BRCA is perhaps the best known DNA repair gene linked with breast cancer risk, and several of these studies have suggested a link between BRCA mutation status and increased susceptibility to breast cancer following ionizing radiation, particularly in women exposed at younger ages (Pijpe et al. 2012).
Outside of the IR literature, multiple studies confirm that mutations can increase proliferation, hyperplasia, and tumors (Podsypanina et al. 2008). Mutations in cell cycle checkpoint, growth factor signaling, and other genes enable mammary epithelial and other cells to proliferate in vitro (Gustin et al. 2009; Higashiguchi et al. 2016; Kouros-Mehr et al. 2006; Shahi et al. 2017; Tao et al. 2011), promote mammary hyperplasia in vivo (Francis et al. 2009), and contribute to tumors when combined with the other mutations in the same pathway (Francis et al. 2011). Restoring function in mutated genes regresses tumors in animals (Martins et al. 2006; Podsypanina et al. 2008). Mutations are common in tumors (CGAN (Cancer Genome Atlas Network) 2012; Garraway and Lander 2013; Greenman et al. 2007; Haag et al. 1996; Stratton et al. 2009; Vandin et al. 2012; Vogelstein et al. 2013; Yang et al. 2015) and tumors are largely clonal, suggesting that individual mutations offer the tumor evolutionary advantages (Begg et al. 2016; Wang et al. 2014c; Yates et al. 2015). GI increases the frequency and duration of DNA damage and mutation after IR, thereby increasing the likelihood of transformation and tumorigenesis (Selvanayagam et al. 1995; Ullrich and Ponnaiya 1998).
The evidence linking DNA damage with epigenetic changes is indirect. The two key events are often observed at the same time or DNA damage precedes epigenetic changes (Koturbash et al. 2016, 2008, 2005, 2006b; Nishida et al. 2013; Pogribny et al. 2005, 2004; Szatmari et al. 2017). However, DNA damage does not always correspond with or precede epigenetic changes (Koturbash et al. 2016, 2006b; Pogribny et al. 2005).
Data gaps and uncertainties
The majority of research on the effects of IR on DNA damage has been performed in tissues other than mammary gland, but several studies suggest that effects in the mammary gland (and its consequences) would be consistent with the other tissues (Haegele et al. 1998; Hernandez et al. 2013; Jones et al. 2007; Snijders et al. 2012; Soler et al. 2009; Wang et al. 2015) and many of the studies in support of the proliferative and tumorigenic role of mutations are in mammary gland or breast cancers (Behjati et al. 2016; CGAN (Cancer Genome Atlas Network) 2012; de Ostrovich et al. 2008; Francis et al. 2009; Francis et al. 2011; Gustin et al. 2009; Haag et al. 1996; Korkola and Archer 1999; Marusyk et al. 2014; Miura et al. 2008; Morganella et al. 2016; Nik-Zainal et al. 2016; Podsypanina et al. 2008; Radice et al. 1997; Tao et al. 2017; Umesako et al. 2005; Wazer et al. 1994; Yang et al. 2015). Recent studies in other tissues have shown a high frequency of cancer-driving mutations in normal tissues, raising questions about factors leading to clonal selection (Martincorena 2019); and further description of these processes in breast represents a data gap.
All cancers have DNA mutations, but mutation is not always the initiating event. Instead mutation can result from other stressors like proliferation or inflammation. Mammary tumor incidence following ionizing radiation treatment of the animal (as discussed above) or transplanted cells (Maffini et al. 2005; McDaniel et al. 2006) varies significantly by sex and depends on the presence of ovarian hormones. Stroma treated with carcinogens or IR supports the growth of tumors from pre-malignant epithelial cells (although these may have pre-existing mutations) (Barcellos-Hoff and Ravani 2000; Maffini et al. 2004; Nguyen et al. 2011b). In these cases, the hormones or inflammation in the mammary gland may modify the stromal environment to increase the likelihood of cellular proliferation, which amplifies the existing mutated cells and generates new mutations.
Some evidence points to a protective effect of lower dose IR on chromosomal damage, mutation, and transformation in vitro (Azzam et al. 2016; Elmore et al. 2008; Ina et al. 2005; Kakinuma et al. 2012; Sakai et al. 2006; Sasaki et al. 2002; Shin et al. 2010; Wolff et al. 1988; Yamauchi et al. 2008) and chromosomal damage, mutation and thymic lymphomas, and skin cancer in vivo (Ina et al. 2005; Kakinuma et al. 2012; Sakai et al. 2006; Shin et al. 2010; Yamauchi et al. 2008). However, the dose range at which protection is observed varies greatly between target tissue, animal models, and other experimental conditions, suggesting that the protection is not a fixed or immutable response (Tang and Loke 2015). Most animal studies (of chemical genotoxicants) are not sufficiently designed or powered to detect low-dose changes in dose–response and are insufficient to reject the linear no threshold model (Guerard et al. 2015), but a large IR animal cancer study and a systematic meta-analysis of IR protection in animals found no convincing evidence of protection against carcinogenesis (Crump et al. 2012; Tanaka et al. 2007). In people, a recent systematic review and meta-analysis of low-dose IR epidemiology concludes that the evidence does not support protection against total solid cancers (Shore et al. 2017), and although the dose–response for epidemiology of leukemia and lymphoma leave open the possibility of a protective response at low doses, a dose–response model without an inflection or protective effect of IR is recommended (Sasaki et al. 2014). No evidence suggests a protective effect of ionizing radiation on breast cancer, and dose–response relationships are further discussed in Sect. 13.2 below.
Proliferation and hyperplasia (KEs)
Evidence of proliferation and hyperplasia following IR
While higher doses of ionizing radiation cause cell death in the short term (especially of dividing cells), IR is associated with proliferation in vitro and in vivo. In vitro, IR promotes proliferation/expansion of p16-suppressed and immortal epithelial populations as well as in bystander CHO cells co-cultured with IR-exposed cells (Han et al. 2010; Mukhopadhyay et al. 2010; Tang et al. 2014). In vivo, IR increases compensatory proliferation in adult rats (Loree et al. 2006), and long-term expression of proliferation in adolescent but not adult mammary gland (Datta et al. 2012a; Snijders et al. 2012; Suman et al. 2012), possibly via the expansion of a population of stem-like cells in vivo (Nguyen et al. 2011b; Tang et al. 2014). This proliferation appears to be associated with TGF-β/Notch activity (Tang et al. 2014) and nitric oxide (Han et al. 2010). Proliferative nodules and hyperplasia appear in mammary terminal-end bud, alveolae, and ducts of rats and mice after exposure to ionizing radiation (Ethier and Ullrich 1982; Faulkin et al. 1967; Imaoka et al. 2006) as well as chemical carcinogens (Beuving et al. 1967a, 1967b; Purnell 1980; Russo et al. 1977).
Essentiality of proliferation
Essentiality is high. Cellular proliferation is a key characteristic of cancer cells (Hanahan and Weinberg 2011). Hyperplasia signals the presence of excess proliferation and represents an intermediate phase in the development of tumorigenesis. Hyperplasia is generally considered to be an adverse outcome in its own right.
Proliferation occurs following the release of inhibitory controls limiting entry into the cell cycle, and oncogenic mutations act via these same pathways to generate abnormal proliferation (Hanahan and Weinberg 2011; Weber et al. 2017), including in mammary gland (Francis et al. 2009, 2011; Garbe et al. 2009; Gustin et al. 2009; Weber et al. 2017). Inhibitory signals such as contact inhibition or TGF-β (Francis et al. 2009; Polyak et al. 1994) stabilize the mechanisms limiting entry into the cell cycle. Proliferative signals such as those following progesterone or estrogen (Croce 2008; Weber et al. 2017) or compensatory proliferation after apoptosis (Fogarty and Bergmann 2017) relieve inhibition and enable cells to enter the cell cycle. Mutations that inactivate inhibitory signals (tumor suppressors) or activate proliferative signals (oncogenes) promote proliferation outside of the normal biological context (Francis et al. 2011; Gustin et al. 2009; Hanahan and Weinberg 2011; Weber et al. 2017). Abnormal proliferation is typically met with apoptosis or senescence, so additional mutations or other mechanisms are required to escape these additional levels of control for proliferation to continue indefinitely (Fernald and Kurokawa 2013; Garbe et al. 2009; Shay and Wright 2011).
Proliferation is required for DNA damage and replication errors to be integrated into the genome as mutations (Kiraly et al. 2015). Proliferation can also promote the expansion and clonal selection of existing cells with proliferative mutations. Genomic mutations favoring further proliferation are positively selected from among the expanded cells, resulting in the accumulation of mutational errors and moving the organism further towards cancer. Different clonal populations can also collaborate to promote growth as observed in mammary gland (Franco et al. 2016; Marusyk et al. 2014).
Increasing proliferation leads to hyperplasia in mammary gland (Korkaya et al. 2009). During mammary tumorigenesis following IR in rodents, proliferating foci precede the development of tumors (Haslam and Bern 1977; Purnell 1980) and form tumors more effectively than non-proliferating tissue (Beuving 1968; Deome et al. 1959; Rivera et al. 1981). Adenocarcinomas in mammary gland appear to preferentially form from terminal-end bud hyperplasia (Haslam and Bern 1977; Purnell 1980; Russo et al. 1977) in rats, similar to the origin of many breast cancers for humans and for some mice after IR (Medina and Thompson 2000).
Supporting the essentiality of these proliferative processes to tumorigenesis, ACI rats that exhibit no mammary proliferation or hyperplasia following IR are resistant to tumors following IR (Kutanzi et al. 2010). Interventions reducing proliferation in susceptible PyVT and BALB/c mice also reduce mammary tumors (Connelly et al. 2011; Luo et al. 2009; Tang et al. 2014).
Data gaps and uncertainties
Some studies report carcinogenesis in the absence of hyperplasia (Sinha and Dao 1974) and others do not find increased tumorigenesis from transplanted hyperplasia (Beuving et al. 1967a; Haslam and Bern 1977; Sinha and Dao 1977).
Inflammation (KEs)
Evidence of activation by IR
Inflammatory pathways are commonly activated in mammary gland (Barcellos-Hoff et al. 1994; Bouchard et al. 2013; Datta et al. 2012a; Dickey et al. 2009; Snijders et al. 2012; Wang et al. 2015) and in breast and mammary cancers following IR (Illa-Bochaca et al. 2014; Nguyen et al. 2013, 2011b).
Essentiality of inflammation
Essentiality is moderate. While inflammation in response to tissue damage or infection is typically resolved without lasting effects, excess initial inflammation, chronic inflammation, and anti-inflammatory signaling (via fibrosis) can contribute to chronic inflammatory diseases including cancer (Perez et al. 2014). Such dysregulation of inflammation may be related to continual competition of inflammatory and anti-inflammatory factors, which can lead to chronic wounds (Eming et al. 2014).
Tumors and tumor cells exhibit features of inflammation, and inflammation is generally understood to promote transformation and tumor progression by supporting multiple hallmarks of cancer including oxidative activity and DNA damage, survival and proliferation, angiogenesis, and invasion and metastasis (Esquivel-Velazquez et al. 2015; Hanahan and Weinberg 2011; Iliopoulos et al. 2009).
Many of these cancer promoting effects of inflammation can be seen following exposure to ionizing radiation (Bisht et al. 2003; Bouchard et al. 2013; Elahi et al. 2009; Illa-Bochaca et al. 2014; Nguyen et al. 2011b, 2013). Polymorphisms in inflammation genes are associated with breast cancer risk from IR in radiation technologists (Schonfeld et al. 2010) and with susceptibility to intestinal adenoma following IR in mice (Elahi et al. 2009). Cytokines TGF-β and IL6 transform primary human mammospheres and pre-malignant mammary epithelial cell lines in vitro and make them tumorigenic in vivo (Iliopoulos et al. 2009; Nguyen et al. 2011b; Sansone et al. 2007), and inflammation-related factors COX2 and TGF-β are required for the full effect of IR on DNA damage and transformation in vitro and mammary tumor growth and invasion in vivo (Bisht et al. 2003; Nguyen et al. 2011b). Further evidence for inflammation comes from tumors arising from mammary epithelial cells transplanted into IR-exposed cleared fat pads: inflammation-related genes and pathways are upregulated or enriched in the gene expression patterns of these indirectly IR-induced tumors (Illa-Bochaca et al. 2014; Nguyen et al. 2013, 2011b).
Activation of an inflammatory NF-kB/IL6/STAT3 signaling pathway generates cancer stem cells in breast cancer cells in vitro (Iliopoulos et al. 2009, 2010, 2011) and is also implicated in colon and other cancers (Iliopoulos et al. 2010). The inflammation-related transcription factor NF-kB also contributes to mammary tumorigenesis and metastasis in PyVt mice, in which mammary tumors are induced by expression of an MMTV-driven oncogene (Connelly et al. 2011).
One mechanism of cancer promotion involves oxidative activity and DNA damage: inflammation in response to IR increases oxidative activity in a positive feedback loop leading to increased DNA lesions, GI, and mutations. Oxidative activity mediates the increase in inflammatory (TNF-α and neutrophil) markers (Ozyurt et al. 2014), and oxidative activity is increased by direct treatment with TNF-α or neutrophils (Jackson et al. 1989; Natarajan et al. 2007; Yan et al. 2006; Yang et al. 2007a) or after IR mediated by Il13, TNF-α, CCL2, COX2, and TLR9 (Ameziane-El-Hassani et al. 2015; Jackson et al. 1989; Kostyuk et al. 2012; Redon et al. 2010; Stevens et al. 1992; Zhang et al. 2017). Cytokines IL13 and TNF-α, neutrophils, and COX2 damage DNA and increase mutations indirectly in IR cells (Ameziane-El-Hassani et al. 2015; Bisht et al. 2003; Burr et al. 2010; Hosseinimehr et al. 2015; Jackson et al. 1989; Ozyurt et al. 2014; Yan et al. 2006), and NO, TGF-β, NF-kβ, and COX2 increase DNA damage and mutations in bystander cells (Chai et al. 2013b; Dickey et al. 2009; Han et al. 2010; Redon et al. 2010; Shao et al. 2008; Zhou et al. 2005; Zhou et al. 2008). The DNA damage and mutations are reduced by blocking the inflammatory factors NF-kβ, COX2, TNF-α, TGF-β, or NO or with antioxidants (Burr et al. 2010; Han et al. 2010; Jackson et al. 1989; Rastogi et al. 2012; Zhang et al. 2017; Zhou et al. 2005, 2008). Inhibiting TNF-α or COX2 or using antioxidants also reduces GI after IR (Lorimore et al. 2008, 2011; Moore et al. 2005; Mukherjee et al. 2012; Natarajan et al. 2007).
Inflammatory pathways activated by IR promote tumor growth and metastasis. Exposure to IR or RONS sensitizes mammary epithelial cells to respond to TGF-β—which is widely activated by IR (Ehrhart et al. 1997). IR and TGF-β signaling leads to an epithelial-to-mesenchymal (EMT)-like transition, which disrupts the expression and distribution of cell adhesion molecules and multicellular organization and promotes invasion (Andarawewa et al. 2011, 2007; Iizuka et al. 2017; Park et al. 2003). This mechanism resembles wound healing (Koh and DiPietro 2011; Landen et al. 2016; Perez et al. 2014), but also resembles malignancy—invasive breast cancer cell lines overexpress TGF-β and respond to TGF-β with increased invasion (Gomes et al. 2012; Kim et al. 2004). The TGF-β-mediated response to IR likely involves an increase in senescence in fibroblasts (Liakou et al. 2016; Perrott et al. 2017; Sourisseau et al. 2011; Tsai et al. 2005).
IL6 is implicated in the carcinogenic response to IR. IL6 is expressed in mouse mammary gland after IR (Bouchard et al. 2013) and is produced by IR-senescent fibroblasts, but may also be expressed by epithelial cells after IR, since primary human mammospheres and pre-malignant mammary epithelial cell lines respond to IL6 with increased IL6 expression (Iliopoulos et al. 2009; Sansone et al. 2007). IL6 promotes the mobility and tumorigenesis of normal and breast cancer epithelial cells (Iliopoulos et al. 2009, 2010; Sansone et al. 2007; Sasser et al. 2007; Studebaker et al. 2008) through a NOTCH dependent pathway. NOTCH supports the renewal of stem-like cell populations (Sansone et al. 2007) and is implicated in the proliferative response to IR in mammary epithelia (Marusyk et al. 2014; Nguyen et al. 2011b; Tang et al. 2014). Interestingly, IL6 is also required for the growth and tumor promoting effects of breast cancer fibroblasts and fibroblasts from common sites of breast cancer metastasis (bone, lung) on ER-positive cancer cells in vitro and in vivo. ER-negative breast epithelial cells release autocrine IL6 and may, therefore, be less dependent on IL6 from fibroblasts, although IL6 also transforms these cells (Iliopoulos et al. 2009; Sasser et al. 2007; Studebaker et al. 2008).
Data gaps and uncertainties
The effects of inflammation can be both pro- and anti-tumorigenic. For example, in addition to TGF-β’s role in EMT, in mammary epithelial cells TGF-β is essential to apoptosis of DNA damaged cells including IR-induced damage (Ewan et al. 2002), thus limiting GI (Maxwell et al. 2008). Inflammatory factors TNF-α and COX2 play a similar role in bone marrow of C57BL/6 mice (Lorimore et al. 2013). By eliminating cells with severe DNA damage and curtailing GI, apoptosis (and therefore TGF-β or TNF-α) limits the appearance of major (possibly carcinogenic) mutations following ionizing radiation. However, apoptosis (and thus TGF-β or TNF-α) can indirectly promote tumorigenesis through compensatory proliferation (Fogarty and Bergmann 2017; Loree et al. 2006).
Genetic background also influences the interaction between inflammation and tumorigenesis. Polymorphisms in inflammatory genes influence susceptibility to intestinal cancer following IR (Elahi et al. 2009). In the SPRET outbred mouse, higher baseline TGF-β during development decreases mammary tumor incidence following lower doses of IR (0.1 Gy), possibly by reducing ductal branching and susceptibility (Zhang et al. 2015). Conversely, the BALB/c mouse susceptible to mammary tumors after IR has a lower baseline TGF-β (and a polymorphism in a DNA damage repair-related gene). Early (4 h) after low dose (0.075 Gy), IR BALB/c mice have suppressed immune pathways and macrophage response but increased IL6, COX2, and TGF-β pathway activation in mammary gland compared to the tumor-resistant C57BL/6 mouse (Bouchard et al. 2013; Snijders et al. 2012). By 1 week after IR, the BALB/c mice show TGF-β-dependent inflammation in the mammary gland, and by 1 month after IR, their mammary glands show proliferation (Nguyen et al. 2011a; Snijders et al. 2012), suggesting that TGF-β is associated with inflammation, proliferation, and mammary tumorigenesis in these mice. Consistent with this pattern, BALB/c mice that are heterozygous for TGF-β are more resistant to mammary tumorigenesis following IR (Nguyen et al. 2011b). However, the BALB/c mouse also has a polymorphism in a DNA repair gene associated with IR-induced GI (Yu et al. 2001), making it difficult to distinguish potentially overlapping mechanisms.
While inflammatory signals are associated with bystander effects including DNA damage, GI, and mutation, these effects vary between organs in vivo (Chai et al. 2013a, b), by genotype (Coates et al. 2008; Lorimore et al. 2008, 2011), and by cell type (Chai et al. 2013a). Further research will be required to identify all the underlying factors determining differences in bystander effects, but one variable is the appearance of a protective apoptotic response to cytokines under some conditions (Lorimore et al. 2011, 2013).
One major piece of conflicting evidence comes from a direct test of the essentiality of inflammation to IR-induced carcinogenesis. In a mouse model of lymphoma, a mutation preventing the PIDD/NEMO-dependent activation of NF-kB blocks the early IR-induced activation of NF-kB (4–24 h) and production of TNF-α (5–48 h) but not lymphoma, suggesting that activation of these inflammatory factors is not essential in this time period (Bock et al. 2013). However, this study examined only day 1 post-IR time points for NF-kB activity, and did not block the production of IL6. Later activation of NF-kB or activation of other inflammation-related factors including IL6 and TGF-β could, therefore, have contributed to lymphoma.
Epigenetic changes (KE)
Evidence of activation by IR
IR produces epigenetic effects including changes in global and gene-specific DNA methylation, histone modification, and expression of methylation-related proteins and miRNA (Cui et al. 2011; Nzabarushimana et al. 2014; Prior et al. 2016; Raiche et al. 2004; Tawa et al. 1998) (see Supplemental Table 1 for more). Epigenetic effects appear in a variety of models within hours after IR and have also been detected days or weeks later (Chaudhry and Omaruddin 2012; Lima et al. 2014; Nzabarushimana et al. 2014; Pogribny et al. 2005).
The mechanism by which IR alters epigenetics is not well established, but RONS are implicated (Bernal et al. 2013; Galligan et al. 2014; Kloypan et al. 2015; Simone et al. 2009; Yang et al. 2014), and DNA damage is suggested (Koturbash et al. 2005, 2006b, 2008, 2016; Nishida et al. 2013; Pogribny et al. 2004, 2005; Szatmari et al. 2017).
Epigenetic changes vary widely with tissue and cell type, target, time after exposure, and sex (Lima et al. 2014; Miousse et al. 2017, 2014; Prior et al. 2016; Raiche et al. 2004), as well as between experiments (e.g., (Pogribny et al. 2004) vs (Raiche et al. 2004), (Miousse et al. 2014) vs (Miousse et al. 2017)), and do not necessarily correlate with expected changes in gene expression (Lima et al. 2014; Luzhna et al. 2015; Miousse et al. 2017, 2014).
Given the variation in epigenetic response in different tissues and sexes, studies in tissue other than female breast and mammary gland may not be informative for this AOP. However, a few studies have examined IR in breast or mammary gland or cells. IR of mammary gland in vivo causes global hypomethylation in the first 4 days and 6 weeks after 3–5 Gy IR (Kutanzi and Kovalchuk 2013; Loree et al. 2006) and hypomethylation of LINE1 in the 4 days after 0.1–2.5 Gy IR (Luzhna et al. 2015). In vitro, IR dose-dependently increased tumor suppressor miR-34a (only miR-34a, let-7a, let-21, and miR-21 were examined) in immortal MCF-10 cells but not MCF-7 cancer cells (Stankevicins et al. 2013). These studies suggest a possible epigenetic response to IR in breast and mammary gland, but the small number of studies and doses prevent drawing conclusions.
Essentiality of epigenetic changes
Essentiality is low. Epigenetic changes in mRNA or miRNA contribute to the short-term DNA-damaging effect of IR on bystander cells. miRNA-containing vesicles from IR-exposed animals promote new DNA damage in unexposed animals (Szatmari et al. 2017) and RNase blocks the short-term DNA-damaging effects of exosome containing media from IR-exposed cells (Al-Mayah et al. 2012, 2015). MiRNA-21 is of particular interest: miRNA-21 in exosomes from exposed cells, miRNA-21 expressing cells, or bystander cells increases DNA damage in bystander cells, while miRNA-21 inhibitors reduce DNA damage from IR in bystander cells (Tian et al. 2015; Xu et al. 2015).
