Abstract
Ethylene is a gaseous phytohormone and the first of this hormone class to be discovered. It is the simplest olefin gas and is biosynthesized by plants to regulate plant development, growth, and stress responses via a well-studied signaling pathway. One of the earliest reported responses to ethylene is the triple response. This response is common in eudicot seedlings grown in the dark and is characterized by reduced growth of the root and hypocotyl, an exaggerated apical hook, and a thickening of the hypocotyl. This proved a useful assay for genetic screens and enabled the identification of many components of the ethylene-signaling pathway. These components include a family of ethylene receptors in the membrane of the endoplasmic reticulum (ER); a protein kinase, called constitutive triple response 1 (CTR1); an ER-localized transmembrane protein of unknown biochemical activity, called ethylene-insensitive 2 (EIN2); and transcription factors such as EIN3, EIN3-like (EIL), and ethylene response factors (ERFs). These studies led to a linear model, according to which in the absence of ethylene, its cognate receptors signal to CTR1, which inhibits EIN2 and prevents downstream signaling. Ethylene acts as an inverse agonist by inhibiting its receptors, resulting in lower CTR1 activity, which releases EIN2 inhibition. EIN2 alters transcription and translation, leading to most ethylene responses. Although this canonical pathway is the predominant signaling cascade, alternative pathways also affect ethylene responses. This review summarizes our current understanding of ethylene signaling, including these alternative pathways, and discusses how ethylene signaling has been manipulated for agricultural and horticultural applications.
Keywords: Arabidopsis thaliana, bioengineering, hormone receptor, phytohormone, plant hormone, signal transduction, signaling, constitutive triple response 1 (CTR1), ethylene, ethylene-insensitive 2 (EIN2)
Introduction
Ethylene (IUPAC name ethene) is the simplest olefin gas and was the first gaseous molecule shown to function as a hormone (1). It is biosynthesized by plants and is well-known to affect various developmental processes, such as seed germination, fruit ripening, senescence, and abscission, as well as responses to various stresses, such as flooding, high salt, and soil compaction (2, 3). The ethylene signal transduction pathway has been extensively studied, in part because ethylene affects so many traits related to plant vigor and post-harvest physiology and storage.
Once biosynthesized, ethylene diffuses throughout the plant and binds to ethylene receptors to stimulate ethylene responses. It can also diffuse to surrounding plants and is the basis of the saying one bad apple spoils the bunch, where ethylene produced by an apple hastens the ripening of bananas. The ethylene-signaling pathway was predominantly delineated with research on Arabidopsis thaliana and is comprised of a combination of components that is not found in other pathways. This review will mainly focus on this research using Arabidopsis. However, it is worth pointing out that similar signaling pathways occur in diverse plants (4–11) so that information from Arabidopsis about ethylene signaling is usually applicable to other species.
Early molecular genetic studies uncovered several key components for ethylene signaling, including a family of receptors; the CTR1 protein kinase; EIN2, which is a transmembrane protein of unknown biochemical activity; and transcription factors, such as EIN3, EILs, and ERFs. This led to a linear, genetic model where, in the absence of ethylene, the receptors activate CTR1, which negatively regulates downstream signaling (Fig. 1). Ethylene functions as an inverse agonist by inhibiting the receptors, leading to release of inhibition by CTR1, resulting in ethylene responses (12). This genetic model provided a general framework that has been refined with further research, resulting in a more complete and detailed model for ethylene signaling, including surprising cases of cross-talk from the receptors to other signaling pathways, details for how a signal perceived at the ER membrane affects transcription in the nucleus, and multiple roles for EIN2. Details from this research have led to various ways to control ethylene signaling. Most of these controls are geared toward inhibiting ethylene responses to prevent post-harvest spoilage. However, there is also a need for stimulating ethylene responses, such as to cause premature germination of parasitic plants so that fields can be cleared of these problematic plants. These discoveries and applications will be summarized in this review.
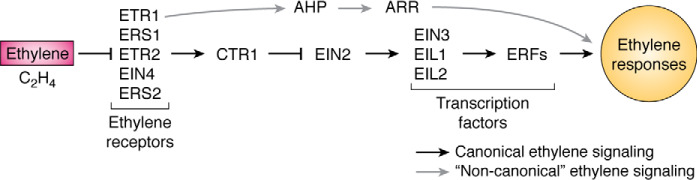
Figure 1.
Simple genetic model of ethylene signaling. In black is shown a model for ethylene signaling based on molecular genetic experiments in Arabidopsis. These experiments showed that ethylene signaling involves ethylene receptors (ETR1, ERS1, ETR2, EIN4, and ERS2), the protein kinase CTR1, and EIN2 that signals to the transcription factors EIN3, EIL1, and EIL2. These, in turn, signal to other transcription factors, such as the ERFs, leading to ethylene responses. This has long been considered the canonical signaling pathway. In this model, CTR1 is a negative regulator of signaling. Ethylene functions as an inverse agonist, where it inhibits the receptors, which leads to lower activity of CTR1 releasing downstream components from inhibition by CTR1. More recent evidence has shown the existence of an alternative, “noncanonical” pathway (in gray), where ETR1 signals to histidine-containing AHPs and then to ARRs to modulate responses to ethylene.
Ethylene-signaling components and the canonical pathway
The first step in ethylene perception is the binding of ethylene to receptors. Ethylene receptors have homology to bacterial two-component receptors that signal via autophosphorylation on a histidine residue followed by phosphotransfer to an aspartate residue in the receiver domain of a response regulator protein (13). Ethylene receptors, as well as other two-component-like receptors, such as the phytochromes and cytokinin receptors, are believed to have been acquired by plants from the cyanobacterium that gave rise to chloroplasts (14–18). Data from a recent phylogenetic analysis suggest a common origin for the ethylene-binding domain in cyanobacteria and plants (19). It is thus interesting to note that ethylene binding has been observed in diverse cyanobacteria, and at least one cyanobacterium, Synechocystis, has a functional ethylene receptor that regulates cell surface properties to affect biofilm formation and phototaxis (20–22). Additionally, ethylene-binding affinities to some of these cyanobacteria and the heterologously expressed Synechocystis ethylene receptor are similar to what has been observed in plants (23), showing a conservation of this domain between these organisms. However, the organism where ethylene receptors first arose remains unknown. The observation that genes encoding for proteins with putative ethylene-binding domains are found in other phyla of bacteria (22) will make answering this question difficult.
By contrast, as will be discussed in more detail below, even though some of the plant ethylene receptor isoforms have retained histidine kinase activity, this activity is not crucial for ethylene perception. This is in contrast to the one cyanobacterial system so far characterized where phosphotransfer is central to the function of the receptor (22, 24, 25). Additionally, some plant ethylene receptor isoforms have serine/threonine kinase activity, indicating that the outputs of these receptors in plants are now diverged from the ancestral proteins. Recent reviews present more information about ethylene receptors in nonplant species (26, 27).
Plants contain multiple ethylene receptor isoforms. Early studies identified ethylene-binding sites in the ER membranes of plants (28, 29), and subsequent research on specific receptor isoforms from various plants confirmed that ethylene receptors are localized to the ER (30–35). In Arabidopsis, five isoforms have been identified and are referred to as ethylene response 1 (ETR1), ethylene response sensor 1 (ERS1), ETR2, ERS2, and EIN4 (36–40). Mutations in any one of these receptors that prevent ethylene binding lead to an ethylene-insensitive plant (12, 20, 36, 37, 41). There are also some mutations in these receptors that have no effect on ethylene binding but prevent signaling through the receptor, which also leads to ethylene insensitivity (20).
The different receptor isoforms in plants have similar domain architecture (Fig. 2) with three transmembrane α-helices at the N terminus, which comprises the ethylene-binding domain, followed by a GAF (cGMP-specific phosphodiesterases, adenylyl cyclases, and FhlA) and kinase domain. Three of the five receptors also contain a receiver domain that is similar to what is found in bacterial two-component receptors (42, 43). The receptors fall into two subfamilies with ETR1 and ERS1 in subfamily 1 and the other three isoforms in subfamily 2 (20). The subfamily 2 receptors contain additional amino acids at the N terminus that are unknown in function. The receptors can be further distinguished by their kinase activity. ETR1 has histidine kinase activity, whereas ETR2, ERS2, and EIN4 have serine/threonine kinase activity, and ERS1 has been documented to have both, depending on assay conditions, although it is believed to be a serine/threonine kinase in vivo (44, 45).
Figure 2.
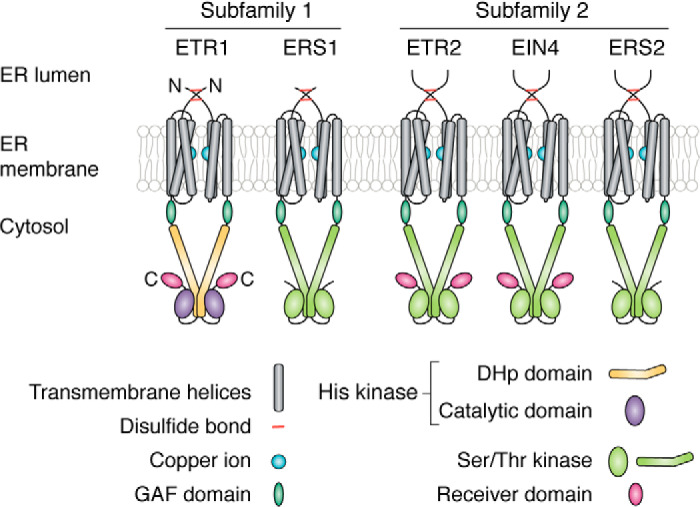
Diagram of domains of receptor isoforms. The receptors are dimers located in the ER membrane. Each dimer is stabilized by two disulfide bonds near the N terminus. All of the receptors contain transmembrane helices that comprise the ethylene-binding domain followed by a GAF and kinase domain. ETR1 is a histidine kinase, and the other four isoforms are serine/threonine kinases. Three of the five contain a receiver domain at the C terminus of the protein. The models for the receptors are based on published structural and computational studies on ETR1 (43, 69), where each monomer coordinates a copper ion required for ethylene binding. In ETR1, the DHp domain of the kinase dimerizes, and a flexible region allows each kinase catalytic domain to associate with the DHp domain. It is unknown whether the kinase domains of the other isoforms also dimerize. The receiver domains are predicted to be orientated away from the central axis of the receptor dimer.
The receptors form homodimers that are stabilized at their N termini by two disulfide bonds (46–48). Nevertheless, these disulfide bonds are necessary neither for binding of ethylene to ETR1 (48) nor for a functional ETR1 receptor in planta (49). In ETR1, it is thought that dimerization between monomers also occurs between the dimerization and histidine phosphotransfer (DHp) domains of each kinase domain (43). It is unclear whether dimerization between kinase domains of the other receptor isoforms occurs. It has also been suggested that heterodimers are possible (35, 50). Evidence that these are receptors is that all of these proteins bind ethylene with high affinity (41, 47, 51, 52), and specific mutations in any one of these proteins lead to ethylene insensitivity (36, 38–40, 53). Similar proteins from tomato also bind ethylene with high affinity and when mutated lead to ethylene insensitivity (51, 54–56).
Ethylene binds to the N-terminal, transmembrane portion of heterologously expressed receptors with Kd values reported in the nanomolar range (21, 41, 52), which corresponds to ethylene-binding affinities reported in plants (57–64). One difference between heterologously expressed receptors and those in planta is that ethylene dissociates from the former with a single, slow rate having a half-time of release of ∼10–12 h (41, 51, 52), whereas there are two rate constants of release in planta (64, 65). In planta, there is an initial, rapid release of ethylene in the first 30 min after ethylene removal, followed by slow release with similar kinetics to the heterologously expressed receptors. Because ethylene can enhance the proteolysis of ethylene receptors (31, 66, 67), this rapid release of ethylene from receptors in plants is likely due to proteolysis of the ethylene-bound receptors.
The cytosolic domains of ETR1 have been structurally characterized (42, 43, 68). This has led to a model of the ETR1 dimer where the DHp domain of the histidine kinase domain dimerizes with the DHp of the other monomer (Fig. 2). In this model, the catalytic domain associates with the DHp domain. The catalytic and receiver domains are modeled to extend outward from the DHp pair. The orientation of the receiver domain in relationship to the remainder of the protein is predicted to be different from prokaryotic histidine kinases, suggesting that this domain may be diverged in function from prokaryotes (68). Additionally, structural studies show that the γ-loop of ETR1, which is part of the catalytic region of receiver domains, is in a different orientation from characterized prokaryote receiver domains (42, 68). No structural information is published characterizing the ethylene-binding domain, but a computational model is available (69). This study coupled with prior research (20) suggests that ethylene binds in the middle of helices 1 and 2 and the signal is transduced via helix 3. The mechanistic details of this transduction through the receptor are unknown.
A key issue in ethylene signaling has been to determine how proteins bind ethylene with high affinity, and mutational studies have identified amino acids in helices 1 and 2 that are important for ethylene binding (20, 21, 41). Based on olefin chemistry, several transition metals were initially suggested as cofactors for binding activity (70–73). It was later determined that ETR1 coordinates copper ions, which act as the cofactor for ethylene binding (21). Cys-65 in helix 2 is required for coordination of copper because the etr1-1 mutant receptor with a C65Y mutation is unable to bind copper or ethylene (21, 36, 37, 41). Mutants such as this render the plant ethylene-insensitive. Additionally, several studies have determined that the ER membrane–localized copper transporter, responsive to antagonist 1 (RAN1), physically interacts with at least some of the receptors and is needed for delivery of copper and proper biogenesis of the ethylene receptors (74–78). Because copper co-purifies with the ETR1 dimer with a 1:1 stoichiometry, it was long thought that each receptor dimer contains one copper ion (21). Recent experimental evidence, however, indicates that there are two copper ions per receptor dimer that are modeled to be coordinated by amino acids in helices 1 and 2 of each monomer (69).
