Abstract
Leishmania is a parasitic protozoon responsible for the neglected tropical disease Leishmaniasis. Approximately, 350 million people are susceptible and close to 70,000 death cases globally are reported annually. The lack of effective leishmanicides, the emergence of drug resistance and toxicity concerns necessitate the pursuit for effective antileishmanial drugs. Natural compounds serve as reservoirs for discovering new drugs due to their chemical diversity. Hardwickiic acid (HA) isolated from the stembark of Croton sylvaticus was evaluated for its leishmanicidal potential against Leishmania donovani and L. major promastigotes. The susceptibility of the promastigotes to HA was determined using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide/phenazine methosulfate colorimetric assay with Amphotericin B serving as positive control. HA showed a significant antileishmanial activity on L. donovani promastigotes with an IC50 value of 31.57± 0.06 µM with respect to the control drug, amphotericin B with IC50 of 3.35 ± 0.14 µM). The cytotoxic activity was observed to be CC50 = 247.83 ± 6.32 µM against 29.99 ± 2.82 µM for curcumin, the control, resulting in a selectivity index of SI = 7.85. Molecular modeling, docking and dynamics simulations of selected drug targets corroborated the observed antileishmanial activity of HA. Novel insights into the mechanisms of binding were obtained for trypanothione reductase (TR), pteridine reductase 1 (PTR1), and glutamate cysteine ligase (GCL). The binding affinity of HA to the drug targets LmGCL, LmPTR1, LdTR, LmTR, LdGCL, and LdPTR1 were obtained as -8.0, -7.8, -7.6, -7.5, -7.4 and -7.1 kcal/mol, respectively. The role of Lys16, Ser111, and Arg17 as critical residues required for binding to LdPTR1 was reinforced. HA was predicted as a Caspase-3 stimulant and Caspase-8 stimulant, implying a possible role in apoptosis, which was shown experimentally that HA induced parasite death by loss of membrane integrity. HA was also predicted as antileishmanial molecule corroborating the experimental activity. Therefore, HA is a promising antileishmanial molecule worthy of further development as a biotherapeutic agent.
Keywords: Croton sylvaticus, hardwickiic acid, leishmaniasis, structural modeling, molecular docking, trypanothione reductase, pteridine reductase 1, glutamate cysteine ligase
Graphical Abstract.
Methodology schema for the evaluation of the leishmanicidal potential of hardwickiic acid.
Introduction
Leishmaniasis which is caused by Leishmania is one of the most neglected tropical diseases needing urgent attention (Feasey et al., 2010; de Vries et al., 2015). It is endemic in about 90 countries with 350 million people at risk of infection and has an annual mortality rate of 70,000 cases globally (Alvar et al., 2012; de Vries et al., 2015; Georgiadou et al., 2016). According to the World Health Organization (WHO), there are about 700,000 to close to a million new cases of both cutaneous leishmaniasis (CL) and visceral leishmaniasis (VL) annually (WHO, 2019). Leishmaniasis is spread to humans after being bitten by infected phlebotomine sand flies (Kamhawi, 2006; Bates, 2007; Corrales et al., 2010; Dostálová and Volf, 2012; Ready, 2013). Generally, leishmaniases are endemic among the under-developed geographic regions and the disease is recognized by the development of single or multiple lesions (WHO, 2018).
The lack of effective and affordable therapeutics is hindering the eradication of leishmaniasis. Currently, therapeutic interventions for controlling leishmaniasis include amphotericin B and its liposomal formulation known as AmBisome, as well as pentavalent antimonial, miltefosine, pentamidine and paromomycin. Also, combination therapy of existing drugs is a preferred treatment option. However, these drugs have been reported to pose serious toxicity challenges and the emergence of drug-resistant strains has become prevalent (Tiwari et al., 2017; Tiwari and Dubey, 2018). The inherent differences in drug sensitivity shown by the different species could be an explanation to the resistance phenomenon (Mandal et al., 2015). Till date, there is no suitable vaccine to entirely eliminate leishmaniasis (Ghorbani and Farhoudi, 2018).
The lack of effective drugs, the emergence of drug resistance, drug toxicity concerns and absence of cost-effectiveness necessitate the search for effective therapeutic molecules and new drug targets (Chawla and Madhubala, 2010). Various efforts have been geared towards finding cures to diseases of parasitic origin (Crowther et al., 2010). The dissimilarities in human and homology amongst parasites have affected the prioritization of drug targets and drug candidates due to reduced interference of these drugs with host systems (Lamotte et al., 2017). Metabolic pathways of the various Leishmania species have been studied in the past decades to elucidate potential targets. Trypanothione reductase (TR), pteridine reductase 1 (PTR1), and glutamate cysteine ligase (GCL) have been identified as plausible targets.
TR is essential for the survival of the parasites in hosts since it is responsible for converting trypanothione disulfide T[S]2 to its dithiol T[SH]2 moiety (Saccoliti et al., 2017), which is required by the tryparedoxin-tryparedoxin peroxidase system for the neutralization of hydrogen peroxide generated during infection by the macrophages of the host (Colotti et al., 2013; Turcano et al., 2018). TR-knockout mutants engineered through gene disruption in L. donovani and L. major strains decreases infectivity thereby stagnating the intracellular survival of the parasites in macrophages (Dumas et al., 1997; Tovar et al., 1998). TR is an attractive drug target for selective inhibition since it plays essential role in immunology mechanisms and pathogenesis.
The growth of the parasite as well as that of trypanosomatid protozoa depends on reduced pteridines especially pterins and folates. PTR1 which is an NADPH-dependent short-chain reductase present in parasitic trypanosomatid protozoans takes part in salvaging pterins (Schüttelkopf et al., 2005). Amplification of PTR1 is critical to the resistance of the parasite to antifolates (Nare et al., 1997b). PTR1 acts as a metabolic bypass for drugs targeting dihydrofolate reductase (DHFR) (Kumar et al., 2008). Since PTR1 has been shown to be less sensitive to antifolates which primarily target DHFR (Nare et al., 1997a), any treatment with antifolate in Leishmania should also target PTR1.
