An unbiased quantitative profiling of the protein expression, phosphorylation, and sialylation events that occur upon β-cell adaptive flexibility during the transition from hyperglycemia to euglycemia was assessed in islets from db/db and WT mice before and after β-cell rest. Our PTMomics data revealed new adaptive proteins, phosphosites, and SA N-linked glycosites involved in the regulation of proinsulin biosynthesis translation, protein folding, not ER stress, proinsulin processing, β-granule biogenesis, vesicular trafficking and the insulin secretory pathway, protein degradation, and endopeptidase activity, associated with β-cell adaptive flexibility.
Keywords: Diabetes, obesity, insulin resistance, signal transduction, protein-protein interactions, phosphoproteome, glycoproteomics, glycoprotein pathways, protein folding, protein synthesis, beta-granule biogenesis, insulin secretory pathway, proinsulin biosynthesis, protein degradation, unfolded protein response
Graphical Abstract
Highlights
db/db β-cells restores appropriate insulin stores and normalize secretory function.
Numerous changes in the phosphorylation and sialylation states by euglycemic rest.
Restoration of numerous dysfunctional biological processes following euglycemic rest.
β-cell adaptive flexibility may lead to improvement in endogenous β-cell function.
Abstract
The onset of obesity-linked type 2 diabetes (T2D) is marked by an eventual failure in pancreatic β-cell function and mass that is no longer able to compensate for the inherent insulin resistance and increased metabolic load intrinsic to obesity. However, in a commonly used model of T2D, the db/db mouse, β-cells have an inbuilt adaptive flexibility enabling them to effectively adjust insulin production rates relative to the metabolic demand. Pancreatic β-cells from these animals have markedly reduced intracellular insulin stores, yet high rates of (pro)insulin secretion, together with a substantial increase in proinsulin biosynthesis highlighted by expanded rough endoplasmic reticulum and Golgi apparatus. However, when the metabolic overload and/or hyperglycemia is normalized, β-cells from db/db mice quickly restore their insulin stores and normalize secretory function. This demonstrates the β-cell's adaptive flexibility and indicates that therapeutic approaches applied to encourage β-cell rest are capable of restoring endogenous β-cell function. However, mechanisms that regulate β-cell adaptive flexibility are essentially unknown. To gain deeper mechanistic insight into the molecular events underlying β-cell adaptive flexibility in db/db β-cells, we conducted a combined proteomic and post-translational modification specific proteomic (PTMomics) approach on islets from db/db mice and wild-type controls (WT) with or without prior exposure to normal glucose levels. We identified differential modifications of proteins involved in redox homeostasis, protein refolding, K48-linked deubiquitination, mRNA/protein export, focal adhesion, ERK1/2 signaling, and renin-angiotensin-aldosterone signaling, as well as sialyltransferase activity, associated with β-cell adaptive flexibility. These proteins are all related to proinsulin biosynthesis and processing, maturation of insulin secretory granules, and vesicular trafficking—core pathways involved in the adaptation of insulin production to meet metabolic demand. Collectively, this study outlines a novel and comprehensive global PTMome signaling map that highlights important molecular mechanisms related to the adaptive flexibility of β-cell function, providing improved insight into disease pathogenesis of T2D.
Diabetes is one of the most common metabolic disorders, and its prevalence in adults is estimated to increase by 69% in developing countries and 20% in developed countries between 2010 and 2030 (1). Notably, type 2 diabetes (T2D)1 accounts for 90–95% of all diabetes cases (2, 3). A key aspect to the development of T2D is an apparently decreased mass and acquired secretory dysfunction of insulin-producing islet β-cells, leading to a relative insulin-deficient diabetic state. T2D risk is strongly associated with obesity and insulin resistance (3, 4), where β-cells initially attempt to compensate to increased metabolic demand, but as this chronically persists, β-cells become overworked, dysfunctional and ultimately succumb to glucolipotoxicity by entering apoptosis (5). However, such β-cell 'dysfunction' could be symptomatic of an inherent adaptive flexibility of β-cells striving to synthesize and secrete sufficient insulin to meet the metabolic demand (6–9). Despite the well-studied pathogenesis of T2D, molecular mechanisms underlying such adaptive flexibility of compensating pancreatic β-cells remains relatively unexplored.
In untreated obesity, chronically increasing insulin resistance and metabolic load raise the demand for insulin which is initially compensated for by increased β-cell functional mass (5, 6). But then, with time and ever-increasing β-cell demand, this compensation eventually fails and a state of relative insulin insufficiency is reached, marking the onset of T2D (5). We have recently shown that β-cells derived from two closely related strains of obese leptin receptor-deficient mice (db/db), one of β-cell compensation (C57BL/6Jdb/db) and the other of β-cell failure (C57BLKS/Jdb/db) (7), both show β-cell adaptive flexibility to markedly increase (pro)insulin production and secretion in a valiant effort to meet metabolic demand in the context of insulin resistance (7, 9). Observations such as these have led to the idea that insulin secretory dysfunction in T2D could be indicative of these efforts toward increased insulin production (6–9). Remarkably, if the glucose concentration is lowered in an insulin-independent manner, and/or insulin resistance is alleviated, the in vivo demand for insulin is lowered and endogenous β-cells recover normal insulin secretory capacity and function (7, 9). These “restored” β-cells are characterized by a restoration of mature insulin secretory granule intracellular stores and insulin secretory capacity, slowing of (pro)insulin production accompanied by normalized endoplasmic reticulum (RER) and Golgi apparatus morphologies, recovered mitochondrial structure and morphology, and re-establishment of normal biphasic insulin secretion in response to glucose (6–9). This illustrates the adaptive flexibility of the pancreatic β-cell and has contributed to the concept of “β-cell rest” as a therapeutic approach to treat T2D (6). However, it remains mechanistically unclear how β-cell adaptive flexibility is regulated at a molecular level.
To characterize the signaling mechanisms underlying β-cell adaptive flexibility in db/db mouse pancreatic islets, we applied a mass spectrometry (MS)-based quantitative proteomics and post-translational modification specific proteomics (PTMomics) approach to both normal wild type (WT) and db/db islets from hyperglycemic and normalized glucose environments. Previously, we focused on β-cell adaptive flexibility related to mechanisms underlying observed morphological changes (e.g. expanded RER and Golgi, vesicular transport, etc.) associated with increased (pro)insulin production (7, 10). Here, we have expanded the analysis of quantitative proteomics and PTMomics (phosphorylation and sialylated N-linked glycosylation) data to elucidate novel molecular processes underlying functional compensation of β-cells in db/db mice and their subsequent recovery following exposure to normal glucose levels to provide deeper insight into the molecular mechanisms underlying regulation of β-cell adaptive flexibility.
Quantitative proteomics and PTMomics were performed on pancreatic islets derived from obese diabetic C57BL/6Jdb/db mice and matching wild-type controls (WT) using isobaric labeling combined with TiO2 enrichment of phosphopeptides and formerly sialylated N-linked glycopeptides (SA glycopeptides) as described (11). We identified functionally unknown proteins and PTM sites (phosphorylation or sialylation) within Gene Ontology biological processes, particularly in regulated proteins with PTMs in isolated db/db mouse islets, which showed a significant restoration after exposure to normal glucose concentrations (5.6 mm) versus WT islets. Several of these biological processes correlated with changes in β-cell biology and function (particularly in the RER and Golgi apparatus) related to (pro)insulin production. We also found significantly altered sialylation changes that were closely related to the ERK1/2 signaling pathway and focal adhesion. Sialic acid content significantly altered SA glycopeptides in db/db mice, and phosphorylation mediated conformational changes on sialyltransferase activities. Characterization of regulated protein kinase pathways unveiled links between cell redox homeostasis and protein refolding that are functionally interrelated and interdependent for proinsulin production. Moreover, twenty proteins that showed recovery in isolated db/db mouse islets after exposure to normal glucose levels, were validated by a targeted MS-based parallel reaction monitoring (PRM) assay. The data presented herein significantly improve our understanding of the cellular signaling mechanisms associated with β-cell adaptive flexibility, both at the protein and PTM level (phosphorylation and sialylation), related to the increased metabolic demand in obesity-linked T2D.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
All experiments were approved by the local Ethics Committee in accordance with Danish legislation. Animal care, use, and experimental protocols were approved by the Institutional Animal and Use Committee (IACUC) of the University of Chicago and MedImmune, LLC. See Fig. 1A for a summary of methods. In this study, we used freshly isolated islets of Langerhans from C57BL/6Jdb/db and WT mice, with 'reduced toward normal' islets cultured at normal 5.6 mm glucose for 12 h in vitro. Four biological replicates (proteomics) and two biological replicates (phosphoproteomics and sialiomics) were performed. Each biological replicate consisted of ∼1,000 islets that were pooled from several mice of each treatment group (n ≥ 8) because the yield of isolating mouse pancreatic islets is limited, especially from diabetic mice. All the quantified proteins including phosphopeptides and SA glycopeptides were overlapped (Proteomics: at least 3 out of the 4 biological replicates, PTMomics: all biological replicates). To further refine the criteria for statistical significance of protein expression, phosphopeptides, SA glycopeptides fold changes, regulated proteins, significantly altered phosphopeptides, or SA glycopeptides were accepted with ±2 fold changes and/or z-test for adjusted p value <0.05 and/or t test for adjusted p value <0.05 with the Benjamini and Hochberg correction for a normal distribution, coefficient of variation (CV%) of 30% or lower, and same direction of change (log2 ratio, positive or negative) in all biological replicates were defined as being significantly regulated. The regulated phosphopeptides or SA glycopeptides were correlated (Pearson correlation of 0.88 to 0.96) between the two biological replicates (supplemental Fig. S1). To identify restored proteins and PTMs in isolated db/db islets in response to glucose, we compared three groups [db/db islets after exposure to normal glucose (DG) versus freshly isolated db/db islets (D) versus normal (WT) islets (N), opposite to D/N; at least log2 fold change ± 0.3 in DG/D] (Fig. 1D and 4B). For instance, if a regulated SA glycopeptide in D/N (e.g. 2/1 = 2-fold) is restored after normal glucose treatment, DG/D (e.g. 1/2 = 0.5 fold) should be regulated opposite to D/N. Of these, we eliminated the differentially expressed proteins or altered phosphorylations at the N versus normal islets after exposure to normal 5.6 mm glucose (NG) comparison as a control for any in vitro tissue culture effect. The remaining regulatory proteins, phosphopeptides and SA glycopeptides in D, that are reduced toward normal at 5.6 mm glucose, were due only to such a “β-cell rest” effect. The PRM assay was performed using islets derived from 8 to 12 mice for each condition. The PRM assay was performed using several technical analyses for significantly regulated proteins, by analyzing a sub-dataset relative to the insulin secretory process in order to reveal new biological insights.
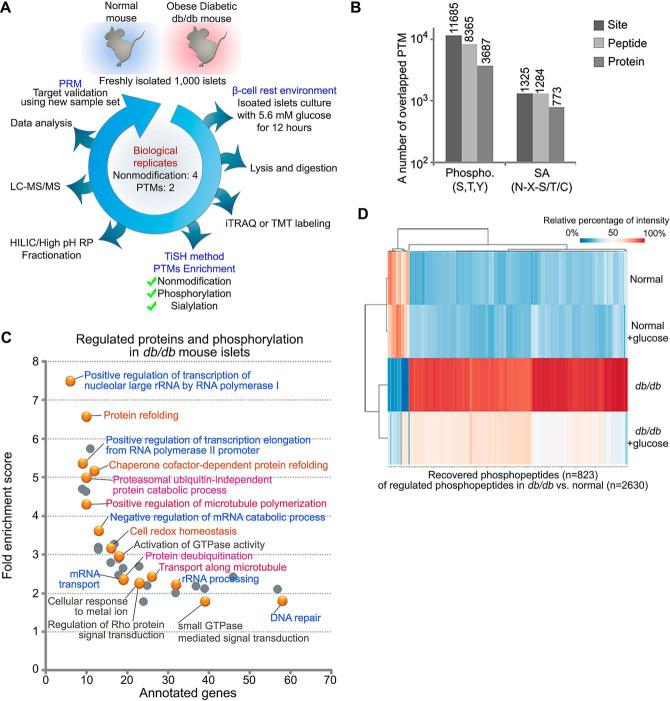
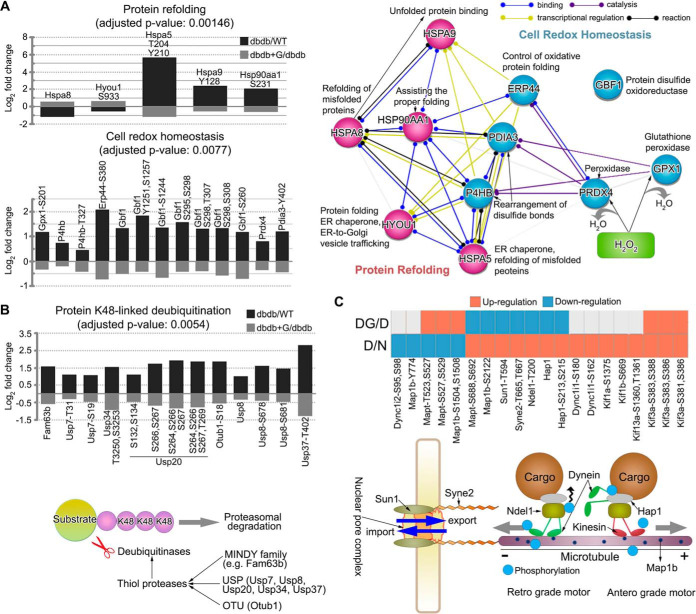
Fig. 1.
Quantitative global phosphoproteomic study. A, islets were isolated from db/db mice and WT mice and incubated with or without euglycemia (5.6 mm, 12 h) after which islets lysates were prepared and digested with trypsin/LysC. Tryptic peptide samples from individual experiments were labeled with isobaric mass tags (iTRAQ/TMT) and the peptides from equal amounts of independent experiments, then phosphopeptides and formerly sialylated N-linked glycopeptides (SA) were enriched by TiSH method. Each sample from modification groups were fractionated using HILIC or high pH reverse phase before being analyzed by LC-MS/MS. Twenty-five differentially regulated peptides were validated using Parallel Reaction Monitoring (PRM) assay with heavy isotope-labeled synthetic peptide. B, Histograms show the overlapping 11,685 and 1325 posttranslationally modified site, 8365 and 1284 peptides, and 3687 and 773 proteins in the phosphorylation group and the SA group, respectively. C, Heatmap of the recovered phosphopeptides (opposite to D/N; at least log2 fold change ± 0.3 in DG/D) ordered by hierarchical clustering. D: db/db mouse islets, N: wild type mouse islets, DG: db/db mouse islets exposed to euglycemia. Values for each phosphopeptide (column) at all analyzed samples (row) are color code based on the relative percentage of normalized intensity, low (blue) and high (red). D, Enriched biological processes for significantly regulated proteins with phosphorylation that exhibited restoration. Panther Gene Ontology enrichment analysis showing the fold enrichment score for the indicated dots (y axis) as well as the number of proteins assigned to classified functions (x axis). Corrected p value < 0.05.