The biochemical output of the receptors has yet to be determined. The GAF, kinase, and receiver domains are the likely output domains, but the specifics of how ethylene signal is transduced are unknown. This is complicated by research showing that even though the receptors have overlapping roles for many traits, for specific traits or under specific conditions, individual receptor isoforms have a role, whereas others do not (52, 79–87). In some cases, individual isoforms display opposite roles from other isoforms. For instance, ETR1 is necessary and sufficient for ethylene-stimulated nutational bending of hypocotyls in dark-grown Arabidopsis seedlings, whereas the other four receptor isoforms inhibit this response (80, 86). Also, loss of ETR1, and to a lesser extent EIN4, results in plants that are less sensitive to the plant hormone abscisic acid (ABA) during seed germination, whereas loss of ETR2 causes plants to be more sensitive to ABA (83, 85). There is recent evidence that ETR1 and ETR2 are signaling independently of CTR1 to cause the changes in ABA responsiveness, but the exact pathway has yet to be determined (84). These observations indicate that there are likely to be differences in the biochemical output between receptor isoforms. Although some of these differences may arise from different kinase specificities (44, 45), this does not easily explain all of these differences.
Ethylene receptors are homologous to bacterial two-component receptors. The simplest bacterial two-component system signals by histidine autophosphorylation followed by relay of the phosphoryl to a conserved aspartate on a receiver domain of a response regulator protein, although more complex variations of this exist (13). Despite the fact that ETR1 possesses histidine kinase activity that is modulated by ethylene (44, 45, 88), this activity is not required for responses to ethylene (89, 90). Rather, it may subtly modulate receptor signaling to downstream components (81, 89, 91–93), including interactions with EIN2 (94). Similarly, receptor serine/threonine kinase activity does not appear to be required for ethylene responses but may have a modulatory role in ethylene receptor signal transduction and responses (95).
Complexes of receptor dimers have been proposed to explain the large range of ethylene concentrations that plants respond to and to explain how one mutant receptor might affect other, nonmutant receptors (48, 49, 96–100). As an example, plants can respond to ethylene at levels down to 0.2 nl/liter (101), which is at least 300-fold below the Kd of binding to the receptors (41). Receptor dimer clusters are proposed as a way for signal amplification to occur, much like how bacterial chemoreceptors function. In chemoreceptors, ligand binding to one receptor dimer can affect the signaling state of neighboring, unbound receptor dimers to increase signal output (102, 103). Structural studies suggest that CTR1 or the receptor receiver domains, or both, may be involved in the formation of ethylene receptor clusters (43, 104). It remains to be determined whether this is important in ethylene signaling.
The receptors also form higher-order complexes with other proteins (48). Specific proteins have been identified as interacting partners with all or a subset of the ethylene receptors. This includes interactions with RAN1 that may be important for correct delivery of copper to the receptors (78). Other interacting partners are less characterized. Reversion to ethylene sensitivity 1 (RTE1) interacts with ETR1 and tetratricopeptide repeat protein 1 (TRP1) with ERS1 to modulate signaling (34, 105–107). A homolog of TRP1 in tomato interacts with both SlETR1 and never ripe (NR or SlETR3) (108). As will be discussed further below, some of the receptors also interact with components of the cytokinin signaling pathway (105, 106, 109–111).
Two proteins, CTR1 and EIN2, are central components of ethylene signaling (112, 113) that physically interact with the receptors (33, 94, 114–118) and each other (119). CTR1 is a serine/threonine protein kinase that functions as a negative regulator of ethylene signaling (113). EIN2 is required for ethylene signaling and is part of the NRAMP (natural resistance-associated microphage protein) family of metal transporters; it is comprised of a large, N-terminal portion containing multiple transmembrane domains in the ER membrane and a cytosolic C-terminal portion (112). In the case of ETR1, the kinase domain of the receptor is required for interactions with both CTR1 and EIN2, although ETR1 histidine kinase activity is only important for modulating interactions with EIN2 (94, 117, 120). These physical interactions appear to be important because mutations in CTR1 that abolish receptor-CTR1 interactions result in a nonfunctional CTR1 (117, 118), and blocking interactions between ETR1 and EIN2 results in ethylene insensitivity (121).
Current models predict that in the absence of ethylene, the ethylene receptors keep CTR1 active (Fig. 3). CTR1 directly phosphorylates EIN2 (119), which may result in EIN2 ubiquitination via an Skp1 Cullen F-box (SCF) E3 ubiquitin ligase complex containing the EIN2-targeting protein 1 (ETP1) and ETP2 F-box proteins and subsequent proteolysis by the 26S proteasome (122), as hypothesized in several studies (119, 123–125). A downstream consequence of this is that the EIN3, EIL1, and EIL2 transcription factors are targeted for ubiquitination by an SCF E3 complex that contains the EBF1 and EBF2 F-box proteins (126–130). The breakdown of these transcription factors prevents ethylene responses. Thus, in the absence of ethylene, signal transduction in the pathway is blocked because EIN2 levels are low.
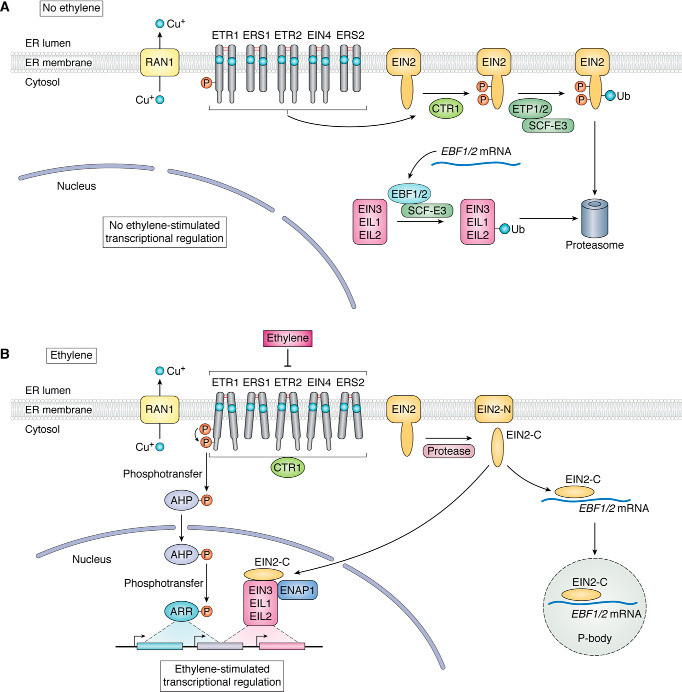
Figure 3.
Model for ethylene signaling. RAN1 is a copper transporter that delivers copper to the lumen of the ER, where it is required for the biogenesis of the receptors and is used as a cofactor by the receptors to bind ethylene. A, in the absence of ethylene, the receptors signal to CTR1, which phosphorylates EIN2. This results in the ubiquitination of EIN2 by an SCF E3 containing the ETP1/2 F-box proteins, leading to EIN2 degradation by the proteasome. Because EIN2 levels are low, an SCF-E3 containing the EBF1/2 F-box proteins ubiquitinates EIN3 and EIL1, leading to their degradation by the proteasome and preventing them from affecting transcription in the nucleus. B, in the presence ethylene, the receptors bind ethylene via a copper cofactor. The binding of ethylene is modeled to cause a conformational change that either reduces CTR1 kinase activity or, as shown, results in CTR1 being sequestered by the receptors so that CTR1 can no longer phosphorylate EIN2. The reduction in EIN2 phosphorylation results in less EIN2 ubiquitination and an increase in EIN2 levels. An unknown protease cleaves EIN2, releasing the C-terminal end (EIN2-C) from the N-terminal end (EIN2-N). One fate of EIN2-C is to bind the RNAs for EBF1 and EBF2 and become sequestered in processing bodies (P-bodies). The reduction of EBF1/2 results in less ubiquitination of EIN3 and EIL1, causing higher EIN3/EIL1 levels. The other fate of EIN2-C is to translocate to the nucleus, where it increases the transcriptional activity of EIN3/EIL1 via ENAP1. This leads to numerous transcriptional changes. In parallel with this pathway, phosphoryl transfer from a conserved histidine in the ETR1 DHp domain to an aspartate in the receiver domain occurs. This is followed by phosphoryl transfer from this residue to AHPs and finally ARRs resulting in transcriptional changes.
In the presence of ethylene, the receptors are inhibited, leading to less phosphorylation of EIN2 by CTR1. Genetic data predict that the binding of ethylene to the receptors should reduce the catalytic activity of CTR1. However, this has not yet been directly tested. Ethylene enhances the interaction between ETR1 and both CTR1 and EIN2 (66, 94, 117). Thus, an alternative explanation for reduced EIN2 phosphorylation by CTR1 is that the binding of ethylene to the receptors results in conformational changes in the receptors that reduces the physical interaction between CTR1 and EIN2, leading to less EIN2 phosphorylation. It is thought that when EIN2 phosphorylation is reduced, there is less EIN2 ubiquitination, resulting in an increase in EIN2 levels and subsequent cleavage of EIN2 by an unknown protease to release the C-terminal portion of EIN2 (EIN2-C) from the membrane-bound N-terminal (EIN2-N) portion (119, 122, 124, 125).
The role of EIN2-N is unknown, but it has diverged from other NRAMP proteins, because no metal transport activity has been detected in heterologously expressed EIN2 and it cannot rescue yeast deficient in metal uptake (107, 112). However, there are hints that EIN2-N has a role in ethylene signaling. In rice, mao huzi 3 (mhz3) mutants are ethylene-insensitive, and the MHZ3 protein physically interacts with OsEIN2-N and regulates OsEIN2 abundance; similar genes have been identified in Arabidopsis that affect ethylene signaling (131, 132). These data indicate the need to further study EIN2-N to delineate the mechanism by which it affects ethylene signaling.
By contrast, EIN2-C has two known roles. One is to bind the mRNAs that encode for EBF1 and EBF2, whereupon this protein/RNA complex associates with processing bodies (133, 134). This results in the degradation of these mRNAs by exoribonuclease 4 (XRN4, also known as EIN5), which is a 5′ → 3′ exoribonuclease known to affect ethylene signaling (133–136). A consequence of the degradation of EBF1 and EBF2 mRNA is that degradation of EIN3 and EIL1 and probably EIL2 is reduced, leading to more ethylene signaling (126, 128, 129). EIN2-C also contains a nuclear localization sequence (NLS). EIN2-C diffuses into the nucleus, where it associates with EIN2 nuclear associated protein 1 (ENAP1), which is required for the ability of EIN2-C to regulate EIN3-dependent transcription (137). Thus EIN2-C provides both transcriptional and translational control to regulate EIN3 and the related EIL1 transcription factor to cause most ethylene responses. This is supported by a recent study where ethylene-stimulated changes in the metabolome did not always correlate with changes in the transcriptome (138). The exception to this model is that short-term, transient responses occur independently of these transcription factors yet require EIN2 (101). Thus, there are more functions for EIN2 that have yet to be discovered.
The increase in EIN3, EIL1, and EIL2 activity caused by EIN2-C leads to changes in the transcription of other ethylene response genes, including other transcription factors, such as the ERFs (139–141). Recent studies have identified histone modifications as having a role in this transcriptional control. Mutational experiments revealed that several histone acetyltransferases and histone deacetylases affect ethylene signaling (142–144). Additionally, research has identified specific histone acetylation marks that are important in ethylene-regulated gene expression by EIN3 (145–147). Even though more details about transcriptional regulation are being discovered, it is also clear from a recent metabolome study that changes in metabolism occur in response to ethylene that are not predicted by changes in the transcriptome (138). This indicates that there is additional regulation for responses to this hormone.
In summary, the model for the canonical ethylene-signaling pathway has developed from a simple genetic model to a more complex model with many more biochemical details. However, there are still gaps in our understanding of this signal transduction pathway.
Noncanonical signaling
The model discussed above is largely linear, and it summarizes the main pathway by which ethylene affects plants. Nonetheless, it is clear from diverse studies that the ethylene-signaling pathway involves feed-forward and feedback regulation leading to sensitization and adaptation (101, 148–160). Most of this research has identified adaptation mechanisms at the level of the receptors. For instance, the levels of the receptors themselves can regulate sensitivity, where higher levels lead to less sensitivity and lower levels to more sensitivity (89, 90, 114, 161–164). However, it is also now clear that other proteins affect sensitivity at the levels of the receptors. This includes negative regulation by RTE1 and the family of proteins called auxin-regulated gene involved in organ size (ARGOS) (149, 154, 165, 166). An RTE1-like protein, green ripe (GR), has a similar role in tomato (166). The exact mechanisms for regulation by these proteins are under investigation. More information about this is contained in a recent review (167).
The existence of nonlinear components to what has been considered the canonical pathway raises the possibility that other ethylene-signaling pathways exist outside of or as branch points from this core pathway. This is an area of active research, and in the cases discussed below, evidence is provided showing that signaling occurs, at least in part, via components not contained in the canonical pathway presented above. These alternative (noncanonical) pathways are not necessary for ethylene responses but appear to have roles in modulating responses to ethylene or in altering responses to other hormones.
