Abstract
Objective
Defects in ion channels and neurotransmitter receptors are implicated in developmental and epileptic encephalopathy (DEE). Metabotropic glutamate receptor 7 (mGluR7), encoded by GRM7, is a presynaptic G‐protein‐coupled glutamate receptor critical for synaptic transmission. We previously proposed GRM7 as a candidate disease gene in two families with neurodevelopmental disorders (NDDs). One additional family has been published since. Here, we describe three additional families with GRM7 biallelic variants and deeply characterize the associated clinical neurological and electrophysiological phenotype and molecular data in 11 affected individuals from six unrelated families.
Methods
Exome sequencing and family‐based rare variant analyses on a cohort of 220 consanguineous families with NDDs revealed three families with GRM7 biallelic variants; three additional families were identified through literature search and collaboration with a clinical molecular laboratory.
Results
We compared the observed clinical features and variants of 11 affected individuals from the six unrelated families. Identified novel deleterious variants included two homozygous missense variants (c.2671G>A:p.Glu891Lys and c.1973G>A:p.Arg685Gln) and one homozygous stop‐gain variant (c.1975C>T:p.Arg659Ter). Developmental delay, neonatal‐ or infantile‐onset epilepsy, and microcephaly were universal. Three individuals had hypothalamic–pituitary–axis dysfunction without pituitary structural abnormality. Neuroimaging showed cerebral atrophy and hypomyelination in a majority of cases. Two siblings demonstrated progressive loss of myelination by 2 years in both and an acquired microcephaly pattern in one. Five individuals died in early or late childhood.
Conclusion
Detailed clinical characterization of 11 individuals from six unrelated families demonstrates that rare biallelic GRM7 pathogenic variants can cause DEEs, microcephaly, hypomyelination, and cerebral atrophy.
Introduction
Defects in ion channels and neurotransmitter receptors are implicated in developmental and epileptic encephalopathies (DEEs) by impairing central nervous system (CNS) neurotransmission. 1 , 2 One such important group of receptors is the glutamate receptors. Glutamate is an excitatory neurotransmitter that has been extensively studied in epilepsy and that acts through two main receptor groups: ionotropic glutamate receptors (iGluRs) and metabotropic glutamate receptors (mGluRs). 3 The iGluRs include N‐methyl‐D‐aspartate receptor (NMDAR), alpha‐amino‐3‐hydroxy‐5‐methyl‐4‐isoxazolepropionic acid receptor (AMPAR), and kainate receptor (KAR). Rare monoallelic and biallelic single nucleotide variants (SNVs) and copy number variants (CNVs) in genes encoding postsynaptic iGluRs subunits such as GRIN1, GRIN2A, GRIN2B, and GRIN2D encoding the NR1, NR2A, NR2B, and NR2D subunits of NMDAR, respectively, GRIA2‐4 encoding the GluR2‐4 subunits of AMPAR, and GRIK2 encoding the GluR6 subunit of KAR have been shown to cause a wide range of neurodevelopmental disorders (NDDs) and DEEs. 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11
The mGluRs consist of eight receptors, mGluRs1‐8, encoded by GRM1‐8, that are classified into three groups based on sequence homology, ligand selectivity, and signal transduction mechanism. 12 Group I consists of mGluR1 and mGluR5, group II consists of mGluR2 and mGluR3, and group III consists of mGluR4, mGluR6, mGluR7, and mGluR8. Groups II and III function presynaptically, whereas group I functions postsynaptically. 12 Recent studies have focused on the role of mGluRs in neurodevelopmental and neuropsychiatric disorders. A genome‐wide CNV study using single nucleotide polymorphism (SNP) array revealed an enrichment of CNVs in GRM1, 5, 7, and 8 in subjects with attention deficit hyperactivity disorders versus apparently healthy controls. 13 Another pilot case–control study supported an association between GRM7 SNPs and autism spectrum disorder. 14 Biallelic rare variants in GRM1 have been shown to cause cerebellar ataxia. 15
mGluRs are also involved in the pathophysiology of epilepsy. 16 Agonists of postsynaptic group I mGluRs have proconvulsive properties while their antagonists have anticonvulsant activity in seizure models, and the opposite has been shown in the presynaptic mGluRs groups II and III. These findings are attributed to the role of group I mGluRs in enhancing neuronal excitability and of groups II and III in inhibiting hyperexcitability. 16
mGluR7, encoded by GRM7, is the most highly conserved mGluR and is exclusively expressed in the CNS. mGluR7 functions as a constitutional dimer and consists of a binding domain, cysteine‐rich domain, transmembrane domain, and intracellular C‐terminus. 12 mGluR7 plays a critical role in synaptic transmission by inhibiting further release of excitatory neurotransmitter glutamate and inhibitory neurotransmitter GABA when they reach high levels at the synapses. 17 mGluR7‐knockout mice develop spontaneous stimulus‐provoked seizures, suggesting that disruption of GRM7 expression may cause epilepsy. 18
We previously proposed GRM7 as a candidate gene for autosomal recessive NDD in four subjects from two unrelated families with NDD and rare biallelic missense variants. 19 One additional family including two siblings with NDD and a rare homozygous stop‐gain allele in GRM7 has been described since. 20 Here, we describe five additional individuals from three unrelated families with biallelic variants in GRM7 and a similar neurological DEE phenotype along with the six individuals previously reported with limited clinical data and provide evidence to support GRM7 biallelic variants as a cause for NDDs, DEE, and microcephaly.
Methods
Participants and ethical approval
This study was approved by the Institutional Review Board (IRB) at Baylor College of Medicine under the Baylor Hopkins Center for Mendelian Genomics Protocol (IRB number H‐29697). All families except for Family 3 (MR005) were enrolled under protocol H‐29697. Family 3 (MR005) was enrolled under an IRB‐approved protocol at the University of Bonn (IRB number 033/08). For all research subjects, informed patient consent was obtained for publication of photographs.
Exome sequencing, linkage analysis, variant interpretation, and phenotyping methods
By performing family‐based exome sequencing and rare variant filtering on a NDD cohort of 220 families from consanguineous populations from Saudi Arabia and Turkey, and using our previously described variant parsing and prioritization workflow, 21 we identified four individuals (BAB6708, BAB6709, BAB10502, and BAB10517) from three unrelated families (Families 1, 5, and 6 in Figs. 1 and 2) with rare homozygous GRM7 variants. Comprehensive analysis of all exome sequencing data was performed to identify rare, potentially damaging variants. No candidate variants in known disease genes were identified in any of the four individuals to provide an alternative genetic etiology.
Figure 1.

Pedigrees, Sanger sequencing, facial features, and brain MRI images of affected individuals from Families 1–3 with biallelic variants in GRM7. (A) Pedigree and Sanger sequencing showing segregation of the variants in GRM7 in Family 1. (B) Facial photograph of individual II‐1 (Family 1) at 13 years showing prominent teeth and everted lower lip. (C‐E) Brain MRI of individual II‐1 (Family 1) at 3 years. T1‐weighted image (midsagittal view) shows moderate to severe thinning of corpus callosum (CC), excess cerebral spinal fluid (CSF) around the falx cerebri indicating severe cerebral volume loss, and minimal cerebellar volume loss (C). T2‐weighted images (axial and coronal views) show hypomyelination, particularly over the anterior temporal horns, with normal fourth ventricle and hippocampi (D and E). (F) Facial photograph of individual II‐2 (Family 1) at 10 years shows high forehead and hypotonic face (G‐I) Brain MRI of individual II‐2 (Family 1) at 2 years. T1‐weighted image (midsagittal view) showing foreshortening of CC, and excess CSF around the falx cerebri indicating severe cerebral volume loss (G). T2‐weighted images (axial and coronal views) show normally myelinated internal capsule, excessive subarachnoid spaces, enlarged lateral ventricles secondary to cerebral volume loss, and T2 hyperintensity in bilateral hippocampi (H and I). (J) Pedigree and Sanger sequencing showing segregation of the variants in GRM7 in Family 2. (K) Facial photograph of individual II‐1 (Family 2) at 15 years shows a wide mouth. (L–N) Brain MRI of individual II‐1 (Family 2) at 2 months. T1‐weighted image (midsagittal view) shows normal CC (L). T2‐weighted images (axial and coronal views) show normal myelination for age (M and N). (O–Q) Brain MRI of individual II‐1 (Family 2) at 7 years. T1‐weighted image (midsagittal view) shows severe thinning of CC and normal cerebellum (O). T2‐weighted images (axial and coronal views) show global hypomyelination and normal hippocampus (P and Q). (R) Facial photograph of individual II‐2 (Family 2) at 10 years shows a wide mouth. (S–U) Brain MRI of individual II‐2 (Family 2) at day of life 2. T1‐weighted image (midsagittal view) shows normal CC (S). T2‐weighted images (axial and coronal views) show normal myelination for age (T‐U). (V‐X) Brain MRI of individual II‐2 (Family 2) at 5 years. T1‐weighted image (midsagittal view) shows severe thinning and foreshortening of CC, normal posterior pituitary and infundibulum, and normal vermis and cerebellum (V). T2‐weighted images (axial and coronal views) show global hypomyelination, small right cerebellar cyst and normal hippocampus (W and X). (Y) Pedigree and Sanger sequencing showing segregation of the variants in GRM7 in Family 3. (Z) Facial features of individual II‐6 (Family 3) at 6 years showing thick lips, crowded teeth, low frontal hairline, remarkable nose, and bulbous nasal tip.