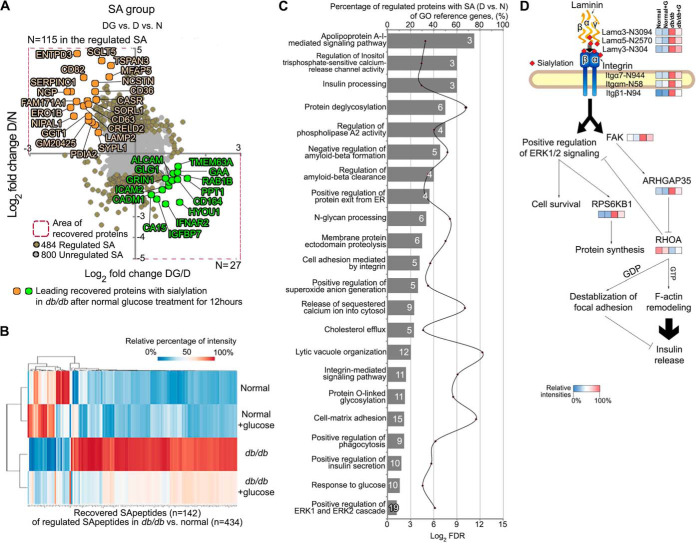
Fig. 4.
Quantitative global sialylomic study. A, Scatter plot of significantly reverted formerly sialylated N-linked glycopeptides (SA) toward baseline (n = 142; opposite to D/N; at least log2 fold change ± 0.3 in DG/D) showing quadrant localization with labels on all significantly regulated proteins (brown; n = 484) and the not significantly regulated proteins (gray; n = 800). D: db/db mouse islets, N: wild type mouse islets, DG: db/db mouse islets exposed to euglycemia. Leading recovered SA shown in light orange or light green are significantly reverted to normal status after exposure to euglycemia in islets of obese diabetic db/db mice. B, Heatmap of the reverted SA (opposite to D/N; at least log2 fold change ± 0.3 in DG/D) ordered by hierarchical clustering. C, Using Panther Gene Ontology analysis, enriched biological process categories showing the percentage of regulated SA in the reference genes of indicated cellular function (left y axis, bars) as well as the false discovery rate (right y axis, line). Note that individual regulated SA can belong to more than one group. The number of proteins assigned to classified functions is indicated by number (white) in a bar. D, Flexibe adaptability of laminin-integrin-mediated signaling show the correlations with inter-related proteins.
Materials
All chemicals were of the highest purity obtainable and purchased from Sigma (Sigma-Aldrich, MO), unless stated otherwise. Titanium dioxide (TiO2) beads were obtained from GL Science, Japan. Poros Oligo R3 reversed phase chromatographic material was obtained from PerSeptive Biosystems.
Animals, Pancreatic Islet Isolation, and Ex Vivo Culture
C57BL/6J wild type (WT) and C57BL/6Jdb/db mice were bred in-house or purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were housed on a 12 h light/dark cycle and were allowed free access to standard mouse food and water. Unless otherwise stated, the mice were sacrificed between 14 and 16 weeks of age. Pancreatic islets were isolated by collagenase digestion as previously described (7). Freshly isolated islets were cultured as previously described (7). Experiments were performed on either fresh islets, immediately after isolation, or on recovered isolated islets after overnight (12 h) culture at 37 °C in RPMI 1640 containing normal 5.6 mm glucose, 50 U/ml penicillin, 50 mg/ml streptomycin and 10% FBS (i.e. euglycemia). Islets were then stored at −80 °C until further processing.
Sample Preparation and iTRAQ Labeling
Proteins were extracted in lysis buffer (starting material: 80 μg of protein lysates from each group) in a total volume of 10 μl containing 6 m Urea, 2 m Thiourea, 10 mm dithiothreitol, phosphatase/protease inhibitor mixture (Roche, Germany)] from ∼1,000 pancreatic islets for each group outlined in Fig. 1A. The samples were mixed and incubated for 2 h at room temperature (RT). After incubation, the samples were diluted 10-fold with 20 mm triethylammonium bicarbonate (TEAB), pH 7.5, and then sonicated for 2 × 20 s on ice. Then, 20 mm iodoacetamide was added and incubated for 20 min in the dark at RT. After incubation, 2% (w/w) trypsin was added to the samples which were then incubated overnight at RT. After this proteolysis, the samples were lyophilized prior to iTRAQ/TMT labeling. Samples from each group were tagged with the iTRAQ 4 plex kit (AB Sciex) or the TMT 10 plex kit (Thermo Fisher Scientific) according to the manufacturer's instructions. After labeling, the samples were combined equally based on the protein quantification achieved from Qubit® Fluorometric Quantitation (Thermo Fisher Scientific) and MALDI-MS/MS analysis of a combined sample (Bruker Daltonics, Billerica, CA, in lift mode).
Enrichment of Phosphorylated Peptides and SA Glycopeptides
Phosphopeptide and SA glycopeptide enrichment were performed from the completely TMT labeled peptide mixture according to a previously published TiSH protocol (11, 12) in which nonmodified peptides are first separated from these two modified peptide species using TiO2. Hereafter, the multi- and monophosphorylated peptides are separated from the formerly SA glycopeptide after a deglycosylation step using the sequential elution from IMAC (SIMAC) procedure. Briefly, the lyophilized labeled sample was adjusted to the TiO2 loading buffer concentration [80% acetonitrile (ACN), 5% Trifluoroacetic acid (TFA), 1 m glycolic acid]. The peptide solution TiO2 beads [0.6 mg/100 μg (bead/peptide)] were added and the solution was mixed at RT for 10 min. After, the sample was centrifuged briefly to pellet the beads. The supernatant was transferred to a new tube labeled as non-modified peptides and 100 μl of TiO2 loading buffer was added to the beads. After mixing, the sample was centrifuged to pellet the beads, and the supernatant then transferred to the non-modified peptide fraction. Subsequently, the TiO2 beads were washed twice with 100 μl of washing buffer 1 (80% ACN, 1% TFA), then dried for 5 min in the vacuum centrifuge to remove all solvent. The bound peptides were eluted with 100 μl of 1% ammonium hydroxide (pH 11.3) for 15 min and then centrifuged at 1000 × g for 1 min. The eluted peptides were passed over a C8 stage tip (3 mTM EmporeTM Bioanalytical Technologies, SigmaAldrich) to retain the TiO2 beads and dried by vacuum centrifugation to produce the enriched phosphopeptide/SA glycopeptide fraction. The flow through from the initial loading buffer (containing non-modified peptides) and washes were combined then dried by vacuum centrifugation to produce the non-modified peptide fraction. The non-modified peptide fraction was acidified (pH < 3) with TFA and desalted on a R3 stage tip column before HILIC fractionation. Non-modified peptides were dried in a vacuum centrifugation and stored at −20 °C.
Deglycosylation
To remove the glycan structures from the enriched SA glycopeptides, the sample was resuspended in 20 mm TEAB, pH 8.0, and treated with 2 μl N-glycosidase F (New England Biolabs) and 0.5 μl Sialidase A (ProZyme, CA) at 37 °C (11).
SIMAC
The SIMAC protocol has been previously described (11). Briefly, the deglycosylated samples were resuspended in SIMAC loading buffer (50% ACN and 0.1% TFA) and the pH was adjusted to 1.6–1.8 by 10% TFA. PhosSelect IMAC beads (Sigma Aldrich) were equilibrated with loading buffer, combined with the samples and incubated for 1 h under gentle rotation. The samples were then centrifuged, and the supernatant recovered. The beads were transferred to a constricted GELoader tip (Eppendorf® AG, Germany) and the column was washed with loading buffer, collecting the flow through in the same tube as the previous supernatant. Mono-phosphorylated and formerly SA glycopeptides were eluted from the column with an acidic solution (20% ACN, 1% TFA). The multi-phosphorylated peptides were subsequently eluted from the column with a basic solution (1% ammonium hydroxide solution, pH 11.3). The basic elution containing multi-phosphopeptides was acidified before desalting on a R3 stage tip column and the eluted peptides lyophilized before LC-MS/MS. The supernatants and acidic eluates collected after SIMAC were subjected to another round of TiO2 purification to separate mono-phosphopeptides from formerly SA glycopeptides. The samples were adjusted to 70% ACN and 2% TFA and the TiO2 beads were added to the solution and incubated on a shaker as described above. The incubation round was performed twice with half the amount of beads the second time. The beads were pelleted, and the supernatant was recovered (containing formerly SA glycopeptides). The two pellets of TiO2 were pooled with 50% ACN, 0.1% TFA, the beads were pelleted, and the supernatant was recovered. The beads were dried and bound phosphopeptides then eluted from the beads with 1% ammonium hydroxide solution (pH 11.3). The phosphopeptide sample was acidified using 100% formic acid (FA) to pH < 3 and subsequently desalted using a R2-R3 stage tip column prior to HILIC. Likewise, the formerly SA glycopeptides were desalted. Finally, all phosphopeptides/formerly SA glycopeptides were dried in a vacuum centrifugation and stored at −20 °C.
HILIC Fractionation
The HILIC fractionation protocol has also been previously described (11). Briefly, all phosphorylated peptides, formerly SA glycosylated peptides, and non-modified peptides were separately resuspended in Buffer B (90% ACN/0.1% TFA). The samples were loaded onto an TSKgel Amide-80 HILIC 320 μm × 170 mm capillary HPLC column using an Agilent 1200 capillary HPLC system. Samples were separated using a gradient from 100% to 60% buffer B (Buffer A: 0.1% TFA) over 35 min, then 60% to 0% over 7 min at a flow rate of 6 μl/min. The fractions were automatically collected in a 96-well plate at 1 min intervals with monitoring by UV detection at 210 nm. Peptide containing fractions were pooled according to UV peak detection. The fractions were then dried by vacuum centrifugation and subsequently stored at −20 °C.
Nanoflow Liquid Chromatography-Mass Spectrometry (nLC-MS/MS) Analysis
All fractions were redissolved in buffer A (0.1% FA) and analyzed using a nLC-MS/MS system consisting of an Easy-nLC (Thermo Fisher Scientific) and an Orbitrap Fusion Lumos (PTMomics) or a Q-exactive HF (proteomics) mass spectrometer (MS) (Thermo Fisher Scientific). The samples were loaded either directly onto a 15 cm fused silica capillary column (75 μm inner diameter) or onto a 2 cm trap column (100 μm inner diameter) and separated on a 15 cm analytical column. All columns were handmade and packed with ReproSil-Pur C18 AQ 3 μm reversed phase material (Dr. Maisch, Germany). The peptides were eluted using 73–133 min gradients from 1 to 40% buffer B (95% ACN, 0.1% FA) and introduced into the MS instrument via nanoelectrospray according to the intensity of each HILIC peptide fraction. A full MS scan in the mass area of 400–1400 Da was performed in the Orbitrap with a resolution of 120,000, an AGC target value of 5 × 105, and a maximum injection time of 60 ms. For each full scan, “Top speed” mode was selected for higher energy collision dissociation (HCD). The settings for the HCD were as follows: AGC target value of 3 × 104, maximum injection time of 60 ms, isolation window of 1.2 Da, and normalized collision energy of 38.
Protein Identification and Quantification
The raw MS data sets were processed for protein identification using the MS-GF+ (v9979, 07/16/2014) combined with the MASIC pipeline with a peptide mass tolerance of 20 ppm, reporter ion m/z tolerance half width of 2 mDa, and a false discovery rate (FDR) of 1% for proteins and peptides. All peak lists were searched against the UniProtKB/Swiss-Prot database (2013_06, 16,613 entries) of mouse sequences with decoy using the parameters as follows: enzyme, trypsin; maximum missed cleavages, 2; fixed modification, carbamidomethylation (C), iTRAQ or TMT tags (K, peptide N termini); variable modifications, oxidation (M), phosphorylation (S,T,Y), and deamidation (N). Datasets with raw MS values were filtered to remove potential errors using the criteria as follows: elimination of contaminants and reversed sequences for each accession number. For sialylated peptides, the Asn-X(except proline)-Ser/Thr/Cys consensus sequence is required to be considered a deglycopeptide. For Phosphopeptides, we assessed the peptide backbone with phosphorylation localization in the MS/MS data sets using Idpicker and PhosphoRS (probability>0.75) (13, 14). For relative protein quantification, the output tsv file from MS-GF+ combined with the MASIC pipeline was imported into Microsoft Excel. Then, protein relative expression values from the respective unique or razor peptides were calculated by summing all of the unique/razor peptide intensities of each protein and normalized to the number of total intensities of each group estimating the relative amounts of the different protein within the relative sample. The resulting ratios were log-transformed (base = 2) to achieve a normal distribution. Ratios were averaged over overlapping proteins, phosphopeptides, or SA peptides.
Functional Analysis of Regulated Proteins
Gene Ontology (GO) annotation enrichment analysis was performed using PANTHER (15) and DAVID (16). These GO analyses allowed the enrichment of biological process, cellular components, and KEGG pathway. The regulated or recovered proteins with PTMs were searched against the STRING database (17) and IntAct database (18) for protein-protein interactions. Ingenuity Pathway Analysis (IPA; Ingenuity Systems) was used to functionally annotate genes implicated in causal biological pathways and functions.
Targeted Quantification Using Parallel-Reaction Monitoring (PRM) and data analysis
Our LC/PRM-MS assay was developed for research use, so the experimental design aligns with the goals of a Tier-2 assay (19). The target peptides were selected and designed based on the proteomics data. They were selected based on the following criteria: they are unique peptides, the most repeatable charge states corresponding to intensity, carry no miss cleavage, and are less than 20 amino acids. We excluded the possible modifications, but artificial modifications were included such as oxidation on Met or carbamidomethylation on Cys. A PRM assay was performed using twenty-five heavy isotope-labeled synthetic peptides (JPT Peptide Technologies) on a Q-Exactive™ HF mass spectrometer (Thermo Scientific) in three technical replicates. All heavy-labeled (K or R) peptides (600 fmol) corresponding to endogenous peptides [aldehyde dehydrogenase family 1 member A3 (ALDH1A3), methionine aminopeptidase 2 (METAP2), mixed lineage kinase 4 (MLK4; MAP3K21), apoptosis-stimulating of p53 protein 1 (PPP1R13B), phosphatase 2A inhibitor I2PP2A (SET), CPE, proSAAS (PCSK1N), protein disulfide-isomerase (P4HB), INS1, somatostatin (SST), melanoma inhibitory activity protein 3 (MIA3), CD63, neudesin (NENF), glycerol-3-phosphate phosphatase (PGP), minor histocompatibility antigen H13 (HM13), polyadenylate-binding protein-interacting protein 2B (PAIP2B), pyruvate kinase (PKM), prominin-1 (PROM1), and mitochondrial import receptor subunit (TOMM22)] were spiked into tryptic peptides originating from 1 μg of islet lysate proteins (∼1,000 islets isolated from 8–12 mice per group) as internal standards. Subsequently, an equal amount of peptide mixture corresponding to each group was desalted using Poros Oligo R3 RP micro-columns (11) prior to PRM analysis. The samples were resuspended in 0.1% FA and loaded onto an EASY-nLC system (Thermo Scientific). The samples were loaded onto a 15 cm analytic column consisting of fused silica capillary (75 m inner diameter) packed with ReproSil-Pur C18 AQ 3 μm reversed-phase material (Dr. Maisch). The peptides were eluted with an organic solvent gradient from 100% phase A (0.1% FA) to 32% phase B (95% ACN, 0.1% FA) at a constant flowrate of 300 nL/min. The PRM method consisted of a full MS scan configured as above followed by targeted MS/MS scans for each selected peptide with a 1 min of retention time window, as defined by a time-scheduled inclusion list. The parameters were set as follows: MS, 120K resolution; MS/MS, 15K resolution; 3e6 AGC target, 15 ms maximum injection, 1.1 m/z isolation window, and normalized collision energy of 28–29. We used Skyline (version 3.5.0.9319) for quantitative analysis of the generated PRM data (11). Briefly, raw MS files were imported into Skyline. And then a FASTA file (from Uniprot) containing targeted proteins was imported to Skyline. We applied the highest confidence of dot-product score (0.95≤) which provides a correlation score between the measured product ion peak areas and the fragment ion intensities for the precursor ions (20). Endogenous peptides relative expression values from the respective group were normalized to standard deviation factors (C, replicates 1,2,3: 1.12, 1.08, 1.05; CG:, replicates 1,2,3: 1.01, 0.97, 1.02; D, replicates 1,2,3: 0.89, 0.94, 0.97; DG, replicates 1, 2, 3: 0.97, 0.92, 0.95) estimating all precursor ion areas of each heavy peptide within relative sample. The peptide quantitation (including peak areas and retention time for all replicates) are provided as supplemental Table S4.
