Abstract
Prion diseases are transmissible, lethal neurodegenerative disorders caused by accumulation of the aggregated scrapie form of the prion protein (PrPSc) after conversion of the cellular prion protein (PrPC). The glycosylphosphatidylinositol (GPI) anchor of PrPC is involved in prion disease pathogenesis, and especially sialic acid in a GPI side chain reportedly affects PrPC conversion. Thus, it is important to define the location and structure of the GPI anchor in human PrPC. Moreover, the sialic acid linkage type in the GPI side chain has not been determined for any GPI-anchored protein. Here we report GPI glycan structures of human PrPC isolated from human brains and from brains of a knock-in mouse model in which the mouse prion protein (Prnp) gene was replaced with the human PRNP gene. LC–electrospray ionization–MS analysis of human PrPC from both biological sources indicated that Gly229 is the ω site in PrPC to which GPI is attached. Gly229 in human PrPC does not correspond to Ser231, the previously reported ω site of Syrian hamster PrPC. We found that ∼41% and 28% of GPI anchors in human PrPCs from human and knock-in mouse brains, respectively, have N-acetylneuraminic acid in the side chain. Using a sialic acid linkage-specific alkylamidation method to discriminate α2,3 linkage from α2,6 linkage, we found that N-acetylneuraminic acid in PrPC's GPI side chain is linked to galactose through an α2,3 linkage. In summary, we report the GPI glycan structure of human PrPC, including the ω-site amino acid for GPI attachment and the sialic acid linkage type.
Keywords: glycosylphosphatidylinositol (GPI), glycosylation, sialic acid, prion, MS, neurodegeneration, Creutzfeldt–Jakob disease, scrapie
Introduction
Glycosylphosphatidylinositol (GPI) anchoring is a posttranslational modification by a glycolipid and is highly conserved among eukaryotes. Precursors of GPI-anchored proteins (GPI-APs) have a GPI attachment signal peptide at the C terminus, which is cleaved off and replaced by a preassembled GPI (1). The amino acid residue to which GPI is attached is termed the ω site (1). The common core structure of GPI is EtNP-6Manα1–2Manα1–6Manα1–4GlcNα1-myoinositol-phospholipid (where EtNP, Man, and GlcN are ethanolamine phosphate, mannose, and glucosamine, respectively; Fig. 1A) (1–3). GPI biosynthesis occurs in the endoplasmic reticulum (ER), followed by attachment to proteins generating nascent GPI-APs. GPI-APs then undergo structural remodeling of the GPI moiety by post-GPI attachment to protein 1 (PGAP1) and PGAP5 in the ER (4, 5). After PGAP1- and PGAP5-dependent reactions, GPI-APs efficiently exit from the ER, mediated by a specific cargo receptor consisting of p24 family proteins (6). They then enter the Golgi apparatus, where they undergo fatty acid remodeling, in which PGAP3 removes an unsaturated fatty acid (usually arachidonic acid) at the sn-2 position, and PGAP2 attaches a saturated fatty acid (usually stearic acid) to the same position, which facilitates association of GPI-APs with lipid rafts (7, 8). In mammalian cells, the GPI core is variously modified by several types of side chains. One is an EtNP side chain linked to the 2 position of the first Man, Man being linked to GlcN (Fig. 1A) (9). This EtNP side chain, which is added during GPI biosynthesis in the ER, is common among mammalian GPI-APs. Another side chain is the fourth Man linked to the third Man via α1,2 linkage (Fig. 1A). The fourth Man is added in the ER immediately before attachment to proteins (10, 11). A third side chain is GalNAc linked to the first Man via β1,4 linkage (Fig. 1A) (11). The GalNAc modification occurs for GPI-APs in the Golgi apparatus after fatty acid remodeling (12). The GalNAc side chain may be further elongated by β1,3-linked galactose (Gal) and then by sialic acid (Sia) (Fig. 1A) (13). The complete chemical structure of mammalian GPI was reported in 1988 (11), but the enzymes responsible for GalNAc side-chain modifications remained unidentified for 30 years. We recently identified PGAP4 (also known as TMEM246 or C9orf125) as a GPI-specific GalNAc transferase (12). More recently, we reported that B3GALT4, which is also known as ganglioside GM1 synthase, also acts as a Gal transferase for the GPI GalNAc side chain (14). Although these achievements have furthered our understanding of the GPI GalNAc side chain, the biosynthesis pathway of the GPI glycan structure is still incompletely understood. A major issue to be resolved is identification the sialyltransferase. Currently, the characteristics of the enzyme are not known because of a lack of information, including the linkage type. Therefore, determination of the Sia linkage type is important.
Figure 1.
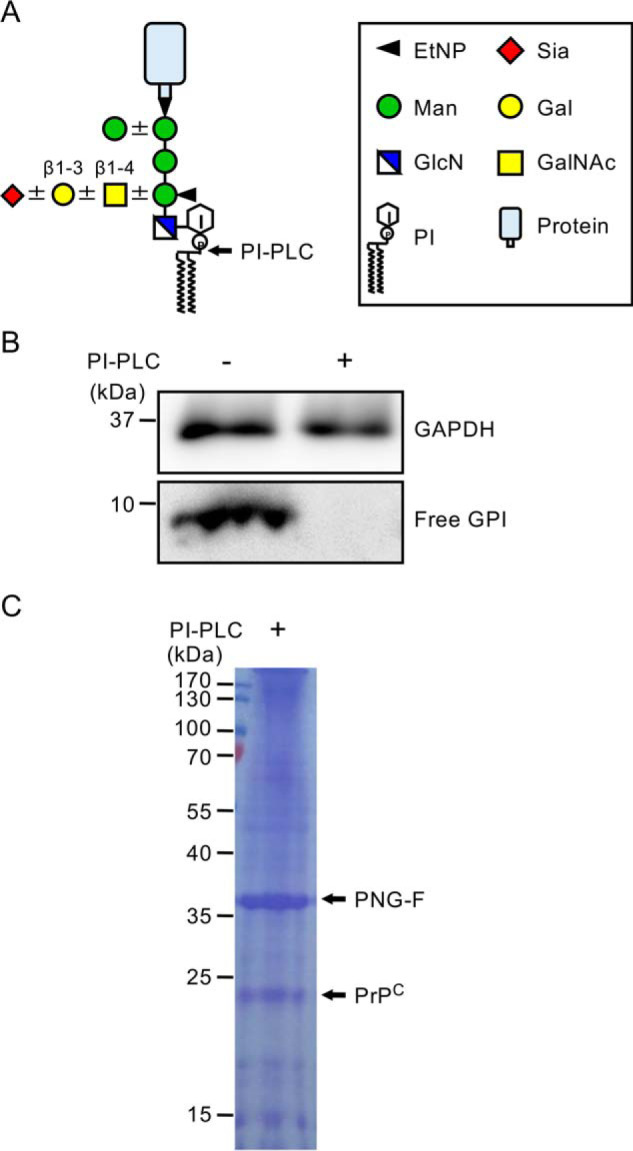
Isolation of human PrPC from KI mouse brains. A, schematic of GPI-AP composition. Core and side-chain structures of mammalian GPI-APs are shown. The cleavage site of PI-PLC is indicated by an arrow. Glycan symbols follow the symbol nomenclature for glycans (44). B, PI-PLC treatment of the mouse medulla oblongata. Medulla oblongata homogenates obtained from mice expressing human PrPC were treated with PI-PLC, and then free GPI-GalNAc was detected by Western blotting. GAPDH was used as a loading control. C, isolation of human PrPC from KI mouse brains. The lipid portion of the GPI anchor of human PrPC was removed by PI-PLC, followed by immunoprecipitation with an anti-prion antibody. N-glycans on isolated PrPC were removed by treatment with PNG-F. Bands corresponding to PNG-F and human PrPC are indicated by arrows.
