Abstract
One long-standing knowledge gap is the role of nuclear proteins in mRNA translation. Nuclear RNA helicase A (DHX9/RHA) is necessary for the translation of the mRNAs of JUND (JunD proto-oncogene AP-1 transcription factor subunit) and HIV-1 genes, and nuclear cap-binding protein 1 (NCBP1)/CBP80 is a component of HIV-1 polysomes. The protein kinase mTOR activates canonical messenger ribonucleoproteins by post-translationally down-regulating the eIF4E inhibitory protein 4E-BP1. We posited here that NCBP1 and DHX9/RHA (RHA) support a translation pathway of JUND RNA that is independent of mTOR. We present evidence from reciprocal immunoprecipitation experiments indicating that NCBP1 and RHA both are components of messenger ribonucleoproteins in several cell types. Moreover, tandem affinity and RT–quantitative PCR results revealed that JUND mRNA is a component of a previously unknown ribonucleoprotein complex. Results from the tandem IP indicated that another component of the JUND-containing ribonucleoprotein complex is NCBP3, a recently identified ortholog of NCBP2/CBP20. We also found that NCBP1, NCBP3, and RHA, but not NCBP2, are components of JUND-containing polysomes. Mutational analysis uncovered two dsRNA-binding domains of RHA that are necessary to tether JUND–NCBP1/NCBP3 to polysomes. We also found that JUND translation is unaffected by inhibition of mTOR, unless RHA was down-regulated by siRNA. These findings uncover a noncanonical cap-binding complex consisting of NCBP1/NCBP3 and RHA substitutes for the eukaryotic translation initiation factors 4E and 4G and activates mTOR-independent translation of the mRNA encoding the tumor suppressor JUND.
Keywords: mammalian target of rapamycin (mTOR), tumor suppressor gene, eukaryotic translation initiation factor 4E (eIF4E), RNA helicase, RNA binding protein, complex 5'-UTR, mTOR, non-canonical cap-binding complex, polyribosome, post-transcriptional control element (PCE)
Introduction
Nuclear proteins engender the translational utilization of mRNAs, but the mechanisms are controversial (1–5). Nuclear proteins are co-transcriptionally bound to nascent RNAs through recognition of the 5′-cap structure. The 5′-cap structure is bound by nuclear cap-binding protein (NCBP)3 2/CBP20 and NCBP1/CBP80 heterodimeric nuclear cap-binding complex (CBC) (6–10). The CBC is necessary for the processing and nuclear export as well as translational utilization of mature transcripts (11–15). The mature transcripts retain the canonical CBC (consisting of NCBP1/NCBP2) to support a pioneer round of ribosome scanning for premature termination codons that trigger nonsense mediated decay (16).
Subsequently, CBC bound at the 5′-RNA cap is exchanged to cytoplasmic cap-binding protein eukaryotic translation initiation factor 4E (eIF4E). eIF4E engages eIF4G and other components of preinitiation complexes (PIC) to initiate canonical cap-dependent translation. The eIF4E-dependent messenger ribonucleoproteins (mRNPs) activate polysome assembly and steady-state protein synthesis (17). These mRNPs are activated by the serine-threonine kinase, mTOR through post-translational down-regulation of 4E-BP1 (18). Treatment with rapamycin or small molecule inhibitors of mTOR (e.g. Torin-1) down-regulates eIF4E activity (18).
The traditional view has been that eIF4E is necessary for cap-dependent protein synthesis (16, 17). Small subsets of cap-dependent mRNAs maintain polysomes during the down-regulation of eIF4E, including JUND (19, 20). Recently, alternatives to eIF4E have been shown to be assembling translation RNPs on select mRNAs. Examples are the DAP5/p97 isoform of eIF4G (21–25), FXR1a/PARN (26, 27), tRNA synthetase (28), and eIF3d (29). NCBP1 is a component of HIV-1 polysomes and polysomes of select histone mRNAs (30–33). Immunoprecipitations (IPs) of HIV-1 polysomes identified that late mRNAs are enriched in NCBP1 co-precipitates while undetected in eIF4E immunoprecipitates. These results posited a non-eIF4E pathway for cap-dependent translation of select mRNAs (31).
Recently an additional cap-binding protein, NCBP3, was identified to serve redundantly with NCBP2 under physiological conditions (9). The observation that co-depletion of NCBP2/NCBP3 curtailed NCBP1 association with 5′-caps demonstrated that NCBP1 alternatively forms CBC with NCBP2 or NCBP3 (9). Further evidence demonstrated that NCBP3 is essential to mount a precise and appropriate stress response (34). NCBP3-deficient mice suffered from severe lung pathology and increased morbidity after influenza A virus challenge, suggesting that NCBP3 supports expression of stress-responsive genes (34). Given that NCBP2 and NCBP3 function redundantly to support nuclear processing of mRNAs and that NCBP3-deficient cells evoke a reduced antiviral response, we posited that the noncanonical CBC (consisting of NCBP1/NCBP3) is important for translation of stress response genes (31). The gap in knowledge remains in determining the involvement of the noncanonical CBC in translation of select mRNAs.
AP-1 transcription factors are stress-response proteins (JUND, JUNB, c-Jun) (35). JUND, a negative regulator of cell proliferation, is among the subset of host mRNAs selectively translated during post-translational down-regulation of eIF4E (19). Previous investigations identified translation of JUND, and retroviral transcripts depend upon nuclear RNA helicase A (DHX9/RHA) (36). DHX9/RHA binds to a specific class of post-transcriptional control element (PCE) in JUND and several retroviral 5′-UTRs to stimulate polysome formation (36–38). RHA down-regulation or ectopic expression of the two N-terminal RNA-binding domains (RBDs) were found to attenuate the synthesis of JUND and HIV-1 virion proteins (36, 38). The RHA co-factors necessary for assembling polysomes remain to be identified. Given the mutual engagement of NCBP1 and RHA to select mRNAs, we posited that NCBP1 and RHA are components of the same RNP that facilitates cap-dependent translation of JUND.
Results
NCBP1-RHA and eIF4E-eIF4G are mutually exclusive RNP
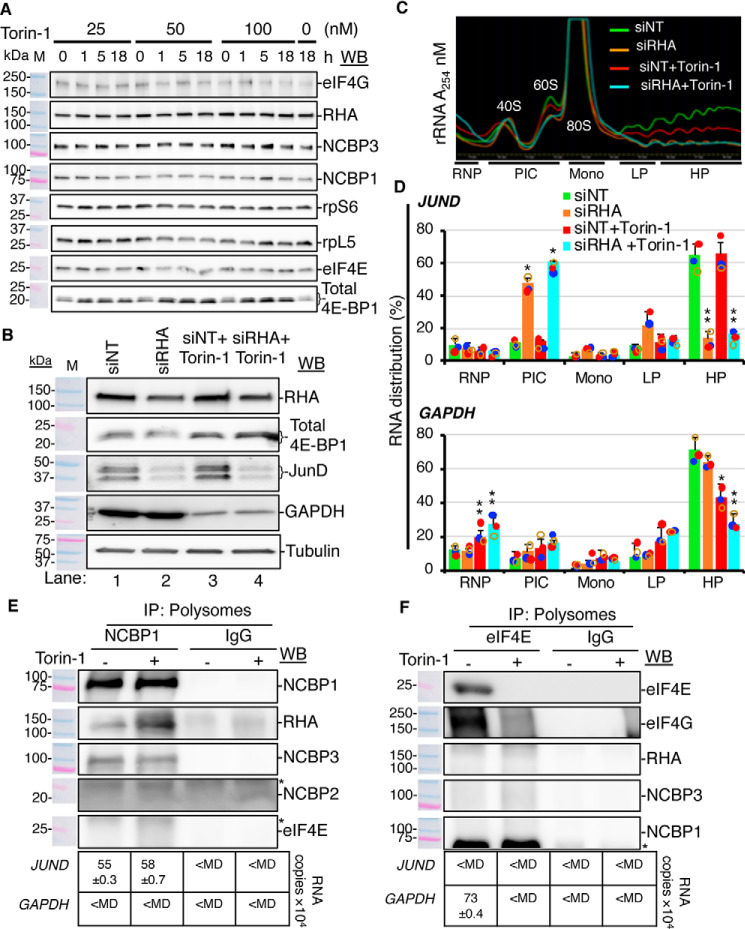
To address the hypothesis that NCBP1 and RHA are components of a mutual RNP, reciprocal co-immunoprecipitations (co-IPs) were performed with specific antisera in three or more replicate experiments. HEK293 cytoplasmic (cyto) lysates were incubated with specific antisera or isotype-specific IgG. Analysis of the immune complexes by Western blots (WB) revealed that the antiserum to NCBP1 co-precipitated RHA, NCBP2, NCBP3, and cytoplasmic poly-A–binding protein (PABPC1). In the course of seven independent experiments, neither eIF4E nor eIF4G were detectable in the NCBP1-RHA immune complexes (Fig. 1A). Antiserum to eIF4E co-precipitated eIF4G, PABPC1, and DDX3 (Fig. 1B) (39, 40). The eIF4E complexes were devoid of detectable NCBP1, RHA, NCBP2, or NCBP3 (Fig. 1B), validating that NCBP1-RHA and eIF4E-DDX3 are components of mutually exclusive mRNPs (Fig. 1).