RESULTS
Quantitative Analysis of the Proteome and PTMome of Islet Cells Derived from WT and db/db Mice
To reveal the cellular mechanism underlying β-cell adaptive flexibility observed in db/db mice, we investigated the proteome and PTMome (phosphorylation and SA N-linked glycosylation) in isolated islets from obese diabetic db/db mice and WT mice using our modified TiSH protocol (11) (workflow in Fig. 1A). We identified a total of 8217 proteins and quantified a total of 4,985 proteins that overlapped in three out of the four performed replicates. Analysis of the phosphopeptides and formerly sialylated N-linked deglycopeptides (SA peptides) (the PTMome) resulted in a total of 8365 phosphopeptides carrying 11,685 phosphosites on 3687 unique proteins and 1284 SA glycopeptides carrying 1325 SA sites on 773 unique proteins (Fig. 1B). To determine whether proteins or modification sites were significantly regulated in freshly isolated db/db mouse islet (D) compared with normal WT (N), we examined regulated proteins, phosphopeptides, or SA glycopeptides between D and N that were significantly increased or decreased (≥ 2.0 fold or ≤ 0.5 fold with CV% ≤ 30% and/or z-test for adjusted p value < 0.05) and altered in the same direction of change (positive or negative) in all of the identified replicates (list of regulated proteins, phosphopeptides, and SA glycopeptides; supplemental Table S1). Our quantitative data analysis revealed that 391 proteins, 2630 phosphopeptides from 1543 proteins, and 434 SA peptides from 339 proteins were significantly changed in D compared with N.
The db/db Islet-associated PTMomic Alterations and Mapping of Key Biological Processes
We further analyzed regulated proteins regarding phosphorylation state and SA modification in D versus N using Panther GO analysis (Fig. 1C and 4C). For phosphopeptides, we found significantly enriched GO categories (FDR < 0.05) that showed strong enrichment in biological processes, such as mRNA/rRNA processing, vesicular transport along microtubules, GTPase-mediated signal transduction, Rho protein signal transduction, protein-folding, and de-ubiquitination, as well as a response to oxidative damage and altered glucose metabolism in D versus N (Fig. 1C). For significantly regulated SA peptides, we identified that twenty-two biological processes linked to the cell surface environment were significantly enriched in D versus N (FDR < 0.05) (Fig. 4C), such as the apolipoprotein A-I-mediated signaling pathway, regulation of inositol trisphosphate-sensitive Ca2+-release channel activity, regulation of phospholipase A2 activity, membrane protein ectodomain proteolysis, cell adhesion mediated by integrins, integrin-mediated signaling pathway, regulation of ERK1 and ERK2 cascades, release of sequestered Ca2+ into cytosol, cholesterol efflux, cell-matrix adhesion, regulation of endocytosis and membrane rearrangement, and regulated insulin secretion (Fig. 4C). The annotated genes are listed in supplemental Table S2.
Upstream Interaction Network of Differentially Expressed Proteins and Altered Phosphorylation in db/db Mouse Islets
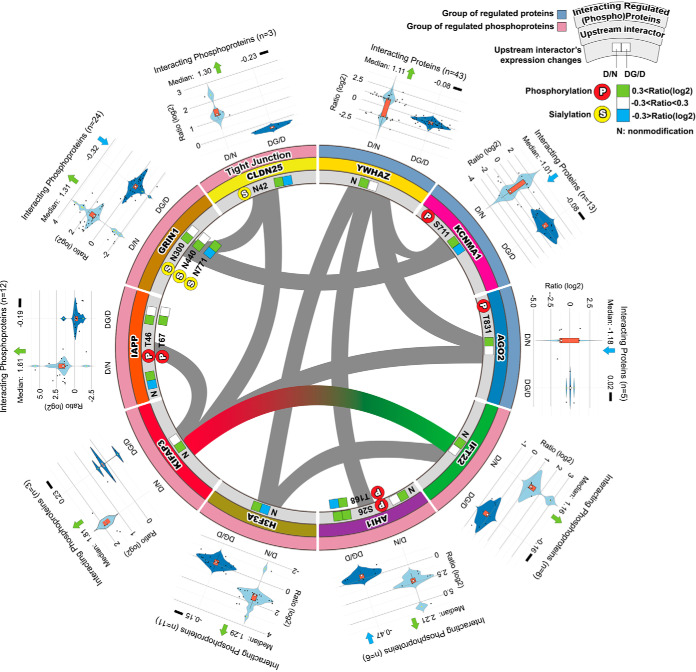
To investigate the protein-protein interactions between proteins that showed a significant regulation on either the protein or phosphosite level, as well as their upstream/downstream relationship between interacting proteins, we used a combination of freely accessible databases programs (IntAct, STRING, DAVID, Pubmed, and UniProt). We determined the ten most qualified upstream interactors by using the p value: [14–3-3 protein zeta, YWHAZ (p value:2.E-04); Ca2+-activated K+ channel subunit alpha-1, KCNMA1 (p value:4.E-02); argonaute RISC catalytic subunit 2, AGO2 (p value:5.E-02); intraflagellar transport 22, IFT22 (p value:8.E-03); abelson helper integration site 1, AHI1 (p value:5.E-02); H3 histone, family 3A, H3F3A (p value:6.E-02); kinesin-associated protein 3, KIFAP3 (p value:7.E-02); islet amyloid polypeptide, IAPP (p value:7.E-02); glutamate receptor ionotropic NMDA1, GRIN1 (p value:1.E-02); and claudin 25, CLDN25 (p value: 7.E-02)], and confirmed probable interactor genes specifically in pancreatic β-cells using Pubmed and UniProt (Fig. 2). The circular interactome map (CIM) of regulated proteins or PTMs in db/db islet β-cells (Fig. 2) represents the ten upstream interactors and their interacting proteins (violin plots of child interactors on each parent interactor were added post-construction). The outer blue or red colored circles indicated the regulated downstream proteins/phosphoproteins as molecular targets of ten upstream interacting proteins in db/db islets compared with controls; up- or downregulation of upstream regulators were colorized in boxes (green: up-regulation, blue: down-regulation, left box: D/N, right box: DG/D) (Fig. 2). These upstream interactors were substantially changed at the protein or PTM levels in db/db mouse islets compared with normal controls. Moreover, such upstream interactors were differentially expressed in parallel with the expression of their interacting protein partners (three upstream interactors in the regulated proteins: YWHAZ, KCNMA1, and AGO2 and seven upstream interactors in the regulated phosphoproteins: IFT22, AHI1, H3F3A, KIFAP3, IAPP, NMDA1, GRIN1, and CLDN25), and were closely associated with interacting proteins/phosphoproteins in db/db mouse islets. If the interacting proteins (not upstream interactor) indirectly associated with another upstream interactor, they were shown by the gray color line (less confidence). If upstream interactor is directly linked to another interactor, the interaction was indicated by its unique color line. Notably, the IFT22-KIFAP3 interaction directly interacted with the highest confidence score (0.9) in Fig. 2. In addition, it has been demonstrated that the interaction of the IFT22 and KIFAP3 plays an important role in nuclear localization signal and cellular regulation (21, 22).
Fig. 2.
Upstream interaction network of regulated proteins or phosphoproteins in db/db mouse islets compared with control using a combination of resources (IntAct, STRING, DAVID, Pubmed, and UniProt).
Selected Phosphoproteins Indicative of Cellular Dysfunction in db/db Mouse Islets
We then examined whether regulated phosphoproteins could influence β-cell function in db/db mice together relative to restoration back to typical levels after exposure to normal (5.6 mm) glucose for 12 h. We performed an analysis to assess the recovered phosphopeptides (opposite to D/N; at least log2 fold change ± 0.3 in DG/D) in isolated db/db mouse islets exposed to normal glucose (DG). For example, if a regulated phosphopeptide in D/N (e.g. 2/1 = 2-fold) was restored after exposure to normal glucose levels, DG/D (e.g. 1/2 = 0.5 fold) should be regulated opposite to D/N. Using these criteria, we identified 823 recovered phosphopeptides that were visualized using a heatmap combined with hierarchical clustering analysis (Fig. 1D). Compared with WT control islets (N and NG), no effect on islet β-cell function was indicated following 12 h culture of WT islets at normal 5.6 mm glucose. Relative to db/db isolated islets exposed to normal (5.6 mm) glucose for 12 h, eleven regulated phosphosites on proteins involved in protein refolding and cell redox homeostasis were recovered back to normal levels found in WT islets [(protein refolding: HSPA8, HYOU1 (GRP170)-S933, HSPA5-T204-Y210, HSPA9-Y128, and HSP90AA1-S231) and (cell redox homeostasis: GPX1-S201, P4HB (PDIA1)-T327, ERP44-S380, GBF1-S260-S295-S298-T307-S308-S1244-Y1251-S1257, PRDX4, and PDIA3-Y402)] (Fig. 3A). To reveal functional connections within protein refolding and cell redox homeostasis, we illustrated relationships between protein refolding and cell redox homeostasis where protein-protein interaction networks could be interconnected. This suggested a potential contribution to systems associated with oxidative protein folding and the stabilization of disulfide bonds of newly synthesized proteins being formed in the endoplasmic reticulum (ER). Interestingly, we observed that the phosphorylation of seven thiol protease proteins [(MINDY deubiquitinase family: FAM63B), (ubiquitin specific peptidase family: USP7-S19-T31, USP8-S678-S681, USP20-S132-S134-S264-S266-S267-T269, USP34-T3250-S3253, and USP37-T402), and (OTU deubiquitinase family: OTUB1-S18)] involved in protein K48-linked de-ubiquitination were recovered upon exposing isolated db/db mouse islets to normal glucose (Fig. 3B). We also identified phosphosites on proteins that connect mRNA transportation (SUN1-T594 and SYNE2-T665-T667) and trafficking along the microtubule (DYNC1I1-S162-S180, DYNC1I2-S95-S98, MAP1B-Y774-S1504-S1508-S2122, MAPT-T523-S527-S529-S688-S692, NDEL1-T200, HAP1-S213-S215, KIF1A-S1375, KIF1B-S669, KIF3A-S381-S383-S386-S388, and KIF13A-S1360-T1361) that were also reverted to normal status after normal (5.6 mm) glucose exposure (Fig. 3C).
Fig. 3.
Phosphoproteome changes in cellular functions in db/db mouse islets. A, Regulated and restored proteins with phosphorylation state that are characterized in protein refolding or cell redox homeostasis (left). The quantitative information for each protein or phosphosite is shown as averaged log2 fold change in a bar chart. STRING subnetworks between protein refolding and cell redox homeostasis show the protein-protein interactions among the restoration of proteins and phosphosites to normal status after exposure to euglycemia in obese diabetic db/db mouse islets (right). B, Proteins with phosphorylation to normal status that are characterized in protein K48-linked deubiquitination (upper). The quantitative information for each protein or phosphosite is shown as averaged log2 fold change in a bar chart. Seven thiol protease proteins (MINDY deubiquitinase family, ubiquitin specific peptidase family, and OTU deubiquitinase family) are involved in protein K48-linked deubiquitination (bottom). C, Proteins with phosphorylation to normal status that are characterized in mRNA export or transport along microtubule (upper). The quantitative information for each protein or phosphosite is shown as up-regulation (red) or down-regulation (blue) in heatmap. Twelve proteins are involved in mRNA export or transport along microtubule (bottom).
Protein Phosphorylation Changes Associated with Islet β-cell Adaptation-A Potential Effect on Disulfide Bridge Formation
We next examined changes in phosphosites which are closely localized to cysteine residues within protein domains. Dynamic phosphorylation or dephosphorylation surrounding a covalent disulfide bond of cysteine within protein domains could be important for protein function, activity or stability and their potential effect on disulfide bond formation or interference (23–28). We identified several new phosphosites in five proteins (LRP1B, IL1R1, NECTIN1, PAPPA, and PCSK2) where the phosphorylation levels were significantly altered in the islets of db/db mice compared with controls (supplemental Fig. S2). In addition, we found that six phosphosites on three proteins (NECTIN1; T171, T173, and S174, PAPPA; S1368/Y1372, and PCSK2; T376) were hyperphosphorylated in db/db islets compared with controls. These were all localized adjacent to cysteine residue important for S-S bridge formation/cleavage required for the structure and function of the proteins of interest (supplemental Fig. S2). In contrast, five phosphosites on two proteins (LRP1B; Y1570 and IL1R1; S104, T105, Y106, and T110) were dephosphorylated in db/db islets, but were similarly localized next to cysteine residues important for S-S bond generation/interference (supplemental Fig. S2). In order to establish possible conformational changes in five proteins, future studies will be needed such as three-dimensional structure analysis of the proteins, providing new insight into redox regulation of protein activity either by phosphorylation or dephosphorylation around cysteine residues.