Several studies suggest that epigenetic changes also contribute to GI after IR. In one experiment, DNMTs (which methylate DNA) appear to be essential to transmission of GI to bystander cells in vitro (Rugo et al. 2011). Global hypomethylation is correlated with chromosomal aberrations in nuclear workers (Lee et al. 2015) and global hypomethylation associated with decreased DNMTs precedes DSBs and chromosomal rearrangements in vivo (Gaudet et al. 2003; Koturbash et al. 2006a). In cell lines with and without GI following IR exposure, the unstable cells had significant changes in repeat element methylation and in mRNA and miRNA associated with instability and mitochondrial respiration (Baulch et al. 2014; Thomas et al. 2012).
Epigenetic mechanisms including miRNAs, DNA, and histone methylation and DNMT expression are also implicated in tumorigenesis and invasion, with several studies in breast and mammary tissue. Invasive breast cancer tissue has more miRNA-21, and mammary adenocarcinomas from IR-exposed rats has enriched areas of both hyper and hypomethylation (Daino et al. 2018; Huang et al. 2009). Most of the genes that were differently methylated and expressed in exposed rats were related to developmental targets of PRC2, a histone methyltransferase (Daino et al. 2018). Mice under-expressing DNMT1 have global hypomethylation preceding thymic tumors (Gaudet et al. 2003), and more malignant breast cancer cells have more extensive hypomethylation and more changes in DNMT and histone methylation (Tryndyak et al. 2006).
Data gaps and uncertainties
The evidence supporting a key role for epigenetic changes in breast cancer from IR is limited in three key areas. First, many studies have established that epigenetic effects vary greatly between organs, cell types, and sex (as well as with the other variables like time after exposure) (Lima et al. 2014; Miousse et al. 2014, 2017; Prior et al. 2016; Raiche et al. 2004). However, only eight studies in breast or mammary gland link IR with epigenetic changes or epigenetic changes with DNA damage or GI, and four of these use breast cancer cells. Given the variation between tissues, additional evidence in the breast or mammary gland is needed. Second, several studies report no link between epigenetic changes and changes in protein expression (Lima et al. 2014; Luzhna et al. 2015; Miousse et al. 2014, 2017), so this link needs to be verified or alternate mechanisms must be identified to assert a mechanistic effect. Finally, the large number of possible epigenetic changes, targets, and outcomes contributes to a general lack of understanding of the downstream effect of observed epigenetic changes.
Additional limitations and conflicting results also undermine confidence in the essentiality of this key event. Limited studies in vivo link epigenetic changes to DNA damage or GI. Furthermore, not all epigenetic changes necessarily increase DNA damage—instead, hypermethylation may protect against DNA damage. Cells exposed to repeated low (0.035 Gy) doses of IR show global hypermethylation but no increase in micronuclei, and blocking hypermethylation reduces the protective effect of low doses against DNA damage from higher (2 Gy) doses (Ye et al. 2013). Epigenetic changes including global methylation, repeat elements (including LINE1) and miRNA can also be seen in the absence of GI (Aypar et al. 2011; Miousse et al. 2014), and global methylation was not associated with GI in unstable cells after IR (Baulch et al. 2014).
Key event measurement
Measurement of these key events is critical both to validate their roles in these and other AOPs, and to test agents for their ability to contribute to these key events, most of which are themselves adverse on the basis of their regulatory relevance. Available methods to measure these events are discussed below and summarized in Table 1. Standardized OECD guideline assays measure several key events and adverse outcomes, and limitations to those are noted where applicable. We also describe non-guideline methods to measure key events. Standardization and adoption into OECD-accepted guideline methods would advance the use of these assays and integration into chemicals testing. Very few of the assays are conducted in breast tissue, so additional validation is needed to ensure that results in other models can be generalized to breast.
There are several guideline assays for measuring breast cancer, DNA damage and mutation, and multiple non-guideline methods for measuring RONS, proliferation, inflammation, and epigenetic changes.
At least one method described for each key event has been applied in mammary glands or mammary cells. However, many of the assays specify a non-mammary tissue and even tissue-agnostic assays are not typically conducted in breast/mammary tissue. For chemical testing work in vitro, it is a priority to develop culture models that reflect the behavior of normal tissue (as opposed to cancer cells), and to establish the relevance of chosen models to the outcome of interest. For some key events, it may be important that models include multiple cell types (Morgan et al. 2020).
Non-guideline methods should be standardized for consistency. In particular GI, proliferation, epigenetic changes, and chronic inflammation all have a strong temporal component, and methods should be developed to measure these key events at multiple time points along with the other standard procedures to improve reproducibility.
Breast cancer
The 2-year rodent carcinogenicity bioassay is the primary assay for breast cancer (Rudel et al. 2007). The assay is included in the OECD Test No. 451 and 453 for carcinogenicity and combined toxicity and carcinogenicity (OECD 2009a, b), and is also used by the US National Toxicology Program (NTP) (Chhabra et al. 1990), and the FDA (2007), and referenced by the EPA (EPA 2005) in guidelines for risk assessments. Other assays from short term (2–4 weeks) and subchronic (90 day) to chronic (1 year) toxicity also call for the documentation of mammary tumors (FDA 2007; OECD 2018), so these assays could capture the early onset of tumors if the methods were sufficiently sensitive to detect small lesions (Makris 2011), and could be modified to report earlier key events like hyperplasia and inflammation. For example, evaluation of breast tissue for hyperplasia or proliferative epithelial lesions could be done at several early time points and could trigger later evaluations. Doing these observations after BrdU injection and staining of tissue sections to evaluate proliferation could also increase sensitivity of the assay.
Several characteristics of classic cancer bioassays limit the sensitivity of these assays to mammary gland carcinogens. First, assays do not require prenatal or early post-natal exposures for carcinogenicity testing. The NTP often starts exposures at 5–6 weeks of age and OECD regulatory assay exposures suggest (but do not require) exposures beginning after weaning and before 8 weeks of age. Since sensitivity appears to peak around or before week 7 for these DNA-damaging agents (around puberty) (Imaoka et al. 2013), studies that start dosing later may have reduced sensitivity. Also, assays that initiate exposure in mature animals have diminished sensitivity to hormonally active agents that act during development to alter breast development and increase future susceptibility to cancer, such as estrogenic hormones, DDT, and dioxins (EPA 2005; Rudel et al. 2011). Second, carcinogenicity assay guidelines do not require the best methods for detecting tumors in mammary gland: whole mount preparations of mammary gland coupled with longitudinal sections (dorsoventral sections parallel to the body) of mammary gland for histology (Tucker et al. 2017). Palpation and transverse sections of mammary gland can easily miss tumors or lesions of interest. The NTP does specify these preferable methods for mammary gland analysis in reproductive toxicity guidelines (NTP 2011a) and an NTP workgroup recommends the early life and in utero dosing for cancer bioassays (Thayer and Foster 2007).
Two additional limitations may reduce the sensitivity of standard carcinogenicity assays. First, benign tumors are not always interpreted as an indicator of carcinogenicity, leading to a possible underestimation of risk. NTP and EPA guidance suggest that benign tumors provide additional weight of evidence if malignant tumors are also present or if studies suggest benign tumors can progress to carcinogenicity. In a short-term study, proliferative epithelial lesions, hyperplasia, or benign tumors may indicate a need for a longer term study. Benign mammary tumors (fibroadenomas) almost always coincide with carcinogenic tumors in mammary gland or other organs, and carcinomas sometimes grow from fibroadenomas (Rudel et al. 2007; Russo 2015), suggesting that benign tumors are an underutilized indicator of mammary carcinogenicity.
Finally, the dose-selection guidance in carcinogenicity testing typically calls for a high dose that is sufficiently toxic to suppress body weight (OECD 2009a). However, body weight in rodent cancer bioassays is correlated with mammary tumors, and so, toxicity-induced weight loss at higher doses may result in fewer mammary tumors, and as a result, observed mammary tumors at the lower doses may be dismissed because of the apparent lack of a dose–response (Haseman et al. 1997; Rudel et al. 2007). This potential for suppression of mammary tumors by toxicity at high dose should be considered in weight of evidence determinations for carcinogenicity.
RONS
RONS is typically measured using fluorescent or other probes that react with RONS to change state, or by measuring the redox state of proteins or DNA (Dickinson and Chang 2011; Griendling et al. 2016; Wang et al. 2013). Optimal methods for RONS detection have high sensitivity, selectivity, and spatiotemporal resolution to distinguish transient and localized activity, but most methods lack one or more of these parameters.
Molecular probes that indicate the presence of RONS species vary in specificity and kinetics (Dickinson and Chang 2011; Griendling et al. 2016; Wang et al. 2013). Small molecule fluorescent probes can be applied to any tissue in vitro, but cannot be finely targeted to different cellular compartments. The non-selective probe DCHF was widely used in the past, but can produce false-positive signals and may not be optimal (Griendling et al. 2016). EPR (electron paramagnetic resonance spectroscopy) provides the most direct and specific detection of free radicals, but requires specialized equipment. Fluorescent protein-based probes can be genetically engineered, expressed in vivo, and targeted to cellular compartments and specific cells. However, these probes are very sensitive to pH in the physiological range and must be carefully controlled. Newer selective small molecule probes such as boronate-based molecules are being developed, but are not yet widely used. EPA’s ToxCast series of high-throughput in vitro assays (Thomas et al. 2019) includes an assay that measures mitochondrial membrane potential in combination with dihydroethidium, a qualitative measure of superoxide formation (Giuliano et al. 2010; Kalyanaraman et al. 2012).
Alternative methods involve the detection of redox-dependent changes to cellular constituents such as proteins, DNA, lipids, or glutathione (Dickinson and Chang 2011; Griendling et al. 2016; Wang et al. 2013). However, these methods cannot generally distinguish between the oxidative species behind the changes, and cannot provide good resolution for kinetics of oxidative activity.
These methods are readily applied to mammary cells. However, measurements in different cell types may vary based on differences in the expression of endogenous antioxidants (Kannan et al. 2014). A standard comparison of response across cell types would be useful.
DNA damage, GI, and mutation
DNA damage can be studied in isolated DNA, fixed cells, or living cells. Types of damage that can be detected include single- and double-strand breaks, nucleotide damage, complex damage, and chromosomal or telomere damage. The OECD test guideline for DNA synthesis Test No. 486 (OECD 1997b) detects nucleotide excision repair, so it will reflect the formation of bulky DNA adducts but not the majority of oxidative damage to nucleotides, which is typically repaired via the Base Excision Repair pathway. This test is not recommended by some agencies, because it is not generalizable beyond the liver (EFSA Scientific Committee et al. 2017). The OECD test guideline alkaline comet assay Test No. 489 (OECD 2016f) detects single- and double-strand breaks, including those arising from repair as well as some (alkali sensitive) nucleotide lesions including some lesions from oxidative damage. OECD tests for chromosomal damage and micronuclei Test Nos. 473, 475, 483, and 487 measure longer term effects of DNA damage, but these tests require the damaged cell to subsequently undergo replication (OECD 2016a, b, d, e). They can, therefore, reflect a wider range of sources of DNA damage, including changes in mitosis. While the comet assay test 489 does not specify a target tissue, it is not typically performed in mammary cells (OECD 2016f) and the other guideline tests are never performed in mammary cells or tissues. Although Rube 2008 reports no difference in degree of DNA damage and repair kinetics in five tissues, expression of DNA repair proteins can vary between tissues (Gottlieb et al. 1997; Gurley and Kemp 2007; Sun et al. 2019) which could lead to a difference in damage observed at various time points. For genotoxicity testing, variations in transport and metabolism can also lead to differences between mammary gland and other tissues (Ding et al. 2014). These tests should, therefore, be generally validated for or performed in mammary gland to address risk in this tissue.
Many other (non-test guideline) techniques have been used to examine specific forms of DNA damage. The comet chip facilitates high-throughput comet assays (Ge et al. 2014). Double-strand breaks are commonly measured microscopically using fluorescently labeled antibodies to H2AX or other labeled probes because of the significant risk attributed to breaks and the relative ease of detecting and quantifying them (Lorat et al. 2016; Nikitaki et al. 2016; Ojima et al. 2008; Rothkamm and Lobrich 2003). Measurement of single-strand break repair is less common but possible by labeling single-strand break repair protein XRCC2 (Lorat et al. 2016; Nikitaki et al. 2016). Base lesions can also be detected using labeled probes for base excision repair enzymes, or by chemical methods such as mass spectroscopy (Madugundu et al. 2014; Nikitaki et al. 2016; Ogawa et al. 2003; Ravanat et al. 2014). Refinements on these methods characterize complex or clustered damage, in which various forms of damage occur in close proximity on a DNA molecule (Lorat et al. 2016; Nikitaki et al. 2016). Some DNA-damaging agents act by directly binding to DNA to form adducts, the detection of which is indicative of the potential for mutation (Rundle 2006). EPA’s ToxCast uses a labeled antibody to p53 to indicate non-specific increases in DNA damage and stress (Giuliano et al. 2010). These methods can be or have been applied to mammary or breast cells (Al-Mayah et al. 2012, 2015; Dutta et al. 2014; Haegele et al. 1998; Hernandez et al. 2013; Jones et al. 2007; Kirshner et al. 2006; Redon et al. 2010; Snijders et al. 2012; Soler et al. 2009; Wang et al. 2015). Since results in other tissue may or may not be applicable to breast tissue, they should be performed in breast to best inform risk of breast cancer.
Certain challenges are common to all methods of detecting DNA damage. In the time required to initiate the detection method, some DNA may already be repaired, leading to undercounting of damage. On the other hand, apoptotic DSBs may be incorrectly included in a measurement of direct (non-apoptotic) induction of DSB damage unless controlled. All methods have difficulty distinguishing individual components of clustered lesions, and microscopic methods may undercount disparate breaks that are processed together in repair centers (Barnard et al. 2013). Methods that use isolated DNA (gel electrophoresis and analytical chemistry) are vulnerable to artifacts and must ensure that the DNA sample is protected from oxidative damage during extraction (Barnard et al. 2013; Pernot et al. 2012; Ravanat et al. 2014).
Finally, tests for mutations reveal past DNA damage that result in a heritable change, including multiple guideline tests (OECD Ames Test No. 471 (OECD 1997a), Hprt or Xprt Test No. 476 (OECD 2016c), Transgenic Rodent Somatic and Germ Cell Gene Mutation Assays Test No. 488 (OECD 2013; White et al. 2019), and Thymidine kinase Test No. 490 (OECD 2016g)) and other assays such as the RaDR-GFP transgenic mouse that detects mutational errors in homologous recombination (Kiraly et al. 2015; Sukup-Jackson et al. 2014). One of the approved TG 488 transgenic mouse lines (Jakubczak et al. 1996) has been used to measure mutations in mammary gland in vivo, and the RaDR-GFP mouse and several TG 488 approved lines have been used for primary or established mammary cell lines (Sukup-Jackson et al. 2014; White et al. 2019). The other tests are limited to specific non-mammary tissues, and should be validated for relevance to mammary gland.
No validated protocols exist specifically to measure GI, but a wide range of the above methods are currently performed at various times after exposure to verify ongoing damage or mutations, including in mammary gland or breast cells (Al-Mayah et al. 2012, 2015; Jakubczak et al. 1996; Maxwell et al. 2008; Ponnaiya et al. 1997a, b; Snijders et al. 2012; Ullrich and Davis 1999; Yu et al. 2001). Since GI can be expressed in a variety of ways, a validated protocol should be developed that measures multiple outcomes and time points.
Proliferation and hyperplasia
Past cellular proliferation can be measured directly using labels that are incorporated into cells upon cell division (BRDU or cytoplasmic proliferation dyes) or indirectly by measuring a change in population size. Ongoing proliferation can be quantified by labeling a protein associated with the cell cycle (e.g., Ki67). Methods have been recently reviewed (Menyhart et al. 2016; Romar et al. 2016) and histopathological assessments for mammary gland hyperplasia in in vivo guideline toxicity studies are reviewed in (Makris 2011).
For in vitro assays, EPA’s ToxCast has a BRDU assay for proliferation in HepG2 (Giuliano et al. 2010) and an assay for estrogen-mediated proliferation in T47D breast cancer cells (Judson et al. 2015). Since many of the cells are proliferating in the sub-confluent cell-culture system, it is not clear how sensitive the assay is to increased proliferation. A large body of work has demonstrated that cancerous and non-cancerous cells grow differently in 3D culture systems and that the differences are not apparent in 2D. Furthermore, the effects of chemical exposure on cell growth and development may only be manifest in tissue that contains multiple cell types together. For example, mammary pre-adipocytes produce estradiol via aromatase and epithelial cells proliferate in response (Morgan et al. 2020). Finally, while many toxicity testing models use cancer cell lines, non-cancerous cells may respond differently. Thus, there is considerable need for investment to strengthen in vitro models.
Hyperplasia is measured histologically based on increased cell numbers leading to increased layers of cells and tissue depth (Collins 2018). Several modifications to guideline in vivo toxicity studies would enhance the detection of hyperplasia and could also detect altered mammary development. These include adding time points and more detailed protocols for evaluating mammary tissue, assessment in male as well as female mammary tissue, longitudinal rather than transverse sectioning, and BrDU injection and staining to better detect proliferation. These assessments could be added to 90-day, 28-day, and 14-day studies, and to studies with developmental exposure such as EPA’s pubertal and the OECD one-generation reproduction study (Makris 2011; Rudel et al. 2011).
In mammary gland, Ki67 or histology is commonly used to characterize proliferation both in vivo and in vitro, and other methods can be easily applied as well. Given the potential variation in endogenous and context-dependent proliferation between mammary and other tissues, findings in non-mammary tissues should be either validated or measured directly in mammary tissues.
Inflammation
Inflammation is commonly measured using cytokine mRNA or extracellular concentrations of cytokines like IL-6, expression of proteins including COX2 or iNOS, activation of key inflammatory signaling molecules MAP kinases and transcription factors NF-kB, and AP1, as well as histological measures like leukocyte infiltration.
Typical assays to detect changes in protein concentration, phosphorylation, or localization include ELISA (El-Saghire et al. 2013; Partridge et al. 2010; Siva et al. 2014), Western Blot (Chai et al. 2013b; Ha et al. 2010), electrophoretic mobility shift assay or EMSA (Haase et al. 2003), or immunostaining (Chai et al. 2013b; Wang et al. 2015). Immunostaining can also detect infiltration of immune cells, a marker for inflammation (Ebrahimian et al. 2018; Monceau et al. 2013; Moravan et al. 2011). Changes in gene transcription are detected using PCR (Azimzadeh et al. 2011; Bouchet et al. 2013; Moravan et al. 2011; Snijders et al. 2012; Wang et al. 2014b). Other methods include histopathological examination of tissue for indicators of inflammation (Haddadi et al. 2017), or measurements of leukocyte adhesion (Arenas et al. 2006; Rodel et al. 2008).
EPA’s ToxCast has a range of assays measuring inflammation-related outcomes (Houck et al. 2009). All of these assays stimulate cells with inflammatory stimuli in addition to the test reagent, so it is possible (though not so reported) that these are more sensitive to reduction than induction of inflammatory pathways.
No specific test is recommended for chronic inflammation. Like GI, the above assays may be applied at various time points after exposure as an indicator of chronic response. We support the development of methods optimized for chronic inflammation. These methods would likely require long-term or repeated exposures. Additionally, since inflammation is a tissue response and not a cell response alone, such an assay would ideally capture tissue interactions.
The above methods are readily applied to mammary tissue (Barcellos-Hoff et al. 1994; Bouchard et al. 2013; Datta et al. 2012a; Snijders et al. 2012; Wang et al. 2015). Since it is not well established how inflammatory responses in a different tissue might apply to mammary tissue, validation studies should be conducted to establish the tissue variation in assay responses to inflammatory stressors, or studies should be conducted in mammary gland.
Epigenetic changes
Epigenetic changes occur and are measured in several different ways. Shifts in global methylation are measured using the [3H]dCTP extension assay (Koturbash et al. 2016), HPLC for 5mdC (Wang et al. 2014a), or quantitative immunoassay for 5-mc (Nzabarushimana et al. 2014). Gene-specific changes in methylation can be measured using MeDIP-on-chip (Hsu et al. 2015), methylation-sensitive/dependent restriction enzymes and qPCR (Nzabarushimana et al. 2014; Oakes et al. 2006a) and bisulfite sequencing and PCR (Wang et al. 2014a). Changes in miRNA expression are measured using qRT-PCR (Stankevicins et al. 2013; Szatmari et al. 2017), hybridization (e.g., microarray, etc.) (Aypar et al. 2011; Jacob et al. 2013), and sequencing (Mestdagh et al. 2014), while histone methylation is measured using ChIP (Prior et al. 2016) or immunohistochemistry (Kutanzi and Kovalchuk 2013).
The most consequential outcome of epigenetic changes is changes in gene expression, which can be measured using western blot (Wang et al. 2014a), qRT-PCR (Prior et al. 2016), or whole transcriptomics, e.g., RNA microarray and RNA-seq (Hrdlickova et al. 2017; Manzoni et al. 2018).
These methods can be and have been applied to breast and mammary gland (Kutanzi and Kovalchuk 2013; Loree et al. 2006; Luzhna et al. 2015; Stankevicins et al. 2013). Considering the tissue, sex, and temporal variability reported in epigenetic effects, experiments that differ in one of these contexts should not be extrapolated to mammary gland and instead the study should directly examine mammary tissue.
Weight of evidence for essentiality of key events
The evidence was evaluated for the essentiality of each key event to downstream events and to breast cancer and is summarized in Table 2. IR appears to be a “complete” carcinogen in the mammary gland in that it acts as an initiator through the formation of oxidative stress and pro-mutagenic DNA damage (the MIEs) and as a promoter through increasing inflammation and proliferation, similar to many chemical carcinogens (Russo and Russo 1996). We have high confidence in the evidence linking stressor (IR) with adverse outcome (breast cancer). The weight of evidence for breast carcinogenesis arising from RONS and DNA damage leading to GI, mutation, proliferation, and hyperplasia is high, while the weight of evidence for the second pathway from RONS and DNA damage to GI, inflammation, proliferation, hyperplasia, and breast cancer is moderate based on the moderate confidence in the inflammation key events. The weight of evidence for epigenetics in these pathways is low based on the current evidence.
Table 2.
