Abstract
Cohesin is a DNA-associated protein complex that forms a tripartite ring controlling sister chromatid cohesion, chromosome segregation and organization, DNA replication, and gene expression. Sister chromatid cohesion is established by the protein acetyltransferase Eco1, which acetylates two conserved lysine residues on the cohesin subunit Smc3 and thereby ensures correct chromatid separation in yeast (Saccharomyces cerevisiae) and other eukaryotes. However, the consequence of Eco1-catalyzed cohesin acetylation is unknown, and the exact nature of the cohesive state of chromatids remains controversial. Here, we show that self-interactions of the cohesin subunits Scc1/Rad21 and Scc3 occur in a DNA replication–coupled manner in both yeast and human cells. Using cross-linking MS-based and in vivo disulfide cross-linking analyses of purified cohesin, we show that a subpopulation of cohesin may exist as dimers. Importantly, upon temperature-sensitive and auxin-induced degron-mediated Eco1 depletion, the cohesin-cohesin interactions became significantly compromised, whereas deleting either the deacetylase Hos1 or the Eco1 antagonist Wpl1/Rad61 increased cohesin dimer levels by ∼20%. These results indicate that cohesin dimerizes in the S phase and monomerizes in mitosis, processes that are controlled by Eco1, Wpl1, and Hos1 in the sister chromatid cohesion-dissolution cycle. These findings suggest that cohesin dimerization is controlled by the cohesion cycle and support the notion that a double-ring cohesin model operates in sister chromatid cohesion.
Keywords: cell cycle, acetylation, DNA replication, dimerization, acetyltransferase, chromatid cohesion, cohesin, double ring model, Eco1, self-interaction, protein acetylation
Introduction
Cohesin is a tripartite ring that controls many, if not all, aspects of chromosome function, including sister chromatid cohesion, chromosome segregation, chromosome condensation, chromosome organization, DNA replication, DNA repair, DNA recombination, and gene expression (1–5). The ring consists of V-shaped heterodimeric SMC proteins Smc1 and Smc3 and an α-kleisin subunit, Scc1/Rad21, bridging their ABC ATPase head domains (6, 7). The fourth subunit, Scc3 (SA1 or SA2 in mammalian cells), binds to the ring through Scc1 (8). The stoichiometry of these subunits is 1:1:1:1 (9–11). Besides such a single-ring model (12–18), higher-order oligomeric cohesin conformations have also been proposed based upon the unusual genetic properties and physical self-interactions of cohesin subunits in yeast (19) and human cells (20), respectively. Therefore, it remains highly debatable whether cohesin functions as single rings, double rings, or clusters (6, 21–23).
As an essential process mediated by cohesin, sister chromatid cohesion is achieved in two steps, cohesin loading and cohesion establishment (24, 25). Cohesin is loaded onto chromosomes with a low affinity in the G1 phase (26). Following DNA replication, cohesion is established, wherein cohesin tightly holds twin chromatids together to resist the pulling force of the spindle before segregation (27, 28). Eco1 plays an essential role in the establishment of cohesion (29–32). It acetylates two conserved lysine residues on Smc3 (Lys-112 and Lys-113 in Saccharomyces cerevisiae) (33–35). Smc3 acetylation appears to counteract an “anti-establishment” activity of Wpl1/Rad61 (36). Wpl1 ablation restores viability and improves sister chromatid cohesion in the absence of Eco1 or Smc3 acetylation (33, 37, 38). Nevertheless, the exact consequences of Smc3 acetylation remain unknown as well.
Results
Self-interactions of cohesin subunits in yeast cells
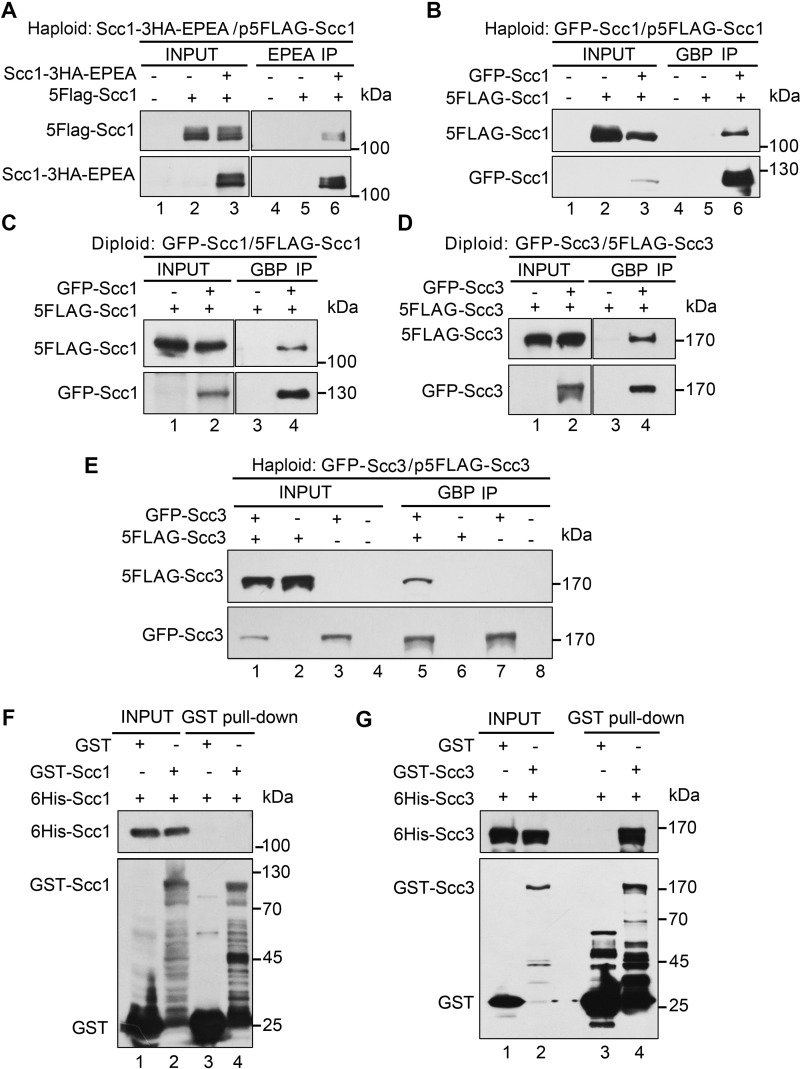
Self-interactions of cohesin subunits have not been detected in yeast (9). In human cells, contradictory observations have been reported (20, 39). To clarify this, we first performed immunoprecipitation (IP)2 experiments using a similar dual-tag strategy. An ectopic copy of SCC1 (pRS-SCC1, Table 1) under the control of its native promoter was introduced into a haploid yeast strain. The two SCC1 alleles were labeled with a pair of orthogonal epitopes (GFP/FLAG or EPEA/FLAG), which have been well-demonstrated to be orthogonal to each other in this and previous studies (Fig. 1) (40, 41). When the epitopes were inserted at the C termini of both copies (Scc1-HA-EPEA/Scc1–5FLAG), after EPEA-IP, no Scc1-Scc1 interaction was virtually detectable (data not shown), in agreement with a previous study from Nasmyth's group (9) in yeast. Epitope tagging may occasionally cause unexpected interference in the structure and function of proteins, which turns out to be true for many cohesin subunits (20, 42). To test this possibility, we switched 5FLAG from the C terminus to the N terminus of Scc1. Although EPEA-IP was performed in the exact same procedure, such a change led to the positive interaction between Scc1-HA-EPEA and 5FLAG-Scc1 (Fig. 1A, lane 6). Because EPEA can only be applied at the C terminus, we then labeled both Scc1 copies at their N termini with another orthogonal epitope pair, GFP/FLAG. A similar intermolecular interaction was observed in GBP-IP (Fig. 1B, lane 6). These results are consistent with the observations in human cells (20), indicating that the self-interaction of Scc1 might be conserved.
Table 1.
Plasmids used in this study
| Plasmid | Base plasmid/Genotype | Source |
|---|---|---|
| pRS315-5FLAG-SCC1 | ampr/LEU2 5FLAG-SCC1 | This study |
| pPADH1-5FLAG-SCC1 | ampr/HIS3 5FLAG-SCC1 | This study |
| pPADH1-5FLAG-SCC3 | ampr/LEU2 5FLAG-SCC3 | This study |
| pPADH1-13MYC-SCC3 | ampr/HIS3 13MYC-SCC3 | This study |
| pPADH1-5FLAG-scc3 K99C | ampr/HIS3 5FLAG-scc3 K99C | This study |
| pPADH1-5FLAG-scc3 K764C | ampr/HIS3 5FLAG-scc3 K764C | This study |
| pPADH1-13MYC-scc3 K1076C | ampr/LEU2 13MYC-scc3 K1076C | This study |
| pPADH1-13MYC-scc3 K1057C | ampr/LEU2 13MYC-scc3 K1057C | This study |
| pRS313-ECO1 | ampr/HIS3 ECO1 | This study |
| pRS313-PCUP1-td-ECO1-13MYC-aid | ampr/HIS3 td-ECO1–13MYC-aid | This study |
| pRK5-FLAG-SMC3 | ampr/FLAG-SMC3 | This study |
| pRK5-MYC-SMC3 | ampr/MYC-SMC3 | This study |
| pGEX-6P-1-SCC1 | ampr/GST-SCC1 | This study |
| pGEX-6P-1-SCC3 | ampr/GST-SCC3 | This study |
| pET28a-SCC1 | kanr/His6-SCC1 | This study |
| pET28a-SCC3 | kanr/His6-SCC3 | This study |
Figure 1.