Modulation of Sialic Acids in db/db Islet
Of the 1284 formerly SA N-linked glycopeptides identified in the analysis, a total of 434 were found to be significantly regulated between normal and db/db islets (Fig. 4A), with 304 being up-regulated and 130 down-regulated. The significantly regulated SA glycopeptides were imported into STRING and the resulting interaction network is shown in Fig. 6 (lower left panel). Here, the modulated SA glycopeptides could be associated with limited proteolysis (29, 30), re-glycosylation with additional sugar residues such as SA in the ER and Golgi apparatus (31, 32), lysosomal function (33), renin-angiotensin system (34), cell adhesion molecules such as integrins and laminin (35–37), PI3K-Akt signaling (38), regulation of the actin cytoskeleton (39), focal adhesion (40), and extracellular matrix-receptor interaction (41). Moreover, ER lectin chaperone-like proteins (e.g. calnexin; CANX) and their associated co-factors (e.g. PDIA3) that recognize N-linked glycans are likely involved (42). In our results, the PTMs of CANX [phosphorylation at T561 (D/N: 12.5 fold, DG/D: −2.5 fold), S569 (D/N: 14.5 fold, DG/D: −3.0 fold); SA glycosylation at N580 (D/N: 2.2 fold, DG/D: −1.4 fold)] and PDIA3 [phosphorylation at Y402 (D/N: 2.3 fold, DG/D: −1.4 fold)] were significantly regulated in D/N and restored toward normal after exposure to normal glucose (supplemental Table S1). The analysis further detected 142 SA glycopeptides out of the 434 regulated SA glycopeptides that reduced toward normal after normal glucose culturing (Fig. 4A), which were also visualized using a heatmap combined with hierarchical clustering analysis (Fig. 4B). To identify the signaling events in recovered islets at the SA level, a GO enrichment and STRING analysis was performed on the proteins with altered SA (Fig 4C and 6). The modulation of SA on cell surface proteins regulates a wide variety of cellular functions and can be a pathophysiological feature of disease (43). Twenty-two biological processes associated with the cell surface environment were significantly enriched using the regulated SA glycoproteins (FDR < 0.05) (Fig. 4C). These signaling events were associated with adaptive compensation in isolated islets of obese diabetic db/db mice when compared with WT mice as revealed by a recovery to normal after euglycemic culturing. They appeared specific to focal adhesion dynamics in pancreatic β-cells and are implicated in insulin processing, Ca2+ release channel activity, apparent amyloid-beta clearance (but the latter likely inappropriately 'called' by the GO/STRING algorithm as islet amyloid does not occur in rodent models of T2D, and more related to the excessive co-secretion of islet-amyloid associated peptide (IAPP) with insulin in the db/db mouse islets), and integrin-mediated signaling pathways, that are all related to the trafficking and secretion of insulin (Fig. 4C). We constructed links between laminin proteins and the integrin-mediated signaling pathway possibly mediated via sialylation that could trigger the ERK1/2 signaling pathway and F-actin remodeling (Fig. 4D). Molecular changes in laminin-integrin-mediated signaling (increased SA levels of laminin and integrin proteins) showed significant correlations with interacting proteins at the expression level [focal adhesion kinase (FAK; up-regulation), Rho GTPase-activating protein 35 (ARHGAP35; up-regulation), transforming protein (RHOA; down-regulation by ARHGAP35), and ribosomal protein S6 kinase beta-1 (RPS6KB1; up-regulation)] possibly involved in the production and release of insulin as part of an adaptive response to obesity-linked T2D.
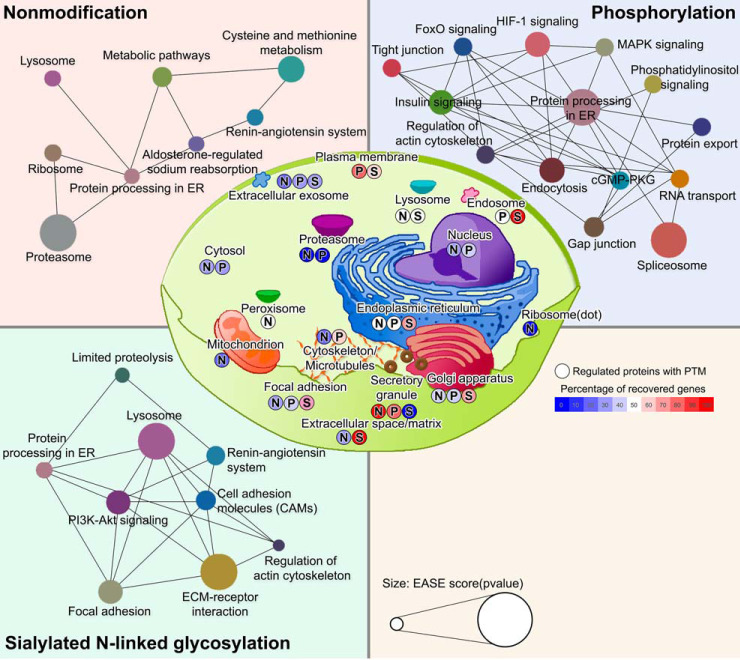
Fig. 6.
Significantly enriched DAVID Gene Ontology categories (p value < 0.05) are classified as cellular component (center) or KEGG pathway network in nonmodification, phosphorylation, or formerly sialylated N-linked glycosylation (SA), respectively. The percentage of recovered genes of each cellular component are indicated by color bar (right in the middle). KEGG pathway nodes are colored according to the type of signaling pathway. The probability (EASE score; log10(p value)) of KEGG pathway annotation are indicated by node size (bottom). N: nonmodification. P: phosphorylation. S: formerly sialylated N-linked glycosylation.
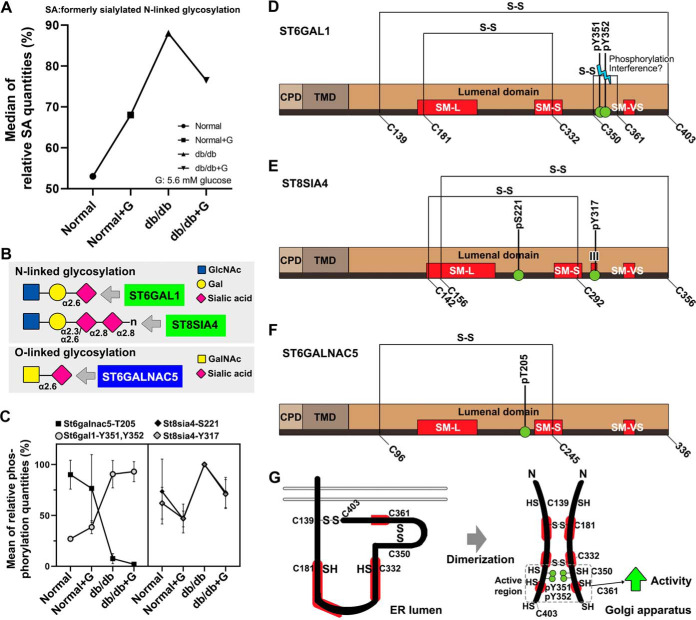
Modulation of Phosphorylation on Sialyltransferases
Sialylation of glycoproteins in db/db islets were associated with certain sialyltransferases and the distribution of the relative intensities of sialylation in db/db mouse islets were compared with controls (Fig. 5A). Similarly, the phosphorylation degree of ST6GAL1 (α2,6-sialyltransferases, Fig. 5B) at Y351 and Y352 was increased in the db/db islet groups compared with controls (left panel, Fig. 5C). In addition, the phosphorylation level of ST8SIA4 (α2,8-polysialyltransferases, Fig. 5B) at S221 and Y317 was elevated in db/db mouse islets (right panel, Fig. 5C). The latter phosphorylation at Y317 (D/N: 1.6 fold, DG/D: −1.5 fold; CV%: 35%) closely returned to baseline after normoglycemic culturing (right panel Fig 5C). Related to sialylated O-linked glycosylation, the phosphorylation of ST6GALNAC5 (α2,6-sialyltransferases, Fig. 5B) at T205 was down-regulated in db/db mouse islets compared with controls. Sialylmotifs L and S in sialyltransferases are related to substrate binding, and sialylmotifs III and VS are connected to catalytic activity (23). Two phosphosites in ST6GAL1 (Y351 and Y352) were closely located to cysteine residues (C350-C361) for potential S-S bond formation/interference environs of the sialylmotif VS (SM-VS) within the catalytic domain (Fig. 5D). In the luminal catalytic domain of ST8SIA4, the phosphorylation at Y317 was identified within sialylmotif III (Fig. 5E). The phosphosite (T205) of ST6GALNAC5 was closely localized to sialylmotif S (SM-S) within the catalytic domain (Fig. 5F) and could potentially influence phosphorylation-induced conformational change related with ST6GAL1 activity (Fig. 5G). Fig. 5G shows the putative effects of phosphorylations at Y351 and Y352 on the conformation and dimerization of the ST6GAL1 monomer (24, 44).
Fig. 5.
Significantly altered phosphorylations of sialyltransferases in db/db islets. A, line plot represents the median of the relative SA percentages in four groups showing the intensity of SA glycopeptides. B, plot chart represents the phosphorylation degree of significantly altered sialyltransferases between db/db islets and normal. Graph shows mean ± standard deviation. C, the N- or O-glycoform can be sialylated by significantly regulated sialyltransferases. N-acetylglucosamine (GlcNac), galactose (Gal), N-acetylgalactosamine (GalNac). D, Schematic representation of ST6GAL1 structure illustrating the cytoplasmic domain (CPD; beige), the plasmamembrane domain (PMD; maroon), the lumenal C-terminal catalytic domain (brown), and three region of sialyltransferase homology termed sialylmotifs (SM-L, SM-S, and SM-VS; red) within the catalytic domain. The S-S bonds and the altered phosphosites (green circle) located in the ST6GAL1 are indicated. E, ST8SIA4. F, ST6GALNAC5. G, Potential influence of phosphorylation- and dimerization-induced conformational alterations on the enzyme activity of ST6GAL1. Putative illustrations are the effects of the phosphorylations at Y351 and Y352 on the conformation and dimerization of ST6GAL1 monomer.
Signaling Pathways Linked to Changes in Protein Processing in the Endoplasmic Reticulum
To generate an overview of potential alterations in the signaling pathways of db/db mouse islet β-cells related to adaptive flexibility, we conducted DAVID GO analysis and KEGG pathway analysis of the regulated proteome, phosphoproteome, and sialiome (FDR < 0.05) (Fig. 6). We identified significant changes in protein processing in the ER across both PTMs (supplemental Fig. S3) linked to protein processing (Fig. 6). For proteome changes, two signaling pathways (aldosterone-regulated sodium reabsorption and metabolic pathway) and three subcellular components (proteasome, ribosome, and lysosome) were potentially linked to protein processing in the ER (upper-left panel, Fig. 6). Of these, ten proteasome proteins (Up-regulation in D/N: PSMB5, PSMB4, PSMB7, PSMB6, PSMB1, PSMA6, PSMA4, PSMA3, PSMD2, and PSMA7) and fourteen ribosome proteins [(Up-regulation: RPS27, MRPL41, GM5239, MRPS25, RPS27L, MRPS31, MRPS2, and MT3) and (Down-regulation: RPL18, RPS19, RPL13, RPL27, FAU, and RPL24)] were significantly changed in db/db mice, but not recovered to normal levels after exposure to euglycemia. For phosphoproteome changes, regulation of protein processing in the ER was interconnected with eleven regulatory signaling pathways (FoxO signaling, insulin signaling, regulation of actin cytoskeleton, endocytosis, cGMP-PKG signaling, spliceosome, RNA transport, protein export, phosphatidylinositol signaling, MAPK signaling, and HIF-1 signaling) (upper-right panel, Fig. 6). For sialiome changes, protein processing in the ER was connected with four signaling pathways (limited proteolysis, regulation of actin cytoskeleton, focal adhesion, and PI3K-Akt signaling) as well as the lysosome (lower-left panel, Fig. 6).
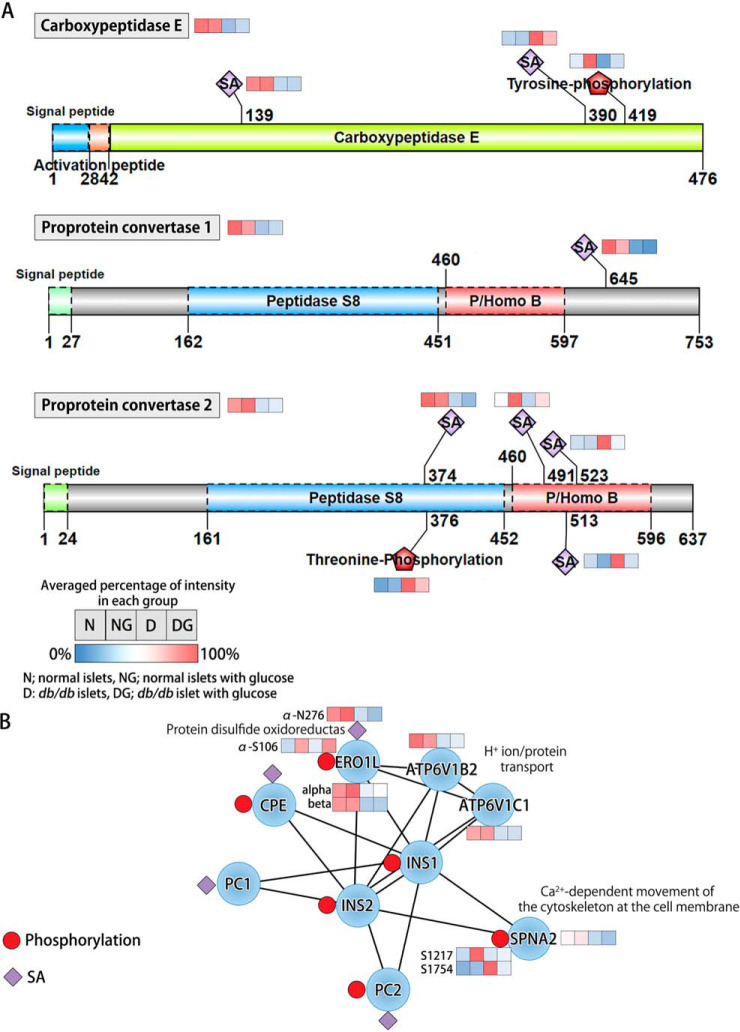
Post-translational Modifications of Proinsulin Processing Enzymes Are Altered in db/db Mouse Islets
The ratio of proinsulin to insulin in isolated db/db mouse islets could reflect accelerated secretory pathway activity or changes in proinsulin processing enzyme activities (7). We examined molecular changes of the three proteolytic enzymes that catalyze proinsulin conversion [Carboxypeptidase E (CPE), proprotein convertase 1 (PCSK1), and proprotein convertase 2 (PCSK2)] (45). PCSK1 (N645; −4.8 fold) and PCSK2 (N374; −2.3 fold) were significantly downregulated at the protein level in db/db islets, but not recovered toward a normal level after culture at normal glucose levels (Fig. 7A), indicating a possible association with the function of these enzymes. We also detected significant alterations of phosphorylation on CPE (Y419) and PCSK2 (T376) in db/db islets, but only CPE (Y419; D/N: −3.5 fold, DG/D: +3.0 fold; ∼86%) was returned to normal after culture at normal glucose (Fig. 7A). Which protein kinase might possibly phosphorylate CPE and PC2 was examined using a kinome analysis that links sequence specificity with cellular context using KinomeXplorer (46). Interestingly, CPE (Y419) and PCSK2 (T376) were most likely phosphorylated by SH3 domain tyrosine phosphorylation [Tensin-3; phosphorylation at S946 (+1.6 fold)] and protein kinase C beta [PRKCB isoform 2; phosphorylation at T641 (+1.6 fold)], respectively (supplemental Information S1). The phosphorylation degree of CPE at Y419 was significantly elevated in normal islets after euglycemic culturing. In addition, DG showed more significant increment in phosphorylation by ∼1.8 fold in the comparison between DG/D and NG/N, that is not necessarily a glucose effect. Furthermore, we observed changes in islet amyloid polypeptide (IAPP) in db/db islets both at the protein (down-regulation) and phosphorylation (up-regulation) levels (Fig. 2). IAPP protein expression was recovered after euglycemic culturing, not surprisingly paralleling the changes in insulin secretory activity because IAPP and insulin are co-secreted. However, the phosphorylation of IAPP at T46 and T67 did not revert to normal after euglycemic culturing (Fig. 2). The T46 and T67 on IAPP may be phosphorylated by the beta-adrenergic receptor kinase 1 [GRK2; protein expression (+1.4 fold); phosphorylation at Y92 (+6.2 fold); phosphorylation at S121 (+2.6 fold)] in db/db β-cells (supplemental Information S1).