GCL is involved in the biosynthesis of trypanothione (TSH) (Mukherjee et al., 2009; Kyriazis et al., 2016). TSH enables the parasite to avoid the deleterious effects of nitric oxide (NO) and reactive oxygen species (ROS), which are generated by the macrophages as part of the host's defense against leishmaniasis (Mukherjee et al., 2009; Kyriazis et al., 2016). An amplification or overexpression of this enzyme has also been shown to increase in vitro resistance of Leishmania to antimonials (Grondin et al., 1997).
Constituents of plants affords limitless opportunities for discovering novel drugs due to the unparalleled diversity of chemical libraries (Carlson, 2010; Lahlou, 2013). Traditional medicine serves as a remedy for over 80% of the global population (Sasidharan et al., 2011). The constituents of plant extracts are natural products for treating plethora of diseases. The phytochemical screening of the constituents serve as the precursor for new therapeutics needed to treat disease (Sasidharan et al., 2011).
The Crotons (Euphorbiaceae) represent around 1300 species of trees, shrubs, and herbs widely distributed in the tropics (Salatino et al., 2007; Xu et al., 2018). They have a rich history of ethnomedicinal uses including malaria, inflammation, tuberculosis, and stomach upset (Salatino et al., 2007). Extensive phytochemical investigations have identified multiple classes of secondary metabolites, predominantly terpenoids and sterols, with diverse pharmacological applications such as antimicrobial, cytotoxicity, anti-inflammatory, antioxidant, antinociceptive, molluscicidal, and wound healing (Jassbi, 2006). Hardwickiic acid (HA) has been isolated from many Croton species including C. sonderianus, C. aromaticus, and C. oblongifolius (Chaichantipyuth et al., 2004). HA also exhibits antimicrobial and insecticidal activities (Bandara et al., 1987; McChesney et al., 1991).
Due to the enormous structural and biological diversity of the chemical constituents, the genus continues to attract remarkable attention (Shi et al., 2008). However, one of the research gaps in existing literature on the Crotons is a lack of correlation between bioactive molecules and their mechanisms of action. In this paper, we present the leishmanicidal potential of hardwickiic acid (HA) isolated from the stembark of Croton sylvaticus by evaluating its biological activity. Also, molecular modeling and docking studies were performed on trypanothione reductase (TR), pteridine reductase 1 (PTR1), and glutamate cysteine ligase (GCL) of both L. donovani and L. major in order to elucidate the binding mechanisms between HA and drug targets. This study also sought to determine novel critical active site residues of LdTR, LmTR, LdGCL, and LmGCL involved in HA binding. More so, sought to predict potential mechanisms of action of HA by using an Open Bayesian-based approach.
Materials and Methods
Plant Material and Isolation
A semi-purified fraction of a previously petroleum ether-extracted stembark of Croton sylvaticus (Mwangi et al., 1998) was analyzed in this work. The plant material was collected at Mombasa in 1989 and identified by Mr. G. Mungai of the East African Herbarium based in Nairobi, Kenya. Voucher specimens were deposited at the Faculty of Pharmacy in the University of Nairobi in Kenya.
The Thin-layer Chromatographic (TLC) profile of the extract, developed in petrol-EtOAc (3:7), indicated one major spot and three others. In order to separate the constituents, 0.535 g of the dried light brown powdery fraction was column chromatographed on silica gel, eluting with petrol and petrol-EtOAc mixtures. A total of fifty-seven 30 ml eluents were collected and pooled into nine fractions F1–F9 based on their TLC profiles. On drying under vacuum, solids precipitated from fractions F1 (235 mg), F2 (0.9 mg), and F6 (0.5 mg). Due to paucity of material, further characterization was conducted on F1 only to afford hardwickiic acid (HA). Analytical liquid chromatography–mass spectrometry (LCMS) analysis was undertaken using an Agilent equipped with Kinetex Core C18, 2.6 µm, 3 x 50 mm, 100 Å maintained at 40 °C and DAD UV-detector (220, 254 and 300 nm). The mobile phase was 10 mM NH4OAc in 90% MeOH in H2O. The ionization technique was Jet Stream-Electrospray in positive mode. Nuclear Magnetic Resonance (NMR) spectra were obtained at 500 MHz on a Brüker Ascend™ 500 Spectrometer in CDCl3 with TMS as the internal standard. Optical rotation was measured on a PerkinElmer141 polarimeter which is equipped with a Na lamp of λ = 589 nm.
Antileishmanial Assay
Parasite Culture
L. donovani (1S MHOM/SD/62/1S strain) and L. major (IFLA/BR/67/PH8 strain) procured from the BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). They were cultured in ME199 medium, pH 7.4, supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin and kept in an air atmosphere at 28°C in 75 cm2 Roux flasks.
Cell Culture
RAW 264.7 cells were procured from the RIKEN BioResource Centre Cell Bank in Japan. They were maintained in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin under 5% CO2 and humidified atmosphere at 37°C.
Cytotoxicity Activity
In vitro evaluation of the cytotoxicity activity was carried out on RAW 264.7 cell line using the resazurin assay, which measures cellular metabolic activity (Süzgeaç-Selaçuk et al., 2011). A sub-confluent cell culture in 75 cm2 culture flask, was trypsinized, and cells were counted and suspended in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were seeded into a 96-well plate (100 μl per well) at concentrations of 1x105 cells per ml and incubated overnight with 5% CO2 at 37°C, to allow cells to attach to the surface of the plate. The cells were then treated in triplicate with increasing concentration of each compound (1.01 to 632.71 μM). After 48 h of incubation, 10 μl of 2.5 mM resazurin solution were added to each well and further incubated for 4 h at 37°C. Fluorescence was measured using the microplate reader (TECAN Infinite M200 Pro Plate Reader, Austria) at excitation and emission wavelengths of 530 nm and 590 nm, respectively. Medium seeded with cells without treatment served as negative control. Curcumin was used as positive control, while medium without cells served as blank. The experiments were done in triplicate and all the data were shown as means ± SD. The percent growth inhibition was computed from the absorbances relative to the negative control, and the concentration of compound that inhibited 50% cell (CC50 values) was calculated with GraphPad Prism 7.0 (GraphPad Prism software Inc. San Diego, CA).