Figure 2.

Pedigrees, Sanger sequencing, facial features, and brain MRI images of affected individuals from Families 4–6 with biallelic variants in GRM7. (A) Pedigree and Sanger sequencing showing segregation of the variants in GRM7 in Family 4. (B) Facial photograph of individual II‐1 (Family 4) in first year of life with normal facial features. (C) Facial photograph of individual II‐4 (Family 4) at 1 month shows a long philtrum. (D and E) Brain MRI of individual II‐4 (Family 4) at 2 weeks. T1‐weighted image (midsagittal view) shows normal cerebrum, CC, and cerebellum (D). T2‐weighted image (axial view) shows under‐opercularization of sylvian fissures (more prominent on the left) (E). (F) Facial photograph of individual II‐6 (Family 4) at 4 years with hypotonic face and tented mouth. (G–I) Brain MRI of individual II‐6 (Family 4) at 4 years. T1‐weighted image (midsagittal view) shows severe cerebral atrophy, severe thinning of CC, and mild cerebellar atrophy (G). T2‐weighted images (axial and coronal views) show severe cerebral atrophy, severe thinning of CC, and bilateral hippocampal atrophy with right hippocampal hyperintensity (H and I). (J) Pedigree and Sanger sequencing showing segregation of the variants in GRM7 in Family 5. (K) Facial photograph of individual II‐1 (Family 5) at 7 months showing normal facial features. Note that individual II‐1 (Family 5) carries a second molecular diagnosis of Klinefelter syndrome as his chromosomal analysis showed 47,XXY. (L‐N) Brain MRI of individual II‐1 (Family 5) at 6 months. T1‐weighted image (midsagittal view) shows thin CC (L). T1‐wieghted and T2‐weighted images (axial views) show simplified gyral pattern (M‐N). (O) B‐allele frequency for individual II‐1 (Family 5) calculated from exome variant data demonstrates a 2.6 Mb block of AOH on chromosome 3, marked by grey zones, and show that GRM7 is located within the AOH block. (P) Pedigree and Sanger sequencing showing segregation of the variants in GRM7 in Family 6. (Q) Facial photograph of individual II‐1 (Family 6) at 20 months of age shows upslanted palpebral fissures. (R‐T) Brain MRI of individual II‐1 (Family 6) at 18 months. T1‐weighted image (midsagittal view) shows severe thinning of CC, moderate cerebral atrophy and mild cerebellar atrophy (R). T2‐weighted images (axial and coronal views) show moderate cerebral atrophy, global hypomyelination for age, and under‐opercularization of sylvian fissures (S and T). (U) B‐allele frequency for individual II‐1 (Family 6) demonstrates that GRM7 is located within a large AOH block (7.3Mb) on chromosome 3 marked by grey zones.
Gene query to the Baylor Genetics diagnostic laboratory revealed five additional individuals (BAB8506, brother of BAB8506, BAB13620, 2 brothers of BAB13620) from two unrelated families (Families 2 and 4 in Figs. 1 and 2) with a similar neurological phenotype and deleterious biallelic GRM7 variants detected on proband exome sequencing. Rudimentary clinical data were previously published on four of these individuals (BAB6708, BAB6709, BAB8506, and brother of BAB8506) from two families (Family 1 and Family 2) as part of a cohort neurological study. 19 Two additional subjects from one family (MR005, Family 3) were identified by literature search, also published with limited phenotypic data in a gene discovery cohort. 20 Gene submission to GeneMatcher re‐captured Family 3 (MR005). 22
In Family 3 (MR005), linkage analysis and homozygosity mapping were performed using HumanCytoSNP‐12 BeadChip of all available members of the family (proband, both parents, and five unaffected siblings) as described in Abou Jamra et al. (2011). 23 In total, five regions of 1Mb or more (between 2.6 and 9.8 Mb, total length of 24.6 Mb) with runs of homozygosity (ROH) resulting in autozygosity were identified. Evaluation of the variants was performed based on zygosity and locus (in linkage regions) as previously described, 24 coupled with variant prevalence in public and private databases and prediction models as described in the cases below.
All seven identified GRM7 variants were absent in the homozygous state from public variant databases including the Genome Aggregation Database (gnomAD), the Exome Aggregation Consortium (ExAC), the Atherosclerosis Risk in Communities Study Database (ARIC), and the National Heart, Lung, and Blood Institute (NHLBI) Grand Opportunity Exome Sequencing Project (ESP) as well as from our in‐house control database. The variants were also absent in heterozygous and homozygous state from the Iranome database (http://www.iranome.com) that contains exomes of 100 healthy individuals from related ethnic groups. 25 Three variants (RefSeq: NM_000844.4; c.461T>C, c.1972C>T, and c.2024C>A) were entered by our group into a public archive of human variation and phenotypes (ClinVar) following the publication of the initial cohort (ClinVar accession numbers VCV000242895, VCV000242900, and VCV000242901, respectively). 21 Bioinformatic analyses (SIFT, PolyPhen2, MutationTaster, CADD, and PhyloP) were used to predict the potential deleterious or pathogenic effect of variants on protein function and also evolutionary conservation. In four subjects (Family 1 individuals II‐1 and II‐2, Family 5 individual II‐1, and Family 6 individual II‐1), absence of heterozygosity (AOH) was determined based on the calculated B‐allele frequency from exome data using an in‐house developed bioinformatic tool, BafCalculator (https://github.com/BCM-Lupskilab/BafCalculator), as previously described. 26 An arbitrary cutoff point or value of 0.5 Mb was used in calculating the size of the AOH, and unphased data or ROH block around the variant as well as the total genomic AOH/ROH. In one subject (Family 4 individual II‐6), ROH was calculated from SNP array data.
Variants in all affected individuals, their parents, and any available unaffected siblings were verified by Sanger sequencing for segregation studies. Molecular diagnosis was confirmed in 8 of the 11 affected individuals that were alive at the time of the study, although all 11 subjects were clinically examined during the course of their illness for the features documented. Referring physicians provided detailed clinical information and assessments on all subjects for deep phenotyping. Updated clinical history and clinical pictures were obtained on two of three previously published families (Families 1 and 2). 19 Brain magnetic resonance images (MRIs) of eight individuals were reviewed by a board‐certified neuroradiologist (JVH). Electroencephalograms (EEGs) of two individuals were reviewed by a board‐certified clinical neurophysiologist (DM).
3D modeling of protein structure
3D protein structure of GRM7 was obtained from SWISS‐MODEL homology modeling server. 27 This crystal structure covers amino acids from 41 to 849. Protein structure was displayed by the PyMol Molecular Graphics System, v.1.5. Schrodinger, LLC. Amino acid conservation was obtained from the Consurf server based on sequence analysis. 28
Results
Clinical findings
We performed a comprehensive retrospective analysis of patients’ clinical data including birth and perinatal history, age of onset of disease, neurological features, seizure types and response to treatment, growth parameters, neurological exam findings as well as diagnostic studies including EEGs and brain MRIs.
Basic clinical information and perinatal history of the 11 affected individuals from six unrelated families with deleterious biallelic GRM7 variants are summarized in Table 1. Subjects were from diverse ethnic backgrounds and countries of origin including Saudi Arabia, Syria, Turkey, United Arab Emirates, and the United States. All individuals were born at term. Prominent perinatal complications included neonatal intensive care unit (NICU) admission for respiratory distress, apnea, desaturation or neonatal seizures in five individuals (Family 1 individual II‐1, Family 2 individual II‐2, and Family 4 individuals II‐1, II‐4, and II‐6), and polyhydramnios in four individuals (Family 4 individuals II‐1, II‐4, and II‐6 and Family 5 individual II‐1). No history of perinatal insult was found in any individuals. Birth weights were available in nine individuals and were normal and within one standard deviation (SD) of the mean for age. Head circumference measurement at birth was only available in one subject (Family 2 individual II‐2) and was also normal while head circumference measurements at birth were not available for the remaining subjects.