Intermolecular interactions of cohesin subunits in yeast. A, self-interaction of Scc1 in the overexpression condition. An extra copy of 5FLAG-SCC1 (Table 1) was introduced into the strain (Table 2, YSD24) carrying a C-terminal 3HA-EPEA-tagged SCC1 at the genomic locus. The lysates (input) were prepared from the cells collected at the exponential phase. Scc1–3HA-EPEA was precipitated via a C-tag affinity matrix. The precipitated proteins were detected via immunoblots against FLAG and HA antibodies, respectively. B, self-interaction of Scc1 detected via another pair of orthogonal epitopes. A plasmid expressing an extra copy of 5FLAG-SCC1 was introduced into the haploid yeast strain carrying an N-terminal GFP-tagged SCC1 at the genomic locus (Table 2, YSD03). GFP-Scc1 was precipitated via GBP beads. The precipitates were analyzed via IB against FLAG and GFP antibodies, respectively. C, self-interaction of Scc1 at physiological protein levels. The two endogenous SCC1 alleles in diploid yeast cells were labeled at their N termini with GFP and 5FLAG, respectively (Table 2, YSD107). The lysates were prepared from the cells collected at the exponential phase. GBP-IP and IB were performed as above. D and E, self-interaction of Scc3 in the physiological (D) or overexpression (E) condition. A diploid yeast strain (Table 2, YSD109) carrying the two endogenous SCC3 alleles with N-terminal GFP and 5FLAG tags was used in D. A haploid strain (Table 2, YSD61) carrying an endogenous N-terminal GFP-tagged SCC3 and an ectopic copy of 5FLAG-SCC3 was used in E. GBP-IP and IB were basically performed as above. F, Scc1 does not bind to itself in vitro. Purified recombinant proteins GST-Scc1 and His6-Scc1 were incubated with GSH Sepharose in the binding buffer. After three washes, the bound proteins were probed against anti-His6 and anti-GST antibodies. As a control, the GST tag alone was mixed with His6-Scc1. G, Scc3 can directly bind itself. GST pulldown and IB were basically conducted as above, using purified recombinant GST-Scc3 and His6-Scc3 proteins in this experiment.
The discrepancy could be explained if Scc1 self-interaction is interrupted by C-terminal tagging. However, it is also possible that self-interaction might be artificially caused by overexpression of cohesin subunits (22). To test the latter possibility, we next labeled two endogenous SCC1 alleles with the same pair of orthogonal epitope tags (i.e. GFP/FLAG) at their genomic loci in the diploid yeast cells. Under the physiological protein levels, Scc1-Scc1 interaction was apparent as well (Fig. 1C, lane 4), consistent with a very recent study in mouse embryonic stem cells (43). Intriguingly, self-interaction was also observed for the fourth cohesin subunit, Scc3, under endogenous and overexpression conditions in diploid (Fig. 1D, lane 4) and haploid (Fig. 1E, lane 5) cells, respectively. Collectively, these data suggest that cohesin is able to form dimers or oligomers. The failure to detect the self-interaction of cohesin is due to inappropriate epitope tagging and/or other experimental conditions.
We then asked which subunit mediates cohesin self-interaction. To answer it, we first examined whether self-interactions of Scc1 and Scc3 are direct or not through GST-pulldown assays. We expressed and purified GST-Scc1 and His6-Scc1 in bacteria. Interestingly, recombinant Scc1 proteins did not bind each other (Fig. 1F, lane 4), indicating that the intermolecular interaction of Scc1 is mediated by other proteins in eukaryotic cells. However, when we repeated the pulldown experiments using purified Scc3 proteins, His6-Scc3 bound with GST-Scc3 (Fig. 1G, lane 4), suggesting a direct physical association.
Isolation and cross-linking analysis of the cohesin complexes
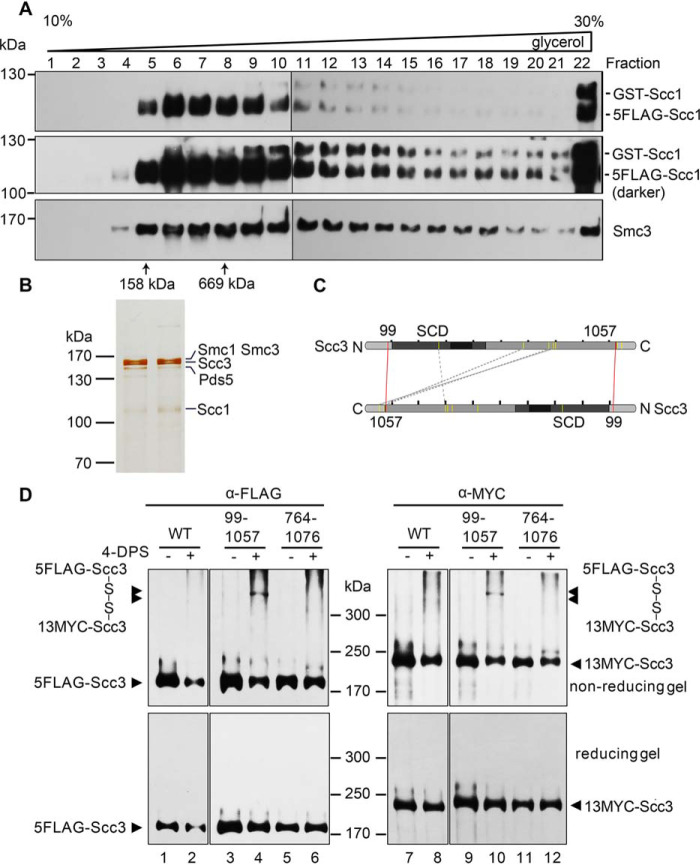
To further address the two different cohesion models, it is critical to test whether cohesin subunits can self-associate in the context of the cohesin complex. For this purpose, we next purified the native cohesin complexes from yeast cells containing two copies of Scc1 with small (5FLAG) and large (GST) epitopes, respectively. This allowed the simultaneous detection of the two Scc1 copies in a single gel by probing with anti-Scc1 antibodies. The lysates were first subjected to anti-FLAG affinity purification and FLAG peptide elution. The eluates were then run on a 10–30% glycerol sedimentation/velocity gradient. After centrifugation, fractions (0.5 ml each, labeled from top to bottom) were collected. After separation by SDS-PAGE, immunoblots revealed the co-purification of Smc3 together with Scc1, suggesting successful isolation of the complex rather than an individual Scc1 subunit (Fig. 2A). The peak of the purified complex (fractions 6–9) contained few GST-Scc1 (i.e. the second copy of Scc1), sedimenting close to the 669 kDa standard (fraction 8). The theoretical molecular weight of the single-ring four-subunit cohesin complex is 478 kDa. The relatively broad distribution of the cohesin complexes in the glycerol gradient might be due to the co-purification of additional factors like Pds5 and Wpl1. However, the cohesin species containing the second Scc1 copy (GST-Scc1) were clearly detected and sedimented much faster than 669 kDa, peaking around fraction 13. These data corroborate the existence of higher-order cohesin complexes in vivo.
Figure 2.
Purification and cross-linking of the cohesin complexes. A and B, purification of the native cohesin complexes. The cohesin complexes were isolated from the yeast cells expressing p5FLAG-SCC1 cells via one-step affinity purification (i.e. anti-FLAG M2 column chromatography and FLAG peptide elution) followed by 10–30% glycerol density gradient centrifugation. The sample was divided into 24 fractions (0.5 ml each). After separation by SDS-PAGE, they were analyzed by IB with the indicated antibodies (A) or silver staining (B). The sedimentation of standard proteins (158 and 669 kDa) is indicated by an arrow. The band of each subunit was validated by MS as well. C, an Scc3-Scc3 connectivity map of the high confidence DSS cross-links detected in CXMS. The purified Scc3-containing complexes were cross-linked by DSS prior to trypsin digestion and LCMS/MS as described under “Experimental procedures.” The cross-linked amino acids were identified using the pLink search engines and labeled by a dashed gray line. The pair of amino acids validated by the following cysteine substitution and in vivo cross-linking are labeled by a red line. D, cysteine substitution of Lys-99 and Lys-1057 at the putative Scc3-Scc3 interface supports the in vivo cross-linking. Cys screening of the putative pairs near the intermolecular interface of Scc3 was conducted. The indicated pairs of amino acid residues (e.g. Lys-99/Lys-1057 and Lys-764/Lys-1076) in two copies of Scc3 were substituted by cysteine. WT or cysteine-substituted mutant cells were grown and treated with 180 μm 4-DPS (+) or DMSO (−) before harvest. The proteins were extracted and analyzed by nonreducing (top) or reducing (bottom) SDS-PAGE followed by anti-FLAG and anti-MYC IBs. The monomeric and dimeric Scc3 are indicated by single and double arrows, respectively.