Figure 1.
NCBP1-RHA and eIF4E-eIF4G are components of mutually exclusive RNPs. Reciprocal co-IP of selected proteins from HEK293 lysates with specific antisera are shown (bold type). Bound proteins were eluted and analyzed by WB with the antisera indicated on the right. Isotype-specific IgG served as the negative control, and input cell lysate served as the positive control. The results are summarized in the tables below each WB. A, IP of NCBP1, RHA, NCBP2, or NCBP3. B, IP of eIF4E, eIF4G, or DDX3. The antiserum detected the specific proteins on the immunoblots, as shown relative to the prestained molecular mass markers (lanes M). The same image of the molecular mass markers was used for each panel. +, positive co-IP. *, nonspecific background. The WBs were subjected to ImageJ densitometry quantification (Table S1).
Antiserum to NCBP2 or NCBP3 independently co-precipitated NCBP1 (Fig. 1A), corroborating the previous finding that NCBP1 independently enriches NCBP2- or NCBP3-bound 5′-capped mRNAs (9). The RHA immune complexes co-precipitated NCBP1 and NCBP2 or NCBP3, and the results of seven replicate experiments confirmed the lack of detectable eIF4E and eIF4G, recapitulating different mRNPs. Reciprocal IPs validated each of the indicated interactions (Fig. 1A). NCBP3 co-precipitated DDX3, but not other components of the eIF4E RNPs. The observed NCBP3-DDX3 interaction may be attributable to DDX3 in transitional interaction between NCBP-bound and eIF4E-bound 5′-caps. In summary, the immune complexes of NCBP1-RHA did not bind DDX3, whereas eIF4E did not bind RHA. The results suggest that there are multiple NCBP3 complexes.
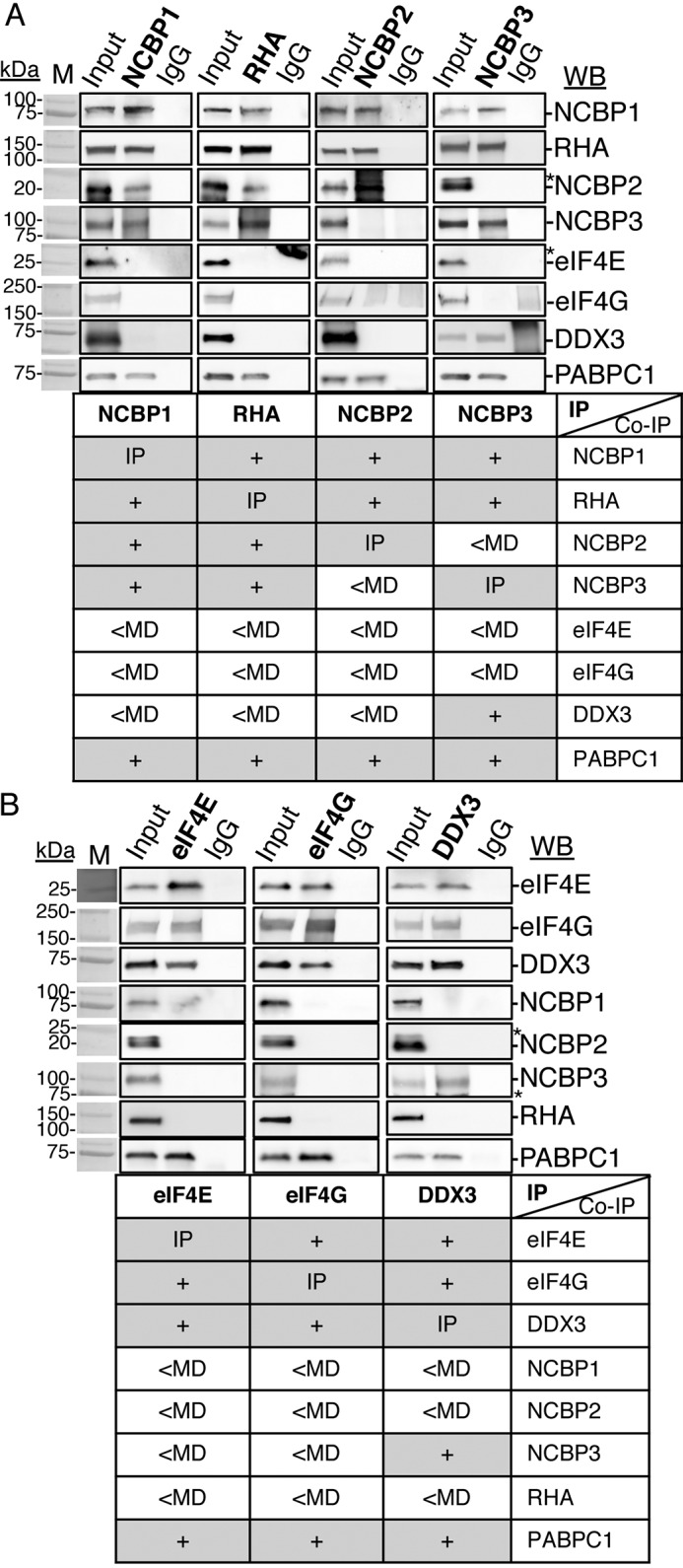
To ascertain whether NCBP1-RHA RNP is conserved in other cell types, we investigated cyto lysates of several other cell lines such as simian fibroblasts (COS), HTLV-1–transformed SLB-1 human T lymphocytes, and CEM×174 human T lymphocytes. The studies assessed co-precipitation of NCBP1 and RHA with or without NCBP2, NCBP3, or eIF4E. The results from three independent experiments demonstrated that NCBP1-RHA complexes selectively interact with NCBP3 or NCBP2 and are mutually exclusive of eIF4E in these cell lines (Fig. 2, A and B). These findings were not surprising because prior studies have demonstrated NCBP1, NCBP3, or NCBP2 and RHA exhibit high-level amino acid sequence conservation across Mammalia and translation of viral mRNAs requires RHA of bovine, feline, human, and simian origin (36).
Figure 2.
NCBP1-RHA complexes exist in several cell types. IP of NCBP1 (A), RHA (B), or isotype IgG from lysates of HEK293, COS7, SLB-1, or CEM×174 cells depleted of nuclei. Immune complexes (bold type) were washed, collected in SDS sample buffer, and analyzed by WB with the antisera indicated on the right. In each case, the input cell lysate served as the positive control, and the isotype-specific IgG served as the negative control for background immunoreactivity. The results are summarized in the table below each WB. The antiserum detected the specific proteins on the immunoblots, as shown relative to the prestained molecular mass markers (M). The same image of the molecular mass markers was used for each panel. +, positive co-IP. *, nonspecific background. The WBs were subjected to ImageJ densitometry quantification (Table S1).
