Clinical samples obtained from 57 glioma patients were analyzed by TOF-SIMS. The analysis allowed to separately cluster normal brain and cancer samples and demonstrated differences between primary and recurrent tumors as well as differences between morphologically distinct regions of the same tumor. Simultaneous detection of proteins and metabolites by TOF-SIMS allowed to visualize Caveolin 1 positive cholesterol rich population of cancer cells that might represent glioma stem cells.
Keywords: Glioblastoma, stem cells, mass spectrometry, cancer biomarker(s), imaging visualization tools, caveolin-1, glioma, TOF-SIMS
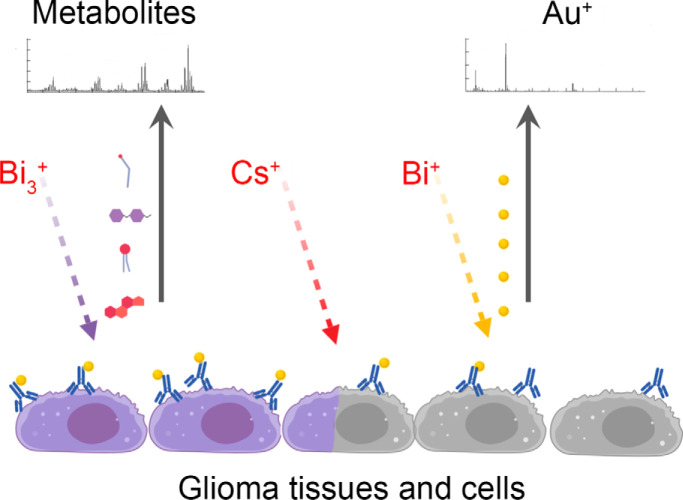
Graphical Abstract
Highlights
TOF-SIMS allows to visualize normal brain and tumor border.
Glioma samples can be subdivided into clinically relevant groups by TOF-SIMS data.
TOF-SIMS allows to simultaneously detect proteins and metabolites in clinical samples.
Abstract
Glioblastoma (GBM) is one of the most aggressive human cancers with a median survival of less than two years. A distinguishing pathological feature of GBM is a high degree of inter- and intratumoral heterogeneity. Intertumoral heterogeneity of GBM has been extensively investigated on genomic, methylomic, transcriptomic, proteomic and metabolomics levels, however only a few studies describe intratumoral heterogeneity because of the lack of methods allowing to analyze GBM samples with high spatial resolution. Here, we applied TOF-SIMS (Time-of-flight secondary ion mass spectrometry) for the analysis of single cells and clinical samples such as paraffin and frozen tumor sections obtained from 57 patients. We developed a technique that allows us to simultaneously detect the distribution of proteins and metabolites in glioma tissue with 800 nm spatial resolution. Our results demonstrate that according to TOF-SIMS data glioma samples can be subdivided into clinically relevant groups and distinguished from the normal brain tissue. In addition, TOF-SIMS was able to elucidate differences between morphologically distinct regions of GBM within the same tumor. By staining GBM sections with gold-conjugated antibodies against Caveolin-1 we could visualize border between zones of necrotic and cellular tumor and subdivide glioma samples into groups characterized by different survival of the patients. Finally, we demonstrated that GBM contains cells that are characterized by high levels of Caveolin-1 protein and cholesterol. This population may partly represent a glioma stem cells. Collectively, our results show that the technique described here allows to analyze glioma tissues with a spatial resolution beyond reach of most of other omics approaches and the obtained data may be used to predict clinical behavior of the tumor.
Glioblastoma (GBM)1 is the most common and aggressive primary brain tumor in adults (1). Despite great efforts to develop new therapeutic strategies, GBM is one of the few types of cancer for which no significant improvement in treatment has been achieved over the past 30 years. The most used glioblastoma chemotherapy drug, Temozolomide, increases the median patient survival from 12 to 14 months (2). One of the reasons underlying the aggressiveness of GBM is the high degree of heterogeneity. Therefore, any tumor will contain a population of cancer cells that are resistant to one or another treatment strategy (3). Over the past 15 years, many studies investigating the inter- and intratumoral heterogeneity of glioblastoma have been published (4–6).
With the help of genome sequencing, it was shown that the presence of a mutation in IDH gene is one of the most important prognostic factors for patients with glioblastoma (7). Transcriptome sequencing allowed to identify mesenchymal and proneuronal subtypes of tumors that are characterized by different sensitivity to therapy (6, 8), these data were also confirmed by the analysis of metabolome (9). Studies of DNA methylation made it possible to understand the origin and evolution of glioblastomas (10), as well as to identify more and less aggressive types of GBM (11). Finally, study of the RNA splicing conducted by our group shed light on the mechanisms of phenotypic plasticity of GBM during tumor growth and radiotherapy (12).
The studies listed above describe intertumoral heterogeneity of glioblastoma. However, recent evidence suggests that intratumoral heterogeneity may play an even more important role (5, 13). By using immunohistological methods, a population of glioblastoma stem cells (GSC) was identified. These cells are in the perivascular region of the tumor and are thought to be responsible for tumor initiation and recurrence (14, 15). Laser microdissection and subsequent RNA sequencing revealed transcriptomic features of GBM cells located in morphological distinct regions of the tumor (16), and as a result showed the corresponding difference in sensitivity to chemotherapy (17) Unfortunately, data describing proteomic and metabolomic differences within the same GBM tumor with good spatial resolution (less than 1 cm) has not yet been published.
Mass spectrometry imaging (MSI) is a set of methods allowing to locate and identify the chemical composition of cells and tissues within specific regions (or microenvironments). The combination of molecular specificity with imaging capabilities has provided a new perspective for biological sample analysis, proteomics, metabolomics and lipidomics (18). Among all MSI methods TOF-SIMS (Time-of-flight secondary ion mass spectrometry) showed the highest lateral resolution (19) which can reach as low as 100 nm in size and 10 nm in depth (20, 21). Frequently TOF-SIMS utilizes two beams option: the analysis beam and the sputter beam. Analysis beam ions collide with molecules of the sample inducing their ionization. Appeared secondary ions are subsequently analyzed with TOF detector. On the other hand, ions of sputter beam (usually Cs+ or Arn+) are used for surface etching, to remove uppermost layers of the material and reach deeper layers of the sample for further analysis (22). So far TOF-SIMS has been successfully used by multiple groups for the detection of overall cancer-related biochemical changes in breast, melanoma, prostate, colon and bladder cancer samples (23, reviewed in 24).
In this study we for the first time applied TOF-SIMS for the analysis of glioma samples and demonstrated that this method allows to produce clinically relevant data that might be used to predict tumor behavior and to elucidate intra- and intertumoral heterogeneity of GBM.
EXPERIMENTAL PROCEDURES
Patients and Specimens
Glioma tissue samples were obtained from N.N. Burdenko National Medical Research Center of Neurosurgery (Moscow, Russia) and processed to the research laboratories after de-identification of the samples. Characteristics of the biological material are given in supplemental Table S1. All diagnoses were confirmed by morphological studies. The study was approved by the ethics committees of the corresponding hospital. The use of the de-identified tissues for the experiments involving isolation of glioma spheres, TOF-SIMS, and/or IHC on human tumors was exempt from requiring consent.
Neurosphere Culture and Sorting by Flow Cytometry
Primary culture of GBM neurospheres were established as described previously (12). Briefly, GBM specimens were collected during surgery under preoperative MRI-guided navigation and mechanically dissociated into pieces with 1–3 mm diameter. The samples were then treated with trypsin for 20 min at +37 °C to obtain single cells. Cell suspensions were run through Lympholyte-H separation (Cedar Lane Labs, Burlington, NC) to remove Red Blood Cells and debris according to manufacturer's specifications. Established cell lines were cultivated for no longer than 10 passages in DMEM/F12 medium containing 2% B27 supplement (Thermo Fisher, Waltham, MA), 1% Penicillin-Streptomycin solution (Thermo Fisher), 2.5 μg/ml heparin (Sigma, St. Louis, MO), 20 ng/ml basic fibroblast growth factor (bFGF; Sigma), and 20 ng/ml epidermal growth factor (EGF; Sigma). bFGF and EGF were added twice a week and the cultural medium was changed every 7 days. For CD133 staining glioma spheres were dissociated into single cells and stained with anti-CD133-APC antibody (Miltenyi Biotec, Bergisch Gladbach, Germany) according to manufacturer's protocol. Stained cells were subsequently sorted with BD FACSAria III cell sorter (BD Biosciences, San Jose, CA) and 10% of cells with the highest and lowest staining intensities were collected and considered as CD133high and CD133low populations respectively. Obtained cells were allowed to attach to laminin coated silicon wafers for 12 h.