Next, we omitted the GST tag, which may cause artificial dimerization. The cohesin complexes at the endogenous level were isolated through 5FLAG-Scc1 or 5FLAG-Scc3. Silver staining showed that the cohesin complex is purified to a nearly homogeneous level (Fig. 2B). Besides all four cohesin subunits, the cellular cohesins often contained other components like Pds5, as validated by LC-MS. To determine how cohesin interacts with itself, we then performed cross-linking MS (CXMS) of the purified cohesin complexes. The representative cross-linked amino acids mapped to Scc3 are shown in Fig. 2C. Although it is challenging to distinguish between intramolecular and intermolecular interfaces of a homodimer, we supposed that the pairs of cross-linked residues apart from each other in the available three-dimensional structure of Scc3 fragment likely represent the intermolecular interface.
To test this, we then substituted these putative pairs by cysteine substitution for the VivosX (in vivo disulfide crosslinking) assay (44). If the two amino acids replaced by cysteine were close enough, a disulfide bond would be introduced by the permeable thiol-specific oxidizing agent 4,4′-dipyridyl disulfide (4-DPS). To simplify the screening and detection, we expressed two Scc3 alleles with a pair of tags (5FLAG-Scc3/13MYC-Scc3) in yeast cells. In WT, Scc3 (either 5FLAG-Scc3 or 13MYC-Scc3) migrated as a monomer (<200 kDa) with or without 4-DPS treatment in nonreducing SDS-PAGE (Fig. 2D, top, lanes 1, 2, 7, and 8). Among all mutated amino acid pairs, a portion of the Scc3-Scc3 cross-linking adducts was only detectable in the Scc3-K99C/Scc3-K1057C pair after 4-DPS treatment (Fig. 2D, compare lanes 3 and 4 and lanes 9 and 10). They migrated more slowly than 300 kDa, close to the expected molecular weight of dimeric Scc3 (∼287 kDa). Importantly, the same band was able to be probed by both anti-FLAG (Fig. 2D, top, lane 4) and anti-MYC (Fig. 2D, top, lane 10), confirming that it is a dimeric Scc3 complex. Moreover, the band was abolished in the reducing gel (Fig. 2D, bottom), further validating that it is formed by disulfide cross-linking of the introduced cysteine pair. Altogether, these results suggest the existence of the Scc3 dimer in vivo, consistent with the direct Scc3-Scc3 association in vitro. Intriguingly, the distal location of Lys-99 and Lys-1057 at the unstructured N and C termini of Scc3 also implicates that the twin Scc3 molecules might bind each other in an antiparallel manner to mediate a double-ring form of the cohesin complex.
Replication-coupled Smc3-Smc3 interaction in human cells
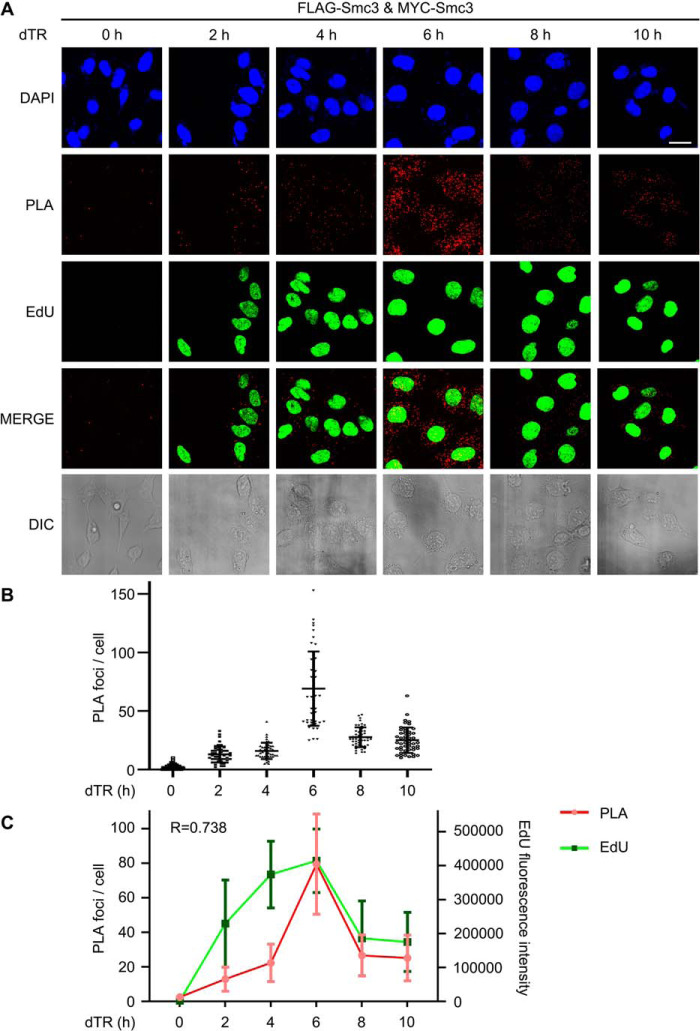
Because the cohesin status is cell cycle–regulated (45), we wanted to know whether cohesin dimerization is similarly controlled. To test this, we applied a proximity ligation assay (PLA) to visualize the cohesin-cohesin interaction in human cells (46). 5FLAG-Smc3 and 13MYC-Smc3 were introduced into HeLa cells. Cells were grown and arrested in G1 (0 h) by double thymidine block, before release into the fresh medium containing EdU for 2, 4, 6, 8, or 10 h. Two Smc3 copies were probed by mouse anti-FLAG and rabbit anti-MYC antibodies, respectively. If two Smc3 proteins are in proximity, their secondary antibodies conjugated to DNA oligonucleotides will bring together another pair of oligonucleotides, which is subsequently ligated and circulated by DNA ligase. The circulated DNA was amplified by rolling circle amplification and finally detected by fluorescence in situ hybridization. In the G1 phase, few fluorescence signals were observed (Fig. 3A), excluding the possible false positives presumably due to the high sensitivity of the PLA method and/or Smc3 overexpression. After G1 release, PLA signals appeared in 2 h and peaked around 6 h (Fig. 3, A and B). These results corroborate the cohesin-cohesin interaction originally discovered by Pati's group in human cells (20). More importantly, these data also demonstrate that cohesin dimerization does not occur in the G1 phase and is regulated in a cell cycle–dependent manner in human cells. Intriguingly, quantification of both PLA and EdU signals revealed a rough correlation between them (r = 0.738). Although PLA signals appeared a little behind EdU during the early S phase (0–4 h), both of them reached the peak at the same time (6 h), followed by a similar decline (Fig. 3C). Because EdU incorporation is an indicator of genome replication progress, it strongly argues that cohesin-cohesin interaction occurs in a DNA replication–coupled manner. The time lag of PLA signals compared with EdU levels during the early replication stage is not surprising, given that cohesin distributes in an average 67-kb distance along the chromosome in HeLa cells (47).
Figure 3.
DNA replication-coupled Smc3-Smc3 interaction in human cells. A, in situ PLA of Smc3-Smc3 in human cells. 293T cells expressing 5FLAG-Smc3 and 13MYC-Smc3 were cultured and synchronized in early S phase by double-thymidine arrest before release (dTR) into the fresh medium containing EdU. Cells were collected at the indicated time points. PLA was performed by proximity probes against FLAG and MYC. EdU was detected via click chemistry. Bar, 20 μm. B, scatter plot of PLA foci per cell throughout the cell cycle. The number of PLA spots within a cell was quantified. At least 50 cells were measured for each time point. The mean ± S.D. values are shown. C, correlation analysis of PLA spots (red) and EdU intensity (green) per cell. The intensity of EdU was measured by ImageJ. The maximal values for in situ PLA and EdU were normalized to allow a comparison between the different assays. The mean ± S.D. (error bars) values and Pearson product-moment correlation coefficient (r) are shown (n = 50 cells).