Prion diseases, including Creutzfeldt–Jakob disease in humans and scrapie or bovine spongiform encephalopathy in animals, are lethal, transmissible, and progressive neurodegenerative disorders. These are designated protein misfolding disorders, similar to other neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, Huntington's disease, and ALS (15). When specific proteins are misfolded because of aging or genetic background, they aggregate and accumulate in cells, leading to cell death. Prion diseases are caused by accumulation of the scrapie form of the prion protein (PrPSc), which is converted from the normal cellular prion protein (PrPC) (16). PrPC has an α helix–rich structure, whereas PrPSc contains more β sheet structures. Prnp (the gene encoding PrPC) knockout mice are resistant to prion disease even after inoculation with PrPSc (17, 18), indicating that the presence of PrPC is critical for the pathogenesis of prion diseases and that prevention of prion conversion can be a therapeutic strategy, although the mechanisms of conversion are still unclear. Because PrPC is anchored on the cell surface via a GPI anchor, previous studies have attempted to uncover the involvement of the GPI anchor in conversion. Although cultured cells expressing GPI anchor–less PrPC were unable to support persistent infection in vitro (19), transgenic mice expressing only GPI anchor–less PrPC showed accumulation of PrPSc and replication of infectivity in the brain after inoculation with PrPSc (20). Transgenic mice had a prolonged incubation period, altered deposition pattern of PrPSc, and distinctive spread pattern of PrPSc in the brain (21). In addition, studies investigating Sia in the GPI anchor strengthen the possibility that the GPI anchor is involved in prion pathogenesis. Desialylated PrPC is resistant to conversion to PrPSc in cultured neurons (22). Moreover, mice expressing PrPC lacking Sia on the GPI anchor have a prolonged incubation time after PrPSc inoculation (23). These studies indicate the significance of the PrPC GPI anchor for the pathogenesis of prion diseases. Therefore, understanding precise structures of human PrPC GPI is important to fully understand the role of the GPI anchor in PrPSc. Nevertheless, elucidation of the human PrPC GPI anchor structure has not been achieved, although an early study determined GPI structures in the Syrian hamster (13).
In this study, we affinity-purified PrPC from knock-in (KI) mouse brains in which Prnp was replaced by human PRNP and from human brains and determined their GPI glycan structure by LC–electrospray ionization (ESI)–MS and the linkage type of Sia using the sialic acid linkage-specific alkylamidation (SALSA) method. We identified the ω site of human PrPC as Gly229 and the Sia linkage type as α2,3 linkage.
Results
Isolation of human PrPC from KI mouse brains
For structural analysis of GPI glycan by MS, the lipid portion of the GPI anchor has to be removed because the presence of lipid interferes with ionization, and, even when ionized, the pattern of product ions is highly complex and confounds analysis. Thus, we first tested phosphatidylinositol-specific phospholipase C (PI-PLC) for removal of the lipid portion of the GPI anchor in mouse brains. The mouse brain (especially the medulla oblongata) contains high levels of free GPIs with the GalNAc side chain that are not associated with proteins and exist as glycolipids on the cell surface (24). We estimated the effect of PI-PLC treatment based on loss of free GPIs. We prepared mouse medulla oblongata homogenates and incubated them with PI-PLC. Samples were subjected to Western blotting with an anti-T5 antibody, which specifically detects free GPIs with GalNAc (12, 25). Free GPIs were detected as reported previously (Fig. 1B) (24). In contrast, no signals were obtained after PI-PLC treatment, indicating that PI-PLC worked well (Fig. 1B). Previously, we generated KI mice expressing human PrPC with the 129M/M genotype (Ki-Hu129M/M) (26). To investigate the structure of the human PrPC GPI glycan, we affinity-purified human PrPC from KI mouse brains after PI-PLC treatment and then treated the isolated PrPC with peptide N-glycosidase F (PNG-F) to remove N-glycans. The prepared sample was separated by SDS-PAGE, and the gel was stained with Coomassie Brilliant Blue to visualize bands of PNG-F and human PrPC lacking N-glycans (Fig. 1C). We excised the band corresponding to de-N-glycosylated human PrPC and subjected the isolated gel fragment to analysis.
Identification of the human PrPC ω site as Gly229
To determine the ω-site amino acid and GPI glycan structures of human PrPC, we first performed LC-ESI-MS analysis. Proteins in the excised gel fragment were digested with trypsin, and the samples were separated by reverse-phase HPLC using a nanoLC system connected to a C18 column. Data were obtained by LTQ Orbitrap Velos MS. A MASCOT search revealed eight peptides of human PrPC, and the coverage was 41% (Fig. 2A and Table S1), indicating that the excised gel fragment mainly contained human PrPC. We searched for peptides with GPI glycan, which contain fragment ions of m/z 422+ and 447+ in the MS/MS profiles (these fragments contain GlcN, which is highly specific to GPI glycan). However, we barely detected peptides containing GPI glycan in this way, probably because of low hydrophobicity of the expected peptides containing a short peptide with large hydrophilic glycans. To overcome this problem, sample separation was performed by forward-phase HPLC with nanoLC using a hydrophilic interaction LC (HILIC) column, which can capture hydrophilic peptides. Using a HILIC column, we found six PrPC peptides (Table S2) and a C-terminal peptide with GPI glycan (total ion chromatogram and MS spectra are shown in Fig. S1, A and B). We initially speculated that the ω site of human PrPC was Ser230 by using PredGPI software to predict GPI attachment sites and because of homology to the Syrian hamster PrPC sequence (whose ω site is Ser231) (Fig. 2B) (27, 28). Unexpectedly, we only identified Gly229 GPI glycan but not Gly229-Ser230 GPI glycan (Table S3; raw data are available from PRIDE). These results strongly indicate that Gly229 and not Ser230 is the bona fide ω site of human PrPC. We compared the sequence of GPI attachment signal among various species, including human and Syrian hamster. Alignment of PrPC peptides indicated that Syrian hamster Ser231 is conserved among rodent PrPC proteins, whereas human Gly229 is conserved among nonrodent species such as cows and sheep (Fig. 2B).
Figure 2.