In vivo Intracranial Xenograft Tumor Models
U87MG cells were cultivated in DMEM/F12 medium containing 10% of fetal bovine serum (Thermo Fisher) and 1% Penicillin-Streptomycin solution. For intracranial transplantation U87MG cells were dissociated into single cells and injected (5 × 105 cells in 3 μl of PBS) into the brains of 6-week-old female nude mice (Foxn1nu) as previously described (25). When neuropathological symptoms developed, mice were sacrificed and perfused with ice-cold PBS and 4% paraformaldehyde (PFA). Mice brains were dissected, fixed in 4% PFA for 24 h and then transferred to 10% formalin. All animal experiments were carried out under an Institutional Animal Care and Use Committee (IACUC) approved protocol according to NIH guidelines.
Immunocytofluorescence Staining
Immunocytofluorescence staining (IF) method was described previously (25). Briefly, U87MG cells were cultivated on laminin coated Lab-Tek II Chambered Coverglass (Nunc) overnight, fixed with 4% PFA, permeabilized with 0.2% Triton-X100, blocked with serum-free protein block solution (Dako, Santa Clara, CA) and incubated with anti-Caveolin-1 primary antibodies (dilution 1:250; ab2910, Abcam, Cambridge, MA) for 1 h at room temperature. Next cells were incubated with Alexa Flour-conjugated secondary antibody for 1 h at room temperature and mounted in Vectashield mounting medium containing DAPI (Vector Laboratories, Burlingame, CA). Images were captured with Nikon DIAPHOT 300 microscope (Nikon, Tokyo, Japan).
Immunohistochemistry
Immunohistochemistry (IHC) was performed as previously described (25). Briefly, GBM frozen sections, were permeabilized with 0.2% Triton-X100, slides were incubated in 0.3% hydrogen peroxide solution in methanol for 15 min at room temperature to inhibit internal peroxidase activity. Next samples were blocked with serum-free protein block solution (Dako) and incubated with anti-Caveolin-1 primary antibodies (dilution 1:250; ab2910, Abcam) overnight at 4 °C. Next day slides were stained with EnVision+ System-HRP labeled Polymer (Dako) and visualized with DAB peroxidase substrate kit (Vector Laboratories).
Conjugation of Antibodies with Gold Nanoparticles
Anti-Caveolin-1 antibodies (ab2910, Abcam) were purified with Pierce Protein A/G Magnetic Beads (Thermo Fisher) according to manufacturer's protocol. Next antibodies were labeled with GOLD Conjugation Kit (20 nm 20OD) (Abcam) as described by the manufacturer.
Preparation of Samples for TOF-SIMS Analysis
Three types of samples have been used for TOF-SIMS. Each of them were processed according to the different protocols.
Cells on silicon wafers were used for both spectroscopy and imaging types of analysis. First, cells were plated on silicon wafers at low density and 12 h later were fixed with 4% PFA in PBS for 15 min at room temperature, then the specimen was washed twice with PBS and three times with water (mQ). After that samples were dried under a weak flow of argon and subjected to TOF-SIMS. For each sample at least 30 single cells were analyzed. For each cell average signal intensity was calculated and normalized to the average total ion count from the background (area of silicon wafers without cells). CD133high and CD133low cells from the same patient were analyzed in parallel during the same run of the TOF-SIMS instrument.
Frozen sections of tissues were used for imaging analysis only. Tissues were fixed in 4% PFA in PBS overnight at +4 C°. Next, samples were incubated in 15% sucrose in PBS overnight at +4 C° and then in 30% sucrose in PBS overnight at +4 C°. After the incubation samples were embedded in OCT medium and frozen in dry ice methanol. Frozen tissues were sectioned, placed on a glass slides and stored at −80 C°. At the day of experiment samples were washed twice with PBS and three times with water (mQ). After that samples were dried under a weak flow of argon and subjected to TOF-SIMS. In some experiments frozen sections were stained with nanogold conjugated anti-Caveolin-1 antibodies. For that reason, at the day of experiment samples were washed twice with PBS, permeabilized with 0.2% Triton-X100 in PBS, blocked with 1% BSA in PBS and incubated with nanogold conjugated anti-Caveolin-1 antibodies overnight at 4 °C. Next day, specimens were subsequently washed twice with 0.1% Triton-X100 in PBS, three times with PBS and three times with water (mQ). After that samples were dried under a weak flow of argon and subjected to TOF-SIMS.
Paraffin embedded tissues were used for spectroscopy analysis only. Tissues were embedded into paraffin and sectioned according to the standard protocol (26). At the day of experiment samples were subsequently washed three times with xylene, three times with 96% ethanol, once with 70% ethanol, twice with PBS and three times with water (mQ). After that samples were dried under a weak flow of argon and subjected to TOF-SIMS. In some experiments, paraffin sections were stained with nanogold conjugated anti-Caveolin-1 antibodies. For that reason, at the day of experiment samples were subsequently washed three times with xylene, three times with 96% ethanol, once with 70% ethanol and twice with PBS. After microwave antigen retrieval in DakoCytomation target retrieval solution pH 6 (Dako), slides were blocked with 1% BSA in PBS and incubated with nanogold conjugated anti-Caveolin-1 antibodies overnight at 4 °C. Next day, specimens were subsequently washed twice with 0.1% Triton-X100 in PBS, three times with PBS and three times with water (mQ). After that samples were dried under a weak flow of argon and subjected to TOF-SIMS.
TOF-SIMS Analysis
Measurements were performed on a TOF-SIMS 5 instrument (ION-TOF Gmbh, Münster, Germany) equipped with a bismuth liquid metal ion gun. Two types of bismuth ions were used: cluster Bi3+ (were used if it is not specified in the text) and monoatomic Bi1+, both types have the energy of 30 keV. The primary ion source was operated at an angle of 45°. Secondary ions were post-accelerated to 10 keV to improve the detection efficiency. Spectra and ion images were recorded for both positive and negative secondary ions. A low-energy electron flood gun was activated to avoid charging effect. Measurements were carried out first with 5 μm lateral resolution (spectroscopy mode) and then in imaging mode with 800 nm resolution. Pulse bunching was applied both in spectroscopy and imaging modes. If analyzed area exceeds 400 × 400 μm stage movement was activated. A 500 eV cesium sputter gun was used for experiments with ion etching. Bombardment duration was 1 min with primary ion dose density ∼ 1015 ions/cm2. ToF-SIMS data were analyzed using SurfaceLab software (ION-TOF GmbH). Peaks on a TOF-SIMS spectra were identified using previously published assignments (27–30) or by ChemCalc software as described previously (31). Principal Component Analysis (PCA) was performed on R using FactoMineR package (32). The confidence ellipse defines the region that contains 95% of all samples that can be drawn from the underlying multivariate normal distribution.
Experimental Design and Statistical Rationale
The primary aims of this study were to test the ability of TOF-SIMS to acquire clinically relevant data from glioma specimens and to investigate heterogeneity of GBM with high spatial resolution. Collection of samples from different patients that represents all major types of glioma in multiple replicates was used: grade II (n = 8 patients), grade III (n = 16 patients), Grade IV (n = 21 patients); tumors from females (n = 21 patients) and males (n = 24 patients); primary (n = 27 patients) and recurrent (n = 18 patients) tumors; age of the patients were from 10 to 85 years with an average age of 47 years. In addition, 12 normal brain samples served as a control. After parafinization sections of the samples listed above were placed on the same glass slide and therefore all treatments were performed simultaneously and with the same reagents for all samples to avoid any variation in preparation and processing. To reduce the potential signal alterations which may occur because of the changes in primary ion current all TOF-SIMS data were normalized to total ion count. To reduce the risk of artifacts in experiments with cells in vitro the study was performed with neurospheres which had been cultivated for only a few passages in serum-free media. This cultivation method allows cells to maintain the phenotype of the original tumor (33). All microscopic and TOF-SIMS images represent data obtained from at least three different samples. All quantitative data are presented as mean ± S.D. We assumed normal distribution based on the appearance of the data and analyzed with Student's tailed t test. The statistical significance of Kaplan-Meier survival plot was determined by log-rank analysis. Statistical analysis was performed by Prism 6 (Graphpad Software). p < 0.05 was considered as statistically significant. No samples, mice or data points were excluded from the reported analyses.