NCBP1-NCBP3-RHA mRNPs exclusive of NCBP2 are components of JUND polysomes
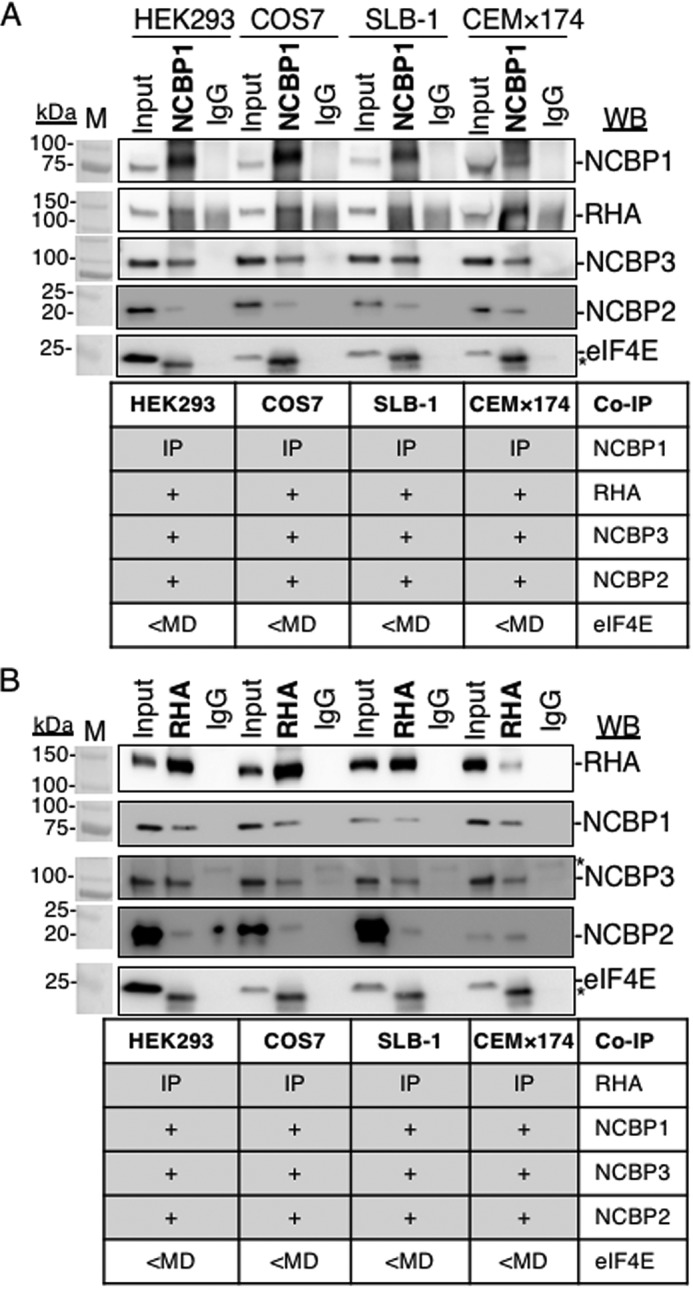
Next, experiments employed tandem IPs to document whether RHA and NCBP1, NCBP3, or NCBP2 are components of the same RNP in polysomes (Fig. 3, A and B). Exogenous expression of pFLAG-RHA was performed, and the HEK293 cells were collected in the lysis buffer and designated as Input cyto lysate (Fig. 3B). Next the Inputs were subjected to sucrose gradient centrifugation, and polysome fractions were collected (Fig. 3A) and subjected to WB (Fig. 3C). WB by several antiserum verified FLAG-RHA, NCBP3, NCBP1, and NCBP2 in the Input cyto lysate (Fig. 3C, lane 1). Noticeably, FLAG-RHA, NCBP3, and NCBP1 were enriched in polysomes, whereas NCBP2 was absent (Fig. 3C, lane 2). These results exposed functional differences between canonical CBC (NCBP1-NCBP2) and noncanonical CBC (NCBP1-NCBP3). The lack of NCBP2 detection in polysomes corroborated previous demonstration of CBC activity in pioneer round, but not steady-state translation (41).
Figure 3.
NCBP1-NCBP3-RHA are components of the same RNP loaded to JUND polysomes. Tandem IPs were employed to isolate components of the same RNP from HEK293 cells transfected with pFLAG-RHA. Input cyto lysates were subjected to sucrose gradient centrifugation, and polysomes were collected. A, A254 spectrometry (red line) of sucrose gradient. B, outline of tandem IP. The polysome fractions were combined, precipitated, and resuspended in low-salt buffer (Input poly). Aliquots of the cyto (white tube) and polysome samples (blue tube) were incubated with FLAG antiserum conjugated to protein G beads. The beads were washed and incubated with 3× FLAG peptides to elute RNPs. A fraction of the eluate was reserved for WB, and the remaining eluate was incubated with NCBP1 antiserum conjugated to protein G beads. The beads were extracted in SDS buffer for WB analysis or with TRIzol to isolate RNA for subsequent RT-qPCR with gene-specific primers. C, WB of Input cyto and polysomes (lanes 1 and 2), eluates of FLAG IP (lanes 3 and 4), eluate of Protein G IP (control for FLAG IP, lane 5), eluates of NCBP1 IP (lanes 6 and 7), and eluates of IgG IP (control for NCBP1 IP; lanes 8 and 9). The antiserum detected the specific proteins on the immunoblots, as shown relative to the prestained molecular mass markers (M). The same image of the molecular mass markers was used for each panel. *, nonspecific band. D, JUND and GAPDH copies by RT-qPCR. The results represent the means of three independent experiments with standard deviation.
Next, the Input and polysome samples were incubated with FLAG antiserum conjugated to protein G magnetic beads. Immune complexes were washed and collected by competitive elution with 3× FLAG peptides (250 μg/ml) as depicted in Fig. 3B. The WB of the cyto IP verified the enrichment of FLAG-RHA and the co-precipitation of NCBP1, NCBP2, and NCBP3 (Fig. 3C, lane 3).
Moreover, the WB of the polysome IP verified the enrichment of FLAG-RHA and the co-precipitation of NCBP1 and NCBP3. Importantly, NCBP2 was not detectable in the polysome IP (Fig. 3C, lane 4), corroborating the lack of NCBP2 in the WB of the collected polysomes (Fig. 3C, lane 2). There was no detectable enrichment of the proteins in the protein G–negative control (Fig. 3, lane 5). Next, the eluates were subjected to tandem IP with NCBP1 antiserum conjugated to protein G magnetic beads and analyzed by WB (Fig. 3B). The results validated that the NCBP1 tandem IP captured FLAG-RHA from cyto and polysome samples (Fig. 3C, lanes 6 and 7, FLAG-RHA panel). The tandem IP ascertained NCBP3 and NCBP2 enrichment in the cyto tandem IP (Fig. 3C, lane 6). However, the polysome tandem IP captured NCBP3 but failed to capture NCBP2 (Fig. 3C, lane 7). There was no enrichment of these proteins in the IgG isotype control (Fig. 3C, lanes 8 and 9). We concluded that NCBP3 is the mutual component of FLAG-RHA-NCBP1 RNPs in polysomes.
To validate that JUND is a component of the FLAG-RHA-NCBP1-NCBP3 RNPs, replicate tandem IP samples were extracted with TRIzol-LS, and the co-precipitating RNA was collected and subjected to RT-qPCR using JUND and GAPDH specific primer pairs. The JUND transcripts were readily detectable in the FLAG-RHA-NCBP1-NCBP3 RNP, whereas GAPDH transcripts were less than the minimum detectable (<MD) (Fig. 3D). Both JUND and GAPDH were <MD in the IgG controls (Fig. 3D). These results validated that FLAG-RHA-NCBP1-NCBP3 are mutual components of JUND polysomes.
RHA tethers NCBP1-NCBP3 to polysomes through RNA
NCBP1-NCBP3 may be tethered to RHA through RNA or via protein–protein interaction, which would be sensitive or resistant to RNase treatment, respectively. To examine the nature of the interaction between RHA and NCBP1-NCBP3, reciprocal IPs were performed using cyto lysates treated with RNase A. The cyto lysate was incubated with RHA or NCBP1 antiserum conjugated to protein G magnetic beads with or without RNase A (1 μg/ml) for 2 h. Immune complexes were washed and collected in SDS sample buffer. The RHA IP and NCBP1 IP co-precipitated RHA, NCBP1, NCPB3, and NCBP2 (Fig. 4A, lanes 1 and 3). However, RNase A treatment of the RHA IP eliminated NCBP1, NCBP3, and NCBP2 (Fig. 4A, lane 2). RNase A treatment of the NCBP1 IP eliminated RHA co-precipitation, establishing that the interaction is RNA-dependent (Fig. 4A, lane 4). We concluded NCBP1 is tethered to RHA through RNA.
Figure 4.