Weight of evidence for essentiality of key events to the breast cancer pathway
| Key events | Essentiality |
|---|---|
| Increase in reactive oxygen and nitrogen species (RONS) | Essentiality is high. A large number of studies using antioxidants or other interventions to reduce RONS show a reduction in DNA damage, GI, mutations, and inflammation. Additional support comes from experiments increasing external oxidants like H2O2, which show that RONS are independently capable of causing DNA damage, mutations inflammation, and epigenetic changes. Uncertainties arise from the smaller effects of RONS on DNA damage compared with ionizing radiation. Mammary gland relevance is less certain due to the relatively few experiments in breast tissue |
| Increase in DNA damage, GI, and mutation | Essentiality is high. The increase in DNA damage and GI precedes mutations, proliferation, and tumorigenesis, while antioxidants that reduce DNA damage and GI also reduce mutations and chromosomal damage. Knock-out and knock-in experiments show that mutations in certain key genes increase tumorigenesis. However, an ongoing debate fueled by transplant studies that show the importance of tissue environment for tumorigenesis questions the necessity of mutations as the singular driver of tumorigenesis, since mutation also follows the other tumor-promoting events like inflammation and proliferation |
| Increase in Proliferation and Hyperplasia | Essentiality is high. Evidence comes from transplant experiments showing that non-proliferating tissue is less tumorigenic than proliferating lesions, and from interventions that reduce both proliferation and tumors. Further evidence comes from animals that are resistant to both mammary gland proliferation and tumors from ionizing radiation. Uncertainty arises from conflicting evidence on the tumorigenicity of hyperplasia, and the absence of hyperplasia observed before some tumors |
| Increase in inflammation | Essentiality is moderate. Evidence comes from using genetic modifications, antibodies, and antioxidants to reduce inflammatory and anti-inflammatory factors. These interventions reduce DNA damage, mutations, and mechanisms contributing to tumorigenesis and invasion. Uncertainty arises from conflicting effects in different genetic backgrounds and in different organs, and the pro-apoptotic effect of TGF-β |
| Epigenetic changes | Essentiality is low. The strongest evidence shows that blocking miRNAs and DNMTs reduces bystander DNA damage and GI. The remainder of the evidence correlates epigenetic changes with downstream effects. Uncertainty arises from the tissue dependence of epigenetic effects combined with the low number of studies in mammary gland, and from the discontinuity between epigenetic changes and changes in gene and protein expression |
Discussion
Context
DNA damage and mutation events correspond to the classical mechanism of carcinogens as mutagens first identified over 50 years ago, while the role of RONS and inflammation is in alignment with the tissue-oriented field theory which elevates the importance of tissue environment (Baker 2015). Modern conceptions for carcinogenesis marry these models and recognize that most carcinogens act on multiple biological targets. For example, IARC’s review of biological activity for known human carcinogens led to the listing of ten “characteristics of carcinogens” that include inducing genotoxicity, GI, oxidative stress, and chronic inflammation, among others (Smith et al. 2016), and the 2012 IARC monograph on carcinogenicity of ionizing radiation reflects many of the same key events which we have highlighted here (IARC 2012). The interaction of key events in this AOP with hormonally driven development and proliferation contributes additional complexity for breast and other hormonally mediated cancers. Because a single agent often has more than one of these characteristics, and host susceptibility and co-exposures also influence these pathways, a simple linear AOP will not adequately represent the underlying processes. For example, experiments suggest that the likelihood of mammary cancer after IR is lower when ovaries are removed and that both mutation and estrogen exposure (via effects on proliferation and other key events) contribute to breast carcinogenesis allowing for additive or synergistic effects (Broerse et al. 1987; Clifton et al. 1985; Cronkite et al. 1960).
Dose–response and low-dose epidemiology and animal studies
Although this paper describes a qualitative AOP, an overview of dose–response observations from low-dose studies provides useful context.
Comprehensive and authoritative reviews of ionizing radiation consistently conclude that the linear no threshold dose–response (sometimes with slope adjustments for low dose or low-dose rate) is the most appropriate and protective model for cancers following exposure to IR (Ruhm et al. 2016; Shore et al. 2018). Low power limits precise estimations of the dose–response in people exposed to low doses (below approximately 0.1 Gy) of mixed or low LET IR like the atomic bomb or medical radiation (Grant et al. 2017; Ozasa et al. 2012; Ruhm et al. 2016; Shore et al. 2018; Suzuki and Yamashita 2012), but solid cancer dose–response in the atomic bomb Life Span Study (LSS) cohort is not significantly different from linear (Furukawa et al. 2016; Grant et al. 2017; Preston et al. 2007). Multiple epidemiological studies of specific cancer sites and total solid cancer following in utero and childhood exposure and in atomic bomb, nuclear worker, and population studies also support significant effects at low doses (Cohen et al. 2018; Little et al. 2014; Mathews et al. 2013; Meulepas et al. 2019; Pearce et al. 2012; Preston et al. 2007; Ruhm et al. 2016; Shore et al. 2017), although a metanalysis of these studies reports that IR does pose a greater cancer risk per unit dose at high-dose rates than at low-dose rates (less than 0.0001 Gy/minute) (Shore et al. 2017).
Although total solid cancers have a linear dose–response, dose–response shape differs by tumor site and sex (Grant et al. 2017; Ozasa et al. 2012; Shore et al. 2018; Suzuki and Yamashita 2012). In males, total tumors and some specific tumor sites have upwardly curving dose–responses with shallower dose–response at low vs high doses, while total tumors in females and sex-specific tumors (combined across sexes) have linear dose–responses (Grant et al. 2017; Sasaki et al. 2014). Studies in animals support overall linearity and show variation in dose–response between affected tissues (Little 2018; Tran and Little 2017).
Available data suggest that breast cancer dose–response is linear. A pooled analysis of eight breast cancer cohorts found no evidence to contradict linear dose–response for breast cancer, although they did find lower excess risk for low-dose rates (Preston et al. 2002). Cancer data from women exposed as infants (mean dose 0.186 Gy) show a linear increased risk of breast cancer (Eidemuller et al. 2015). More recent examinations of LSS data using updated dose estimates support this conclusion, finding no significant non-linearity of breast cancer over the whole or 0–2 Gy dose range (Preston et al. 2007), but the most recent dose and incident LSS data have not yet been reported for breast cancer.
Like breast cancer in people, the dose–response for mammary gland tumors in rodents is linear (Gragtmans et al. 1984; Imaoka et al. 2013), but has not been widely investigated in the low-dose range (< 0.1 Gy). One low-dose experiment reporting mammary tumors did not find a significant change (increase or decrease) in mammary tumors at chronic low doses, all of which were below 0.021 Gy/day (Tanaka et al. 2007). Although each group contained 500 animals, this study was performed in B6C3F1 mice which do not carry the mouse mammary tumor virus and are not otherwise particularly susceptible to mammary tumors, so the power to detect changes may still have been insufficient (Bennett and Davis 2002).
Animal and in vitro studies suggest that high LET radiation (alpha particles from cosmic radiation and radioactive decay of isotopes such as plutonium from nuclear materials or radon gas) has a linear or even supralinear dose–response at low doses (Azzam et al. 2016; Little 2018; Tran and Little 2017).
Generalizability to other agents
Breast carcinogenesis from IR and other DNA-damaging agents has more similarities than differences (Imaoka et al. 2009). Both IR and other DNA-damaging agents form adenocarcinomas in rodents with similar pathology and gene expression, although IR also creates a much larger fraction of fibroadenomas than the other DNA-damaging agents (Imaoka et al. 2009). Carcinogenicity for IR and chemical mammary carcinogens NMU and DMBA varies consistently with age and exposure to ovarian hormones (Imaoka et al. 2009; Medina 2007; Russo 2015). Breast carcinogenesis from IR and chemical carcinogens depends strongly on developmental or ongoing exposure to ovarian hormones (Nandi et al. 1995; Russo 2015), and estrogen status of IR and chemical carcinogen-induced tumors increases with ovarian hormone exposure in rats (Imaoka et al. 2009; Nandi et al. 1995). The mammary gland is especially susceptible to both IR and mammary carcinogens DMBA and NMU around puberty. This is presumably because puberty is when undifferentiated cells are present in large numbers and poised to undergo subsequent proliferative expansion, although additional factors including metabolism and expression of DNA damage repair genes contribute to variations in the age of maximal susceptibility between agents (Imaoka et al. 2009, 2013, 2011; Medina 2007). IR has an additive effect in combination with NMU (Imaoka et al. 2014), consistent with general accepted risk assessment assumptions of additivity in carcinogenesis (National Research Council 2009). Some differences between mammary carcinogens appear around the protective role of breast maturation: pregnancy appears to be more protective in rats exposed to chemical carcinogens than in rats exposed to IR (Imaoka et al. 2009).
The role of DNA damage, mutation, and proliferation outlined in this AOP would presumably apply to other DNA-damaging agents, while the role of RONS and inflammation is more likely to vary between DNA-damaging and other agents based on their ability to induce these key events. DNA-damaging agents differ in the degree, type, and reparability of the DNA damage which they cause. Mammary carcinogens NMU, DMBA, PhIP, and urethane mostly cause adducts with single-nucleotide substitutions (Committee to Assess Health Risks from Exposure to Low Levels of Ionizing Radiation 2006; Imaoka et al. 2009; Nik-Zainal et al. 2015; Sherborne et al. 2015; Westcott et al. 2014). Like ionizing radiation, mammary carcinogen PhIP can cause amplifications and NMU can cause GI (Goepfert et al. 2007; Imaoka et al. 2009). While IR also induces adducts, it characteristically generates complex damage and double-strand breaks leading to deletions and inversions as well as amplification and GI (Behjati et al. 2016; Datta et al. 2012b; Mavragani et al. 2017; Mukherjee et al. 2012; Pazhanisamy et al. 2011; Snijders et al. 2012; Yang et al. 2015). The prevalence of complex damage and double-strand breaks is likely due to the density of damage delivered by ionizing radiation, but is also attributable to oxidative activity, since IR creates an oxidative state and H2O2 and other oxidizing agents can also cause complex damage, double-strand breaks and mutations (Cadet et al. 2017; Seager et al. 2012; Sharma et al. 2016). Radiomimetic compounds (used in chemotherapy) also cause double-strand breaks and simple complex damage. Agents like bleomycin cause double-strand breaks through oxidized lesions (Regulus et al. 2007), while agents like etoposide and cisplatin cause double-strand breaks by interfering with DNA replication forks (Kawashima et al. 2017).
Proliferation and inflammation are also implicated in chemical carcinogenicity. The aforementioned pubertal susceptibility to carcinogen exposure implies an important role for proliferation, as does the fact that tumorigenesis following NMU depends on proliferation during treatment (Medina 2007). Like IR, NMU and DMBA promote hyperplasia in terminal-end buds and ducts and ductal carcinoma in situ leading to carcinogenesis (Goepfert et al. 2007; Imaoka et al. 2009; Medina 2007; Russo 2015). In terms of inflammation, like IR, some chemical carcinogens increase inflammatory reactions in mammary stroma and also show a tumor-promoting effect of exposed stroma (Barcellos-Hoff and Ravani 2000; Maffini et al. 2004; Nguyen et al. 2011b; Russo and Russo 1996) and although bleomycin has not been characterized for its effects on mammary stroma or mammary carcinogenesis, it causes lung fibrosis (an inflammatory reaction) so consistently that it is used as a research model for that endpoint (Moeller et al. 2008).
Other key events
This review does not address a large number of biological processes that would be expected to interact with the key events described here. For example, changes in the tissue microenvironment such as increased breast density are associated with increased risk of cancer. Multiple changes occur in the tissue microenvironment following IR and inflammation that may increase density, but insufficient data are available to characterize these changes as a separate key event after IR. Subsequent experiments should examine the time course, dose-dependence, and essentiality of changes to the breast microenvironment including breast density after IR and consider whether these factors should be considered a separate key event. Other important biological effects of IR that would be expected to interact with this pathway include immune surveillance which may change with the inflammatory environment after IR (Barcellos-Hoff 2013; Lumniczky and Safrany 2015; Schreiber et al. 2011); IR effect on survival/apoptosis and interactions of apoptosis with inflammation, mutation, compensatory proliferation, and selection process; changes to DNA repair or the many possible influences on those events. The interaction of these key events with hormonally driven development and proliferation represent a critical aspect of breast carcinogenesis with significant implications for sensitivity of experimental models as well as dose–response.
Data gaps
Guideline tests are not established for important cancer-related endpoints such as RONS, GI, proliferation, hyperplasia, and inflammation. In addition, assessment of mammary gland effects in guideline studies is limited, and so, important changes can be missed (Makris 2011; Rudel et al. 2011). Adopting standardized methods for these key events and ensuring that the methods are either applied in mammary gland or that results in different tissues are relevant to mammary gland would better capture the potential of a chemical to act as a breast carcinogen. Improved assays are needed to measure genomic instability and chronic inflammation, as well as the interaction with hormonally driven development and proliferation.
An important gap relates to knowledge of the interactions between the key events in this AOP and hormonally driven development and proliferation in the breast, and this gap is a particular barrier to developing a quantitative AOP. Because stressors often activate multiple parts of this pathway, we caution against generalizing the dose–response across different stressors. We expect a different dose–response relationship depending on if a stressor only increases one part of the pathway (such as increasing mutations) or affects multiple parts (such as RONS, mutation and proliferation). In addition, individuals live in a complex mixture of environmental and genetic influences, some of which act on one or more events in the same carcinogenesis pathway to increase the overall risk of breast cancer. As a result, dose–response relationships for a particular stressor will also vary depending on host susceptibilities and co-exposures.
Evidence suggests that breast cancer increases linearly with dose even at lower doses, and that breast cancer incidence is elevated at doses relevant to diagnostic radiation (0.001 to 0.1 Gy), but greater power is needed to determine the precise shape of the breast cancer dose–response at these doses. DNA damage appears to increase linearly even at low doses, but even for this well-studied key event, there are insufficient studies with multiple low doses and sufficient power to enable the estimate of the lower end of the dose–response relationship in mammary gland. For other key events, a few studies inform the dose–response in any tissue. For example, although many studies support the link between IR, RONS, and DNA damage, a few RONS studies use mammary tissue, multiple doses, doses below 1 Gy, or explicitly measure RNS presenting an addressable data gap. Given these uncertainties, a better characterization of the low-dose end of the dose–response for the key events would give a more complete picture of the relative contribution of these different events and pathways to the breast cancer outcome across a range of doses. If more evidence is needed, it would be helpful to measure the dose–response of DNA damage and other key events in breast tissue using the highest possible power in this lower dose range and test multiple doses.
Conclusions
This paper extends the characteristics of mammary carcinogens beyond DNA damage, highlighting the important role in breast cancer of agents that increase RONS, GI, cell proliferation, and inflammation. The interaction of these key events with hormonally driven development and proliferation represent a critical aspect of breast carcinogenesis. The AOPs which we have described for ionizing radiation leading to breast cancer are consistent with recent enumeration of characteristics of many known human carcinogens by IARC and other scientists (IARC 2012; Smith et al. 2016). Standardized assays are needed to measure RONS, GI, proliferation, and chronic inflammation, and breast cancer-related effects may be missed unless mammary gland is more consistently included in testing and assays in other tissues are evaluated to see if they can be generalized to breast. Adopting standardized methods for these key events would better capture the potential of a chemical to act as a breast carcinogen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to Laura Borth for her assistance in proofreading this manuscript, and to Paul Yaswen for his helpful comments on an earlier version of this manuscript.
Abbreviations
- IR
Ionizing radiation
- RONS
Reactive oxygen and nitrogen species
- AOP
Adverse outcome pathway
Author contributions
JH reviewed articles and extracted relevant information, wrote the initial draft, drafted the OECD AOP, and edited the manuscript; RAR conceived the project, contributed to conceptualizing the AOP, and edited the manuscript, contributing certain content.
Funding
Financial support was provided by grants to Silent Spring Institute from the California Breast Cancer Research Program (17UB-8708, 21UB-8012, and 21UB-8100), the Avon Foundation for Women, the Cedar Tree Foundation, and Silent Spring Institute’s charitable gifts Innovation Fund. RAR and JH are or were (JH) employed at the Silent Spring Institute, a scientific research organization dedicated to studying environmental factors in women’s health. The Institute is a 501(c)3 public charity funded by federal grants and contracts, foundation grants, and private donations, including from breast cancer organizations. Study funders had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
This manuscript is an original work, has not been previously published in whole or in part in a peer-reviewed journal, and is not under consideration for publication in another peer-reviewed journal. A more extensive description of this AOP containing over 30,000 words and 500 references has been entered into the OECD's online AOP wiki (aopwiki.org) and will be submitted to review by the Extended Advisory Group on Molecular Screening and Transcriptomics for publication in the OECD series on adverse outcome pathways. Both of the authors have read and approved the paper, and it has not been published previously nor is it being considered by any other peer-reviewed journal.
Availability of data and material
Supplemental files to the manuscript contain key data extracted from the 467 articles that we reviewed.
Footnotes
Biological systems studied
This review article summarizes and synthesizes data from cell-culture, experimental animals, and human epidemiology.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adams LM, Ethier SP, Ullrich RL. Enhanced in vitro proliferation and in vivo tumorigenic potential of mammary epithelium from BALB/c mice exposed in vivo to gamma-radiation and/or 7,12-dimethylbenz[a]anthracene. Cancer Res. 1987;47:4425–4431. [PubMed] [Google Scholar]
- Al-Mayah A, et al. The non-targeted effects of radiation are perpetuated by exosomes. Mutat Res. 2015;772:38–45. doi: 10.1016/j.mrfmmm.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Al-Mayah AH, Irons SL, Pink RC, Carter DR, Kadhim MA. Possible role of exosomes containing RNA in mediating nontargeted effect of ionizing radiation. Radiat Res. 2012;177:539–545. doi: 10.1667/rr2868.1. [DOI] [PubMed] [Google Scholar]
- Alkner S, Ehinger A, Bendahl PO, Ryden L, Ferno M. Prognosis, stage and oestrogen receptor status of contralateral breast cancer in relation to characteristics of the first tumour, prior endocrine treatment and radiotherapy. Eur J Cancer. 2015;51:2304–2313. doi: 10.1016/j.ejca.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Ameziane-El-Hassani R, et al. Role of H2O2 in RET/PTC1 chromosomal rearrangement produced by ionizing radiation in human thyroid cells. Cancer Res. 2010;70:4123–4132. doi: 10.1158/0008-5472.can-09-4336. [DOI] [PubMed] [Google Scholar]
- Ameziane-El-Hassani R, et al. NADPH oxidase DUOX1 promotes long-term persistence of oxidative stress after an exposure to irradiation. Proc Natl Acad Sci USA. 2015;112:5051–5056. doi: 10.1073/pnas.1420707112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andarawewa KL, Costes SV, Fernandez-Garcia I, Chou WS, Ravani SA, Park H, Barcellos-Hoff MH. Lack of radiation dose or quality dependence of epithelial-to-mesenchymal transition (EMT) mediated by transforming growth factor beta. Int J Radiat Oncol Biol Phys. 2011;79:1523–1531. doi: 10.1016/j.ijrobp.2010.11.058. [DOI] [PubMed] [Google Scholar]
- Andarawewa KL, et al. Ionizing radiation predisposes nonmalignant human mammary epithelial cells to undergo transforming growth factor beta induced epithelial to mesenchymal transition. Cancer Res. 2007;67:8662–8670. doi: 10.1158/0008-5472.CAN-07-1294. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Stevens DL, Goodhead DT. M-FISH analysis shows that complex chromosome aberrations induced by alpha -particle tracks are cumulative products of localized rearrangements. Proc Natl Acad Sci USA. 2002;99:12167–12172. doi: 10.1073/pnas.182426799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SM, Rudolph MC, McManaman JL, Neville MC. Key stages in mammary gland development. Secretory activation in the mammary gland: it's not just about milk protein synthesis. Breast Cancer Res. 2007;9:204. doi: 10.1186/bcr1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrieu N, et al. Effect of chest X-rays on the risk of breast cancer among BRCA1/2 mutation carriers in the international BRCA1/2 carrier cohort study: a report from the EMBRACE GENEPSO, GEO-HEBON, and IBCCS Collaborators'. Group J Clin Oncol. 2006;24:3361–3366. doi: 10.1200/JCO.2005.03.3126. [DOI] [PubMed] [Google Scholar]
