Abstract
Objective
CircRNAs are emerging as vital regulators in a variety of cancers. However, the expression pattern and potential mechanism of circRNAs in triple-negative breast cancer remain unclear. In this study, we aim to systematically investigate circRNAs alteration in triple-negative breast cancer tissues.
Methods
Microarray and bioinformatics analyses were used to identify circRNAs expression in cancer tissues. qRT-PCR was conducted to measure the expression of RNAs. Cell Counting Kit‐8, wound-healing and transwell assays were conducted to investigate the function of circRNAs. Dual-luciferase reporter assay was performed to validate target binding.
Results
Hsa_circ_0131242 was highly expressed in both cancer tissues and cell lines compared to control. Subsequently, statistical analyses revealed that high expression of hsa_circ_0131242 was positively correlated with advanced tumor stages and poorer clinical features in cancer patients. Hsa_circ_0131242 knockdown could suppress the progression of breast cancer cells. Bioinformatics prediction and luciferase reporter assay showed that hsa_circ_0131242 acted as a sponge for hsa-miR-2682. Moreover, co-transfection of hsa-miR-2682 inhibitor and si-hsa_circ_0131242 rescued cell proliferation and migration in BT549 and MDA-MB-468 cell lines.
Conclusion
Our study identified hsa_circ_0131242 expression in TNBC for the first time and found that hsa_circ_0131242 may promote triple-negative breast cancer progression by sponging hsa-miR-2682.
Keywords: breast cancer, hsa_circ_0131242, proliferation, sponge, biomarker
Introduction
Breast cancer is one of the most frequently diagnosed cancers among women in the world.1 Triple-negative breast cancer (TNBC) represents an aggressive subtype of breast cancer, accounts for 15–20% of clinical cases. TNBC is characterized by lack of estrogen receptor (ER), progesterone receptor (PR) and HER2 receptor and thus resistance to conventional chemotherapy for breast cancer.2–4 Therefore, it is essential to better understand TNBC progression and identify more prognostic and therapeutic molecules to promote TNBC diagnosis and treatment.
Circular RNAs (circRNAs) are a novel class of non-coding RNAs characterized by the lack of 3ʹpolyA tail and a special loop structure.5 With the development of microarray and high-throughput sequencing, the expression profile and potential function of circRNAs were investigated in a variety of human tissues.6,7 Accumulating evidences indicated that circRNAs may play pivotal roles in disease progression by acting as miRNAs sponges,8 regulating splicing events and interacting with RNA-binding proteins.9 The sponge mechanism of circRNAs indicated that they may bind to miRNAs and consequently repress function of miRNAs.10 For instance, Zhou et, al found that circSMARCA5 could inhibit progression of hepatocellular carcinoma by acting as sponges of miR-17 and miR-181b;11 CircLMTK2 was proved to promote gastric cancer proliferation through sponging miR-150.12 However, the expression profile and function of circRNAs in TNBC progression have not been clearly elucidated.
In this study, we performed microarray and bioinformatics analyses to investigate circRNAs alteration in TNBC tissues compared to control. Among the differentially expressed circRNAs, hsa_circ_0131242 was validated to be significantly up-regulated in both TNBC samples and cell lines by qRT-PCR experiment. Subsequently, the correlation between hsa_circ_0131242 and clinical outcomes of TNBC patients was calculated. Finally, the function and potential mechanism of hsa_circ_0131242 in breast cancer cells were investigated, which provides a new insight into the diagnosis and treatment of TNBC.
Materials and Methods
Cell Lines, Patient Samples and Ethical Statement
Human breast cancer cell lines including BT549, MDA-MB-231, MDA-MB-468, T47D and HCC1806, normal mammary epithelial cell line (MCF-10A) were obtained from Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and American Type Culture Collection (ATCC, USA). Cells were cultured following the instruction of supplier. The breast cancer tissues and adjacent non-tumorigenesis tissue were collected from 120 TNBC patients aged from 18 to 70 years old that received therapy in Cancer Hospital of China Medical University between 2009 and 2016 (Supplementary Table 1). Patients enrolled in this study did not receive neoadjuvant chemotherapy before. The written informed consents were obtained from all participants. This study was approved by Ethics Committee of Cancer Hospital of China Medical University and was in accordance with Helsinki Declaration.