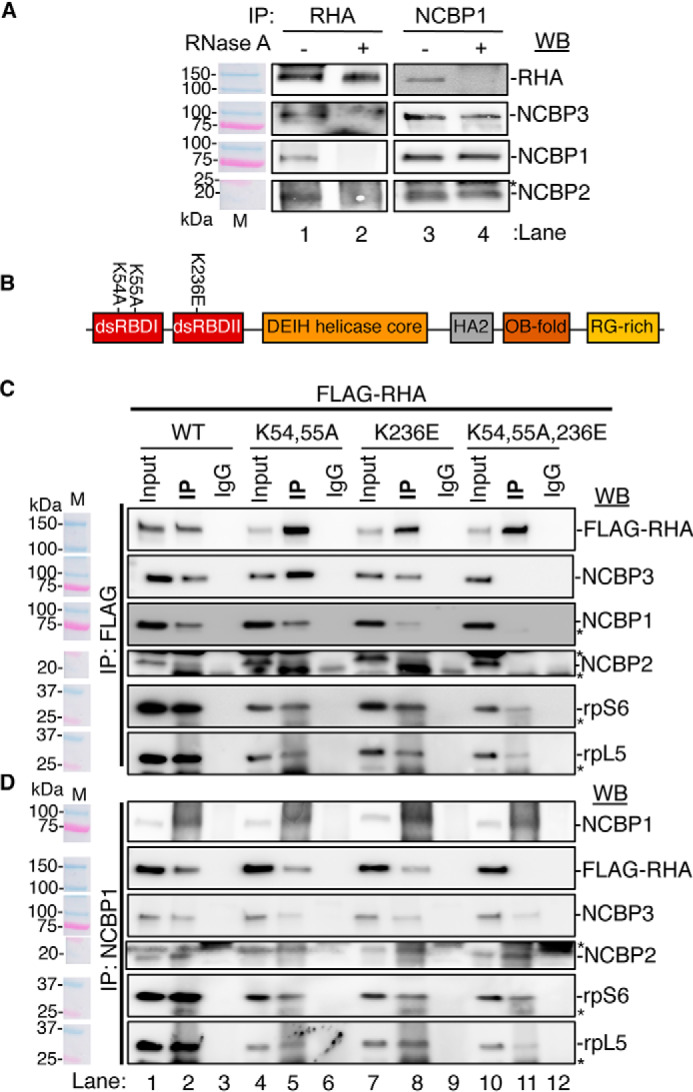
NCBP1 interaction with RHA requires N-terminal dsRBDs. A, RHA or NCBP1 IP of CEM×174 cyto lysates treated with RNase A (or left untreated). Co-precipitates were analyzed by WB using antiserum against RHA, NCBP3, NCBP1, or NCBP2. B, depiction of RHA with annotation of the domain structure and position of mutations K54A,K55A and K236E. Red, dsRBD I and II; tangerine, DEIH helicase core; gray, helicase-associated 2 (HA2); brown, OB fold; gold, arginine-glycine–rich (RG-rich). C and D, FLAG IP (C) and NCBP1 IP (D) of cyto lysates of HEK293 cells transfected with FLAG-tagged WT RHA or mutant RHA (K54A,K55A [KK], K236E, or K54A,K55A,K236E [KKK]) for 24 h followed by WB for the indicated proteins. The antiserum detected the specific proteins on the immunoblots, as shown relative to the prestained molecular mass markers (M). The same image of the molecular mass markers was used for each panel. *, nonspecific band. The WBs were subjected to ImageJ densitometry quantification (Table S1).
Because RHA binding to PCE RNA requires the N-terminal double-stranded (ds) RBDs and is eliminated by mutation of surface-exposed lysine residues (K54A, K55A, and K236E) (42) (Fig. 4B), we postulated that the same residues are critical for RHA interaction with NCBP1-NCBP3 or NCBP1-NCBP2. To examine this, HEK293 cells were transfected with FLAG-RHA-WT or FLAG-RHA modified by the following substitutions: K54A,K55A (KK), K236E, or K54A,K55A,K236E (KKK) and subjected to reciprocal IPs using antiserum to FLAG or NCBP1.
The antiserum to FLAG identified WT, KK and K236E co-precipitated NCBP1 (Fig. 4C, lanes 2, 5, and 8), whereas KKK was not detectable (Fig. 4C, lane 11). The longer exposure of the blot identified NCBP1 was barely detected in the KKK co-precipitate (3% of WT) (Fig. S1 and Table S1). Likewise, the co-precipitation of NCBP2 and NCBP3 was eliminated by the KKK mutation (Fig. 4C, lane 11).
Reciprocal IP was performed using antiserum to NCBP1. NCBP1 co-precipitated FLAG-WT, KK or K236E, but not KKK (Fig. 4D). NCBP3 and NCBP2 interactions were maintained. The co-precipitation of ribosomal proteins rpS6 and rpL5 by FLAG or NCBP1 was diminished but not abrogated by the lysine mutations (Fig. 4, C and D). The results demonstrated that NCBP1-NCBP3 and NCBP1-NCBP2 are tethered through RNA to the N-terminal dsRBDs of RHA.
RHA is necessary for the assembly of NCBP1-NCBP3-JUND polysomes and mTOR-resistant JUND translation
Next, we sought to establish whether RHA-PCE interaction is necessary for the assembly of NCBP1-NCBP3 polysomes. HEK293 cells were transfected with siRNAs targeting NCBP3 or RHA or with nontargeting (NT) control siRNA. 24 h post-transfection, the cell lysates were analyzed by WB, and total cellular RNA was analyzed by RT-qPCR using gene-specific primers. WB showed that NCBP3 and RHA proteins were down-regulated by the specific siRNA, whereas NCBP1 and GAPDH protein expression was unaffected (Fig. 5A). The RT-qPCR recapitulated the down-regulation of ncbp3 and dhx9/rha by the specific siRNAs relative to GAPDH (Fig. 5B). The cyto lysates were subjected to sucrose density sedimentation and A254 spectrometry to identify the rRNA profile. The results of three independent experiments identified that the magnitude of polysomes was modestly diminished by RHA down-regulation compared with the siNT control (Fig. 5C). The siNCBP3 treatment did not diminish polysomes and reproducibly increased the robustness of the 60S and 80S peaks (Fig. 5C). The results indicate that NCBP3 has a generalized effect on ribosome recruitment.
Figure 5.

RHA down-regulation reduces JUND polysomes. Cyto lysates of HEK293 cells transfected with siRNA targeting NCBP3 (siNCBP3), RHA (siRHA), or nontargeting siRNA (siNT) were subjected to WB and RT-qPCR. A, representative immunoblot of the cyto lysates with antiserum against RHA, NCBP3, NCBP1, or the loading control GAPDH. The antiserum detected the specific proteins on the immunoblots, as shown relative to the prestained molecular mass markers (M). The same image of the molecular mass markers was used for each panel. B, dhx9/rha and ncbp3 expression in cells treated with the indicated siRNA by RT-qPCR. The bar graph represents the expression of dhx9/rha and ncbp3 relative to GAPDH. C, A254 spectrometry of sucrose gradient showing rRNA distribution and designation of the fractions. PIC, preinitiation complex composed of 40S and 60S RNPs; Mono, monosome (80S); LP, light polysome (two or three polysomes); HP, heavy polysome (4 or more polysomes). D, RT-qPCR of RNA extracted from the fractions identified in C for expression of JUND and GAPDH. Copies were calculated relative to standard curves. The graphs present the distribution of the RNA copies across the gradients. The results represent the means of three independent experiments (bars) with standard deviation. The colored circles indicate the values from the individual experiments. Statistical significance is indicated with asterisks: *, p ≤ 0.05; **, p ≤ 0.005.
The distribution of mRNAs in the gradient fractions was determined by RT-qPCR. The down-regulation of RHA significantly reduced JUND polysomes, whereas no change was observed in GAPDH polysomes (Fig. 5D). The results recapitulated previous metabolic labeling assays showing that RHA down-regulation reduces the synthesis of JUND, but not GAPDH protein (36). The down-regulation of NCBP3 significantly increased PIC (48S and 60S mRNPs) and diminished polysomes (Fig. 5D). The JUND PIC increased from 2% to 15%, whereas heavy polysomes decreased from 69% to 58% of the RNA copies, accounting for the redistribution to PIC. The GAPDH PIC increased from 12% to 22%, and light polysomes decreased from 20% to 10% of the RNA copies, accounting for the redistribution in RNA copies. The increase in PIC was 4-fold greater for JUND than GAPDH polysomes (Fig. 5D). The results do not exclude the possibility of redundant activity by RHA-NCBP1-NCBP2 as a result of NCBP3 down-regulation. Indeed, NCBP2 and NCBP3 were shown to function redundantly to support nuclear processing of mRNAs (9). We concluded that RHA-NCBP1-NCBP3 activity is important for JUND polysome assembly at steady state. The prospect of RHA-NCBP1-NCBP3 RNP activity during mTOR down-regulation was an important open issue.
Therefore, we evaluated the effect of mTOR down-regulation on the assembly of JUND polysome components, RHA-NCBP1-NCBP3. The small molecule mTOR inhibitor, Torin-1 activates the allosteric inhibitor of eIF4E, hypophosphorylated 4E-BP1. Torin-1 titration experiments were first carried out to establish treatment time and optimal dose to activate hypophosphorylated 4E-BP1 while maintaining ribosome profiles and no change in cell viability. The titration experiments identified cells exposed to 50 nm Torin-1 for 18 h maintained consistent polysome profiles and up-regulated hypophosphorylated 4E-BP1 in several replicate experiments with the cell viability remaining similar to mock-treated cells. Immunoblotting of the cell lysates documented that RHA, NCBP1, NCBP3, and other proteins maintained steady state at 50 nm Torin-1 for 18 h (Fig. 6A and Fig. S2).
Figure 6.