Fig. 7.
Proinsulin processing. A, Dysregulation and recovery of proinsulin processing enzymes as proprotein convertase1, proprotein convertase 2, and carboxypeptidase E. B, Schematic representation of the relationships between insulin proteins and inter-connected proteins in the protein interaction network using STRING database.
CPE and PC1/2 were also identified to be associated with three insulin-modulating proteins [ERO1 like protein α/β (ERO1Lα/β), v-type proton ATPase subunit B (ATP6V1B2), and v-type proton ATPase subunit C (ATP6V1C1)] and a calcium-dependent calmodulin binding protein, spectrin alpha chain (SPNA2; Sptan1), all of which were decreased in db/db islets and did not recover upon euglycemic culturing (Fig 6B). Changes in the phosphorylation (S106) and sialylation (N276) on ERO1Lα were also observed. After exposure to euglycemia, phosphorylation (S106) was recovered, but sialylation (N276) did not return to normal, paralleling protein expression (Fig. 7B).
Kinome Profiling of Protein Kinases Related to Adaptive Flexibility of Islets
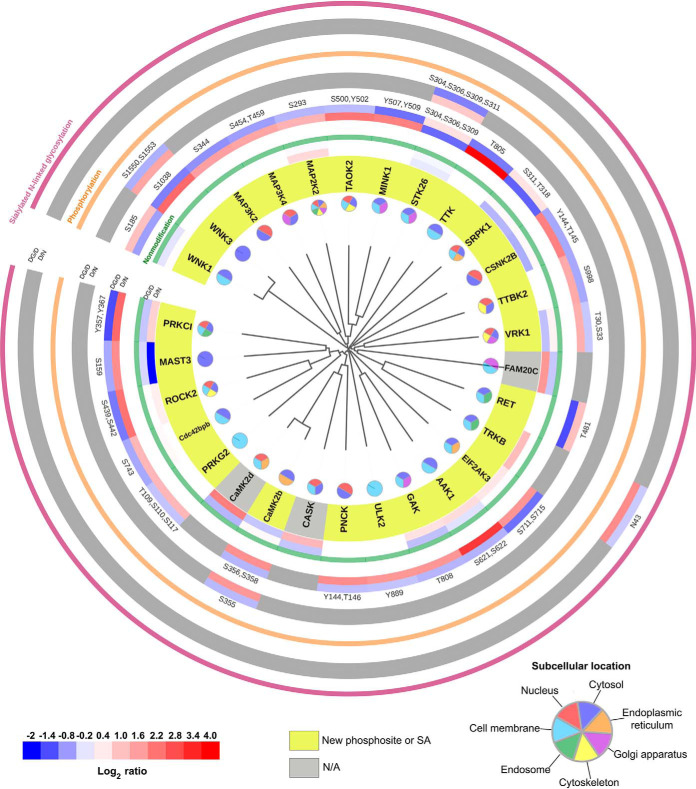
In a total of 378 kinases, we identified 162 kinases associated with changes between D and N pancreatic islets. We investigated 29 classified protein kinases that were significantly increased or decreased at either the protein expression or PTM level (Fig. 8 and supplemental Table S3). We used the FASTA sequences of the kinase domains retrieved from the KinBase resource and aligned them by ClustalX2.1 using default parameters for multiple alignments and bootstrapping N-J tree. Kinase sequences were visualized by phylogenetic distances using the Interactive Tree of Life (ITOL) tool (47). We presented the expression changes of 29 protein kinases according to the homology of the kinase domains at the protein and PTM levels (Fig. 8). Euglycemic recovered protein kinases were mostly found to be associated with the cell membrane, nucleus, ER or Golgi apparatus.
Fig. 8.
Significantly restored 29 kinases (opposite to D/N; at least log2 fold change ± 0.3 in DG/D) to normal status showing the homology of the kinase domains and protein posttranslational modifications. The rims indicate, the ratio of D/N (inner rim) or DG/D (outer rim) by color bar (bottom left) on recovered kinase in the respective PTM group such as nonmodification (green rim), phosphorylation (orange rim), or formerly sialylated N-linked glycosylation (SA, purple rim). D: db/db mouse islets, N: wild type mouse islets, DG: db/db mouse islets exposed to euglycemia. Among 29 restored kinases in phosphorylation or SA group, light yellow marked gene symbol indicate the newly identified phosphosite or SA site in PubMed. Pie chart for the restored kinase classification according to cellular component by Uniprot search (bottom right).
Significance of the Canonical Pathway for β-cell Dysfunction and Rest after Euglycemic Culture
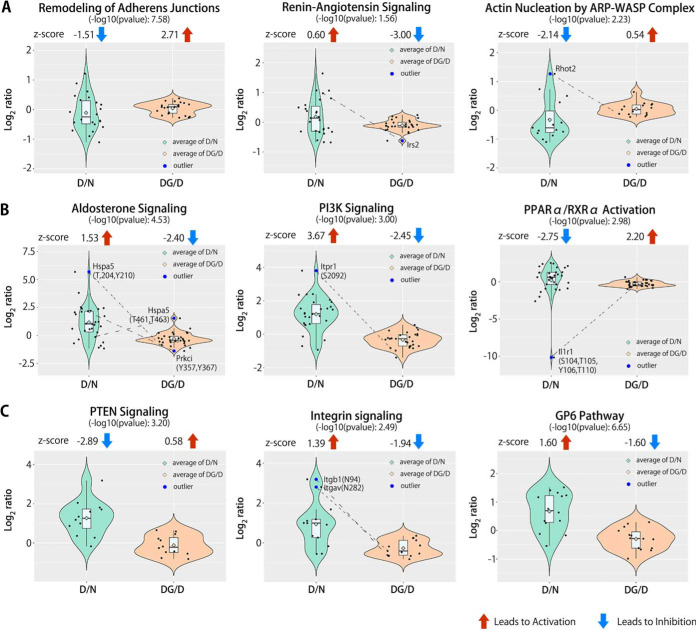
To reveal which canonical signaling pathways were activated or inhibited in db/db islets and those that were modulated by euglycemic rest, we conducted IPA analysis on all of the regulated proteins, phosphopeptides, or SA glycopeptides based on the statistical significance of the pattern match (11). In the protein group, we identified three overrepresented pathways (Fig. 9A). Remodeling of adherence junctions and actin nucleation by the ARP-WASP complex showed significant inhibition by z-scores (based on p values, as an index of confidence in the overlap) in isolated db/db islets compared with WT. After euglycemic culture, the two signaling pathways were inversely activated (Fig. 9A). In contrast, the renin-angiotensin signaling pathway showed activation by z-scores in isolated db/db islets, and returned to normal after euglycemic rest (Fig. 9A). From the phosphoproteome data, three significantly overrepresented signaling network pathways were observed (Fig. 9B). The aldosterone signaling pathway and PI3K signaling pathway showed activation by z-scores in isolated db/db mouse islets when compared with controls, and subsequently returned to normal after exposure to basal glucose levels (Fig. 9B). PPARα/RXRα activation showed significant inhibition by z-scores in isolated db/db mouse islets, and was activated after ex vivo culture at euglycemia (Fig. 9B). From the sialiome data, three overrepresented canonical pathways were apparent. The integrin signaling pathway and GP6 signaling pathway indicated substantial activation by z-scores and the PTEN signaling pathway showed predominant inhibition in isolated db/db mouse islets (Fig. 9C). In recovered db/db islets, the three signaling pathways tended to revert to normal versus WT controls (Fig 9C). We next investigated the crucial regulators in the representative signaling pathways using the inter-quartile range (IQR) test (less than quartile 1–1.5 × IQR, or greater than quartile 3 + 1.5 × IQR), which are either highly up-regulated or downregulated and thus may influence the overall observation from the overrepresented signaling pathways (11). Interestingly, two proteins [insulin receptor substrate 2 (IRS2), mitochondrial Rho GTPase 2 (RHOT2)], four phosphoproteins [endoplasmic reticulum chaperone BiP (HSPA5)-T204-Y210-T461-T463, protein kinase C iota type (PRKCI)-Y357-Y367, inositol 1,4,5-trisphosphate receptor type 1 (ITPR1)-S2092, and interleukin-1 receptor type 1 (IL1R1)-S104-T105-Y106-T110)], and two SA glycoproteins [integrin beta-1 (ITGB1)-N94 and integrin alpha-V (ITGAV)-N282] were identified in the overrepresented signaling pathways (Fig. 9). Furthermore, these proteins with PTMs were significantly regulated in isolated db/db mouse islets and recovered to normal following ex vivo euglycemic culture.
Fig. 9.
Dynamics of functional activity for canonical signaling pathways in the Ingenuity Pathway Analysis (IPA). A, For the protein group, combined violin plot with box plot shows significant inhibition or activation of z-scores in 3 overrepresented pathways based on p value (as an index of confidence in the overlap) as recovered canonical pathways to normal status in obese diabetic db/db mouse islets exposed to normal glucose concentrations. B, The phosphopeptides group. C, The SA peptides group. Significantly restored proteins or PTMs shown in box plot as outlier based on statistical significance of IQR test.
Validation of Some Regulated and Recovered Proteins using Parallel Reaction Monitoring (PRM)
Our global proteomics and PTMomics elucidated molecular and cellular signatures of islet dysfunction related to the production and secretion of insulin in db/db mouse islet β-cells, including the restoration of β-cell pathophysiology after exposure to euglycemic conditions for 12 h. We selected twenty regulated proteins and validated them in isolated islets from db/db and WT mice either fresh or after euglycemic rest using the PRM approach. Synthetic heavy isotope-labeled peptides were generated and used for internal quantitation standards for PRM validation of all four sample groups. We confirmed a high correlation for retention time (Pearson correlation of 0.999) between endogenous and heavy labeled peptides in all groups. By using the PRM strategy, we confirmed the expression levels of the 25 endogenous peptides, which correlated with the iTRAQ/TMT analysis (supplemental Fig. S4 and supplemental Table S4).
DISCUSSION
In this study, proteome and PTMome (sialylation and phosphorylation) analyses were used to map dynamic molecular changes in islets isolated from obese diabetic db/db mice. These changes were then related to the adaptive cellular response to compensate for the increased metabolic load triggered by obesity and an insulin-resistant state (7). A comprehensive analysis enabled us to explore the expression level of many proteins in islets from db/db and WT mice before and after β-cell rest, in conjunction with monitoring more than 11,685 phosphosites and 1325 formerly SA N-linked glycosites. In addition, our PTMomics data revealed dramatic multiple changes associated with previously identified notable biological and morphological alterations (e.g. enlarged RER and Golgi, (pro)insulin production and processing, insulin secreoty granule maturation, vesicle/granule trafficking and secretion, and controlled protein degradation) (6–9), which are related to β-cell adaptive flexibility. Some of these molecular events, especially in the phosphorylation data, will be casual/control of elements of β-cell adaptive flexibility and some will be consequential, but future studies will be needed to substantiate this differentiation. We hope the dataset presented here as well as find it useful resource for biologists in the diabetes area.
There are two closely related leptin receptor-deficient obese T2D db/db mouse models, C57BLKSdb/db and C57BL/6Jdb/db (7). The major difference between these models is that C57BL/6Jdb/db mice have an increased compensatory β-cell mass and that C57BLKSdb/db mice have a markedly decreased β-cell mass and thus more severe T2D despite a similar degree of obesity (7). However, at the level of β-cell biology, C57BLKSdb/db and C57BL/6Jdb/db mice are phenotypically similar, with both exhibiting a marked increase in proinsulin biosynthesis and processing, an almost total reduction of mature β-granules (the intracellular store of insulin), and a consequential large increase in insulin secretory pathway activity and exocytosis. This adaptability gives these animals a greater than 10-fold increase in basal hyperinsulinemia, an attempt to compensate for the inherent insulin resistance, hyperglycemia, and increased metabolic load related to obesity (6–9). In other words, the β-cells adapt to produce and rapidly secrete substantial amounts of insulin for this compensation (6, 7). Upon reducing glucose levels to normal and/or alleviating the insulin resistance in vivo, pancreatic β-cells rapidly restore their normal function and insulin secretory capacity (6, 7, 9). This result calls into question whether β-cell “dysfunction” in T2D might be symptomatic of this adaptive compensation and not necessarily be indicative of dysfunction (6). In this study, we used islets from C57BL/6Jdb/db mice because they contain many more β-cells versus C57BLKSdb/db mice and provide a better opportunity to observe changes in protein expression and PTMs specifically related to β-cell adaptive flexibility.
Nonetheless, there could be subtle differences between C57BLKSdb/db and C57BL/6Jdb/db mice, which may be related to differences in β-cell mass adaptation. For example, in C57BLKSdb/db mouse islets, phosphorylation of glycogen synthase kinase-3β (GSK3B) activity at S9 is decreased, and activation of phosphorylation of GSK3B activity at Y216 increases (48, 49). Our data on C57BL/6Jdb/db mouse islets indicate the dephosphorylation of GSK3A/B at Y216 and new phosphosites of GSK3A/B at S215 and S219, which themselves are increased. However, in relation to β-cell adaptive flexibility, similar mechanistic changes at the molecular level are likely to occur in both C57BLKSdb/db and C57BL/6Jdb/db mouse models. We previously demonstrated that the ER protein chaperone, BiP (also known as GPR78 or HSPA5), is elevated in isolated islets from both mouse models relative to the marked increase in proinsulin biosynthesis, but without inducing ER stress (7). Here, we have furthered those observations by identifying changes in proteins involved in unfolded protein-binding proteins, ER chaperones, effectors of proper protein folding, regulators of oxidative folding, oxidoreductases, and peroxidases in the C57BL/6Jdb/db islets that parallel β-cell adaptive flexibility. As such, our PTMome data reveal previously unknown molecular events related to this functional β-cell compensation in obesity and T2D.
Furthermore, our PTMomic and bioinformatic analyses complemented increased proinsulin biosynthesis translation in β-cell adaptive flexibility C57BL/6Jdb/db islets; protein synthesis, protein folding, cell redox homeostasis, and the UPR (not ER stress) are adjusted to meet metabolic demand mediated by glucose (6–9). In diabetes, overwhelming ER stress has been related to β-cell death as a result of abnormal protein folding in the ER (50). However, it is the UPR which contributes to the adaptive pathway processes, such as ER redox homeostasis, and improves the misfolded/unfolded protein response to facilitate functional and mature insulin production (50). In addition, the UPR can be activated because of an enhanced protein translation level (6). Correspondingly, in our comprehensive PTMomic map of cellular components we identified dynamic plasticity in the phosphorylation (50% of the regulated genes) and SA (70% of the regulated genes) states of proteins that regulate the UPR (not ER stress), biosynthesis and trafficking of proteins, cellular redox homeostasis, and protein folding in both the ER and Golgi apparatus after euglycemic rest in parallel (Fig. 3, 6, supplemental Fig. S3). The β-cell adaptive flexibility falls into translational control of proinsulin biosynthesis as a beneficial mechanism, implying a critical role for ER protein phosphorylation and SA in adjusting to ER redox homeostasis and appropriate protein folding to produce more insulin from C57BL/6Jdb/db islets to meet the metabolic demand.