Antipromastigote Assay
The susceptibility of promastigotes to HA was determined using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide/phenazine methosulfate (MTS/PMS, Promega) colorimetric assay (Wong et al., 2014). Glucose-6-phosphate dehydrogenase which takes part in the pentose phosphate pathway of Leishmania parasites are needed for the reduction of MTS to formazan. Briefly, stationary-phase promastigotes were seeded into 96-well flat-bottomed microtiter plates at 2x107 parasites per well and incubated at 28°C in the absence or presence of different concentrations of HA (39.54 to 632.71 μM). After 72 h of incubation, 10 μl of MTS/PMS solution were added to each well of the microtiter plate. The plates were further incubated at 28°C for colour development. After 4 h of incubation, the optical densities were read at 490 nm using an automated microtiter plate reader (TECAN Infinite M200 Pro Plate Reader, Austria). The negative control consisted of medium with untreated parasites, while medium without parasites served as blank. Amphotericin B was used as positive control. All the experiments were done in triplicate and the data were shown as means ± SD. The percent growth inhibition was computed from the optical densities relative to the negative control, and the concentration of compound that inhibited the parasite growth by 50% (IC50 values) were calculated with GraphPad Prism 7.0.
Determination of Phosphatidylserine Externalization
Externalization of the Phosphatidylserine was done using Annexin–V FLUOS staining kit (Roche), whilst Fluorescein-conjugated Annexin-V was used to detect the externalized phosphatidylserine due to very high binding affinity to the phospholipid portion. Additionally, annexin-V FLUOS distinguishes between surviving, apoptotic and necrotic cells. The stationary phase promastigotes (1×107 cells/ml) were either treated or untreated with HA and amphotericin B at IC50 for 72 h at 28°C. The downstream processing complied with the instructions of the manufacturer. Parasites were washed in cold phosphate-buffered saline (PBS) by centrifuging at 200 g for 5 min and the pellet was incubated with Annexin-V-FLOUS and PI (20 μl each) togther with sample buffer (100 μl) for 15 min at room temperature in the dark. After incubation, the samples were acquired on a flow cytometer (BD LSRFortessa II-x20), FlowJo software program was used for analysis and the percent of positive parasites was calculated per sample.
Preparation of the Structures of Pteridine Reductase 1 (PTR1) of L. donovani (Ld) and L. major (Lm) for In Silico Analysis
The X-ray crystallographic structures of Pteridine reductase 1 (PTR1) of L. major (Lm) and L. donovani (Ld) were obtained from protein databank (PDB) (Berman et al., 2000; Rose et al., 2017) with IDs 2XOX and 2BFO at resolutions of 2.5 Å and 2.6 Å, respectively. Water molecules, ligands and other hetero atoms, and all chains except chain A were removed using PyMOL (DeLano, 2002).
Homology Modeling of the Structures of LdGCL, LmGCL, LdTR, and LmTR
Due to the absence of experimentally solved structures of Trypanothione reductase (TR) and Glutamate cysteine ligase (GCL) of L. donovani as well as L. major in the RCSB PDB database, the primary sequences of Gamma-Glutamylcysteine synthetase (previous name for GCL) of L. donovani and Trypanothione reductase (TR) of L. donovani as well as L. major were obtained from UniProtKB (Magrane and Consortium, 2011) with IDs Q67BG3, C6GKV5, and Q4QJG7, respectively in fasta formats. The sequence of the putative gamma-glutamyl cysteine synthetase of L. major (strain Friedlin) with corresponding ID Q4QDM2 was also retrieved from UniProtKB. SWISS-MODEL (accessible via https://swissmodel.expasy.org) was used to search for templates of the LdGCL, LmGCL, LdTR, and LmTR (Kiefer et al., 2009).
Modeller 9.2 was then employed to build homology models of LdGCL, LmGCL, LdTR, and LmTR using the most reasonably identical templates with high sequence identity and maximum coverage to the queried sequences (Fiser and Šali, 2003). Homology models were built using pairwise alignment of target and template sequences utilizing spatial restraint techniques with default parameters. The generated models were ranked based on the DOPE scores.
The quality of the modelled structures was assessed using SAVES v5.0 (accessible via http://servicesn.mbi.ucla.edu/SAVES/). ERRAT, PROVE, VERIFY, and PROCHECK. ProSA-web were utilised in SAVES v5.0 for quality assessment. Also, Ramachandra plots were generated using Rampage (Sippl, 1993; Wiederstein and Sippl, 2007).
Binding Site Characterization
The binding pockets were predicted using CASTp (Binkowski et al., 2003; Dundas et al., 2006) and the results were visualized with Chimera 1.12 (Pettersen et al., 2004). The dimensions of the areas and volumes were taken into consideration when selecting the most plausible binding cavities. The active site of the PTR1 proteins was obtained from previous studies (Kaur et al., 2010). Prediction of residues involved in intermolecular bonding was done using LigPLOT+ v1.4.5 (Laskowski and Swindells, 2011).
Preparation of Ligands and Molecular Docking
The 3D structure of HA was retrieved from PubChem (Wang et al., 2009) with ID 15559629. Docking experiments were performed using AutoDock Vina embedded in PyRx 0.8 (Trott and Olson, 2010; Dallakyan and Olson, 2015). The “make macromolecule” option embedded within PyRx was used to convert the structure into a pdbqt format. The MMFF94 force field and Conjugate Gradients algorithms were used for energy minimization of ligands in 200 steps via OpenBabel embedded in PyRx 0.8. Blind docking was carried out on all structures used in this study. The docked structures were visualized and saved as pdb complexes using PyMOL. The resulting docked complexes were imported into LigPLOT+ v1.4.5 (Laskowski and Swindells, 2011) and ligand interaction 2D diagrams were generated for each complex.