RESULTS
Validation of TOF-SIMS Analysis for Glioblastoma Samples
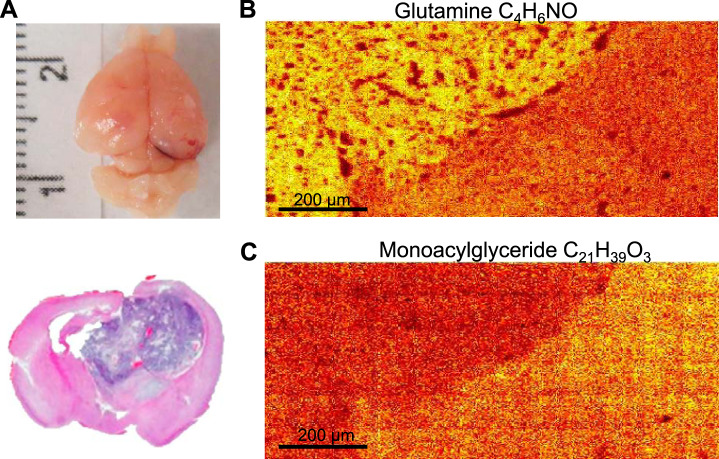
Silicon wafers and conductive indium tin oxide glass slides are mainly used as a substrate for cells and tissue sections for TOF-SIMS investigations (34). These substrates can be used in small scale laboratory studies but not in clinic practice. To apply TOF-SIMS for the analysis of glioma samples obtained from patients, we first tested if this method allows to acquire data from the samples most often created in clinic - frozen and paraffin sections of tissues located on glass slides. To verify the capabilities of TOF-SIMS, we used mice intracranial glioblastoma xenografts. These samples have easily visible boundary between the tumor and the normal brain. First, U87MG glioblastoma cells were injected into the brain of immunocompromised mice, and after tumor formation, frozen brain sections were obtained according to the standard protocol (Fig. 1A). Next, these sections were washed to remove remaining freezing media and analyzed by TOF-SIMS. As a glass slide is an insulator, the sample surface becomes positively charged during TOF-SIMS analysis because of primary ion bombardment. Such charging distorts trajectory of primary and secondary ions and decreases ion signal. To compensate this effect a low energy electron flood gun was used to reduce charging of the surface. The results of this experiment demonstrated that TOF-SIMS can clearly visualize the boundary between GBM and the normal brain. Our data showed an increased amount of glutamine (m/z = 84,04 (27, 35)) in the tumor (Fig. 1B). This result correlates well with previous reports indicating that glutamine accumulation is one of the characteristic features of GBM (36). In addition, we have also observed a reduced amount of monoacylglyceride C18:1 (m/z = 339,29 (28, 37)) in tumor tissue, which also made it possible to clearly distinguish GBM from the normal brain (Fig. 1C).
Fig. 1.
TOF-SIMS analysis of GBM xenografts in mouse brain. A, Mouse brain one month after intracranial transplantation of U87MG cells (top). H&E staining of mouse brain section containing tumor (bottom). B, Representative pseudocoloured image demonstrating the distribution of glutamine fragment (m/z = 84,04) detected by TOF-SIMS near the border of normal mouse brain and a tumor. C, The same as in B for the monoacylglycerol ion (m/z = 339.29).
Next, we compared TOF-SIMS spectra obtained from frozen and paraffin sections of human GBM tissues. As expected, the process of paraffinization/deparaffinization significantly decreased the amount and the intensity of the recorded peaks (supplemental Fig. S1A), however, most of the peaks with m/z below 100 and some of the higher molecular mass peaks were still present in the spectra. Therefor our data indicate that TOF-SIMS can be used for the analysis of conventionally prepared clinical glioma samples. It is important to note, that despite the extensive washing procedure of the samples there was a significant amount of material left from paraffin embedding medium as can be seen from the representative TOF-SIMS spectra obtained from the same glass slide right next to the tissue slice (supplemental Fig. S1B). To overcome this issue for further analysis we used only the peaks from the sample that did not match peaks in the embedding medium.
Investigation of Intertumoral Heterogeneity of Gliomas Using TOF-SIMS
Having confirmed that TOF-SIMS is capable of detecting the previously described metabolic features of GBM in clinical samples, we proceeded to the analysis of a large panel of samples from patients with various types of gliomas, as well as a non-tumor controls. Obtained mass spectra revealed more than 200 peaks corresponding to various fragments of organic compounds with m/z of up to 500. Fifty peaks demonstrating the largest differences between samples were used for further analysis. Clusterization of data allowed us to divide the samples into three groups (Fig. 2A, supplemental Table S2). By comparing these groups with the etiology of the studied tumors, we found that the groups created on the basis of TOF-SIMS data reflects the clinical characteristics of the samples. All normal brain samples were clustered together and represented the majority of group I. Group II included mainly primary tumors, and group III consists of predominantly recurrent tumors after therapy (Fig. 2B). Interestingly, we did not find any noticeable correlation of TOF-SIMS groups with the grade of glioma tumors as well as with the gender of the patients (supplemental Fig. S2). Next we tested the contribution of each peak to the separation of glioma samples into either TOF-SIMS signature groups (Group I/Group II/Group III) or groups created based on the clinical characteristics of the tumors (Normal brain/Primary glioma/Recurrent glioma). Supplemental Fig. S3 demonstrates that most of the peaks were significantly different between all TOF-SIMS groups and between normal brain and the rest of the glioma samples. Interestingly, differences between primary and recurrent gliomas were much less pronounced and observed only for 3 peaks.
Fig. 2.
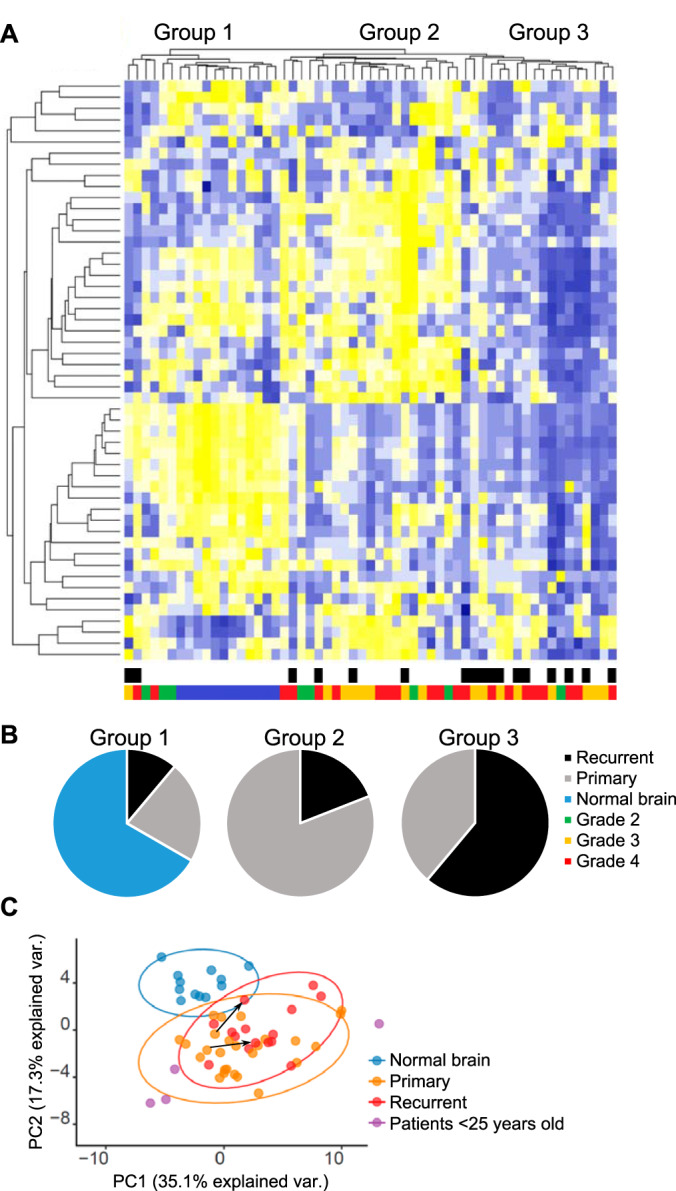
Levels of metabolites' fragments detected by TOF-SIMS in glioma (n = 45) and normal brain (n = 12) sections. A, Heatmap demonstrating relative amounts of detected molecules in each sample and sample clusterization into the groups. Color scales below the heatmap represent clinical characteristics of the samples (black - recurrent tumor, gray - primary tumor, blue - normal brain, green - glioma II grade, yellow - glioma III grade, red - GBM). B, Amount of samples with different clinical features in each group (black - recurrent tumor, gray - primary tumor, blue - normal brain). C, The principal component analysis of TOF-SIMS data (blue - normal brain, yellow - primary tumors, red - recurrent tumors, violet - tumors obtained from patients less than 25 years old). Ovals represent confident interval of 95%. Arrows show changes in tumors obtained from the same patients before and after therapy.