Results from several studies have led to the suggestion that the ethylene receptors signal independently of CTR1 or EIN2 (44, 80, 82–85, 168–170). For instance, epistasis analysis has shown that the role of ETR1 and ETR2 in the control of seed germination by ABA is, at least in part, independent of CTR1 (84). It is possible that such alternative signaling occurs via CTR1 homologues, but so far no CTR1 homologue has been identified as being involved in this. Even though ETR1 histidine kinase activity is not required for ethylene signaling, this activity does modulate sensitivity to ethylene, growth recovery kinetics when ethylene is removed, growth of root apical meristem, seed germination under stress conditions or in response to ABA, and interactions with EIN2 (81, 83, 84, 89–91, 93, 94, 111). Likely targets for phosphorelay from ETR1 are components of the cytokinin signaling pathway (Fig. 1). The cytokinin receptors are two-component receptors in plants that, unlike the ethylene receptors, use phosphorelay as the primary route for signaling (18, 171). In this pathway, the phosphoryl is transferred from the cytokinin receptors to histidine-containing phosphotransfer proteins (AHP family in Arabidopsis) and finally to response regulator proteins (ARR family in Arabidopsis) that function as transcription factors. Various studies have demonstrated that ETR1 physically interacts with ARR and AHP proteins (109–111, 172). This interaction involves the C-terminal portion of ETR1 (109, 111). The affinity between ETR1 and AHP1 is altered by their phosphorylation state, where it is highest if one protein is phosphorylated and the other is not (172).
In support of interactions between ETR1 and the cytokinin pathway having functional consequences, mutational analyses revealed that the ARRs are involved in ethylene responses such as sensitivity to ethylene, recovery kinetics after ethylene is removed, stomatal aperture control, and the regulation of root apical meristem (92, 93, 111, 173). Null mutants of ARR1 are less responsive to ethylene, and this appears to depend upon ETR1 histidine kinase activity (93). Similarly, null mutants in several AHPs and ARRs prolong growth recovery when ethylene is removed, similar to what is observed in plants deficient in ETR1 histidine kinase activity (81, 92). Additionally, ETR1 histidine kinase activity is involved in both ethylene- and cytokinin-induced changes in root apical meristem (111). Together, these results are consistent with a model where ETR1 histidine kinase activity is directly involved in affecting components of the cytokinin pathway, resulting in changes in transcription that modulate ethylene responses (Fig. 3). There is some overlap between transcriptional changes caused by ethylene and cytokinin (174), raising the possibility that there are both overlapping and nonoverlapping targets of transcriptional control from this signaling pathway involving ETR1 histidine kinase and the well-known pathway involving EIN3 and EILs. It is interesting to note that in rice, a histidine kinase (MHZ1/OsHK1) that may have a role in cytokinin signaling functions downstream of the OsERS2 ethylene receptor and signals independently of OsEIN2 (175). Thus, our model for canonical ethylene signaling probably needs to be expanded to include secondary pathways such as phosphorelay from some of the ethylene receptors to the AHPs and ARRs.
It should be noted that biochemical experiments show that ETR1 histidine autophosphorylation decreases upon binding of ethylene or ethylene receptor agonists (88, 94), whereas genetic experiments suggest that ethylene leads to more phosphotransfer (81, 89). Histidine kinases can carry out multiple enzymatic reactions, including kinase, phosphatase, and phosphotransfer reactions, and receiver domains can catalyze both phosphotransfer and autodephosphorylation reactions (13, 176). Given this complexity, one possible resolution to this discrepancy between biochemical and genetic data is that histidine autophosphorylation occurs in the absence of ethylene, but phosphotransfer to the receiver domain does not occur until ethylene binds to the receptor to bring the DHp (site of histidine phosphorylation) and receiver domains into the correct orientation. Thus, ethylene may be increasing phosphotransfer through the pathway, causing the steady-state level of ETR1 histidine phosphorylation to decrease. This will only be answered conclusively when we have structural data.
Noncanonical signaling is also likely to occur downstream of the receptors. For instance, PpCTR1 in Physcomitrella patens has a role in both ethylene and ABA signal transduction, raising the possibility that CTR1 has more functions than simply phosphorylating EIN2 (177). Also, mutants of EIN2 have altered responses to various hormones (reviewed in Ref. 178), but whether this reflects alternative signaling from EIN2 or is due to many pathways converging on EIN2 has yet to be completely explored.
The signaling pathway downstream of EIN2 is complex because it involves at least two levels of transcriptional regulation. Because of this, it is harder to distinguish “canonical” from “noncanonical” signaling. EIN3 is the transcription factor with the largest role in ethylene signaling (128, 139), and it homodimerizes to interact with its target DNA (141). However, environmental factors such as dark versus light or the presence of other hormones can affect this so that, depending on conditions, EIN3 interacts with other transcription factors, leading to outputs not predicted by the common ethylene-signaling models (179–181). As an example, ethylene is well-known for inhibiting hypocotyl growth in dark-grown eudicot seedlings (36, 182) and stimulating hypocotyl growth in the light (183–187). In the dark, EIN3 directly interacts with another transcription factor, phytochrome-interacting factor 3 (PIF3), forming an output module distinct from either transcription factor alone (181). A recent meta-analysis of transcriptomic data sets comparing ethylene-responsive genes in the light versus the dark uncovered a set of genes that were similarly regulated in both conditions, but also many that were differentially regulated (188). It will be interesting to determine which of these differentially regulated transcripts are controlled by this EIN3/PIF3 module.
The above summarizes evidence that specific ethylene receptor isoforms signal to affect other hormone pathways, such as cytokinin and ABA. The exact pathways for this have yet to be delineated, but it appears that at least some of these roles are independent of CTR1. This also raises the interesting possibility that the ethylene receptors are affecting other signaling pathways via yet to be discovered mechanisms. Additionally, environmental factors can affect the output of this pathway, adding an additional layer of complexity to understanding all of the nuances of how ethylene signaling occurs and how we can manipulate this signaling.
Regulating ethylene signal transduction for agricultural and horticultural uses
As can be seen from the information provided above, our understanding about the signaling pathways for the perception of ethylene has grown and become increasingly complicated. This increased complexity provides challenges in determining how to modulate responses to ethylene for commercial purposes, but it also provides opportunities to perhaps modulate specific responses without off-target outcomes.
It is likely that signaling pathways comparable with those outlined above in Arabidopsis also occur in most land plant species because similar genes have been uncovered in diverse plants, including rice, tomato, strawberry, the clubmoss Selaginella moellendorffi, and the moss P. patens (4–11). However, it is important to keep in mind that there are also likely to be variations on this general signaling pathway that occur from species to species that need to be taken into account when trying to manipulate ethylene responses. Because ethylene affects many processes that are important in horticulture and agriculture, a great deal of research has used the information outlined above to develop ways to regulate ethylene signal transduction. Even though ethylene itself is used for some applications, such as to cause uniform fruit ripening, most applications involve minimizing ethylene signaling. These approaches have generally been either genetic or chemical in nature.
Early attempts to bioengineer plants that do not respond to ethylene involved the heterologous expression of the Arabidopsis etr1-1 gene, which, as mentioned above, contains a mutation that leads to ethylene insensitivity in Arabidopsis. Heterologous etr1-1 expression leads to ethylene insensitivity and reduced flower senescence and longer vase life in several plant species, delayed fruit ripening in tomato and melon, and altered regeneration in lettuce leaf explants (189–197). It is likely that any ethylene-insensitive receptor transgene will have similar outcomes because Nemesia strumosa flower life was extended when heterologously expressing a cucumber etr1-1 homolog (198). A drawback of constitutive expression of ethylene receptor mutants that cause ethylene insensitivity is unintended effects that can have adverse agricultural and horticultural outcomes. These adverse outcomes include increased stress in tomato plants; increased pathogen susceptibility in tobacco; reduction in seed germination, pollen viability, number of adventitious roots, and root performance in petunias; and reduced femaleness in melon flowers (194, 195, 199–203). These unwanted effects reduce the efficacy of this approach for commercial use.
One potential way around this is to target etr1-1 or another similar receptor mutant that causes ethylene insensitivity to tissues of interest. For instance, flower-specific expression of etr1-1 reduced flower senescence and increased flower life of two plant species (189, 190, 204). A potential problem with this approach is that ethylene insensitivity can lead to increased biosynthesis of ethylene (36), which in turn could affect tissues not expressing the mutant receptor (194). Another way to address these issues is to use an inducible promoter for heterologous expression of the mutant receptor. Relevant to this is the observation that some ethylene receptors are ethylene-inducible, including ETR2 from Arabidopsis and NR from tomato (51, 54). Both etr2-1 and nr mutants contain point mutations that result in ethylene-insensitive plants with long-term ethylene treatments (40, 54). However, both show a transient response to ethylene and only become insensitive to ethylene when levels of the mutant receptor increase due to increased ethylene levels (205). Thus, controlling mutant receptor expression with inducible heterologous gene expression could provide control over both the timing and amount of expression. This has been used in tomato to delay ripening (206), but it remains to be determined whether or not this reduces the severity of unwanted effects from the transgene.
Another alternative is to find mutants in other genes that affect ethylene signaling. For instance, down-regulation of SlEIN2 in tomato results in inhibition of ripening (207, 208). One ethylene-signaling mutant that alters ripening is in the GR gene in tomato, which has homology to RTE1 in Arabidopsis (165, 166). A drawback is that it requires overexpression of GR to inhibit ripening in tomato (209), leading to issues similar to those outlined above for heterologous expression of genes. Fruit ripening, like other developmental processes, is complex and is regulated by a network of transcription factors (210). Thus, to avoid unwanted effects of mutations, it may be necessary to target specific transcription factors for mutagenesis to regulate specific traits affected by ethylene, without altering other responses to this hormone. For instance, virus-induced gene silencing of SlEIN3 leads to delayed tomato fruit ripening, but no other traits were analyzed to determine whether there were detrimental outcomes (211). This will require more research to link specific transcription factors with specific ethylene-related traits.
Research has also focused on developing chemicals that can regulate ethylene signal transduction. Silver has long been known to block ethylene responses in plants (73). Silver ions are larger than copper ions (212–214) yet support ethylene binding to heterologously expressed ETR1 (21, 215). This led to an early hypothesis that silver ions replace the copper in the ethylene binding site of the receptors, allowing for ethylene binding but preventing stimulus-response coupling through the receptors because of steric effects (21, 99, 215, 216). This model may be incorrect because silver largely functions via the subfamily I receptors, in particular ETR1 (49, 52, 114). Also, silver functions as a noncompetitive inhibitor, suggesting that it binds to a site other than the ethylene-binding site to inhibit the receptor (52, 73), although it is possible that silver has the characteristics of a noncompetitive inhibitor yet acts at the ethylene-binding site (217). Even though silver ions are effective at blocking ethylene responses in plants, the adverse human health and environmental effects of silver limit its use. Additionally, silver has off-target effects, such as altering auxin transport (218).
Because of this, other compounds have been developed. Strained alkenes such as cyclic olefins can inhibit ethylene binding and action (219), and they have been studied for commercial use (for examples, see Fig. 4). They have also been used to characterize the ethylene-binding site of the receptors. For instance, even though ethylene is a symmetric molecule, the use of different enantiomers of trans-cyclooctene, a competitive antagonist of ethylene receptors, showed that the ethylene-binding site is asymmetric (220). Of these cyclic olefins, 1-methylcyclopropene (1-MCP) has a high binding affinity to the ethylene receptors and has been patented (221–223). Even though it is gaseous, it has become commercially successful because a solid formulation was developed where 1-MCP is released when the formulation is dissolved in water. This effectively blocks ethylene responses and is currently used to prolong the storage life of a variety of produce (224). Because the active component is a gas, its use is generally limited to enclosed spaces, such as for post-harvest storage.
Figure 4.
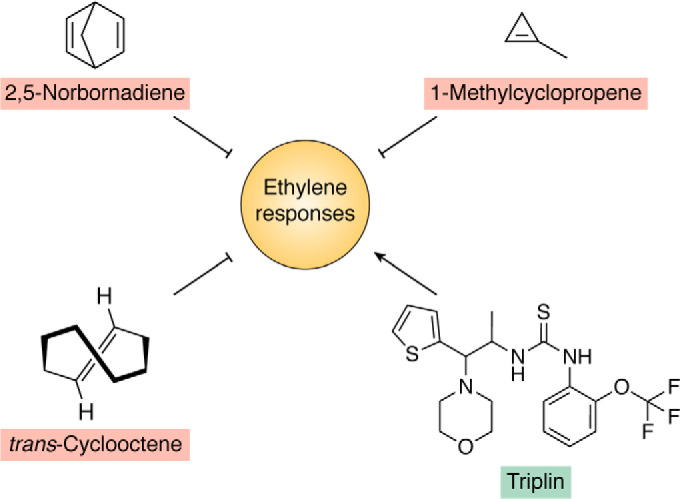
Chemicals that affect ethylene responses in plants. Many strained alkenes, such as 2,5-norbornadiene, trans-cyclooctene, and 1-methylcyclopropene, have been demonstrated to be effective antagonists of ethylene responses that function on the ethylene receptors. Other compounds, such as triplin, are agonists of ethylene responses. Triplin is believed to function by altering the delivery of copper ions to the receptors.
Because gaseous compounds cannot easily be used in open space applications, such as open fields, research has focused on finding liquid agonists and antagonists of ethylene receptors that can be used in open locations. Using a chemical genetics approach, several such compounds have been identified (225–227). One of these compounds, triplin (Fig. 4), mimics the effects of ethylene and was used to help identify the protein antioxidant protein 1 (ATX1) as a key transporter of copper to RAN1 (227).
Investigations using details about ethylene signaling, such as receptor-protein interactions and copper as a cofactor for ethylene binding, have also resulted in interesting compounds. One such compound is NOP-1, a synthetic octapeptide that was developed based on details about ETR1-EIN2 interactions (121). This peptide corresponds to the NLS in EIN2-C and disrupts the interaction between EIN2 and ETR1 in Arabidopsis as well as interactions between SlETR1 and SlEIN2 in tomato (121, 228). NOP-1 binds to various ethylene receptors, including ETR1 from Arabidopsis, NR and SlETR4 from tomato, and DcETR1 from carnation (229–231). Importantly, NOP-1 leads to reduced ethylene sensitivity in various plants and has been shown to delay tomato fruit ripening and carnation flower senescence (121, 230). Also important is that it can achieve these effects by surface application to the plants. Because the NLS in EIN2s is conserved across many flowering ornamental species (229), it is very likely that NOP-1 and derivatives will be effective at blocking ethylene responses in most, if not all, plants used in agriculture and horticulture.