- Arenas M, et al. Anti-inflammatory effects of low-dose radiotherapy in an experimental model of systemic inflammation in mice. Int J Radiat Oncol Biol Phys. 2006;66:560–567. doi: 10.1016/j.ijrobp.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Asaithamby A, Chen DJ. Cellular responses to DNA double-strand breaks after low-dose gamma-irradiation. Nucleic Acids Res. 2009;37:3912–3923. doi: 10.1093/nar/gkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atashgaran V, Wrin J, Barry SC, Dasari P, Ingman WV. Dissecting the biology of menstrual cycle-associated breast cancer risk front. Oncol. 2016;6:267. doi: 10.3389/fonc.2016.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aypar U, Morgan WF, Baulch JE. Radiation-induced epigenetic alterations after low and high LET irradiations. Mutat Res. 2011;707:24–33. doi: 10.1016/j.mrfmmm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Azimzadeh O, et al. Rapid proteomic remodeling of cardiac tissue caused by total body ionizing radiation. Proteomics. 2011;11:3299–3311. doi: 10.1002/pmic.201100178. [DOI] [PubMed] [Google Scholar]
- Azzam EI, Colangelo NW, Domogauer JD, Sharma N, de Toledo SM. Is ionizing radiation harmful at any exposure? An echo that continues to vibrate. Health Phys. 2016;110:249–251. doi: 10.1097/HP.0000000000000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam EI, De Toledo SM, Spitz DR, Little JB. Oxidative metabolism modulates signal transduction and micronucleus formation in bystander cells from alpha-particle-irradiated normal human fibroblast cultures. Can Res. 2002;62:5436–5442. [PubMed] [Google Scholar]
- Baker SG. A cancer theory kerfuffle can lead to new lines of research. J Natl Cancer Inst. 2015 doi: 10.1093/jnci/dju405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff MH. Does microenvironment contribute to the etiology of estrogen receptor-negative breast cancer? Clin Cancer Res. 2013;19:541–548. doi: 10.1158/1078-0432.CCR-12-2241. [DOI] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Derynck R, Tsang ML, Weatherbee JA. Transforming growth factor-beta activation in irradiated murine mammary gland. J Clin Invest. 1994;93:892–899. doi: 10.1172/JCI117045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Kleinberg DL. Breast cancer risk in BRCA1 mutation carriers: insight from mouse models. Ann Oncol. 2013;24(Suppl 8):8–12. doi: 10.1093/annonc/mdt305. [DOI] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Mao JH. HZE radiation non-targeted effects on the microenvironment that mediate mammary carcinogenesis. Front Oncol. 2016;6:57. doi: 10.3389/fonc.2016.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Nguyen DH. Radiation carcinogenesis in context: how do irradiated tissues become tumors? Health Phys. 2009;97:446–457. doi: 10.1097/HP.0b013e3181b08a10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Ravani SA. Irradiated mammary gland stroma promotes the expression of tumorigenic potential by unirradiated epithelial cells. Cancer Res. 2000;60:1254–1260. [PubMed] [Google Scholar]
- Barnard S, Bouffler S, Rothkamm K. The shape of the radiation dose response for DNA double-strand break induction and repair. Genome Integr. 2013;4:1. doi: 10.1186/2041-9414-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartstra RW, Bentvelzen PA, Zoetelief J, Mulder AH, Broerse JJ, van Bekkum DW. Induction of mammary tumors in rats by single-dose gamma irradiation at different ages. Radiat Res. 1998;150:442–450. [PubMed] [Google Scholar]
- Baulch JE, Aypar U, Waters KM, Yang AJ, Morgan WF. Genetic and epigenetic changes in chromosomally stable and unstable progeny of irradiated cells. PLoS ONE. 2014;9:e107722. doi: 10.1371/journal.pone.0107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RA, et al. Increasing scientific confidence in adverse outcome pathways: application of tailored bradford-hill considerations for evaluating weight of evidence. Regul Toxicol Pharmacol. 2015;72:514–537. doi: 10.1016/j.yrtph.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Beels L, Werbrouck J, Thierens H. Dose response and repair kinetics of gamma-H2AX foci induced by in vitro irradiation of whole blood and T-lymphocytes with X- and gamma-radiation. Int J Radiat Biol. 2010;86:760–768. doi: 10.3109/09553002.2010.484479. [DOI] [PubMed] [Google Scholar]
- Begg CB, et al. Clonal relationships between lobular carcinoma in situ and other breast malignancies. Breast Cancer Res. 2016;18:66. doi: 10.1186/s13058-016-0727-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, et al. Mutational signatures of ionizing radiation in second malignancies. Nat Commun. 2016;7:12605. doi: 10.1038/ncomms12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LM, Davis BJ. Identification of mammary carcinogens in rodent bioassays. Environ Mol Mutagen. 2002;39:150–157. doi: 10.1002/em.10068. [DOI] [PubMed] [Google Scholar]
- Bensimon J, et al. Forced extinction of CD24 stem-like breast cancer marker alone promotes radiation resistance through the control of oxidative stress. Mol Carcinog. 2016;55:245–254. doi: 10.1002/mc.22273. [DOI] [PubMed] [Google Scholar]
- Berdelle N, Nikolova T, Quiros S, Efferth T, Kaina B. Artesunate induces oxidative DNA damage, sustained DNA double-strand breaks, and the ATM/ATR damage response in cancer cells. Mol Cancer Ther. 2011;10:2224–2233. doi: 10.1158/1535-7163.MCT-11-0534. [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Dolinoy DC, Huang D, Skaar DA, Weinhouse C, Jirtle RL. Adaptive radiation-induced epigenetic alterations mitigated by antioxidants. Faseb J. 2013;27:665–671. doi: 10.1096/fj.12-220350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JL, et al. Radiation exposure, the ATM Gene, and contralateral breast cancer in the women's environmental cancer and radiation epidemiology study. J Natl Cancer Inst. 2010;102:475–483. doi: 10.1093/jnci/djq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JL, et al. Contralateral breast cancer after radiotherapy among BRCA1 and BRCA2 mutation carriers: a WECARE study report. Eur J Cancer. 2013;49:2979–2985. doi: 10.1016/j.ejca.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrington de Gonzalez A, Curtis RE, Gilbert E, Berg CD, Smith SA, Stovall M, Ron E. Second solid cancers after radiotherapy for breast cancer in SEER cancer registries. Br J Cancer. 2010;102:220–226. doi: 10.1038/sj.bjc.6605435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruyer C, et al. Vanin-1-/- mice exhibit a glutathione-mediated tissue resistance to oxidative stress. Mol Cell Biol. 2004;24:7214–7224. doi: 10.1128/mcb.24.16.7214-7224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuving LJ. Mammary tumor formation within outgrowths of transplanted hyperplastic nodules from carcinogen-treated rats. J Natl Cancer Inst. 1968;40:1287–1291. [PubMed] [Google Scholar]
- Beuving LJ, Bern HA, DeOme KB. Occurrence and transplantation of carcinogen-induced hyperplastic nodules in fischer rats2. JNCI J Natl Cancer Inst. 1967;39:431–447. doi: 10.1093/jnci/39.3.431. [DOI] [Google Scholar]
- Beuving LJ, Faulkin JLJ, DeOme KB, Bergs VV. Hyperplastic lesions in the mammary glands of sprague-dawley rats after 7,12-dimethylbenz[a]anthracene treatment2. JNCI J Natl Cancer Inst. 1967;39:423–429. doi: 10.1093/jnci/39.3.423. [DOI] [PubMed] [Google Scholar]
- Bijwaard H, Brenner A, Dekkers F, van Dillen T, Land CE, Boice JD., Jr Breast cancer risk from different mammography screening practices. Radiat Res. 2010;174:367–376. doi: 10.1667/RR2067.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht KS, Bradbury CM, Zoberi I, Curry HA, Kaushal A, Roti Roti JL, Gius D. Inhibition of cyclooxygenase-2 with NS-398 and the prevention of radiation-induced transformation, micronuclei formation and clonogenic cell death in C3H 10T1/2 cells. Int J Radiat Biol. 2003;79:879–888. doi: 10.1080/09553000310001621400. [DOI] [PubMed] [Google Scholar]
- Bock FJ, et al. Loss of PIDD limits NF-kappaB activation and cytokine production but not cell survival or transformation after DNA damage. Cell Death Differ. 2013;20:546–557. doi: 10.1038/cdd.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boice JD, Jr, Preston D, Davis FG, Monson RR. Frequent chest X-ray fluoroscopy and breast cancer incidence among tuberculosis patients in Massachusetts. Radiat Res. 1991;125:214–222. [PubMed] [Google Scholar]
- Bonnet F, et al. An array CGH based genomic instability index (G2I) is predictive of clinical outcome in breast cancer and reveals a subset of tumors without lymph node involvement but with poor prognosis. BMC Med Genom. 2012;5:54. doi: 10.1186/1755-8794-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard G, Bouvette G, Therriault H, Bujold R, Saucier C, Paquette B. Pre-irradiation of mouse mammary gland stimulates cancer cell migration and development of lung metastases. Br J Cancer. 2013;109:1829–1838. doi: 10.1038/bjc.2013.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet A, et al. Early gene expression analysis in 9L orthotopic tumor-bearing rats identifies immune modulation in molecular response to synchrotron microbeam radiation therapy. PLoS ONE. 2013;8:e81874. doi: 10.1371/journal.pone.0081874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady NJ, Chuntova P, Schwertfeger KL. Macrophages: regulators of the inflammatory microenvironment during mammary gland development and breast cancer. Mediat Inflamm. 2016;2016:4549676. doi: 10.1155/2016/4549676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken C, Hess K, Jeitziner R. Progesterone and overlooked endocrine pathways in breast cancer pathogenesis. Endocrinology. 2015;156:3442–3450. doi: 10.1210/en.2015-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt K, Ashworth A, Smalley M. Pregnancy and the risk of breast cancer. Endocr Relat Cancer. 2007;14:907–933. doi: 10.1677/ERC-07-0137. [DOI] [PubMed] [Google Scholar]
- Broeks A, et al. Identification of women with an increased risk of developing radiation-induced breast cancer: a case only study. Breast Cancer Res. 2007;9:R26. doi: 10.1186/bcr1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broerse JJ, Hennen LA, Klapwijk WM, Solleveld HA. Mammary carcinogenesis in different rat strains after irradiation and hormone administration. Int J Radiat Biol Relat Stud Phys Chem Med. 1987;51:1091–1100. doi: 10.1080/09553008714551381. [DOI] [PubMed] [Google Scholar]
- Brooks JD, et al. Reproductive status at first diagnosis influences risk of radiation-induced second primary contralateral breast cancer in the WECARE study. Int J Radiat Oncol Biol Phys. 2012;84:917–924. doi: 10.1016/j.ijrobp.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JD, et al. Variants in activators and downstream targets of ATM, radiation exposure, and contralateral breast cancer risk in the WECARE study. Hum Mutat. 2012;33:158–164. doi: 10.1002/humu.21604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno M, de Toledo SM, Pain D, Azzam EI. Long-term consequences of radiation-induced bystander effects depend on radiation quality and dose and correlate with oxidative stress. Radiat Res. 2011;175:405–415. doi: 10.1667/RR2461.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr KL, Robinson JI, Rastogi S, Boylan MT, Coates PJ, Lorimore SA, Wright EG. Radiation-induced delayed bystander-type effects mediated by hemopoietic cells. Radiat Res. 2010;173:760–768. doi: 10.1667/RR1937.1. [DOI] [PubMed] [Google Scholar]
- Cadet J, Davies KJA, Medeiros MH, Di Mascio P, Wagner JR. Formation and repair of oxidatively generated damage in cellular DNA. Free Radic Biol Med. 2017;107:13–34. doi: 10.1016/j.freeradbiomed.2016.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcerrada P, Peluffo G, Radi R. Nitric oxide-derived oxidants with a focus on peroxynitrite: molecular targets, cellular responses and therapeutic implications. Curr Pharm Des. 2011;17:3905–3932. doi: 10.2174/138161211798357719. [DOI] [PubMed] [Google Scholar]
- Castiglioni F, et al. Radiation effects on development of HER2-positive breast carcinomas. Clin Cancer Res. 2007;13:46–51. doi: 10.1158/1078-0432.CCR-06-1490. [DOI] [PubMed] [Google Scholar]
- CGAN (Cancer Genome Atlas Network) Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, et al. Radiation induced COX-2 expression and mutagenesis at non-targeted lung tissues of gpt delta transgenic mice. Br J Cancer. 2013;108:91–98. doi: 10.1038/bjc.2012.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Lam RK, Calaf GM, Zhou H, Amundson S, Hei TK. Radiation-induced non-targeted response in vivo: role of the TGFbeta-TGFBR1-COX-2 signalling pathway. Br J Cancer. 2013;108:1106–1112. doi: 10.1038/bjc.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Zhang M, Rajapakshe K, Coarfa C, Edwards D, Huang S, Rosen JM. Mammary stem cells and tumor-initiating cells are more resistant to apoptosis and exhibit increased DNA repair activity in response to DNA damage. Stem Cell Rep. 2015;5:378–391. doi: 10.1016/j.stemcr.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry MA, Omaruddin RA. Differential DNA methylation alterations in radiation-sensitive and -resistant cells. DNA Cell Biol. 2012;31:908–916. doi: 10.1089/dna.2011.1509. [DOI] [PubMed] [Google Scholar]
- Chhabra RS, Huff JE, Schwetz BS, Selkirk J. An overview of prechronic and chronic toxicity/carcinogenicity experimental study designs and criteria used by the national toxicology program. Environ Health Perspect. 1990;86:313–321. doi: 10.1289/ehp.9086313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KM, Kang CM, Cho ES, Kang SM, Lee SB, Um HD. Ionizing radiation-induced micronucleus formation is mediated by reactive oxygen species that are produced in a manner dependent on mitochondria, Nox1, and JNK. Oncol Rep. 2007;17:1183–1188. [PubMed] [Google Scholar]
- Clifton KH, Yasukawa-Barnes J, Tanner MA, Haning RV., Jr Irradiation and prolactin effects on rat mammary carcinogenesis: intrasplenic pituitary and estrone capsule implants. J Natl Cancer Inst. 1985;75:167–175. [PubMed] [Google Scholar]
- Coates PJ, Rundle JK, Lorimore SA, Wright EG. Indirect macrophage responses to ionizing radiation: implications for genotype-dependent bystander signaling. Cancer Res. 2008;68:450–456. doi: 10.1158/0008-5472.CAN-07-3050. [DOI] [PubMed] [Google Scholar]
- Cohen S, et al. Exposure to low-dose ionizing radiation from cardiac procedures and malignancy risk in adults with congenital heart disease. Circulation. 2018;137:1334–1345. doi: 10.1161/CIRCULATIONAHA.117.029138. [DOI] [PubMed] [Google Scholar]
- Colditz GA, Wei EK. Preventability of cancer: the relative contributions of biologic and social and physical environmental determinants of cancer mortality. Annu Rev Public Health. 2012;33:137–156. doi: 10.1146/annurev-publhealth-031811-124627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins LC. Precursor lesions of the low-grade breast neoplasia pathway. Surg Pathol Clin. 2018;11:177–197. doi: 10.1016/j.path.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Committee to Assess Health Risks from Exposure to Low Levels of Ionizing Radiation Health risks from exposure to low levels of ionizing radiation: BEIR VII, Phase 2. Natl Res Council Natl Acad. 2006 doi: 10.17226/11340. [DOI] [PubMed] [Google Scholar]
- Connelly L, Barham W, Onishko HM, Sherrill T, Chodosh LA, Blackwell TS, Yull FE. Inhibition of NF-kappa B activity in mammary epithelium increases tumor latency and decreases tumor burden. Oncogene. 2011;30:1402–1412. doi: 10.1038/onc.2010.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- Cronkite EP, Shellabarger CJ, Bond VP, Lippincott A. Studies on radiation-induced mammary gland neoplasia in the rat. I. The role of the ovary in the neoplastic response of the breast tissue to total- or partial-body x-irradiation. Radiat Res. 1960;12:81–93. [PubMed] [Google Scholar]
- Crump KS, Duport P, Jiang H, Shilnikova NS, Krewski D, Zielinski JM. A meta-analysis of evidence for hormesis in animal radiation carcinogenesis, including a discussion of potential pitfalls in statistical analyses to detect hormesis. J Toxicol Environ Health B Crit Rev. 2012;15:210–231. doi: 10.1080/10937404.2012.659140. [DOI] [PubMed] [Google Scholar]
- Cui W, Ma J, Wang Y, Biswal S. Plasma miRNA as biomarkers for assessment of total-body radiation exposure dosimetry. PLoS ONE. 2011;6:e22988. doi: 10.1371/journal.pone.0022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm-Daphi J, Sass C, Alberti W. Comparison of biological effects of DNA damage induced by ionizing radiation and hydrogen peroxide in CHO cells. Int J Radiat Biol. 2000;76:67–75. doi: 10.1080/095530000139023. [DOI] [PubMed] [Google Scholar]
- Daino K, et al. Epigenetic dysregulation of key developmental genes in radiation-induced rat mammary carcinomas. Int J Cancer. 2018;143:343–354. doi: 10.1002/ijc.31309. [DOI] [PubMed] [Google Scholar]
- Dall G, Risbridger G, Britt K. Mammary stem cells and parity-induced breast cancer protection- new insights. J Steroid Biochem Mol Biol. 2017;170:54–60. doi: 10.1016/j.jsbmb.2016.02.018. [DOI] [PubMed] [Google Scholar]
- Das U, et al. Role of ferulic acid in the amelioration of ionizing radiation induced inflammation: a murine model. PLoS ONE. 2014;9:e97599. doi: 10.1371/journal.pone.0097599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Hyduke DR, Suman S, Moon BH, Johnson MD, Fornace AJ., Jr Exposure to ionizing radiation induced persistent gene expression changes in mouse mammary gland. Radiat Oncol. 2012;7:205. doi: 10.1186/1748-717X-7-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Suman S, Kallakury BV, Fornace AJ., Jr Exposure to heavy ion radiation induces persistent oxidative stress in mouse intestine. PLoS ONE. 2012;7:e42224. doi: 10.1371/journal.pone.0042224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayal D, Martin SM, Limoli CL, Spitz DR. Hydrogen peroxide mediates the radiation-induced mutator phenotype in mammalian cells. Biochem J. 2008;413:185–191. doi: 10.1042/bj20071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayal D, et al. Mitochondrial complex II dysfunction can contribute significantly to genomic instability after exposure to ionizing radiation. Radiat Res. 2009;172:737–745. doi: 10.1667/rr1617.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruin ML, et al. Breast cancer risk in female survivors of Hodgkin's lymphoma: lower risk after smaller radiation volumes. J Clin Oncol. 2009;27:4239–4246. doi: 10.1200/JCO.2008.19.9174. [DOI] [PubMed] [Google Scholar]
- de Ostrovich KK, et al. Paracrine overexpression of insulin-like growth factor-1 enhances mammary tumorigenesis in vivo. Am J Pathol. 2008;173:824–834. doi: 10.2353/ajpath.2008.071005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissova NG, Nasello CM, Yeung PL, Tischfield JA, Brenneman MA. Resveratrol protects mouse embryonic stem cells from ionizing radiation by accelerating recovery from DNA strand breakage. Carcinogenesis. 2012;33:149–155. doi: 10.1093/carcin/bgr236. [DOI] [PubMed] [Google Scholar]
- Deome KB, Faulkin LJ, Jr, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- Dickey JS, Baird BJ, Redon CE, Sokolov MV, Sedelnikova OA, Bonner WM. Intercellular communication of cellular stress monitored by gamma-H2AX induction. Carcinogenesis. 2009;30:1686–1695. doi: 10.1093/carcin/bgp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol. 2011;7:504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Bishop ME, Pearce MG, Davis KJ, White GA, Lyn-Cook LE, Manjanatha MG. Sex-specific dose-response analysis of genotoxicity in cyproterone acetate-treated F344 rats. Mutat Res Genet Toxicol Environ Mutagen. 2014;774:1–7. doi: 10.1016/j.mrgentox.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Dong C, et al. Role of the MAPK pathway in the observed bystander effect in lymphocytes co-cultured with macrophages irradiated with gamma-rays or carbon ions. Life Sci. 2015;127:19–25. doi: 10.1016/j.lfs.2015.02.017. [DOI] [PubMed] [Google Scholar]
- Dores GM, Anderson WF, Beane Freeman LE, Fraumeni JF, Jr, Curtis RE. Risk of breast cancer according to clinicopathologic features among long-term survivors of Hodgkin's lymphoma treated with radiotherapy. Br J Cancer. 2010;103:1081–1084. doi: 10.1038/sj.bjc.6605877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douki T, Ravanat JL, Pouget JP, Testard I, Cadet J. Minor contribution of direct ionization to DNA base damage induced by heavy ions. Int J Radiat Biol. 2006;82:119–127. doi: 10.1080/09553000600573788. [DOI] [PubMed] [Google Scholar]
- Driessens N, et al. Hydrogen peroxide induces DNA single- and double-strand breaks in thyroid cells and is therefore a potential mutagen for this organ. Endocr Relat Cancer. 2009;16:845–856. doi: 10.1677/ERC-09-0020. [DOI] [PubMed] [Google Scholar]
- Du C, et al. Mitochondrial ROS and radiation induced transformation in mouse embryonic fibroblasts. Cancer Biol Ther. 2009;8:1962–1971. doi: 10.4161/cbt.8.20.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Warshall C, Bandyopadhyay C, Dutta D, Chandran B. Interactions between exosomes from breast cancer cells and primary mammary epithelial cells leads to generation of reactive oxygen species which induce DNA damage response, stabilization of p53 and autophagy in epithelial cells. PLoS ONE. 2014;9:e97580. doi: 10.1371/journal.pone.0097580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimian TG, et al. Chronic exposure to external low-dose gamma radiation induces an increase in anti-inflammatory and anti-oxidative parameters resulting in atherosclerotic plaque size reduction in ApoE(-/-) Mice Radiat Res. 2018;189:187–196. doi: 10.1667/RR14823.1. [DOI] [PubMed] [Google Scholar]
- EFSA Scientific Committee et al. Clarification of some aspects related to genotoxicity assessment. EFSA J. 2017;15:e05113. doi: 10.2903/j.efsa.2017.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea J, et al. European contribution to the study of ROS: a summary of the findings and prospects for the future from the COST action BM1203 (EU-ROS) Redox Biol. 2017;13:94–162. doi: 10.1016/j.redox.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhart EJ, Segarini P, Tsang ML, Carroll AG, Barcellos-Hoff MH. Latent transforming growth factor beta1 activation in situ: quantitative and functional evidence after low-dose gamma-irradiation. Faseb J. 1997;11:991–1002. doi: 10.1096/fasebj.11.12.9337152. [DOI] [PubMed] [Google Scholar]
- Eidemuller M, Holmberg E, Jacob P, Lundell M, Karlsson P. Breast cancer risk and possible mechanisms of radiation-induced genomic instability in the Swedish hemangioma cohort after reanalyzed dosimetry. Mutat Res. 2015;775:1–9. doi: 10.1016/j.mrfmmm.2015.03.002. [DOI] [PubMed] [Google Scholar]
- El-Saghire H, Michaux A, Thierens H, Baatout S. Low doses of ionizing radiation induce immune-stimulatory responses in isolated human primary monocytes. Int J Mol Med. 2013;32:1407–1414. doi: 10.3892/ijmm.2013.1514. [DOI] [PubMed] [Google Scholar]
- Elahi E, et al. Five quantitative trait loci control radiation-induced adenoma multiplicity in Mom1R Apc Min/+ mice. PLoS ONE. 2009;4:e4388. doi: 10.1371/journal.pone.0004388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore E, Lao XY, Kapadia R, Giedzinski E, Limoli C, Redpath JL. Low doses of very low-dose-rate low-LET radiation suppress radiation-induced neoplastic transformation in vitro and induce an adaptive response. Radiat Res. 2008;169:311–318. doi: 10.1667/RR1199.1. [DOI] [PubMed] [Google Scholar]
- Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med. 2014 doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA U . Guidelines for carcinogen risk assessment. Washington: Environmental Protection Agency, Risk Assessment Forum; 2005. [Google Scholar]
- Ervik M, Lam F, Ferlay J, Mery L, Soerjomataram I, Bray F (2016) Cancer today. International Agency for Research on Cancer. https://gco.iarc.fr/today. Accessed 23 Mar 2018
- Eskiocak U, et al. CDDO-Me protects against space radiation-induced transformation of human colon epithelial cells. Radiat Res. 2010;174:27–36. doi: 10.1667/RR2155.1. [DOI] [PubMed] [Google Scholar]
- Esquivel-Velazquez M, Ostoa-Saloma P, Palacios-Arreola MI, Nava-Castro KE, Castro JI, Morales-Montor J. The role of cytokines in breast cancer development and progression. J Interferon Cytokine Res. 2015;35:1–16. doi: 10.1089/jir.2014.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier SP, Ullrich RL. Detection of ductal dysplasia in mammary outgrowths derived from carcinogen-treated virgin female BALB/c mice. Cancer Res. 1982;42:1753–1760. [PubMed] [Google Scholar]
- Ewan KB, et al. Transforming growth factor-beta1 mediates cellular response to DNA damage in situ. Cancer Res. 2002;62:5627–5631. [PubMed] [Google Scholar]
- Ezz MK, Ibrahim NK, Said MM, Farrag MA. The beneficial radioprotective effect of tomato seed oil against gamma radiation-induced damage in male rats. J Diet Suppl. 2018;15:923–938. doi: 10.1080/19390211.2017.1406427. [DOI] [PubMed] [Google Scholar]
- Faulkin JLJ, Shellabarger CJ, DeOme KB. Hyperplastic lesions of sprague-dawley rat mammary glands after x irradiation2. JNCI. 1967;39:449–459. doi: 10.1093/jnci/39.3.449. [DOI] [PubMed] [Google Scholar]
- FDA U . Redbook 2000: Guidance for industry and other stakeholders. Toxicological principles for the safety assessment of food ingredients. Silver Spring: US Department of Health and Human Services Food and Drug Administration; 2007. [Google Scholar]
- Fernald K, Kurokawa M. Evading apoptosis in cancer. Trends Cell Biol. 2013;23:620–633. doi: 10.1016/j.tcb.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibach E, Rachmilewitz EA. The effect of fermented papaya preparation on radioactive exposure. Radiat Res. 2015;184:304–313. doi: 10.1667/rr14000.1. [DOI] [PubMed] [Google Scholar]
- Fogarty CE, Bergmann A. Killers creating new life: caspases drive apoptosis-induced proliferation in tissue repair and disease. Cell Death Differ. 2017;24:1390–1400. doi: 10.1038/cdd.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJL, Naghavi M. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–1484. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- Francis SM, et al. A functional connection between pRB and transforming growth factor beta in growth inhibition and mammary gland development. Mol Cell Biol. 2009;29:4455–4466. doi: 10.1128/MCB.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SM, Chakrabarti S, Dick FA. A context-specific role for retinoblastoma protein-dependent negative growth control in suppressing mammary tumorigenesis. PLoS ONE. 2011;6:e16434. doi: 10.1371/journal.pone.0016434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco OE, et al. Altered TGF-alpha/beta signaling drives cooperation between breast cancer cell populations. Faseb J. 2016;30:3441–3452. doi: 10.1096/fj.201500187RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasor J, Weaver AE, Pradhan M, Mehta K. Synergistic up-regulation of prostaglandin E synthase expression in breast cancer cells by 17beta-estradiol and proinflammatory cytokines. Endocrinology. 2008;149:6272–6279. doi: 10.1210/en.2008-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Misumi M, Cologne JB, Cullings HM. A bayesian semiparametric model for radiation dose–response estimation. Risk Anal. 2016;36:1211–1223. doi: 10.1111/risa.12513. [DOI] [PubMed] [Google Scholar]
- Galligan JJ, Rose KL, Beavers WN, Hill S, Tallman KA, Tansey WP, Marnett LJ. Stable histone adduction by 4-oxo-2-nonenal: a potential link between oxidative stress and epigenetics. J Am Chem Soc. 2014;136:11864–11866. doi: 10.1021/ja503604t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Schottker B. Reduction–oxidation pathways involved in cancer development: a systematic review of literature reviews. Oncotarget. 2017;8:51888–51906. doi: 10.18632/oncotarget.17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe JC, et al. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Can Res. 2009;69:7557–7568. doi: 10.1158/0008-5472.CAN-09-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Gaudet F, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Ge J, et al. CometChip: a high-throughput 96-well platform for measuring DNA damage in microarrayed human cells. J Vis Exp. 2014 doi: 10.3791/50607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano KA, Gough AH, Taylor DL, Vernetti LA, Johnston PA. Early safety assessment using cellular systems biology yields insights into mechanisms of action. J Biomol Screen. 2010;15:783–797. doi: 10.1177/1087057110376413. [DOI] [PubMed] [Google Scholar]
- Goepfert TM, Moreno-Smith M, Edwards DG, Pathak S, Medina D, Brinkley WR. Loss of chromosomal integrity drives rat mammary tumorigenesis. Int J Cancer. 2007;120:985–994. doi: 10.1002/ijc.22420. [DOI] [PubMed] [Google Scholar]
- Gomes LR, Terra LF, Wailemann RA, Labriola L, Sogayar MC. TGF-beta1 modulates the homeostasis between MMPs and MMP inhibitors through p38 MAPK and ERK1/2 in highly invasive breast cancer cells. BMC Cancer. 2012;12:26. doi: 10.1186/1471-2407-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodhead DT. Spatial and temporal distribution of energy. Health Phys. 1988;55:231–240. doi: 10.1097/00004032-198808000-00015. [DOI] [PubMed] [Google Scholar]
- Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int J Radiat Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- Gottlieb E, Haffner R, King A, Asher G, Gruss P, Lonai P, Oren M. Transgenic mouse model for studying the transcriptional activity of the p53 protein: age- and tissue-dependent changes in radiation-induced activation during embryogenesis. Embo J. 1997;16:1381–1390. doi: 10.1093/emboj/16.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradzka I, Iwanenko T. A non-radioactive, PFGE-based assay for low levels of DNA double-strand breaks in mammalian cells. DNA Repair (Amst) 2005;4:1129–1139. doi: 10.1016/j.dnarep.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Gragtmans NJ, Myers DK, Johnson JR, Jones AR, Johnson LD. Occurrence of mammary tumors in rats after exposure to tritium beta rays and 200-kVp X rays. Radiat Res. 1984;99:636–650. [PubMed] [Google Scholar]
- Grant EJ, et al. Solid cancer incidence among the life span study of atomic bomb survivors: 1958–2009. Radiat Res. 2017;187:513–537. doi: 10.1667/RR14492.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant EJ, et al. Bioavailable serum estradiol may alter radiation risk of postmenopausal breast cancer: a nested case-control study. Int J Radiat Biol. 2018;94:97–105. doi: 10.1080/09553002.2018.1419303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griendling KK, et al. Measurement of reactive oxygen species, reactive nitrogen species, and redox-dependent signaling in the cardiovascular system: a scientific statement from the American heart association. Circ Res. 2016;119:e39–75. doi: 10.1161/RES.0000000000000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerard M, et al. Assessment of mechanisms driving non-linear dose-response relationships in genotoxicity testing. Mutat Res Rev Mutat Res. 2015;763:181–201. doi: 10.1016/j.mrrev.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Gurley KE, Kemp CJ. Ataxia-telangiectasia mutated is not required for p53 induction and apoptosis in irradiated epithelial tissues. Mol Cancer Res. 2007;5:1312–1318. doi: 10.1158/1541-7786.MCR-07-0223. [DOI] [PubMed] [Google Scholar]
- Gustin JP, et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci USA. 2009;106:2835–2840. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha YM, et al. Molecular activation of NF-kappaB, pro-inflammatory mediators, and signal pathways in gamma-irradiated mice. Biotechnol Lett. 2010;32:373–378. doi: 10.1007/s10529-009-0165-4. [DOI] [PubMed] [Google Scholar]
- Haag JD, Hsu LC, Newton MA, Gould MN. Allelic imbalance in mammary carcinomas induced by either 7,12-dimethylbenz[a]anthracene or ionizing radiation in rats carrying genes conferring differential susceptibilities to mammary carcinogenesis. Mol Carcinog. 1996;17:134–143. doi: 10.1002/(SICI)1098-2744(199611)17:3<134::AID-MC5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Haase MG, et al. Sustained elevation of NF-kappaB DNA binding activity in radiation-induced lung damage in rats. Int J Radiat Biol. 2003;79:863–877. doi: 10.1080/09553000310001632903. [DOI] [PubMed] [Google Scholar]
- Haddadi GH, Rezaeyan A, Mosleh-Shirazi MA, Hosseinzadeh M, Fardid R, Najafi M, Salajegheh A. Hesperidin as radioprotector against radiation-induced lung damage in rat: a histopathological study. J Med Phys. 2017;42:25–32. doi: 10.4103/jmp.JMP_119_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegele AD, Wolfe P, Thompson HJ. X-radiation induces 8-hydroxy-2'-deoxyguanosine formation in vivo in rat mammary gland DNA. Carcinogenesis. 1998;19:1319–1321. doi: 10.1093/carcin/19.7.1319. [DOI] [PubMed] [Google Scholar]
- Han W, Chen S, Yu KN, Wu L. Nitric oxide mediated DNA double strand breaks induced in proliferating bystander cells after alpha-particle irradiation. Mutat Res. 2010;684:81–89. doi: 10.1016/j.mrfmmm.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Haseman JK, Young E, Eustis SL, Hailey JR. Body weight-tumor incidence correlations in long-term rodent carcinogenicity studies. Toxicol Pathol. 1997;25:256–263. doi: 10.1177/019262339702500302. [DOI] [PubMed] [Google Scholar]
- Haslam SZ, Bern HA. Histopathogenesis of 7,12-diemthylbenz(a)anthracene-induced rat mammary tumors. Proc Natl Acad Sci USA. 1977;74:4020–4024. doi: 10.1073/pnas.74.9.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaki S, Kotsinas A, Chronopoulos E, Kletsas D, Georgakilas A, Gorgoulis VG. The role of oxidative DNA damage in radiation induced bystander effect. Cancer Lett. 2015;356:43–51. doi: 10.1016/j.canlet.2014.01.023. [DOI] [PubMed] [Google Scholar]
- Henderson TO, et al. Systematic review: surveillance for breast cancer in women treated with chest radiation for childhood, adolescent, or young adult cancer. Ann Intern Med. 2010;152:444–455. doi: 10.1059/0003-4819-152-7-201004060-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L, Terradas M, Martin M, Feijoo P, Soler D, Tusell L, Genesca A. Increased mammogram-induced DNA damage in mammary epithelial cells aged in vitro. PLoS ONE. 2013;8:e63052. doi: 10.1371/journal.pone.0063052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiguchi M, et al. Clarifying the biological significance of the CHK2 K373E somatic mutation discovered in the Cancer genome Atlas database. FEBS Lett. 2016;590:4275–4286. doi: 10.1002/1873-3468.12449. [DOI] [PubMed] [Google Scholar]
- Hinck L, Silberstein GB. Key stages in mammary gland development: the mammary end bud as a motile organ. Breast Cancer Res. 2005;7:245–251. doi: 10.1186/bcr1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson LJ, Chua AC, Evdokiou A, Robertson SA, Ingman WV. Macrophage phenotype in the mammary gland fluctuates over the course of the estrous cycle and is regulated by ovarian steroid hormones. Biol Reprod. 2013;89:65. doi: 10.1095/biolreprod.113.109561. [DOI] [PubMed] [Google Scholar]
- Holtzman S, Stone JP, Shellabarger CJ. Synergism of diethylstilbestrol and radiation in mammary carcinogenesis in female F344 rats. J Natl Cancer Inst. 1979;63:1071–1074. [PubMed] [Google Scholar]
- Holtzman S, Stone JP, Shellabarger CJ. Synergism of estrogens and X-rays in mammary carcinogenesis in female ACI rats. J Natl Cancer Inst. 1981;67:455–459. [PubMed] [Google Scholar]
- Holtzman S, Stone JP, Shellabarger CJ. Radiation-induced mammary carcinogenesis in virgin, pregnant, lactating, and postlactating rats. Cancer Res. 1982;42:50–53. [PubMed] [Google Scholar]
- Horst KC, et al. Histologic subtypes of breast cancer following radiotherapy for Hodgkin lymphoma. Ann Oncol. 2014;25:848–851. doi: 10.1093/annonc/mdu017. [DOI] [PubMed] [Google Scholar]
- Hosseinimehr SJ, Nobakht R, Ghasemi A, Pourfallah TA. Radioprotective effect of mefenamic acid against radiation-induced genotoxicity in human lymphocytes. Radiat Oncol J. 2015;33:256–260. doi: 10.3857/roj.2015.33.3.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck KA, Dix DJ, Judson RS, Kavlock RJ, Yang J, Berg EL. Profiling bioactivity of the ToxCast chemical library using BioMAP primary human cell systems. J Biomol Screen. 2009;14:1054–1066. doi: 10.1177/1087057109345525. [DOI] [PubMed] [Google Scholar]
- Hrdlickova R, Toloue M, Tian B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip Rev RNA. 2017 doi: 10.1002/wrna.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YW, Huang RL, Lai HC. MeDIP-on-chip for methylation profiling methods. Mol Biol. 2015;1249:281–290. doi: 10.1007/978-1-4939-2013-6_21. [DOI] [PubMed] [Google Scholar]
- Huang GL, et al. Clinical significance of miR-21 expression in breast cancer: SYBR-Green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep. 2009;21:673–679. [PubMed] [Google Scholar]
- Huang WY, Newman B, Millikan RC, Schell MJ, Hulka BS, Moorman PG. Hormone-related factors and risk of breast cancer in relation to estrogen receptor and progesterone receptor status. Am J Epidemiol. 2000;151:703–714. doi: 10.1093/oxfordjournals.aje.a010265. [DOI] [PubMed] [Google Scholar]
- IARC (2012) Radiation volume 100D: a review of human carcinogens vol 100D. International Agency for Research on Cancer, Lyon
- Iizuka D, Sasatani M, Barcellos-Hoff MH, Kamiya K. Hydrogen peroxide enhances TGFbeta-mediated epithelial-to-mesenchymal transition in human mammary epithelial MCF-10A cells. Anticancer Res. 2017;37:987–995. doi: 10.21873/anticanres.11408. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci USA. 2011;108:1397–1402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illa-Bochaca I, et al. Densely ionizing radiation acts via the microenvironment to promote aggressive Trp53-null mammary carcinomas. Cancer Res. 2014;74:7137–7148. doi: 10.1158/0008-5472.CAN-14-1212. [DOI] [PubMed] [Google Scholar]
- Imaoka T, et al. Age modifies the effect of 2-MeV fast neutrons on rat mammary carcinogenesis. Radiat Res. 2017;188:419–425. doi: 10.1667/RR14829.1. [DOI] [PubMed] [Google Scholar]
- Imaoka T, et al. Influence of age on the relative biological effectiveness of carbon ion radiation for induction of rat mammary carcinoma. Int J Radiat Oncol Biol Phys. 2013;85:1134–1140. doi: 10.1016/j.ijrobp.2012.08.035. [DOI] [PubMed] [Google Scholar]
- Imaoka T, et al. Molecular characterization of cancer reveals interactions between ionizing radiation and chemicals on rat mammary carcinogenesis. Int J Cancer. 2014;134:1529–1538. doi: 10.1002/ijc.28480. [DOI] [PubMed] [Google Scholar]
- Imaoka T, et al. Radiation-induced mammary carcinogenesis in rodent models: what's different from chemical carcinogenesis? J Radiat Res. 2009;50:281–293. doi: 10.1269/jrr.09027. [DOI] [PubMed] [Google Scholar]
- Imaoka T, Nishimura M, Iizuka D, Nishimura Y, Ohmachi Y, Shimada Y. Pre- and postpubertal irradiation induces mammary cancers with distinct expression of hormone receptors ErbB ligands, and developmental genes in rats. Mol Carcinog. 2011;50:539–552. doi: 10.1002/mc.20746. [DOI] [PubMed] [Google Scholar]
- Imaoka T, et al. High relative biologic effectiveness of carbon ion radiation on induction of rat mammary carcinoma and its lack of H-ras and Tp53 mutations. Int J Radiat Oncol Biol Phys. 2007;69:194–203. doi: 10.1016/j.ijrobp.2007.05.026. [DOI] [PubMed] [Google Scholar]
- Imaoka T, Nishimura M, Nishimura Y, Kakinuma S, Shimada Y. Persistent cell proliferation of terminal end buds precedes radiation-induced rat mammary carcinogenesis. In Vivo. 2006;20:353–358. [PubMed] [Google Scholar]
- Ina Y, Tanooka H, Yamada T, Sakai K. Suppression of thymic lymphoma induction by life-long low-dose-rate irradiation accompanied by immune activation in C57BL/6 mice. Radiat Res. 2005;163:153–158. doi: 10.1667/rr3289. [DOI] [PubMed] [Google Scholar]
- Inano H, et al. Chemoprevention by curcumin during the promotion stage of tumorigenesis of mammary gland in rats irradiated with gamma-rays. Carcinogenesis. 1999;20:1011–1018. doi: 10.1093/carcin/20.6.1011. [DOI] [PubMed] [Google Scholar]
- Inano H, Suzuki K, Ishii-Ohba H, Ikeda K, Wakabayashi K. Pregnancy-dependent initiation in tumorigenesis of Wistar rat mammary glands by 60Co-irradiation. Carcinogenesis. 1991;12:1085–1090. doi: 10.1093/carcin/12.6.1085. [DOI] [PubMed] [Google Scholar]
- Inano H, Suzuki K, Onoda M, Wakabayashi K. Relationship between induction of mammary tumors and change of testicular functions in male rats following gamma-ray irradiation and/or diethylstilbestrol. Carcinogenesis. 1996;17:355–360. doi: 10.1093/carcin/17.2.355. [DOI] [PubMed] [Google Scholar]
- Inano H, Suzuki K, Onoda M, Yamanouchi H. Susceptibility of fetal, virgin, pregnant and lactating rats for the induction of mammary tumors by gamma rays. Radiat Res. 1996;145:708–713. [PubMed] [Google Scholar]
- Inano H, Yamanouchi H, Suzuki K, Onoda M, Wakabayashi K. Estradiol-17 beta as an initiation modifier for radiation-induced mammary tumorigenesis of rats ovariectomized before puberty. Carcinogenesis. 1995;16:1871–1877. doi: 10.1093/carcin/16.8.1871. [DOI] [PubMed] [Google Scholar]
- Inskip PD, et al. Radiation dose and breast cancer risk in the childhood cancer survivor study. J Clin Oncol. 2009;27:3901–3907. doi: 10.1200/JCO.2008.20.7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Nystrom S, Nygren J, Hammarsten O. Activation of ataxia telangiectasia mutated by DNA strand break-inducing agents correlates closely with the number of DNA double strand breaks. J Biol Chem. 2005;280:4649–4655. doi: 10.1074/jbc.M411588200. [DOI] [PubMed] [Google Scholar]
- Iwasaki T, Takashima Y, Suzuki T, Yoshida MA, Hayata I. The dose response of chromosome aberrations in human lymphocytes induced in vitro by very low-dose gamma rays. Radiat Res. 2011;175:208–213. doi: 10.1667/rr2097.1. [DOI] [PubMed] [Google Scholar]
- Jackson JH, Gajewski E, Schraufstatter IU, Hyslop PA, Fuciarelli AF, Cochrane CG, Dizdaroglu M. Damage to the bases in DNA induced by stimulated human neutrophils. J Clin Invest. 1989;84:1644–1649. doi: 10.1172/JCI114342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob NK, et al. Identification of sensitive serum microRNA biomarkers for radiation biodosimetry. PLoS ONE. 2013;8:e57603. doi: 10.1371/journal.pone.0057603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczak JL, Merlino G, French JE, Muller WJ, Paul B, Adhya S, Garges S. Analysis of genetic instability during mammary tumor progression using a novel selection-based assay for in vivo mutations in a bacteriophage lambda transgene target. Proc Natl Acad Sci USA. 1996;93:9073–9078. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerry DJ, Dunphy KA, Hagen MJ. Estrogens, regulation of p53 and breast cancer risk: a balancing act. Cell Mol Life Sci. 2010;67:1017–1023. doi: 10.1007/s00018-009-0244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JA, Riggs PK, Yang TC, Pedemonte CH, Clarke MS, Feeback DL, Au WW. Ionizing radiation-induced bioeffects in space and strategies to reduce cellular injury and carcinogenesis. Aviat Space Environ Med. 2007;78:A67–78. [PubMed] [Google Scholar]
- Judson RS, et al. Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol Sci. 2015;148:137–154. doi: 10.1093/toxsci/kfv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadhim M, Salomaa S, Wright E, Hildebrandt G, Belyakov OV, Prise KM, Little MP. Non-targeted effects of ionising radiation–implications for low dose risk. Mutat Res. 2013;752:84–98. doi: 10.1016/j.mrrev.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakinuma S, et al. Combined exposure to X-irradiation followed by N-ethyl-N-nitrosourea treatment alters the frequency and spectrum of Ikaros point mutations in murine T-cell lymphoma. Mutat Res. 2012;737:43–50. doi: 10.1016/j.mrfmmm.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Kalyanaraman B, et al. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, et al. Glutathione-dependent and -independent oxidative stress-control mechanisms distinguish normal human mammary epithelial cell subsets. Proc Natl Acad Sci USA. 2014;111:7789–7794. doi: 10.1073/pnas.1403813111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, Lim PX, Helgadottir HR, Moynahan ME, Jasin M. Robust homology-directed repair within mouse mammary tissue is not specifically affected by Brca2 mutation. Nat Commun. 2016;7:13241. doi: 10.1038/ncomms13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y, et al. Detection of DNA double-strand breaks by pulsed-field gel electrophoresis. Genes Cells. 2017;22:84–93. doi: 10.1111/gtc.12457. [DOI] [PubMed] [Google Scholar]
- Khan A, et al. Gossypetin ameliorates ionizing radiation-induced oxidative stress in mice liver—a molecular approach. Free Radic Res. 2015;49:1173–1186. doi: 10.3109/10715762.2015.1053878. [DOI] [PubMed] [Google Scholar]
- Kim ES, Kim MS, Moon A. TGF-beta-induced upregulation of MMP-2 and MMP-9 depends on p38 MAPK, but not ERK signaling in MCF10A human breast epithelial cells. Int J Oncol. 2004;25:1375–1382. [PubMed] [Google Scholar]
- Kim GJ, Fiskum GM, Morgan WF. A role for mitochondrial dysfunction in perpetuating radiation-induced genomic instability. Cancer Res. 2006;66:10377–10383. doi: 10.1158/0008-5472.CAN-05-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiraly O, Gong G, Olipitz W, Muthupalani S, Engelward BP. Inflammation-induced cell proliferation potentiates DNA damage-induced mutations in vivo. PLoS Genet. 2015;11:e1004901. doi: 10.1371/journal.pgen.1004901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshner J, et al. Inhibition of transforming growth factor-beta1 signaling attenuates ataxia telangiectasia mutated activity in response to genotoxic stress. Cancer Res. 2006;66:10861–10869. doi: 10.1158/0008-5472.CAN-06-2565. [DOI] [PubMed] [Google Scholar]
- Kloypan C, Srisa-art M, Mutirangura A, Boonla C. LINE-1 hypomethylation induced by reactive oxygen species is mediated via depletion of S-adenosylmethionine. Cell Biochem Funct. 2015;33:375–385. doi: 10.1002/cbf.3124. [DOI] [PubMed] [Google Scholar]
- Koh TJ, DiPietro LA. Inflammation and wound healing: the role of the macrophage. Expert Rev Mol Med. 2011;13:e23. doi: 10.1017/S1462399411001943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H, et al. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 2009;7:e1000121. doi: 10.1371/journal.pbio.1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkola JE, Archer MC. Resistance to mammary tumorigenesis in Copenhagen rats is associated with the loss of preneoplastic lesions. Carcinogenesis. 1999;20:221–227. doi: 10.1093/carcin/20.2.221. [DOI] [PubMed] [Google Scholar]