Intermolecular cohesin interaction is cell cycle–regulated
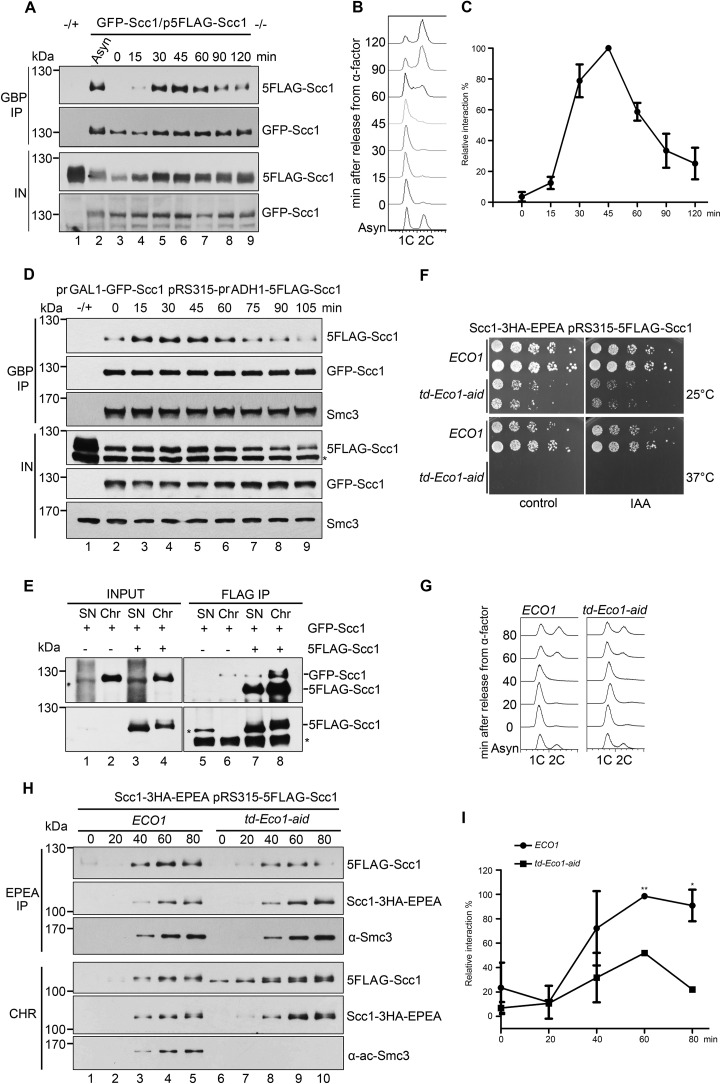
To further elucidate how cohesin dimerization is regulated, we investigated it in the synchronized yeast cells. For this purpose, a strain carrying FLAG-Scc1 and GFP-Scc1 was grown at 30 °C and arrested in G1 by α-factor (0 min). After release into S phase, cells were collected at different time points. Then we carried out GBP-IP of whole-cell extracts. Although Scc1 is expressed in G1 (Fig. 4A, lane 3, input (IN)), few Scc1 proteins co-precipitated with themselves (Fig. 4A, top panel). If we normalized the precipitated GFP-Scc1 (second panel), the co-precipitated 5FLAG-Scc1 gradually increased and peaked around 45 min (S phase), followed by a decline in M phase (Fig. 4, A (first panel) and B). The relative Scc1-Scc1 interaction was quantified as the 5FLAG-Scc1/GFP-Scc1 ratio in the precipitates, which clearly fluctuated with the cell cycle (Fig. 4C). Consistent with the results above in human cells, these data suggest that cohesin dimerization occurs exclusively in S phase in a cell cycle–regulated fashion.
Figure 4.
Cohesin-cohesin interaction is regulated by Eco1 during the cell cycle. A, the GFP-SCC1/p5FLAG-SCC1 cells were grown, synchronized in G1 by α-factor (0 min), and released into S phase at 25 °C for the indicated time. The cell lysates were subjected to GBP-IP and IB against anti-FLAG and anti-GFP antibodies. B, a representative cell cycle profile analyzed by flow cytometry of the samples used in A. C, quantification of the relative intermolecular interaction of cohesin during the cell cycle. The densities of the FLAG-Scc1 and GFP-Scc1 bands in the precipitates were quantified. The ratio of FLAG-Scc1/GFP-Scc1 was calculated to indicate the relative cohesin-cohesin interaction in each sample. The maximum percentage among all samples was normalized to 100%. To ensure that the signals were within the linear range, immunoblots with appropriate exposure were quantified by Quantity One (Bio-Rad). Data shown are the mean ± S.D. (error bars) of three biological replicates. D, both GFP-SCC1 and 5FLAG-SCC1 under control of the GAL1 promoter were overexpressed in α-factor–arrested cells by galactose. All other experimental conditions were the same as described in A. E, the exponentially grown GFP-SCC1/p5FLAG-SCC1 cells were collected and fractionated into native Chr and nonchromatin-bound SN as described under “Experimental procedures.” Both SN and Chr fractions were subjected to FLAG-IP and IB using anti-Scc1 antibodies and anti-FLAG antibodies. 5FLAG-Scc1 cannot be well-detected in INPUT fraction using anti-Scc1 antibodies. Bottom, 5FLAG-Scc1 detected by anti-FLAG antibodies. F, efficient depletion of Eco1 via combined td and aid degrons leads to cell death. The growth of WT (ECO1) and Eco1 depletion strains (td-ECO1-aid) was examined by spotting on the medium with or without IAA at either 25 or 37 °C. G, Eco1 depletion causes only subtle changes in the cell-cycle progression. Shown are representative cell-cycle profiles of WT and Eco1 depletion strains used in H. After release from G1 arrest, cells were collected at the indicated time at 37 °C and analyzed by flow cytometry. H, Eco1 depletion interferes with cohesin-cohesin interaction on chromatin. Synchronized cells were prepared as in G. Native Chr was prepared as described under “Experimental procedures.” Scc1–3HA-EPEA was then precipitated via a C-tag affinity matrix and probed with the indicated antibodies. I, the relative cohesin-cohesin interaction in the presence or absence of Eco1 was quantified as described in C. Data shown are the mean ± S.D. of three biological replicates. *, p < 0.05; **, p < 0.01.
Notably, there was increased Scc1 expression during S phase (Fig. 4A). To test whether cohesin dimerization in the S phase is due to the increased Scc1 protein level, we overexpressed both GFP-Scc1 and 5FLAG-Scc1 by strong promoters. This resulted in a very high level of both versions of Scc1 in G1 (Fig. 4D, lane 2, bottom). However, the Scc1-Scc1 interaction remained very weak at that time and augmented in S phase (Fig. 4D, top), similar to that in WT (Fig. 4A, top). Meanwhile, the amounts of Smc3 in the precipitates were not significantly changed (Fig. 4D, third panel), indicating a constant Scc1-Smc3 interaction throughout the cell cycle. These data suggest that cell cycle–regulated cohesin dimerization is not merely due to the fluctuation of the Scc1 protein level.
Cohesin dimerization shares common factors with sister chromatid cohesion
The above results suggest a similar cell cycle–regulated pattern between cohesin-cohesin interaction and sister chromatid cohesion. Notably, both of them occur concomitantly with DNA replication. These facts prompted us to speculate on a functional relationship between the two critical events. To test this notion, we carried out five sets of experiments.
First, given that sister chromatid cohesion occurs on chromatin, we asked how cohesin-cohesin interaction is controlled spatially. To address this question, we prepared native chromatin-bound fraction (Chr) and nonchromatin-bound fraction (supernatant (SN)) before FLAG-IPs. Cohesin existed in both fractions (Fig. 4E, lanes 3 and 4). However, the intermolecular interaction of Scc1 was detectable exclusively in the Chr fraction (Fig. 4E, compare lanes 7 and 8). This suggests that cohesin is dimerized in the context of chromatin as well.
Second, we examined whether the vital cohesion establishment factor, Eco1, is required for the cohesin-cohesin interaction. Because Eco1 is essential for cell viability, we combined both temperature-sensitive (td) and auxin-induced (aid) degrons to deplete cellular Eco1 protein. The Ubr1 and Tir1 ubiquitin E3 ligases were induced by galactose. The td and aid degrons were turned on by switching from 25 to 37 °C and adding indole-3-acetic acid (IAA), respectively. These led to cell death (Fig. 4F) and abolished S phase Smc3 acetylation (Fig. 4H, bottom, lanes 7–10), indicating effective Eco1 depletion. However, the first S phase progression right after Eco1 depletion was only subtly affected (Fig. 4G). Meanwhile, we monitored the intermolecular cohesin interaction during the cell cycle through EPEA-IP and immunoblotting in the Chr. In WT, the cohesin-cohesin interaction displayed a cell-cycle pattern (Fig. 4, H and I) as shown in Fig. 4C, but relatively slow, which is in accord with the slower cell-cycle progression under this condition (Fig. 4G). When Eco1 was depleted, co-precipitated 5FLAG-Scc1 was largely decreased (Fig. 4, H and I), whereas the chromatin-associated Scc1 levels of both versions were not much affected (Fig. 4H, bottom). Meanwhile, Scc1-Smc3 interaction was not affected either. These data suggest that Eco1 is required for cohesin dimerization, but not for chromatin association of single rings.