There is also interest in applying ethylene or ethylene response agonists to open fields. Even though this may seem counterintuitive because of the adverse agricultural effects this could have (such as increased senescence and abscission), there is strong interest in such compounds as a way to control parasitic weeds, such as species of Striga. Striga is an obligate parasitic plant that is estimated to cause billions of dollars of crop damage annually and can result in 100% crop loss in many parts of sub-Saharan Africa (232, 233). Striga germinates when other plants germinate nearby, and one of the major cues for this is ethylene produced by the host plant, although it is unclear whether this is true for all parasitic weeds (234–240). A strategy being explored to control this weed is to stimulate seed germination in the absence of a host in a process termed suicidal germination, because the parasite cannot survive without a host plant (233, 235, 238, 241–244). Ethylene gas was successfully used for this purpose in the United States in the 1960s, where soil contaminated with Striga seeds was fumigated with ethylene to stimulate germination of the Striga seeds in the absence of a host needed for survival (235). This has also been shown to work to varying degrees in Africa (245, 246). Unfortunately, fumigating with ethylene is not a good solution in sub-Saharan Africa where this weed is a severe problem, because the farmers cannot afford the expensive equipment needed for fumigating soil. Therefore, alternative, less expensive, and more easily deployed approaches need to be developed. One approach that was developed is the use of ethylene-producing bacteria to stimulate germination of Striga (247). Alternatively, application of ethylene-releasing agents or compounds that stimulate ethylene biosynthesis by Striga seeds have been shown to increase Striga seed germination (239, 248, 249). However, these approaches are either cost-prohibitive or less effective, so low-cost and effective measures still need to be developed to control parasitic weeds.
Concluding remarks
The details about ethylene signal transduction provided in this review illustrate that we now know many important aspects of how plants perceive ethylene. This includes a new appreciation that the ethylene receptors signal via alternative pathways, in addition to the canonical pathway that was originally delineated in genetic screens. Nonetheless, there are clearly gaps in our understanding of this pathway with unanswered questions.
Despite decades of research on the ethylene receptors, it is still not known what conformational changes occur when the receptors bind ethylene and what enzymatic activity or receptor-protein interaction is modulated by this binding event. Determining the output of the receptors is complicated by the fact that the receptor isoforms have both overlapping and nonoverlapping roles, indicating that output from the receptors is not entirely redundant. We also do not know whether we have uncovered all instances of cross-talk from the receptors to other pathways, and we still lack fundamental details about the cross-talk that has been discovered. These details may prove crucial to develop better-targeted control of ethylene responses. Despite recent advances in understanding EIN2, there are still open questions about this protein: What is the role of the N-terminal, transmembrane portion of EIN2? What additional roles does EIN2-C have? Given that EIN2 is a central regulator for ethylene signal transduction, such details are also important as we develop new methods of controlling ethylene responses. It is clear that transcriptional regulation by application of ethylene is influenced by environmental conditions. Thus, there is still a great deal we need to examine regarding transcriptional networks that are influenced by ethylene and the factors that affect these networks. Without this information, targeting specific transcription factors may have unintended outcomes, depending on environmental conditions.
No doubt, as we obtain answers to these and other questions, we will develop new methods to control responses to ethylene for agricultural and horticultural uses that will have fewer unwanted, off-target effects that decrease plant vigor and post-harvest storage. Such an improvement in methods will require more specific targeting (with either chemicals or genetic modification) that will only come with further research on this pathway.
Acknowledgments
I thank Caren Chang, Dan Roberts, Anna Stepanova, and Gyeongmee Yoon for helpful conversations and insights.
Funding and additional information—This work was supported by National Science Foundation Grants MCB-1716279, MCB-1817304, and IOS-1855066.
Conflict of interest—The author declares that he has no conflicts of interest with the contents of this article.
- CTR
- constitutive triple response
- ABA
- abscisic acid
- AHP
- Arabidopsis histidine-containing phosphotransfer protein
- ARGOS
- auxin-regulated gene involved in organ size
- ARR
- Arabidopsis response regulator
- ATX1
- antioxidant protein 1
- DHp
- dimerization and histidine phosphotransfer
- EBF
- EIN3-binding F-box
- EIL
- EIN3-like
- EIN
- ethylene-insensitive
- ENAP1
- EIN2 nuclear associated protein 1
- ERF
- ethylene response factor
- ERS
- ethylene response sensor
- ETP
- EIN2-targeting protein
- ETR
- ethylene response
- GAF
- cGMP-specific phosphodiesterases adenylyl cyclases, and FhlA
- GR
- green ripe
- 1-MCP
- 1-methylcyclopropene
- NLS
- nuclear localization sequence
- NR
- never ripe
- NRAMP
- natural resistance-associated macrophage proteins
- PIF3
- phytochrome-interacting factor 3
- RAN1
- responsive to antagonist 1
- RTE1
- reversion to ethylene sensitivity 1
- SCF
- Skp1 Cullen F-box
- TRP1
- tetratricopeptide repeat protein 1
- XRN4
- exoribonuclease 4.
References
- 1. Bakshi A., Shemansky J. M., Chang C., and Binder B. M. (2015) History of research on the plant hormone ethylene. J. Plant Growth Regul. 34, 809–827 10.1007/s00344-015-9522-9 [DOI] [Google Scholar]
- 2. Abeles F., Morgan P., and Saltveit M. J. (1992) Ethylene in Plant Biology, 2nd Ed., Academic Press, San Diego, CA [Google Scholar]
- 3. Mattoo A. K., and Suttle J. C. (1991) The Plant Hormone Ethylene, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 4. Banks J. A., Nishiyama T., Hasebe M., Bowman J. L., Gribskov M., dePamphilis C., Albert V. A., Aono N., Aoyama T., Ambrose B. A., Ashton N. W., Axtell M. J., Barker E., Barker M. S., Bennetzen J. L., et al. (2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 10.1126/science.1203810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rensing S. A., Lang D., Zimmer A. D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P. F., Lindquist E. A., Kamisugi Y., Tanahashi T., Sakakibara K., Fujita T., Oishi K., Shin-I T., et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 10.1126/science.1150646 [DOI] [PubMed] [Google Scholar]
- 6. Ju C., Van de Poel B., Cooper E. D., Thierer J. H., Gibbons T. R., Delwiche C. F., and Chang C. (2015) Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants 1, 14004 10.1038/nplants.2014.4 [DOI] [PubMed] [Google Scholar]
- 7. Gallie D. (2015) Appearance and elaboration of the ethylene receptor family during land plant evolution. Plant Mol. Biol. 87, 521–539 10.1007/s11103-015-0296-z [DOI] [PubMed] [Google Scholar]
- 8. Rzewuski G., and Sauter M. (2008) Ethylene biosynthesis and signaling in rice. Plant Sci. 175, 32–42 10.1016/j.plantsci.2008.01.012 [DOI] [Google Scholar]
- 9. Ma B., Chen S. Y., and Zhang J. S. (2010) Ethylene signaling in rice. Chin. Sci. Bull. 55, 2204–2210 10.1007/s11434-010-3192-2 [DOI] [Google Scholar]
- 10. Klee H. J., and Giovannoni J. J. (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59 10.1146/annurev-genet-110410-132507 [DOI] [PubMed] [Google Scholar]
- 11. Shulaev V., Sargent D. J., Crowhurst R. N., Mockler T. C., Folkerts O., Delcher A. L., Jaiswal P., Mockaitis K., Liston A., Mane S. P., Burns P., Davis T. M., Slovin J. P., Bassil N., Hellens R. P., et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43, 109–116 10.1038/ng.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hall A. E., Chen Q. G., Findell J. L., Schaller G. E., and Bleecker A. B. (1999) The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol. 121, 291–300 10.1104/pp.121.1.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao R., and Stock A. M. (2009) Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63, 133–154 10.1146/annurev.micro.091208.073214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kehoe D. M., and Grossman A. R. (1996) Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273, 1409–1412 10.1126/science.273.5280.1409 [DOI] [PubMed] [Google Scholar]
- 15. Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., Leister D., Stoebe B., Hasegawa M., and Penny D. (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12246–12251 10.1073/pnas.182432999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mount S. M., and Chang C. (2002) Evidence for a plastid origin of plant ethylene receptor genes. Plant Physiol. 130, 10–14 10.1104/pp.005397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Timmis J. N., Ayliffe M. A., Huang C. Y., and Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 10.1038/nrg1271 [DOI] [PubMed] [Google Scholar]
- 18. Schaller G. E., Shiu S.-H., and Armitage J. P. (2011) Two-component systems and their co-option for eukaryotic signal transduction. Curr. Biol. 21, R320–R330 10.1016/j.cub.2011.02.045 [DOI] [PubMed] [Google Scholar]
- 19. Hérivaux A., Dugé de Bernonville T., Roux C., Clastre M., Courdavault V., Gastebois A., Bouchara J.-P., James T. Y., Latgé J.-P., Martin F., and Papon N. (2017) The identification of phytohormone receptor homologs in early diverging fungi suggests a role for plant sensing in land colonization by fungi. mBio 8, e01739–16 10.1128/mBio.01739-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang W., Esch J. J., Shiu S. H., Agula H., Binder B. M., Chang C., Patterson S. E., and Bleecker A. B. (2006) Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell 18, 3429–3442 10.1105/tpc.106.044537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodríguez F. I., Esch J. J., Hall A. E., Binder B. M., Schaller G. E., and Bleecker A. B. (1999) A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283, 996–998 10.1126/science.283.5404.996 [DOI] [PubMed] [Google Scholar]
- 22. Lacey R. F., and Binder B. M. (2016) Ethylene regulates the physiology of the cyanobacterium Synechocystis sp. PCC 6803 via an ethylene receptor. Plant Physiol 171, 2798–2809 10.1104/pp.16.00602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allen C. J., Lacey R. F., Binder Bickford A. B., Beshears C. P., Gilmartin C. J., and Binder B. M. (2019) Cyanobacteria respond to low levels of ethylene. Front. Plant Sci. 10, 950 10.3389/fpls.2019.00950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song J.-Y., Cho H. S., Cho J.-I., Jeon J.-S., Lagarias J. C., and Park Y.-I. (2011) Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. U.S.A. 108, 10780–10785 10.1073/pnas.1104242108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Narikawa R., Suzuki F., Yoshihara S., Higashi S., Watanabe M., and Ikeuchi M. (2011) Novel photosensory two-component system (PixA-NixB-NixC) involved in the regulation of positive and negative phototaxis of cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 52, 2214–2224 10.1093/pcp/pcr155 [DOI] [PubMed] [Google Scholar]
- 26. Carlew T. S., Allen C. J., and Binder B. M. (2019) Ethylene receptors in nonplant species. Small Methods 10.1002/smtd.201900266 [DOI] [Google Scholar]
- 27. Papon N., and Binder B. M. (2019) An Evolutionary perspective on ethylene sensing in microorganisms. Trends Microbiol. 27, 193–196 10.1016/j.tim.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 28. Evans D. E., Bengochea T., Cairns A. J., Dodds J. H., and Hall M. A. (1982) Studies on ethylene binding by cell-free preparations from cotyledons of Phaseolus vulgaris L.: subcellular localization. Plant Cell Environ. 5, 101–107 10.1111/1365-3040.ep11588010 [DOI] [Google Scholar]
- 29. Evans D. E., Dodds J. H., Lloyd P. C., Apgwynn I., and Hall M. A. (1982) A study of the subcellular localisation of an ethylene binding site in developing cotyledons of Phaseolus vulgaris L. by high resolution autoradiography. Planta 154, 48–52 10.1007/BF00385495 [DOI] [PubMed] [Google Scholar]
- 30. Chen Y.-F., Randlett M. D., Findell J. L., and Schaller G. E. (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J. Biol. Chem. 277, 19861–19866 10.1074/jbc.M201286200 [DOI] [PubMed] [Google Scholar]
- 31. Chen Y.-F., Shakeel S. N., Bowers J., Zhao X.-C., Etheridge N., and Schaller G. E. (2007) Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. J. Biol. Chem. 282, 24752–24758 10.1074/jbc.M704419200 [DOI] [PubMed] [Google Scholar]
- 32. Ma B., Cui M.-L., Sun H.-J., Takada K., Mori H., Kamada H., and Ezura H. (2006) Subcellular localization and membrane topology of the melon ethylene receptor CmERS1. Plant Physiol. 141, 587–597 10.1104/pp.106.080523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhong S., Lin Z., and Grierson D. (2008) Tomato ethylene receptor-CTR interactions: visualization of NEVER-RIPE interactions with multiple CTRs at the endoplasmic reticulum. J. Exp. Bot. 59, 965–972 10.1093/jxb/ern021 [DOI] [PubMed] [Google Scholar]