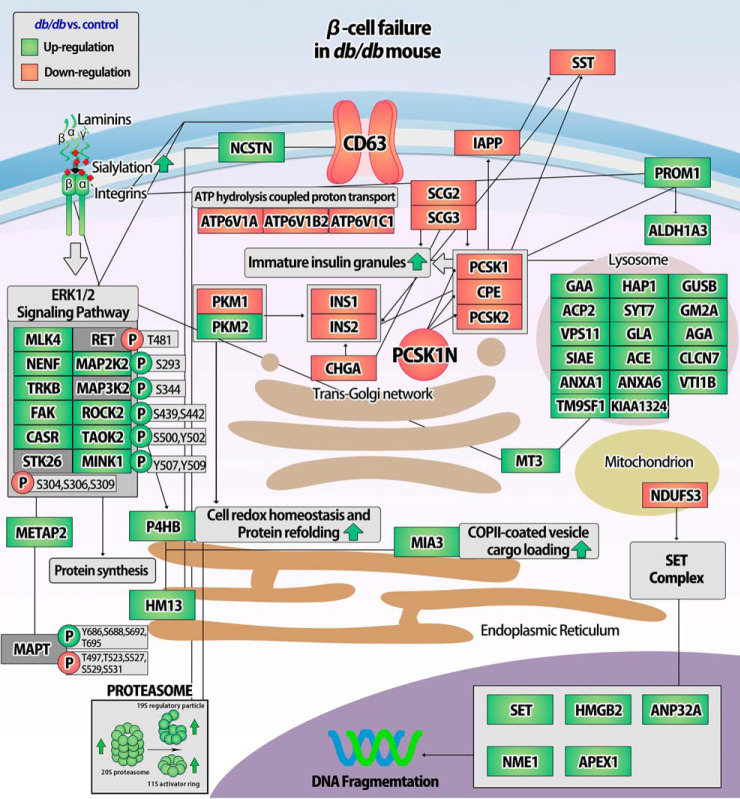
Proinsulin processing is complex and occurs at multiple stages of protein folding in the ER, including disulfide bond formation, ER to Golgi translocation through the secretory pathway, and β-granule biogenesis in parallel to proteolytic proinsulin-to-insulin conversion (6, 50, 51). Disulfide bond generation in proinsulin is necessary for the (pro)insulin maturation process, catalyzed by protein disulfide isomerases (PDIs) in the ER and the secretory pathway (52). Consequently, PDIs induce the ER export of properly folded proinsulin (53). For instance, P4HB (also known as PDIA1) is important for the oxidative maturation of proinsulin in the ER and insulin secretory granules (54). In the current study, we observed that the restoration of PDIs together with chaperones correlated with increased insulin production and mature insulin secretory granules (Fig. 3 and 7A) (7). Our MS-based targeted PRM approach shows that several key proteins are involved in proinsulin processing and β-granule biogenesis (Fig. 10 and supplemental Fig. S4). Of these proteins, CPE, PCSK1N, P4HB, INS1 (beta-chain), MIA3, and SST (SST-28) are involved in the conversion of proinsulin-to-insulin together with the maturation of insulin secretory granules (45, 53–55). Our PTMomics and PRM data demonstrate that the expression of proteins related to proinsulin processing and the maturation of insulin secretory granules is coordinately regulated and hence involved in the adaptive signaling mechanisms underlying these processes.
Fig. 10.
Schematic overview of regulated and validated proteins associated with β-cell failure in db/db mouse islets using PRM with synthetic peptides. (Green: Up-regulation, Red: Down-regulation).
Consistent with regulating the mature insulin granule population in β-cells (7), our integrated analysis indicates the complementary changes of the secretory pathway to facilitate vesicular trafficking and exocytosis in response to euglycemic and metabolic demand. We previously demonstrated that FAM20C is a key component in the secretory pathway and facilitates the preservation of ER redox homeostasis and the regulation of oxidative protein folding by ERO1Lα phosphorylation in the Golgi apparatus and retrograde transport back to the ER (Fig. 7B and 8) (10, 56, 57). Furthermore, we illustrated the relationship among the nuclear lamina, the microtubules mediated by the Linker of Nucleoskeleton and Cytoskeleton (LINC) protein complex, and the dynein and kinesin motor proteins involved in the trafficking of proteins or mRNAs. We previously demonstrated the dynamic organization of the nuclear pore complexes and protein/mRNA translocation in β-cells exposed to high glucose concentrations (58). Although this trafficking mechanism is not well understood in β-cells, the LINC complex proteins have been identified in cardiac and skeletal muscle diseases to play an important role in the LINC complex and motor protein function of trafficking proteins and mRNAs along microtubules (59). In general, dynein and kinesin proteins are the essential molecular motors involved in the intracellular transport of organelles, proteins, and mRNAs along microtubules (60, 61). In this study, we observed dynamic changes in protein-mRNA transport and vesicular trafficking along the microtubules in C57BL/6Jdb/db islets. This adaptability suggests that the LINC complex and motor proteins may regulate the trafficking of proinsulin and other insulin secretory granule proteins from the ER to the trans-Golgi network in β-cells. In addition, the modulation of focal adhesion contributes to the regulation of cytoskeletal structure, vesicle trafficking, and cell movement (62).
Thiol proteases involved in protein K48-linked deubiquitination are restored upon β-cell rest. The K48-linked polyubiquitin chains are a clear signal of protein degradation by the 26S proteasome (63). Deubiquitinating enzymes (DUBs) switch off the proteolytic system by deubiquitinating proteins to prevent chronic cellular stress (63). The K48-linked ubiquitination of proteins by DUBs is reversible and modulates protein degradation, DNA repair, chromatin remodeling, endocytosis, and the activity of the NF-κB and AMPK-related kinase signaling pathways (64). Insufficient UPR and protein degradation are implicated in T2D (65, 66). In the present study, we identified new phosphosites in seven DUBs that are dephosphorylated in C57BL/6Jdb/db islets after euglycemic rest. This may suggest that the protein quality control system differs between obese diabetic db/db and WT mice, with a tendency for protein degradation to be decreased after euglycemic rest.
Our PTMomic analysis allowed for an in-depth bioinformatic analysis identifying both known and unknown molecular events related to β-cell adaptive flexibility. The primary function of KCNMA1 (also known as SLO1) is the repolarization of action potentials in mouse pancreatic β-cells (67), and affects insulin secretion as well as the sensitivity of pancreatic β-cells to oxidative stress (68, 69). We identified that the phosphorylation of S711 in KCNMA1 and its 13 known interacting proteins are restored after euglycemic rest. These results highlight a previously unknown phosphosite of KCNMA1 associated with T2D. KCNMA1's interacting proteins in the CIM analysis may be involved in β-cell secretory function, but how phosphorylation of KCNMA1 contributes to β-cell adaptive flexibility requires further studies.
A previous genome-wide association study (GWAS) on T2D identified a specific relationship with a polymorphism (rs1535435 and rs9494266) for a gene encoding a unique AHI1 (Jouberin) protein (70), which was previously unknown in pancreatic β-cells. AHI1 is localized to the cytoskeleton and adherent junctions and is involved in vesicle trafficking and the regulation of protein secretion in neurons (71, 72). In our results, we have demonstrated that AHI1 recovers to a normal state after euglycemic rest. All downstream interacting proteins for AHI1 in the CIM analysis also correlated with β-cell adaptive flexibility. Although the role of AHI1 in β-cells has not yet been defined, our data could suggest that up-regulation of AHI1 or its phosphosites may be implicated in the trafficking and secretion of insulin in relation to adaptive compensation during T2D. Additional studies are required to better understand the exact roles of AHI1 and its protein-protein interactions related to β-cell adaptive flexibility.
Furthermore, our large-scale and targeted quantification analyses allowed the identification of twelve new proteins (CD63, NENF, PGP, HM13, PAIP2B, PKM [isotopes M1 and M2], PROM1, TOMM22, SET, PPP1R13B, and METAP2) likely related to β-cell adaptive flexibility in T2D (Fig. 10 and supplemental Fig. S4). These proteins are involved in the transport from the ER to the Golgi apparatus, transport from the trans-Golgi network to the plasma membrane, protein synthesis, protein transportation, vesicular trafficking, PI3K-AKT and ERK1/2 signaling pathways, and endopeptidase activity as annotated in the UniProt database. For instance, METAP2 is a metalloaminopeptidase associated with the co-translational cleavage of N-terminal methionine from nascent proteins, implying that it is involved in most cellular functions, including metabolism, protein synthesis, protein degradation, growth, and proliferation through binding to ERK1/2 and elongation initiation factor 2α (73–75). METAP2 inhibition has also been recently emphasized as a therapeutic strategy for treating diabetes, obesity, and associated metabolic disorders (76, 77). Thus, our data provide new insights into potential molecular targets that have beneficial effects on β-cell adaptability.
A marked increase in sialic acid on the cell membrane proteins found in C57BL/6Jdb/db islets correlates with the significant modulation of phosphorylation of sialyltransferases (78, 79). Alteration of ST8SIA4 phosphorylation at Y317 is restored toward a normal state in response to euglycemic rest, also correlating with β-cell adaptive flexibility. The sialylation of proteins may be associated with aberrant signaling in obesity and T2D (79–81). In addition, an expanded GWAS identified a T2D-specific association with a polymorphism (rs9820223) of a gene encoding a unique ST6GAL1 protein (82). We identified several significantly altered new phosphosites within the lumenal C-terminal catalytic domain in three sialyltransferases (ST6GAL1, ST8SIA4, and ST6GALNAC5), which could be related to abnormal sialic acid addition; the enzymatic activity of these sialyltransferases may be regulated by the phosphorylation- or dephosphorylation-induced S-S bond formation and dimerization or interference (24, 83, 84). Interestingly, Qian et al. have demonstrated that the disulfide-bonded dimer formed with ST6GAL1 (C139-C403 and C350-C361) decreases enzyme activity in the ER, whereas mutagenesis of C181 and C332 of ST6GAL1 causes predominant localization in the ER (24). In addition, mutation of C350S or C361S provokes the inactivation of ST6GAL1 without causing protein targeting to the ER or dimerization processing; these cysteine residues are a key component of the sialyltransferase's active site (24). In this study, the hyperphosphorylation of ST6GAL1 at Y351 and Y352 is localized near cysteine residues (C350-C361) for potential S-S bond generation or interference adjacent to the sialylmotif VS (SM-VS). Sialylmotifs L and S are involved in substrate binding, whereas sialylmotifs III and VS are associated with catalytic activity (23). However, future studies will be required to substantiate the idea of how the phosphorylation or dephosphorylation-induced catalytic activity of sialyltransferases contribute to the pathogenesis of T2D. ST6GAL1 and ST8SIA4 (phosphorylation at Y317 within SM-III) can catalyze α2,6/α2,8-sialylation of β1 integrin (85), consistent with elevated sialylation of β1 integrin in C57BL/6Jdb/db islets, suggesting that the elevated phosphorylation of ST6GAL1 and ST8SIA4 within SM-III and -VS could inhibit β1 integrin binding to matrix proteins, such as laminin. Integrin-mediated signaling is activated upon the binding of extracellular proteins, such as laminin, promoting activation of focal adhesion and the ERK1/2 signaling pathway (86). Our results also suggest that the hyperphosphorylation of ST6GAL1 and ST8SIA4 near the SM-VS/III sialylmotif could increase α2,6/α2,8-sialylation levels in the laminin-integrin signaling pathway to repress the ERK1/2 signaling pathway and focal adhesion, consequently causing defective insulin secretion in β-cells. Altered glycosylation of integrins, especially sialylation, is associated with structural changes that reduce cell adhesion to laminin and other ligands (85). Increased sialylation of β1 integrin is related to increased Ras activity and altered cell adhesion to collagen (87). In pancreatic β-cells, specific blocking of β1 integrin-laminin binding interferes with cell spreading and reduces the release of insulin in response to glucose (88). A laminin-integrin interaction may be required normal for glucose-stimulated insulin secretion in healthy β-cells (89). Consistent with the retrievability of sialylation on the laminin and integrin proteins, our results suggest that the sialylation levels in the laminin-integrin signaling pathway modulate the interplay between the ERK1/2 signaling pathway and focal adhesion, which impact correct insulin granule trafficking in β-cells. These results illustrate the potential influence of phosphorylation- and dephosphorylation-induced conformational changes on sialyltransferases in T2D, implying that sialylation is a key PTM in β-cell adaptive flexibility, capable of cross-talking with phosphorylation as we previously demonstrated (11).
Several overrepresented signaling pathways were highlighted in C57BL/6Jdb/db islets. Notably, the renin-angiotensin-aldosterone system (RAAS) is a biological process well associated with the development of obesity-linked T2D (90–92). Malfunctioning of the RAAS has previously been suggested to lead to insulin resistance by increasing cellular oxidative stress and impaired glucose metabolism in obesity and T2D. Activation of aldosterone signaling suppresses glucose-stimulated insulin secretion in vivo and in vitro (93). In the current study, we identified that renin-angiotensin signaling and aldosterone signaling are activated in C57BL/6Jdb/db islets and inhibited after euglycemic rest. These data suggest that restoration of the RAAS may play a protective role in preventing T2D progression.
In summary, we performed integrated large-scale, targeted, and bioinformatics analyses using our novel enrichment strategy of proteins, phosphopeptides, and SA N-linked glycopeptides, thereby identifying numerous dysfunctional biological processes related to the restoration of β-cell function to a normal state following euglycemic rest. By similarly treating WT islets in parallel, the effects of tissue culture could be mostly eliminated and only those changes associated with resolution by normal glucose in db/db islets were assessed (7). The scope of the study was to reveal changes in the proteome and PTMome of molecular pathways between db/db and normal islets as well as what happen to the β-cells when they undergo “β-cell-rest”. Keeping db/db islets and/or exposing WT islets to hyperglycemic conditions may yield additional interesting proteome/PTMome insight in β-cell biology, however, this was not within the scope of the study. Overall, in addition to identifying pathways for β-cell adaptive flexibility, we identified new adaptive proteins, phosphosites, and SA N-linked glycosites involved in the regulation of proinsulin biosynthesis translation, protein folding, not ER stress, proinsulin processing, β-granule biogenesis, vesicular trafficking and the insulin secretory pathway, protein degradation, and endopeptidase activity. These results demonstrate that β-cell adaptive flexibility may lead to improvement in endogenous β-cell function in C57BL/6Jdb/db islets. Our sialiomics analysis offers more in-depth insight into the laminin-integrin-mediated pathways between the ERK1/2 signaling pathway and focal adhesion. However, additional challenges remain regarding implementing our findings in diabetes studies. Although we observed β-cell adaptability as a positive effect in C57BL/6Jdb/db islets, observation of similar protein changes in diabetic human subjects would greatly support our findings. Our in-depth exploration of β-cell adaptive flexibility provided herein may contribute to the discovery of other potential β-cell specific therapeutic targets for diabetes.