- Kostyuk SV, et al. Role of extracellular DNA oxidative modification in radiation induced bystander effects in human endotheliocytes. Mutat Res. 2012;729:52–60. doi: 10.1016/j.mrfmmm.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Koturbash I, et al. Epigenetic dysregulation underlies radiation-induced transgenerational genome instability in vivo. Int J Radiat Oncol Biol Phys. 2006;66:327–330. doi: 10.1016/j.ijrobp.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Jadavji NM, Kutanzi K, Rodriguez-Juarez R, Kogosov D, Metz GAS, Kovalchuk O. Fractionated low-dose exposure to ionizing radiation leads to DNA damage, epigenetic dysregulation, and behavioral impairment. Environ Epigenet. 2016;2:25. doi: 10.1093/eep/dvw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Kutanzi K, Hendrickson K, Rodriguez-Juarez R, Kogosov D, Kovalchuk O. Radiation-induced bystander effects in vivo are sex specific. Mutat Res. 2008;642:28–36. doi: 10.1016/j.mrfmmm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Pogribny I, Kovalchuk O. Stable loss of global DNA methylation in the radiation-target tissue—a possible mechanism contributing to radiation carcinogenesis? Biochem Biophys Res Commun. 2005;337:526–533. doi: 10.1016/j.bbrc.2005.09.084. [DOI] [PubMed] [Google Scholar]
- Koturbash I, et al. Irradiation induces DNA damage and modulates epigenetic effectors in distant bystander tissue in vivo. Oncogene. 2006;25:4267–4275. doi: 10.1038/sj.onc.1209467. [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhne M, Rothkamm K, Lobrich M. No dose-dependence of DNA double-strand break misrejoining following alpha-particle irradiation. Int J Radiat Biol. 2000;76:891–900. doi: 10.1080/09553000050050909. [DOI] [PubMed] [Google Scholar]
- Kushman ME, Kraft AD, Guyton KZ, Chiu WA, Makris SL, Rusyn I. A systematic approach for identifying and presenting mechanistic evidence in human health assessments. Regul Toxicol Pharmacol. 2013;67:266–277. doi: 10.1016/j.yrtph.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutanzi K, Kovalchuk O. Exposure to estrogen and ionizing radiation causes epigenetic dysregulation, activation of mitogen-activated protein kinase pathways, and genome instability in the mammary gland of ACI rats. Cancer Biol Ther. 2013;14:564–573. doi: 10.4161/cbt.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutanzi KR, Koturbash I, Bronson RT, Pogribny IP, Kovalchuk O. Imbalance between apoptosis and cell proliferation during early stages of mammary gland carcinogenesis in ACI rats. Mutat Res. 2010;694:1–6. doi: 10.1016/j.mrfmmm.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Lambe M, Hsieh C, Trichopoulos D, Ekbom A, Pavia M, Adami HO. Transient increase in the risk of breast cancer after giving birth. N Engl J Med. 1994;331:5–9. doi: 10.1056/NEJM199407073310102. [DOI] [PubMed] [Google Scholar]
- Land CE, Hayakawa N, Machado SG, Yamada Y, Pike MC, Akiba S, Tokunaga M. A case–control interview study of breast cancer among Japanese A-bomb survivors. II. Interactions with radiation dose. Cancer Causes Control. 1994;5:167–176. doi: 10.1007/BF01830263. [DOI] [PubMed] [Google Scholar]
- Land CE, Tokunaga M, Koyama K, Soda M, Preston DL, Nishimori I, Tokuoka S. Incidence of female breast cancer among atomic bomb survivors, Hiroshima and Nagasaki, 1950–1990. Radiat Res. 2003;160:707–717. doi: 10.1667/rr3082. [DOI] [PubMed] [Google Scholar]
- Land CE, Tokunaga M, Tokuoka S, Nakamura N. Early-onset breast cancer in A-bomb survivors. Lancet. 1993;342:237. doi: 10.1016/0140-6736(93)92324-m. [DOI] [PubMed] [Google Scholar]
- Landen NX, Li D, Stahle M. Transition from inflammation to proliferation: a critical step during wound healing. Cell Mol Life Sci. 2016;73:3861–3885. doi: 10.1007/s00018-016-2268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach JK, Black SM, Schmidt-Ullrich RK, Mikkelsen RB. Activation of constitutive nitric-oxide synthase activity is an early signaling event induced by ionizing radiation. J Biol Chem. 2002;277:15400–15406. doi: 10.1074/jbc.M110309200. [DOI] [PubMed] [Google Scholar]
- Leach JK, Van Tuyle G, Lin PS, Schmidt-Ullrich R, Mikkelsen RB. Ionizing radiation-induced, mitochondria-dependent generation of reactive oxygen/nitrogen. Cancer Res. 2001;61:3894–3901. [PubMed] [Google Scholar]
- Lee Y, Kim YJ, Choi YJ, Lee JW, Lee S, Cho YH, Chung HW. Radiation-induced changes in DNA methylation and their relationship to chromosome aberrations in nuclear power plant workers. Int J Radiat Biol. 2015;91:142–149. doi: 10.3109/09553002.2015.969847. [DOI] [PubMed] [Google Scholar]
- Lemon HM, Kumar PF, Peterson C, Rodriguez-Sierra JF, Abbo KM. Inhibition of radiogenic mammary carcinoma in rats by estriol or tamoxifen. Cancer. 1989;63:1685–1692. doi: 10.1002/1097-0142(19900501)63:9<1685::aid-cncr2820630907>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. Cyclin D1 integrates estrogen-mediated DNA damage repair signaling. Cancer Res. 2014;74:3959–3970. doi: 10.1158/0008-5472.CAN-13-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakou E, et al. Ionizing radiation-mediated premature senescence and paracrine interactions with cancer cells enhance the expression of syndecan 1 in human breast stromal fibroblasts: the role of TGF-beta. Aging (Albany NY) 2016;8:1650–1669. doi: 10.18632/aging.100989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima F, Ding D, Goetz W, Yang AJ, Baulch JE. High LET (56)Fe ion irradiation induces tissue-specific changes in DNA methylation in the mouse. Environ Mol Mutagen. 2014;55:266–277. doi: 10.1002/em.21832. [DOI] [PubMed] [Google Scholar]
- Limoli CL, Giedzinski E. Induction of chromosomal instability by chronic oxidative stress. Neoplasia. 2003;5:339–346. doi: 10.1016/S1476-5586(03)80027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli CL, Giedzinski E, Morgan WF, Swarts SG, Jones GD, Hyun W. Persistent oxidative stress in chromosomally unstable cells. Cancer Res. 2003;63:3107–3111. [PubMed] [Google Scholar]
- Limoli CL, Kaplan MI, Corcoran J, Meyers M, Boothman DA, Morgan WF. Chromosomal instability and its relationship to other end points of genomic instability. Cancer Res. 1997;57:5557–5563. [PubMed] [Google Scholar]
- Lindstrom LS, et al. Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J Clin Oncol. 2012;30:2601–2608. doi: 10.1200/JCO.2011.37.2482. [DOI] [PubMed] [Google Scholar]
- Little MP. Heterogeneity of variation of relative risk by age at exposure in the Japanese atomic bomb survivors. Radiat Environ Biophys. 2009;48:253–262. doi: 10.1007/s00411-009-0228-x. [DOI] [PubMed] [Google Scholar]
- Little MP. Evidence for dose and dose rate effects in human and animal radiation studies. Ann ICRP. 2018;47:97–112. doi: 10.1177/0146645318756235. [DOI] [PubMed] [Google Scholar]
- Little MP, et al. Association of chromosome translocation rate with low dose occupational radiation exposures in U.S. radiologic technologists. Radiat Res. 2014;182:1–17. doi: 10.1667/RR13413.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little MP, McElvenny DM. Male breast cancer incidence and mortality risk in the Japanese atomic bomb survivors—differences in excess relative and absolute risk from female breast cancer. Environ Health Perspect. 2017;125:223–229. doi: 10.1289/EHP151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobrich M, Rief N, Kuhne M, Heckmann M, Fleckenstein J, Rube C, Uder M. vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc Natl Acad Sci USA. 2005;102:8984–8989. doi: 10.1073/pnas.0501895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorat Y, Brunner CU, Schanz S, Jakob B, Taucher-Scholz G, Rube CE. Nanoscale analysis of clustered DNA damage after high-LET irradiation by quantitative electron microscopy—the heavy burden to repair. DNA Repair (Amst) 2015;28:93–106. doi: 10.1016/j.dnarep.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Lorat Y, Timm S, Jakob B, Taucher-Scholz G, Rube CE. Clustered double-strand breaks in heterochromatin perturb DNA repair after high linear energy transfer irradiation. Radiother Oncol. 2016;121:154–161. doi: 10.1016/j.radonc.2016.08.028. [DOI] [PubMed] [Google Scholar]
- Loree J, Koturbash I, Kutanzi K, Baker M, Pogribny I, Kovalchuk O. Radiation-induced molecular changes in rat mammary tissue: possible implications for radiation-induced carcinogenesis. Int J Radiat Biol. 2006;82:805–815. doi: 10.1080/09553000600960027. [DOI] [PubMed] [Google Scholar]
- Lorimore SA, Chrystal JA, Robinson JI, Coates PJ, Wright EG. Chromosomal instability in unirradiated hemaopoietic cells induced by macrophages exposed in vivo to ionizing radiation. Cancer Res. 2008;68:8122–8126. doi: 10.1158/0008-5472.CAN-08-0698. [DOI] [PubMed] [Google Scholar]
- Lorimore SA, Mukherjee D, Robinson JI, Chrystal JA, Wright EG. Long-lived inflammatory signaling in irradiated bone marrow is genome dependent. Cancer Res. 2011;71:6485–6491. doi: 10.1158/0008-5472.CAN-11-1926. [DOI] [PubMed] [Google Scholar]
- Lorimore SA, Rastogi S, Mukherjee D, Coates PJ, Wright EG. The influence of p53 functions on radiation-induced inflammatory bystander-type signaling in murine bone marrow. Radiat Res. 2013;179:406–415. doi: 10.1667/RR3158.2. [DOI] [PubMed] [Google Scholar]
- Lumniczky K, Safrany G. The impact of radiation therapy on the antitumor immunity: local effects and systemic consequences. Cancer Lett. 2015;356:114–125. doi: 10.1016/j.canlet.2013.08.024. [DOI] [PubMed] [Google Scholar]
- Luo M, et al. Mammary epithelial-specific ablation of the focal adhesion kinase suppresses mammary tumorigenesis by affecting mammary cancer stem/progenitor cells. Can Res. 2009;69:466–474. doi: 10.1158/0008-5472.CAN-08-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzhna L, Ilnytskyy Y, Kovalchuk O. Mobilization of LINE-1 in irradiated mammary gland tissue may potentially contribute to low dose radiation-induced genomic instability. Genes Cancer. 2015;6:71–81. doi: 10.18632/genesandcancer.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyng FM, Seymour CB, Mothersill C. Oxidative stress in cells exposed to low levels of ionizing radiation. Biochem Soc Trans. 2001;29:350–353. doi: 10.1042/0300-5127:0290350. [DOI] [PubMed] [Google Scholar]
- Ma H, Hill CK, Bernstein L, Ursin G. Low-dose medical radiation exposure and breast cancer risk in women under age 50 years overall and by estrogen and progesterone receptor status: results from a case-control and a case–case comparison. Breast Cancer Res Treat. 2008;109:77–90. doi: 10.1007/s10549-007-9625-5. [DOI] [PubMed] [Google Scholar]
- Madugundu GS, Cadet J, Wagner JR. Hydroxyl-radical-induced oxidation of 5-methylcytosine in isolated and cellular DNA. Nucleic Acids Res. 2014;42:7450–7460. doi: 10.1093/nar/gku334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffini MV, Calabro JM, Soto AM, Sonnenschein C. Stromal regulation of neoplastic development: age-dependent normalization of neoplastic mammary cells by mammary stroma. Am J Pathol. 2005;167:1405–1410. doi: 10.1016/S0002-9440(10)61227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffini MV, Soto AM, Calabro JM, Ucci AA, Sonnenschein C. The stroma as a crucial target in rat mammary gland carcinogenesis. J Cell Sci. 2004;117:1495–1502. doi: 10.1242/jcs.01000. [DOI] [PubMed] [Google Scholar]
- Mahdi AH, et al. Evidence of intertissue differences in the DNA damage response and the pro-oncogenic role of NF-kappaB in mice with disengaged BRCA1–PALB2 interaction. Cancer Res. 2018;78:3969–3981. doi: 10.1158/0008-5472.CAN-18-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailander PC, Meza JL, Higginbotham S, Chakravarti D. Induction of A.T to G.C mutations by erroneous repair of depurinated DNA following estrogen treatment of the mammary gland of ACI rats. J Steroid Biochem Mol Biol. 2006;101:204–215. doi: 10.1016/j.jsbmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Makris SL. Current assessment of the effects of environmental chemicals on the mammary gland in guideline rodent studies by the U.S. Environmental Protection Agency (U.S. EPA), Organisation for Economic Co-operation and Development (OECD), and National Toxicology Program (NTP) Environ Health Perspect. 2011;119:1047–1052. doi: 10.1289/ehp.1002676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna K, Das U, Das D, Kesh SB, Khan A, Chakraborty A, Dey S. Naringin inhibits gamma radiation-induced oxidative DNA damage and inflammation, by modulating p53 and NF-kappaB signaling pathways in murine splenocytes. Free Radic Res. 2015;49:422–439. doi: 10.3109/10715762.2015.1016018. [DOI] [PubMed] [Google Scholar]
- Manzoni C, Kia DA, Vandrovcova J, Hardy J, Wood NW, Lewis PA, Ferrari R. Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences. Brief Bioinform. 2018;19:286–302. doi: 10.1093/bib/bbw114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NT, et al. Homozygous mutation of MTPAP causes cellular radiosensitivity and persistent DNA double-strand breaks. Cell Death Dis. 2014;5:e1130. doi: 10.1038/cddis.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I. Somatic mutation and clonal expansions in human tissues. Genome Med. 2019;11:35. doi: 10.1186/s13073-019-0648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514:54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews JD, et al. Cancer risk in 680,000 people exposed to computed tomography scans in childhood or adolescence: data linkage study of 11 million. Aust Bmj. 2013;346:f2360. doi: 10.1136/bmj.f2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavragani IV, et al. Complex DNA damage: a route to radiation-induced genomic instability and carcinogenesis. Cancers (Basel) 2017 doi: 10.3390/cancers9070091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell CA, et al. Targeted and nontargeted effects of ionizing radiation that impact genomic instability. Cancer Res. 2008;68:8304–8311. doi: 10.1158/0008-5472.CAN-08-1212. [DOI] [PubMed] [Google Scholar]
- McDaniel SM, Rumer KK, Biroc SL, Metz RP, Singh M, Porter W, Schedin P. Remodeling of the mammary microenvironment after lactation promotes breast tumor cell metastasis. Am J Pathol. 2006;168:608–620. doi: 10.2353/ajpath.2006.050677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measurements IICoRUa . ICRU report 16: linear energy transfer. Washington: International Commission on Radiation Units and Measurements; 1970. [Google Scholar]
- Medina D. Chemical carcinogenesis of rat and mouse mammary glands. Breast Dis. 2007;28:63–68. doi: 10.3233/bd-2007-28107. [DOI] [PubMed] [Google Scholar]
- Medina D, Thompson HJ. A comparison of the salient features of mouse, rat, and human mammary tumorigenesis. In: Ip MM, Asch BB, editors. Methods in mammary gland biology and breast cancer research. Boston: Springer; 2000. pp. 31–36. [Google Scholar]
- Menyhart O, Harami-Papp H, Sukumar S, Schafer R, Magnani L, de Barrios O, Gyorffy B. Guidelines for the selection of functional assays to evaluate the hallmarks of cancer. Biochim Biophys Acta. 2016;1866:300–319. doi: 10.1016/j.bbcan.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Mestdagh P, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11:809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- Meulepas JM, et al. Radiation exposure from pediatric CT scans and subsequent cancer risk in the Netherlands. J Natl Cancer Inst. 2019;111:256–263. doi: 10.1093/jnci/djy104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen RB, Wardman P. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene. 2003;22:5734–5754. doi: 10.1038/sj.onc.1206663. [DOI] [PubMed] [Google Scholar]
- Miller AB, et al. Mortality from breast cancer after irradiation during fluoroscopic examinations in patients being treated for tuberculosis. N Engl J Med. 1989;321:1285–1289. doi: 10.1056/NEJM198911093211902. [DOI] [PubMed] [Google Scholar]
- Millikan RC, Player JS, Decotret AR, Tse CK, Keku T. Polymorphisms in DNA repair genes, medical exposure to ionizing radiation, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:2326–2334. doi: 10.1158/1055-9965.EPI-05-0186. [DOI] [PubMed] [Google Scholar]
- Miousse IR, et al. Inter-strain differences in LINE-1 DNA methylation in the mouse hematopoietic system in response to exposure to ionizing radiation. Int J Mol Sci. 2017 doi: 10.3390/ijms18071430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, et al. Exposure to low-dose (56)Fe-ion radiation induces long-term epigenetic alterations in mouse bone marrow hematopoietic progenitor and stem cells. Radiat Res. 2014;182:92–101. doi: 10.1667/RR13580.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, et al. Significance of HER2 and C-MYC oncogene amplifications in breast cancer in atomic bomb survivors: associations with radiation exposure and histologic grade. Cancer. 2008;112:2143–2151. doi: 10.1002/cncr.23414. [DOI] [PubMed] [Google Scholar]
- Moeller A, Ask K, Warburton D, Gauldie J, Kolb M. The bleomycin animal model: a useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int J Biochem Cell Biol. 2008;40:362–382. doi: 10.1016/j.biocel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller S, et al. The heritability of breast cancer among women in the nordic twin study of cancer. Cancer Epidemiol Biomarkers Prev. 2016;25:145–150. doi: 10.1158/1055-9965.EPI-15-0913. [DOI] [PubMed] [Google Scholar]
- Monceau V, et al. Enhanced sensitivity to low dose irradiation of ApoE-/- mice mediated by early pro-inflammatory profile and delayed activation of the TGFbeta1 cascade involved in fibrogenesis. PLoS ONE. 2013;8:e57052. doi: 10.1371/journal.pone.0057052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SR, et al. Genomic instability in human lymphocytes irradiated with individual charged particles: involvement of tumor necrosis factor alpha in irradiated cells but not bystander cells. Radiat Res. 2005;163:183–190. doi: 10.1667/rr3298. [DOI] [PubMed] [Google Scholar]
- Moravan MJ, Olschowka JA, Williams JP, O'Banion MK. Cranial irradiation leads to acute and persistent neuroinflammation with delayed increases in T-cell infiltration and CD11c expression in C57BL/6 mouse brain. Radiat Res. 2011;176:459–473. doi: 10.1667/RR2587.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MM, Schuler LA, Ciciliano JC, Johnson BP, Alarid ET, Beebe DJ. Modeling chemical effects on breast cancer: the importance of the microenvironment in vitro. Integr Biol (Camb) 2020;12:21–33. doi: 10.1093/intbio/zyaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morganella S, et al. The topography of mutational processes in breast cancer genomes. Nat Commun. 2016;7:11383. doi: 10.1038/ncomms11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita M, et al. Chromothripsis-like chromosomal rearrangements induced by ionizing radiation using proton microbeam irradiation system. Oncotarget. 2016;7:10182–10192. doi: 10.18632/oncotarget.7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz CS, et al. Breast cancer after chest radiation therapy for childhood cancer. J Clin Oncol. 2014;32:2217–2223. doi: 10.1200/JCO.2013.54.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Coates PJ, Lorimore SA, Wright EG. The in vivo expression of radiation-induced chromosomal instability has an inflammatory mechanism. Radiat Res. 2012;177:18–24. doi: 10.1667/RR2793.1. [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Coates PJ, Lorimore SA, Wright EG. Responses to ionizing radiation mediated by inflammatory mechanisms. J Pathol. 2014;232:289–299. doi: 10.1002/path.4299. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Costes SV, Bazarov AV, Hines WC, Barcellos-Hoff MH, Yaswen P. Promotion of variant human mammary epithelial cell outgrowth by ionizing radiation: an agent-based model supported by in vitro studies. Breast Cancer Res. 2010;12:R11. doi: 10.1186/bcr2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J, Purvis ER, Swenberg JA. Micromolar concentrations of hydrogen peroxide induce oxidative DNA lesions more efficiently than millimolar concentrations in mammalian cells. Nucleic Acids Res. 2003;31:1790–1795. doi: 10.1093/nar/gkg263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao N, Kurokawa T, Nonami T, Tumurkhuu G, Koide N, Yokochi T. Hydrogen peroxide induces the production of tumor necrosis factor-alpha in RAW 264.7 macrophage cells via activation of p38 and stress-activated protein kinase. Innate Immun. 2008;14:190–196. doi: 10.1177/1753425908093932. [DOI] [PubMed] [Google Scholar]
- Nandi S, Guzman RC, Yang J. Hormones and mammary carcinogenesis in mice, rats, and humans: a unifying hypothesis. Proc Natl Acad Sci USA. 1995;92:3650–3657. doi: 10.1073/pnas.92.9.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan PK, Goodwin EH, Lehnert BE. Alpha particles initiate biological production of superoxide anions and hydrogen peroxide in human cells. Can Res. 1997;57:3963–3971. [PubMed] [Google Scholar]
- Natarajan M, Gibbons CF, Mohan S, Moore S, Kadhim MA. Oxidative stress signalling: a potential mediator of tumour necrosis factor alpha-induced genomic instability in primary vascular endothelial cells. Br J Radiol. 2007;1:S13–22. doi: 10.1259/bjr/15316848. [DOI] [PubMed] [Google Scholar]
- National Research Council . Science and decisions: advancing risk assessment. Washington: The National Academies Press; 2009. [PubMed] [Google Scholar]
- Neta G, Anderson WF, Gilbert E, Berrington A. Variation in the risk of radiation-related contralateral breast cancer by histology and estrogen receptor expression in SEER. Breast Cancer Res Treat. 2012;131:1021–1027. doi: 10.1007/s10549-011-1820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, Fredlund E, Zhao W, Perou CM, Balmain A, Mao JH, Barcellos-Hoff MH. Murine microenvironment metaprofiles associate with human cancer etiology and intrinsic subtypes. Clin Cancer Res. 2013;19:1353–1362. doi: 10.1158/1078-0432.CCR-12-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, Martinez-Ruiz H, Barcellos-Hoff MH. Consequences of epithelial or stromal TGFbeta1 depletion in the mammary gland. J Mammary Gland Biol Neoplasia. 2011;16:147–155. doi: 10.1007/s10911-011-9218-0. [DOI] [PubMed] [Google Scholar]
- Nguyen DH, et al. Radiation acts on the microenvironment to affect breast carcinogenesis by distinct mechanisms that decrease cancer latency and affect tumor type. Cancer Cell. 2011;19:640–651. doi: 10.1016/j.ccr.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, et al. The genome as a record of environmental exposure. Mutagenesis. 2015;30:763–770. doi: 10.1093/mutage/gev073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitaki Z, Hellweg CE, Georgakilas AG, Ravanat JL. Stress-induced DNA damage biomarkers: applications and limitations. Front Chem. 2015;3:35. doi: 10.3389/fchem.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitaki Z, et al. Measurement of complex DNA damage induction and repair in human cellular systems after exposure to ionizing radiations of varying linear energy transfer (LET) Free Radic Res. 2016;50:S64–S78. doi: 10.1080/10715762.2016.1232484. [DOI] [PubMed] [Google Scholar]
- Nishida N, et al. Reactive oxygen species induce epigenetic instability through the formation of 8-hydroxydeoxyguanosine in human hepatocarcinogenesis. Dig Dis. 2013;31:459–466. doi: 10.1159/000355245. [DOI] [PubMed] [Google Scholar]
- NTP U (2011a) SPECIFICATIONS FOR THE CONDUCT OF STUDIES TO EVALUATE THE REPRODUCTIVE AND DEVELOPMENTAL TOXICITY OF CHEMICAL, BIOLOGICAL AND PHYSICAL AGENTS IN LABORATORY ANIMALS FOR THE NATIONAL TOXICOLOGY PROGRAM (NTP). https://ntp.niehs.nih.gov/ntp/test_info/finalntp_reprospecsmay2011_508.pdf.