Table 1.
Basic clinical information and perinatal history of families with biallelic GRM7 variants.
| Individual | Country of origin | Sex | Current age | Gestational age at birth/mode of delivery | Perinatal complications | OFC‐birth/cm (z‐score) | Birth weight/gram (z‐score) |
|---|---|---|---|---|---|---|---|
| Family 1, II‐1 (BAB6709)* | Saudi Arabia | M | 13 years | FT (39 w)/CS | Decreased fetal movements, oligohydramnios, and fetal bradycardia requiring emergency CS; NICU admission for respiratory distress (on mechanical ventilation) | N/A | 3000 g (−0.9 SD) |
| Family 1, II‐2 (BAB6708)* | Saudi Arabia | M | 10 years | FT (39 w)/CS | None | N/A | 3000 g (−0.9 SD) |
| Family 2, II‐1 (BAB8506)* | USA | F | 15 years | FT (38 w)/SVD | None; failed hearing screen one side | N/A | 3800 g (+0.82SD) |
| Family 2, II‐2 (BAB8506's brother)* | USA | M | 10 years | FT (39 w)/SVD | NICU admissions neonatal seizures at 12 HOL | 36 cm (+0.06SD) | 3700 g (+0.28 SD) |
| Family 3, II‐2 (MR005‐1)* | Syria | F | Died at 13 years (poor nutritional status) | FT/SVD | None | N/A | N/A |
| Family 3, II‐6 (MR005‐2)* | Syria | M | N/A | FT/SVD | None | N/A | N/A |
| Family 4, II‐1 | UAE | M | Died at 13 months (respiratory failure) | FT/CS | Polyhydramnios, NICU admission for respiratory distress and desaturations | N/A | 3500 g (−0.10 SD) |
| Family 4, II‐4 | UAE | M | Died at 45 days (SIDS) | FT (41 w)/SVD | Polyhydramnios, NICU admission for respiratory distress and seizures | N/A | 3700 g (+0.28 SD) |
| Family 4, II‐6 (BAB13620) | UAE | F | Died at 4 years (aspiration PNA) | FT (41 w)/CS | Polyhydramnios and GDM, maternal influenza, and NICU admission for apnea, desaturations, hypoglycemia, and jaundice | N/A | 3200 g (−0.43 SD) |
| Family 5, II‐1 (BAB10502) | Turkey | M | 2 years 2 months | FT/SVD | Polyhydramnios and GDM | N/A | 3700 g (+0.28 SD) |
| Family 6, II‐1 (BAB10517) | Turkey | M | Died at 5 years (aspiration PNA) | FT/SVD | None | N/A | 3300 g (−0.42 SD) |
CS, C‐section; F, female; FT, full term; g, grams; GDM, gestational diabetes mellitus; HOL, hours of life; M, male; N/A, not available; NICU, neonatal intensive care unit; OFC, occipital frontal circumference; PNA, pneumonia; SIDS, sudden infant death syndrome; SD, standard deviation; SVD, spontaneous vaginal delivery; w, weeks; UAE, United Arab Emirates; USA, United States of America.
Previously published with limited clinical data.
Neurological and other clinical features are detailed in Table 2. Shared clinical features in all 11 subjects include severe to profound global developmental delays (GDD), intellectual disability (ID), and early‐onset seizures within the first year of life. Head circumference measurements at time of last visit were available in eight individuals and were consistent with microcephaly (−3.8 to −2.7 SD from mean for age). Review of the head circumference chart from birth to current age in one individual (Family 2 individual II‐2) revealed an acquired microcephaly pattern with stagnation of head growth and crossing of centiles between 4 and 14 months (Fig. 3A and B). Other common neurological features included axial hypotonia (8/8), peripheral hypertonia (7/8), hyper‐reflexia (4/6), and drug‐resistant epilepsy (6/9). Status epilepticus was reported in four affected individuals (4/8). Additional clinical features include failure to thrive (6/11), recurrent infections (6/11), short stature (5/11), dysphagia (4/11), and cortical visual impairment (3/11). Hormonal problems were present in 4/11 individuals in the form of panhypopituitarism (1/11), growth hormone deficiency (1/11), hypothyroidism (1/11), or unspecified hormonal deficiency (1/11). Review of brain MRI images confirmed the absence of a structural pituitary abnormality in 3/4 of these subjects. The brain MRI images were not available for review in the subject with the unspecific hormonal deficiency. Occasionally reported medical problems include iron or B12 deficiency anemia (3/11), lipodystrophy (2/11), and sensory or conductive hearing loss (2/11). One subject had a nasogastric tube for feeding during infancy due to poor feeding and evidence of aspiration on modified barium swallow assessment. Self‐mutilation was reported in one subject. Four subjects died in infancy or early childhood due to either aspiration pneumonia (2/4), respiratory failure (1/4), or sudden infant death syndrome (1/4), and one individual died in late childhood (at 13 years) from poor nutritional status and lack of access to advanced medical care (Table 1).
Table 2.
Neurological, developmental, and other clinical features of families with biallelic GRM7 variants.
| Individual | Age at last exam | OFC‐last exam cm (z‐score) | Microcephaly | Axial hypo‐tonia | Peripheral hyper‐tonia | Hyper‐reflexia | DD/ID | Seizures (onset) | DRE (Current AEDs) | Seizure types | SE | Other clinical features |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family 1, II‐1 (BAB6709)* | 13 yrs. | 50 cm (−2.9SD) | + | + | + | + | + | + (4 mo.) | − (LEV and RFM‐effective) | Myoclonic | − | FTT, RI, dysphagia, anemia (B12 & iron deficiency), hypothyroidism, bilateral hyperopia, and occasional self‐mutilation |
| Family 1, II‐2 (BAB6708)* | 10 yrs. | N/A | N/A | + | + | + | + | + (8 mo.) | − (none) | GTCs (seizure free off AEDs for 5 yrs with subsequent recurrence) | − | N/A |
| Family 2, II‐1 (BAB8506)* | 15 yrs. | 50.5 cm (−3.31SD) | + | + | + | − | + | + (1 mo.) | + (LEV and VNS) | Myoclonic & GTCs | + (one admission) | FTT, SS, dysphagia, cortical visual impairment, GH deficiency, hypotonic facies, vitamin D deficiency, low bone density, and stool incontinence |
| Family 2, II‐2 (BAB8506's brother)* | 10 yrs. | 48.6 cm (−3.29SD) | + | + | + | N/A | + | + (DOL1) | + (LEV, ZNS, and VNS) | Myoclonic & GTCs | − | FTT, SS, dysphagia, cortical visual impairment, hypotonic facies, and micropenis |
| Family 3, II‐2 (MR005‐1)* | N/A | N/A (<−2 SD) | + | N/A | N/A | N/A | + | + | N/A | N/A | N/A | FTT, RI, SS, and lipodystrophy |
| Family 3, II‐6 (MR005‐2)* | 5 yrs. 6 mo. | N/A (<−2 SD) | + | N/A | + | N/A | + | + (3 w) | N/A | N/A | N/A | FTT, RI, SS, “hormonal deficiency,” and lipodystrophy |
| Family 4, II‐1 | 13 mo. | N/A | N/A | + | N/A | N/A | + | + (3 mo.) | + (VPA and PB) | N/A | + (ICU admission at 8 mo) | Dysphagia (nasogastric tube feeding), hepatomegaly, and an episode of hypoglycemia |
| Family 4, II‐4 | 1 mo. | N/A | N/A | N/A | N/A | N/A | + | + (1 w) | − | N/A | N/A | N/A |
| Family 4, II‐6 (BAB13620) | 3 yrs. 3 mo. | 42.5 (−3.8 SD) | + | + | − | − | + | + (5 mo.) | + (VPA, LEV, and CLB) | GTCs & focal | − | RI, SS, cortical visual impairment, panhypopituitarism on hydrocortisone and thyroid hormone replacement, mild peripheral pulmonary stenosis, conductive hearing loss, CSA, ASD, concern for long QT syndrome, and HIE following aspiration at 3y 3m |
| Family 5, II‐1 (BAB10502) | 7 mo. | 40 cm (−3.2 SD) | + | + | + | + | + | + (2 mo.) | + (PB, CLB, LEV) | Myoclonic & GTCs | + (2 admissions) | RI, OSA, anemia, and unilateral hearing loss |
| Family 6, II‐1 (BAB10517) | 20 mo. | 44.5 cm (−2.7 SD) | + | + | + | + | + | + (2d) | + (TPM, LMG, VPA, and CLB) | Multifocal | + (multiple ICU admissions) | FTT, RI, and anemia |
| Total | 8/8 | 8/8 | 7/8 | 4/6 | 11/11 | 11/11 | 6/9 | 4/8 |
ADHD, attention deficit hyperactivity disorder; AED, antiepileptic drug; ASD, atrial septal defect; CLB, clobazam; CSA, central sleep apnea; DD, developmental delay; DOL, date of life; DRE, drug‐resistant epilepsy; FTT, failure to thrive; GH, growth hormone; GTC, generalized tonic‐clonic; HIE, hypoxic ischemic injury; ICU, intensive care unit; ID, intellectual disability; mo., months; LEV, levetiracetam; LMG, lamotrigine; N/A, not available; OSA, obstructive sleep apnea; PB, phenobarbital; RI, recurrent infections; SE, status epilepticus; SS, short stature; TPM, topiramate; VNS, vagal nerve stimulator; VPA, valproic acid, w, weeks; yrs., years; ZNS, zonisamide.