Finally, we applied principal component analysis (PCA) to the data. The graph on Fig. 2C demonstrates the significant difference between normal brain samples and gliomas. In addition, it is interesting to note that the data of mass spectra obtained from tumors of young patients (less than 25 years) were clustered separately from the other tumors. This result is in good agreement with the data described previously (38) and may indicate a different type of the genetic alterations underlying carcinogenesis in young patients and, therefore, a different metabolic profile of these tumors. The differences between the primary and secondary tumors from the PCA analysis were less pronounced as opposed to sample clustering, however, a comparison of pairs of samples obtained from the same patient before and after therapy shows a tendency that treatment causes similar changes in the glioblastoma metabolome. It is important to mention that for some peaks on the TOF-SIMS spectra the signal intensity was evenly distributed over the sample, whereas for others the separate groups of cells were clearly visible, showing higher or lower value for this parameter within the sample. This result indicates a significant intratumoral heterogeneity of the molecules which correspond to these peaks (supplemental Fig. S4).
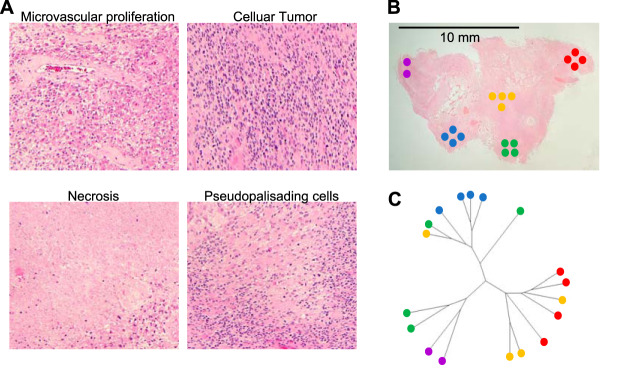
Investigation of Intratumoral Heterogeneity of GBM
To study the intratumoral heterogeneity of glioblastoma in more detail, we obtained a section of a patient's tumor with a relatively large spatial size (more than 1 cm2). It contained all morphologically distinct zones of GBM described in the literature (39, 16), namely, the necrosis zone, the pseudopalisade region, the cellular tumor and microvascular proliferation zone (Fig. 3A). In each of these regions, we selected 4 sites for TOF-SIMS analysis (Fig. 3B). Similarly, to the previous experiment, we used 50 differential peaks for cauterization of the data (supplemental Table S3). The diagram in Fig. 3C demonstrates that in most cases samples from the same morphological region have similar TOF-SIMS signatures. Therefore, spatially distant but morphologically similar areas of microvascular proliferation are like each other according to TOF-SIMS data. On the other hand, samples of the pseudopalisade and necrotic regions are also clustered next to each other. Interestingly, the areas of the cellular tumor were more scattered, possibly because these regions are relatively small in size and located closely to the areas of necrosis and pseudopalisades. Thus, TOF-SIMS analysis makes it possible to distinguish morphologically different parts of glioblastomas that otherwise can only be distinguished by H&E staining.
Fig. 3.
Analysis of intratumoral heterogeneity of GBM using TOF-SIMS. A, Microscopic images of morphologically distinct regions of GBM located within one section of the tumor. The section was stained with H&E. B, Low magnification image of GBM tumor section. Circles represent regions selected for TOF-SIMS analysis, colors indicate morphologically distinct zones of GBM (microvascular proliferation - red and yellow, cellular tumor - green, pseudopalisading area - blue, necrotic zone - violet). C, Diagram demonstrating phylogenetic relationships of the tumor regions calculated according to the TOF-SIMS data.
Simultaneous Detection of Caveolin-1 Protein and Cholesterol in Glioma Tissues by TOF-SIMS
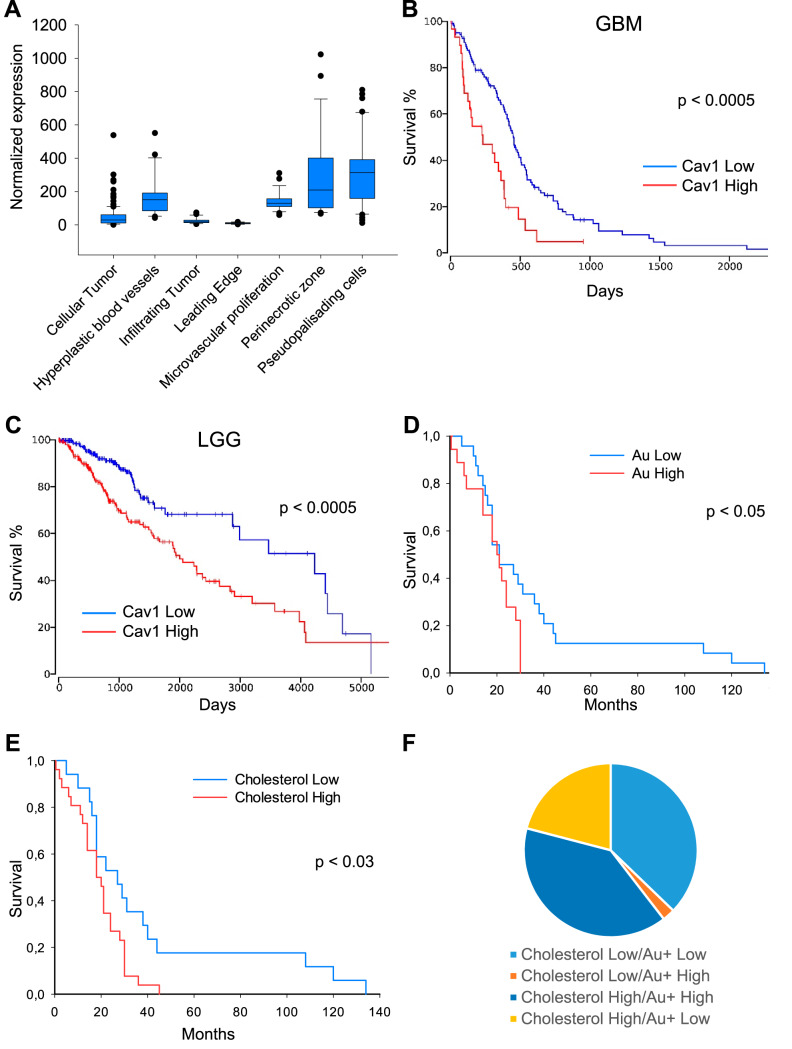
A major drawback of TOF-SIMS is the inability to detect specific proteins in the samples. To overcome this deficiency, we attempted to visualize the distribution of the protein of interest by staining sections of glioblastoma with antibodies conjugated to gold nanoparticles. We hypothesized that the amount of secondary Au+ ions detected by TOF-SIMS should reflect both the distribution and the amount of protein to which the corresponding antibodies were raised. To select the most suitable protein for the study, we conducted a bioinformatic analysis of the IVY and TCGA databases. According to the results of the analysis, we selected the protein Caveolin-1. This choice was made on the basis of several factors: first, the amount of this protein correlates well with the amount of cholesterol (40), which is reliably detected by TOF-SIMS (29). Second, Caveolin-1 demonstrated an uneven distribution within glioblastoma (Fig. 4A) and therefore, it can potentially serve as a marker of intratumoral heterogeneity. Third, the gene expression of Caveolin-1 is statistically significant correlated with survival of patients both with low grade glioma and glioblastoma and therefore its' protein level may also reflect the survival of patients (Fig. 4B and 4C).