Molecular Dynamics Simulations
The protein-hardwickiic acid complexes were subjected to molecular dynamics simulations (MDs). PRODRG (Schüttelkopf and Van Aalten, 2004) (http://davapc1.bioch.dundee.ac.uk/prodrg/) was used to prepare HA topology prior to the molecular dynamics simulation. Chirality, charges and energy minimization parameters were set to “yes”, “full,” and “no” respectively. MD simulation was conducted with GROMACS 2018 (Van Der Spoel et al., 2005; Abraham et al., 2015) under the GROMOS96 43A1 force field. The box boundary for solvation of the protein-ligand complexes was 1.0 nm. Ions of sodium and chloride were used to neutralize the charges on the complexes. The Steepest Descent was used to minimize the complexes over 1,000 steps.
Thereafter, the complexes were restrained as well as relaxed via equilibration. Also, the PME adopted for the simulation was over 100 ns. Finally, graphs were generated using the Xmgrace (Turner, 2005).
In Silico Pharmacokinetic Properties Prediction
HA, miltefosine, amphotericin B and curcumin were subjected to pharmacokinetics profiling using SwissADME. The canonical SMILES of HA (ID: 15559629), miltefosine (ID: 3599), amphotericin B (ID: 5280965), and curcumin (ID: 969516) were retrieved from PubChem and used as inputs for predicting the pharmacokinetic properties via SwissADME (Daina et al., 2017).
Prediction of Leishmanicidal Potential of Hardwickiic Acid
The SMILES file of HA was used to predict the biological activity of the compound (Lagunin et al., 2000; Parasuraman, 2011).
Results and Discussion
Structural Elucidation of Hardwickiic Acid (HA)
Hardwickiic acid (HA) was obtained as a white powder with specific rotation, -0.773 (c 6.49 x 10-3, CHCl3). The structure of HA (Figure 1) was elucidated from its 1H and 13C-NMR spectral and MS data as follows: 1H NMR (500 MHz, CDCl3): 0.77 (3H, Me-20), 0.84 (3H, d, J = 6.6 Hz, Me-17), 1.18 (1H, td, J = 12.9, 4.0 Hz, H-6a), 1.27 (3H, s, Me-19), 1.39 (1H, d, J = 12.1 Hz, 10-H), 1.44 (1H, dd, J = 10.1, 4.2 Hz, 7-Ha), 1.47 (1H, m, H-1a), 1.57 (1H, m, 11-Ha), 1.60 (1H, dd, J = 7.5, 3.4 Hz, 8-H), 1.66 (1H, dd, J = 13.9, 6.3 Hz, 11-Hb), 2.17 (2H, dd, J = 13.8, 5.3 Hz, 2-H), 2.23 (1H, m, 12-Ha), 2.31 (1H, t, J = 4.9 Hz, 12-Hb), 2.35 (1H, dd, J = 10.1, 4.2 Hz, 7-Hb), 2.45 (1H, dt, J = 12.9, 3.0 Hz, 6-Hb), 6.26 (1H, s, 14-H), 6.87 (1H, m, 3-H), 7.21 (1H, s, 16-H), 7.35 (1H, s, 15-H); 13C NMR (126 MHz, CDCl3): 16.1 (C-17, CH3), 17.6 (C-1, CH3), 18.2 (C-20, CH3), 18.3 (C-12, CH2), 18.4 (C-20, CH3), 20.7 (C-19, CH3), 27.4 (C-7, CH2), 27.6 (C-2, CH2), 36.0 (C-6, CH2), 37.7 (C-5, C), 38.8 (C-9, C), 39.0 (C-11, CH2), 46.8 (C-10, CH), 111.1 (C-14, CH), 125.7 (C-13, C), 138.5 (C-16, CH), 140.5 (C-3, C), 141.7 (C-4, C), 142.9 (C-15, CH),173.2 (C-18, C); ESI-MS [M+H]+ m/z: 317.1. The MS and 13C-NMR spectra of HA are shown in Supplementary Figures S1 and S2, respectively.
Figure 1.
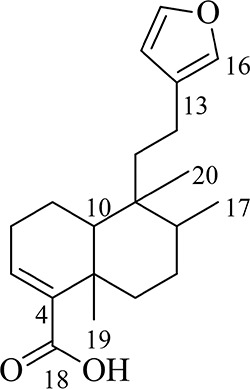
The structure of hardwickiic acid (HA).
Antipromastigote and Cytotoxicity Activities of HA
The antileishmanial assay of HA against promastigotes of L. major and L. donovani gave IC50 values of 62.82 and 31.57 μM, respectively. Moreover, its cytotoxicity on RAW cells was low with a CC50 value of 247.83 µM (SI values of 3.94 and 7.85, respectively for L. major and L. donovani), indicating hardwickiic acid to be a potential antileishmanial compound (Table 1, Figure 2). There are no previous reports about antileishmanial potential of HA. However, natural products from Croton species have been reported to have activity against Leishmania parasites. A MeOH : DCM (1:1 v/v) leaf extract from C. alienus showed activity on L. donovani parasite with an IC50 value of 80 μg/ml (Ndunda, 2014). Lima et al. (2015) reported the antileishmanial activity of clerodane diterpenes from C. cajucara on L. amazonensis parasites (Lima et al., 2015). Trans-dehydrocrotonin was the most potent having IC50 of 6.30, 19.18 and 0.47 μg/ml, respectively against promastigotes, axenic and intracellular amastigote forms without toxic effect (CC50 >100 μg/ml).
Table 1.
Antileishmanial and cytotoxicity activities.
| Cytotoxicity on RAW cells | L. major promastigotes | L. donovani promastigotes | |||
|---|---|---|---|---|---|
| CC50 (μM) | IC50 (μM) | SI | IC50 (μM) | SI | |
| Hardwickiic acid | 247.83 ± 6.32 | 62.82 ± 2.53 | 3.94 | 31.57 ± 0.14 | 7.85 |
| Curcumin | 29.99 ± 2.82 | ND | ND | ND | ND |
| Amphotericin B | ND | 3.67 ± 0.14 | ND | 3.35 ± 0.14 | ND |
Concentration of compounds which inhibited parasite proliferation by 50% (IC50); concentration of compounds which inhibited normal mammalian cells proliferation by 50% (CC50); and not determined (ND).