- 34. Dong C.-H., Rivarola M., Resnick J. S., Maggin B. D., and Chang C. (2008) Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 53, 275–286 10.1111/j.1365-313X.2007.03339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grefen C., Städele K., Rz̊icka K., Obrdlik P., Harter K., and Horák J. (2008) Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol. Plant 1, 308–320 10.1093/mp/ssm015 [DOI] [PubMed] [Google Scholar]
- 36. Bleecker A. B., Estelle M. A., Somerville C., and Kende H. (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089 10.1126/science.241.4869.1086 [DOI] [PubMed] [Google Scholar]
- 37. Chang C., Kwok S. F., Bleecker A. B., and Meyerowitz E. M. (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262, 539–544 10.1126/science.8211181 [DOI] [PubMed] [Google Scholar]
- 38. Hua J., Chang C., Sun Q., and Meyerowitz E. M. (1995) Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269, 1712–1714 10.1126/science.7569898 [DOI] [PubMed] [Google Scholar]
- 39. Hua J., Sakai H., Nourizadeh S., Chen Q. G., Bleecker A. B., Ecker J. R., and Meyerowitz E. M. (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10, 1321–1332 10.1105/tpc.10.8.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sakai H., Hua J., Chen Q. G., Chang C., Medrano L. J., Bleecker A. B., and Meyerowitz E. M. (1998) ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 5812–5817 10.1073/pnas.95.10.5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schaller G. E., and Bleecker A. B. (1995) Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270, 1809–1811 10.1126/science.270.5243.1809 [DOI] [PubMed] [Google Scholar]
- 42. Müller-Dieckmann H.-J., Grantz A. A., and Kim S.-H. (1999) The structure of the signal receiver domain of the Arabidopsis thaliana ethylene receptor ETR1. Structure 7, 1547–1556 10.1016/S0969-2126(00)88345-8 [DOI] [PubMed] [Google Scholar]
- 43. Mayerhofer H., Panneerselvam S., Kaljunen H., Tuukkanen A., Mertens H. D. T., and Mueller-Dieckmann J. (2015) Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1). J. Biol. Chem. 290, 2644–2658 10.1074/jbc.M114.587667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gamble R. L., Coonfield M. L., and Schaller G. E. (1998) Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 7825–7829 10.1073/pnas.95.13.7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moussatche P., and Klee H. J. (2004) Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J. Biol. Chem. 279, 48734–48741 10.1074/jbc.M403100200 [DOI] [PubMed] [Google Scholar]
- 46. Schaller G. E., Ladd A. N., Lanahan M. B., Spanbauer J. M., and Bleecker A. B. (1995) The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J. Biol. Chem. 270, 12526–12530 10.1074/jbc.270.21.12526 [DOI] [PubMed] [Google Scholar]
- 47. Hall A. E., Findell J. L., Schaller G. E., Sisler E. C., and Bleecker A. B. (2000) Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 123, 1449–1458 10.1104/pp.123.4.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen Y.-F., Gao Z., Kerris R. J. 3rd, Wang W., Binder B. M., and Schaller G. E. (2010) Ethylene receptors function as components of high-molecular-mass protein complexes in Arabidopsis. PLoS ONE 5, e8640 10.1371/journal.pone.0008640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xie F., Liu Q., and Wen C.-K. (2006) Receptor signal output mediated by the ETR1 N-terminus is primarily subfamily I receptor dependent. Plant Physiol. 142, 492–508 10.1104/pp.106.082628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berleth M., Berleth N., Minges A., Hänsch S., Burkart R. C., Stork B., Stahl Y., Weidtkamp-Peters S., Simon R., and Groth G. (2019) Molecular analysis of protein-protein interactions in the ethylene pathway in the different ethylene receptor subfamilies. Front. Plant Sci. 10, 726 10.3389/fpls.2019.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. O'Malley R. C., Rodriguez F. I., Esch J. J., Binder B. M., O'Donnell P., Klee H. J., and Bleecker A. B. (2005) Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J. 41, 651–659 10.1111/j.1365-313X.2004.02331.x [DOI] [PubMed] [Google Scholar]
- 52. McDaniel B. K., and Binder B. M. (2012) Ethylene receptor 1 (ETR1) is sufficient and has the predominant role in mediating inhibition of ethylene responses by silver in Arabidopsis thaliana. J. Biol. Chem. 287, 26094–26103 10.1074/jbc.M112.383034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen Q. G., and Bleecker A. B. (1995) Analysis of ethylene signal-transduction kinetics associated with seedling-growth response and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiol. 108, 597–607 10.1104/pp.108.2.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wilkinson J. Q., Lanahan M. B., Yen H.-C., Giovannoni J. J., and Klee H. J. (1995) An ethylene-inducible component of signal transduction encoded by Never-ripe. Science 270, 1807–1809 10.1126/science.270.5243.1807 [DOI] [PubMed] [Google Scholar]
- 55. Okabe Y., Asamizu E., Ariizumi T., Shirasawa K., Tabata S., and Ezura H. (2012) Availability of Micro-Tom mutant library combined with TILLING in molecular breeding of tomato fruit shelf-life. Breed. Sci. 62, 202–208 10.1270/jsbbs.62.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen Y., Hu G., Rodriguez C., Liu M., Binder B. M., and Chervin C. (2020) Role of SlETR7, a newly discovered ethylene receptor, in tomato plant and fruit development. Hortic. Res. 7, 17 10.1038/s41438-020-0239-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sisler E. C. (1979) Measurement of ethylene binding in plant tissue. Plant Physiol. 64, 538–542 10.1104/pp.64.4.538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blankenship S. M., and Sisler E. C. (1989) Ethylene binding changes in apple and morning glory during ripening and senescence. J. Plant Growth Reg. 8, 37–44 10.1007/BF02024924 [DOI] [Google Scholar]
- 59. Blankenship S. M., and Sisler E. C. (1993) Ethylene binding site affinity in ripening apples. J. Amer. Soc. Hortic. Sci. 118, 609–612 10.21273/JASHS.118.5.609 [DOI] [Google Scholar]
- 60. Sanders I. O., Ishizawa K., Smith A. R., and Hall M. A. (1990) Ethylene binding and action in rice seedlings. Plant Cell Physiol. 31, 1091–1099 10.1093/oxfordjournals.pcp.a078021 [DOI] [Google Scholar]
- 61. Smith A. R., Robertson D., Sanders I. O., Williams R. A. N., and Hall M. A. (1987) Ethylene binding sites. in Plant Hormone Receptors (Klambt D., ed) pp. 229–238, Springer-Verlag, Berlin [Google Scholar]
- 62. Sisler E. C., Reid M. S., and Yang S. F. (1986) Effect of antagonists of ethylene action on binding of ethylene in cut carnations. Plant Growth Reg. 4, 213–218 10.1007/BF00028164 [DOI] [Google Scholar]
- 63. Goren R., and Sisler E. C. (1986) Ethylene-binding characteristics in phaseolus, citrus, and ligustrum plants. Plant Growth Regul. 4, 43–54 10.1007/BF00025348 [DOI] [Google Scholar]
- 64. Sanders I. O., Harpham N. V. J., Raskin I., Smith A. R., and Hall M. A. (1991) Ethylene binding in wild type and mutant Arabidopsis thaliana (L.) Heynh. Ann. Bot. 68, 97–103 10.1093/oxfordjournals.aob.a088242 [DOI] [Google Scholar]
- 65. Sisler E. C. (1991) Ethylene-binding components in plants. in The Plant Hormone Ethylene (Mattoo A. K., and Suttle J. C. eds) pp. 81–99, CRC Press, Inc., Baca Raton, FL [Google Scholar]
- 66. Shakeel S. N., Gao Z., Amir M., Chen Y.-F., Rai M. I., Haq N. U., and Schaller G. E. (2015) Ethylene regulates levels of ethylene receptor/CTR1 signaling complexes in Arabidopsis thaliana. J. Biol. Chem. 290, 12415–12424 10.1074/jbc.M115.652503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kevany B. M., Tieman D. M., Taylor M. G., Cin V. D., and Klee H. J. (2007) Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 51, 458–467 10.1111/j.1365-313X.2007.03170.x [DOI] [PubMed] [Google Scholar]
- 68. Hung Y.-L., Jiang I., Lee Y.-Z., Wen C.-K., and Sue S.-C. (2016) NMR study reveals the receiver domain of Arabidopsis ETHYLENE RESPONSE1 ethylene receptor as an atypical type response regulator. PLoS ONE 11, e0160598 10.1371/journal.pone.0160598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schott-Verdugo S., Müller L., Classen E., Gohlke H., and Groth G. (2019) Structural model of the ETR1 ethylene receptor transmembrane sensor domain. Sci. Rep. 9, 8869 10.1038/s41598-019-45189-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thompson J., Harlow R., and Whitney J. (1983) Copper(I)-olefin complexes: support for the proposed role of copper in the ethylene effect in plants. J. Am. Chem. Soc. 105, 3522–3527 10.1021/ja00349a026 [DOI] [Google Scholar]
- 71. Burg S. P., and Burg E. A. (1967) Molecular requirements for the biological activity of ethylene. Plant Physiol. 42, 144–152 10.1104/pp.42.1.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sisler E. C. (1976) Ethylene activity of some π-acceptor compounds. Tob. Sci. 21, 43–45 [Google Scholar]
- 73. Beyer E. M. (1976) A potent inhibitor of ethylene action in plants. Plant Physiol. 58, 268–371 10.1104/pp.58.3.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hirayama T., Kieber J. J., Hirayama N., Kogan M., Guzman P., Nourizadeh S., Alonso J. M., Dailey W. P., Dancis A., and Ecker J. R. (1999) RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97, 383–393 10.1016/S0092-8674(00)80747-3 [DOI] [PubMed] [Google Scholar]
- 75. Woeste K. E., and Kieber J. J. (2000) A strong loss-of-function mutation in RAN1 results in constitutive activation of the ethylene response pathway as well as a Rosette-lethal phenotype. Plant Cell 12, 443–455 10.1105/tpc.12.3.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Himelblau E., and Amasino R. M. (2001) Nutrients mobilized from leaves during leaf senescence. J. Plant Physiol. 158, 1317–1323 10.1078/0176-1617-00608 [DOI] [Google Scholar]
- 77. Binder B. M., Rodríguez F. I., and Bleecker A. B. (2010) The copper transporter RAN1 is essential for biogenesis of ethylene receptors in Arabidopsis. J. Biol. Chem. 285, 37263–37270 10.1074/jbc.M110.170027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hoppen C., Müller L., Hänsch S., Uzun B., Milić D., Meyer A. J., Weidtkamp-Peters S., and Groth G. (2019) Soluble and membrane-bound protein carrier mediate direct copper transport to the ethylene receptor family. Sci. Rep. 9, 10715 10.1038/s41598-019-47185-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu Q., Xu C., and Wen C.-K. (2010) Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Biol. 10, 60 10.1186/1471-2229-10-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Binder B. M., O'Malley R. C., Wang W., Zutz T. C., and Bleecker A. B. (2006) Ethylene stimulates nutations that are dependent on the ETR1 receptor. Plant Physiol. 142, 1690–1700 10.1104/pp.106.087858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Binder B. M., O'Malley R. C., Wang W., Moore J. M., Parks B. M., Spalding E. P., and Bleecker A. B. (2004) Arabidopsis seedling growth response and recovery to ethylene: a kinetic analysis. Plant Physiol. 136, 2913–2920 10.1104/pp.104.050369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wilson R. L., Bakshi A., and Binder B. M. (2014) Loss of the ETR1 ethylene receptor reduces the inhibitory effect of far-red light and darkness on seed germination of Arabidopsis thaliana. Front. Plant Sci. 5, 433 10.3389/fpls.2014.00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wilson R. L., Kim H., Bakshi A., and Binder B. M. (2014) The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol. 165, 1353–1366 10.1104/pp.114.241695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bakshi A., Piya S., Fernandez J. C., Chervin C., Hewezi T., and Binder B. M. (2018) Ethylene receptors signal via a non-canonical pathway to regulate abscisic acid responses. Plant Physiol. 176, 910–929 10.1104/pp.17.01321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bakshi A., Wilson R. L., Lacey R. F., Kim H., Wuppalapati S. K., and Binder B. M. (2015) Identification of regions in the receiver domain of the ETHYLENE RESPONSE1 ethylene receptor of Arabidopsis important for functional divergence. Plant Physiol 169, 219–232 10.1104/pp.15.00626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kim H., Helmbrecht E. E., Stalans M. B., Schmitt C., Patel N., Wen C.-K., Wang W., and Binder B. M. (2011) Ethylene receptor ETR1 domain requirements for ethylene responses in Arabidopsis seedlings. Plant Physiol. 156, 417–429 10.1104/pp.110.170621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Harkey A. F., Watkins J. M., Olex A. L., DiNapoli K. T., Lewis D. R., Fetrow J. S., Binder B. M., and Muday G. K. (2018) Identification of transcriptional and receptor networks that control root responses to ethylene. Plant Physiol. 176, 2095–2118 10.1104/pp.17.00907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Voet-van-Vormizeele J., and Groth G. (2008) Ethylene controls autophosphorylation of the histidine kinase domain in ethylene receptor ETR1. Mol. Plant 1, 380–387 10.1093/mp/ssn004 [DOI] [PubMed] [Google Scholar]