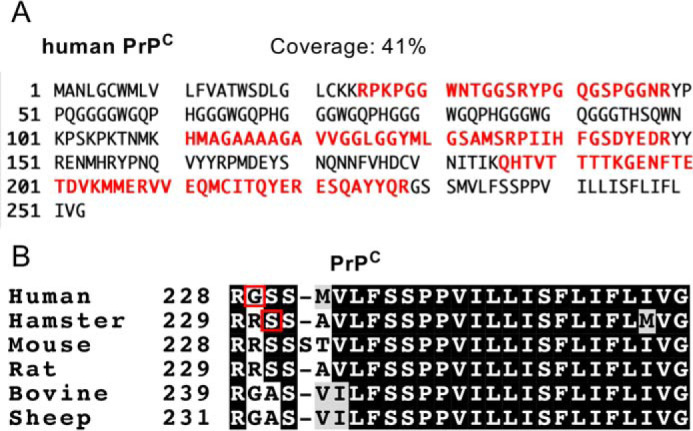
Identification of the ω site in human PrPC from KI mouse brains. A, peptide coverage of purified human PrPC from KI mouse brains. Trypsinized peptides were analyzed by LC-ESI-MS, and the obtained data were analyzed using the MASCOT database. B, alignment of PrPC sequences. Sequence alignment was performed using ClustalW and BoxShade. Residues conserved among species are shaded in gray or black. Red boxes indicate the ω sites determined in this and previous studies (28).
Determination of GPI glycan structures of human PrPC from KI mice
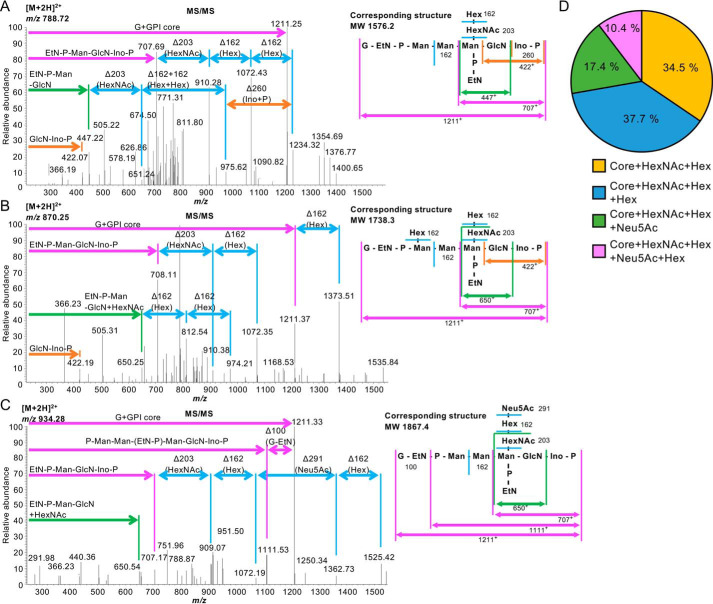
Based on the known structures of the mammalian GPI anchors, there are eight possible GPI glycan species: core only (m/z 1211.3+), core + fourth Man (m/z 1373.4+), core + GalNAc (m/z 1414.4+), core + GalNAc + fourth Man (m/z 1576.4+), core + GalNAc + Gal (m/z 1576.4+), core + GalNAc + Gal + fourth Man (m/z 1738.5+), core + GalNAc + Gal + Sia (m/z 1867.5+ when Sia is N-acetylneuraminic acid (Neu5Ac)), and core + GalNAc + Gal + Sia + fourth Man (m/z 2029.6+ when Sia is Neu5Ac). Because MS analysis provides m/z only, peptides with the same m/z, such as core + GalNAc + fourth Man and core + GalNAc + Gal cannot be distinguished, and the precise sugar species are not determined. Thus, there are seven possible species in MS analysis. We found four species of GPI glycans and verified them by MS/MS spectra (Fig. 3, A–C, and Table S3). Next we investigated the percentage of each GPI glycan species. For this, peak intensities of each fragment were calculated from the MS spectra (Fig. S1B). We revealed that core + N-acetylhexosamine (HexNAc) + hexose (Hex), core + HexNAc + Hex + Hex, core + HexNAc + Hex + Neu5Ac, and core + HexNAc + Hex + Neu5Ac + Hex were present at 34.5%, 37.7%, 17.4%, and 10.4%, respectively (Fig. 3D and Table S3; HexNAc and Hex are GalNAc and Gal or fourth Man, respectively). The peptides corresponding to core only, core + HexNAc, and core + Hex were not detected (Fig. 3D and Table S3). These results indicate that almost all of the GPI anchor of human PrPC is modified by a GalNAc side chain and that at least 60% of the GPI anchor is elongated by Gal and 25% is further elongated by Neu5Ac in the mouse brain. We did not find N-glycolylneuraminic acid–containing GPI, probably because of very low levels of N-glycolylneuraminic acid in vertebrate brains (29).
Figure 3.
Determination of GPI glycan structures in human PrPC from KI mice by LC-ESI-MS/MS analysis. A, left panel, the MS/MS spectra of the m/z 788.722+ ion shown in Fig. S1B and the identities of fragment ions. Right panel, the corresponding structure (MW 1576.2) of a C-terminal peptide with GPI containing a HexNAc-Hex side chain. B, left panel, the MS/MS spectra of the m/z 870.252+ ion. Right panel, the corresponding structure (MW 1738.3) of a C-terminal peptide with GPI containing Hex and HexNAc-Hex side chains. C, left panel, the MS/MS spectra of the m/z 934.32+ ion. Right panel, the corresponding structure (MW 1867.4) of a C-terminal peptide with GPI containing a HexNAc-Hex-Neu5Ac side chain. In A–C, all annotated fragment ions are monovalent cations. D, percentages of GPI glycan species in KI mouse brains. Core only, Core + Hex (fourth Man), and Core + HexNAc (GalNAc) were not detected.
Neu5Ac in the GPI anchor is attached by α2,3 linkage
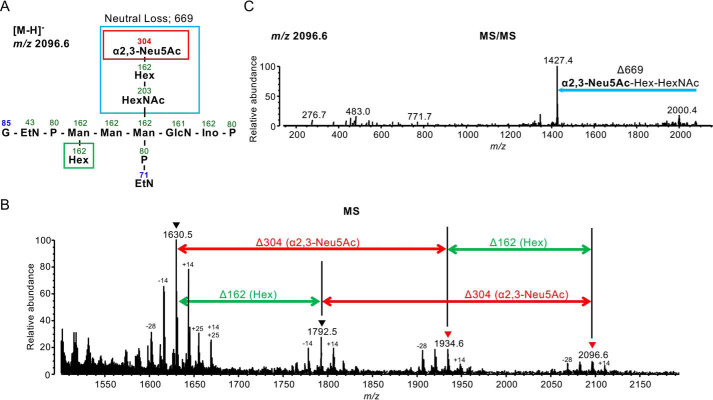
Next, to determine the linkage type of a Neu5Ac in the human PrPC GPI anchor, we used a linkage-specific chemical derivatization approach, SALSA (30, 31). SALSA converts carboxyl groups in Sia residues into alkylamide forms with different alkyl chain lengths in a linkage-specific manner, allowing direct discrimination of the α2,3 linkage type from the α2,6 linkage type by MS. To suppress unfavorable by-products during SALSA reactions, free amine groups were capped by dimethyl labeling (DML) prior to SALSA reactions. After DML and SALSA treatments, the derivatized sample was measured in negative ion mode MALDI-MS. There are mainly four glycoforms at m/z 1630.5−, m/z 1792.5−, m/z 1934.6−, and m/z 2096.6− in human PrPC (Fig. 4, A and B, and Fig. S2, A–C). The peaks at m/z 1630.5− and 1792.5− correspond to nonsialylated GPI anchor glycopeptides (Fig. 4B). The peaks at m/z 1934.6− and 2096.6− correspond to sialylated GPI anchor glycopeptides, which include a methylamidated Neu5Ac residue (304 Da) (Fig. 4B). This indicates that the linkage type of a Neu5Ac residue is α2,3-linked, not α2,6-linked. Structural analysis by MS/MS supports these assignments (see Fig. 4C and Fig. S2, A–C for MS/MS spectra). We could not identify GPI species without HexNAc (m/z 1427−) or Hex and HexNAc (m/z 1265−) because of dense peaks of foreign substances (HexNAc, GalNAc; Hex, fourth Man). Accompanying signals, including −28, −14, and +14 Da, were by-products attributable to incomplete or additional DML reactions (MS/MS data were not shown). Signals of +25 Da also seemed to be by-products, but their structures and origins were not identified because of unexpected reactions. Although these by-product signals were detected, the main signals with Δm = 304 Da clearly indicated the presence of α2,3-linked Neu5Ac.