RNA Extraction and circRNAs Microarray
Total RNA was isolated from five pairs of breast cancer tissues and adjacent non-tumorigenesis tissue using TRIzol (Invitrogen, CA, USA) following the instructions of manufacturer. RNA quality and integrity were examined by a NanoDrop ND-1000 (Thermo, MA, USA) and denaturing agarose gel electrophoresis. CircRNA microarray was conducted in CapitalBio company (CapitalBio, Beijing, China) using Human CircRNA Array v2.0. The raw expression data were processed and normalized by GeneSpring software.
Bioinformatics and Statistical Analyses
The annotations and sequences of circRNAs were obtained from circBase.13 miRNA expression data of human breast cancer were downloaded from GEO database (GSE28969). Differentially expressed circRNAs and miRNAs were analyzed by EdgeR14 and setting the threshold as |fold change| ≥2 and P-value <0.05. Pearson’s correlation analysis was conducted to search the expression tendency between hsa_circ_0131242 and miRNAs. The target binding of miRNAs to circRNAs was predicted by TargetScan and Miranda algorithms.15 Pathway enrichment was performed by KOBAS software.16 The Kaplan–Meier analysis was conducted to evaluate the overall survival rate. Correlation between clinical features and survival was calculated by Cox regression analysis. All statistical tests were two sided and conducted by R software (https://www.r-project.org); moreover, all experiments were performed at least three times and performed in triplicate. P-value <0.05 was considered statistically significant.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNAs from cancer tissues or cell lines were isolated using TRIzol (Invitrogen, CA, USA) following the instructions. RNAs were reverse-transcribed into complementary cDNAs by Prime Script RT reagent kit (Takara, Japan). The quantitative real-time PCR was conducted with SYBR Taq (Takara, Japan) at an Applied Real-Time PCR System following the instruction of manufacturer. U6 and GAPDH were used as controls for miRNAs and circRNAs, respectively. The relative expression of RNAs was calculated by 2−ΔΔCt method. Primer sequences are listed in Supplementary Table 2.
Cell Transfection
MDA-MB-468 and BT549 cells were transfected in six-well plates with small interfering RNA for hsa_circ_0131242 and hsa-miR-2682 inhibitor using Lipofectamine 2000 (4μL/well, Invitrogen, USA) following the manufacture’s guidance. The siRNA and miRNA inhibitor were synthesized by RiboBio company (Guangzhou, China).
Cell Counting Kit‐8 (CCK-8) and Colony Formation Assays
Cells (1×103) were seeded in a 96-well plate. After 48 h of transfection, CCK-8 solution (10 μL) was added and then cells were incubated for 2 h (37°C). The absorbance at 450 nM was observed. For colony formation assay, cells were plated into 6-well plates (500 cells/well) followed by 2-week incubation at 37°C. The colonies were treated with 4% formaldehyde and then stained with crystal violet (1%) for 5 min. Finally, colonies were counted under a microscope. All experiments were performed in triplicate.
Wound-Healing and Transwell Assays
The transfected cells were seeded in six-well cell culture plates, grown to confluence overnight. The wound was scratched using 200μL pipette tip and were then imaged after 0 and 48h. For transwell assay, cells (2×104) in serum-free medium (200μL) were added into transwell chamber (Corning, NY, USA), 20% FBS medium was added into the lower chamber. Twenty-four hours later, cells on the upper chamber were removed and the remaining cells were treated with paraformaldehyde (4%) and stained with crystal violet after washing. Cells were quantified using Image J software.
Dual-Luciferase Reporter Assay
Bioinformatics algorithms predicted the target binding of miRNA and circRNA. To verify the target binding, dual-luciferase reporter assay was conducted. To construct plasmid of circRNA, the conserved sequence of hsa_circ_0131242 was cloned into the pGL3-Basic luciferase vectors. Cells (3×104) were seeded in 24-well plates and transiently transfected with pGL3- hsa_circ_0131242 and miRNA mimics for 24 h. Luciferase activity was obtained by dual-luciferase reporter assay system (Promega, USA) following the instruction of manufacturer. Independent experiments were performed in triplicate.
RNA-Binding Protein Immunoprecipitation (RIP)
The MS2bp-MS2bs based RIP experiment was conducted to verify the interaction between circRNA and miRNA. Cancer cells were co‐transfected with vectors of MS2bs-Rluc (control), MS2bs‐hsa_circ_0131242 and MS2bs‐hsa_circ_0131242mt (complementarity to hsa-miR-2682 was removed) by Lipofectamine 2000 (Invitrogen, CA, USA). RIP experiment was performed after 48 h using Magna RIP Kit (Millipore) according to the manufacturer’s guidance.