Fig. 4.
Association between Caveolin-1 gene expression and characteristics of glioma tumors. A, Analysis of RNA sequencing data from IVY database demonstrating expression of Cav1 gene by GBM cells located in morphologically different regions of the tumors. B, Kaplan-Meier curve showing the overall survival of glioblastoma patients (n = 152) subdivided based on Caveolin-1 expression levels. Data were obtained from TCGA database. C, Same as B for low grade glioma patients (n = 510). D, Kaplan-Meier curve showing the overall survival of glioma patients (n = 45) subdivided into two groups based on the intensity of immunogold staining detected by TOF-SIMS. E, Same as D for glioma samples subdivided by cholesterol content detected by TOF-SIMS. F, Diagram demonstrating the amount of glioma samples with low and high cholesterol and Caveolin-1 content.
First, we conjugated antibodies against Caveolin-1 with gold (Au+) nanoparticles and confirmed that this modification did not affect the specificity of the antibodies (supplemental Fig. S5A). Low Au ionization efficiency and superposition of Au peak with organic components with similar mass complicates the detection of Au+ secondary ions localization in sections stained by conjugated antibodies. Therefore, optimized multi-step procedure was designed to obtain reliable results. First, Bi3+ scanning was employed for metabolites detection in glioblastoma cells. Then the same area was treated by cesium sputter gun to uncover Au+ signal. The sputter gun parameters were adjusted to minimize layer mixing and gold atom relocation. Finally, the same area was scanned with Bi1+ ions. Bi1+ provides lower secondary ion yield compare with Bi3+ especially for organic molecules, which was favorable in this case. Using this protocol, we detected the accumulation of cholesterol and gold in 45 glioma samples stained with anti-Caveolin-1 antibodies conjugated with gold nanoparticles. Ion 147.12 was chosen for cholesterol detection as it provided more than 10-fold higher signal intensity as compared with standard 369.35 cholesterol ion (supplemental Fig. S5B and S5C) (29). Comparison of the Au+ intensities with the survival of the patients demonstrated that the high level of gold staining detected using TOF-SIMS correlates with a poor prognosis for the patients (Fig. 4D). These data are in good agreement with the results of our bioinformatics analysis (Fig. 4B). Importantly, we detected a similar trend for the cholesterol content, which also correlated with an unfavorable prognosis for patients (Fig. 4E). Of note, in most samples, high cholesterol content was accompanied by a high amount of Caveolin-1, and vice versa, low amount of Caveolin-1 corresponded to low cholesterol levels (Fig. 4F).
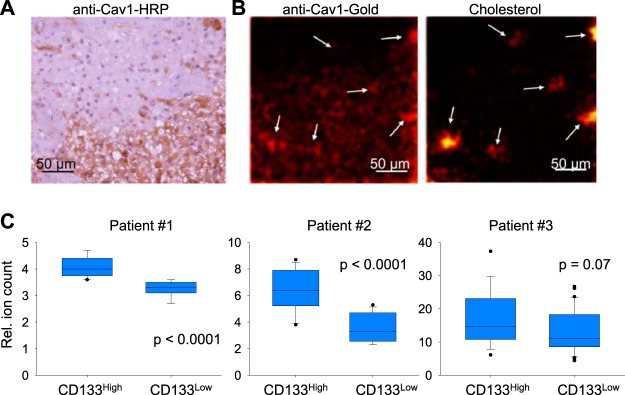
Detection of Caveolin-1 Protein and Cholesterol in GBM Cells by TOF-SIMS
Because the total amount of Caveolin-1 in glioma sections correlates well with the amount of cholesterol, we next investigated the distribution of cholesterol and Caveolin-1 in GBM sections with high spatial resolution. We first showed that the distribution of Caveolin-1 visualized by standard staining with primary antibodies and secondary antibodies conjugated to horseradish peroxidase looks similar to the distribution of Caveolin-1 detected by the accumulation of gold nanoparticles detected by TOF-SIMS (Fig. 5A and Fig. 5B). Based on the results of H&E staining, we selected the border of the cellular tumor and perinecrotic zone for the subsequent TOF-SIMS analysis. Fig. 5B shows that cells in the necrotic zone contain significantly more Caveolin-1 than cells in the cellular tumor zone. This result is in good agreement with our bioinformatics analysis (Fig. 4A). In addition, we were able to observe cells or small groups of cells that simultaneously contain an increased amount of cholesterol and Caveolin-1. We hypothesized that these cholesterol-rich cells may represent glioblastoma stem cells (GSC) because one of the well characterized GSC markers is the cholesterol-binding protein CD133 (41). To test this hypothesis, we isolated glioblastoma cells from the tumors of three different patients, and after short-term in vitro cultivation populations of CD133high and CD133low cells were obtained by flow cytometry sorting. Sorted cells were allowed to attach to silicon wafers coated with laminin, and 12 h later samples were analyzed by TOF-SIMS. The results of this experiment showed that for all three patients CD133high cells contained significantly more cholesterol than CD133low population (Fig. 5C). These data are in good agreement with our hypothesis that the cholesterolhigh/Caveolin-1high population of GBM cells found using TOF-SIMS may represent GSC. This hypothesis is also indirectly supported by the fact that cancer stem cells are characterized by an increased level of cholesterol biosynthesis (42).
Fig. 5.
Distribution of Caveolin-1 and cholesterol in GBM cells and tissues. A, Section of GBM tumor stained with primary antibodies against Caveolin-1 and secondary antibodies conjugated with horseradish peroxidase. B, Representative pseudocolored images demonstrating intensity of signal corresponding to cholesterol fragment (m/z = 147.12) and Au+ which were detected by TOF-SIMS in GBM section stained with antibodies against Caveolin-1 conjugated with gold nanoparticles. C, Signal of cholesterol fragment (m/z = 147.12) detected by TOF-SIMS in GBM neurosphere primary cultures obtained from three different patients and divided into CD133high and CD133low populations by staining with anti-CD133 antibodies conjugated with APC and subsequent FACS sorting.
DISCUSSION
In this study, we for the first time applied TOF-SIMS for the analysis of glioblastoma cells and tissues. We showed that this method can simultaneously detect fragments from several hundreds of molecules with a spatial resolution like that in a fluorescence microscopy. A significant drawback of TOF-SIMS is excessive molecules fragmentation during the ionization process and inability to detect ions with a molecular mass of more than 2000 Da. This complicates or even makes impossible the precise identification of the original molecule that gave rise to the corresponding secondary ion. For example, it was previously shown that an ion with a mass of 127.04 can arise during ionization of at least five different metabolites and may have a different chemical structure (43). To overcome this issue, we used two complementary approaches. First, instead of focusing only on those peaks on mass spectra that can be uniquely identified, we analyzed a large set of peaks for all gliomas and used these peaks for samples clusterisation into the groups according to the TOF-SIMS signature. Second, simultaneously with the detection of low molecular weight compounds, we detected the protein which is functionally associated with cell metabolism. To the best of our knowledge, this is the first attempt to simultaneously determine the spatial distribution and the relative amount of protein and lipid in the same sample using mass spectrometry imaging. Such simultaneous detection allows to acquire considerably more information about the functional differences of the tumor regions, as well as to additionally verify the obtained metabolic results by comparing them with the distribution of the protein related to the metabolite. In this study, we used only antibodies conjugated to gold particles, however, by the similarity with the recently established CyTOF method (44), it can be assumed that the simultaneous use of antibodies conjugated with various metals (such as cadmium, neodymium, terbium and etc.) will enable TOF-SIMS to detect tens of proteins in one sample in addition to a standard set of metabolites.