Figure 2.
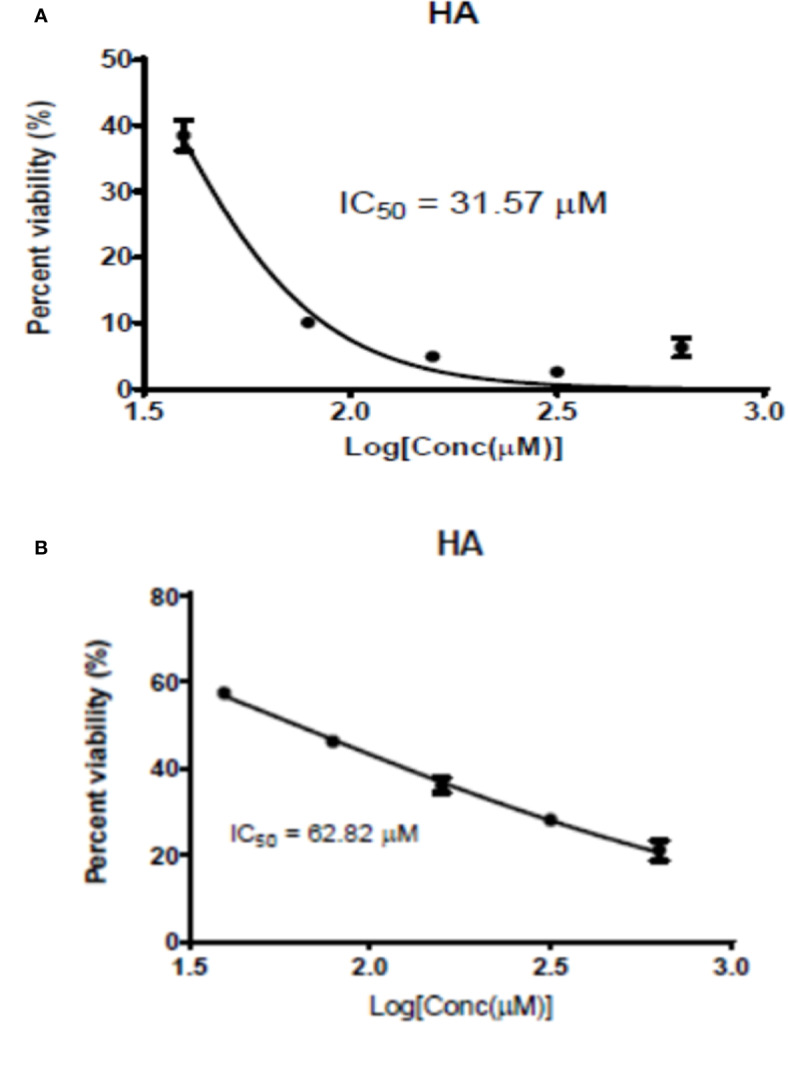
Graphic 1: Dose-response activity of HA on L. major (A) and on L. donovani (B).
Externalization of Phosphatidyl Serine by HA
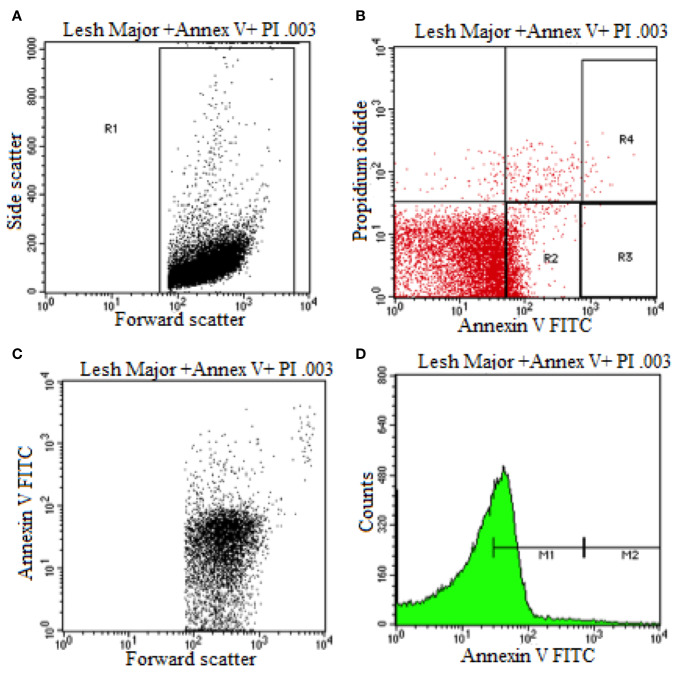
Flow cytometry was used to elucidate the Leishmania promastigotes parasite death mechanism triggered by HA at its IC50 values (Figure 3). Untreated control parasites died due to apoptosis or necrosis. Co-staining with annexin V and PI distinguishes between live (PI−/annexin V−), necrotic or late apoptotic (PI+/annexin V−) and early apoptotic (annexin V+/PI−) cells.
Figure 3.
Flow cytometry analysis of promastigotes following treatment with HA and after labeling with annexin-v and pi (untreated Leishmania major promastigote). (A) Dot Plot analysis of gated L. Major (R1) showing its size (Forward Scatter) and internal granularity (Side Scatter). (B) Dot Plot analysis of cellular apoptotic (Annexin V) and necrotic cells (Propidium Iodide) where L. Major expresses early cellular apoptosis (R2) and necrotic and apoptotic cells (R4). (C) Dot Plot analysis of L. Major being expressed in size (Forward Scatter) and cellular apoptosis (Annexin V). (D) Histogram plot analysis of L. Major showing low apoptotic cell expression (M1) and high apoptotic cell expression.