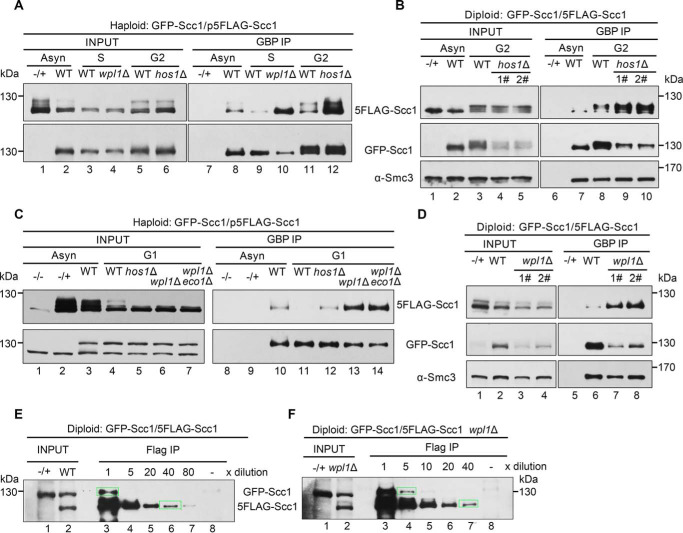
Third, Smc3 acetylation is erased by deacetylase Hos1 in anaphase and recycled in the subsequent cell cycle, so we examined the change of cohesin-cohesin interaction in the absence of Hos1. In the GFP-Scc1/p5FLAG-Scc1 dual-tagged haploid background, WT or mutant cells were cultured and arrested in G2 by nocodazole. Although the amounts of both GFP-Scc1 and 5FLAG-Scc1 were nearly equal in WT and mutant cells (Fig. 5A, lanes 5 and 6), the hos1Δ cells showed a significant augment of Scc1-Scc1 interaction (Fig. 5A, compare lanes 11 and 12). Similar results were obtained from the diploid cells in which two endogenous Scc1 copies carry the same pair of orthogonal epitopes (Fig. 5B, compare lanes 8–10). These results suggest that Hos1 either partially relieves cohesin-cohesin interaction in the M phase or prevents precocious cohesin-cohesin interaction before the S phase.
Figure 5.
The dimerized cohesin increases once wpl1 or hos1 is depleted. A, the haploid WT (GFP-Scc1/p5FLAG-Scc1), wpl1Δ or hos1Δ cells were grown with or without synchronization. The S phase cells were obtained by α-factor arrest and release for 60 min at 30 °C, whereas the G2 cells were arrested by nocodazole. GFP-Scc1 was precipitated via GBP beads from WCE. The proteins were detected via IBs against the indicated antibodies. −/+, control strain that does not harbor the GFP-tagged version of Scc1. B, the diploid WT (GFP-Scc1/5FLAG-Scc1) or hos1Δ cells were grown with or without G2 arrest. The lysates were subjected to GBP-IP and IB as above. 1# and 2# denote the biological repeats. C, the haploid WT (GFP-Scc1/p5FLAG-Scc1), hos1Δ, wpl1Δ, or wpl1Δeco1Δ cells were grown with or without G1 arrest. GBP-IPs and IBs were performed as above. D, the diploid WT (GFP-Scc1/5FLAG-Scc1) or wpl1Δ cells were grown exponentially. The lysates were subjected to GBP-IP and IB. 1# and 2# denote the biological repeats. E and F, the diploid WT (GFP-Scc1/5FLAG-Scc1) (E) or wpl1Δ (F) cells were grown exponentially. The lysates were precipitated by anti-FLAG M2 beads. A series of dilutions (5×, 10×, 20×, 40×, and 80×) of the samples were probed by anti-Scc1 antibodies. The indicated relative density of the band in a rectangular marquee was measured by Quantity One (Bio-Rad). The percentage of cohesin dimers was calculated as described under “Experimental procedures.” −/+, control strain that does not harbor the 5FLAG-tagged version of Scc1.
Fourth, prior to Eco1-dependent cohesion establishment, cohesin remains dynamic on chromatin due to the destabilized activity of Wpl1. The essential function of ECO1 can be bypassed by WPL1 deletion (33, 34), so we next compared the cohesin-cohesin interaction in the presence or absence of Wpl1. The experiments were basically conducted as described for hos1Δ. When WPL1 was deleted, cohesin oligomerization increased prominently in both G1 (Fig. 5A, compare lanes 11 and 13) and S (Fig. 5A, compare lanes 9 and 10). Consistently, in asynchronized diploid cells, a dramatic increase was observed in the absence of Wpl1 as well (Fig. 5D, compare lanes 6–8). These data indicate that Wpl1 prevents the cohesin-cohesin interaction, correlating with a loose and dynamic association of cohesin with chromatin in G1.
Fifth, based on the ratio of GFP-Scc1/5FLAG-Scc1 in the cell extracts and precipitates, we estimated the percentage of cohesin dimers at the endogenous protein levels in diploid cells using a method published recently (41). The measured cellular levels of GFP-Scc1 and 5FLAG-Scc1 were nearly identical (Fig. 5E). Through serial dilutions of the precipitates, we quantified the band densities of GFP-Scc1 and 5FLAG-Scc1 probed by anti-Scc1 antibodies in the same gel. The percentage of cohesin dimers was roughly estimated through the following formula,
| (Eq. 1) |
In asynchronized WT cells, ∼20% of cohesins could be detected in the dimeric state (Fig. 5E). Intriguingly, ∼20–30% of Smc3 is acetylated in budding yeast in a previous report (35). Using a similar method, Cattoglio et al. (43) recently reported that cohesin dimers occupy at least ∼8% of mouse embryonic stem cells. When WPL1 was deleted, cohesin dimers increased up to 40% in yeast (Fig. 5F). This result suggests that Wpl1, the anti-establishment factor of cohesion, acts as a negative regulator in cohesin dimerization as well. Taken together, all of these lines of evidence suggest that cohesin dimerization is cell cycle–regulated as the sister chromatid cohesion cycle by the same mechanisms (i.e. Wpl1/Eco1/Hos1).
Discussion
Here, we show that cohesin is dimerized in S phase and monomerized again in mitosis and G1, which is controlled by the same regulators (Eco1, Wpl1, and Hos1) as the sister chromatid cohesion/dissolution cycle. Besides this biochemical evidence described here and literature (20, 43), genetic interactions also support cohesin-cohesin interactions (19). Both yeast cohesin and prokaryotic SMC condensin have been proposed to act as dimers in extruding DNA loops (48, 49).
Notably, through bifluorescent complementation assays, Zhang et al. (20) also showed a similar antiparallel orientation of Scc1-Scc1 in human cells, generally in agreement with the results shown in this study. The only difference lies in the number of Scc3 subunits in the cohesin dimer. According to a very recent study from the Peters group, the stoichiometry of the four cohesin subunits remains 1:1:1:1 (monomer) or 2:2:2:2 (dimer) throughout the cell cycle in human (47). Therefore, the discrepancy might be due to the different experimental procedures/conditions.
Although the exact interface of such a cohesin handcuff remains to be solved, we speculate that it might be mediated via self-association of Scc3 rather than Scc1 for at least two reasons. First, both pulldown and VivosX data show a direct physical association of two Scc3 molecules. Second, lack of Scc3 or Eco1 causes a defect in sister chromatid cohesion but not in cohesin binding to DNA, whereas Scc1 is indispensable for both (29, 30, 50). Besides the canonical single-ring structure, dimerization may provide an additional mechanism for cohesin to execute various functions in sister chromatid cohesion, DNA repair, chromatin loop extrusion, and high-order chromatin organization (21). It will also be very interesting to investigate how Eco1-catalyzed acetylation and Wpl1 regulate the structural/conformational change of cohesin in the future.
Experimental procedures
Strain and plasmid construction
Strains and plasmids used in this study are listed in Tables 1 and 2, respectively.