Results
Identification and Characterization of hsa_circ_0131242
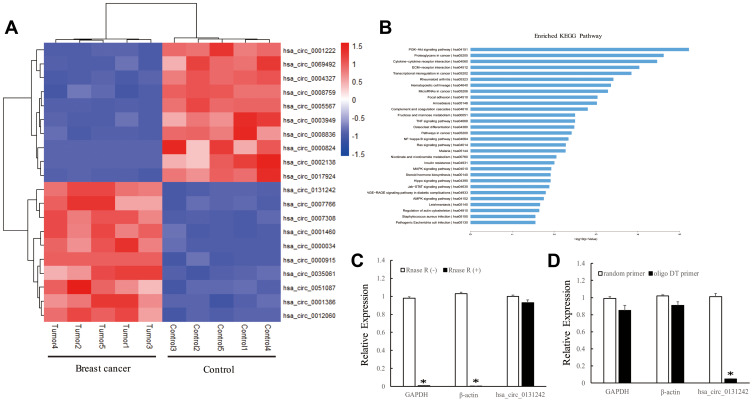
To identify potential circRNAs that may involve in breast cancer progression, we performed microarray and bioinformatics analyses. A total of 624 differentially expressed circRNAs were identified between TNBC tissues and adjacent normal tissues. Figure 1A presents the top 20 differentially expressed circRNAs (fold change ≥2 and P-value <0.05) based on high expression level and fold change. Screening the Kyoto Encyclopedia of Genes and Genomes (KEGG) database enables us to find cancer-related pathways of top 20 circRNAs. As shown in Figure 1B, host genes of circRNAs were mainly enriched in ECM−receptor interaction (hsa04512), transcriptional misregulation in cancer (hsa05202) and MicroRNAs in cancer (hsa05206), indicating their potential roles in cancer pathogenesis. Among the top 20 differentially expressed circRNAs, hsa_circ_0131242 had the highest expression in TNBC tissues, which is 6.85 times higher than that in normal control tissues. Hsa_circ_0131242 (chr6:161022009−161034449) is partially derived from mitogen-activated protein kinase 4 (MAP3K4) with genomic sequence of 12441nt. To verify the loop structure of hsa_circ_0131242, total RNA was treated with RNase R. As shown in Figure 1C, hsa_circ_0131242 was resistant to RNase R digestion compared with linear GAPDH or β-actin mRNAs. Moreover, hsa_circ_0131242 was successfully reversed by the random primer but resistant to oligo DT primer (Figure 1D). The above findings indicated the circular structure of hsa_circ_0131242.
Figure 1.
Identification and characterization of hsa_circ_0131242. (A) Top 20 differentially expressed circRNAs in TNBC breast cancer tissues compared to control. (B) KEGG analysis of host genes corresponding to top 20 circRNAs. (C) qRT-PCR analysis of GAPDH, β-actin and hsa_circ_0131242 after RNase R treatment. (D) Poly A-tailed mRNAs can be synthesized to cDNA using random primer or oligo DT primer, non-poly A-tailed hsa_circ_0131242 cannot be reversed to cDNA by using oligo DT primer. *P < 0.01.
hsa_circ_0131242 Was Associated with Clinical Features in TNBC Patients
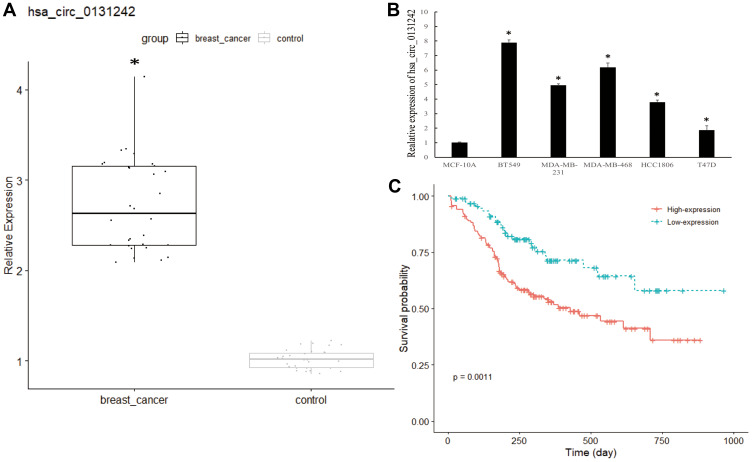
We validated the expression level of hsa_circ_0131242 in 30 pairs of TNBC tissues and control tissues. As shown in Figure 2A, hsa_circ_0131242 was remarkably up-regulated in TNBC samples compared to control as identified by qRT-PCR. Moreover, high expression level of hsa_circ_0131242 was also detected in breast cancer cell lines (BT549, MDA-MB-231, MDA-MB-468, HCC1806 and T47D) (Figure 2B). The above results were consistent with microarray expression data. We then measured the association between hsa_circ_0131242 expression and clinical features in 120 TNBC tissues. As shown in Table 1, hsa_circ_0131242 expression was significantly associated with tumor size and TNM stages (P < 0.001). Besides, Kaplan–Meier survival analysis showed that patients with high hsa_circ_0131242 level had a poorer overall survival (Figure 2C).