We investigated the localization and relative amount of Caveolin-1 protein in tissue sections of human gliomas. Caveolin-1 is a 22 kDa integral, cholesterol binding protein, predominantly localized on the inner surface of the plasma membrane inside lipid rafts and caveoles (45). This protein plays a crucial role in the organization and functioning of receptor complexes located on the membrane. For example, Caveolin-1 interacts with important tumor-associated proteins such as PDGFRα, EGFR, LYN, PKA, Hedgehog receptor, Estrogen and Androgen receptors (46). Interestingly, Caveolin-1 can both activate and inhibit the activity of the receptors (47, 48). Accordingly, in some types of cancer Caveolin-1 was characterized as an oncosuppressor (breast cancer (49)), and in others as an oncogene (colorectal and prostate cancer (50, 51)). Controversial data were also obtained for GBM, where in some experimental models Caveolin-1 promoted tumor growth (52, 53), whereas in others, it slowed down proliferation and increased sensitivity to the therapy (54, 55). In this study, using GBM patient clinical samples, we demonstrated Caveolin-1 expression varies across the tumor. We can propose that in the necrotic region, which is characterized by a lack of nutrition and hypoxia, Caveolin-1 is normally up-regulated and it plays an oncogenic role. On the other hand, in areas of actively proliferating tumor cells which are rich with blood supply and oxygen, Caveolin-1 may act as an oncosuppressor and therefore its level in these regions is markedly reduced. This hypothesis may explain the controversial data published in recent years about the role of Caveolin-1 in GBM.
In addition to these data, we showed that GBM contains a small population of cells characterized by high levels of cholesterol and Caveolin-1. These cells theoretically may represent a glioblastoma stem cell population. It is important to note that no universal protein marker for GSC has been described so far. It was shown that for GBM with proneuronal phenotype, the main GSC marker is CD133 protein (6). On the other hand, for GBM with mesenchymal phenotype, as well as for GBM exposed to therapy, the major GSC marker is CD109 protein (56). Although the functions of CD133 and CD109 are unclear, it is known that both proteins are associated with cholesterol and lipid rafts (41, 57). In addition, recent data revealed the activation of cholesterol biosynthesis pathways in cancer stem cells (42). Thus, it is possible that cholesterol may serve as a lipid marker for GSC. Indirectly, this assumption is supported by our findings that increased cholesterol level correlates with a poor prognosis for patient survival. However, most likely, a single protein or lipid marker is not enough to determine GSC, and therefore further studies are needed to find a universal panel of markers of various nature for identification and visualization of GSC. From this point of view, application of TOF-SIMS can be particularly beneficial because of its high spatial resolution and the ability to simultaneously detect a wide variety of molecules.
Using TOF-SIMS to analyze many glioma samples, we showed that this method allows to divide the samples into the clinically relevant groups. We were able to clearly separate normal brain samples from glioma tumors. This is consistent with previous reports that have demonstrated differences in TOF-SIMS signatures between breast, bladder and prostate cancers and corresponding normal cells (58–61). Our results indicate that subgrouping of gliomas according to TOF-SIMS signature partly but not completely matches separation of samples into primary or recurrent tumors. This observation is in good agreement with the previous publications, which indicate that therapy frequently causes phenotypic transformations of gliomas and significantly alters cell metabolism toward aerobic glycolysis (mesenchymal phenotype), however, a fraction of primary tumors also have mesenchymal properties (6, 12, 56). Therefore, it is possible that TOF-SIMS allows to detect the metabolic changes associated with mesenchymal transformation, but additional experiments are needed to confirm this hypothesis.
Collectively, our study demonstrated the applicability of TOF-SIMS for the analysis of various types of conventionally prepared glioma samples, such as frozen and paraffin tumor sections on glass substrate. We showed that the differences detected by TOF-SIMS correlate with the clinical and histological features of the tumor, and in some cases, it allows to subdivide patients into groups characterized by more favorable and less favorable survival. Because TOF-SIMS does not require specific sample preparation procedures, and the analysis of one sample takes less than 10 min, it is possible that after further development this technology can be used for the rapid identification of GBM cells. This procedure will allow more accurate removal of a tumor and may help to determine the optimal postoperative therapy, ultimately improving the treatment efficiency of patients with GBM.
DATA AVAILABILITY
The processed data can be found in supplemental Tables S2 and S3. Unprocessed TOF-SIMS spectra have been deposited to Mendeley Data (Project: Investigation of inter- and intra-tumoral heterogeneity of glioblastoma using TOF-SIMS; http://dx.doi.org/10.17632/ssgyybm6d3.2).
Supplementary Material
Acknowledgments
We thank all our respective lab mates for their helpful discussion.
Footnotes
* This work was supported by The Russian Foundation for Basic Research grants 16-04-00660 (S.K.G.), 17-29-06056 (M.S.P.), 18-29-01027 (M.I.S.), 20-04-00804 (M.S.P.), 20-34-70147 (M.S.P.), Russian Science Foundation grant 19-44-02027 (M.S.P.), Ministry of Science and Higher Education of the Russian Federation grant 075-15-2019-1669 (V.O.S.), and the Program of the Presidium of the Russian Academy of Sciences “Molecular and Cellular Biology” project no. 0101-2018-0012 (N.V.A.). Investigations were performed using the facilities of Semenov FRCCP RAS CCE no. 506694. The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- GBM
- glioblastoma multiforme
- GSC
- glioma stem cells
- H&E
- Hematoxylin & Eosin
- MSI
- mass spectrometry imaging
- m/z
- mass-to-charge ratio
- PCA
- principal component analysis
- PBS
- phosphate buffered saline
- TOF-SIMS
- time-of-flight secondary ion mass spectrometry.
REFERENCES
- 1. Walid M. S. (2008) Prognostic factors for long-term survival after glioblastoma. Perm. J. 12, 45–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R., Mason W. P., van den Bent M. J., Weller M., Fisher B., Taphoorn M. J., Belanger K., Brandes A. A., Marosi C., Bogdahn U., Curschmann J., Janzer R. C., Ludwin S. K., Gorlia T., Allgeier A., Lacombe D., Cairncross J. G., Eisenhauer E., and Mirimanoff R. O. (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 [DOI] [PubMed] [Google Scholar]
- 3. Aum D. J., Kim D. H., Beaumont T. L., Leuthardt E. C., Dunn G. P., and Kim A. H. (2014) Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg. Focus. 37, E11. [DOI] [PubMed] [Google Scholar]