Figure 4.
Linkage analysis of Sia in GPI anchors using the SALSA method. A, structure of a peptide with the GPI glycan displayed in B and C. Numbers indicate m/z values. Boxes show the neutral loss after MS/MS analysis. B, MS spectra of peptides containing the GPI glycan shown in A. A 304-Da difference indicates the presence of α2,3-linked Neu5Ac. C, the MS/MS spectrum of a C-terminal peptide with GPI containing Hex and HexNAc-Hex-Neu5Ac. A 669-Da loss indicates loss of α2,3-linked Neu5Ac, Hex (Gal), and HexNAc (GalNAc).
The ω site and GPI glycan structures of PrPC in the human brain
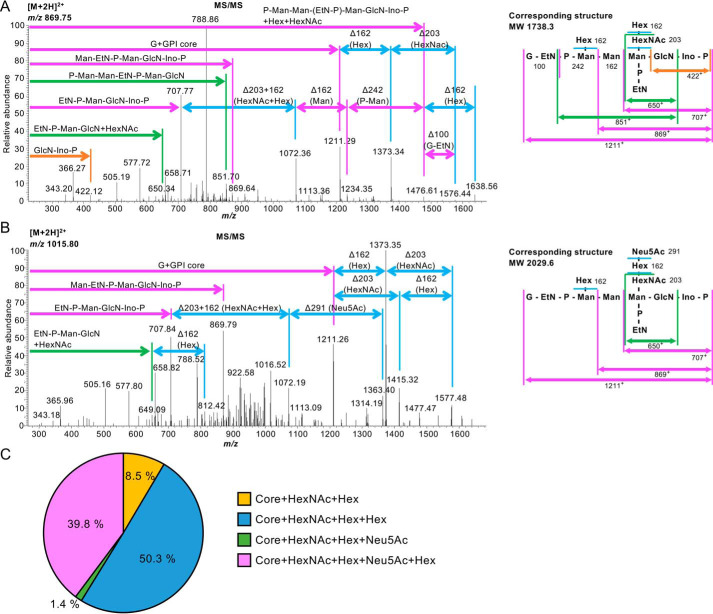
Finally, we determined GPI-glycan structures of PrPC isolated from a human brain. Deglycosylated and delipidated PrPC was prepared by PNG-F and PI-PLC treatments, followed by affinity purification as described above. Trypsinized samples were analyzed by LC-ESI-MS (Table S4). Similar to the results for human PrPC from KI mice, the C-terminal glycopeptide corresponding to Gly229 GPI glycan, but not Gly229-Ser230 GPI glycan, was detected, confirming that Gly229 was indeed the ω site of human PrPC (Table S5; raw data are available from PRIDE). We again detected four kinds of GPI glycans, core + HexNAc + Hex, core + HexNAc + Hex + Hex, core + HexNAc + Hex + Neu5Ac, and core + HexNAc + Hex + Neu5Ac + Hex, in the human brain sample (see Fig. 5, A and B, for MS/MS spectra). The percentages of these species were 8.5%, 50.3%, 1.4%, and 39.8%, respectively (Fig. 5C and Table S5). This indicates that almost all GPI in human brain PrPC is modified by a GalNAc side chain; 90% of the GalNAc side chain is further elongated by Gal, and 40% of the GalNAc side chain has Gal-Neu5Ac elongation. These results also indicate that 90% of GPI in the human brain PrPC contains a fourth Man. Taken together, we conclude that the ω site of human PrPC is Gly229 and that the majority of human brain PrPC contains a fourth Man and a galactosylated GalNAc side chain with or without Neu5Ac modification.
Figure 5.
Determination of GPI glycan structures of human PrPC from human brain by LC-ESI-MS/MS analysis. A, left panel, the MS/MS spectra of the m/z 869.752+ ion. All annotated fragment ions are monovalent cations. Right panel, the corresponding structure (MW 1738.3) of a C-terminal peptide with GPI containing Hex and HexNAc-Hex side chains. B, left panel, the MS/MS spectra of the m/z 1015.802+ ion. All annotated fragment ions are monovalent cations. Right panel, the corresponding structure (MW 2029.6) of a C-terminal peptide with GPI containing Hex and HexNAc-Hex-Neu5Ac side chains. C, percentages of GPI glycan species in the human brain. Core only, Core + Hex (fourth Man), and Core + HexNAc (GalNAc) were not detected.
Discussion
Here we determined the GPI glycan structures of human PrPC, including the Sia linkage type. The GPI Sia linkage type was unknown for any GPI-APs. We took advantage of KI mice expressing human PrPC in place of mouse PrPC and the SALSA method. In general, analysis of Sia-containing glycans by MS is difficult because of low ionization and chemical instability. We employed the SALSA method to enhance detection of Sia and to determine the linkage types. We demonstrated that Sia in the human PrPC GPI anchor is Neu5Ac linked to Gal via α2,3 linkage (Fig. 4). Intriguingly, the GPI side-chain structure, NeuNAcα2–3Galβ1–3GalNAcβ1-, is exactly the same as a partial structure of GD1a, a product of sialylation of GM1a ganglioside (32). Because B3GALT4 is a common Gal transferase for GPI GalNAc modification and GM1 ganglioside synthesis (33, 34), it seems possible that sialyltransferase for the GPI anchor is the same as GD1a synthase. This possibility is being tested.
The profile of GPI glycan species identified in human PrPC is similar to that of Syrian hamster PrPC (13), indicating that GPI glycan structures of PrPC are conserved among species. Conservation of GPI glycan structures might suggest the significance of these structures for PrPC functions. Consistent with this notion, sialylation of PrPC might be involved in the pathology of prion diseases (22, 23). Elucidation of the physiological significance of the GPI GalNAc side chain is an issue to be addressed.