Previously published with limited clinical data.
Figure 3.

Head circumference charts and EEG epochs. (A) Head circumference‐for‐age chart (boys, birth to 2 years) of individual II‐2 from Family 2 showing normal head circumference at closer to 50th centile with crossing of centiles to less than 2nd centile starting at around 4 months of age through 14 months of age. The pattern is consistent with an acquired microcephaly. (B) Head circumference‐for‐age chart (boys, 2–18 years) of same individual (individual II‐2 from Family 2) showing that despite steady head growth, head circumference remains consistently under the 2nd centile from 2 years to 9 years of age. (C) 10‐sec epoch from a routine EEG of individual II‐1 from Family 1 (anterior–posterior bipolar montage, sensitivity of 15 μV/Sec) showing diffuse background slowing and multifocal epileptiform activity most prominent at 6–7 sec from start of epoch. (D) 10‐se epoch from a routine EEG of Individual II‐1 Family 6 (anterior–posterior bipolar montage, sensitivity of 7 μV/Sec) showing diffuse background slowing and abundant multifocal epileptiform activity throughout the epoch.
Neuroimaging studies of 10 affected individuals are summarized in Table 3, and representative images are displayed in Figures 1 and 2. Neuroimaging included brain MRIs in nine subjects and a head CT in one additional individual. Prominent neuroimaging findings included cerebral atrophy (8/10) that ranged from mild (2/8) to moderate/severe (6/8), global hypomyelination in 8/10 individuals, and moderate or severe thinning of corpus callosum (CC) in 7/8. Additional findings included mild cerebellar atrophy in 4/10 individuals, under‐opercularization of the Sylvian fissures in 3/10, and hippocampal atrophy or signal abnormality in 2/9, and brainstem volume loss in one individual (Family 4 individual II‐6).
Table 3.
Radiological, electrographic, and other significant laboratory findings in families with biallelic GRM7 variants.
| Individual | Family 1, II‐1 (BAB6709)* | Family 1, II‐2 (BAB6708)* | Family 2, II‐1 (BAB8506)* | Family 2, II‐2 (BAB8506's brother)* | Family 3, II‐2 (MR005‐1)* | Family 3, II‐6 (MR005‐2)* | Family 4, II‐1 | Family 4, II‐4 | Family 4, II‐6 (BAB13620) | Family 5, II‐1 (BAB10502) | Family 6, II‐1 (BAB10517) | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Electroencephalogram (EEG) findings | |||||||||||||
| Diffuse background slowing | + | N/A | − | + (also disorganized with lack of AP gradient) | N/A | N/A | + (also attenuated) | − | + | + | + | 6/8 | |
| Epileptiform activity | + (frequent to abundant, multifocal over LPO, L hemispheric, LFC, LF, LT> R hemispheric) | N/A | + (occasional, sleep‐activated, focal over LT and RF regions | + (frequent, multifocal over RF, LF, and BiO regions) | N/A | N/A | + (occasional, multifocal) | − | + (frequent, high voltage 1‐2 Hz GSSW) | + (focal, LFC with generalized spread) | + (frequent, multifocal over RFC, RT, LF, and LP) | 7/8 | |
| Ictal findings | + (2 clinical generalized myoclonic seizures with generalized electro decrement; 6 electrographic seizures) | N/A | − | − | N/A | N/A | − | − | + (7 clinical focal tonic seizures with generalized electro decrement; 1 electrographic seizure) | − | − | 2/8 | |
| Type of neuroimaging (age) | Brain MRI (3 years) | Brain MRI (2 years) | Serial brain MRIs (2 mo, 18 mo, 3 yrs, 7 yrs & 10 yrs) | Serial brain MRIs (DOL2, 2 yrs & 5 yrs) | N/A | Brain MRI (N/A) | CT head (2 mo) | Brain MRI (2 w) | Brain MRI (3 yrs) | Brain MRI (6 mo) | Brain MRI (18 mo) | ||
| Neuroimaging findings | |||||||||||||
| Cerebral atrophy | ++/+++ | ++/+++ | + | + | N/A | +++ | +++ | − | +++ | − (simplified gyri) | ++ | 8/10 | |
| Hypomyelination | + | − | ++ | ++ | N/A | + | + | − | + | + | + | 8/10 | |
| CC thinning | ++/+++ | +++ | +++ | +++ | N/A | N/A | N/A | − | ++/+++ | +++ | +++ | 7/8 | |
| Cerebellar atrophy | + | − | − | − | N/A | + | − | − | + | − | + | 4/10 | |
| Under‐opercularization of sylvian fissures | − | − | − | − | N/A | − | − | + | − | + | + | 3/10 | |
| Hippocampal abnormality | − | + (b/l T2 hyper‐intensity) | − | − | N/A | N/A | − | + (b/l hippocampal atrophy with R hippocampal T2 hyperintensity) | − | − | 2/9 | ||
AP, anterior–posterior; BiO, Bi‐occipital; b/l, bilateral; CC, corpus callosum; DOL, day of life; GSSW, generalized slow spike wave; indiv., individual; L, left; LF, left frontal; LFC, left frontocentral; LP, left parietal; LPO, left parieto‐occipital; LT, left temporal; mo, months; MRI, magnetic resonance imaging; N/A, not available; R, right; RF, right frontal; RT, right temporal; w, weeks; yrs, years. Note that (− and +) is used to indicate the absence or presence of the finding, respectively. Degree of presence is reflected by number of (+) signs, where one is used for mild, (++) is used for moderate and (+++) is used for severe.
Previously published with limited clinical data.
One subject (Family 4 individual II‐6) had a complicated medical course and developed hypoxic ischemic encephalopathy (HIE) at age 3 years and 3 months following an aspiration event during oral feeding that led to cardiac asystole and respiratory failure with return of circulation after 30 min of cardiopulmonary resuscitation. A brain MRI obtained 1 week following the aspiration event showed interval worsening of the previously identified cerebral volume loss and atrophy.
Serial brain MRIs were available in two siblings (Family 2 Individuals II‐1 and II‐2) from birth or 2 weeks of age until the ages of 10 and 5 years, respectively. Of note, in both siblings, the myelination and CC were normal in the first 2 months of life but showed marked CC atrophy and loss of myelination at follow‐up brain MRI during the second year of life with plateauing of myelination and an apparent static myelin state on brain MRIs obtained following the second year of life (Fig. 1L–Q and S–X). One of these siblings (Family 2 Individual II‐2) had documented acquired microcephaly with onset of 4 months as discussed above (Fig. 3A and B). These findings suggest that the natural course of disease is progressive in the first 2 years of life followed by a static course.
Brain magnetic resonance spectroscopy (MRS) was performed in only one individual (Family 4 individual II‐6) and was obtained at 6 months of age revealing a markedly reduced N‐acetyl aspartate level and relatively high choline peak. No glutamate peak was noted. Additionally, cerebrospinal fluid (CSF), glucose, lactate, and neurotransmitter metabolites were performed in two affected individuals (Family 2 individual II‐2 and Family 4 individual II‐6) and were all within normal limits. One of these subjects (Family 2 individual II‐2) also had a normal CSF amino acids profile.