RHA is essential for mTOR-resistant translation of JUND. A, HEK293 cells were treated with mTOR inhibitor Torin-1 (0–100 nm) for 0–18 h, and the cell lysates were analyzed by WB with the indicated antisera. B, cells transfected with siRNA targeting RHA (siRHA) or nontargeting control (siNT) and treated with 50 nm Torin-1 or 0.2% DMSO for 18 h were analyzed by WB with antiserum to RHA, 4E-BP1, JUND, GAPDH, or tubulin (loading control). C, A254 spectrometry of the sucrose gradient showing rRNA distribution in the fractions. PIC, preinitiation complex composed of 40S and 60S RNPs; Mono, monosome (80S); LP, light polysome (two or three polysomes); HP, heavy polysome (four or more polysomes). D, JUND and GAPDH RNA copies were analyzed by RT-qPCR and calculated relative to standard curves. The graphs present the distribution of the RNA across the gradients. The results represent the means of three independent experiments (bars) with standard deviation. The colored circles indicate the values from the individual experiments. Statistical significance is indicated with asterisks: *, p ≤ 0.05; **, p ≤ 0.005. E and F, the light and heavy polysome fractions were combined, and proteins were precipitated and subjected to IP with NCBP1 or eIF4E antiserum. Co-precipitates were subjected to WB with the indicated antiserum or RNA extraction followed by RT-qPCR. Isotype-specific IgG were used as negative controls. The antiserum detected the specific proteins on the immunoblots, as shown relative to the prestained molecular mass markers (M). The same image of the molecular mass markers was used for each panel. *, nonspecific band. The table below each WB panel shows the number of copies of JUND and GAPDH relative to IgG control detected by RT-qPCR.
To evaluate the capacity for cells to recover from the Torin-1 treatment, the medium was exchanged, and cells were cultured without Torin-1 for 1 or 18 h. Immunoblotting validated recovery of hyperphosphorylated 4E-BP1 in the cells subject to short-term (1 h) and long-term (18 h) culture (Fig. S3). We concluded the treatment conditions employed were appropriate to maintain polysome profiles and cell viability for 18 h.
Next, HEK293 cells were transfected with RHA-specific or NT siRNAs (mock) for 24 h and then exposed to 50 nm Torin-1 for 18 h. The WB on cyto lysates validated that RHA was down-regulated by siRNA (Fig. 6B, lanes 2 and 4), and hypophosphorylated 4E-BP1 was up-regulated by Torin-1 treatment (Fig. 6B, lanes 3 and 4). Tubulin was unaffected by the treatments. The results identified JUND proteins reduced in response to the RHA down-regulation (Fig. 6B, compare lanes 1, 2, and 4) but not by Torin-1 treatment (compare lanes 1 and 3). Conversely, GAPDH protein was reduced by Torin-1 but not by RHA down-regulation. The results verified JUND translation is resistant to mTOR inhibition.
The cyto lysates were subjected to sucrose density sedimentation and A254 spectrometry. As expected, ribosomal profiles were diminished by the Torin-1 treatment (Fig. 6C). Next, RT-qPCR was performed on the input lysate, and the RNA was isolated from the gradient fractions. The results verified that RHA down-regulation significantly reduces JUND polysomes (Fig. 6D, upper panel) but not GAPDH polysomes (Fig. 6D, lower panel). Torin-1 treatment significantly down-regulated GAPDH polysomes (Fig. 6D, lower panel) but not JUND polysomes (Fig. 6D, upper panel). As expected, the combination of Torin-1 treatment and RHA down-regulation significantly reduced both JUND and GAPDH polysomes (Fig. 6D). The distribution across the sucrose gradients of RHA, NCBP1, NCBP3, and other proteins was evaluated by immunoblotting (Fig. S4). The results validated that Torin-1 eliminated eIF4E from polysome fractions, whereas RHA, NCBP3, and NCBP1 remained.
Next, replicate polysome fractions were combined, and proteins were co-precipitated by NCBP1 or eIF4E antiserum. The immunoblots validated NCBP1 co-precipitated RHA and NCBP3 polysomes in the presence or absence of the Torin-1 treatment, whereas neither NCBP2 nor eIF4E were detectable (Fig. 6E). The eIF4E immunoprecipitates enriched eIF4G, and these polysomes were eliminated by Torin-1 (Fig. 6F). None of the candidate proteins were detected in the IgG isotype controls (Fig. 6, E and F).
The IP samples were extracted with TRIzol-LS, and the co-precipitating RNAs were collected and subjected to the RT-qPCR. In results of three independent experiments, JUND transcripts were readily detectable in immunoprecipitated NCBP1 polysomes, whereas GAPDH transcripts were not (<MD) (Fig. 6E, below WB). In contrast, the eIF4E polysomes enriched GAPDH transcripts, but not JUND (<MD) (Fig. 6F, below WB). The IgG IPs did not enrich either JUND or GAPDH (<MD) (Fig. 6, E and F). These findings demonstrated the assembly of the JUND polysome components, RHA-NCBP1-NCBP3, is mTOR-independent.
Discussion
This study has identified a specialized translation pathway for cap-dependent translation of JUND that does not utilize eIF4E or eIF4G nor NCBP2. Strong evidence is provided that NCBP1 and RHA are components of the same RNP that is facilitating cap-dependent translation of JUND. Moreover, JUND polysomes are composed of RHA-NCBP1-NCBP3 and assemble at steady state and during mTOR inhibition. Current evidence indicates that the RHA-NCBP1-NCBP3 cap-dependent translation mRNP is not regulated by eIF4E levels and appears to be a constitutively active pathway rather than a rescue pathway.
In addition to JUND, several retroviruses contain the RHA-responsive element; PCE and RHA-NCBP1-NCBP3 are likely involved in the translation control of these mRNAs. PCE does not support internal ribosome entry, and PCE translation activity has been shown to be cap-dependent (37). Prior research established the RHA-PCE RNA interaction neutralizes structural barriers within the 5′-UTR that repress ribosome scanning to promote efficient translation (36). Our new findings provide strong evidence that RHA is necessary for the assembly of JUND polysomes at steady state and during mTOR inhibition (Fig. 7). Moreover, NCBP1-NCBP3 and NCBP1-NCBP2 are tethered to RHA through JUND by the N-terminal dsRBDs of RHA. The molecular basis by which RHA-PCE interaction subverts canonical exchange of the nuclear cap-binding proteins to eIF4E remains an open issue.
Figure 7.
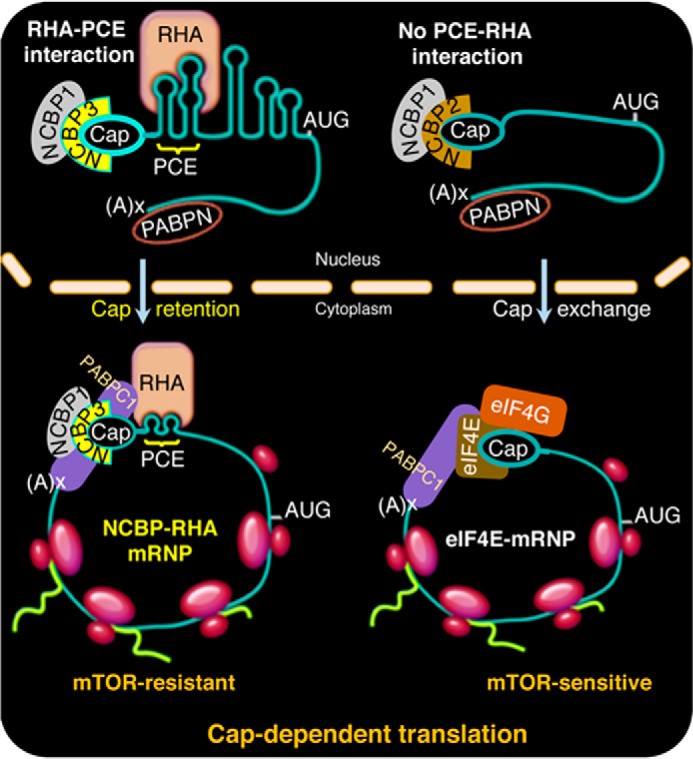
Model of the specialized translation of PCE-containing mRNAs and assembly of mTOR-resistant RNP complexes. Model comparing nuclear assembly of cap-dependent mRNPs. Left, PCE-bearing mRNAs bind NCBP1-NCBP3 at the 5′-cap and RHA at the PCE and experience the exchange of PABPN to PABPC1. RHA-NCBP1-NCBP3 mRNPs facilitate assembly of JUND polysomes whether the eIF4E-mRNPs are active or inhibited by 4E-BP1. Right, canonical engagement of NCBP1-NCBP2 at the 5′-cap is depicted by mRNA lacking PCE. Consequential to nuclear export, CBC undergoes exchange to eIF4E, which engages eIF4G and other factors to assemble polysomes. In common, both mRNPs experience the exchange of PABPN to PABPC1. The eIF4E-eIF4G–dependent polysome assembly is abrogated upon the inhibition of mTOR because of down-regulated eIF4E activity.