Figure 2.
Hsa_circ_0131242 was associated with clinical features in TNBC patients. (A and B) Hsa_circ_0131242 was highly expressed in breast cancer tissues and cell lines. (C) Overall survival for breast cancer patients with high or low hsa_circ_0131242 expression. *P < 0.01.
Table 1.
Hsa_circ_0131242 Expression and Clinical Features of TNBC Patients (n = 120)
| Clinical Index | hsa_circ_0131242 Expression | P-value | |
|---|---|---|---|
| High (n = 86) | Low (n = 34) | ||
| Age (years) | 0.847 | ||
| <50 | 27 | 10 | |
| ≥50 | 59 | 24 | |
| TNM stage | <0.001 | ||
| I II–III |
10 76 |
15 19 |
|
| Tumor size (cm) | <0.001 | ||
| ≤2 | 9 | 13 | |
| >2 | 77 | 21 | |
Note: Patients were divided into high/low expression groups based on the mean expression of hsa_circ_0131242.
hsa_circ_0131242 Knockdown Inhibited the Progression of TNBC Cells
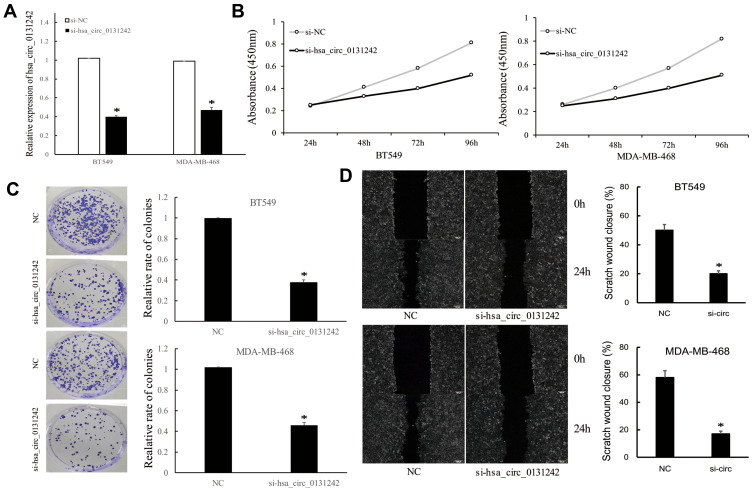
To gain insights into the cellular effects of hsa_circ_0131242 in TNBC, BT549 and MDA-MB-468 cell lines were used to knock down hsa_circ_0131242, as shown in Figure 3A. CCK-8 and colony formation assays revealed that hsa_circ_0131242 knockdown significantly inhibited cell proliferation (Figure 3B) and colony formation (Figure 3C). We further examined the effects of hsa_circ_0131242 on migration of cancer cells. As shown in Figure 3D, wound-healing assays indicated that hsa_circ_0131242 knockdown significantly inhibited the migration of BT549 and MDA-MB-468 cells (Figure 3D).
Figure 3.
Hsa_circ_0131242 knockdown inhibited the progression of TNBC cells. (A) Transfection of si-hsa_circ_0131242 in BT549 and MDA-MB-468 cell lines. (B–D) CCK-8, colony formation and wound-healing assays revealed that hsa_circ_0131242 knockdown inhibited cell proliferation, colony formation and migration of BT549 and MDA-MB-468 cell lines. *P < 0.01.