- 89. Qu X., and Schaller G. E. (2004) Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol. 136, 2961–2970 10.1104/pp.104.047126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang W., Hall A. E., O'Malley R., and Bleecker A. B. (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc. Natl. Acad. Sci. U.S.A. 100, 352–357 10.1073/pnas.0237085100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hall B. P., Shakeel S. N., Amir M., Ul Haq N., Qu X., and Schaller G. E. (2012) Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol. 159, 682–695 10.1104/pp.112.196790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Binder B. M., Kim H. J., Mathews D. E., Hutchison C. E., Kieber J. J., and Schaller G. E. (2018) A role for two-component signaling elements in the Arabidopsis growth recovery response to ethylene. Plant Direct 2, e00058 10.1002/pld3.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Street I. H., Aman S., Zubo Y., Ramzan A., Wang X., Shakeel S. N., Kieber J. J., and Schaller G. E. (2015) Ethylene inhibits cell proliferation of the Arabidopsis root meristem. Plant Physiol. 169, 338–350 10.1104/pp.15.00415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Bisson M. M. A., and Groth G. (2010) New insight in ethylene signaling: autokinase activity of ETR1 modulates the interaction of receptors and EIN2. Mol. Plant 3, 882–889 10.1093/mp/ssq036 [DOI] [PubMed] [Google Scholar]
- 95. Chen T., Liu J., Lei G., Liu Y.-F., Li Z.-G., Tao J.-J., Hao Y.-J., Cao Y.-R., Lin Q., Zhang W.-K., Ma B., Chen S.-Y., and Zhang J.-S. (2009) Effects of tobacco ethylene receptor mutations on receptor kinase activity, plant growth and stress responses. Plant Cell Physiol. 50, 1636–1650 10.1093/pcp/pcp107 [DOI] [PubMed] [Google Scholar]
- 96. Gao Z., Wen C.-K., Binder B. M., Chen Y.-F., Chang J., Chiang Y.-H., Kerris R. J. 3rd, Chang C., and Schaller G. E. (2008) Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J. Biol. Chem. 283, 23801–23810 10.1074/jbc.M800641200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gamble R. L., Qu X., and Schaller G. E. (2002) Mutational analysis of the ethylene receptor ETR1: role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol. 128, 1428–1438 10.1104/pp.010777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gao Z., and Schaller G. E. (2009) The role of receptor interactions in regulating ethylene signal transduction. Plant Signal. Behav. 4, 1152–1153 10.4161/psb.4.12.9943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Binder B. M., and Bleecker A. B. (2003) A model for ethylene receptor function and 1-methylcyclopropene action. Acta Hortic. 628, 177–187 10.17660/ActaHortic.2003.628.21 [DOI] [Google Scholar]
- 100. Liu Q., and Wen C.-K. (2012) Arabidopsis ETR1 and ERS1 differentially repress the ethylene response in combination with other ethylene receptor genes. Plant Physiol. 158, 1193–1207 10.1104/pp.111.187757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Binder B. M., Mortimore L. A., Stepanova A. N., Ecker J. R., and Bleecker A. B. (2004) Short-term growth responses to ethylene in Arabidopsis seedlings are EIN3/EIL1 independent. Plant Physiol. 136, 2921–2927 10.1104/pp.104.050393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bray D., Levin M. D., and Morton-Firth C. J. (1998) Receptor clustering as a cellular mechanism to control sensitivity. Nature 393, 85–88 10.1038/30018 [DOI] [PubMed] [Google Scholar]
- 103. Parkinson J. S., Hazelbauer G. L., and Falke J. J. (2015) Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 23, 257–266 10.1016/j.tim.2015.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Mayerhofer H., Panneerselvam S., and Mueller-Dieckmann J. (2012) Protein kinase domain of CTR1 from Arabidopsis thaliana promotes ethylene receptor cross talk. J. Mol. Biol. 415, 768–779 10.1016/j.jmb.2011.11.046 [DOI] [PubMed] [Google Scholar]
- 105. Lin Z., Ho C. W., and Grierson D. (2009) AtTRP1 encodes a novel TPR protein that interacts with the ethylene receptor ERS1 and modulates development in Arabidopsis. J. Exp. Bot. 60, 3697–3714 10.1093/jxb/erp209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Dong C.-H., Jang M., Scharein B., Malach A., Rivarola M., Liesch J., Groth G., Hwang I., and Chang C. (2010) Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. J. Biol. Chem. 285, 40706–40713 10.1074/jbc.M110.146605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Thomine S., Wang R., Ward J. M., Crawford N. M., and Schroeder J. I. (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. U.S.A. 97, 4991–4996 10.1073/pnas.97.9.4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lin Z., Arciga-Reyes L., Zhong S., Alexander L., Hackett R., Wilson I., and Grierson D. (2008) SlTPR1, a tomato tetratricopeptide repeat protein, interacts with the ethylene receptors NR and LeETR1, modulating ethylene and auxin responses and development. J. Exp. Bot. 59, 4271–4287 10.1093/jxb/ern276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Scharein B., Voet-van Vormizeele J., Harter K., and Groth G. (2008) Ethylene signaling: identification of a putative ETR1-AHP1 phosphorelay complex by fluorescence spectroscopy. Anal. Biochem. 377, 72–76 10.1016/j.ab.2008.03.015 [DOI] [PubMed] [Google Scholar]
- 110. Urao T., Miyata S., Yamaguchi-Shinozaki K., and Shinozaki K. (2000) Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 478, 227–232 10.1016/S0014-5793(00)01860-3 [DOI] [PubMed] [Google Scholar]
- 111. Zdarska M., Cuyacot A. R., Tarr P. T., Yamoune A., Szmitkowska A., Hrdinová V., Gelová Z., Meyerowitz E. M., and Hejátko J. (2019) ETR1 integrates response to ethylene and cytokinins into a single multistep phosphorelay pathway to control root growth. Mol. Plant 12, 1338–1352 10.1016/j.molp.2019.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Alonso J. M., Hirayama T., Roman G., Nourizadeh S., and Ecker J. R. (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152 10.1126/science.284.5423.2148 [DOI] [PubMed] [Google Scholar]
- 113. Kieber J. J., Rothenberg M., Roman G., Feldmann K. A., and Ecker J. R. (1993) CTR1, A negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441 10.1016/0092-8674(93)90119-B [DOI] [PubMed] [Google Scholar]
- 114. Cancel J. D., and Larsen P. B. (2002) Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol. 129, 1557–1567 10.1104/pp.003780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Bisson M. M. A., Bleckmann A., Allekotte S., and Groth G. (2009) EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem. J. 424, 1–6 10.1042/BJ20091102 [DOI] [PubMed] [Google Scholar]
- 116. Clark K. L., Larsen P. B., Wang X., and Chang C. (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. U.S.A. 95, 5401–5406 10.1073/pnas.95.9.5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Gao Z., Chen Y. F., Randlett M. D., Zhao X. C., Findell J. L., Kieber J. J., and Schaller G. E. (2003) Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J. Biol. Chem. 278, 34725–34732 10.1074/jbc.M305548200 [DOI] [PubMed] [Google Scholar]
- 118. Huang Y. F., Li H., Hutchison C. E., Laskey J., and Kieber J. J. (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 33, 221–233 10.1046/j.1365-313x.2003.01620.x [DOI] [PubMed] [Google Scholar]
- 119. Ju C., Yoon G. M., Shemansky J. M., Lin D. Y., Yin Z. I., Chang J., Garrett W. M., Kessenbrock M., Groth G., Tucker M. L., Cooper B., Kieber J. J., and Chang C. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 19486–19491 10.1073/pnas.1214848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Bisson M. M., and Groth G. (2011) New paradigm in ethylene signaling: EIN2, the central regulator of the signaling pathway, interacts directly with the upstream receptors. Plant Signal. Behav. 6, 164–166 10.4161/psb.6.1.14034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bisson M. M. A., and Groth G. (2015) Targeting plant ethylene responses by controlling essential protein-protein interactions in the ethylene pathway. Mol. Plant 8, 1165–1174 10.1016/j.molp.2015.03.014 [DOI] [PubMed] [Google Scholar]
- 122. Qiao H., Chang K. N., Yazaki J., and Ecker J. R. (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 23, 512–521 10.1101/gad.1765709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Chen R., Binder B. M., Garrett W. M., Tucker M. L., Chang C., and Cooper B. (2011) Proteomic responses in Arabidopsis thaliana seedlings treated with ethylene. Mol. Biosyst. 7, 2637–2650 10.1039/c1mb05159h [DOI] [PubMed] [Google Scholar]
- 124. Wen X., Zhang C., Ji Y., Zhao Q., He W., An F., Jiang L., and Guo H. (2012) Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 22, 1613–1616 10.1038/cr.2012.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Qiao H., Shen Z., Huang S.-S. C., Schmitz R. J., Urich M. A., Briggs S. P., and Ecker J. R. (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338, 390–393 10.1126/science.1225974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., and Genschik P. (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689 10.1016/S0092-8674(03)00968-1 [DOI] [PubMed] [Google Scholar]
- 127. Guo H., and Ecker J. R. (2003) Plant responses to ethylene gas are mediated by SCF (EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677 10.1016/S0092-8674(03)00969-3 [DOI] [PubMed] [Google Scholar]
- 128. Binder B. M., Walker J. M., Gagne J. M., Emborg T. J., Hemmann G., Bleecker A. B., and Vierstra R. D. (2007) The Arabidopsis EIN3-binding F-box proteins, EBF1 and 2 have distinct but overlapping roles in regulating ethylene signaling. Plant Cell 19, 509–523 10.1105/tpc.106.048140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. An F., Zhao Q., Ji Y., Li W., Jiang Z., Yu X., Zhang C., Han Y., He W., Liu Y., Zhang S., Ecker J. R., and Guo H. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22, 2384–2401 10.1105/tpc.110.076588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Gagne J. M., Smalle J., Gingerich D. J., Walker J. M., Yoo S. D., Yanagisawa S., and Vierstra R. D. (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 6803–6808 10.1073/pnas.0401698101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ma B., Zhou Y., Chen H., He S.-J., Huang Y.-H., Zhao H., Lu X., Zhang W.-K., Pang J.-H., Chen S.-Y., and Zhang J.-S. (2018) Membrane protein MHZ3 stabilizes OsEIN2 in rice by interacting with its Nramp-like domain. Proc. Natl. Acad. Sci. U.S.A. 115, 2520–2525 10.1073/pnas.1718377115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ma B., He S.-J., Duan K.-X., Yin C.-C., Chen H., Yang C., Xiong Q., Song Q.-X., Lu X., Chen H.-W., Zhang W.-K., Lu T.-G., Chen S.-Y., and Zhang J.-S. (2013) Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant 6, 1830–1848 10.1093/mp/sst087 [DOI] [PubMed] [Google Scholar]
- 133. Merchante C., Brumos J., Yun J., Hu Q., Spencer K. R., Enríquez P., Binder B. M., Heber S., Stepanova A. N., and Alonso J. M. (2015) Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163, 684–697 10.1016/j.cell.2015.09.036 [DOI] [PubMed] [Google Scholar]
- 134. Li W., Ma M., Feng Y., Li H., Wang Y., Ma Y., Li M., An F., and Guo H. (2015) EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163, 670–683 10.1016/j.cell.2015.09.037 [DOI] [PubMed] [Google Scholar]
- 135. Olmedo G., Guo H., Gregory B. D., Nourizadeh S. D., Aguilar-Henonin L., Li H., An F., Guzman P., and Ecker J. R. (2006) ETHYLENE-INSENSITIVE5 encodes a 5′ → 3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl. Acad. Sci. U.S.A. 103, 13286–13293 10.1073/pnas.0605528103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Potuschak T., Vansiri A., Binder B. M., Lechner E., Vierstra R. D., and Genschik P. (2006) The exonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell 18, 3047–3057 10.1105/tpc.106.046508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Zhang F., Wang L., Qi B., Zhao B., Ko E. E., Riggan N. D., Chin K., and Qiao H. (2017) EIN2 mediates direct regulation of histone acetylation in the ethylene response. Proc. Natl. Acad. Sci. U.S.A. 114, 10274–10279 10.1073/pnas.1707937114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hildreth S. B., Foley E. E., Muday G. K., Helm R. F., and Winkel B. S. J. (2020) The dynamic response of the Arabidopsis root metabolome to auxin and ethylene is not predicted by changes in the transcriptome. Sci. Rep. 10, 679 10.1038/s41598-019-57161-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Alonso J. M., Stepanova A. N., Solano R., Wisman E., Ferrari S., Ausubel F. M., and Ecker J. R. (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 2992–2997 10.1073/pnas.0438070100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., and Ecker J. R. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144 10.1016/S0092-8674(00)80300-1 [DOI] [PubMed] [Google Scholar]
- 141. Solano R., Stepanova A., Chao Q., and Ecker J. R. (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714 10.1101/gad.12.23.3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Poulios S., and Vlachonasios K. E. (2016) Synergistic action of histone acetyltransferase GCN5 and receptor CLAVATA1 negatively affects ethylene responses in Arabidopsis thaliana. J. Exp. Bot. 67, 905–918 10.1093/jxb/erv503 [DOI] [PubMed] [Google Scholar]
- 143. Li C., Xu J., Li J., Li Q., and Yang H. (2014) Involvement of Arabidopsis histone acetyltransferase HAC family genes in the ethylene signaling pathway. Plant Cell Physiol. 55, 426–435 10.1093/pcp/pct180 [DOI] [PubMed] [Google Scholar]