Galactosylated and sialylated GPI anchors are more abundant in human brain PrPC than in human PrPC in KI mouse brain (compare Fig. 3D with Fig. 5C) and Syrian hamster PrPC (13). This implies that enzyme activities for GPI anchor galactosylation and sialylation are diverse in brain tissues of different organisms. We recently reported that the percentages of Gal-modified and Sia-Gal–modified GPI-glycans in CD59, a GPI-AP, were only 10% and 1% respectively, in HEK293 cells (14). Overexpression of B3GALT4 in HEK293 cells increased these percentages, indicating that B3GALT4-mediated Gal transfer was rate-limiting. We found that ∼90% and ∼40% of GPI glycans in human brain PrPC are modified by Gal and Sia-Gal, respectively, indicating that the brain has high B3GALT4 activity for GPI side-chain modification (Figs. 3D and 5C). The adult mammalian brain has high GM1 content (35, 36) and B3GALT4 transcript levels (37). Therefore, the activity of B3GALT4 in ganglioside biosynthesis and GPI-AP maturation is high in the brain. In contrast to PrPC, GPI in Thy-1 from rat brain always contains a GalNAc side chain without Gal modification (11). This indicates that the GPI glycan structures are different among proteins, even in the same tissue. What determines the different GPI structures is currently unknown. Recently, it has been reported that GPI glycan structures are dependent on their GPI signal peptides; replacement of the PrPC GPI signal peptide with that of Thy-1 caused loss of Sia modification in PrPC (23). Because loss of Sia modification was examined only by lectin blotting using Sambucus nigra lectin, which preferentially binds to α2,6-linked Sia and, to a lesser degree, α2,3-linked Sia, structural determination is needed for a solid conclusion.
We showed that the ω site of human brain PrPC is Gly229. The ω site of Syrian hamster PrPC is Ser231 (28), and that of mouse PrPC is Ser230 (38), both of which correspond to Ser230 of human PrPC. In addition, the PredGPI software, used to predict GPI attachment sites, indicated the ω site of human PrPC to be Ser230. Therefore, it was believed that the ω site of human PrPC is Ser230. However, LC-ESI-MS analysis demonstrated that the bona fide ω site of human brain PrPC is Gly229 (Tables S3 and S5). Thus, the ω-site amino acid could be different among species, and experimental validation of GPI anchor structures, including the ω site in various species, is important. It is unknown whether the difference in ω sites has any effect on protein function because several studies investigating the function of GPI attachment signal peptides (23, 39) indicate that a difference in the ω site might cause a functional difference in the same protein from different species.
In conclusion, we report a bona fide ω-site amino acid and precise GPI glycan structures, including the Sia linkage type, of human PrPC in the brain. Our study will further our understanding of PrPC physiology, the pathology of prion diseases, and the complete pathway of GPI-AP biosynthesis.
Experimental procedures
Ethics statement
This study was approved by the Institutional Ethics Committee of Hokkaido University Graduate School of Veterinary Medicine (VET27-1). All experiments using human materials were in compliance with the Helsinki Declaration. Animal experiments were performed in strict accordance with the Regulations for Animal Experiments and Related Activities at Osaka University and Hokkaido University, and Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions by the Ministry of Education, Culture, Sports, Science, and Technology in Japan, Notice 71. The protocol was approved by the Institutional Animal Care and Use Committee of Hokkaido University (14-0170).
Animals
Nine-week-old C57BL/6 mice were used to estimate the effect of PI-PLC treatment. Production of knock-in mice expressing human PrPC with the 129M/M genotype (Ki-Hu129M/M) has been reported previously (26). Two-month-old Ki-Hu129M/M mice were sacrificed, and the brains were frozen immediately until purification of PrPC.
Patient
A patient included in this study was clinically, genetically, and histopathologically diagnosed as having nonprion disease. Brain tissue was obtained at autopsy after receiving written informed consent for research use. The genotype and mutations in the ORF of the PRNP gene were determined by sequence analysis as described previously (40). The PRNP genotypes at polymorphic codon 129 and 219 were methionine homozygosity and glutamine homozygosity, respectively.
Antibodies and reagents
The mAb against GAPDH (clone 6c5) was purchased from Thermo Fisher (Waltham, MA). The T5_4E10 mAb against free GPI GalNAc (mouse IgM) was kindly provided by Dr. Jean François Dubremetz (Montpellier University) (41). The T5_4E10 mAb is available from BEI Resources (ATCC, catalog no. NR-50267). The mAb against PrP (clone 3F4) was purchased from BioLegend (San Diego, CA). The secondary antibodies were HRP-conjugated goat anti-mouse IgG (GE Healthcare) and goat anti-mouse IgM (Thermo Fisher). PI-PLC was purchased from Thermo Fisher. All reagents for DML, such as 37% formaldehyde, 1 m triethylammonium bicarbonate buffer (pH 8.5), sodium cyanoborohydride, 28% ammonium hydroxide solution, and formic acid, were purchased from Sigma-Aldrich (St. Louis, MO). All reagents for Sia derivatization, such as 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride, 1-hydroxybenzotriazole monohydrate, isopropylamine hydrochloride, 40% methylamine in water, and DMSO, were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). MALDI matrix chemicals including 3-aminoquinoline, p-coumaric acid, and ammonium dihydrogen phosphate, were purchased from Sigma-Aldrich. GL-Tip amide for HILIC solid phase extraction was from GL Science Inc. (Tokyo, Japan).
PI-PLC treatment of mouse brain homogenates
Nine-week-old male mice were sacrificed, and brains were eviscerated. To detect free GPI, the medulla oblongata was isolated. Tissue homogenates were obtained as described previously (24). Briefly, tissues were washed with PBS and homogenized in 30 volumes of lysis buffer (20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 60 mm n-octyl-β-d-glucoside, and 1× protease inhibitor mixture) using a hand-held microhomogenizer (Microtec Co. Ltd.) and rotated at 4 °C for 2 h, followed by centrifugation at 17,900 × g at 4 °C for 15 min. The protein concentrations of samples were measured using a Pierce BCA Protein Assay Kit (Thermo Fisher). 5 μg of protein samples was treated with or without PI-PLC (1 unit) at 37 °C for 1 h. Samples were mixed with 4× SDS sample buffer and boiled at 95 °C for 5 min. 5 μg of protein samples was run on SDS-PAGE, followed by Western blotting.
Western blotting
Samples were run on 10%–20% gradient SDS-PAGE gels, followed by transfer to PVDF membranes. The PVDF membranes were blocked with 5% nonfat milk in Tris buffered saline containing Tween 20 at room temperature for 1 h. Free GPI GalNAc and GAPDH were detected using anti-T5_4E10 (1:500) and anti-GAPDH (1:4000), followed by detection with anti-mouse IgM HRP (1:1000) and anti-mouse IgG HRP (1:4000).
Purification of PrPC
PrPC was purified from human or mouse brains using Protein G Mag–Sepharose (GE Healthcare) according to the manufacturer's instructions. Briefly, 10% brain homogenates were prepared with binding buffer (PBS, 0.5% Triton X-100, 0.5% sodium deoxycholate, and 1× protease inhibitor mixture Complete (Merck)). After removal of cell debris by centrifugation at 1000 × g for 10 min at 4 °C, the supernatants were treated with PI-PLC (1 units/200 mg of tissue) for 30 min at 37 °C. The digested homogenates were incubated with anti-PrP mAb 3F4 for 3 h at 4 °C and then incubated with Protein G Mag–Sepharose beads overnight at 4 °C. The beads were collected by a magnetic stand and washed with binding buffer three times. The washed beads were resuspended in Laemmli's sample buffer (60 mm Tris-HCl (pH 6.8), 5% glycerol, 2% SDS, and 0.01% bromphenol blue) and boiled at 100 °C for 10 min. After removal of the beads, the supernatants were digested with PNG-F prime (1000 units/200 mg of tissue) (N-Zyme Scientifics) overnight at 37 °C to remove N-linked glycans.