hsa_circ_0131242 Regulated Breast Cancer Progression by Sponging hsa-miR-2682
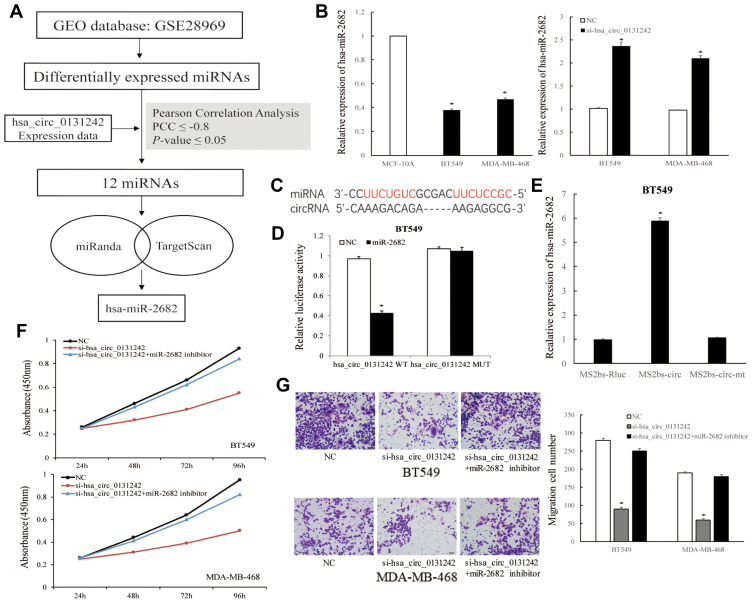
Considering circRNAs could act as miRNAs sponges to participate in biological process, we analyzed the miRNA expression data from human breast cancer database (GEO: GSE28969). Twelve differentially expressed miRNAs were found to be significantly correlated with hsa_circ_0131242 based on Pearson Correlation Coefficient (PCC) ≤ −0.8 and P-value ≤0.05. Among these miRNAs, hsa-miR-2682-3p was predicted to be a potential target of hsa_circ_0131242 by two algorithms (Figure 4A). QRT-PCR showed that hsa-miR-2682 was down-regulated in breast cancer cell lines and knockdown of hsa_circ_0131242 enhanced hsa-miR-2682 expression (Figure 4B). Luciferase reporter assays were conducted to verify the target binding of hsa-miR-2682 and hsa_circ_0131242 (Figure 4C). As shown in Figure 4D, hsa-miR-2682 significantly reduced the luciferase activity of hsa_circ_0131242 wild type while had no effect on mutant type. Besides, RIP assay showed an enrichment of hsa-miR-2682 in RNAs retrieved from MS2bs-hsa_circ_0131242 (Figure 4E), suggested that hsa_circ_0131242 could act as a sponge for hsa-miR-2682. Moreover, co-transfection of hsa-miR-2682 inhibitor and si-hsa_circ_0131242 rescued cell proliferation and migration in BT549 and MDA-MB-468 cell lines (Figure 4F and G).
Figure 4.
Hsa_circ_0131242 regulated breast cancer progression by sponging hsa-miR-2682. (A) Workflow of screening candidate miRNAs that targeted by hsa_circ_0131242. (B) Hsa-miR-2682 was down-regulated in breast cancer cell lines and knockdown of hsa_circ_0131242 enhanced hsa-miR-2682 expression. (C) Predicted binding sites of hsa-miR-2682 and hsa_circ_0131242. (D) Luciferase reporter assay of BT549 cells co-transfected with hsa-miR-2682 mimics and wild‐type (WT) or mutant (Mut) reporter. (E) MS2‐based RIP assay was conducted in BT549 cells. (F and G) Cell proliferation and migration were reversed by co-transfection of hsa-miR-2682 inhibitor and si-hsa_circ_0131242, as detected by CCK-8 and transwell assays. *P < 0.01.
Discussion
Triple-negative breast cancer is an aggressive subtype of breast cancer with rapid tumor growth and chemotherapy resistance.17,18 To explore potential therapeutic targets for TNBC diagnosis and treatment, we performed genome-wide circRNA microarray analyses in TNBC tissues and control tissues. CircRNAs are a novel class of endogenous ncRNAs with covalently closed loop structure.5 This property makes circRNAs more stable than linear RNAs and is expected to serve as diagnostic biomarkers for a variety of human cancers.19 Liu et, al identified certain circRNAs were abnormally expressed in human laryngeal squamous cell carcinomas and hsa_circ:chr20:31876585–31897648 may be a candidate biomarker for disease treatment.20 By analyzing microarray data, we noticed that hsa_circ_0131242 was highly expressed in breast cancer tissues and the up-regulation was validated by qRT-PCR experiment in 30 TNBC tissues and breast cancer cell lines. High expression level of hsa_circ_0131242 in breast cancer deserves us to further explore its clinical significance in TNBC patients. The Kaplan–Meier results showed that high expression level of hsa_circ_0131242 in TNBC patients was correlated with poorer prognosis.