JUND is a member of the Jun family of transcription factors that dimerize with c-Jun, Fos, or other family members to form AP-1 (29, 35). Recently, the 5′-UTR of c-jun was shown to assemble non-eIF4E translation initiation complexes (29). In preliminary studies, we observed c-jun RNA copies are enriched in RHA-NCBP1-NCBP3 immune complexes (5.3 × 104 and 3.1 × 104, minus and plus Torin-1, respectively) at levels similar to JUND in Fig. 6E. Next, we replicated the tandem IP experiment and identified the enrichment of c-jun-RHA-NCBP1 mRNPs in cyto and polysomes (11 × 104 and 39 × 104 copies). Further experimentation is warranted to establish that the c-jun 5′-UTR recapitulates activity of JUND PCE and possibly retroviral PCEs, which is necessary for RHA interaction (36, 43–45) and mTOR-resistant translation.
AP-1 is a critical regulator of nuclear gene expression during T-cell activation, innate response to viral infections, and anti-tumor immune response through type I interferons and proinflammatory cytokines (46). Dysregulation of AP-1 is hallmark of viral pathogenesis, neoplastic transformation, and tumor progression (35, 46). Likewise, dysregulation of dhx9/rha is associated with productive viral infection (31, 37, 38, 45, 47–50) and tumor survival (51–54). The recent finding that NCBP3 is essential to mount a precise antiviral response (34) posits dysfunction in a RHA-NCBP1-NCBP3 translation pathway that contributes to deficient innate response. The DHX9, DExH-box helicase 9 (alias RNA helicase A) gene is within the 1q25 prostate/lung/breast cancer locus, and gene mutation or overexpression is pervasive in human clinical samples (55). We speculate that mutations in PCE or RHA residues identified in this report provide biomarkers for neoplastic transformation.
Experimental procedures
Cell culture and transfection
Dulbecco's modified Eagle's medium or RMPI culture medium were supplemented with 10% fetal bovine serum and 1× antibiotic–antimycotic solution (Gibco) and used to culture HEK293 and COS7 cells or SLB-1 and CEM×174 cells, respectively. The cell lines were obtained from ATCC, and low-passage cultures were used in the experiments. In addition, mycoplasma testing was performed periodically and on a weekly basis. The cells were authenticated and documented free of mycoplasma based on morphological evaluation of cells plated at high and low culture densities under a microscope, and the cell morphology images were maintained for comparisons.
Transfection of HEK293 cells (1 × 106/35-mm well) with siRNA (50 nm) targeting NCBP3 (9), RHA, or NT (36) siRNA used Lipofectamine 2000 (1 μl/10 nm siRNA) (Invitrogen) and Opti-MEM medium (500 μl). Fresh medium was exchanged 6 and 24 h later, supplemented with Torin-1 (50 nm) for 18 h. Plasmid transfections were performed using X-tremeGENE (Roche) (3 or 1.5 μl), plasmids (1 or 0.5 μg) in OptiMEM (500 μl), and cells that had been cultured overnight in 6-well (1 × 106/well) or 12-well (2 × 105/well) plates. After 24 h, the transfected cells were washed twice in ice-cold 1× PBS and collected by 3-min low-speed centrifugation. Total cellular proteins and RNA were isolated in cell lysis buffer and TRIzol-LS (Ambion), respectively (56). Equivalent (20 μg) amounts of cytoplasmic protein were subjected to WB with specific antibodies, and protein–antibody complexes were detected by enhanced chemiluminescence (GE Biosciences) and quantified by ImageJ (National Institutes of Health), and densitometry results were compiled in Table S1. The antibodies used in this study are listed in Tables S2 and S3.
As described previously, cDNA was generated using Omniscript (Qiagen), random primers (Invitrogen), cellular RNA (2 μg), co-precipitated RNA samples (56), or tandem IP samples (15, 57), followed by RT-qPCR using gene-specific primers, which are provided in Table S4. All experiments were performed in three or more biological replicates.
HEK293 cells (1 × 106/35-mm well) were cultured in medium containing different concentrations (0, 25, 50, or 100 nm) of Torin-1 or 0.2% DMSO for 0, 1, 5, 18, or 24 h. The cells treated with 50 nm Torin-1 for 18 h were washed twice and cultured in Dulbecco's modified Eagle's medium for 1 or 18 h. The cells were washed once with ice-cold 1× PBS and lysed in radioimmune precipitation assay buffer. The soluble lysates were collected by centrifugation at 12,000 rpm for 2 min.
Density sedimentation and analysis of RNPs
Published protocols were used to perform density sedimentation and A254 spectrometry (58). Briefly, the cells were transfected for 24 h and treated with Torin-1 (50 nm) for 18 h. The culture medium was supplemented with 0.1 mg/ml cycloheximide (CHX) for 10 min. The cells were washed twice with ice-cold 1× PBS containing 0.1 mg/ml CHX and collected in 5 ml of ice-cold 1× PBS with CHX by scraping. The cells were subjected to centrifugation at 1500 rpm for 4 min at 4 °C and resuspended in 0.75 ml of low salt buffer (20 mm Tris-HCl, pH 7.5, 3 mm MgCl2, 10 mm NaCl, 2 mm DTT, 1× protease inhibitor mixture EDTA-free, 5 μl/ml RNase Out) and allowed to swell on ice for 5 min. The cells were lysed on ice by the addition of 0.25 ml of lysis buffer (0.2 m sucrose, 0.5% Nonidet P-40, and 0.1% Triton X-100 prepared in low salt buffer) and 10 strokes in a Dounce homogenizer (Kimble Chase). The lysates were depleted of nuclei by centrifugation at 13,000 rpm for 1 min at 4 °C to collect cytoplasm. The lysates were used for the sucrose gradients and IPs. Equivalent amounts of RNA OD units across samples were layered on the top of 10–50% sucrose prepared in 10 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 100 mm KCl, 2 mm DTT, 0.1 mg/ml CHX and centrifuged using a SW41 rotor (Beckman Coulter) for 2 h 40 min at 35,000 rpm at 4 °C. Polysome profiles were generated by continuous monitoring of RNA absorbance at 254 nm by the ISCO UA-6 absorbance detector unit (Brandel) and fractionated into 22 equivalent volume (0.5 ml) fractions by the ISCO Foxy R1 fraction collector. Brandel Peak Trace software was used to generate the corresponding profile traces. The proteins were precipitated from the collected fractions by ProteoExtract protein precipitation kit (EMD Millipore) according to the manufacturer's instructions or by TCA precipitation. The fractions were supplemented with ice-cold TCA (final concentration, 20%), vortexed, and incubated overnight at −20 °C. The samples were centrifuged at 12,000 rpm for 15 min at room temperature, and the pellets were washed twice with three volumes of ice-cold acetone. The precipitated proteins were resuspended in equivalent volume of low salt buffer and used for WB or tandem IP. The even fractions were used for protein precipitation, and the odd fractions were used for RNA extraction in TRIzol-LS (Ambion).
IP and tandem IP
Dynabeads Protein G (30 μl) (Invitrogen) were washed two times in 10 bed volumes of IP lysis buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40). The beads were incubated with the antiserum in 10 bed volumes of IP lysis buffer containing 1 mm BSA for 45 min at room temperature. The antibodies used for IP are listed in Table S2. The bead–antibody complexes were washed once in 10 bed volumes of IP wash buffer (20 mm Tris-HCl, pH 7.4, 300 mm NaCl, 0.5% Nonidet P-40) and incubated with 300 μg of cell lysate at 4 °C for 2 h with rotation. The enriched immune complexes were washed four times in IP wash buffer and collected by boiling with 1× SDS sample buffer.