EEG findings are summarized in Table 3 with available EEG traces shown in Figure 3. The majority of the subjects (6/8) had diffuse background slowing indicating cerebral dysfunction. Epileptiform activity was also present in almost all subjects (7/8), with focal or multifocal epileptiform activity (6/8) seen more frequently than a generalized slow spike and slow wave pattern (1/8). Seizures captured on EEGs in two subjects included generalized myoclonic seizures with generalized electro‐decrement and superimposed paroxysmal fast activity in one subject, and focal tonic with generalized electro‐decrement in the second subject. The presence of severe developmental impairment, early‐onset seizures and frequent epileptiform activity with background slowing on EEG in a majority of subjects is consistent with DEE.
Molecular characterization of rare GRM7 biallelic variants
Table 4 contains the summary of the variants observed: seven variants from six unrelated families that included five homozygous variants and one family with compound heterozygous variants. The pedigree diagrams, Sanger sequencing data and segregation studies of the variants, and facial morphological features of the affected individuals are shown in Figures 1 and 2. mGluR7 protein model structure, localization of variants and implicated amino acids, and variant conservation are displayed in Figure 4. The majority of the identified variants cluster within the transmembrane domain, except for two variants that are located at the ligand‐binding or intracellular domains (Fig. 4A and C). All variants were located within highly conserved regions in vertebrates and higher species (Fig. 4B and C). In general, subjects with the stop‐gain or missense variants within the transmembrane domain (Families 2–4 and 6) showed a more severe phenotype compared to those with missense variants located within the ligand‐binding (Family 1) or intracellular (Family 5) domains. Compound heterozygous variants were present in one family (Family 2) with no reported history of consanguinity, while homozygous variants were present in the other five families (Families 1 and 3–6) with self‐reported history of consanguinity (1o cousin marriages). All GRM7 homozygous variants were located within large blocks of AOH ranging from 2.6 to 9.7Mb. Total AOH in these affected individuals ranged from 24.6 to 575Mb (Table 4). One subject (Family 5 individual II‐1) carried a second molecular diagnosis consistent with Klinefelter syndrome based on a clinical G‐banded chromosomal analysis showing 47,XXY. The diagnosis of Klinefelter syndrome in this subject may contribute to the NDD phenotype but does not fully explain the severe neurological features in isolation.
Table 4.
Summary of GRM7 variants.
| Individual | Position (GRCh37\hg19) | Nucleotide (Protein) | Zygosity | Allele count/Zygosity (gnomAD) | CADD Score (PHRED) | Conservation (phyloP) | Conservation (ConSurf algorithm) | AOH region around gene (Mb) | Total AOH (Mb) |
|---|---|---|---|---|---|---|---|---|---|
| Family 1, II‐1 (BAB6709)* | Chr3:6903536; T>C | c.461T>C (p.I154T) | Hmz | 0 Htz–0 Hmz | 28.9 | 1.978 | 7 | 8.7 | 575 |
| Family 1, II‐2 (BAB6708)* | Chr3:6903536; T>C | c.461T>C (p.I154T) | Hmz | 0 Htz–0 Hmz | 28.9 | 1.978 | 7 | 9.7 | 411 |
| Family 2, II‐1 (BAB8506)* | Chr3:7620565; C>T | c.1972C>T (p.R658W) | Comp Htz | 0 Htz–0 Hmz | 26.2 | 0.962 | 7 | – | – |
| Chr3:7620617; C>A | c.2024C>A (p.T675K) | Comp Htz | 0 Htz–0 Hmz | 26.1 | 6.104 | 9 | – | – | |
| Family 2, II‐2 (BAB8506's brother)* | Chr3:7620565; C>T | c.1972C>T (p.R658W) | Comp Htz | 0 Htz–0 Hmz | 26.2 | 0.962 | 7 | – | – |
| Chr3:7620617; C>A | c.2024C>A (p.T675K) | Comp Htz | 0 Htz–0 Hmz | 26.1 | 6.104 | 9 | – | – | |
| Family 3, II‐2 (MR005‐1)* | – | – | – | – | – | – | |||
| Family 3, II‐6 (MR005‐2)* | Chr3:7620350; G>A | c.1757G>A (p.W586*) | Hmz | 0 Htz–0 Hmz | 51 | N/A | 3 | 3.7 | 24.6 |
| Family 4, II‐1 | – | – | – | – | – | – | – | – | – |
| Family 4, II‐4 | – | – | – | – | – | – | – | – | – |
| Family 4, II‐6 (BAB13620) | Chr3:7620566; G>A | c.1973G>A (p.R658Q) | Hmz | 5 Htz–0 Hmz | 24.9 | 0.962 | 7 | 6 | 113 |
| Family 5, II‐1 (BAB10502) | Chr3:7721955; G>A | c.2671G>A (p.E891K) | Hmz | 5 Htz–0 Hmz | 33 | 0.902 | 8 | 2.6 | 545 |
| Family 6, II‐1 (BAB10517) | Chr3:7620568; C>T | c.1975C>T (p.R659*) | Hmz | 1 Htz–0 Hmz | 37 | ND | 8 | 7.3 | 361.2 |
AOH, absence of heterozygosity; CADD, Combined Annotation Dependent Depletion; Comp.Htz, compound heterozygous; Hmz, homozygous; Htz, heterozygous; N/A, not available; ND, not determined.
Previously published with limited clinical data.
Figure 4.
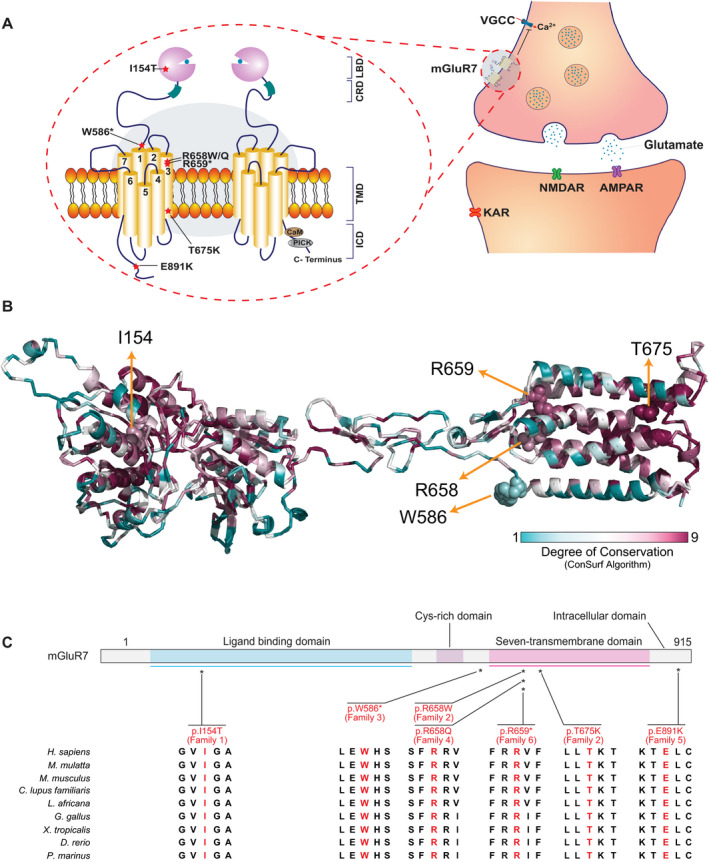
Variant locations on mGluR7 schematic and protein model and their conservation. (A) Schematic diagram of the neuronal synapse showing the presynaptic location of mGluR7 and the postsynaptic location of the iGluR7 including NMDAR, AMPAR, and KAR. A magnified illustration of mGluR7 shows that mGluR7s function as constitutional dimers and consist of four regions: N‐terminal ligand‐binding domain (LBD), cysteine‐rich domain (CRD), transmembrane domain (TMD), and intracellular domain (ICD). The approximate location of the altered amino acid residues described in this paper is displayed on the mGluR7 on the left. (B) Model protein structure of GRM7. The model structure covers amino acids from 41 to 849. The locations of the altered amino acids are represented by spheres on the protein cartoon model. I154 is buried on a loop between highly conserved alpha helices and beta sheets. Amino acid residues R658, R659, and T675 are highly conserved and located on the same helix of on transmembrane domain, while W586 is less conserved and located on the loop of the transmembrane domain. Note that E891 is not shown here, since the model structure does not include the intracellular region much. (C) Structure of the mGluR7 protein and the position of the altered amino acid residues (I154T, W586*, R658W/Q, R659*, T675K, and E891K) and the evolutionary conservation of the altered amino acids is also displayed.