- 144. Zhou C., Zhang L., Duan J., Miki B., and Wu K. (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17, 1196–1204 10.1105/tpc.104.028514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wang F., Wang L., Qiao L., Chen J., Pappa M. B., Pei H., Zhang T., Chang C., and Dong C. H. (2017) Arabidopsis CPR5 regulates ethylene signaling via molecular association with the ETR1 receptor. J. Integr. Plant Biol. 59, 810–824 10.1111/jipb.12570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Zhang F., Qi B., Wang L., Zhao B., Rode S., Riggan N. D., Ecker J. R., and Qiao H. (2016) EIN2-dependent regulation of acetylation of histone H3K14 and non-canonical histone H3K23 in ethylene signalling. Nat. Commun. 7, 13018 10.1038/ncomms13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zhang F., Wang L., Ko E. E., Shao K., and Qiao H. (2018) Histone deacetylases SRT1 and SRT2 interact with ENAP1 to mediate ethylene-induced transcriptional repression. Plant Cell 30, 153–166 10.1105/tpc.17.00671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kim J., Wilson R. L., Case J. B., and Binder B. (2012) A comparative study of ethylene growth response kinetics in eudicots and monocots reveals a role for gibberellin in growth inhibition and recovery. Plant Physiol. 160, 1567–1580 10.1104/pp.112.205799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Rai M. I., Wang X., Thibault D. M., Kim H. J., Bombyk M. M., Binder B. M., Shakeel S. N., and Schaller G. E. (2015) The ARGOS gene family functions in a negative feedback loop to desensitize plants to ethylene. BMC Plant Biol. 15, 157 10.1186/s12870-015-0554-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Prescott A. M., McCollough F. W., Eldreth B. L., Binder B. M., and Abel S. M. (2016) Analysis of network topologies underlying ethylene growth response kinetics. Front. Plant Sci. 7, 1308 10.3389/fpls.2016.01308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Shi J., Drummond B. J., Wang H., Archibald R. L., and Habben J. E. (2016) Maize and Arabidopsis ARGOS proteins interact with ethylene receptor signaling complex, supporting a regulatory role for ARGOS in ethylene signal transduction. Plant Physiol. 171, 2783–2797 10.1104/pp.16.00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Deslauriers S. D., Alvarez A. A., Lacey R. F., Binder B. M., and Larsen P. B. (2015) Dominant gain-of-function mutations in transmembrane domain III of ERS1 and ETR1 suggest a novel role for this domain in regulating the magnitude of ethylene response in Arabidopsis. New Phytol. 208, 442–455 10.1111/nph.13466 [DOI] [PubMed] [Google Scholar]
- 153. Konishi M., and Yanagisawa S. (2008) Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 55, 821–831 10.1111/j.1365-313X.2008.03551.x [DOI] [PubMed] [Google Scholar]
- 154. Shi J., Habben J. E., Archibald R. L., Drummond B. J., Chamberlin M. A., Williams R. W., Lafitte H. R., and Weers B. P. (2015) Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiol. 169, 266–282 10.1104/pp.15.00780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Robles L. M., Wampole J. S., Christians M. J., and Larsen P. B. (2007) Arabidopsis enhanced ethylene response 4 encodes an EIN3-interacting TFIID transcription factor required for proper ethylene response, including ERF1 induction. J. Exp. Bot. 58, 2627–2639 10.1093/jxb/erm080 [DOI] [PubMed] [Google Scholar]
- 156. Christians M. J., and Larsen P. B. (2007) Mutational loss of the prohibitin AtPHB3 results in an extreme constitutive ethylene response phenotype coupled with partial loss of ethylene-inducible gene expression in Arabidopsis seedlings. J. Exp. Bot. 58, 2237–2248 10.1093/jxb/erm086 [DOI] [PubMed] [Google Scholar]
- 157. Christians M. J., Robles L. M., Zeller S. M., and Larsen P. B. (2008) The eer5 mutation, which affects a novel proteasome-related subunit, indicates a prominent role for the COP9 signalosome in resetting the ethylene-signaling pathway in Arabidopsis. Plant J. 55, 467–477 10.1111/j.1365-313X.2008.03521.x [DOI] [PubMed] [Google Scholar]
- 158. Kim H. G., Kwon S. J., Jang Y. J., Nam M. H., Chung J. H., Na Y.-C., Guo H., and Park O. K. (2013) GDSL LIPASE1 modulates plant immunity through feedback regulation of ethylene signaling. Plant Physiol. 163, 1776–1791 10.1104/pp.113.225649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kim H. G., Kwon S. J., Jang Y. J., Chung J. H., Nam M. H., and Park O. K. (2014) GDSL lipase 1 regulates ethylene signaling and ethylene-associated systemic immunity in Arabidopsis. FEBS Lett. 588, 1652–1658 10.1016/j.febslet.2014.02.062 [DOI] [PubMed] [Google Scholar]
- 160. Kwon S. J., Jin H. C., Lee S., Nam M. H., Chung J. H., Kwon S. I., Ryu C.-M., and Park O. K. (2009) GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis. Plant J. 58, 235–245 10.1111/j.1365-313X.2008.03772.x [DOI] [PubMed] [Google Scholar]
- 161. Hua J., and Meyerowitz E. M. (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94, 261–271 10.1016/S0092-8674(00)81425-7 [DOI] [PubMed] [Google Scholar]
- 162. Ciardi J. A., Tieman D. M., Lund S. T., Jones J. B., Stall R. E., and Klee H. J. (2000) Response to Xanthomonas campestris pv. vesicatoria in tomato involves regulation of ethylene receptor gene expression. Plant Physiol. 123, 81–92 10.1104/pp.123.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Tieman D. M., Taylor M. G., Ciardi J. A., and Klee H. J. (2000) The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc. Natl. Acad. Sci. U.S.A. 97, 5663–5668 10.1073/pnas.090550597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Hall A. E., and Bleecker A. B. (2003) Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell 15, 2032–2041 10.1105/tpc.013060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Resnick J. S., Wen C.-K., Shockey J. A., and Chang C. (2006) REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 7917–7922 10.1073/pnas.0602239103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Barry C. S., and Giovannoni J. J. (2006) Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc. Natl. Acad. Sci. U.S.A. 103, 7923–7928 10.1073/pnas.0602319103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Azhar B. J., Zulfiqar A., Shakeel S. N., and Schaller G. E. (2019) Amplification and adaptation in the ethylene signaling pathway. Small Methods 10.1002/smtd.201900452 [DOI] [Google Scholar]
- 168. Desikan R., Hancock J. T., Bright J., Harrison J., Weir I., Hooley R., and Neill S. J. (2005) A role for ETR1 in hydrogen peroxide signaling in stomatal guard cells. Plant Physiol. 137, 831–834 10.1104/pp.104.056994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Qiu L., Xie F., Yu J., and Wen C.-K. (2012) Arabidopsis RTE1 is essential to ethylene receptor ETR1 amino-terminal signaling independent of CTR1. Plant Physiol. 159, 1263–1276 10.1104/pp.112.193979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Kim J., Patterson S. E., and Binder B. M. (2013) Reducing jasmonic acid levels causes ein2 mutants to become ethylene responsive. FEBS Lett. 587, 226–230 10.1016/j.febslet.2012.11.030 [DOI] [PubMed] [Google Scholar]
- 171. Kieber J. J., and Schaller G. E. (2018) Cytokinin signaling in plant development. Development 145, dev149344 10.1242/dev.149344 [DOI] [PubMed] [Google Scholar]
- 172. Scharein B., and Groth G. (2011) Phosphorylation alters the interaction of the Arabidopsis phosphotransfer protein AHP1 with its sensor kinase ETR1. PLoS ONE 6, e24173 10.1371/journal.pone.0024173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Mira-Rodado V., Veerabagu M., Witthöft J., Teply J., Harter K., and Desikan R. (2012) Identification of two-component system elements downstream of AHK5 in the stomatal closure response of Arabidopsis thaliana. Plant Signal. Behav. 7, 1467–1476 10.4161/psb.21898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Nemhauser J. L., Hong F., and Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126, 467–475 10.1016/j.cell.2006.05.050 [DOI] [PubMed] [Google Scholar]
- 175. Zhao H., Duan K.-X., Ma B., Yin C.-C., Hu Y., Tao J.-J., Huang Y.-H., Cao W.-Q., Chen H., Yang C., Zhang Z.-G., He S.-J., Zhang W.-K., Wan X.-Y., Lu T.-G., et al. (2020) Histidine kinase MHZ1/OsHK1 interacts with ethylene receptors to regulate root growth in rice. Nat. Commun. 11, 518 10.1038/s41467-020-14313-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Bourret R. B. (2010) Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 13, 142–149 10.1016/j.mib.2010.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Yasumura Y., Pierik R., Kelly S., Sakuta M., Voesenek L. A. C. J., and Harberd N. P. (2015) An ancestral role for CONSTITUTIVE TRIPLE RESPONSE1 proteins in both ethylene and abscisic acid signaling. Plant Physiol. 169, 283–298 10.1104/pp.15.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Van de Poel B., Smet D., and Van Der Straeten D. (2015) Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol. 169, 61–72 10.1104/pp.15.00724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Zhu Z., An F., Feng Y., Li P., Xue L., Jiang A. M. Z., Kim J.-M., To T. K., Li W., Zhang X., Yu Q., Dong Z., Chen W.-Q., Seki M., Zhou J.-M., and Guo H. (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 12539–12544 10.1073/pnas.1103959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. He X., Jiang J., Wang C.-Q., and Dehesh K. (2017) ORA59 and EIN3 interaction couples jasmonate-ethylene synergistic action to antagonistic salicylic acid regulation of PDF expression. J. Integr. Plant Biol. 59, 275–287 10.1111/jipb.12524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Liu X., Liu R., Li Y., Shen X., Zhong S., and Shi H. (2017) EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. Plant Cell 29, 3051–3067 10.1105/tpc.17.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Guzmán P., and Ecker J. R. (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523 10.1105/tpc.2.6.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Smalle J., Haegman M., Kurepa J., Van Montagu M., and Straeten D. V. D. (1997) Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc. Natl. Acad. Sci. U.S.A. 94, 2756–2761 10.1073/pnas.94.6.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Das D., St Onge K. R., Voesenek L. A. C. J., Pierik R., and Sasidharan R. (2016) Ethylene- and shade-induced hypocotyl elongation share transcriptome patterns and functional regulators. Plant Physiol. 172, 718–733 10.1104/pp.16.00725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Le J., Vandenbussche F., De Cnodder T., Van Der Straeten D., and Verbelen J.-P. (2005) Cell elongation and microtubule behavior in the Arabidopsis hypocotyl: responses to ethylene and auxin. J. Plant Growth Regul. 24, 166–178 10.1007/s00344-005-0044-8 [DOI] [Google Scholar]
- 186. Seo D. H., and Yoon G. M. (2019) Light-induced stabilization of ACS contributes to hypocotyl elongation during the dark-to-light transition in Arabidopsis seedlings. Plant J. 98, 898–911 10.1111/tpj.14289 [DOI] [PubMed] [Google Scholar]
- 187. Vandenbussche F., Vancompernolle B., Rieu I., Ahmad M., Phillips A., Moritz T., Hedden P., and Van Der Straeten D. (2007) Ethylene-induced Arabidopsis hypocotyl elongation is dependent on but not mediated by gibberellins. J. Exp. Bot. 58, 4269–4281 10.1093/jxb/erm288 [DOI] [PubMed] [Google Scholar]
- 188. Harkey A. F., Yoon G. M., Seo D. H., DeLong A., and Muday G. K. (2019) Light modulates ethylene synthesis, signaling, and downstream transcriptional networks to control plant development. Front. Plant. Sci. 10, 1094 10.3389/fpls.2019.01094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Bovy A., Angenent G., Dons H. M., and van Altvorst A.-C. (1999) Heterologous expression of the Arabidopsis etr1-1 allele inhibits the senescence of carnation flowers. Mol. Breed. 5, 301–308 10.1023/A:1009617804359 [DOI] [Google Scholar]
- 190. Sriskandarajah S., Mibus H., and Serek M. (2007) Transgenic Campanula carpatica plants with reduced ethylene sensitivity. Plant Cell Rep. 26, 805–813 10.1007/s00299-006-0291-6 [DOI] [PubMed] [Google Scholar]
- 191. Wang H., Stier G., Lin J., Liu G., Zhang Z., Chang Y., Reid M. S., and Jiang C.-Z. (2013) Transcriptome changes associated with delayed flower senescence on transgenic petunia by inducing expression of etr1-1, a mutant ethylene receptor. PLoS ONE 8, e65800 10.1371/journal.pone.0065800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Wilkinson J. Q., Lanahan M. B., Clark D. G., Bleecker A. B., Chang C., Meyerowitz E. M., and Klee H. J. (1997) A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat. Biotechnol. 15, 444–447 10.1038/nbt0597-444 [DOI] [PubMed] [Google Scholar]
- 193. Langston B. J., Bai S., and Jones M. L. (2005) Increases in DNA fragmentation and induction of a senescence-specific nuclease are delayed during corolla senescence in ethylene-insensitive (etr1-1) transgenic petunias. J. Exp. Bot. 56, 15–23 10.1093/jxb/eri002 [DOI] [PubMed] [Google Scholar]
- 194. Little H., Papadopoulou E., Hammar S., and Grumet R. (2007) The influence of ethylene perception on sex expression in melon (Cucumis melo L.) as assessed by expression of the mutant ethylene receptor, At-etr1–1, under the control of constitutive and floral targeted promoters. Sex Plant Reprod. 20, 123–136 10.1007/s00497-007-0049-5 [DOI] [Google Scholar]
- 195. Gubrium E. K., Clevenger D. J., Clark D. G., Barrett J. E., and Nell T. A. (2000) Reproduction and horticultural performance of transgenic ethylene-insensitive petunias. J. Am. Soc. Hortic. Sci. 125, 277–281 10.21273/JASHS.125.3.277 [DOI] [Google Scholar]
- 196. Hee Kim J., and Botella J. R. (2004) Etr1-1 gene expression alters regeneration patterns in transgenic lettuce stimulating root formation. Plant Cell Tiss. Org. 78, 69–73 10.1023/B:TICU.0000020396.64257.c1 [DOI] [Google Scholar]