Tandem IP was performed on the cyto lysates from pFLAG-RHA-transfected HEK293 cell cultured in three 15-cm plates. Dynabeads Protein G (100 μl) (Invitrogen) was washed twice in 10 bed volumes of lysis buffer (20 mm Tris-HCl, pH 7.5, 3 mm MgCl2, 10 mm NaCl, 2 mm DTT, 1× protease inhibitor mixture EDTA-free, 5 μl/ml RNase Out, 0.2 m sucrose, 0.5% Nonidet P-40, and 0.1% Triton X-100) and incubated with 10 μl of FLAG antiserum (1 mg/ml; Sigma) in 10 bed volumes of lysis buffer containing 1 mm BSA for 45 min at room temperature. 500 μg of cyto lysate was diluted to 1 ml in the lysis buffer, and the final concentration of NaCl was adjusted to 300 mm. The diluted lysate was mixed with protein G–anti-FLAG complex and incubated at 4 °C for 2 h. The RNPs captured by the anti-FLAG–conjugated protein G beads were subsequently washed twice (1 ml each) in ice-cold wash buffer I (20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 0.1% Nonidet P-40) followed by two washes in ice-cold wash buffer II (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Nonidet P-40). The samples were incubated with one bed volume of wash buffer II containing 250 μg/ml 3× FLAG peptide and 5 μl/ml RNase Out with gentle shaking at 4 °C overnight. The suspension was cleared by magnetic force followed by centrifugation at 12,000 rpm for 5 min at 4 °C. The cleared suspension was incubated with protein G (50 μl)–anti-NCBP1 (4 μl) complex for 2 h at 4 °C. The captured RNP complexes were washed four times in wash buffer II and divided into two equal parts for WB and RNA isolation. The proteins precipitated from polysome fractions were processed in a similar manner. 10% of each sample volume was reserved from each step of the tandem IP for WB. The isolated proteins were resolved using commercial 4–15% gradient SDS-PAGE gels (Bio-Rad) and transferred to nitrocellulose membrane and immunoblotted.
Statistical data analysis
Three or more independent experiments were performed for each assay and results were combined to define the means ± S.D. Statistical significance was assessed using a two-tailed Student's t test, and a p value (*) of < 0.05 was considered to be significant. Bar graphs present mean and S.D. of three independent experiments, and the precise values from the individual experiments are denoted by small dots. Statistical significance is denoted with asterisks: *, p ≤ 0.05; **, p ≤ 0.005.
Data availability
The raw RNA copies from the polysome analysis are available from the corresponding author upon request. All other data are contained within the manuscript.
Author contributions
G. S., S. E. F., and K. B.-L. conceptualization; G. S., S. E. F., and B. S. data curation; G. S. and S. E. F. formal analysis; G. S. supervision; G. S. validation; G. S. and K. B.-L. investigation; G. S. methodology; G. S. writing-original draft; G. S. and K. B.-L. writing-review and editing; K. B.-L. funding acquisition.
Supplementary Material
This work was supported by National Institutes of Health Grants P50 NIGMS0103297, T32GM068412, and U54AI50470. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S4 and Figs. S1–S4.
- NCBP
- nuclear cap-binding protein
- AP-1
- activator protein-1
- CBC
- cap-binding complex
- IP
- immunoprecipitation
- dsRBD
- double-stranded RNA-binding domains
- PIC
- preinitiation complex
- mRNP
- messenger ribonucleoprotein
- PABP
- poly-A–binding protein
- PCE
- post-transcriptional control element
- RHA
- RNA helicase A
- WB
- Western blot
- qPCR
- quantitative PCR
- eIF
- eukaryotic translation initiation factor
- cyto
- cytoplasmic
- <MD
- less than the minimum detectable
- NT
- nontargeting
- CHX
- cycloheximide.
References
- 1. Braddock M., Muckenthaler M., White M. R., Thorburn A. M., Sommerville J., Kingsman A. J., and Kingsman S. M. (1994) Intron-less RNA injected into the nucleus of Xenopus oocytes accesses a regulated translation control pathway. Nucleic Acids Res. 22, 5255–5264 10.1093/nar/22.24.5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matsumoto K., Wassarman K. M., and Wolffe A. P. (1998) Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 17, 2107–2121 10.1093/emboj/17.7.2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim V. N., Kataoka N., and Dreyfuss G. (2001) Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293, 1832–1836 10.1126/science.1062829 [DOI] [PubMed] [Google Scholar]
- 4. Nott A., Meislin S. H., and Moore M. J. (2003) A quantitative analysis of intron effects on mammalian gene expression. RNA 9, 607–617 10.1261/rna.5250403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu S., and Cullen B. R. (2003) Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9, 618–630 10.1261/rna.5260303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., and Mattaj I. W. (1994) A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78, 657–668 10.1016/0092-8674(94)90530-4 [DOI] [PubMed] [Google Scholar]
- 7. Mazza C., Segref A., Mattaj I. W., and Cusack S. (2002) Large-scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21, 5548–5557 10.1093/emboj/cdf538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choe J., Kim K. M., Park S., Lee Y. K., Song O. K., Kim M. K., Lee B. G., Song H. K., and Kim Y. K. (2013) Rapid degradation of replication-dependent histone mRNAs largely occurs on mRNAs bound by nuclear cap-binding proteins 80 and 20. Nucleic Acids Res. 41, 1307–1318 10.1093/nar/gks1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gebhardt A., Habjan M., Benda C., Meiler A., Haas D. A., Hein M. Y., Mann A., Mann M., Habermann B., and Pichlmair A. (2015) mRNA export through an additional cap-binding complex consisting of NCBP1 and NCBP3. Nat. Commun. 6, 8192 10.1038/ncomms9192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ryu I., and Kim Y. K. (2017) Translation initiation mediated by nuclear cap-binding protein complex. BMB Rep. 50, 186–193 10.5483/BMBRep.2017.50.4.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le Hir H., Nott A., and Moore M. J. (2003) How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28, 215–220 10.1016/S0968-0004(03)00052-5 [DOI] [PubMed] [Google Scholar]
- 12. Moore M. J. (2005) From birth to death: the complex lives of eukaryotic mRNAs. Science 309, 1514–1518 10.1126/science.1111443 [DOI] [PubMed] [Google Scholar]
- 13. Moore M. J., and Proudfoot N. J. (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 10.1016/j.cell.2009.02.001 [DOI] [PubMed] [Google Scholar]
- 14. Topisirovic I., Svitkin Y. V., Sonenberg N., and Shatkin A. J. (2011) Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA 2, 277–298 10.1002/wrna.52 [DOI] [PubMed] [Google Scholar]
- 15. Singh G., Kucukural A., Cenik C., Leszyk J. D., Shaffer S. A., Weng Z., and Moore M. J. (2012) The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151, 750–764; Correction (2012) Cell151, 915–916 10.1016/j.cell.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Isken O., and Maquat L. E. (2008) The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 9, 699–712 10.1038/nrg2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sonenberg N., and Hinnebusch A. G. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santulli G., and Totary-Jain H. (2013) Tailoring mTOR-based therapy: molecular evidence and clinical challenges. Pharmacogenomics 14, 1517–1526 10.2217/pgs.13.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Short J. D., and Pfarr C. M. (2002) Translational regulation of the JunD messenger RNA. J. Biol. Chem. 277, 32697–32705 10.1074/jbc.M204553200 [DOI] [PubMed] [Google Scholar]
- 20. Hsieh A. C., Liu Y., Edlind M. P., Ingolia N. T., Janes M. R., Sher A., Shi E. Y., Stumpf C. R., Christensen C., Bonham M. J., Wang S., Ren P., Martin M., Jessen K., Feldman M. E., et al. (2012) The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henis-Korenblit S., Strumpf N. L., Goldstaub D., and Kimchi A. (2000) A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol. 20, 496–506 10.1128/MCB.20.2.496-506.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nevins T. A., Harder Z. M., Korneluk R. G., and Holcík M. (2003) Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 278, 3572–3579 10.1074/jbc.M206781200 [DOI] [PubMed] [Google Scholar]