Table 2.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 lys2Δ0 | In stock |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | In stock |
| YSD03 | BY4741 KanMX6::PGAL1-GFP-SCC1 (p315-5FLAG-SCC1::LEU2) | This study (Fig. 1B) |
| YSD24 | BY4741 SCC1-3HA-EPEA::HygR (p315-5FLAG-SCC1::LEU2) | This study (Fig. 1A) |
| YSD107 | BY4743 5FLAG-SCC1::HIS3/GFP-SCC1::LEU2 | This study (Fig. 1C) |
| ZSJ22 | BY4741 KanMX6::PADH1-GST-SCC1 wpl1Δ::HygR (p315-5FLAG-SCC1::LEU2) | This study (Fig. 2A) |
| YSD61 | BY4741 KanMX6::PGAL1-GFP-SCC3 (pPADH1-5FLAG-SCC3::LEU2) | This study (Fig. 1E) |
| YSD109 | BY4743 5FLAG-SCC3::HIS3/GFP-SCC3::LEU2 | This study (Fig. 1D) |
| YSD83 | BY4741 scc3Δ::NatMX (pScc3::URA3 pPADH1-5FLAG-SCC3::HIS3 pPADH1-13MYC-SCC3::LEU2) | This study (Fig. 3, C and D) |
| YSD87 | BY4741 scc3Δ::NatMX (pScc3::URA3 pPADH1-5FLAG- scc3 K99C::HIS3 pPADH1-13MYC-scc3 K1057C::LEU2) | This study (Fig. 3, C and D) |
| YSD88 | BY4741 scc3Δ::NatMX (pScc3::URA3 pPADH1-5FLAG- scc3 K764C::HIS3 pPADH1-13MYC-scc3 K1076C::LEU2) | This study (Fig. 3, C and D) |
| YSD17 | BY4741 KanMX6::PGAL1-GFP-SCC1 wpl1Δ::HygR (p315-5FLAG-SCC1::LEU2) | This study (Fig. 5, A and C) |
| YSD08 | BY4741 KanMX6::PGAL1-GFP-SCC1 hos1Δ::HygR (p315-5FLAG-SCC1::LEU2) | This study (Fig. 5, A and C) |
| YSD159 | BY4743 5FLAG-SCC1::HIS3/GFP-SCC1::LEU2 hos1Δ::HygR/hos1Δ::NatMX | This study (Fig. 5B) |
| YSD141 | BY4743 5FLAG-SCC1::HIS3 / GFP-SCC1::LEU2 wpl1Δ::HygR/wpl1Δ::NatMX | This study (Fig. 5D) |
| YSD33 | BY4741 SCC1-3HA-EPEA::HygR eco1Δ::NatMX ubr1::PGal1-UBR1-PGAP-OsTIR1-9MYC-URA3 (p315-5FLAG-SCC1::LEU2 p313-ECO1::HIS3) | This study (Fig. 3, E–H) |
| YSD35 | BY4741 SCC1-3HA-EPEA::HygR eco1Δ::NatMX ubr1::PGal1-UBR1-PGAP-OsTIR1-9MYC-URA3 (p315-5FLAG-SCC1::LEU2 p313-PCUP1-td-ECO1-13MYC-aid::HIS3) | This study (Fig. 3, E–H) |
Preparation of antibodies
To raise polyclonal antibodies specific to Scc1 and Smc3, purified Scc1N (amino acids 1–333) and Smc3 hinge domain were used to immunize rabbits. Polyclonal antibodies were affinity-purified. Scc1 and Smc3 beads were prepared by immobilizing purified Scc1N and Smc3 proteins to NHS-activated agarose beads as recommended by the manufacturer (GE Healthcare).
Cell synchronization and flow cytometry analysis
Cells were grown to logarithmic phase, and 7.5 μg/ml α-factor was added for cell synchronization in G1 phase. After washing twice, G1-arrested cells were released in fresh medium and continued growth for the indicated time. Cells were collected and fixed with 70% ethanol and then processed for flow cytometry using a BECKMAN Cytoflex-S flow cytometer.
Conditional depletion of cellular Eco1 protein
The efficient depletion of endogenous Eco1 protein was achieved through a two-degron strategy. td and aid degrons were added to the N and C terminus of Eco1, respectively. The corresponding two ubiquitin ligases (E3), UBR1 and OsTIR1, were integrated into the genomic UBR1 locus under control of the galactose-inducible Gal1-10 promoter. Cells were first grown at 25 °C in rich medium containing 0.1 mm Cu2+ supplemented with 2% raffinose before transferring to 2% galactose to induce the expression of two E3s. Two degrons were turned on by adding 1 mm IAA (for aid) and switching to 37 °C (for td) for 2 h. The protein level of Eco1-MYC was measured by immunoblotting (IB) with anti-MYC and anti-Smc3ac antibodies.
Whole-cell extracts (WCE) and IB
WCE of 100 OD600 units of asynchronized or synchronized cells were prepared by glass bead beating (Mini-Beadbeater-16, Biospec) in lysis buffer (50 mm HEPES/KOH, pH 7.4, 150 mm NaCl, 1 mm EDTA, 10% glycerol, 1 mm DTT, 1 mm PMSF, protease inhibitor tablets (EDTA-free; Roche Applied Science)). Protein samples were separated by SDS-PAGE and immunoblotted with the antibodies specifically indicated in each figure. Antibodies used in this study are as follows: mouse anti-FLAG M2-specific mAb (1:1000; Sigma), rabbit polyclonal anti-GFP (1:500; GeneScript), mouse anti-HA 16B12 (1:1000; Millipore), anti-ac-Smc3, polyclonal anti-Smc3 (1:1000), and polyclonal anti-Scc1 (1:1000). Horseradish peroxidase–conjugated anti-rabbit or anti-mouse IgG was used as the secondary antibody (1:10,000; Sigma).
IP
Monoclonal GBP agarose, monoclonal anti-EPEA agarose (Thermo Fisher), and monoclonal anti-FLAG M2 affinity gel (Sigma–Aldrich) were used for IP. IP was performed using strains co-expressing the tagged versions of each protein as indicated in each figure. After three washes, the proteins specifically associated with beads were boiled and analyzed by SDS-PAGE and IB using the indicated antibodies.
Glycerol gradient centrifugation
The native protein complexes in the peptide eluates after FLAG-IPs were concentrated and applied to the top of a 10–30% glycerol gradient in EBX-3 buffer (50 mm HEPES/KOH, pH 7.5, 150 mm KCl, 2.5 mm MgOAc, 0.1 mm ZnOAc, 2 mm NaF, 0.5 mm spermidine, 20 mm glycerophosphate, 1 mm ATP, 1 mm DTT, 1 mm PMSF, protease inhibitor tablets (EDTA-free; Roche Applied Science)). The gradients were centrifuged in a P55ST2 swinging bucket rotor (Hitachi CP100NX ultracentrifuge) at 120,000 × g for 9 h using slow deceleration. After centrifugation, the fractions were collected from the top of the gradient and subjected to SDS-PAGE and immunoblots. Aldlase (158 kDa) and thyroglobulin (669 kDa) were used as size markers.
CXMS
5FLAG-Scc3 was prepared by FLAG-IP of yeast WCE and FLAG peptide elution. About 15 μg of purified Scc3 in a volume of 15 μl was cross-linked through incubation with the lysine cross-linker disuccinimidyl suberate (DSS) at a final concentration of 0.5 mm for 1 h at room temperature. The final concentration of 20 mm NH4HCO3 was added to quench the reaction. The cross-linked proteins were precipitated with ice-cold acetone of 4–5-fold volume at −20 °C overnight, resuspended in 8 m urea, 100 mm Tris, pH 8.5. After trypsin digestion, the LC-MS/MS analysis was performed on an Easy-nLC 1000 UHPLC (Thermo Fisher Scientific) coupled to a Q Exactive HF Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides were loaded on a precolumn (75-μm inner diameter, 4 cm long, packed with ODS-AQ 12 nm–10 mm beads from YMC Co., Ltd.) and separated on an analytical column (75-μm inner diameter, 13 cm long, packed with ReproSil-Pur C18-AQ 1.9-μm 120-Å resin from Dr. Maisch GmbH) using a linear gradient of 0–35% buffer B (100% acetonitrile and 0.1% formic acid) at a flow rate of 250 nl/min over 73 min. The top 20 most intense precursor ions from each full scan (resolution 60,000) were isolated for HCD MS2 (resolution 15,000; NCE 27) with a dynamic exclusion time of 30 s. Precursors with 1+ or unassigned charge states were excluded. pLink was used to identify cross-linked peptides with the cutoffs of false discovery rate of 5% and E value of 0.001.
Disulfide cross-linking to capture site-specific protein-protein interactions in vivo
The Scc3-Scc3 interaction was captured by a disulfide cross-linking method in yeast cells (44). The indicated pairs of amino acid residues in Scc3 were substituted by cysteine. WT or mutant cells were cultured in 5 ml of CSM medium (without cysteine) at 30 °C to OD600 of 0.5 before the addition of 180 μm 4-DPS (Sigma–Aldrich). The cultures were resumed for 20 min and then quenched with 20% TCA. The cells were pelleted and washed with 20% TCA before homogenization in the presence of 400 μl of 20% TCA and ∼450 μl of glass beads using Mini-Beadbeater-16 (Biospec). N-Ethyl maleimide was added to prevent any free thiol groups from cross-linking after cell lysis. Proteins were extracted and separated by nonreducing and reducing SDS-PAGE for immunoblots against the indicated antibodies as described previously (44).