- 4. Verhaak R. G., Hoadley K. A., Purdom E., Wang V., Qi Y., Wilkerson M. D., Miller C. R., Ding L., Golub T., Mesirov J. P., Alexe G., Lawrence M., O'Kelly M., Tamayo P., Weir B. A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H. S., Hodgson J. G., James C. D., Sarkaria J. N., Brennan C., Kahn A., Spellman P. T., Wilson R. K., Speed T. P., Gray J. W., Meyerson M., Getz G., Perou C. M., and Hayes D. N. (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patel A. P., Tirosh I., Trombetta J. J., Shalek A. K., Gillespie S. M., Wakimoto H., Cahill D. P., Nahed B. V., Curry W. T., Martuza R. L., Louis D. N., Rozenblatt-Rosen O., Suvà M. L., Regev A., and Bernstein B. E. (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mao P., Joshi K., Li J., Kim S. H., Li P., Santana-Santos L., Luthra S., Chandran U. R., Benos P. V., Smith L., Wang M., Hu B., Cheng S. Y., Sobol R. W., and Nakano I. (2013) Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl. Acad. Sci. U.S.A. 110, 8644–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cancer Genome Atlas Research Network Brat D. J., Verhaak R. G., Aldape K. D., Yung W. K., Radenbaugh A. J., and Zhang J. (2015) Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 372, 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhat K. P. L., Balasubramaniyan V., Vaillant B., Ezhilarasan R., Hummelink K., Hollingsworth F., Wani K., Heathcock L., James J. D., Goodman L. D., Conroy S., Long L., Lelic N., Wang S., Gumin J., Raj D., Kodama Y., Raghunathan A., Olar A., Joshi K., Pelloski C. E., Heimberger A., Kim S. H., Cahill D. P., Rao G., Den Dunnen W. F. A., Boddeke H. W., Phillips H. S., Nakano I., Lang F. F., Colman H., Sulman E. P., and Aldape K. (2013) Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 24, 331–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cuperlovic-Culf M., Ferguson D., Culf A., Morin P. Jr, and Touaibia M. (2012) 1H NMR metabolomics analysis of glioblastoma subtypes: correlation between metabolomics and gene expression characteristics. J. Biol. Chem. 287, 20164–20175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ozawa T., Riester M., Cheng Y. K., Huse J. T., Squatrito M., Helmy K., Charles N., Michor F., and Holland E. C. (2014) Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 26, 288–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Noushmehr H., Weisenberger D., Diefes K., Phillips H. S., Pujara K., Berman B. P., Pan F., Pelloski C. E., Sulman E. P., Bhat K. P., Verhaak R. G., Hoadley K. A., Hayes D. N., Perou C. M., Schmidt H. K., Ding L., Wilson R. K., Van Den Berg D., Shen H., Bengtsson H., Neuvial P., Cope L. M., Buckley J., Herman J. G., Baylin S. B., Laird P. W., and Aldape K. (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17, 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pavlyukov M. S., Yu H., Bastola S., Minata M., Shender V. O., Lee Y., Zhang S., Wang J., Komarova S., Wang J., Yamaguchi S., Alsheikh H. A., Shi J., Chen D., Mohyeldin A., Kim S. H., Shin Y. J., Anufrieva K., Evtushenko E. G., Antipova N. V., Arapidi G. P., Govorun V., Pestov N. B., Shakhparonov M. I., Lee L. J., Nam D. H., and Nakano I. (2018) Apoptotic Cell-Derived Extracellular Vesicles Promote Malignancy of Glioblastoma Via Intercellular Transfer of Splicing Factors. Cancer Cell 34, 119–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qazi M. A., Vora P., Venugopal C., Sidhu S. S., Moffat J., Swanton C., and Singh S. K. (2017) Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann. Oncol. 28, 1448–1456 [DOI] [PubMed] [Google Scholar]
- 14. Bao S., Wu Q., McLendon R. E., Hao Y., Shi Q., Hjelmeland A. B., Dewhirst M. W., Bigner D. D., and Rich J. N. (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760 [DOI] [PubMed] [Google Scholar]
- 15. Bao S., Wu Q., Sathornsumetee S., Hao Y., Li Z., Hjelmeland A. B., Shi Q., McLendon R. E., Bigner D. D., and Rich J. N. (2006) Stem Cell-like Glioma Cells Promote Tumor Angiogenesis through Vascular Endothelial Growth Factor. Cancer Res. 66, 7843–7848 [DOI] [PubMed] [Google Scholar]
- 16. Puchalski R. B., Shah N., Miller J., Dalley R., Nomura S. R., Yoon J. G., Smith K. A., and Lankerovich M. (2018) An anatomic transcriptional atlas of human glioblastoma. Science 360, 660–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fidoamore A., Cristiano L., Antonosante A., d'Angelo M., Di Giacomo E., Astarita C., Giordano A., Ippoliti R., Benedetti E., and Cimini A. (2016) Glioblastoma stem cells microenvironment: the paracrine roles of the niche in drug and radioresistance. Stem Cells Int. 10.1155/2016/6809105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bluestein B. M., Morrish F., Graham D. J., Guenthoer J., Hockenbery D., Porter P. L., and Gamble L. J. (2016) An unsupervised MVA method to compare specific regions in human breast tumor tissue samples using ToF-SIMS. Analyst. 141, 1947–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H. W., Hua X., and Long Y. T. (2019) Graphene quantum dots enhanced ToF-SIMS for single-cell imaging. Anal. Bioanal. Chem. 411, 4025–4030 [DOI] [PubMed] [Google Scholar]
- 20. Draude F., Körsgen M., Pelster A., Schwerdtle T., Müthing J., and Arlinghaus H. F. (2015) Characterization of freeze-fractured epithelial plasma membranes on nanometer scale with ToF-SIMS. Anal. Bioanal. Chem. 407, 2203–2211 [DOI] [PubMed] [Google Scholar]
- 21. Jungnickel H., Laux P., and Luch A. (2016) Time-of-flight secondary ion mass spectrometry (ToF-SIMS): a new tool for the analysis of toxicological effects on single cell level. Toxics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robinson M. A., Graham D., and Castner D. G. (2012) ToF-SIMS depth profiling of cells: z-correction, 3D imaging, and sputter rate of individual NIH/3T3 fibroblasts. Anal. Chem. 84, 4880–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bobrowska J., Awsiuk K., Pabijan J., Bobrowski P., Lekki J., Sowa K. M., Rysz J., Budkowski A., and Lekka M. (2019) Biophysical and biochemical characteristics as complementary indicators of melanoma progression. Anal. Chem. 91, 9885–9892 [DOI] [PubMed] [Google Scholar]
- 24. Denbigh J. L., and Lockyer N. P. (2015) ToF-SIMS as a tool for profiling lipids in cancer and other diseases. Materials Sci. Technol. 31, 137–147 [Google Scholar]
- 25. Wang J., Cheng P., Pavlyukov M. S., Yu H., Zhang Z., Kim S. H., Minata M., Mohyeldin A., Xie W., Chen D., Goidts V., Frett B., Hu W., Li H., Shin Y. J., Lee Y., Nam D. H., Kornblum H. I., Wang M., and Nakano I. (2017) Targeting NEK2 attenuates glioblastoma growth and radioresistance by destabilizing histone methyltransferase EZH2. J. Clin. Invest. 127, 3075–3089 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Canene-Adams K. (2013) Preparation of formalin-fixed paraffin-embedded tissue for immunohistochemistry. Methods Enzymol. 533, 225–233 [DOI] [PubMed] [Google Scholar]
- 27. Kaweckia M. (2018) Database of proteinogenic amino acid reference spectra for Bismuth-cluster ToF-SIMS. II. Positive polarity. Surface Sci. Spectra. 25, 015002 [Google Scholar]
- 28. Debois D., Bralet M. P., Le Naour F., Brunelle A., and Laprévote O. (2009) In situ lipidomic analysis of nonalcoholic fatty liver by cluster TOF-SIMS imaging. Anal. Chem. 81, 2823–2831 [DOI] [PubMed] [Google Scholar]
- 29. Piehowski P. D., Carado A. J., Kurczy M. E., Ostrowski S. G., Heien M. L., Winograd N., and Ewing A. G. (2008) MS/MS methodology to improve subcellular mapping of cholesterol using TOF-SIMS. Anal. Chem. 80, 8662–8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang H. J., Ishizaki I., Sanada N., Zaima N., Sugiura Y., Yao I., Ikegami K., and Setou M. (2010) Detection of characteristic distributions of phospholipid head groups and fatty acids on neurite surface by time-of-flight secondary ion mass spectrometry. Med. Mol. Morphol. 43, 158–164 [DOI] [PubMed] [Google Scholar]
- 31. Dimzon I. K., Trier X., Frömel T., Helmus R., Knepper T. P., and de Voogt P. (2016) High Resolution Mass Spectrometry of Polyfluorinated Polyether-Based Formulation. J. Am. Soc. Mass Spectrom. 27, 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lê S., Josse J., and Husson F. (2008) FactoMineR: An R package for multivariate analysis. J. Statistical Software. 25 [Google Scholar]
- 33. Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N. M., Pastorino S., Purow B. W., Christopher N., Zhang W., Park J. K., and Fine H. A. (2006) Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 [DOI] [PubMed] [Google Scholar]