Discussion
We comprehensively characterized the clinical features and variants in six families with biallelic variants in GRM7 and showed that rare GRM7 biallelic variants can cause a severe neurological phenotype characterized by microcephaly, DEE, hypomyelination, and cerebral atrophy. Overall, the findings of both congenital neuroimaging features such as under‐opercularization and thin corpus callosum and progressive features such as cerebral atrophy suggest that GRM7‐related disorders are both congenital and progressive in nature similar to that seen in GRIA2‐related NDDs. 7 Additionally, other disorders such as CDKL5‐related and FOXG1‐related disorders cause a wide and heterogeneous range of NDDs. 29 , 30 It is possible that the phenotype‐first approach of our cohort has introduced an ascertainment bias toward enrolling subjects with more severe, and potentially homogenous, phenotype. Identification of more cases using a gene‐first approach will likely reveal the entire and broad spectrum of GRM7‐related disorders. 31 , 32
Genotype–phenotype correlation of our limited data suggests that stop‐gain and missense variants affecting the transmembrane domain result in a more severe phenotype compared to missense variants located in ligand‐binding or intracellular domains. We hypothesize that the reported biallelic variants are loss‐of‐function (LoF) alleles, with the missense variants causing partial LoF (hypomorphic) while the stop‐gain variants result in complete LoF (amorphic). Both stop‐gain variants identified (Family 3 and Family 6) were in exon 8 of total 10 exons in GRM7. The resultant transcripts with these premature termination codons will either be degraded by nonsense‐mediated decay or result in a truncated protein, both of which are expected to result in complete LoF. 33 This LoF of mGluR7 is expected to result in loss of inhibition in the glutamatergic pathway, excess glutamate release, and subsequently hyperexcitability.
Glutamate is an excitatory neurotransmitter that is robustly implicated in the pathophysiology of epilepsy, a disorder characterized by aberrant neuronal hyperexcitability. Excessive and abnormal neuronal firing in seizures leads to depolarization shift, increase in extracellular glutamate, and subsequent neuronal excitotoxicity. 3 This results in a vicious cycle with aberrant glutamate release contributing to further epileptogenicity, and seizure maintenance and progression. 3
Hyperexcitability of the glutamatergic pathway could lead to the severe epilepsy phenotype as noted in all affected individuals. All our subjects had the onset of their seizures in infancy, with seizures starting in the neonatal period in nearly half of them (Table 2). Additionally, the significant history of early postnatal apneas and desaturations requiring NICU admission may suggest the possibility of missed seizures with earlier onset than reported in some individuals. Two of our subjects showed early signs of early hippocampal sclerosis (HS) with hippocampal atrophy and signal abnormality. While it remains unclear if HS is a cause or a sequela of frequent seizures, the aberrant expression of mGluR7 in the hippocampus may be a contributing factor. 34 , 35
Excessive glutamate is also neurotoxic, potentially explaining the marked and early progressive cerebral atrophy and axonal loss observed in our subjects, particularly the affected siblings in Family 2 with the observations of longitudinal MRI data and the acquired microcephaly pattern in one of them. Interestingly, postnatal deceleration of head growth has been observed in 5/28 subjects with deleterious monoallelic variants in GRIA2, a gene encoding a subunit of another glutamate receptor, AMPAR. 7 However, our ability to further assess or quantify this finding is limited due to the lack of longitudinal data in other subjects. One caveat is that one of our subjects (Family 4 individual II‐6) had an MRS that did not show a glutamate peak, but this observation cannot be generalized either, as it was a one‐time observation in a single subject without longitudinal data.
At the neuronal synapses, glutamate signals through two groups of receptors, iGluRs and mGluRs. AMPAR is the most abundant iGluR in the mammalian brain and is responsible for fast excitatory neurotransmission while NMDAR is important for slow synaptic potential and information processing. 3 KAR is thought to play a role in presynaptic and postsynaptic modulation of neurotransmission. 36 De novo and inherited pathogenic variations in genes encoding postsynaptic iGluRs subunits cause a wide range of NDDs and DEEs. 4 , 5 , 6 , 7 , 8 , 9 , 10 Interestingly, biochemical and in vitro functional assays show that variants in the same gene encoding some NMDAR and AMPAR subunits (e.g. GRIN2A, GRIN2B, GRIN2D, and GRIA2) may act as both gain‐of‐function (GoF) and LoF and may result in indistinguishable neurological phenotypes. 5 , 6 , 7 This alludes to the complexity of the glutaminergic pathway with both enhanced and reduced function leading to NDDs and DEEs. Patho‐mechanisms include alterations in any of the multiple functional parameters including agonist potency, sensitivity to negative allosteric modulators, channel opening probability, surface expression, and current amplitude response. 6 , 7
mGluR7, encoded by GRM7, is the most abundant CNS mGluR and is only activated by high glutamate and GABA concentrations due to its low affinity for these neurotransmitters. This allows it to function as an auto or hetero‐receptor to downregulate further calcium‐dependent glutamate release and thus prevent the neurotoxic effect of extracellular glutamate accumulation. 37 mGluR7 is evolutionarily conserved and is widely distributed across the CNS including the hippocampus, hypothalamus, and thalamo‐cortical circuitry synapses. 35
Activation of mGluR7 produces a cascade of events starting with the liberation of Gβγ subunits to inhibition of adenylyl cyclase and reduced cAMP production and ending with the downregulation of voltage‐gated calcium channels (VGCC). 38 mGluR7 also interacts with several intracellular and scaffolding proteins including PKC (protein kinase C) and PDZ domain‐containing protein PICK1 (protein interacting with C kinase 1), providing further complexity to the mechanisms by which it regulates synaptic transmission. 39 The mGluR7‐PICK interaction is critical for receptor function, thus raising the possibility that the intracellular variant (p.Glu891Lys) in one of our subjects causes disease by disrupting this PICK‐mGlu7 interaction.
Rodent models of mGluR7 and its scaffolding (PICK1) or interacting proteins (ELFN1) show that mGluR7 reduced expression or impairment of its function or recruitment to synaptic sites lead to symptoms overlapping with NDDs and DEEs. mGluR7‐knockout mice exhibit spontaneous sensory‐provoked seizures; increased seizure vulnerability to two proconvulsant agents; reduced fear learning; signs of impaired learning, short‐term and spatial memory, and synaptic plasticity; and dysregulation of the hypothalamic–pituitary–adrenal (HPA) axis. 18 , 40 , 41 Of note, HPA axis dysregulation was present in at least three of our subjects. Additionally, mice lacking transmembrane protein ELFN1 (extracellular‐leucine‐rich repeat fibronectin domain 1) show early postnatal deficits in recruiting mGluR7 to synaptic site and display a remarkably similar neurological phenotype to mGluR7‐deficient mice with late‐onset sensory‐triggered epileptic seizures and motor and behavioral abnormalities. 42 Furthermore, impaired mGluR7 interaction with PICK1 by knockin PDZ‐recognition binding motif of mGluR7 or by pharmacological uncoupling in rodent models results in low seizure threshold to proconvulsive agents and produces behavioral changes and EEG discharges consistent with absence‐like seizures. 43 , 44 These animal models demonstrate that abnormal mGluR7 expression or function consistently produce an epilepsy phenotype.
Apart from one subject with a reported favorable response to rufinamide, we could not assess the response to each antiepileptic drug (AED) in retrospect, and our subjects had poor responses to AEDs overall (Table 2). However, several US Food and Drug administration (FDA)‐approved AEDs that reduce the glutaminergic pathway hyperexcitability already exist and show potential for use in such conditions. These AEDs exert their effect either directly or indirectly and may possess additional neuroprotective effect against glutamate excitotoxicity. Topiramate selectively inhibits the excitatory neurotransmission mediated by KAR and partially depresses AMPAR‐mediated excitatory current. 45 Topiramate also shows a neuroprotective effect against glutamate excitotoxicity in rodent hippocampal neurons. 46 Felbamate blocks NMDAR particularly at excessive exposure, thus inhibiting seizure discharges while preserving normal neuronal firing. 47 Perampanel is a first‐in‐class noncompetitive selective AMPAR antagonist that blocks AMPAR by acting on an allosteric site rather than the glutamate recognition site. 48 Voltage‐gated sodium channel blockers such as lamotrigine, phenytoin, carbamazepine, oxcarbazepine, lacosamide, rufinamide, and eslicarbazepine can also reduce glutamate release indirectly by blocking sodium‐evoked calcium influx. 49 Gabapentin and pregabalin reduce the calcium‐dependent glutamate release by selectively binding at the α2δ‐1 subunit of VGCC. 50 In addition, cannabinoids, a newly emerging therapy in treatment of refractory epilepsy, can indirectly modulate glutaminergic neurotransmission through neuronal cannabinoid receptor 1 (CB1). 51 Whether any of the above‐mentioned AEDs that target the glutaminergic pathway prove to be particularly effective in treating epilepsy due to deleterious variants in GRM7 is an area for further investigation.