Accumulating evidences demonstrated that circRNAs play important roles in regulating cellular physiology processes,9 for example, circPRKCI was identified to act as a tumor promoter in lung adenocarcinoma.21 We investigated the cellular function of hsa_circ_0131242 in BT549 and MDA-MB-468 cell lines. The results indicated that knockdown of hsa_circ_0131242 could suppress proliferation, colony formation and migration of breast cancer cells. Currently, the mechanism of circRNAs remains to be explored, previous studies have shown that circRNAs could function as miRNAs sponges to regulate gene expression.8,9 For example, circRNA circMTO1 was found to suppress the progression of hepatocellular carcinoma by sponging miR-9 and thus promoted p21 expression.22 By means of bioinformatics prediction and experimental validation, we verified that hsa_circ_0131242 could act as a sponge for hsa-miR-2682. Further co-transfection of hsa-miR-2682 inhibitor and si-hsa_circ_0131242 in cells indicated that hsa_circ_0131242 regulated breast cancer progression by sponging hsa-miR-2682.
Collectively, our study showed that hsa_circ_0131242 was highly expressed in TNBC and its expression was positively correlated with advanced tumor stages and poorer clinical features of TNBC patients. Furthermore, hsa_circ_0131242 could regulate breast cancer progression by sponging hsa-miR-2682, which may serve as a potential therapeutic target for breast cancer.
Data Sharing Statement
The microarray data used to support the findings of this study are available from the corresponding author upon reasonable request.
Author Contributions
Yueting Li and Daqing Jiang designed this research and wrote the paper. Yueting Li and Pengxu Shi performed data statistics. Tianzhi Zheng and Ziwei Ying performed experiments. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare there are no competing financial interests.
References
- 1.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. doi: 10.3322/caac.21412 [DOI] [PubMed] [Google Scholar]
- 2.Jayasekera J, Schechter CB, Sparano JA, et al. Effects of radiotherapy in early-stage, low-recurrence risk, hormone-sensitive breast cancer. J Natl Cancer Inst. 2018;110(12):1370–1379. doi: 10.1093/jnci/djy128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalimutho M, Parsons K, Mittal D, Lopez JA, Srihari S, Khanna KK. Targeted therapies for triple-negative breast cancer: combating a stubborn disease. Trends Pharmacol Sci. 2015;36(12):822–846. doi: 10.1016/j.tips.2015.08.009 [DOI] [PubMed] [Google Scholar]
- 4.Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA. New strategies for triple-negative breast cancer–deciphering the heterogeneity. Clin Cancer Res. 2014;20(4):782–790. doi: 10.1158/1078-0432.CCR-13-0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034 [DOI] [PubMed] [Google Scholar]
- 6.Li S, Teng S, Xu J, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. 2019;20(4):1420–1433. doi: 10.1093/bib/bby006 [DOI] [PubMed] [Google Scholar]
- 7.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692. doi: 10.1038/nrg.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 9.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(5):309–316. doi: 10.1016/j.tig.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi: 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Tang D, Wang W, et al. circLMTK2 acts as a sponge of miR-150-5p and promotes proliferation and metastasis in gastric cancer. Mol Cancer. 2019;18(1):162. doi: 10.1186/s12943-019-1081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glazar P, Papavasileiou P, Rajewsky N. CircBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. Common features of microRNA target prediction tools. Front Genet. 2014;5:23. doi: 10.3389/fgene.2014.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–322. doi: 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YR, Jiang YZ, Xu XE, Hu X, Yu KD, Shao ZM. Comprehensive transcriptome profiling reveals multigene signatures in triple-negative breast cancer. Clin Cancer Res. 2016;22(7):1653–1662. doi: 10.1158/1078-0432.CCR-15-1555 [DOI] [PubMed] [Google Scholar]
- 18.Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017;389(10087):2430–2442. doi: 10.1016/S0140-6736(16)32454-0 [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu C, Shi X, Wang AY, et al. RNA-Seq profiling of circular RNAs in human laryngeal squamous cell carcinomas. Mol Cancer. 2018;17(1):86. doi: 10.1186/s12943-018-0833-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu M, Xia W, Chen R, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78(11):2839–2851. doi: 10.1158/0008-5472.CAN-17-2808 [DOI] [PubMed] [Google Scholar]
- 22.Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi: 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]