During early apoptosis, phosphatidylserine is translocated from the cytosolic face of the plasma membrane to the external face, which can be detected using Annexin V Fluos (Sousa et al., 2014). As shown in Figure 3 and Supplementary Figures S3B, S4, and S5B, 67.19% and 52.58% of hardwickiic acid-treated L. major and L. donovani promastigotes, respectively, became annexin positive, indicating early apoptotic phase. In early apoptotic phase, numerous changes occur in the parasite's plasma membrane. Translocation of phophatidylserine from the inside of the parasite membrane to the outside causes changes (Khademvatan et al., 2011). Consequently, 18.84% and 32.52% of a double positive were observed in hardwickiic acid treated-L. major and L. donovani promastigotes, respectively (Supplementary Figures S3B and S5B). In contrast, only 1.82% and 16.80% double positive were observed in untreated controls, respectively for L. major and L. donovani (Figure 3B and Supplementary Figure S3B). Therefore, HA triggered cell death via late apoptotic phase due to membrane integrity loss resulting from the binding of annexin-V and increment in PI incorporation.
Homology Modeling
The X-ray crystallographic structures of L. donovani (Ld) and L. major (Lm) Pteridine reductase 1 (PTR1) with IDs 2XOX and 2BFO, were retrieved from RCSB PDB with resolutions of 2.5 Å and 2.6 Å, respectively (Schüttelkopf et al., 2005; Barrack et al., 2011).
Since there are no solved structures, the sequences of LdTR, LmTR, LdGCL, and LmGCL with corresponding UNIPROT IDs C6GKV5, Q4QJG7, Q67BG3, and Q4QDM2 were retrieved in fasta format, respectively.
PTR1 receptors of L. major and L. donovani share high sequence conservation with sequence identity of 91% with structurally overlapping active site cavities (Kaur et al., 2010). Using LALIGN (Huang and Miller, 1991) (https://www.ebi.ac.uk/Tools/psa/lalign/), it was shown that LdTR and LmTR shared 95.5% sequence identity and 99.2% similarity, while LdGCL and LmGCL shared 94.8% identity and 98.3% similarity.
A BLAST search for homologues of LdTR, LmTR, and LdGCL was carried out via SWISS-MODEL (Schwede et al., 2003; Bienert et al., 2017). The search revealed a total of 5,445 and 5,452 templates that matched the LdTR and LmTR sequences, respectively. The structure with PDB ID: 1TYT was chosen as the reasonable template for comparative modelling of both receptors. 1TYT which is the structure of TR from Crithidia fasciculata, is a homo-dimer with a resolution of a 2.6 Å and has sequence similarity of 78.44% and 79.09% to LdTR and LmTR, respectively. C. fasciculata is a non-human infective trypanosomatid which is related to Trypanosoma brucei and Leishmania spp. (Rojas et al., 2014). The BLAST search also revealed a total of 16 and 11 templates as homologues to LdGCL and LmGCL, respectively. The X-ray crystallographic structure of Glutamate cysteine ligase (GCL) from Saccharomyces cerevisiae (Sc) at resolution of 2.1Å (Biterova and Barycki, 2009) with PDB ID 3IG5 was selected as the reasonably best homologue of both LdGCL and LmGCL with similarity of 39.42% and 39.01%, respectively. The protein encoded by the isolated cDNA of Lm is a homologue to those of S. cerevisiae and silent information regulator 2 (SIR2) (Yahiaoui et al., 1996).
Modeller 9.2 was used to generate five structures each of LdTR, LmTR, LdGCL, and LmGCL using the selected templates. The most suitable models were selected based on the DOPE scores (Supplementary Table S1 and Supplementary Figure S6). Predicted structures are evaluated using the DOPE score, which is a statistical potential. The model with the least DOPE value can be selected as the best model for the same target (Eswar et al., 2006; Shen and Sali, 2006). The least DOPE scores obtained for the best models of LdTR, LmTR, LdGCL, and LmGCL were -53014.125, -53008.44531, -73083.05469, and -73102.125, respectively.
Model Quality Assessment
The quality of the selected models of LdTR, LmTR, LdGCL, and LmGCL were assessed using SAVESv5.0. The summary of the quality assessment scores are shown in Table 2. ERRAT, PROVE, VERIFY, and PROCHECK were used to assess the quality of the structures. From Table 2, LdTR, LmTR, and LmGCL passed VERIFY validation since more than 80% of the amino acids had scores >= 0.2 in the 3D/1D profile, while LdGCL had 77.58%. PROVE showed that LdTR had 83 (4.8%), LmTR had 88 (5.0%), LdGCL had 193 (7.4%), and LmGCL had 210 (8%) buried outlier protein atoms. The overall quality factor obtained via ERRAT (Supplementary Figure S6) showed that LdTR, LmTR, LdGCL, and LmGCL had overall quality factors of 87.7847, 85.7143, 54.2474, and 57.037%, respectively. LdTR had ERRAT error values at residues 34, 162, 218, 222, 99, 170, 172, and 182. LmTR also had error values at residues 149 to 155, 168 to 173, 175, and 182, while LdGCL and LmGCL showed the most error values, and similar error trends among the four predicted models (Supplementary Figures S6C, D).
Table 2.
Model Evaluation of the Selected Models using various tools.
| TOOL | MODEL SCORE | |||
|---|---|---|---|---|
| LdTR | LmTR | LdGCL | LmGCL | |
| VERIFY | 90.02% | 93.08% | 77.58% | 80.35% |
| ERRAT (Quality Factor) | 87.7847 | 85.7143 | 54.2474 | 57.037 |
| PROVE | 4.8% | 5.0% | 7.4% | 8.0% |
| PROCHECK | 3 Errors, 3 Warnings and 3 Passes. | 3 Errors, 3 Warnings and 3 Passes. | 4 Errors, 2 Warnings and 3 Passes. | 4 Errors, 2 Warnings and 3 Passes. |
ProSA-web predicted the z-scores of LdTR, LmTR, LdGCL, and LmGCL as -11.31, -11.81, -7.12, and -8.26, respectively. All four modeled structures were predicted to lie within the z-scores of X-ray determined proteins. The overall quality of the model is shown by the z-score falling within the range known for native proteins of comparable size (Sippl, 1993; Wiederstein and Sippl, 2007).