In silico analyses
Prediction of the ω site of human PrPC was carried out using PredGPI software (27). An amino acid sequence of human PrPC (reference sequence NP_000302.1) was obtained from the NCBI and submitted to PredGPI (RRID: SCR_018363) for ω site prediction using a general model. Alignment of PrPC sequences was constructed using ClustalW and BoxShade software. For this, amino acid sequences of human, hamster, mouse, rat, bovine, and sheep PrPC (reference sequences NP_000302.1, XP_012967855.1, NP_035300.1, NP_036763.1, NP_001258555.1, and NP_001009481.1, respectively) were obtained from the NCBI and submitted to ClustalW (RRID: SCR_017277). Alignment was performed in protein alignment mode with the default setting except for output options, ORDER: input. The data were downloaded as an aln file and opened with a text editing application. Sequence alignment with a shaded background was created with the default setting and put out in an EPS_portrait using BoxShade software (RRID: SCR_007165).
LC-ESI-MS analysis
To analyze the GPI structures of human PrPC, bands corresponding to PrPC were excised, reduced with 10 mm DTT, and alkylated with 55 mm iodoacetamide. After in-gel digestion with trypsin, samples were subjected to LC-ESI-MS/MS analysis. To obtain peptide coverage for human PrPC, mass spectrometry analysis was performed using a hydrophobic column as described previously (12). The sample was purified using Zip-Tipμ-C18 pipette tips (Millipore, Burlington, MA) before analysis. Data were collected in positive ion mode on a nanoLC system (Advance, Michrom BioResources) using a C18 column (0.1 × 150 mm) coupled to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher). The mass tolerance for precursor ions was ±10 ppm. The mass tolerance for fragment ions was ±0.8 Da. The threshold score/expectation value for accepting individual spectra was p < 0.05. The mobile phase consisted of water containing 0.1% formic acid (solvent A) and acetonitrile (solvent B). Peptides were eluted by a gradient of 5%–35% solvent B over 45 min. The mass scanning range was set at m/z 350–1500. The ion spray voltage was set at 1.8 kV. The MS/MS analysis was performed by automatic switching between MS and MS/MS modes at a collision energy of 35%. To detect peptides containing a GPI anchor, mass spectrometry analysis was performed using a hydrophilic column. The samples were purified with Nu-Tip polyHydroxyethylA (HILIC) tips (GlySci) according to the manufacturer's protocol before measurements. Purified peptides were then dissolved in 0.1% formic acid for measurement. Data were collected in positive ion mode on a nanoLC system using an EX-Nano Inert Sustain Amide capillary (0.1 × 150 mm; particle size, 3 μm) coupled to the same mass spectrometer. Measurement samples were injected onto the column by loop injection method. The mobile phase was the same as above, and peptides were eluted by a gradient of 90%–5% solvent B over 24 min or 67 min for samples from KI mouse brains or human brains, respectively. The same MS parameters were used as described above. The obtained mass data were analyzed by Xcalibur (Proteome Discoverer1.4 v. 1.4.0.288), and peptides were identified by MASCOT v. 2.6(2.6.0) (Matrix Science) using the SwissProt_2020_01 database (561,911 sequences; 202,173,710 residues; taxonomy: Homo sapiens (human) (20,366 sequences)). Protease specificity was set for trypsin (C-term, KR; Restrict, P; Independent, no; Semispecific, no; two missed and/or nonspecific cleavages permitted). Fixed modification considered was carbamidomethyl cysteine, and variable modifications considered were acetyl N terminus, N-terminal Gln to pyro-Glu, and oxidation of methionine. For quantification, the intensities of the top three peaks (monoisotopic peak + 0.5 or 0.33 + 1.0 or 0.66 Da for divalent or trivalent ions, respectively) corresponding to GPI-containing peptides in the MS spectra were calculated.
Chemical derivatization for prion GPI-anchor glycopeptides
In-gel tryptic digest of human PrPC was collected to a tube, and solvents were removed in vacuo. The digest was derivatized by DML according to the literature with some modifications (42). Briefly, the digest was redissolved with 20 μl of 100 mm triethylammonium bicarbonate buffer. The mixture was vortexed and then mixed with 1.6 μl of 2% formaldehyde and 1.6 μl of 300 mm sodium cyanoborohydride. The mixture was gently vortexed for 1 h at 25 °C (room temperature). After the reaction, 3.2 μl of 1% ammonium hydroxide was added and vortexed for quenching. To acidify, 1.6 μl of formic acid was then added, and the mixture was diluted with 170 μl of acetonitrile (ACN).
Excess reagents were removed by HILIC solid phase extraction microtip (GL-Tip Amide). The ACN-diluted reaction mixture was loaded into an equilibrated microtip. The microtip was then washed three times with 100 μl of 90% ACN containing 0.1% TFA. The DML-derivatized samples were eluted using 20 μl of H2O. To ensure elution from styrene-divinylbenzene frit located under amide particles, the microtip were additionally washed with 20 μl of 90% ACN containing 0.1% TFA. The eluents were mixed, and the solvents were then removed in vacuo.
DML-derivatized samples were further derivatized in a sialyl linkage–specific manner by SALSA. The dried sample was redissolved with 20 μl of the linkage-specific alkylamidation solution (500 mm 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride, 500 mm 1-hydroxybenzotriazole monohydrate, and 2 m isopropylamine hydrochloride in DMSO) and then incubated at 25 °C (room temperature) for 1 h. In this reaction step, α2,6-linked sialic acids are amidated by isopropylamine hydrochloride, whereas α2,3-linked sialic acids were converted to lactone forms. Lactone forms were then converted to methylamide forms via ring-opening aminolysis by adding 20 μl of 10% methylamine solution. The reaction mixture was diluted by adding 160 μl of 2.5% TFA in ACN. The excess reagents were removed by the HILIC microtip, and then the solvent was removed in vacuo.
MALDI-MS analysis
Mass spectra of DML and SALSA-derivatized GPI anchor glycopeptides were obtained by MALDI-quadrupole ion trap-TOF-MS (AXIMA-Resonance, Shimadzu/Kratos) in negative ion mode using 3-aminoquinoline/p-coumaric acid liquid matrix doped with 2 mm ammonium dihydrogen phosphate.
Data availability
The mass spectrometric proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (43) partner repository with the dataset identifiers PXD017655 (project name “GPI-glycan structures of human Prion from knock-in mice_LC-MSMS”) and PXD017656 (project name “GPI-glycan structures of human Prion from a human brain”).
Supplementary Material
Acknowledgments
We thank Dr. J. F. Dubremetz for the T5_4E10 mAb, Dr. Yoshiko Murakami and Dr. Yicheng Wang for discussions, and Keiko Kinoshita for technical help. We thank Jeremy Allen, Ph.D., from the Edanz Group for editing a draft of this manuscript.
This article contains supporting information.