DATA AVAILABILITY
Proteomics data was previously published (10) and is available via ProteomeXchange with identifier PXD010265. Phosphoroteomics and Sialiomics data are available via ProteomeXchange with identifier PXD017469, allow viewing of results (e.g., all spectra, chromatograms, identification, and quantification) using PRIDE Inspector (94). Skyline files from the PRM analysis were uploaded into Panorama Public (https://panoramaweb.org/piX2gg.url).
Supplementary Material
Acknowledgments
We thank Vibeke Jørgensen and Lene Jakobsen for their excellent technical support at SDU.
Footnotes
* This study was supported by the Danish Medical Research Council (grant number: 1331–00338B), AstraZeneca BioPharmaceuticals R&D, The Novo Nordisk Foundation (grant number: NNF16OC0023448) and the Villum Center for Bioanalytical Sciences at SDU, the Danish Diabetes Academy, and from the National Institutes of Health (NIH/NIDDK) R01DK50610 (C.J.R.). The authors have declared a conflict of interest. C.J.R., B.B.B., and J.S.G. are or were employees of AstraZeneca, LLC. T.K., P.J., C.A., A.N., and M.R.L. declare no competing interests.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- T2D
- type 2 diabetes
- PTMomics
- post-translational modification specific proteomic
- db/db
- obese leptin receptor-deficient mice
- WT
- C57BL/6J wild type
- SA
- formerly sialylated N-linked glycosylation
- PRM
- parallel reaction monitoring
- CV
- coefficient of variation
- Tio2
- titanium dioxide
- SIMAC
- deglycosylation step using the sequential elution from IMAC
- nLC-MS/MS
- nanoflow liquid chromatography-tandem mass spectrometry
- TMT
- tandem mass tags
- FDR
- false discovery rate
- GO
- gene ontology
- N
- normal wild type mouse islet
- NG
- normal islets with 5.6mM glucose
- D
- freshly isolated db/db mouse islet
- DG
- db/db mouse islet with 5.6mM glucose
- CIM
- circular interactome map
- UPR
- unfolded protein response.
REFERENCES
- 1. Shaw J. E., Sicree R. A., and Zimmet P. Z. (2010) Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res. Clin. Pract. 87, 4–14 [DOI] [PubMed] [Google Scholar]
- 2. Moller D. E. (2001) New drug targets for type 2 diabetes and the metabolic syndrome. Nature 414, 821–827 [DOI] [PubMed] [Google Scholar]
- 3. American Diabetes, A. (2005) Diagnosis and classification of diabetes mellitus. Diabetes care 28, S37–S42 [DOI] [PubMed] [Google Scholar]
- 4. Kahn S. E., Hull R. L., and Utzschneider K. M. (2006) Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444, 840–846 [DOI] [PubMed] [Google Scholar]
- 5. Rhodes C. J. (2005) Type 2 diabetes-a matter of beta-cell life and death? Science 307, 380–384 [DOI] [PubMed] [Google Scholar]
- 6. Boland B. B., Rhodes C. J., and Grimsby J. S. (2017) The dynamic plasticity of insulin production in beta-cells. Mol. Metab. 6, 958–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alarcon C., Boland B. B., Uchizono Y., Moore P. C., Peterson B., Rajan S., Rhodes O. S., Noske A. B., Haataja L., Arvan P., Marsh B. J., Austin J., and Rhodes C. J. (2016) Pancreatic beta-cell adaptive plasticity in obesity increases insulin production but adversely affects secretory function. Diabetes 65, 438–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boland B. B., Brown C. Jr, Alarcon C., Demozay D., Grimsby J. S., and Rhodes C. J. (2018) beta-Cell control of insulin production during starvation-refeeding in male rats. Endocrinology 159, 895–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boland B. B., Brown C. Jr, Boland M. L., Cann J., Sulikowski M., Hansen G., Gronlund R. V., King W., Rondinone C., Trevaskis J., Rhodes C. J., and Grimsby J. S. (2019) Pancreatic beta-cell rest replenishes insulin secretory capacity and attenuates diabetes in an extreme model of obese type 2 diabetes. Diabetes 68, 131–140 [DOI] [PubMed] [Google Scholar]
- 10. Kang T., Boland B. B., Alarcon C., Grimsby J. S., Rhodes C. J., and Larsen M. R. (2019) Proteomic analysis of restored insulin production and trafficking in obese diabetic mouse pancreatic islets following euglycemia. J. Proteome Res. [DOI] [PubMed] [Google Scholar]
- 11. Kang T., Jensen P., Huang H., Lund Christensen G., Billestrup N., and Larsen M. R. (2018) Characterization of the molecular mechanisms underlying glucose stimulated insulin secretion from isolated pancreatic beta-cells using post-translational modification specific proteomics (PTMomics). Mol. Cell Proteomics 17, 95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Engholm-Keller K., Birck P., Storling J., Pociot F., Mandrup-Poulsen T., and Larsen M. R. (2012) TiSH–a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J. Proteomics 75, 5749–5761 [DOI] [PubMed] [Google Scholar]
- 13. Taus T., Kocher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 14. Ma Z. Q., Dasari S., Chambers M. C., Litton M. D., Sobecki S. M., Zimmerman L. J., Halvey P. J., Schilling B., Drake P. M., Gibson B. W., and Tabb D. L. (2009) IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. Journal of Proteome Research 8, 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mi H. Y., Muruganujan A., Casagrande J. T., and Thomas P. D. (2013) Large-scale gene function analysis with the PANTHER classification system. Nat. Protocols 8, 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Szklarczyk D., Morris J. H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N. T., Roth A., Bork P., Jensen L. J., and von Mering C. (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N. H., Chavali G., Chen C., del-Toro N., Duesbury M., Dumousseau M., Galeota E., Hinz U., Iannuccelli M., Jagannathan S., Jimenez R., Khadake J., Lagreid A., Licata L., Lovering R. C., Meldal B., Melidoni A. N., Milagros M., Peluso D., Perfetto L., Porras P., Raghunath A., Ricard-Blum S., Roechert B., Stutz A., Tognolli M., van Roey K., Cesareni G., and Hermjakob H. (2014) The MIntAct project-IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, D358–D363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carr S. A., Abbatiello S. E., Ackermann B. L., Borchers C., Domon B., Deutsch E. W., Grant R. P., Hoofnagle A. N., Huttenhain R., Koomen J. M., Liebler D. C., Liu T., MacLean B., Mani D., Mansfield E., Neubert H., Paulovich A. G., Reiter L., Vitek O., Aebersold R., Anderson L., Bethem R., Blonder J., Boja E., Botelho J., Boyne M., Bradshaw R. A., Burlingame A. L., Chan D., Keshishian H., Kuhn E., Kinsinger C., Lee J. S. H., Lee S. W., Moritz R., Oses-Prieto J., Rifai N., Ritchie J., Rodriguez H., Srinivas P. R., Townsend R. R., Van Eyk J., Whiteley G., Wiita A., and Weintraub S. (2014) Targeted peptide measurements in biology and medicine: best practices for mass spectrometry- based assay development using a fit- for- purpose approach. Mol. Cell Proteomics 13, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schilling B., Rardin M. J., MacLean B. X., Zawadzka A. M., Frewen B. E., Cusack M. P., Sorensen D. J., Bereman M. S., Jing E. X., Wu C. C., Verdin E., Kahn C. R., MacCoss M. J., and Gibson B. W. (2012) Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline application to protein acetylation and phosphorylation. Mol. Cell Proteomics 11, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuan X., Serra R. A., and Yang S. Y. (2015) Function and regulation of primary cilia and intraflagellar transport proteins in the skeleton. Marrow 1335, 78–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Funabashi T., Katoh Y., Michisaka S., Terada M., Sugawa M., and Nakayama K. (2017) Ciliary entry of KIF17 is dependent on its binding to the IFT-B complex via IFT46-IFT56 as well as on its nuclear localization signal. Mol. Biol. Cell 28, 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takashima S., Matsumoto T., Tsujimoto M., and Tsuji S. (2013) Effects of amino acid substitutions in the sialylmotifs on molecular expression and enzymatic activities of alpha 2,8-sialyltransferases ST8Sia-I and ST8Sia-VI. Glycobiology 23, 603–612 [DOI] [PubMed] [Google Scholar]
- 24. Qian R., Chen C., and Colley K. J. (2001) Location and mechanism of alpha 2,6-sialyltransferase dimer formation. Role of cysteine residues in enzyme dimerization, localization, activity, and processing. J. Biol. Chem. 276, 28641–28649 [DOI] [PubMed] [Google Scholar]
- 25. Lu X. M., Tompkins R. G., and Fischman A. J. (2013) Nitric oxide activates intradomain disulfide bond formation in the kinase loop of Akt1/PKBalpha after burn injury. Int. J. Mol. Med. 31, 740–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang W. Y., and Linsdell P. (2012) Relative movements of transmembrane regions at the outer mouth of the cystic fibrosis transmembrane conductance regulator channel pore during channel gating. Journal of Biol. Chem. 287, 32136–32146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tobiume K., Saitoh M., and Ichijo H. (2002) Activation of apoptosis signal-regulating kinase 1 by the stress-induced activating phosphorylation of pre-formed oligomer. J. Cell Physiol. 191, 95–104 [DOI] [PubMed] [Google Scholar]
- 28. Salmeen A., and Barford D. (2005) Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid. Redox. Signal 7, 560–577 [DOI] [PubMed] [Google Scholar]
- 29. Zhang Z. Q., Shah B., and Richardson J. (2019) Impact of Fc N-glycan sialylation on IgG structure. Mabs-Austin 11, 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McGrath R. T., McKinnon T. A., Byrne B., O'Kennedy R., Terraube V., McRae E., Preston R. J., Laffan M. A., and O'Donnell J. S. (2010) Expression of terminal alpha2–6-linked sialic acid on von Willebrand factor specifically enhances proteolysis by ADAMTS13. Blood 115, 2666–2673 [DOI] [PubMed] [Google Scholar]
- 31. Bieberich E. (2014) Synthesis, processing, and function of N-glycans in N-glycoproteins. Adv. Neurobiol. 9, 47–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petit D., Teppa E., Cenci U., Ball S., and Harduin-Lepers A. (2018) Reconstruction of the sialylation pathway in the ancestor of eukaryotes. Sci. Rep. 8, 2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winchester B. (2005) Lysosomal metabolism of glycoproteins. Glycobiology 15:1R–15R, [DOI] [PubMed] [Google Scholar]
- 34. Gimenez-Roqueplo A. P., Celerier J., Lucarelli G., Corvol P., and Jeunemaitre X. (1998) Role of N-glycosylation in human angiotensinogen. J. Biol. Chem. 273, 21232–21238 [DOI] [PubMed] [Google Scholar]
- 35. Seales E. C., Jurado G. A., Brunson B. A., Wakefield J. K., Frost A. R., and Beilis S. L. (2005) Hypersialylation of beta(1) integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up-regulating cell motility. Cancer Res. 65, 4645–4652 [DOI] [PubMed] [Google Scholar]
- 36. Bellis S. L. (2004) Variant glycosylation: an underappreciated regulatory mechanism for beta1 integrins. Biochim. Biophys. Acta 1663, 52–60 [DOI] [PubMed] [Google Scholar]
- 37. Hart G. W. (1992) Glycosylation. Curr. Opin. Cell Biol. 4, 1017–1023 [DOI] [PubMed] [Google Scholar]
- 38. Wei A., Fan B., Zhao Y., Zhang H., Wang L., Yu X., Yuan Q., Yang D., and Wang S. (2016) ST6Gal-I overexpression facilitates prostate cancer progression via the PI3K/Akt/GSK-3beta/beta-catenin signaling pathway. Oncotarget 7, 65374–65388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hino M., Kijima-Suda I., Nagai Y., and Hosoya H. (2003) Glycosylation of the alpha and beta tubulin by sialyloligosaccharides. Zool. Sci. 20, 709–715 [DOI] [PubMed] [Google Scholar]
- 40. Bassaganas S., Perez-Garay M., and Peracaula R. (2014) Cell surface sialic acid modulates extracellular matrix adhesion and migration in pancreatic adenocarcinoma cells. Pancreas 43, 109–117 [DOI] [PubMed] [Google Scholar]
- 41. Ohtsubo K., Takamatsu S., Minowa M. T., Yoshida A., Takeuchi M., and Marth J. D. (2005) Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell 123, 1307–1321 [DOI] [PubMed] [Google Scholar]
- 42. Lamriben L., Graham J. B., Adams B. M., and Hebert D. N. (2016) N-glycan-based ER molecular chaperone and protein quality control system: the calnexin binding cycle. Traffic 17, 308–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Varki A. (2008) Sialic acids in human health and disease. Trends Mol. Med. 14, 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim J. W., Tang Q. Q., Li X., and Lane M. D. (2007) Effect of phosphorylation and S-S bond-induced dimerization on DNA binding and transcriptional activation by C/EBPbeta. Proc. Natl. Acad. Sci. U.S.A. 104, 1800–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guest P. C., Abdel-Halim S. M., Gross D. J., Clark A., Poitout V., Amaria R., Ostenson C. G., and Hutton J. C. (2002) Proinsulin processing in the diabetic Goto-Kakizaki rat. J. Endocrinol. 175, 637–647 [DOI] [PubMed] [Google Scholar]
- 46. Horn H., Schoof E. M., Kim J., Robin X., Miller M. L., Diella F., Palma A., Cesareni G., Jensen L. J., and Linding R. (2014) KinomeXplorer: an integrated platform for kinome biology studies. Nat. Methods 11, 603–604 [DOI] [PubMed] [Google Scholar]
- 47. Letunic I., and Bork P. (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bhat R. V., Shanley J., Correll M. P., Fieles W. E., Keith R. A., Scott C. W., and Lee C. M. (2000) Regulation and localization of tyrosine216 phosphorylation of glycogen synthase kinase-3beta in cellular and animal models of neuronal degeneration. Proc. Natl. Acad. Sci. U.S.A. 97, 11074–11079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sacco F., Seelig A., Humphrey S. J., Krahmer N., Volta F., Reggio A., Marchetti P., Gerdes J., and Mann M. (2019) Phosphoproteomics reveals the GSK3-PDX1 axis as a key pathogenic signaling node in diabetic islets. Cell Metabolism 29, 1422–1432.e3 [DOI] [PubMed] [Google Scholar]
- 50. Scheuner D., and Kaufman R. J. (2008) The unfolded protein response: A pathway that links insulin demand with beta-cell failure and diabetes. Endocr. Rev. 29, 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davidson H. W., Rhodes C. J., and Hutton J. C. (1988) Intraorganellar calcium and pH control proinsulin cleavage in the pancreatic beta cell via two distinct site-specific endopeptidases. Nature 333, 93–96 [DOI] [PubMed] [Google Scholar]