PLA
The PLA was performed as described previously (51). Briefly, HeLa cells were fixed with 4% paraformaldehyde (Sigma–Aldrich) in PBS for 15 min, permeabilized with 0.1% Triton X-100 (Sigma) for 5 min, and blocked for 1 h with a blocking solution (250 μg/ml BSA, 2.5 μg/ml sonicated salmon sperm DNA, 5 mm EDTA, 0.05% Tween 20 in PBS). Cells were washed with PBS and incubated in two primary antibodies. The primary antibodies used were as follows: mouse monoclonal anti-FLAG (1:100; Sigma) and rabbit polyclonal anti-MYC (1:100; 16286-1-AP, Proteintech). After washing with PBS, samples were incubated with secondary antibodies conjugated with PLA probes for 1 h at 37 °C. After washing with PBS, samples were incubated with ligation-ligase solution for 30 min at 37 °C. Then the samples were washed with PBS and continued with amplification-polymerase solution incubation for 90 min at 37 °C. The detection mix was added, followed by incubation for 30 min at 37 °C. Then the slide was kept in the dark. After washing three times for 5 min each with PBS, slides were mounted using Duolink in situ Mounting Medium with 4′,6-diamidino-2-phenylindole. Pictures were taken using a fluorescent microscope (Leica DIM8).
Native chromatin fractionation
Native chromatin fractionation was performed as described (40, 52) with minor modifications. Yeast cells of 200 OD600 units were spheroplasted by 75 units/ml lyticase. Crude extracts were prepared by Triton X-100 treatment and fractionated via sucrose cushion in 500 μl of EBX-3 buffer (50 mm HEPES/KOH, pH 7.4, 150 mm NaCl, 2.5 mm MgCl2, 0.1 mm ZnOAc, 5 mm NaF, 1 mm NaVO4, 10 mm β-glycerophosphate, 1 mm ATP, 1 mm DTT, 1 mm PMSF, protease inhibitor tablets (EDTA-free; Roche Applied Science). The supernatant contained non-chromatin-bound proteins. Chromatin-bound proteins in the pellet were released by incubation in EBX-3 buffer containing 500 units/ml of benzonase (Sigma) for 60 min at 4 °C.
In vitro pulldown assay
An approximately 10 pm concentration of each protein was mixed with GSH Sepharose 4B (GE Healthcare Life Sciences) in 100 μl of binding buffer (50 mm HEPES-NaOH, pH 7.6, 150 mm NaCl, 10% glycerol, 1 mm EDTA, 1 mm PMSF, 1 μg/μl BSA, and 0.1% Triton X-100) and incubated for 1 h at 4 °C. The beads were washed at least three times prior to Western blotting.
Statistical analyses
Statistical analyses were performed using GraphPad Prism software. The data are presented as the mean ± S.D. Student's t test was employed to determine the statistical significance of the differences. A value of p < 0.05 was regarded as statistically significant.
Data availability
All data are contained within the article.
Author contributions
D. S., S. Z., Q. C., and H. L. conceptualization; D. S., S. Z., J. Z., W. H., Q. C., and H. L. resources; D. S., S. Z., M.-Q. Z., J. Z., W. H., M.-Q. D., Q. C., and H. L. data curation; D. S., S. Z., M.-Q. Z., J. Z., W. H., M.-Q. D., Q. C., and H. L. software; D. S., S. Z., M.-Q. Z., J. Z., W. H., M.-Q. D., Q. C., and H. L. formal analysis; M.-Q. D., Q. C., and H. L. supervision; D. S., S. Z., J. Z., W. H., Q. C., and H. L. validation; D. S., S. Z., M.-Q. Z., J. Z., W. H., M.-Q. D., Q. C., and H. L. investigation; D. S., S. Z., M.-Q. Z., J. Z., W. H., M.-Q. D., Q. C., and H. L. visualization; D. S., S. Z., M.-Q. Z., M.-Q. D., Q. C., and H. L. methodology; D. S. and H. L. writing-original draft; W. H., M.-Q. D., Q. C., and H. L. project administration; D. S. and H. L. writing-review and editing; J. Z., M.-Q. D., Q. C., and H. L. funding acquisition.
Acknowledgments
We thank Dr. Katsuhiko Shirahige (Tokyo Institute of Technology) for anti-Smc3ac antibodies and members of the Lou laboratory for helpful discussion and comments on the manuscript.
This work was supported by the National Natural Science Foundation of China Grants 31630005, 31770084, and 31771382; National Basic Research Program (973 Program) of China Grant 2014CB849801; and Program for Extramural Scientists of the State Key Laboratory of Agrobiotechnology Grant 2018SKLAB6-5. The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as one of our Editors' Picks.
- IP
- immunoprecipitation
- CXMS
- cross-linking mass spectrometry
- VivosX
- in vivo disulfide cross-linking
- 4-DPS
- 4,4′-dipyridyl disulfide
- PLA
- proximity ligation assay
- Chr
- chromatin-bound fraction
- WCE
- whole-cell extracts
- GBP
- GFP-binding protein
- SN
- supernatant
- td
- temperature-dependent
- aid
- auxin-induced
- IAA
- indole-3-acetic acid
- IB
- immunoblotting
- OD
- optical density
- PMSF
- phenylmethylsulfonyl fluoride
- DSS
- disuccinimidyl suberate.
References
- 1. Onn I., Heidinger-Pauli J. M., Guacci V., Unal E., and Koshland D. E. (2008) Sister chromatid cohesion: a simple concept with a complex reality. Annu. Rev. Cell Dev. Biol. 24, 105–129 10.1146/annurev.cellbio.24.110707.175350 [DOI] [PubMed] [Google Scholar]
- 2. Peters J.-M., Tedeschi A., and Schmitz J. (2008) The cohesin complex and its roles in chromosome biology. Genes Dev. 22, 3089–3114 10.1101/gad.1724308 [DOI] [PubMed] [Google Scholar]
- 3. Nasmyth K., and Haering C. H. (2009) Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- 4. Jeppsson K., Kanno T., Shirahige K., and Sjögren C. (2014) The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell Biol. 15, 601–614 10.1038/nrm3857 [DOI] [PubMed] [Google Scholar]
- 5. Uhlmann F. (2016) SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399 10.1038/nrm.2016.30 [DOI] [PubMed] [Google Scholar]
- 6. Nasmyth K. (2011) Cohesin: a catenase with separate entry and exit gates? Nat. Cell Biol. 13, 1170–1177 10.1038/ncb2349 [DOI] [PubMed] [Google Scholar]
- 7. Gligoris T., and Löwe J. (2016) Structural insights into ring formation of cohesin and related Smc complexes. Trends Cell Biol. 26, 680–693 10.1016/j.tcb.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Y., Muir K. W., Bowler M. W., Metz J., Haering C. H., and Panne D. (2018) Structural basis for Scc3-dependent cohesin recruitment to chromatin. Elife 7, e38356 10.7554/eLife.38356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haering C. H., Löwe J., Hochwagen A., and Nasmyth K. (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788 10.1016/S1097-2765(02)00515-4 [DOI] [PubMed] [Google Scholar]
- 10. Holzmann J., Fuchs J., Pichler P., Peters J. M., and Mechtler K. (2011) Lesson from the stoichiometry determination of the cohesin complex: a short protease mediated elution increases the recovery from cross-linked antibody-conjugated beads. J. Proteome Res. 10, 780–789 10.1021/pr100927x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holzmann J., Politi A. Z., Nagasaka K., Hantsche-Grininger M., Walther N., Koch B., Fuchs J., Dürnberger G., Tang W., Ladurner R., Stocsits R. R., Busslinger G. A., Novák B., Mechtler K., Davidson I. F., et al. (2019) Absolute quantification of cohesin, CTCF and their regulators in human cells. Elife 8, e46269 10.7554/eLife.46269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gruber S., Haering C. H., and Nasmyth K. (2003) Chromosomal cohesin forms a ring. Cell 112, 765–777 10.1016/S0092-8674(03)00162-4 [DOI] [PubMed] [Google Scholar]
- 13. Ivanov D., and Nasmyth K. (2005) A topological interaction between cohesin rings and a circular minichromosome. Cell 122, 849–860 10.1016/j.cell.2005.07.018 [DOI] [PubMed] [Google Scholar]
- 14. Chan K. L., Roig M. B., Hu B., Beckouët F., Metson J., and Nasmyth K. (2012) Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell 150, 961–974 10.1016/j.cell.2012.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murayama Y., and Uhlmann F. (2014) Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505, 367–371 10.1038/nature12867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huis in 't Veld P. J., Herzog F., Ladurner R., Davidson I. F., Piric S., Kreidl E., Bhaskara V., Aebersold R., and Peters J.-M. (2014) Characterization of a DNA exit gate in the human cohesin ring. Science 346, 968–972 10.1126/science.1256904 [DOI] [PubMed] [Google Scholar]