Additionally, there are ample neuropharmacological studies focusing on the potential for using allosteric modulators of mGluRs in the treatment of neurological diseases and epilepsy. 52 , 53 Selective positive allosteric modulators (PAMs) for group III mGluRs have been recently developed, potentially representing novel targeted and personalized therapy in partial mGluR7 LoF (hypomorph alleles). PAMs are noncompetitive agonists that bind to a site other than the ligand‐binding site to potentiate their effect. 12 N,N′‐dibenzhydrylethane‐1,2‐diamine dihydrochloride (AMN082) is the first selective mGluR7 PAM to be discovered. 54 AMN082 binds to an allosteric site at the transmembrane domain to fully activate mGluR7. 54 Thus, the potential use of AMN082 would be predicted to be dependent on the variant location. However, AMN082 shows scarce selectivity in vivo, off‐target effect, and mixed proconvulsive and anticonvulsive profile in pentylenetetrazol‐treated rats, limiting its potential for clinical use and underscoring a need for preclinical in vivo knockout studies. 55 , 56
We show here that rare biallelic variants in GRM7 cause a severe neurological phenotype characterized by microcephaly, DEE, hypomyelination, and cerebral atrophy. Functional studies at the variant level would provide better insight into the patho‐mechanism of the disorder and evaluate the potential for targeted therapy.
Conflict of Interest
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals and Novartis, and is a co‐inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics (BG) Laboratories; JRL is a member of the Scientific Advisory Board of BG Laboratories. Other authors have no potential conflicts to report. J.V.H receives royalties from chapter in UpToDate on pediatric neuroimaging.
Authors’ Contribution
D.M., T.M., D.P., J.F, J.E.P., and J.R.L. contributed to the conception and design of the study. All authors contributed to the acquisition and analysis of data. D.M., T.M, D.P., J.F., H.D., S.E., J.V.H, J.E.P., and J.R.L drafted a significant portion of the manuscript or contributed to the design of the figures.
Acknowledgments
We would like to thank all families for their participation in this study. We would also like to thank Julia Hentschel for her continuous support. This study was supported in part by the US National Human Genome Research Institute (NHGRI) and National Heart Lung and Blood Institute (NHBLI) to the Baylor‐Hopkins Center for Mendelian Genomics (BHCMG, UM1 HG006542, J.R.L); NHGRI grant to Baylor College of Medicine Human Genome Sequencing Center (U54HG003273 to R.A.G.), US National Institute of Neurological Disorders and Stroke (NINDS) (R35NS105078 to J.R.L.) and Muscular Dystrophy Association (MDA) (512848 to J.R.L.). D.M. is supported by a Medical Genetics Research Fellowship Program through the United States National Institute of Health (T32 GM007526‐42). T.M. is supported by the Uehara Memorial Foundation. R.A.J is supported by Deutsche Forschungsgemeinschaft (AB393/1‐1 and AB393/1‐2). D.P. is supported by a Clinical Research Training Scholarship in Neuromuscular Disease partnered by the American Academy of Neurology (AAN), American Brain Foundation (ABF) and Muscle Study Group (MSG), NIH – Brain Disorders and Development Training Grant (T32 NS043124‐17), and International Rett Syndrome Foundation (IRSF grant #3701‐1). J.E.P. was supported by NHGRI K08 HG008986.
Funding Information
This study was supported in part by the US National Human Genome Research Institute (NHGRI) and National Heart Lung and Blood Institute (NHBLI) to the Baylor‐Hopkins Center for Mendelian Genomics (BHCMG, UM1 HG006542, J.R.L); NHGRI grant to Baylor College of Medicine Human Genome Sequencing Center (U54HG003273 to R.A.G.), US National Institute of Neurological Disorders and Stroke (NINDS) (R35NS105078 to J.R.L.) and Muscular Dystrophy Association (MDA) (512848 to J.R.L.). D.M. is supported by a Medical Genetics Research Fellowship Program through the United States National Institute of Health (T32 GM007526‐42). T.M. is supported by the Uehara Memorial Foundation. R.A.J is supported by Deutsche Forschungsgemeinschaft (AB393/1‐1 and AB393/1‐2). D.P. is supported by a Clinical Research Training Scholarship in Neuromuscular Disease partnered by the American Academy of Neurology (AAN), American Brain Foundation (ABF) and Muscle Study Group (MSG), NIH – Brain Disorders and Development Training Grant (T32 NS043124‐17), and International Rett Syndrome Foundation (IRSF grant #3701‐1). J.E.P. was supported by NHGRI K08 HG008986.
Funding Statement
This work was funded by Muscle Study Group grant ; Deutsche Forschungsgemeinschaft grants AB393/1‐1 and AB393/1‐2; National Heart Lung and Blood Institute grant UM1 HG006542; Muscular Dystrophy Association grant 512848; National Institute of Neurological Disorders and Stroke grants R35NS105078 and K08 HG008986; Uehara Memorial Foundation grant ; National Human Genome Research Institute grants K08 HG008986, U54HG003273, and UM1 HG006542; American Academy of Neurology grant ; National Institutes of Health grants T32 GM007526‐42 and T32 NS043124‐17; American Brain Foundation grant ; Baylor College of Medicine Human Genome Sequencing Center grant U54HG003273; International Rett Syndrome Foundation grant #3701‐1.
Contributor Information
Dana Marafi, Email: dana.marafi@bcm.edu.
Jennifer E. Posey, Email: Jennifer.Posey@bcm.edu.
James R. Lupski, Email: jlupski@bcm.edu.