Author contributions—A. K. and T. Kinoshita conceptualization; A. K. and T. Kitamoto resources; A. K. and T. Kinoshita funding acquisition; A. K., T. H., T. N., A. N., Y. M., and Y. T. investigation; A. K., T. H., T. N., A. N., and T. Kinoshita writing-original draft; A. K. and T. Kinoshita project administration; T. H. data curation; T. H. and T. N. methodology; T. H., Y. M., and T. Kinoshita writing-review and editing; T. Kinoshita supervision.
Funding and additional information—This work was supported by Japan Society for the Promotion of Science and MEXT KAKENHI grants JP16H04753 and 17H06422 (to T. Kinoshita), a grant from The Kato Memorial Trust for Nambyo Research (to A. K.), and Japan Society for the Promotion of Science KAKENHI Grant 18K05963 (to A. K.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- GPI
- glycosylphosphatidylinositol
- GPI-AP
- GPI-anchored protein
- EtNP
- ethanolamine phosphate
- Man
- mannose
- GlcN
- glucosamine
- ER
- endoplasmic reticulum
- Gal
- galactose
- Sia
- sialic acid
- KI
- knock-in
- ESI
- electrospray ionization
- SALSA
- sialic acid linkage-specific alkylamidation
- PI-PLC
- phosphatidylinositol-specific phospholipase C
- PNG-F
- peptide N-glycosidase F
- HILIC
- hydrophilic interaction LC
- Neu5Ac
- N-acetylneuraminic acid
- HexNAc
- N-acetylhexosamine
- Hex
- hexose
- DML
- dimethyl labeling
- ACN
- acetonitrile
- PrPC
- normal cellular prion protein
- PrPSc
- scrapie form of the prion protein
- MW
- molecular weight.
References
- 1. Kinoshita T. (2020) Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol. 10, 190290 10.1098/rsob.190290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kinoshita T., and Fujita M. (2016) Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling. J. Lipid Res. 57, 6–24 10.1194/jlr.R063313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ferguson M. A. J., Hart G. W., and Kinoshita T. (2015) in Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L., and Seeberger P. H., eds.), 3rd Ed., pp. 137–150, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 4. Tanaka S., Maeda Y., Tashima Y., and Kinoshita T. (2004) Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 279, 14256–14263 10.1074/jbc.M313755200 [DOI] [PubMed] [Google Scholar]
- 5. Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., and Kinoshita T. (2009) GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell 139, 352–365 10.1016/j.cell.2009.08.040 [DOI] [PubMed] [Google Scholar]
- 6. Fujita M., Watanabe R., Jaensch N., Romanova-Michaelides M., Satoh T., Kato M., Riezman H., Yamaguchi Y., Maeda Y., and Kinoshita T. (2011) Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J. Cell Biol. 194, 61–75 10.1083/jcb.201012074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maeda Y., Tashima Y., Houjou T., Fujita M., Yoko-o T., Jigami Y., Taguchi R., and Kinoshita T. (2007) Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol. Biol. Cell 18, 1497–1506 10.1091/mbc.e06-10-0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tashima Y., Taguchi R., Murata C., Ashida H., Kinoshita T., and Maeda Y. (2006) PGAP2 is essential for correct processing and stable expression of GPI-anchored proteins. Mol. Biol. Cell 17, 1410–1420 10.1091/mbc.e05-11-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hong Y., Maeda Y., Watanabe R., Ohishi K., Mishkind M., Riezman H., and Kinoshita T. (1999) Pig-n, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J. Biol. Chem. 274, 35099–35106 10.1074/jbc.274.49.35099 [DOI] [PubMed] [Google Scholar]
- 10. Taron B. W., Colussi P. A., Wiedman J. M., Orlean P., and Taron C. H. (2004) Human Smp3p adds a fourth mannose to yeast and human glycosylphosphatidylinositol precursors in vivo. J. Biol. Chem. 279, 36083–36092 10.1074/jbc.M405081200 [DOI] [PubMed] [Google Scholar]
- 11. Homans S. W., Ferguson M. A., Dwek R. A., Rademacher T. W., Anand R., and Williams A. F. (1988) Complete structure of the glycosyl phosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature 333, 269–272 10.1038/333269a0 [DOI] [PubMed] [Google Scholar]
- 12. Hirata T., Mishra S. K., Nakamura S., Saito K., Motooka D., Takada Y., Kanzawa N., Murakami Y., Maeda Y., Fujita M., Yamaguchi Y., and Kinoshita T. (2018) Identification of a Golgi GPI-N-acetylgalactosamine transferase with tandem transmembrane regions in the catalytic domain. Nat. Commun. 9, 405 10.1038/s41467-017-02799-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stahl N., Baldwin M. A., Hecker R., Pan K. M., Burlingame A. L., and Prusiner S. B. (1992) Glycosylinositol phospholipid anchors of the scrapie and cellular prion proteins contain sialic acid. Biochemistry 31, 5043–5053 10.1021/bi00136a600 [DOI] [PubMed] [Google Scholar]
- 14. Wang Y., Maeda Y., Liu Y. S., Takada Y., Ninomiya A., Hirata T., Fujita M., Murakami Y., and Kinoshita T. (2020) Cross-talks of glycosylphosphatidylinositol biosynthesis with glycosphingolipid biosynthesis and ER-associated degradation. Nat. Commun. 11, 860 10.1038/s41467-020-14678-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aguzzi A., and Lakkaraju A. K. K. (2016) cell biology of prions and prionoids: a status report. Trends Cell Biol. 26, 40–51 10.1016/j.tcb.2015.08.007 [DOI] [PubMed] [Google Scholar]
- 16. Prusiner S. B., Scott M. R., DeArmond S. J., and Cohen F. E. (1998) Prion protein biology. Cell 93, 337–348 10.1016/S0092-8674(00)81163-0 [DOI] [PubMed] [Google Scholar]
- 17. Büeler H., Aguzzi A., Sailer A., Greiner R. A., Autenried P., Aguet M., and Weissmann C. (1993) Mice devoid of PrP are resistant to scrapie. Cell 73, 1339–1347 10.1016/0092-8674(93)90360-3 [DOI] [PubMed] [Google Scholar]
- 18. Prusiner S. B., Groth D., Serban A., Koehler R., Foster D., Torchia M., Burton D., Yang S. L., and DeArmond S. J. (1993) Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc. Natl. Acad. Sci. U.S.A. 90, 10608–10612 10.1073/pnas.90.22.10608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McNally K. L., Ward A. E., and Priola S. A. (2009) Cells expressing anchorless prion protein are resistant to scrapie infection. J. Virol. 83, 4469–4475 10.1128/JVI.02412-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., Raymond L., Favara C., Baron G., Priola S., Caughey B., Masliah E., and Oldstone M. (2005) Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 308, 1435–1439 10.1126/science.1110837 [DOI] [PubMed] [Google Scholar]