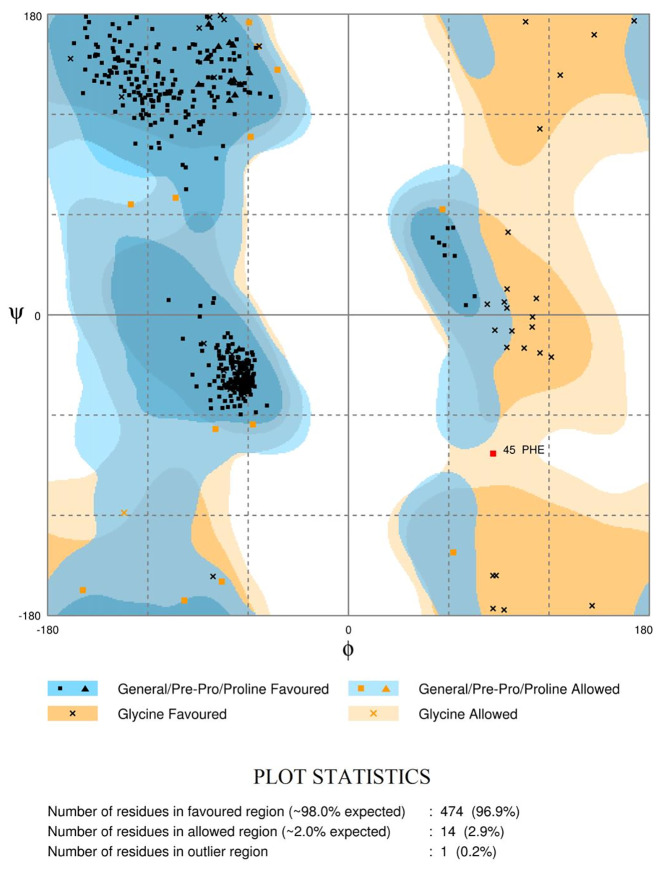
The Ramachandran plots (Figure 4 and Supplementary Figure S7) evaluate the structural quality of the models using the Rampage (Sippl, 1993; Wiederstein and Sippl, 2007). LdTR had 474 residues (96.9%) in the favoured region, 14 (2.9%) in the allowed region, and only 1 residue (0.2%) in the outlier region. LmTR had 476 residues (97.3%) in the favoured region, 11 (2.2%) in the allowed region and 2 residues (0.4%) in the outlier region. LdGCL had 635 residues (92.7%) in the favoured region, 34 (5.0%) in the allowed region and 16 (2.3%) in the outlier region. LmGCL had 630 residues (92.0%) in the favoured region, 35 (5.1%) in the allowed region and 20 residues (2.9%) in the outlier region. The expected threshold for Rampage is approximately 98% in the favoured region and 2% in the allowed region. All four models had almost all their residues in the favoured regions and were quite close to the expected values. The modeled LdTR had only 1 residue in the outlier region while LmTR had 2 residues in the outlier region. The results from the study showed that the models were of very high quality.
Figure 4.
Ramachandran plot of the LdTR modeled receptor obtained via RAMPAGE. The plots evaluate the residues in the outlier, allowed and favoured regions.
Binding Site Predictions and Molecular Docking
The active sites of LdPTR1 and LmPTR1 have been determined in previous studies. The active site cavity of LdPTR1 comprise key amino acid residues Arg17, Asn109, Ser111, Asp181, Tyr191, Tyr194, Lys198, Leu226, and Ala230 (Kaur et al., 2010). The receptors of Lm and Ld PTR1 are highly conserved and have structurally similar active site regions (Cavazzuti et al., 2008; Kaur et al., 2010; Ferrari et al., 2011). The active sites of LdTR, LmTR, LdGCL, and LmGCL were determined using the CASTp. The results were visually analyzed, and the areas and volumes were considered when selecting the most plausible binding sites (Supplementary Table S2 and Figure S8).
HA was docked against six Leishmania receptors comprising LdPTR1, LmPTR1, LdTR, LmTR, LdGCL, and LmGCL. From the docking results (Table 3), the binding energies of all the six models docked with HA exceeded the threshold of −7.0 kcal/mol (Chang et al., 2007).
Table 3.
Binding affinities and protein-ligand interactions of HA docked with the six receptors, namely LdPTR1, LmPTR1, LdTR, LmTR, LdGCL, and LmGCL.