References
- 1. McTague A, Howell KB, Cross JH, et al. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol 2016;15:304–316. [DOI] [PubMed] [Google Scholar]
- 2. Klassen T, Davis C, Goldman A, et al. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell 2011;145:1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barker‐Haliski M, White HS. Glutamatergic mechanisms associated with seizures and epilepsy. Cold Spring Harb Perspect Med 2015;5:a022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen W, Shieh C, Swanger SA, et al. GRIN1 mutation associated with intellectual disability alters NMDA receptor trafficking and function. J Hum Genet 2017;62:589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Swanger SA, Chen W, Wells G, et al. Mechanistic insight into NMDA receptor dysregulation by rare variants in the GluN2A and GluN2B agonist binding domains. Am J Hum Genet 2016;99:1261–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. XiangWei W, Kannan V, Xu Y, et al. Heterogeneous clinical and functional features of GRIN2D‐related developmental and epileptic encephalopathy. Brain 2019;142(10):3009–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salpietro V, Dixon CL, Guo H, et al. AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat Commun 2019;10:3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu Y, Arai AC, Rumbaugh G, et al. Mutations in ionotropic AMPA receptor 3 alter channel properties and are associated with moderate cognitive impairment in humans. Proc Natl Acad Sci USA 2007;104:18163–18168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martin S, Chamberlin A, Shinde DN, et al. De novo variants in GRIA4 lead to intellectual disability with or without seizures and gait abnormalities. Am J Hum Genet 2017;101:1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Motazacker MM, Rost BR, Hucho T, et al. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am J Hum Genet 2007;81:792–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sajan SA, Jhangiani SN, Muzny DM, et al. Enrichment of mutations in chromatin regulators in people with Rett syndrome lacking mutations in MECP2 . Genet Med 2017;19:13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niswender C, Conn P. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol 2010;50:295–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elia J, Glessner JT, Wang K, et al. Genome‐wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet 2011;44:78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang Y, Pan C. Role of metabotropic glutamate receptor 7 in autism spectrum disorders: a pilot study. Life Sci 2013;92:149–153. [DOI] [PubMed] [Google Scholar]
- 15. Guergueltcheva V, Azmanov DN, Angelicheva D, et al. Autosomal‐recessive congenital cerebellar ataxia is caused by mutations in metabotropic glutamate receptor 1. Am J Hum Genet 2012;91:553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ure J, Baudry M, Perassolo M. Metabotropic glutamate receptors and epilepsy. J Neurol Sci 2006;247:1–9. [DOI] [PubMed] [Google Scholar]
- 17. Palazzo E, Marabese I, de Novellis V, et al. Metabotropic glutamate receptor 7: from synaptic function to therapeutic implications. Curr Neuropharmacol 2016;14:504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sansig G, Bushell TJ, Clarke VR, et al. Increased seizure susceptibility in mice lacking metabotropic glutamate receptor 7. J Neurosci 2001;21:8734–8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Charng WL, Karaca E, Coban Akdemir Z, et al. Exome sequencing in mostly consanguineous Arab families with neurologic disease provides a high potential molecular diagnosis rate. BMC Med Genomics 2016;9:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reuter MS, Tawamie H, Buchert R, et al. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiatry 2017;74:293–299. [DOI] [PubMed] [Google Scholar]
- 21. Karaca E, Harel T, Pehlivan D, et al. Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron 2015;88:499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 2015;36:928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abou Jamra R, Wohlfart S, Zweier M, et al. Homozygosity mapping in 64 Syrian consanguineous families with non‐specific intellectual disability reveals 11 novel loci and high heterogeneity. Eur J Hum Genet 2011;19:1161–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tawamie H, Martianov I, Wohlfahrt N, et al. Hypomorphic pathogenic variants in TAF13 are associated with autosomal‐recessive intellectual disability and microcephaly. Am J Hum Genet 2017;100:555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fattahi Z, Beheshtian M, Mohseni M, et al. Iranome: a catalog of genomic variations in the Iranian population. Hum Mutat 2019;40:1968–1984. [DOI] [PubMed] [Google Scholar]
- 26. Karaca E, Posey JE, Coban Akdemir Z, et al. Phenotypic expansion illuminates multilocus pathogenic variation. Genet Med 2018;20:1528–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Waterhouse A, Bertoni M, Bienert S, et al. SWISS‐MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 2018;46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ashkenazy H, Abadi S, Martz E, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 2016;44:W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vegas N, Cavallin M, Maillard C, et al. Delineating FOXG1 syndrome: From congenital microcephaly to hyperkinetic encephalopathy. Neurol Genet 2018;4:e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kilstrup‐Nielsen C, Rusconi L, La Montanara P, et al. What we know and would like to know about CDKL5 and its involvement in epileptic encephalopathy. Neural Plast 2012;2012:728267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. White J, Beck CR, Harel T, et al. POGZ truncating alleles cause syndromic intellectual disability. Genome Med 2016;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bostwick BL, McLean S, Posey JE, et al. Phenotypic and molecular characterisation of CDK13‐related congenital heart defects, dysmorphic facial features and intellectual developmental disorders. Genome Med 2017;9:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coban‐Akdemir Z, White JJ, Song X, et al. Identifying genes whose mutant transcripts cause dominant disease traits by potential gain‐of‐function alleles. Am J Hum Genet 2018;103:171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walker MC. Hippocampal sclerosis: causes and prevention. Semin Neurol 2015;35:193–200. [DOI] [PubMed] [Google Scholar]
- 35. Ohishi H, Akazawa C, Shigemoto R, et al. Distributions of the mRNAs for L‐2‐amino‐4‐phosphonobutyrate‐sensitive metabotropic glutamate receptors, mGluR4 and mGluR7, in the rat brain. J Comp Neurol 1995;360:555–570. [DOI] [PubMed] [Google Scholar]
- 36. Lerma J, Marques JM. Kainate receptors in health and disease. Neuron 2013;80:292–311. [DOI] [PubMed] [Google Scholar]
- 37. Lafon‐Cazal M, Fagni L, Guiraud MJ, et al. mGluR7‐like metabotropic glutamate receptors inhibit NMDA‐mediated excitotoxicity in cultured mouse cerebellar granule neurons. Eur J Neurosci 1999;11:663–672. [DOI] [PubMed] [Google Scholar]
- 38. Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol 1997;37:205–237. [DOI] [PubMed] [Google Scholar]
- 39. Suh YH, Pelkey KA, Lavezzari G, et al. Corequirement of PICK1 binding and PKC phosphorylation for stable surface expression of the metabotropic glutamate receptor mGluR7. Neuron 2008;58:736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Holscher C, Schmid S, Pilz PK, et al. Lack of the metabotropic glutamate receptor subtype 7 selectively impairs short‐term working memory but not long‐term memory. Behav Brain Res 2004;154:473–481. [DOI] [PubMed] [Google Scholar]
- 41. Masugi M, Yokoi M, Shigemoto R, et al. Metabotropic glutamate receptor subtype 7 ablation causes deficit in fear response and conditioned taste aversion. J Neurosci 1999;19:955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tomioka NH, Yasuda H, Miyamoto H, et al. Elfn1 recruits presynaptic mGluR7 in trans and its loss results in seizures. Nat Commun 2014;5:4501. [DOI] [PubMed] [Google Scholar]
- 43. Bertaso F, Zhang C, Scheschonka A, et al. PICK1 uncoupling from mGluR7a causes absence‐like seizures. Nat Neurosci 2008;11:940–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang CS, Bertaso F, Eulenburg V, et al. Knock‐in mice lacking the PDZ‐ligand motif of mGluR7a show impaired PKC‐dependent autoinhibition of glutamate release, spatial working memory deficits, and increased susceptibility to pentylenetetrazol. J Neurosci 2008;28:8604–8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gryder DS, Rogawski MA. Selective antagonism of GluR5 kainate‐receptor‐mediated synaptic currents by topiramate in rat basolateral amygdala neurons. J Neurosci 2003;23:7069–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mao XY, Cao YG, Ji Z, et al. Topiramate protects against glutamate excitotoxicity via activating BDNF/TrkB‐dependent ERK pathway in rodent hippocampal neurons. Prog Neuropsychopharmacol Biol Psychiatry 2015;60:11–17. [DOI] [PubMed] [Google Scholar]
- 47. Kuo CC, Lin BJ, Chang HR, Hsieh CP. Use‐dependent inhibition of the N‐methyl‐D‐aspartate currents by felbamate: a gating modifier with selective binding to the desensitized channels. Mol Pharmacol 2004;65:370–380. [DOI] [PubMed] [Google Scholar]
- 48. Rogawski MA, Hanada T. Preclinical pharmacology of perampanel, a selective non‐competitive AMPA receptor antagonist. Acta Neurol Scand Suppl 2013;127:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vohora D, Saraogi P, Yazdani MA, et al. Recent advances in adjunctive therapy for epilepsy: focus on sodium channel blockers as third‐generation antiepileptic drugs. Drugs Today 2010;46:265–277. [DOI] [PubMed] [Google Scholar]
- 50. Dooley DJ, Taylor CP, Donevan S, Feltner D. Ca2+ channel alpha2delta ligands: novel modulators of neurotransmission. Trends Pharmacol Sci 2007;28:75–82. [DOI] [PubMed] [Google Scholar]
- 51. Monory K, Blaudzun H, Massa F, et al. Genetic dissection of behavioural and autonomic effects of Delta(9)‐tetrahydrocannabinol in mice. PLoS Biol 2007;5:e269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tang FR. Agonists and antagonists of metabotropic glutamate receptors: anticonvulsants and antiepileptogenic agents? Curr Neuropharmacol 2005;3:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fisher NM, Seto M, Lindsley CW, Niswender CM. Metabotropic glutamate receptor 7: a new therapeutic target in neurodevelopmental disorders. Front Mol Neurosci 2018;11:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mitsukawa K, Yamamoto R, Ofner S, et al. A selective metabotropic glutamate receptor 7 agonist: activation of receptor signaling via an allosteric site modulates stress parameters in vivo. Proc Natl Acad Sci USA 2005;102:18712–18717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ahnaou A, Raeyemaekers L, Huysmans H, Drinkenburg W. Off‐target potential of AMN082 on sleep EEG and related physiological variables: evidence from mGluR7 (‐/‐) mice. Behav Brain Res 2016;311:287–297. [DOI] [PubMed] [Google Scholar]
- 56. Mares P. AMN 082, an agonist of mGluR7, exhibits mixed anti‐ and proconvulsant effects in developing rats. Physiol Res 2008;57:969–972. [DOI] [PubMed] [Google Scholar]