- 52. Weiss M., Steiner D. F., and Philipson L. H. (2000) Insulin Biosynthesis, Secretion, Structure, and Structure-Activity Relationships. In: Feingold K. R., Anawalt B., Boyce A., Chrousos G., Dungan K., Grossman A., Hershman J. M., Kaltsas G., Koch C., Kopp P., Korbonits M., McLachlan R., Morley J. E., New M., Perreault L., Purnell J., Rebar R., Singer F., Trence D. L., Vinik A., and Wilson D. P., eds. Endotext, South Dartmouth (MA) [PubMed] [Google Scholar]
- 53. Rajpal G., Schuiki I., Liu M., Volchuk A., and Arvan P. (2012) Action of protein disulfide isomerase on proinsulin exit from endoplasmic reticulum of pancreatic beta-cells. J. Biol. Chem. 287, 43–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jang I., Pottekat A., Poothong J., Yong J., Lagunas-Acosta J., Charbono A., Chen Z., Scheuner D. L., Liu M., Itkin-Ansari P., Arvan P., and Kaufman R. J. (2019) PDIA1/P4HB is required for efficient proinsulin maturation and ss cell health in response to diet induced obesity. Elife 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Strowski M. Z., Parmar R. M., Blake A. D., and Schaeffer J. M. (2000) Somatostatin inhibits insulin and glucagon secretion via two receptors subtypes: an in vitro study of pancreatic islets from somatostatin receptor 2 knockout mice. Endocrinology 141, 111–117 [DOI] [PubMed] [Google Scholar]
- 56. Pollak A. J., Liu C., Gudlur A., Mayfield J. E., Dalton N. D., Gu Y., Chen J., Heller Brown J., Hogan P. G., Wiley S. E., Peterson K. L., and Dixon J. E. (2018) A secretory pathway kinase regulates sarcoplasmic reticulum Ca(2+) homeostasis and protects against heart failure. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang J., Zhu Q., Wang X., Yu J., Chen X., Wang J., Wang X., Xiao J., Wang C. C., and Wang L. (2018) Secretory kinase Fam20C tunes endoplasmic reticulum redox state via phosphorylation of Ero1alpha. EMBO J. 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kang T., Jensen P., Solovyeva V., Brewer J. R., and Larsen M. R. (2018) Dynamic changes in the protein localization in the nuclear environment in pancreatic beta-cell after brief glucose stimulation. J. Proteome Res. 17, 1664–1676 [DOI] [PubMed] [Google Scholar]
- 59. Stroud M. J., Banerjee I., Veevers J., and Chen J. (2014) Linker of nucleoskeleton and cytoskeleton complex proteins in cardiac structure, function, and disease. Circ. Res. 114, 538–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hirokawa N., Noda Y., Tanaka Y., and Niwa S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Bio. 10, 682–696 [DOI] [PubMed] [Google Scholar]
- 61. Caviston J. P., and Holzbaur E. L. F. (2006) Microtubule motors at the intersection of trafficking and transport. Trends in Cell Biol. 16, 530–537 [DOI] [PubMed] [Google Scholar]
- 62. Parsons J. T., Horwitz A. R., and Schwartz M. A. (2010) Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Welchman R. L., Gordon C., and Mayer R. J. (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Bio. 6, 599–609 [DOI] [PubMed] [Google Scholar]
- 64. Reyes-Turcu F. E., Ventii K. H., and Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bugliani M., Liechti R., Cheon H., Suleiman M., Marselli L., Kirkpatrick C., Filipponi F., Boggi U., Xenarios I., Syed F., Ladriere L., Wollheim C., Lee M. S., and Marchetti P. (2013) Microarray analysis of isolated human islet transcriptome in type 2 diabetes and the role of the ubiquitin-proteasome system in pancreatic beta cell dysfunction. Mol. Cell Endocrinol. 367, 1–10 [DOI] [PubMed] [Google Scholar]
- 66. Quan W., Hur K. Y., Lim Y., Oh S. H., Lee J. C., Kim K. H., Kim G. H., Kim S. W., Kim H. L., Lee M. K., Kim K. W., Kim J., Komatsu M., and Lee M. S. (2012) Autophagy deficiency in beta cells leads to compromised unfolded protein response and progression from obesity to diabetes in mice. Diabetologia 55, 392–403 [DOI] [PubMed] [Google Scholar]
- 67. Smith P. A., Bokvist K., Arkhammar P., Berggren P. O., and Rorsman P. (1990) Delayed rectifying and calcium-activated K+ channels and their significance for action-potential repolarization in mouse pancreatic beta-cells. J. Gen. Physiol. 95, 1041–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dufer M., Neye Y., Horth K., Krippeit-Drews P., Hennige A., Widmer H., McClafferty H., Shipston M. J., Haring H. U., Ruth P., and Drews G. (2011) BK channels affect glucose homeostasis and cell viability of murine pancreatic beta cells. Diabetologia 54, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rorsman P., and Braun M. (2013) Regulation of insulin secretion in human pancreatic islets. Annu. Rev. Physiol. 75, 155–179 [DOI] [PubMed] [Google Scholar]
- 70. Salonen J. T., Uimari P., Aalto J. M., Pirskanen M., Kaikkonen J., Todorova B., Hypponen J., Korhonen V. P., Asikainen J., Devine C., Tuomainen T. P., Luedemann J., Nauck M., Kerner W., Stephens R. H., New J. P., Ollier W. E., Gibson J. M., Payton A., Horan M. A., Pendleton N., Mahoney W., Meyre D., Delplanque J., Froguel P., Luzzatto O., Yakir B., and Darvasi A. (2007) Type 2 diabetes whole-genome association study in four populations: The DiaGen consortium. Am. J. Hum. Genet. 81, 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hsiao Y. C., Tong Z. J., Westfall J. E., Ault J. G., Page-McCaw P. S., and Ferland R. J. (2009) Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking. Human Mol. Gen. 18, 3926–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sheng G., Xu X., Lin Y. F., Wang C. E., Rong J., Cheng D., Peng J., Jiang X., Li S. H., and Li X. J. (2008) Huntingtin-associated protein 1 interacts with Ahi1 to regulate cerebellar and brainstem development in mice. J. Clin. Invest. 118, 2785–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bennett B., and Holz R. C. (1997) EPR studies on the mono- and dicobalt(II)-substituted forms of the aminopeptidase from Aeromonas proteolytica. Insight into the catalytic mechanism of dinuclear hydrolases. J. Am. Chem. Soc. 119, 1923–1933 [Google Scholar]
- 74. Leopoldini M., Russo N., and Toscano M. (2007) Which one among Zn(II), Co(II), Mn(II), and Fe(II) is the most efficient ion for the methionine aminopeptidase catalyzed reaction? J. Am. Chem. Soc. 129, 7776–7784 [DOI] [PubMed] [Google Scholar]
- 75. Datta B., Majumdar A., Datta R., and Balusu R. (2004) Treatment of cells with the angiogenic inhibitor fumagillin results in increased stability of eukaryotic initiation factor 2-associated glycoprotein, p67, and reduced phosphorylation of extracellular signal-regulated kinases. Biochemistry 43, 14821–14831 [DOI] [PubMed] [Google Scholar]
- 76. Burkey B. F., Hoglen N. C., Inskeep P., Wyman M., Hughes T. E., and Vath J. E. (2018) Preclinical efficacy and safety of the novel antidiabetic, antiobesity MetAP2 inhibitor ZGN-1061. J. Pharmacol. Exp. Ther. 365, 301–313 [DOI] [PubMed] [Google Scholar]
- 77. Malloy J., Zhuang D. L., Kim T., Inskeep P., Kim D., and Taylor K. (2018) Single and multiple dose evaluation of a novel MetAP2 inhibitor: Results of a randomized, double-blind, placebo-controlled clinical trial. Diabetes Obesity Metabolism 20, 1878–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Breen K. C., and Georgopoulou N. (2003) The role of protein phosphorylation in alpha2,6(N)-sialyltransferase activity. Biochem. Biophys. Res. Commun. 309, 32–35 [DOI] [PubMed] [Google Scholar]
- 79. Gu X. B., Preuss U., Gu T. J., and Yu R. K. (1995) Regulation of sialyltransferase activities by phosphorylation and dephosphorylation. J. Neurochem. 64, 2295–2302 [DOI] [PubMed] [Google Scholar]
- 80. Reily C., Stewart T. J., Renfrow M. B., and Novak J. (2019) Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee C. L., Chiu P. C. N., Pang P. C., Chu I. K., Lee K. F., Koistinen R., Koistinen H., Seppala M., Morris H. R., Tissot B., Panico M., Dell A., and Yeung W. S. B. (2011) Glycosylation failure extends to glycoproteins in gestational diabetes mellitus evidence from reduced alpha 2–6 sialylation and impaired immunomodulatory activities of pregnancy-related glycodelin-A. Diabetes 60, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Scott R. A., Scott L. J., Maegi R., Marullo L., Gaulton K. J., Kaakinen M., Pervjakova N., Pers T. H., Johnson A. D., Eicher J. D., Jackson A. U., Ferreira T., Lee Y., Ma C., Steinthorsdottir V., Thorleifsson G., Qi L., Van Zuydam N. R., Mahajan A., Chen H., Almgren P., Voight B. F., Grallert H., Mueller-Nurasyid M., Ried J. S., Rayner N. W., Robertson N., Karssen L. C., Van Leeuwen E. M., Willems S. M., Fuchsberger C., Kwan P., Teslovich T. M., Chanda P., Li M., Lu Y., Dina C., Thuillier D., Yengo L., Jiang L., Sparso T., Kestler H. A., Chheda H., Eisele L., Gustafsson S., Franberg M., Strawbridge R. J., Benediktsson R., Hreidarsson A. B., Kong A., Sigurdsson G., Kerrison N. D., Luan J., Liang L., Meitinger T., Roden M., Thorand B., Esko T., Mihailov E., Fox C., Liu C. T., Rybin D., Isomaa B., Lyssenko V., Tuomi T., Couper D. J., Pankow J. S., Grarup N., Have C. T., Jorgensen M. E., Jorgensen T., Linneberg A., Cornelis M. C., Van Dam R. M., Hunter D. J., Kraft P., Sun Q., Edkins S., Owen K. R., Perry J. R. B., Wood A. R., Zeggini E., Tajes-Fernandes J., Abecasis G. R., Bonnycastle L. L., Chines P. S., Stringham H. M., Koistinen H. A., Kinnunen L., Sennblad B., Muehleisen T. W., Noethen M. M., Pechlivanis S., Baldassarre D., Gertow K., Humphries S. E., Tremoli E., Klopp N., Meyer J., Steinbach G., Wennauer R., Eriksson J. G., Mannisto S., Peltonen L., Tikkanen E., Charpentier G., Eury E., Lobbens S., Gigante B., Leander K., McLeod O., Bottinger E. P., Gottesman O., Ruderfer D., Blueher M., Kovacs P., Tonjes A., Maruthur N. M., Scapoli C., Erbel R., Joeckel K. H., Moebus S., De Faire U., Hamsten A., Stumvoll M., Deloukas P., Donnelly P. J., Frayling T. M., Hattersley A. T., Ripatti S., Salomaa V., Pedersen N. L., Boehm B. O., Bergman R. N., Collins F. S., Mohlke K. L., Tuomilehto J., Hansen T., Pedersen O., Barroso I., Lannfelt L., Ingelsson E., Lind L., Lindgren C. M., Cauchi S., Froguel P., Loos R. J. F., Balkau B., Boeing H., Franks P. W., Gurrea A. B., Palli D., Van der Schouw Y. T., Altshuler D., Groop L. C., Langenberg C., Wareham N. J., Sijbrands E., Van Duijn C. M., Florez J. C., Meigs J. B., Boerwinkle E., Gieger C., Strauch K., Metspalu A., Morris A. D., Palmer C. N. A., Hu F. B., Thorsteinsdottir U., Stefansson K., Dupuis J., Morris A. P., Boehnke M., McCarthy M. I., Prokopenko I., and Replicat D. G. (2017) An expanded genome-wide association study of Type 2 diabetes in Europeans. Diabetes 66, 2888–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sato C., and Kitajima K. (2013) Impact of structural aberrancy of polysialic acid and its synthetic enzyme ST8SIA2 in schizophrenia. Front. Cell Neurosci. 7, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bhide G. P., and Colley K. J. (2017) Sialylation of N-glycans: mechanism, cellular compartmentalization and function. Histochem. Cell Biol. 147, 149–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chammas R., Veiga S. S., Travassos L. R., and Brentani R. R. (1993) Functionally distinct roles for glycosylation of alpha-integrin and beta-integrin chains in cell matrix interactions. Proc. Natl. Acad. Sci. U.S.A. 90, 1795–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Stupack D. G., and Cheresh D. A. (2002) Get a ligand, get a life: integrins, signaling and cell survival. J. Cell Sci. 115, 3729–3738 [DOI] [PubMed] [Google Scholar]
- 87. Seales E. C., Jurado G. A., Singhal A., and Bellis S. L. (2003) Ras oncogene directs expression of a differentially sialylated, functionally altered beta 1 integrin. Oncogene 22, 7137–7145 [DOI] [PubMed] [Google Scholar]
- 88. Parnaud G., Hammar E., Rouiller D. G., Armanet M., Halban P. A., and Bosco D. (2006) Blockade of beta 1 integrin-laminin-5 interaction affects spreading and insulin secretion of rat beta-cells attached on extracellular matrix. Diabetes 55, 1413–1420 [DOI] [PubMed] [Google Scholar]
- 89. Bosco D., Meda P., Halban P. A., and Rouiller D. G. (2000) Importance of cell-matrix interactions in rat islet beta-cell secretion in vitro - Role of alpha 6 beta 1 integrin. Diabetes 49, 233–243 [DOI] [PubMed] [Google Scholar]
- 90. Cooper M. E. (2004) The role of the renin-angiotensin-aldosterone system in diabetes and its vascular complications. Am. J. Hypertens. 17, 16S–20S; quiz A12–14 [DOI] [PubMed] [Google Scholar]
- 91. Sauter N. S., Thienel C., Plutino Y., Kampe K., Dror E., Traub S., Timper K., Bedat B., Pattou F., Kerr-Conte J., Jehle A. W., Boni-Schnetzler M., and Donath M. Y. (2015) Angiotensin II induces interleukin-1beta-mediated islet inflammation and beta-cell dysfunction independently of vasoconstrictive effects. Diabetes 64, 1273–1283 [DOI] [PubMed] [Google Scholar]
- 92. Chu K. Y., Lau T., Carlsson P. O., and Leung P. S. (2006) Angiotensin II type 1 receptor blockade improves beta-cell function and glucose tolerance in a mouse model of type 2 diabetes. Diabetes 55, 367–374 [DOI] [PubMed] [Google Scholar]
- 93. Luther J. M., and Brown N. J. (2011) The renin-angiotensin-aldosterone system and glucose homeostasis. Trends Pharmacol. Sci. 32, 734–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Perez-Riverol Y., Xu Q. W., Wang R., Uszkoreit J., Griss J., Sanchez A., Reisinger F., Csordas A., Ternent T., Del-Toro N., Dianes J. A., Eisenacher M., Hermjakob H., and Vizcaino J. A. (2016) PRIDE inspector toolsuite: moving toward a universal visualization tool for proteomics data standard formats and quality assessment of ProteomeXchange datasets. Mol. Cell Proteomics 15, 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data was previously published (10) and is available via ProteomeXchange with identifier PXD010265. Phosphoroteomics and Sialiomics data are available via ProteomeXchange with identifier PXD017469, allow viewing of results (e.g., all spectra, chromatograms, identification, and quantification) using PRIDE Inspector (94). Skyline files from the PRM analysis were uploaded into Panorama Public (https://panoramaweb.org/piX2gg.url).