- NTP U (2011b) SPECIFICATIONS FOR THE CONDUCT OF STUDIES TO EVALUATE THE TOXIC AND CARCINOGENIC POTENTIAL OF CHEMICAL, BIOLOGICAL AND PHYSICAL AGENTS IN LABORATORY ANIMALS FOR THE NATIONAL TOXICOLOGY PROGRAM (NTP). https://ntp.niehs.nih.gov/ntp/test_info/finalntp_toxcarspecsjan2011.pdf.
- Nzabarushimana E, et al. Long-term epigenetic effects of exposure to low doses of 56Fe in the mouse lung. J Radiat Res. 2014;55:823–828. doi: 10.1093/jrr/rru010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, La Salle S, Robaire B, Trasler JM. Evaluation of a quantitative DNA methylation analysis technique using methylation-sensitive/dependent restriction enzymes and real-time. PCR Epigenet. 2006;1:146–152. doi: 10.4161/epi.1.3.3392. [DOI] [PubMed] [Google Scholar]
- Oakes SR, Hilton HN, Ormandy CJ. The alveolar switch: coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res. 2006;8:207. doi: 10.1186/bcr1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD (1997a) Test No. 471: bacterial reverse mutation test. doi:10.1787/9789264071247-en
- OECD (1997b) Test No. 486: Unscheduled DNA Synthesis (UDS) Test with Mammalian Liver Cells in vivo. doi:10.1787/9789264071520-en [DOI] [PubMed]
- OECD (2009a) Test No. 451: Carcinogenicity Studies. doi:10.1787/9789264071186-en
- OECD (2009b) Test No. 453: Combined Chronic Toxicity/Carcinogenicity Studies. doi:10.1787/9789264071223-en
- OECD (2013) Test No. 488: Transgenic Rodent Somatic and Germ Cell Gene Mutation Assays. doi:10.1787/9789264203907-en [DOI] [PubMed]
- OECD (2016a) Test No. 473: In Vitro Mammalian Chromosomal Aberration Test. doi:10.1787/9789264264649-en
- OECD (2016b) Test No. 475: Mammalian Bone Marrow Chromosomal Aberration Test. doi:10.1787/9789264264786-en
- OECD (2016c) Test No. 476: In Vitro Mammalian Cell Gene Mutation Tests using the Hprt and xprt genes. doi:10.1787/9789264264809-en
- OECD (2016d) Test No. 483: Mammalian Spermatogonial Chromosomal Aberration Test. doi:10.1787/9789264264847-en
- OECD (2016e) Test No. 487: In Vitro Mammalian Cell Micronucleus Test. doi:10.1787/9789264264861-en
- OECD (2016f) Test No. 489: In Vivo Mammalian Alkaline Comet Assay. doi:10.1787/9789264264885-en
- OECD (2016g) Test No. 490: In Vitro Mammalian Cell Gene Mutation Tests Using the Thymidine Kinase Gene. doi:10.1787/9789264264908-en
- OECD (2017) Revised guidance document on developing and assessing adverse outcome pathways.
- OECD OECD guidelines for the testing of chemicals Section 4. OECD, Paris. 2018 doi: 10.1787/20745788. [DOI] [Google Scholar]
- User's handbook supplement to the guidance document for developing and assessing adverse outcome pathways. doi:10.1787/5jlv1m9d1g32-en
- Ogawa Y, Kobayashi T, Nishioka A, Kariya S, Hamasato S, Seguchi H, Yoshida S. Radiation-induced oxidative DNA damage, 8-oxoguanine, in human peripheral T cells. Int J Mol Med. 2003;11:27–32. [PubMed] [Google Scholar]
- Ojima M, Ban N, Kai M. DNA double-strand breaks induced by very low X-ray doses are largely due to bystander effects. Radiat Res. 2008;170:365–371. doi: 10.1667/RR1255.1. [DOI] [PubMed] [Google Scholar]
- Olipitz W, et al. Integrated molecular analysis indicates undetectable change in DNA damage in mice after continuous irradiation at ~ 400-fold natural background radiation. Environ Health Perspect. 2012;120:1130–1136. doi: 10.1289/ehp.1104294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolenghi A, Merzagora M, Paretzke HG. DNA complex lesions induced by protons and alpha-particles: track structure characteristics determining linear energy transfer and particle type dependence. Radiat Environ Biophys. 1997;36:97–103. doi: 10.1007/s004110050060. [DOI] [PubMed] [Google Scholar]
- Owens KM, Aykin-Burns N, Dayal D, Coleman MC, Domann FE, Spitz DR. Genomic instability induced by mutant succinate dehydrogenase subunit D (SDHD) is mediated by O2(-*) and H2O2. Free Radic Biol Med. 2012;52:160–166. doi: 10.1016/j.freeradbiomed.2011.10.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oya Y, Yamamoto K, Tonomura A. The biological activity of hydrogen peroxide. I. Induction of chromosome-type aberrations susceptible to inhibition by scavengers of hydroxyl radicals in human embryonic fibroblasts. Mutat Res. 1986;172:245–253. doi: 10.1016/0165-1218(86)90062-5. [DOI] [PubMed] [Google Scholar]
- Ozasa K, et al. Studies of the mortality of atomic bomb survivors, Report 14, 1950–2003: an overview of cancer and noncancer diseases. Radiat Res. 2012;177:229–243. doi: 10.1667/rr2629.1. [DOI] [PubMed] [Google Scholar]
- Ozyurt H, et al. Quercetin protects radiation-induced DNA damage and apoptosis in kidney and bladder tissues of rats. Free Radic Res. 2014;48:1247–1255. doi: 10.3109/10715762.2014.945925. [DOI] [PubMed] [Google Scholar]
- Padula G, Ponzinibbio MV, Seoane AI. Possible radioprotective effect of folic acid supplementation on low dose ionizing radiation-induced genomic instability in vitro. Indian J Exp Biol. 2016;54:537–543. [PubMed] [Google Scholar]
- Park CC, Henshall-Powell RL, Erickson AC, Talhouk R, Parvin B, Bissell MJ, Barcellos-Hoff MH. Ionizing radiation induces heritable disruption of epithelial cell interactions. Proc Natl Acad Sci USA. 2003;100:10728–10733. doi: 10.1073/pnas.1832185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge MA, Chai Y, Zhou H, Hei TK. High-throughput antibody-based assays to identify and quantify radiation-responsive protein biomarkers. Int J Radiat Biol. 2010;86:321–328. doi: 10.3109/09553000903564034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhanisamy SK, Li H, Wang Y, Batinic-Haberle I, Zhou D. NADPH oxidase inhibition attenuates total body irradiation-induced haematopoietic genomic instability. Mutagenesis. 2011;26:431–435. doi: 10.1093/mutage/ger001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MS, et al. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet. 2012;380:499–505. doi: 10.1016/S0140-6736(12)60815-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Evinger AJ, Lee E, Levin ER. Estrogen inhibits ATR signaling to cell cycle checkpoints and DNA repair. Mol Biol Cell. 2009;20:3374–3389. doi: 10.1091/mbc.E09-01-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DA, Vago JP, Athayde RM, Reis AC, Teixeira MM, Sousa LP, Pinho V. Switching off key signaling survival molecules to switch on the resolution of inflammation. Mediators Inflamm. 2014;2014:829851. doi: 10.1155/2014/829851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernot E, et al. Ionizing radiation biomarkers for potential use in epidemiological studies. Mutat Res. 2012;751:258–286. doi: 10.1016/j.mrrev.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Perrott KM, Wiley CD, Desprez PY, Campisi J. Apigenin suppresses the senescence-associated secretory phenotype and paracrine effects on breast cancer cells. Geroscience. 2017;39:161–173. doi: 10.1007/s11357-017-9970-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson NC, et al. Tamoxifen resistance and Her2/neu expression in an aged, irradiated rat breast carcinoma model. Carcinogenesis. 2005;26:1542–1552. doi: 10.1093/carcin/bgi103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijpe A, et al. Exposure to diagnostic radiation and risk of breast cancer among carriers of BRCA1/2 mutations: retrospective cohort study (GENE-RAD-RISK) BMJ. 2012;345:e5660. doi: 10.1136/bmj.e5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto M, Prise KM, Michael BD. Evidence for complexity at the nanometer scale of radiation-induced DNA DSBs as a determinant of rejoining kinetics. Radiat Res. 2005;164:73–85. doi: 10.1667/rr3394. [DOI] [PubMed] [Google Scholar]
- Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci USA. 2008;105:5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny I, et al. Fractionated low-dose radiation exposure leads to accumulation of DNA damage and profound alterations in DNA and histone methylation in the murine thymus. Mol Cancer Res. 2005;3:553–561. doi: 10.1158/1541-7786.MCR-05-0074. [DOI] [PubMed] [Google Scholar]
- Pogribny I, Raiche J, Slovack M, Kovalchuk O. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Biophys Res Commun. 2004;320:1253–1261. doi: 10.1016/j.bbrc.2004.06.081. [DOI] [PubMed] [Google Scholar]
- Poirier MC, Beland FA. DNA adduct measurements and tumor incidence during chronic carcinogen exposure in rodents. Environ Health Perspect. 1994;102(Suppl 6):161–165. doi: 10.1289/ehp.94102s6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- Ponnaiya B, Cornforth MN, Ullrich RL. Induction of chromosomal instability in human mammary cells by neutrons and gamma rays. Radiat Res. 1997;147:288–294. [PubMed] [Google Scholar]
- Ponnaiya B, Cornforth MN, Ullrich RL. Radiation-induced chromosomal instability in BALB/c and C57BL/6 mice: the difference is as clear as black and white. Radiat Res. 1997;147:121–125. [PubMed] [Google Scholar]
- Pouget JP, Frelon S, Ravanat JL, Testard I, Odin F, Cadet J. Formation of modified DNA bases in cells exposed either to gamma radiation or to high-LET particles. Radiat Res. 2002;157:589–595. doi: 10.1667/0033-7587(2002)157[0589:fomdbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Preston DL, Mattsson A, Holmberg E, Shore R, Hildreth NG, Boice JD., Jr Radiation effects on breast cancer risk: a pooled analysis of eight cohorts. Radiat Res. 2002;158:220–235. doi: 10.1667/0033-7587(2002)158[0220:reobcr]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Preston DL, et al. Solid cancer incidence in atomic bomb survivors: 1958–1998. Radiat Res. 2007;168:1–64. doi: 10.1667/RR0763.1. [DOI] [PubMed] [Google Scholar]
- Prior S, et al. Densely ionizing radiation affects DNA methylation of selective LINE-1 elements. Environ Res. 2016;150:470–481. doi: 10.1016/j.envres.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell DM. The relationship of terminal duct hyperplasia to mammary carcinoma in 7,12-dimethylbenz(alpha)anthracene-treated LEW/Mai rats. Am J Pathol. 1980;98:311–324. [PMC free article] [PubMed] [Google Scholar]
- Radice GL, Ferreira-Cornwell MC, Robinson SD, Rayburn H, Chodosh LA, Takeichi M, Hynes RO. Precocious mammary gland development in P-cadherin-deficient mice. J Cell Biol. 1997;139:1025–1032. doi: 10.1083/jcb.139.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiche J, Rodriguez-Juarez R, Pogribny I, Kovalchuk O. Sex- and tissue-specific expression of maintenance and de novo DNA methyltransferases upon low dose X-irradiation in mice. Biochem Biophys Res Commun. 2004;325:39–47. doi: 10.1016/j.bbrc.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Rajkumar L, Kittrell FS, Guzman RC, Brown PH, Nandi S, Medina D. Hormone-induced protection of mammary tumorigenesis in genetically engineered mouse models. Breast Cancer Res. 2007;9:R12. doi: 10.1186/bcr1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Coates PJ, Lorimore SA, Wright EG. Bystander-type effects mediated by long-lived inflammatory signaling in irradiated bone marrow. Radiat Res. 2012;177:244–250. doi: 10.1667/RR2805.1. [DOI] [PubMed] [Google Scholar]
- Ravanat JL. Chromatographic methods for the analysis of oxidatively damaged DNA. Free Radic Res. 2012;46:479–491. doi: 10.3109/10715762.2011.623161. [DOI] [PubMed] [Google Scholar]
- Ravanat JL, Breton J, Douki T, Gasparutto D, Grand A, Rachidi W, Sauvaigo S. Radiation-mediated formation of complex damage to DNA: a chemical aspect overview. Br J Radiol. 2014;87:20130715. doi: 10.1259/bjr.20130715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon CE, et al. Tumors induce complex DNA damage in distant proliferative tissues in vivo. Proc Natl Acad Sci USA. 2010;107:17992–17997. doi: 10.1073/pnas.1008260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulus P, Duroux B, Bayle PA, Favier A, Cadet J, Ravanat JL. Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc Natl Acad Sci USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera EM, Hill SD, Taylor M. Organ culture passage enhances the oncogenicity of carcinogen-induced hyperplastic mammary nodules. In Vitro. 1981;17:159–166. doi: 10.1007/BF02618074. [DOI] [PubMed] [Google Scholar]
- Rivina L, Davoren MJ, Schiestl RH. Mouse models for radiation-induced cancers. Mutagenesis. 2016;31:491–509. doi: 10.1093/mutage/gew019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodel F, Hofmann D, Auer J, Keilholz L, Rollinghoff M, Sauer R, Beuscher HU. The anti-inflammatory effect of low-dose radiation therapy involves a diminished CCL20 chemokine expression and granulocyte/endothelial cell adhesion. Strahlenther Onkol. 2008;184:41–47. doi: 10.1007/s00066-008-1776-8. [DOI] [PubMed] [Google Scholar]
- Romar GA, Kupper TS, Divito SJ. Research techniques made simple: techniques to assess cell proliferation. J Invest Dermatol. 2016;136:e1–7. doi: 10.1016/j.jid.2015.11.020. [DOI] [PubMed] [Google Scholar]
- Ronckers CM, Erdmann CA, Land CE. Radiation and breast cancer: a review of current evidence. Breast Cancer Res. 2005;7:21–32. doi: 10.1186/bcr970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roots R, Holley W, Chatterjee A, Irizarry M, Kraft G. The formation of strand breaks in DNA after high-LET irradiation: a comparison of data from in vitro and cellular systems. Int J Radiat Biol. 1990;58:55–69. doi: 10.1080/09553009014551431. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci USA. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel RA, Attfield KR, Schifano JN, Brody JG. Chemicals causing mammary gland tumors in animals signal new directions for epidemiology, chemicals testing, and risk assessment for breast cancer prevention. Cancer. 2007;109:2635–2666. doi: 10.1002/cncr.22653. [DOI] [PubMed] [Google Scholar]
- Rudel RA, Fenton SE, Ackerman JM, Euling SY, Makris SL. Environmental exposures and mammary gland development: state of the science, public health implications, and research recommendations. Environ Health Perspect. 2011;119:1053–1061. doi: 10.1289/ehp.1002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugo RE, Mutamba JT, Mohan KN, Yee T, Chaillet JR, Greenberger JS, Engelward BP. Methyltransferases mediate cell memory of a genotoxic insult. Oncogene. 2011;30:751–756. doi: 10.1038/onc.2010.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhm W, Azizova TV, Bouffler SD, Little MP, Shore RE, Walsh L, Woloschak GE. Dose-rate effects in radiation biology and radiation protection. Ann ICRP. 2016;45:262–279. doi: 10.1177/0146645316629336. [DOI] [PubMed] [Google Scholar]
- Rundle A. Carcinogen-DNA adducts as a biomarker for cancer risk. Mutat Res. 2006;600:23–36. doi: 10.1016/j.mrfmmm.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Russo IH, Russo J. Mammary gland neoplasia in long-term rodent studies. Environ Health Perspect. 1996;104:938–967. doi: 10.1289/ehp.96104938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo J. Significance of rat mammary tumors for human risk assessment. Toxicol Pathol. 2015;43:145–170. doi: 10.1177/0192623314532036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo J, Saby J, Isenberg WM, Russo IH. Pathogenesis of mammary carcinomas induced in rats by 7, 12-dimethylbenz[a]anthracene2. JNCI. 1977;59:435–445. doi: 10.1093/jnci/59.2.435. [DOI] [Google Scholar]
- Rydberg B, Cooper B, Cooper PK, Holley WR, Chatterjee A. Dose-dependent misrejoining of radiation-induced DNA double-strand breaks in human fibroblasts: experimental and theoretical study for high- and low-LET radiation. Radiat Res. 2005;163:526–534. doi: 10.1667/rr3346. [DOI] [PubMed] [Google Scholar]
- Rydberg B, Heilbronn L, Holley WR, Lobrich M, Zeitlin C, Chatterjee A, Cooper PK. Spatial distribution and yield of DNA double-strand breaks induced by 3–7 MeV helium ions in human fibroblasts. Radiat Res. 2002;158:32–42. doi: 10.1667/0033-7587(2002)158[0032:sdayod]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ryu TH, Kim JH, Kim JK. Chromosomal aberrations in human peripheral blood lymphocytes after exposure to ionizing radiation. Genome Integr. 2016;7:5. doi: 10.4103/2041-9414.197172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenko Y, Cieslar-Pobuda A, Skonieczna M, Rzeszowska-Wolny J. Changes of reactive oxygen and nitrogen species and mitochondrial functioning in human K562 and HL60 cells exposed to ionizing radiation. Radiat Res. 2013;180:360–366. doi: 10.1667/RR3247.1. [DOI] [PubMed] [Google Scholar]
- Sakai K, Nomura T, Ina Y. Enhancement of bio-protective functions by low dose/dose-rate radiation. Dose Response. 2006;4:327–332. doi: 10.2203/dose-response.06-115.Sakai. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama SA, et al. Effect of tumor necrosis factor-alpha on estrogen metabolism and endometrial cells: potential physiological and pathological relevance. J Clin Endocrinol Metab. 2009;94:285–293. doi: 10.1210/jc.2008-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu JK, Birnboim HC. Mutagenicity and cytotoxicity of reactive oxygen and nitrogen species in the MN-11 murine tumor cell line. Mutat Res. 1997;379:241–252. doi: 10.1016/s0027-5107(97)00140-1. [DOI] [PubMed] [Google Scholar]
- Sansone P, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci-Pereira J, et al. Mimicking pregnancy as a strategy for breast cancer prevention. Breast Cancer Manag. 2013;2:283–294. doi: 10.2217/bmt.13.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki MS, Ejima Y, Tachibana A, Yamada T, Ishizaki K, Shimizu T, Nomura T. DNA damage response pathway in radioadaptive response. Mutat Res. 2002;504:101–118. doi: 10.1016/s0027-5107(02)00084-2. [DOI] [PubMed] [Google Scholar]
- Sasaki MS, Tachibana A, Takeda S. Cancer risk at low doses of ionizing radiation: artificial neural networks inference from atomic bomb survivors. J Radiat Res. 2014;55:391–406. doi: 10.1093/jrr/rrt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasser AK, Sullivan NJ, Studebaker AW, Hendey LF, Axel AE, Hall BM. Interleukin-6 is a potent growth factor for ER-alpha-positive human breast cancer. Faseb J. 2007;21:3763–3770. doi: 10.1096/fj.07-8832com. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Khogali F, Carls N. Reversion of the mouse pink-eyed unstable mutation induced by low doses of x-rays. Science. 1994;266:1573–1576. doi: 10.1126/science.7985029. [DOI] [PubMed] [Google Scholar]
- Schonfeld SJ, et al. Polymorphisms in oxidative stress and inflammation pathway genes, low-dose ionizing radiation, and the risk of breast cancer among US radiologic technologists. Cancer Causes Control. 2010;21:1857–1866. doi: 10.1007/s10552-010-9613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- Seager AL, et al. Pro-oxidant induced DNA damage in human lymphoblastoid cells: homeostatic mechanisms of genotoxic tolerance. Toxicol Sci. 2012;128:387–397. doi: 10.1093/toxsci/kfs152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segaloff A, Maxfield WS. The synergism between radiation and estrogen in the production of mammary cancer in the rat. Cancer Res. 1971;31:166–168. [PubMed] [Google Scholar]
- Selvanayagam CS, Davis CM, Cornforth MN, Ullrich RL. Latent expression of p53 mutations and radiation-induced mammary cancer. Cancer Res. 1995;55:3310–3317. [PubMed] [Google Scholar]
- Shahi P, et al. GATA3 targets semaphorin 3B in mammary epithelial cells to suppress breast cancer progression and metastasis. Oncogene. 2017;36:5567–5575. doi: 10.1038/onc.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Folkard M, Prise KM. Role of TGF-beta1 and nitric oxide in the bystander response of irradiated glioma cells. Oncogene. 2008;27:434–440. doi: 10.1038/sj.onc.1210653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, et al. Oxidative stress at low levels can induce clustered DNA lesions leading to NHEJ mediated mutations. Oncotarget. 2016;7:25377–25390. doi: 10.18632/oncotarget.8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Semin Cancer Biol. 2011;21:349–353. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shellabarger CJ. Mammary neoplastic response of Lewis and Sprague-Dawley female rats to 7,12-dimethylbenz(a)anthracene or x-ray. Cancer Res. 1972;32:883–885. [PubMed] [Google Scholar]
- Shellabarger CJ, Stone JP, Holtzman S. Synergism between neutron radiation and diethylstilbestrol in the production of mammary adenocarcinomas in the rat. Can Res. 1976;36:1019–1022. [PubMed] [Google Scholar]
- Sherborne AL, Davidson PR, Yu K, Nakamura AO, Rashid M, Nakamura JL. Mutational analysis of ionizing radiation induced neoplasms. Cell Rep. 2015;12:1915–1926. doi: 10.1016/j.celrep.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SC, Kang YM, Kim HS. Life span and thymic lymphoma incidence in high- and low-dose-rate irradiated AKR/J mice and commonly expressed genes. Radiat Res. 2010;174:341–346. doi: 10.1667/RR1946.1. [DOI] [PubMed] [Google Scholar]
- Shore R, Walsh L, Azizova T, Ruhm W. Risk of solid cancer in low dose-rate radiation epidemiological studies and the dose-rate effectiveness factor. Int J Radiat Biol. 2017;93:1064–1078. doi: 10.1080/09553002.2017.1319090. [DOI] [PubMed] [Google Scholar]
- Shore RE, et al. Implications of recent epidemiologic studies for the linear nonthreshold model and radiation protection. J Radiol Prot. 2018;38:1217–1233. doi: 10.1088/1361-6498/aad348. [DOI] [PubMed] [Google Scholar]
- Sigurdson AJ, et al. Polymorphisms in estrogen biosynthesis and metabolism-related genes, ionizing radiation exposure, and risk of breast cancer among US radiologic technologists. Breast Cancer Res Treat. 2009;118:177–184. doi: 10.1007/s10549-009-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone NL, et al. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS ONE. 2009;4:e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha D, Dao TL. A direct mechanism of mammary carcinogenesis induced by 7,12-dimethylbenz[a]anthracene2. JNCI. 1974;53:841–846. doi: 10.1093/jnci/53.3.841. [DOI] [PubMed] [Google Scholar]
- Sinha D, Dao TL. Hyperplastic alveolar nodules of the rat mammary gland: tumor-producing capability in vivo and in vitro. Cancer Lett. 1977;2:153–160. doi: 10.1016/s0304-3835(77)80005-0. [DOI] [PubMed] [Google Scholar]
- Sinha M, Das DK, Bhattacharjee S, Majumdar S, Dey S. Leaf extract of Moringa oleifera prevents ionizing radiation-induced oxidative stress in mice. J Med Food. 2011;14:1167–1172. doi: 10.1089/jmf.2010.1506. [DOI] [PubMed] [Google Scholar]