- 34. Gamble L. J., Graham D. J., Bluestein B., Whitehead N. P., Hockenbery D., Morrish F., and Porter P. (2015) ToF-SIMS of tissues: “lessons learned” from mice and women. Biointerphases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tyler B. J., Bruening C., Rangaranjan S., and Arlinghaus H. F. (2011) TOF-SIMS imaging of adsorbed proteins on topographically complex surfaces with Bi3 + primary ions. Biointerphases. 6, 135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Venneti S., Dunphy M. P., Zhang H., Pitter K. L., Zanzonico P., Campos C., Carlin S. D., La Rocca G., Lyashchenko S., Ploessl K., Rohle D., Omuro A. M., Cross J. R., Brennan C. W., Weber W. A., Holland E. C., Mellinghoff I. K., Kung H. F., Lewis J. S., and Thompson C. B. (2015) Glutamine-based PET imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Sci. Transl. Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gulin A. A., Pavlyukov M. S., Gularyan S. K., and Nadtochenko V. A. (2015) Visualization of the spatial distribution of Pt+ ions in cisplatin-treated glioblastoma cells by time-of-flight secondary ion mass spectrometry. Biochem. Moscow Suppl. 9, 202–209 [Google Scholar]
- 38. Ferguson S. D., Xiu J., Weathers S. P., Zhou S., Kesari S., Weiss S. E., Verhaak R. G., Hohl R. J., Barger G. R., Reddy S. K., and Heimberger A. B. (2016) GBM-associated mutations and altered protein expression are more common in young patients. Oncotarget. 7, 69466–69478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hambardzumyan D., and Bergers G. (2015) Glioblastoma: Defining Tumor Niches. Trends Cancer 1, 252–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lange Y., and Steck T. L. (2016) Active membrane cholesterol as a physiological effector. Chem. Phys. Lipids. 199, 74–93 [DOI] [PubMed] [Google Scholar]
- 41. Rappa G., Fargeas C. A., Le T. T., Corbeil D., and Lorico A. (2015) Letter to the editor: An intriguing relationship between lipid droplets, cholesterol-binding protein CD133 and Wnt/β-catenin signaling pathway in carcinogenesis. Stem Cells 33, 1366–1370 [DOI] [PubMed] [Google Scholar]
- 42. Ehmsen S., Pedersen M. H., Wang G., Terp M. G., Arslanagic A., Hood B. L., Conrads T. P., Leth-Larsen R., and Ditzel H. J. (2019) Increased cholesterol biosynthesis is a key characteristic of breast cancer stem cells influencing patient outcome. Cell Rep. 27, 3927–3938 [DOI] [PubMed] [Google Scholar]
- 43. Yun S. J., Park J. W., Choi I. J., Kang B., Kim H. K., Moon D. W., Lee T. G., and Hwang D. (2011) TOFSIMS-P: a web-based platform for analysis of large-scale TOF-SIMS data. Anal. Chem. 83, 9298–92305 [DOI] [PubMed] [Google Scholar]
- 44. Kay A. W., Strauss-Albee D. M., and Blish C. A. (2016) Application of mass cytometry (CyTOF) for functional and phenotypic analysis of natural killer cells. Methods Mol. Biol. 1441, 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Di Vizio D., Adam R. M., Kim J., Kim R., Sotgia F., Williams T., Demichelis F., Solomon K. R., Loda M., Rubin M. A., Lisanti M. P., and Freeman M. R. (2008) Caveolin-1 interacts with a lipid raft-associated population of fatty acid synthase. Cell Cycle 7, 2257–2267 [DOI] [PubMed] [Google Scholar]
- 46. Liu P., Rudick M., and Anderson R. G. (2002) Multiple functions of caveolin-1. J. Biol. Chem. 277, 41295–41298 [DOI] [PubMed] [Google Scholar]
- 47. Fang K., Fu W., Beardsley A. R., Sun X., Lisanti M. P., and Liu J. (2007) Overexpression of caveolin-1 inhibits endothelial cell proliferation by arresting the cell cycle at G0/G1 phase. Cell Cycle 6, 199–204 [DOI] [PubMed] [Google Scholar]
- 48. Syme C. A., Zhang L., and Bisello A. (2006) Caveolin-1 regulates cellular trafficking and function of the glucagon-like Peptide 1 receptor. Mol. Endocrinol. 20, 3400–3411 [DOI] [PubMed] [Google Scholar]
- 49. Williams T. M., Sotgia F., Lee H., Hassan G., Di Vizio D., Bonuccelli G., Capozza F., Mercier I., Rui H., Pestell R. G., and Lisanti M. P. (2006) Stromal and epithelial caveolin-1 both confer a protective effect against mammary hyperplasia and tumorigenesis: Caveolin-1 antagonizes cyclin D1 function in mammary epithelial cells. Am. J. Pathol. 169, 1784–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kitowska A., Wesserling M., Seroczynska B1, Szutowicz A., Ronowska A., Peksa R., and Pawelczyk T. (2015) Differentiation of high-risk stage I and II colon tumors based on evaluation of CAV1 gene expression. J. Surg. Oncol. 112, 408–414 [DOI] [PubMed] [Google Scholar]
- 51. Yang G., Truong L. D., Wheeler T. M., and Thompson T. C. (1999) Caveolin-1 expression in clinically confined human prostate cancer: a novel prognostic marker. Cancer Res. 59, 5719–5723 [PubMed] [Google Scholar]
- 52. Eser Ocak P., Ocak U., Tang J., and Zhang J. H. (2019) The role of caveolin-1 in tumors of the brain - functional and clinical implications. Cell Oncol 42, 423–447 [DOI] [PubMed] [Google Scholar]
- 53. Martin S., Cosset E. C., Terrand J., Maglott A., Takeda K., and Dontenwill M. (2009) Caveolin-1 regulates glioblastoma aggressiveness through the control of alpha(5)beta(1) integrin expression and modulates glioblastoma responsiveness to SJ749, an alpha(5)beta(1) integrin antagonist. Biochim. Biophys. Acta. 1793, 354–367 [DOI] [PubMed] [Google Scholar]
- 54. Quann K., Gonzales D. M., Mercier I., Wang C., Sotgia F., Pestell R. G., Lisanti M. P., and Jasmin J. F. (2013) Caveolin-1 is a negative regulator of tumor growth in glioblastoma and modulates chemosensitivity to temozolomide. Cell Cycle. 12, 1510–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barresi V., Buttarelli F. R., Vitarelli E. E., Arcella A., Antonelli M., and Giangaspero F. (2009) Caveolin-1 expression in diffuse gliomas: correlation with the proliferation index, epidermal growth factor receptor, p53, and 1p/19q status. Hum. Pathol. 40, 1738–1746 [DOI] [PubMed] [Google Scholar]
- 56. Minata M., Audia A., Shi J., Lu S., Bernstock J., Pavlyukov M. S., Das A., Kim S. H., Shin Y. J., Lee Y., Koo H., Snigdha K., Waghmare I., Guo X., Mohyeldin A., Gallego-Perez D., Wang J., Chen D., Cheng P., Mukheef F., Contreras M., Reyes J. F., Vaillant B., Sulman E. P., Cheng S. Y., Markert J. M., Tannous B. A., Lu X., Kango-Singh M., Lee L. J., Nam D. H., Nakano I., and Bhat K. P. (2019) Phenotypic plasticity of invasive edge glioma stem-like cells in response to ionizing radiation. Cell Rep. 26, 1893–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hagiwara S., Murakumo Y., Mii S., Shigetomi T., Yamamoto N., Furue H., Ueda M., and Takahashi M. (2010) Processing of CD109 by furin and its role in the regulation of TGF-beta signaling. Oncogene. 29, 2181–2191 [DOI] [PubMed] [Google Scholar]
- 58. Kulp K. S., Berman E. S., Knize M. G., Shattuck D. L., Nelson E. J., Wu L., Montgomery J. L., Felton J. S., and Wu K. J. (2006) Chemical and biological differentiation of three human breast cancer cell types using time-of-flight secondary ion mass spectrometry. Anal. Chem. 78, 3651–3658 [DOI] [PubMed] [Google Scholar]
- 59. Robinson M. A., Graham D. J., Morrish F., Hockenbery D., and Gamble L. J. (2015) Lipid analysis of eight human breast cancer cell lines with ToF-SIMS. Biointerphases. 11, 02A303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Baker M. J., Brown M. D., Gazi E., Clarke N. W., Vickerman J. C., and Lockyer N. P. (2008) Discrimination of prostate cancer cells and non-malignant cells using secondary ion mass spectrometry. Analyst 133, 175–179 [DOI] [PubMed] [Google Scholar]
- 61. Gostek J., Awsiuk K., Pabijan J., Rysz J., Budkowski A., and Lekka M. (2015) Differentiation between single bladder cancer cells using principal component analysis of time-of-flight secondary ion mass spectrometry. Anal. Chem. 87, 3195–3201 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed data can be found in supplemental Tables S2 and S3. Unprocessed TOF-SIMS spectra have been deposited to Mendeley Data (Project: Investigation of inter- and intra-tumoral heterogeneity of glioblastoma using TOF-SIMS; http://dx.doi.org/10.17632/ssgyybm6d3.2).