- 197. Khatami F., Najafi F., Yari F., and Khavari-Nejad R. A. (2020) Expression of etr1-1 gene in transgenic Rosa hybrida L. increased postharvest longevity through reduced ethylene biosynthesis and perception. Sci. Hortic. 263, 109103 10.1016/j.scienta.2019.109103 [DOI] [Google Scholar]
- 198. Cui M.-L., Takada K., Ma B., and Ezura H. (2004) Overexpression of a mutated melon ethylene receptor gene Cm-ETR1/H69A confers reduced ethylene sensitivity in a heterologous plant, Nemesia strumosa. Plant Sci. 167, 253–258 10.1016/j.plantsci.2004.03.024 [DOI] [Google Scholar]
- 199. Mubarok S., Okabe Y., Fukuda N., Ariizumi T., and Ezura H. (2015) Potential use of a weak ethylene receptor mutant, Sletr1-2, as breeding material to extend fruit shelf life of tomato. J. Agric. Food Chem. 63, 7995–8007 10.1021/acs.jafc.5b02742 [DOI] [PubMed] [Google Scholar]
- 200. Switzenberg J. A., Beaudry R. M., and Grumet R. (2015) Effect of CRC::etr1–1 transgene expression on ethylene production, sex expression, fruit set and fruit ripening in transgenic melon (Cucumis melo L.). Transgenic Res. 24, 497–507 10.1007/s11248-014-9853-5 [DOI] [PubMed] [Google Scholar]
- 201. Clark D. G., Gubrium E. K., Barrett J. E., Nell T. A., and Klee H. J. (1999) Root formation in ethylene-insensitive plants. Plant Physiol. 121, 53–60 10.1104/pp.121.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Clevenger D. J., Barrett J. E., Klee H. J., and Clark D. G. (2004) Factors affecting seed production in transgenic ethylene-insensitive petunias. J. Am. Soc. Hortic. Sci. 129, 401–406 10.21273/JASHS.129.3.0401 [DOI] [Google Scholar]
- 203. Knoester M., van Loon L. C., van den Heuvel J., Hennig J., Bol J. F., and Linthorst H. J. (1998) Ethylene-insensitive tobacco lacks nonhost resistance against soil-borne fungi. Proc. Natl. Acad. Sci. U.S.A. 95, 1933–1937 10.1073/pnas.95.4.1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Sanikhani M., Mibus H., Stummann B. M., and Serek M. (2008) Kalanchoe blossfeldiana plants expressing the Arabidopsis etr1-1 allele show reduced ethylene sensitivity. Plant Cell Rep. 27, 729–737 10.1007/s00299-007-0493-6 [DOI] [PubMed] [Google Scholar]
- 205. Chen Y., Rofidal V., Hem S., Gil J., Nosarzewska J., Berger N., Demolombe V., Bouzayen M., Azhar B. J., Shakeel S. N., Schaller G. E., Binder B. M., Santoni V., and Chervin C. (2019) Targeted proteomics allows quantification of ethylene receptors and reveals SlETR3 accumulation in Never-Ripe tomatoes. Front. Plant Sci. 10, 1054 10.3389/fpls.2019.01054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Gallie D. (2010) Regulated ethylene insensitivity through the inducible expression of the Arabidopsis etr1-1 mutant ethylene receptor in tomato. Plant Physiol. 152, 1928–1939 10.1104/pp.109.151688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Fu D.-Q., Zhu B.-Z., Zhu H.-L., Jiang W.-B., and Luo Y.-B. (2005) Virus-induced gene silencing in tomato fruit. Plant J. 43, 299–308 10.1111/j.1365-313X.2005.02441.x [DOI] [PubMed] [Google Scholar]
- 208. Hu Z. L., Deng L., Chen X. Q., Wang P. Q., and Chen G. P. (2010) Co-suppression of the EIN2-homology gene LeEIN2 inhibits fruit ripening and reduces ethylene sensitivity in tomato. Russ. J. Plant Physiol. 57, 554–559 10.1134/S102144371004014X [DOI] [Google Scholar]
- 209. Barry C. S., McQuinn R. P., Thompson A. J., Seymour G. B., Grierson D., and Giovannoni J. J. (2005) Ethylene insensitivity conferred by the Green-ripe and Never-ripe 2 ripening mutants of tomato. Plant Physiol. 138, 267–275 10.1104/pp.104.057745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Liu M., Pirrello J., Chervin C., Roustan J.-P., and Bouzayen M. (2015) Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol. 169, 2380–2390 10.1104/pp.15.01361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Zhang Y. P., Huang Y. Q., Wang C., Mu Q., Jiu S. T., Zhu X. D., Zheng T., Zhang K. K., Jia H. F., Tariq P., and Fang J. G. (2019) Characterization and identification of PpEIN3 during the modulation of fruit ripening process by ectopic expressions in tomato. Plant Genome 12, 10.3835/plantgenome2018.11.0089 [DOI] [PubMed] [Google Scholar]
- 212. Liao M. S., and Schwarz W. H. E. (1994) Effective radii of the monovalent coin metals. Acta Crystallogr. B Struct. Sci. 50, 9–12 10.1107/S0108768193007864 [DOI] [Google Scholar]
- 213. Bayler A., Schier A., Bowmaker G. A., and Schmidbaur H. (1996) Gold is smaller than silver: crystal structures of [bis(trimesitylphosphine)gold(I)] and [bis(trimesitylphosphine)silver(I)] tetrafluoroborate. J. Am. Chem. Soc. 118, 7006–7007 10.1021/ja961363v [DOI] [Google Scholar]
- 214. Pyykkö P. (1988) Relativistic effects in structural chemistry. Chem. Rev. 88, 563–594 10.1021/cr00085a006 [DOI] [Google Scholar]
- 215. Binder B. M., Rodriguez F. I., Bleecker A. B., and Patterson S. E. (2007) The effects of group 11 transition metals, including gold, on ethylene binding to the ETR1 receptor and growth of Arabidopsis thaliana. FEBS Lett. 581, 5105–5109 10.1016/j.febslet.2007.09.057 [DOI] [PubMed] [Google Scholar]
- 216. Zhao X. C., Qu X., Mathews D. E., and Schaller G. E. (2002) Effect of ethylene pathway mutations upon expression of the ethylene receptor ETR1 from Arabidopsis. Plant Physiol. 130, 1983–1991 10.1104/pp.011635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Blat Y. (2010) Non-competitive inhibition by active site binders. Chem. Biol. Drug Des. 75, 535–540 10.1111/j.1747-0285.2010.00972.x [DOI] [PubMed] [Google Scholar]
- 218. Strader L. C., Beisner E. R., and Bartel B. (2009) Silver ions increase auxin efflux independently of effects on ethylene response. Plant Cell 21, 3585–3590 10.1105/tpc.108.065185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Sisler E. C., and Pian A. (1973) Effect of ethylene and cyclic olefins on tobacco leaves. Tob. Sci. 17, 68–72 [Google Scholar]
- 220. Pirrung M. C., Bleecker A. B., Inoue Y., Rodríguez F. I., Sugawara N., Wada T., Zou Y., and Binder B. M. (2008) Ethylene receptor antagonists: strained alkenes are necessary but not sufficient. Chem. Biol. 15, 313–321 10.1016/j.chembiol.2008.02.018 [DOI] [PubMed] [Google Scholar]
- 221. Sisler E. C., and Blankenship S. M. (May 21, 1996) Method of counteracting an ethylene response in plants. U. S. Patent 5,518,988
- 222. Sisler E. C., Dupille E., and Serek M. (1996) Effect of 1-methylcyclopropene and methylenecyclopropane on ethylene binding and ethylene action on cut carnations. Plant Growth Regul. 18, 79–86 10.1007/BF00028491 [DOI] [Google Scholar]
- 223. Sisler E. C., Serek M., and Dupille E. (1996) Comparison of cyclopropene, 1-methylcyclopropene, and 3,3-dimethylcyclopropene as ethylene antagonists in plants. Plant Growth Regul. 18, 169–174 10.1007/BF00024378 [DOI] [Google Scholar]
- 224. Watkins C. B. (2006) The use of 1-methylcyclopropene (1-MCP) on fruits and vegetables. Biotechnol. Adv. 24, 389–409 10.1016/j.biotechadv.2006.01.005 [DOI] [PubMed] [Google Scholar]
- 225. Oh K., Hoshi T., Tomio S., Ueda K., and Hara K. (2017) A chemical genetics strategy that identifies small molecules which induce the triple response in Arabidopsis. Molecules 22, 2270 10.3390/molecules22122270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Hu Y., Callebert P., Vandemoortel I., Nguyen L., Audenaert D., Verschraegen L., Vandenbussche F., and Van Der Straeten D. (2014) TR-DB: an open-access database of compounds affecting the ethylene-induced triple response in Arabidopsis. Plant Physiol. Biochem. 75, 128–137 10.1016/j.plaphy.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 227. Li W., Lacey R. F., Ye Y., Lu J., Yeh K.-C., Xiao Y., Li L., Wen C.-K., Binder B. M., and Zhao Y. (2017) Triplin, a small molecule, reveals copper ion transport in ethylene signaling from ATX1 to RAN1. PLoS Genet. 13, e1006703 10.1371/journal.pgen.1006703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Bisson M. M. A., Kessenbrock M., Müller L., Hofmann A., Schmitz F., Cristescu S. M., and Groth G. (2016) Peptides interfering with protein-protein interactions in the ethylene signaling pathway delay tomato fruit ripening. Sci. Rep. 6, 30634 10.1038/srep30634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Hoppen C., Müller L., Albrecht A. C., and Groth G. (2019) The NOP-1 peptide derived from the central regulator of ethylene signaling EIN2 delays floral senescence in cut flowers. Sci. Rep. 9, 1287–1287 10.1038/s41598-018-37571-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Kessenbrock M., Klein S. M., Müller L., Hunsche M., Noga G., and Groth G. (2017) Novel protein-protein inhibitor based approach to control plant ethylene responses: synthetic peptides for ripening control. Front. Plant Sci. 8, 1528 10.3389/fpls.2017.01528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Milić D., Dick M., Mulnaes D., Pfleger C., Kinnen A., Gohlke H., and Groth G. (2018) Recognition motif and mechanism of ripening inhibitory peptides in plant hormone receptor ETR1. Sci. Rep. 8, 3890–3901 10.1038/s41598-018-21952-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Runo S., and Kuria E. K. (2018) Habits of a highly successful cereal killer, Striga. PLoS Pathol. 14, e1006731 10.1371/journal.ppat.1006731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Kountche B. A., Jamil M., Yonli D., Nikiema M. P., Blanco-Ania D., Asami T., Zwanenburg B., and Al-Babili S. (2019) Suicidal germination as a control strategy for Striga hermonthica (Benth.) in smallholder farms of sub-Saharan Africa. Plants People Planet 1, 107–118 10.1002/ppp3.32 [DOI] [Google Scholar]
- 234. Logan D. C., and Stewart G. R. (1991) Role of ethylene in the germination of the hemiparasite Striga hermonthica. Plant Physiol. 97, 1435–1438 10.1104/pp.97.4.1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Eplee R. E. (1975) Ethylene: a witchweed seed germination stimulant. Weed Sci. 23, 433–436 10.1017/S0043174500062822 [DOI] [Google Scholar]
- 236. Egley G. H., and Dale J. E. (1970) Ethylene, 2-chloroethylphosphonic acid, and witchweed germination. Weed Sci. 18, 586–589 10.1017/S0043174500034263 [DOI] [Google Scholar]
- 237. Babiker A. G. T., Ejeta G., Butler L. G., and Woodson W. R. (1993) Ethylene biosynthesis and strigol-induced germination of Striga asiatica. Physiol. Plant 88, 359–365 10.1111/j.1399-3054.1993.tb05510.x [DOI] [Google Scholar]
- 238. Zwanenburg B., Mwakaboko A. S., and Kannan C. (2016) Suicidal germination for parasitic weed control. Pest Manag. Sci. 72, 2016–2025 10.1002/ps.4222 [DOI] [PubMed] [Google Scholar]
- 239. Logan D. C., and Stewart G. R. (1995) Thidiazuron stimulates germination and ethylene production in Striga hermonthica—comparison with the effects of GR-24, ethylene and 1-aminocyclopropane-1-carboxylic acid. Seed Sci. Res. 5, 99–108 10.1017/S0960258500002671 [DOI] [Google Scholar]
- 240. Zehhar N., Ingouff M., Bouya D., and Fer A. (2002) Possible involvement of gibberellins and ethylene in Orobanche ramosa germination. Weed Res. 42, 464–469 10.1046/j.1365-3180.2002.00306.x [DOI] [Google Scholar]
- 241. Samejima H., Babiker A. G., Takikawa H., Sasaki M., and Sugimoto Y. (2016) Practicality of the suicidal germination approach for controlling Striga hermonthica. Pest Manag. Sci. 72, 2035–2042 10.1002/ps.4215 [DOI] [PubMed] [Google Scholar]
- 242. Kgosi R. L., Zwanenburg B., Mwakaboko A. S., and Murdoch A. J. (2012) Strigolactone analogues induce suicidal seed germination of Striga spp. in soil. Weed Res. 52, 197–203 10.1111/j.1365-3180.2012.00912.x [DOI] [Google Scholar]
- 243. Nefkens G. H. L., Thuring J. W. J. F., Beenakkers M. F. M., and Zwanenburg B. (1997) Synthesis of a phthaloylglycine-derived strigol analogue and its germination stimulatory activity toward seeds of the parasitic weeds Striga hermonthica and Orobanche crenata. J. Agric. Food Chem. 45, 2273–2277 10.1021/jf9604504 [DOI] [Google Scholar]
- 244. Wigchert S. C. M., Kuiper E., Boelhouwer G. J., Nefkens G. H. L., Verkleij J. A. C., and Zwanenburg B. (1999) Dose-response of seeds of the parasitic weeds Striga and Orobanche toward the synthetic germination stimulants GR 24 and Nijmegen 1. J. Agric. Food Chem. 47, 1705–1710 10.1021/jf981006z [DOI] [PubMed] [Google Scholar]
- 245. Bebawi F. F., and Eplee R. E. (1986) Efficacy of ethylene as a germination stimulant of Striga hermonthica seed. Weed Sci. 34, 694–698 10.1017/S0043174500067709 [DOI] [Google Scholar]
- 246. Bebawi F. F., Eplee R. E., and Norris R. S. (1985) The dispersion of backpack-applied ethylene in soil. Weed Sci. 33, 74–77 10.1017/S0043174500083958 [DOI] [Google Scholar]
- 247. Berner D. K., Schaad N. W., and Völksch B. (1999) Use of ethylene-producing bacteria for stimulation of Striga spp. seed germination. Biol. Control 15, 274–282 10.1006/bcon.1999.0718 [DOI] [Google Scholar]
- 248. Sugimoto Y., Ali A. M., Yabuta S., Kinoshita H., Inanaga S., and Itai A. (2003) Germination strategy of Striga hermonthica involves regulation of ethylene biosynthesis. Physiol. Plant 119, 137–145 10.1034/j.1399-3054.2003.00162.x [DOI] [Google Scholar]
- 249. Babiker A. G. T., and Hamdoun A. M. (1983) Factors affecting the activity of ethephon in stimulating seed germination of Striga hermonthica (Del.) Benth. Weed Res. 23, 125–131 10.1111/j.1365-3180.1983.tb00530.x [DOI] [Google Scholar]