- Sinha M, Das DK, Manna K, Datta S, Ray T, Sil AK, Dey S. Epicatechin ameliorates ionising radiation-induced oxidative stress in mouse liver. Free Radic Res. 2012;46:842–849. doi: 10.3109/10715762.2012.684245. [DOI] [PubMed] [Google Scholar]
- Sishc BJ, et al. Telomeres and telomerase in the radiation response: implications for instability, reprograming, and carcinogenesis. Front Oncol. 2015;5:257. doi: 10.3389/fonc.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva S, et al. A pattern of early radiation-induced inflammatory cytokine expression is associated with lung toxicity in patients with non-small cell lung cancer. PLoS ONE. 2014;9:e109560. doi: 10.1371/journal.pone.0109560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MT, et al. Key characteristics of carcinogens as a basis for organizing data on mechanisms of carcinogenesis. Environ Health Perspect. 2016;124:713–721. doi: 10.1289/ehp.1509912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders AM, et al. Genetic differences in transcript responses to low-dose ionizing radiation identify tissue functions associated with breast cancer susceptibility. PLoS ONE. 2012;7:e45394. doi: 10.1371/journal.pone.0045394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler D, Pampalona J, Tusell L, Genesca A. Radiation sensitivity increases with proliferation-associated telomere dysfunction in nontransformed human epithelial cells. Aging Cell. 2009;8:414–425. doi: 10.1111/j.1474-9726.2009.00488.x. [DOI] [PubMed] [Google Scholar]
- Solleveld HA, van Zwieten MJ, Broerse JJ, Hollander CF. Effects of X-irradiation, ovariohysterectomy and estradiol-17 beta on incidence, benign/malignant ratio and multiplicity of rat mammary neoplasms—a preliminary report. Leuk Res. 1986;10:755–759. doi: 10.1016/0145-2126(86)90292-4. [DOI] [PubMed] [Google Scholar]
- Sourisseau T, Harrington KJ, Muller WJ, Marshall CJ, White DE. Changes in tumor tissue organization in collagen-I sensitize cells to ionizing radiation in an ex vivo model of solid mammary tumor growth and local invasion. Cell Cycle. 2011;10:3979–3981. doi: 10.4161/cc.10.22.18291. [DOI] [PubMed] [Google Scholar]
- Sridharan DM, et al. Understanding cancer development processes after HZE-particle exposure: roles of ROS DNA damage repair and inflammation. Radiat Res. 2015;183:1–26. doi: 10.1667/rr13804.1. [DOI] [PubMed] [Google Scholar]
- Stanicka J, Russell EG, Woolley JF, Cotter TG. NADPH oxidase-generated hydrogen peroxide induces DNA damage in mutant FLT3-expressing leukemia cells. J Biol Chem. 2015;290:9348–9361. doi: 10.1074/jbc.M113.510495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankevicins L, et al. MiR-34a is up-regulated in response to low dose, low energy X-ray induced DNA damage in breast cells. Radiat Oncol. 2013;8:231. doi: 10.1186/1748-717X-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenerlow B, Hoglund E, Carlsson J, Blomquist E. Rejoining of DNA fragments produced by radiations of different linear energy transfer. Int J Radiat Biol. 2000;76:549–557. doi: 10.1080/095530000138565. [DOI] [PubMed] [Google Scholar]
- Sternlicht MD, Sunnarborg SW, Kouros-Mehr H, Yu Y, Lee DC, Werb Z. Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development. 2005;132:3923–3933. doi: 10.1242/dev.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CR, Bucurenci N, Abbot SE, Sahinoglu T, Blake DR, Naughton D, Grootveld MC. Application of methionine as a detector molecule for the assessment of oxygen radical generation by human neutrophils and endothelial cells. Free Radic Res Commun. 1992;17:143–154. doi: 10.3109/10715769209082271. [DOI] [PubMed] [Google Scholar]
- Stovall M, et al. Dose to the contralateral breast from radiotherapy and risk of second primary breast cancer in the WECARE study. Int J Radiat Oncol Biol Phys. 2008;72:1021–1030. doi: 10.1016/j.ijrobp.2008.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studebaker AW, et al. Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res. 2008;68:9087–9095. doi: 10.1158/0008-5472.CAN-08-0400. [DOI] [PubMed] [Google Scholar]
- Sukup-Jackson MR, et al. Rosa26-GFP direct repeat (RaDR-GFP) mice reveal tissue- and age-dependence of homologous recombination in mammals in vivo. PLoS Genet. 2014;10:e1004299. doi: 10.1371/journal.pgen.1004299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suman S, Johnson MD, Fornace AJ, Jr, Datta K. Exposure to ionizing radiation causes long-term increase in serum estradiol and activation of PI3K-Akt signaling pathway in mouse mammary gland. Int J Radiat Oncol Biol Phys. 2012;84:500–507. doi: 10.1016/j.ijrobp.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Osterman MD, Li M. Tissue specificity of DNA damage response and tumorigenesis. Cancer Biol Med. 2019;16:396–414. doi: 10.20892/j.issn.2095-3941.2019.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland BM, Bennett PV, Sutherland JC, Laval J. Clustered DNA damages induced by X-rays in human cells. Radiat Res. 2002;157:611–616. doi: 10.1667/0033-7587(2002)157[0611:cddibx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Sutherland BM, Georgakilas AG, Bennett PV, Laval J, Sutherland JC. Quantifying clustered DNA damage induction and repair by gel electrophoresis, electronic imaging and number average length analysis. Mutat Res. 2003;531:93–107. doi: 10.1016/j.mrfmmm.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Ishii-Ohba H, Yamanouchi H, Wakabayashi K, Takahashi M, Inano H. Susceptibility of lactating rat mammary glands to gamma-ray-irradiation-induced tumorigenesis. Int J Cancer. 1994;56:413–417. doi: 10.1002/ijc.2910560321. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kashino G, Kodama S, Watanabe M. Long-term persistence of X-ray-induced genomic instability in quiescent normal human diploid cells. Mutat Res. 2009;671:33–39. doi: 10.1016/j.mrfmmm.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Yamashita S. Low-dose radiation exposure and carcinogenesis. Jpn J Clin Oncol. 2012;42:563–568. doi: 10.1093/jjco/hys078. [DOI] [PubMed] [Google Scholar]
- Sykora P, et al. Next generation high throughput DNA damage detection platform for genotoxic compound screening. Sci Rep. 2018;8:2771. doi: 10.1038/s41598-018-20995-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari T, et al. Extracellular vesicles mediate radiation-induced systemic bystander signals in the bone marrow and spleen. Front Immunol. 2017;8:347. doi: 10.3389/fimmu.2017.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostakowska M, Trebinska-Stryjewska A, Grzybowska EA, Fabisiewicz A. Resistance to endocrine therapy in breast cancer: molecular mechanisms and future goals. Breast Cancer Res Treat. 2019;173:489–497. doi: 10.1007/s10549-018-5023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabatake M, et al. Differential effect of parity on rat mammary carcinogenesis after pre- or post-pubertal exposure to radiation. Sci Rep. 2018;8:14325. doi: 10.1038/s41598-018-32406-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka IB, 3rd, et al. Cause of death and neoplasia in mice continuously exposed to very low dose rates of gamma rays. Radiat Res. 2007;167:417–437. doi: 10.1667/RR0728.1. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kohda A, Satoh K, Toyokawa T, Ichinohe K, Ohtaki M, Oghiso Y. Dose-rate effectiveness for unstable-type chromosome aberrations detected in mice after continuous irradiation with low-dose-rate gamma rays. Radiat Res. 2009;171:290–301. doi: 10.1667/RR1238.1. [DOI] [PubMed] [Google Scholar]
- Tang FR, Loke WK. Molecular mechanisms of low dose ionizing radiation-induced hormesis, adaptive responses, radioresistance, bystander effects, and genomic instability. Int J Radiat Biol. 2015;91:13–27. doi: 10.3109/09553002.2014.937510. [DOI] [PubMed] [Google Scholar]
- Tang J, et al. Irradiation of juvenile, but not adult, mammary gland increases stem cell self-renewal and estrogen receptor negative tumors. Stem Cells. 2014;32:649–661. doi: 10.1002/stem.1533. [DOI] [PubMed] [Google Scholar]
- Tao L, Roberts AL, Dunphy KA, Bigelow C, Yan H, Jerry DJ. Repression of mammary stem/progenitor cells by p53 is mediated by Notch and separable from apoptotic activity. Stem Cells. 2011;29:119–127. doi: 10.1002/stem.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Xiang D, Xie Y, Bronson RT, Li Z. Induced p53 loss in mouse luminal cells causes clonal expansion and development of mammary tumours. Nat Commun. 2017;8:14431. doi: 10.1038/ncomms14431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartier L, Gilchrist S, Burdak-Rothkamm S, Folkard M, Prise KM. Cytoplasmic irradiation induces mitochondrial-dependent 53BP1 protein relocalization in irradiated and bystander cells. Cancer Res. 2007;67:5872–5879. doi: 10.1158/0008-5472.CAN-07-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawa R, et al. Effects of X-ray irradiation on genomic DNA methylation levels in mouse tissues. J Radiat Res. 1998;39:271–278. doi: 10.1269/jrr.39.271. [DOI] [PubMed] [Google Scholar]
- Thayer KA, Foster PM. Workgroup report: national toxicology program workshop on hormonally induced reproductive tumors—relevance of rodent bioassays. Environ Health Perspect. 2007;115:1351–1356. doi: 10.1289/ehp.10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS, et al. The next generation blueprint of computational toxicology at the U.S Environmental Protection Agency. Toxicol Sci. 2019;169:317–332. doi: 10.1093/toxsci/kfz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SN, Waters KM, Morgan WF, Yang AJ, Baulch JE. Quantitative proteomic analysis of mitochondrial proteins reveals prosurvival mechanisms in the perpetuation of radiation-induced genomic instability. Free Radic Biol Med. 2012;53:618–628. doi: 10.1016/j.freeradbiomed.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Yin X, Wang L, Wang J, Zhu W, Cao J, Yang H. The key role of miR-21-regulated SOD2 in the medium-mediated bystander responses in human fibroblasts induced by alpha-irradiated keratinocytes. Mutat Res. 2015;780:77–85. doi: 10.1016/j.mrfmmm.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017;355:1330–1334. doi: 10.1126/science.aaf9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V, Little MP. Dose and dose rate extrapolation factors for malignant and non-malignant health endpoints after exposure to gamma and neutron radiation. Radiat Environ Biophys. 2017;56:299–328. doi: 10.1007/s00411-017-0707-4. [DOI] [PubMed] [Google Scholar]
- Travis LB, et al. Breast cancer following radiotherapy and chemotherapy among young women with Hodgkin disease. JAMA. 2003;290:465–475. doi: 10.1001/jama.290.4.465. [DOI] [PubMed] [Google Scholar]
- Tryndyak VP, Kovalchuk O, Pogribny IP. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4–20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol Ther. 2006;5:65–70. doi: 10.4161/cbt.5.1.2288. [DOI] [PubMed] [Google Scholar]
- Tsai KK, Chuang EY, Little JB, Yuan ZM. Cellular mechanisms for low-dose ionizing radiation-induced perturbation of the breast tissue microenvironment. Cancer Res. 2005;65:6734–6744. doi: 10.1158/0008-5472.can-05-0703. [DOI] [PubMed] [Google Scholar]
- Tucker DK, Foley JF, Bouknight SA, Fenton SE. Sectioning mammary gland whole mounts for lesion identification. J Vis Exp. 2017 doi: 10.3791/55796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich RL, Bowles ND, Satterfield LC, Davis CM. Strain-dependent susceptibility to radiation-induced mammary cancer is a result of differences in epithelial cell sensitivity to transformation. Radiat Res. 1996;146:353–355. [PubMed] [Google Scholar]
- Ullrich RL, Davis CM. Radiation-induced cytogenetic instability in vivo. Radiat Res. 1999;152:170–173. [PubMed] [Google Scholar]
- Ullrich RL, Ponnaiya B. Radiation-induced instability and its relation to radiation carcinogenesis. Int J Radiat Biol. 1998;74:747–754. doi: 10.1080/095530098141023. [DOI] [PubMed] [Google Scholar]
- Umesako S, et al. Atm heterozygous deficiency enhances development of mammary carcinomas in p53 heterozygous knockout mice. Breast Cancer Res. 2005;7:R164–170. doi: 10.1186/bcr968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger K, et al. Novel gene rearrangements in transformed breast cells identified by high-resolution breakpoint analysis of chromosomal aberrations. Endocr Relat Cancer. 2010;17:87–98. doi: 10.1677/erc-09-0065. [DOI] [PubMed] [Google Scholar]
- Vandin F, Upfal E, Raphael BJ. De novo discovery of mutated driver pathways in cancer. Genome Res. 2012;22:375–385. doi: 10.1101/gr.120477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, et al. Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol Sci. 2014;142:312–320. doi: 10.1093/toxsci/kfu199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HH, Jr, Turner JE. Genetic component in rat mammary carcinogenesis. Radiat Res. 1982;89:264–273. [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VoPham T, et al. Environmental radon exposure and breast cancer risk in the Nurses' Health Study II. Environ Health. 2017;16:97. doi: 10.1186/s12940-017-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KU. Models of breast cancer: quo vadis, animal modeling? Breast Cancer Res. 2004;6:31–38. doi: 10.1186/bcr723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Su F, Smilenov LB, Zhou L, Hu W, Ding N, Zhou G. Mechanisms of increased risk of tumorigenesis in Atm and Brca1 double heterozygosity. Radiat Oncol. 2011;6:96. doi: 10.1186/1748-717X-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. Genome-wide screen of DNA methylation changes induced by low dose X-ray radiation in mice. PLoS ONE. 2014;9:e90804. doi: 10.1371/journal.pone.0090804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Guo F, Han L, Wang X, Li J, Guo Y, Lu Y. X-ray-induced changes in the expression of inflammation-related genes in human peripheral blood. Int J Mol Sci. 2014;15:19516–19534. doi: 10.3390/ijms151119516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, et al. Induction of non-targeted stress responses in mammary tissues by heavy ions. PLoS ONE. 2015;10:e0136307. doi: 10.1371/journal.pone.0136307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fang H, Huang Z, Shang W, Hou T, Cheng A, Cheng H. Imaging ROS signaling in cells and animals. J Mol Med (Berl) 2013;91:917–927. doi: 10.1007/s00109-013-1067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- Watanabe R, Rahmanian S, Nikjoo H. Spectrum of radiation-induced clustered non-DSB damage—a monte carlo track structure modeling and calculations. Radiat Res. 2015;183:525–540. doi: 10.1667/RR13902.1. [DOI] [PubMed] [Google Scholar]
- Wazer DE, Chu Q, Liu XL, Gao Q, Safaii H, Band V. Loss of p53 protein during radiation transformation of primary human mammary epithelial cells. Mol Cell Biol. 1994;14:2468–2478. doi: 10.1128/mcb.14.4.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RJ, Desai TA, Gartner ZJ. Non-autonomous cell proliferation in the mammary gland and cancer. Curr Opin Cell Biol. 2017;45:55–61. doi: 10.1016/j.ceb.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch CW, Goodrich-Smith M, Brown CK, Miglorie N, Clifton KH. Effect of an estrogen antagonist (tamoxifen) on the initiation and progression of gamma-irradiation-induced mammary tumors in female Sprague-Dawley rats. Eur J Cancer Clin Oncol. 1981;17:1255–1258. doi: 10.1016/0014-2964(81)90004-9. [DOI] [PubMed] [Google Scholar]
- Werner E, Wang H, Doetsch PW. Opposite roles for p38MAPK-driven responses and reactive oxygen species in the persistence and resolution of radiation-induced genomic instability. PLoS ONE. 2014;9:e108234. doi: 10.1371/journal.pone.0108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott PMK, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2014;517:489. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyemi U, et al. Inactivation of NADPH oxidases NOX4 and NOX5 protects human primary fibroblasts from ionizing radiation-induced DNA damage. Radiat Res. 2015;183:262–270. doi: 10.1667/RR13799.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PA, Luijten M, Mishima M, Cox JA, Hanna JN, Maertens RM, Zwart EP. In vitro mammalian cell mutation assays based on transgenic reporters: a report of the International Workshop on genotoxicity testing (IWGT) Mutat Res. 2019;847:403039. doi: 10.1016/j.mrgentox.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Williams ES, et al. Telomere dysfunction and DNA-PKcs deficiency: characterization and consequence. Cancer Res. 2009;69:2100–2107. doi: 10.1158/0008-5472.CAN-08-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winyard PG, Faux SP, Smith AJ, Davies JE, Chipman JK. Bleomycin-induced unscheduled DNA synthesis in non-permeabilized human and rat hepatocytes is not paralleled by 8-oxo-7,8-dihydrodeoxyguanosine formation. Biochem Pharmacol. 1992;44:1255–1260. doi: 10.1016/0006-2952(92)90523-l. [DOI] [PubMed] [Google Scholar]
- Wolff S, Afzal V, Wiencke JK, Olivieri G, Michaeli A. Human lymphocytes exposed to low doses of ionizing radiations become refractory to high doses of radiation as well as to chemical mutagens that induce double-strand breaks in DNA. Int J Radiat Biol Relat Stud Phys Chem Med. 1988;53:39–47. doi: 10.1080/09553008814550401. [DOI] [PubMed] [Google Scholar]
- Xu S, et al. Exosome-mediated microRNA transfer plays a role in radiation-induced bystander effect. RNA Biol. 2015;12:1355–1363. doi: 10.1080/15476286.2015.1100795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi H, Ishii-Ohba H, Suzuki K, Onoda M, Wakabayashi K, Inano H. Relationship between stages of mammary development and sensitivity to gamma-ray irradiation in mammary tumorigenesis in rats. Int J Cancer. 1995;60:230–234. doi: 10.1002/ijc.2910600217. [DOI] [PubMed] [Google Scholar]
- Yamauchi K, et al. Differential effects of low- and high-dose X-rays on N-ethyl-N-nitrosourea-induced mutagenesis in thymocytes of B6C3F1 gpt-delta mice. Mutat Res. 2008;640:27–37. doi: 10.1016/j.mrfmmm.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Yan B, et al. Tumor necrosis factor-alpha is a potent endogenous mutagen that promotes cellular transformation. Cancer Res. 2006;66:11565–11570. doi: 10.1158/0008-5472.CAN-06-2540. [DOI] [PubMed] [Google Scholar]
- Yang B, et al. A novel pathway links oxidative stress to loss of insulin growth factor-2 (IGF2) imprinting through NF-kappaB activation. PLoS ONE. 2014;9:e88052. doi: 10.1371/journal.pone.0088052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Elner SG, Bian ZM, Till GO, Petty HR, Elner VM. Pro-inflammatory cytokines increase reactive oxygen species through mitochondria and NADPH oxidase in cultured RPE cells. Exp Eye Res. 2007;85:462–472. doi: 10.1016/j.exer.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Anzenberg V, Held KD. The time dependence of bystander responses induced by iron-ion radiation in normal human skin fibroblasts. Radiat Res. 2007;168:292–298. doi: 10.1667/RR0864.1. [DOI] [PubMed] [Google Scholar]
- Yang H, Asaad N, Held KD. Medium-mediated intercellular communication is involved in bystander responses of X-ray-irradiated normal human fibroblasts. Oncogene. 2005;24:2096–2103. doi: 10.1038/sj.onc.1208439. [DOI] [PubMed] [Google Scholar]
- Yang T-H, Craise LM, Prioleau JC, Stampfer MR, Rhim JS. Chromosomal changes in cultured human epithelial cells transformed by low- and high-LET radiation. Adv Space Res. 1992;12:127–136. doi: 10.1016/0273-1177(92)90099-j. [DOI] [PubMed] [Google Scholar]
- Yang TC, Georgy KA, Craise LM, Durante M. Initiation of oncogenic transformation in human mammary epithelial cells by charged particles. Radiat Oncol Investig. 1997;5:134–138. doi: 10.1002/(SICI)1520-6823(1997)5:3<134::AID-ROI10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Yang XR, et al. Characterization of genomic alterations in radiation-associated breast cancer among childhood cancer survivors, using comparative genomic hybridization (CGH) arrays. PLoS ONE. 2015;10:e0116078. doi: 10.1371/journal.pone.0116078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Cell cycle stage-specific roles of Rad18 in tolerance and repair of oxidative DNA damage. Nucleic Acids Res. 2013;41:2296–2312. doi: 10.1093/nar/gks1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21:751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Yuan D, Xie Y, Pan Y, Shao C. Role of DNA methylation in long-term low-dose gamma-rays induced adaptive response in human B lymphoblast cells. Int J Radiat Biol. 2013;89:898–906. doi: 10.3109/09553002.2013.806832. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Goto S, Kawakatsu M, Urata Y, Li TS. Mitochondrial dysfunction, a probable cause of persistent oxidative stress after exposure to ionizing radiation. Free Radic Res. 2012;46:147–153. doi: 10.3109/10715762.2011.645207. [DOI] [PubMed] [Google Scholar]
- Yu Y, et al. Elevated breast cancer risk in irradiated BALB/c mice associates with unique functional polymorphism of the Prkdc (DNA-dependent protein kinase catalytic subunit) gene. Cancer Res. 2001;61:1820–1824. [PubMed] [Google Scholar]
- Yuan L, Dietrich AK, Ziegler YS, Nardulli AM. 17beta-Estradiol alters oxidative damage and oxidative stress response protein expression in the mouse mammary gland. Mol Cell Endocrinol. 2016;426:11–21. doi: 10.1016/j.mce.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetner D, Andersen LP, Rosenberg J. Melatonin as protection against radiation injury: a systematic review. Drug Res (Stuttg) 2016;66:281–296. doi: 10.1055/s-0035-1569358. [DOI] [PubMed] [Google Scholar]
- Zhang P, et al. Identification of genetic loci that control mammary tumor susceptibility through the host microenvironment. Sci Rep. 2015;5:8919. doi: 10.1038/srep08919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhu L, Wang G, Zhao Y, Xiong N, Bao H, Jin W. Ionizing radiation promotes CCL27 secretion from keratinocytes through the cross talk between TNF-alpha and ROS. J Biochem Mol Toxicol. 2017 doi: 10.1002/jbt.21868. [DOI] [PubMed] [Google Scholar]
- Zhou H, et al. Mechanism of radiation-induced bystander effect: role of the cyclooxygenase-2 signaling pathway. Proc Natl Acad Sci USA. 2005;102:14641–14646. doi: 10.1073/pnas.0505473102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Ivanov VN, Lien YC, Davidson M, Hei TK. Mitochondrial function and nuclear factor-kappaB-mediated signaling in radiation-induced bystander effects. Cancer Res. 2008;68:2233–2240. doi: 10.1158/0008-5472.CAN-07-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.