- 21. Rangel A., Race B., Phillips K., Striebel J., Kurtz N., and Chesebro B. (2014) Distinct patterns of spread of prion infection in brains of mice expressing anchorless or anchored forms of prion protein. Acta Neuropathol. Commun. 2, 8 10.1186/2051-5960-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bate C., Nolan W., and Williams A. (2016) Sialic acid on the glycosylphosphatidylinositol anchor regulates PrP-mediated cell signaling and prion formation. J. Biol. Chem. 291, 160–170 10.1074/jbc.M115.672394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Puig B., Altmeppen H. C., Linsenmeier L., Chakroun K., Wegwitz F., Piontek U. K., Tatzelt J., Bate C., Magnus T., and Glatzel M. (2019) GPI-anchor signal sequence influences PrPC sorting, shedding and signalling, and impacts on different pathomechanistic aspects of prion disease in mice. PLoS Pathog. 15, e1007520 10.1371/journal.ppat.1007520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y., Hirata T., Maeda Y., Murakami Y., Fujita M., and Kinoshita T. (2019) Free, unlinked glycosylphosphatidylinositols on mammalian cell surfaces revisited. J. Biol. Chem. 294, 5038–5049 10.1074/jbc.RA119.007472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Striepen B., Zinecker C. F., Damm J. B., Melgers P. A., Gerwig G. J., Koolen M., Vliegenthart J. F., Dubremetz J. F., and Schwarz R. T. (1997) Molecular structure of the “low molecular weight antigen” of Toxoplasma gondii: a glucose α1–4 N-acetylgalactosamine makes free glycosyl-phosphatidylinositols highly immunogenic. J. Mol. Biol. 266, 797–813 10.1006/jmbi.1996.0806 [DOI] [PubMed] [Google Scholar]
- 26. Asano M., Mohri S., Ironside J. W., Ito M., Tamaoki N., and Kitamoto T. (2006) vCJD prion acquires altered virulence through trans-species infection. Biochem. Biophys. Res. Commun. 342, 293–299 10.1016/j.bbrc.2006.01.149 [DOI] [PubMed] [Google Scholar]
- 27. Pierleoni A., Martelli P. L., and Casadio R. (2008) PredGPI: a GPI-anchor predictor. BMC Bioinformatics 9, 392 10.1186/1471-2105-9-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stahl N., Baldwin M. A., Burlingame A. L., and Prusiner S. B. (1990) Identification of glycoinositol phospholipid linked and truncated forms of the scrapie prion protein. Biochemistry 29, 8879–8884 10.1021/bi00490a001 [DOI] [PubMed] [Google Scholar]
- 29. Davies L. R., Pearce O. M., Tessier M. B., Assar S., Smutova V., Pajunen M., Sumida M., Sato C., Kitajima K., Finne J., Gagneux P., Pshezhetsky A., Woods R., and Varki A. (2012) Metabolism of vertebrate amino sugars with N-glycolyl groups: resistance of α2–8-linked N-glycolylneuraminic acid to enzymatic cleavage. J. Biol. Chem. 287, 28917–28931 10.1074/jbc.M112.365056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nishikaze T., Tsumoto H., Sekiya S., Iwamoto S., Miura Y., and Tanaka K. (2017) Differentiation of sialyl linkage isomers by one-pot sialic acid derivatization for mass spectrometry-based glycan profiling. Anal. Chem. 89, 2353–2360 10.1021/acs.analchem.6b04150 [DOI] [PubMed] [Google Scholar]
- 31. Hanamatsu H., Nishikaze T., Miura N., Piao J., Okada K., Sekiya S., Iwamoto S., Sakamoto N., Tanaka K., and Furukawa J. I. (2018) Sialic acid linkage specific derivatization of glycosphingolipid glycans by ring-opening aminolysis of lactones. Anal. Chem. 90, 13193–13199 10.1021/acs.analchem.8b02775 [DOI] [PubMed] [Google Scholar]
- 32. Schnaar R. L. (2016) Gangliosides of the vertebrate nervous system. J. Mol. Biol. 428, 3325–3336 10.1016/j.jmb.2016.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amado M., Almeida R., Carneiro F., Levery S. B., Holmes E. H., Nomoto M., Hollingsworth M. A., Hassan H., Schwientek T., Nielsen P. A., Bennett E. P., and Clausen H. (1998) A family of human β3-galactosyltransferases: characterization of four members of a UDP-galactose:β-N-acetyl-glucosamine/β-N-acetyl-galactosamine β-1,3-galactosyltransferase family. J. Biol. Chem. 273, 12770–12778 10.1074/jbc.273.21.12770 [DOI] [PubMed] [Google Scholar]
- 34. Miyazaki H., Fukumoto S., Okada M., Hasegawa T., and Furukawa K. (1997) Expression cloning of rat cDNA encoding UDP-galactose:GD2 β1,3-galactosyltransferase that determines the expression of GD1b/GM1/GA1. J. Biol. Chem. 272, 24794–24799 10.1074/jbc.272.40.24794 [DOI] [PubMed] [Google Scholar]
- 35. Svennerholm L., Boström K., Fredman P., Månsson J. E., Rosengren B., and Rynmark B. M. (1989) Human brain gangliosides: developmental changes from early fetal stage to advanced age. Biochim. Biophys. Acta 1005, 109–117 10.1016/0005-2760(89)90175-6 [DOI] [PubMed] [Google Scholar]
- 36. Olsen A. S. B., and Faergeman N. J. (2017) Sphingolipids: membrane microdomains in brain development, function and neurological diseases. Open Biol. 7, 170069 10.1098/rsob.170069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shiina T., Kikkawa E., Iwasaki H., Kaneko M., Narimatsu H., Sasaki K., Bahram S., and Inoko H. (2000) The β-1,3-galactosyltransferase-4 (B3GALT4) gene is located in the centromeric segment of the human MHC class II region. Immunogenetics 51, 75–78 10.1007/s002510050012 [DOI] [PubMed] [Google Scholar]
- 38. Hizume M., Kobayashi A., Mizusawa H., and Kitamoto T. (2010) Amino acid conditions near the GPI anchor attachment site of prion protein for the conversion and the GPI anchoring. Biochem. Biophys. Res. Commun. 391, 1681–1686 10.1016/j.bbrc.2009.12.128 [DOI] [PubMed] [Google Scholar]
- 39. Miyagawa-Yamaguchi A., Kotani N., and Honke K. (2015) Each GPI-anchored protein species forms a specific lipid raft depending on its GPI attachment signal. Glycoconj. J. 32, 531–540 10.1007/s10719-015-9595-5 [DOI] [PubMed] [Google Scholar]
- 40. Kitamoto T., Ohta M., Doh-ura K., Hitoshi S., Terao Y., and Tateishi J. (1993) Novel missense variants of prion protein in Creutzfeldt-Jakob disease or Gerstmann-Straussler syndrome. Biochem. Biophys. Res. Commun. 191, 709–714 10.1006/bbrc.1993.1275 [DOI] [PubMed] [Google Scholar]
- 41. Tomavo S., Couvreur G., Leriche M. A., Sadak A., Achbarou A., Fortier B., and Dubremetz J. F. (1994) Immunolocalization and characterization of the low molecular weight antigen (4–5 kDa) of Toxoplasma gondii that elicits an early IgM response upon primary infection. Parasitology 108, 139–145 [DOI] [PubMed] [Google Scholar]
- 42. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., and Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 10.1038/nprot.2009.21 [DOI] [PubMed] [Google Scholar]
- 43. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–456 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., et al. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 10.1093/glycob/cwv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometric proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (43) partner repository with the dataset identifiers PXD017655 (project name “GPI-glycan structures of human Prion from knock-in mice_LC-MSMS”) and PXD017656 (project name “GPI-glycan structures of human Prion from a human brain”).