- 23. Nousch M., Reed V., Bryson-Richardson R. J., Currie P. D., and Preiss T. (2007) The eIF4G-homolog p97 can activate translation independent of caspase cleavage. RNA 13, 374–384 10.1261/rna.372307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lewis S. M., Cerquozzi S., Graber T. E., Ungureanu N. H., Andrews M., and Holcik M. (2008) The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Res. 36, 168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoffe Y., David M., Kalaora R., Povodovski L., Friedlander G., Feldmesser E., Ainbinder E., Saada A., Bialik S., and Kimchi A. (2016) Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes Dev. 30, 1991–2004 10.1101/gad.285239.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bukhari S. I. A., Truesdell S. S., Lee S., Kollu S., Classon A., Boukhali M., Jain E., Mortensen R. D., Yanagiya A., Sadreyev R. I., Haas W., and Vasudevan S. (2016) A specialized mechanism of translation mediated by FXR1a-associated microRNP in cellular quiescence. Mol. Cell. 61, 760–773 10.1016/j.molcel.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bukhari S. I., and Vasudevan S. (2017) FXR1a-associated microRNP: a driver of specialized non-canonical translation in quiescent conditions. RNA Biol. 14, 137–145 10.1080/15476286.2016.1265197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jeong S. J., Park S., Nguyen L. T., Hwang J., Lee E. Y., Giong H. K., Lee J. S., Yoon I., Lee J. H., Kim J. H., Kim H. K., Kim D., Yang W. S., Kim S. Y., Lee C. Y., et al. (2019) A threonyl-tRNA synthetase-mediated translation initiation machinery. Nat. Commun. 10, 1357 10.1038/s41467-019-09086-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee A. S., Kranzusch P. J., Doudna J. A., and Cate J. H. (2016) EIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536, 96–99 10.1038/nature18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim K. M., Cho H., Choi K., Kim J., Kim B. W., Ko Y. G., Jang S. K., and Kim Y. K. (2009) A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev. 23, 2033–2045 10.1101/gad.1823409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharma A., Yilmaz A., Marsh K., Cochrane A., and Boris-Lawrie K. (2012) Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 8, e1002612 10.1371/journal.ppat.1002612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choe J., Ryu I., Park O. H., Park J., Cho H., Yoo J. S., Chi S. W., Kim M. K., Song H. K., and Kim Y. K. (2014) EIF4AIII enhances translation of nuclear cap-binding complex-bound mRNAs by promoting disruption of secondary structures in 5′ UTR. Proc. Natl. Acad. Sci. U.S.A. 111, E4577–E4586 10.1073/pnas.1409695111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Toro-Ascuy D., Rojas-Araya B., García-de-Gracia F., Rojas-Fuentes C., Pereira-Montecinos C., Gaete-Argel A., Valiente-Echeverría F., Ohlmann T., and Soto-Rifo R. (2018) A Rev–CBP80–eIF4AI complex drives Gag synthesis from the HIV-1 unspliced mRNA. Nucleic Acids Res. 46, 11539–11552 10.1093/nar/gky851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gebhardt A., Bergant V., Schnepf D., Moser M., Meiler A., Togbe D., MacKowiak C., Reinert L. S., Paludan S. R., Ryffel B., Stukalov A., Staeheli P., and Pichlmair A. (2019) The alternative cap-binding complex is required for antiviral defense in vivo. PLoS Pathog. 15, e1008155 10.1371/journal.ppat.1008155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hernandez J. M., Floyd D. H., Weilbaecher K. N., Green P. L., and Boris-Lawrie K. (2008) Multiple facets of junD gene expression are atypical among AP-1 family members. Oncogene 27, 4757–4767 10.1038/onc.2008.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hartman T. R., Qian S., Bolinger C., Fernandez S., Schoenberg D. R., and Boris-Lawrie K. (2006) RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 13, 509–516 10.1038/nsmb1092 [DOI] [PubMed] [Google Scholar]
- 37. Bolinger C., Yilmaz A., Hartman T. R., Kovacic M. B., Fernandez S., Ye J., Forget M., Green P. L., and Boris-Lawrie K. (2007) RNA helicase A interacts with divergent lymphotropic retroviruses and promotes translation of human T-cell leukemia virus type 1. Nucleic Acids Res. 35, 2629–2642 10.1093/nar/gkm124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bolinger C., Sharma A., Singh D., Yu L., and Boris-Lawrie K. (2010) RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 38, 1686–1696 10.1093/nar/gkp1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soto-Rifo R., Rubilar P. S., and Ohlmann T. (2013) The DEAD-box helicase DDX3 substitutes for the cap-binding protein eIF4E to promote compartmentalized translation initiation of the HIV-1 genomic RNA. Nucleic Acids Res. 41, 6286–6299 10.1093/nar/gkt306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shih J. W., Tsai T. Y., Chao C. H., and Wu Lee Y. H. (2008) Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27, 700–714 10.1038/sj.onc.1210687 [DOI] [PubMed] [Google Scholar]
- 41. Ishigaki Y., Li X., Serin G., and Maquat L. E. (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617 10.1016/S0092-8674(01)00475-5 [DOI] [PubMed] [Google Scholar]
- 42. Ranji A., Shkriabai N., Kvaratskhelia M., Musier-Forsyth K., and Boris-Lawrie K. (2011) Features of double-stranded RNA-binding domains of RNA helicase A are necessary for selective recognition and translation of complex mRNAs. J. Biol. Chem. 286, 5328–5337 10.1074/jbc.M110.176339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roberts T. M., and Boris-Lawrie K. (2000) The 5′ RNA terminus of spleen necrosis virus stimulates translation of nonviral mRNA. J. Virol. 74, 8111–8118 10.1128/JVI.74.17.8111-8118.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roberts T. M., and Boris-Lawrie K. (2003) Primary sequence and secondary structure motifs in spleen necrosis virus RU5 confer translational utilization of unspliced human immunodeficiency virus type 1 reporter RNA. J. Virol. 77, 11973–11984 10.1128/JVI.77.22.11973-11984.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brady S., Singh G., Bolinger C., Song Z., Boeras I., Weng K., Trent B., Brown W. C., Singh K., Boris-Lawrie K., and Heng X. (2019) Virion-associated, host-derived DHX9/RNA helicase A enhances the processivity of HIV-1 reverse transcriptase on genomic RNA. J. Biol. Chem. 294, 11473–11485 10.1074/jbc.RA119.007679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Atsaves V., Leventaki V., Rassidakis G. Z., and Claret F. X. (2019) AP-1 transcription factors as regulators of immune responses in cancer. Cancers (Basel) 11, E1037 10.3390/cancers11071037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Childs-Disney J. L., Tran T., Vummidi B. R., Velagapudi S. P., Haniff H. S., Matsumoto Y., Crynen G., Southern M. R., Biswas A., Wang Z. F., Tellinghuisen T. L., and Disney M. D. (2018) A massively parallel selection of small molecule-RNA motif binding partners informs design of an antiviral from sequence. Chem. 4, 2384–2404 10.1016/j.chempr.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ng Y. C., Chung W. C., Kang H. R., Cho H. J., Park E. B., Kang S. J., and Song M. J. (2018) A DNA-sensing–independent role of a nuclear RNA helicase, DHX9, in stimulation of NF-B–mediated innate immunity against DNA virus infection. Nucleic Acids Res. 46, 9011–9026 10.1093/nar/gky742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shen B., Chen Y., Hu J., Qiao M., Ren J., Hu J., Chen J., Tang N., Huang A., and Hu Y. (2020) Hepatitis B virus X protein modulates upregulation of DHX9 to promote viral DNA replication. Cell. Microbiol. 22, e13148 [DOI] [PubMed] [Google Scholar]
- 50. Boeras I., Song Z., Moran A., Franklin J., Brown W. C., Johnson M., Boris-Lawrie K., and Heng X. (2016) DHX9/RHA binding to the PBS-segment of the genomic RNA during HIV-1 assembly bolsters virion infectivity. J. Mol. Biol. 428, 2418–2429 10.1016/j.jmb.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee T., Paquet M., Larsson O., and Pelletier J. (2016) Tumor cell survival dependence on the DHX9 DExH-box helicase. Oncogene 35, 5093–5105 10.1038/onc.2016.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee T., and Pelletier J. (2016) The biology of DHX9 and its potential as a therapeutic target. Oncotarget 7, 42716–42739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng D. D., Zhang H. Z., Yuan J. Q., Li S. J., Yang Q. C., and Fan C. Y. (2017) Minichromosome maintenance protein 2 and 3 promote osteosarcoma progression via DHX9 and predict poor patient prognosis. Oncotarget 8, 26380–26393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cao S., Sun R., Wang W., Meng X., Zhang Y., Zhang N., and Yang S. (2017) RNA helicase DHX9 may be a therapeutic target in lung cancer and inhibited by enoxacin. Am. J. Transl. Res. 9, 674–682 [PMC free article] [PubMed] [Google Scholar]
- 55. Lee C. G., Eki T., Okumura K., Nogami M., Soares V., Murakami Y., Hanaoka F., and Hurwitz J. (1999) The human RNA helicase a (DDX9) gene maps to the prostate cancer susceptibility locus at chromosome band Iq25 and its pseudogene (DDX9P) to 13q22, respectively. Somat. Cell Mol. Genet. 25, 33–39 10.1023/B:SCAM.0000007138.44216.3a [DOI] [PubMed] [Google Scholar]
- 56. Singh G., Rife B. D., Seufzer B., Salemi M., Rendahl A., and Boris-Lawrie K. (2018) Identification of conserved, primary sequence motifs that direct retrovirus RNA fate. Nucleic Acids Res. 46, 7366–7378 10.1093/nar/gky369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Singh G., Fritz S. M., Ranji A., Singh D., and Boris-Lawrie K. (2017) Isolation of cognate RNA-protein complexes from cells using oligonucleotide-directed elution. J. Vis. Exp. 119, 54391 10.3791/54391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Boeras I., Seufzer B., Brady S., Rendahl A., Heng X., and Boris-Lawrie K. (2017) The basal translation rate of authentic HIV-1 RNA is regulated by 5′UTR nt-pairings at junction of R and U5. Sci. Rep. 7, 6902 10.1038/s41598-017-06883-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA copies from the polysome analysis are available from the corresponding author upon request. All other data are contained within the manuscript.