- 17. Srinivasan M., Scheinost J. C., Petela N. J., Gligoris T. G., Wissler M., Ogushi S., Collier J. E., Voulgaris M., Kurze A., Chan K.-L., Hu B., Costanzo V., and Nasmyth K. A. (2018) The cohesin ring uses its hinge to organize DNA using non-topological as well as topological mechanisms. Cell 173, 1508–1519.e18 10.1016/j.cell.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murayama Y., Samora C. P., Kurokawa Y., Iwasaki H., and Uhlmann F. (2018) Establishment of DNA-DNA interactions by the cohesin ring. Cell 172, 465–477.e15 10.1016/j.cell.2017.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eng T., Guacci V., and Koshland D. (2015) Interallelic complementation provides functional evidence for cohesin-cohesin interactions on DNA. Mol. Biol. Cell 26, 4224–4235 10.1091/mbc.e15-06-0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang N., Kuznetsov S. G., Sharan S. K., Li K., Rao P. H., and Pati D. (2008) A handcuff model for the cohesin complex. J. Cell Biol. 183, 1019–1031 10.1083/jcb.200801157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skibbens R. V. (2016) Of rings and rods: regulating cohesin entrapment of DNA to generate intra- and intermolecular tethers. PLoS Genet. 12, e1006337 10.1371/journal.pgen.1006337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hansen A. S., Pustova I., Cattoglio C., Tjian R., and Darzacq X. (2017) CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 6, e25776 10.7554/eLife.25776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hirano T., Nishiyama T., and Shirahige K. (2017) Hot debate in hot springs: report on the second international meeting on SMC proteins. Genes Cells 22, 934–938 10.1111/gtc.12539 [DOI] [PubMed] [Google Scholar]
- 24. Peters J.-M., and Nishiyama T. (2012) Sister chromatid cohesion. Cold Spring Harb. Perspect. Biol. 4, a011130 10.1101/cshperspect.a011130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morales C., and Losada A. (2018) Establishing and dissolving cohesion during the vertebrate cell cycle. Curr. Opin. Cell Biol. 52, 51–57 10.1016/j.ceb.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 26. Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A., and Nasmyth K. (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5, 243–254 10.1016/S1097-2765(00)80420-7 [DOI] [PubMed] [Google Scholar]
- 27. Sherwood R., Takahashi T. S., and Jallepalli P. V. (2010) Sister acts: coordinating DNA replication and cohesion establishment. Genes Dev. 24, 2723–2731 10.1101/gad.1976710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Makrantoni V., and Marston A. L. (2018) Cohesin and chromosome segregation. Curr. Biol. 28, R688–R693 10.1016/j.cub.2018.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skibbens R. V., Corson L. B., Koshland D., and Hieter P. (1999) Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13, 307–319 10.1101/gad.13.3.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tóth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., and Nasmyth K. (1999) Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13, 320–333 10.1101/gad.13.3.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skibbens R. V. (2009) Establishment of sister chromatid cohesion. Curr. Biol. 19, R1126–R1132 10.1016/j.cub.2009.10.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Uhlmann F. (2009) A matter of choice: the establishment of sister chromatid cohesion. EMBO Rep. 10, 1095–1102 10.1038/embor.2009.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rolef Ben-Shahar T., Heeger S., Lehane C., East P., Flynn H., Skehel M., and Uhlmann F. (2008) Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321, 563–566 10.1126/science.1157774 [DOI] [PubMed] [Google Scholar]
- 34. Unal E., Heidinger-Pauli J. M., Kim W., Guacci V., Onn I., Gygi S. P., and Koshland D. E. (2008) A molecular determinant for the establishment of sister chromatid cohesion. Science 321, 566–569 10.1126/science.1157880 [DOI] [PubMed] [Google Scholar]
- 35. Zhang J., Shi X., Li Y., Kim B.-J., Jia J., Huang Z., Yang T., Fu X., Jung S. Y., Wang Y., Zhang P., Kim S.-T., Pan X., and Qin J. (2008) Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 31, 143–151 10.1016/j.molcel.2008.06.006 [DOI] [PubMed] [Google Scholar]
- 36. Sutani T., Kawaguchi T., Kanno R., Itoh T., and Shirahige K. (2009) Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr. Biol. 19, 492–497 10.1016/j.cub.2009.01.062 [DOI] [PubMed] [Google Scholar]
- 37. Tanaka K., Yonekawa T., Kawasaki Y., Kai M., Furuya K., Iwasaki M., Murakami H., Yanagida M., and Okayama H. (2000) Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 20, 3459–3469 10.1128/MCB.20.10.3459-3469.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Feytout A., Vaur S., Genier S., Vazquez S., and Javerzat J. P. (2011) Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1. Mol. Cell. Biol. 31, 1771–1786 10.1128/MCB.01284-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hauf S., Roitinger E., Koch B., Dittrich C. M., Mechtler K., and Peters J. M. (2005) Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 3, e69 10.1371/journal.pbio.0030069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quan Y., Xia Y., Liu L., Cui J., Li Z., Cao Q., Chen X. S., Campbell J. L., and Lou H. (2015) Cell-cycle-regulated interaction between Mcm10 and double hexameric Mcm2-7 is required for helicase splitting and activation during S phase. Cell Rep. 13, 2576–2586 10.1016/j.celrep.2015.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu L., Zhang Y., Zhang J., Wang J.-H., Cao Q., Li Z., Campbell J. L., Dong M.-Q., and Lou H. (2019) Characterization of the dimeric CMG/pre-initiation complex and its transition into DNA replication forks. Cell. Mol. Life Sci. 10.1007/s00018-019-03333-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maradeo M. E., and Skibbens R. V. (2010) Epitope tag-induced synthetic lethality between cohesin subunits and Ctf7/Eco1 acetyltransferase. FEBS Lett. 584, 4037–4040 10.1016/j.febslet.2010.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cattoglio C., Pustova I., Walther N., Ho J. J., Hantsche-Grininger M., Inouye C. J., Hossain M. J., Dailey G. M., Ellenberg J., Darzacq X., Tjian R., and Hansen A. S. (2019) Determining cellular CTCF and cohesin abundances to constrain 3D genome models. eLife 8, e40164 10.7554/eLife.40164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mohan C., Kim L. M., Hollar N., Li T., Paulissen E., Leung C. T., and Luk E. (2018) VivosX, a disulfide crosslinking method to capture site-specific, protein-protein interactions in yeast and human cells. eLife 7, e36654 10.7554/eLife.36654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uhlmann F., and Nasmyth K. (1998) Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 8, 1095–1101 10.1016/S0960-9822(98)70463-4 [DOI] [PubMed] [Google Scholar]
- 46. Söderberg O., Leuchowius K. J., Gullberg M., Jarvius M., Weibrecht I., Larsson L. G., and Landegren U. (2008) Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45, 227–232 10.1016/j.ymeth.2008.06.014 [DOI] [PubMed] [Google Scholar]
- 47. Holzmann J., Politi A. Z., Nagasaka K., Hantsche-Grininger M., Walther N., Koch B., Fuchs J., Dürnberger G., Tang W., Ladurner R., Stocsits R. R., Busslinger G. A., Novák B., Mechtler K., Davidson I. F., et al. (2019) Absolute quantification of cohesin, CTCF and their regulators in human cells. eLife 8, e46269 10.7554/eLife.46269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim Y., Shi Z., Zhang H., Finkelstein I. J., and Yu H. (2019) Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang X., Brandão H. B., Le T. B. K., Laub M. T., and Rudner D. Z. (2017) Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355, 524–527 10.1126/science.aai8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kulemzina I., Schumacher M. R., Verma V., Reiter J., Metzler J., Failla A. V., Lanz C., Sreedharan V. T., Rätsch G., and Ivanov D. (2012) Cohesin rings devoid of Scc3 and Pds5 maintain their stable association with the DNA. PLoS Genet. 8, e1002856 10.1371/journal.pgen.1002856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weibrecht I., Leuchowius K. J., Clausson C. M., Conze T., Jarvius M., Howell W. M., Kamali-Moghaddam M., and Söderberg O. (2010) Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev. Proteomics 7, 401–409 10.1586/epr.10.10 [DOI] [PubMed] [Google Scholar]
- 52. Sheu Y. J., and Stillman B. (2006) Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell 24, 101–113 10.1016/j.molcel.2006.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the article.