| Receptor | Binding energy (kcal/mol) | Number of hydrogen bonds | Hydrogen Bond Residues | Hydrogen Bond Length (Å) | Hydrophobic Contacts |
|---|---|---|---|---|---|
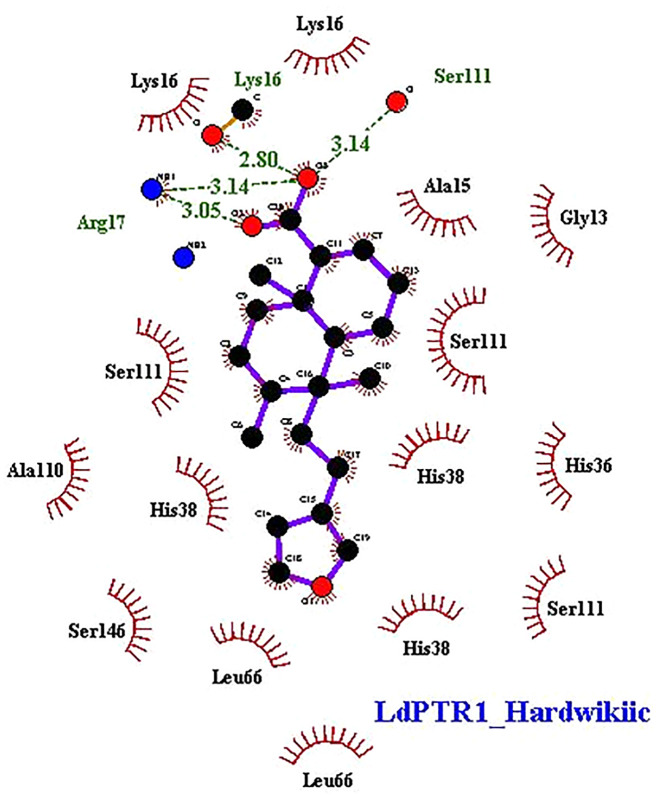
| LdPTR1 | -7.1 | 4 | Lys16, Arg17, Arg17, Ser111. | 2.8, 3.05, 3.14, 3.14. | Gly13, Ala15, Lys16, His36, His38, Leu66, Ala110, Ser146, Ser111. |
| LmPTR1 | -7.8 | 3 | Asn109, Gly225, Ser227. | 2.9, 2.78, 2.98. | Arg17, Leu18, Gly19, Asn109, Ser111, Phe113, Asp181, Tyr194, Lys198, Pro224, Leu226. |
| LdTR | -7.6 | 0 | – | – | Gly66, Tyr69, Met70, Leu88, Pro90, Asn91, Thr94, Leu95, Tyr210. |
| LmTR | -7.5 | 1 | Met70 | 3.09 | Gly66, Ala67, Tyr69, Met70, Ile73, Pro90, Asn91, Thr94, Leu95, Gly209, Tyr210. |
| LdGCL | -7.4 | 4 | Met1, Gly51, Glu52, Thr330. | 3.07, 3.24, 3.05, 3.01. | Met1, Gly51, Glu52, Glu53, Thr101, Pro102, Asp103, Pro105, Thr330, Ile491, Arg494. |
| LmGCL | -8.0 | 4 | Phe151, Val152, Val152, Cys154. | 3.0, 2.99, 3.09, 2.98. | Gln148, Gly149, Asn150, Phe151, Val152, Cys154, Ser155, Asp156, Ser159, Ser164, Leu165, Phe166, Val167, Pro168, Val253, Ser256, Ser257, Arg261. |
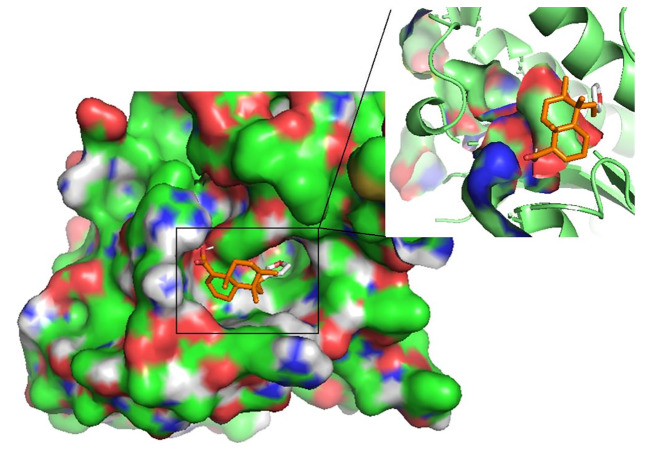
HA demonstrated the strongest binding affinity to LmGCL (-8.0kcal/mol), followed by LmPTR1 (-7.8kcal/mol), LdTR (-7.6 kcal/mol), LmTR (-7.5 kcal/mol), LdGCL (-7.4 kcal/mol), and then LdPTR1 (-7.1 kcal/mol). HA is a potential antileishmanial compound based on the docking results which supports the in vitro studies conducted herein. The best binding pose of HA in the binding cavities of the 6 proteins are provided (Figure 5 and Supplementary Figure S8). HA docked firmly into the predicted LmTR pocket 4, LdGCL pocket 2, and LmGCL pocket 2.
Figure 5.
The pose of hardwickiic acid within the binding cavity of LdPTR1. The receptor is represented as a surface while hardwickiic acid as sticks.
Characterization of Protein-Ligand Interactions
The interaction profiles of the receptors and HA complexes have been elucidated (Table 3 and Supplementary Table S2; and Figure 6, Supplementary Figures S8 and S9). HA was predicted not to interact via H-bonds with LdTR. HA interacted with LdPTR1 via hydrogen bonds with Lys16 (2.8 Å bond length), Ser111 (3.14 Å bond length) and Arg17 (bond lengths of 3.05 Å and 3.14 Å). Arg17 and Ser111 were identified as key residues in the active site of PTR1 (Bernal and Coy-Barrera, 2014). HA also interacted with LmPTR1 via 3 hydrogen bonds with Asn109, Gly225, and Ser227 with bond lengths of 2.9 Å, 2.78 Å, and 2.98 Å, respectively. Some dimeric xanthanolide compounds were shown to have the best binding affinity when docked against PTR1. Also, the dimeric xanthanolide compounds formed hydrogen bond interactions with Ser111, Ser227, and Arg17, which are critical amino acid residues required for stability (Bernal and Coy-Barrera, 2014). These two compounds were reported to be reasonably active against L. donovani amastigotes (IC50:22–27 µM) (Schmidt et al., 2009). Herein, hardwickiic acid is a promising lead which warrants further structural optimization studies for PTR1 inhibition.
Figure 6.
2D representations of protein-ligand interaction between hardwickiic acid and the receptors LdPTR1. Hydrogen bonds are denoted as green dash lines while red spike arcs represent hydrophobic contacts.
HA also interacted with LmTR via a hydrogen bond with Met70 with bond length of 3.09 Å. Similarly, HA interacted with Met1, Gly51, Glu52, and Thr330 of LdGCL with bond lengths of 3.07 Å, 3.24 Å, 3.05 Å, and 3.01 Å, respectively. It was also observed that HA interacted with LmGCL via 4 hydrogen bonds with Phe151, Val152, Val152, and Cys154 of lengths 3.0 Å, 2.99 Å, 3.09 Å, and 2.98 Å, respectively. The high numbers of hydrogen bonds with relatively short bond distances indicate a strong binding which thereby positions HA as a promising leishmanicide. HA shared a single hydrophobic contact with Ser111 in LdPTR1, and two with Arg17 and Ser111 in LmPTR1.
Molecular Dynamics Simulation of Complexes
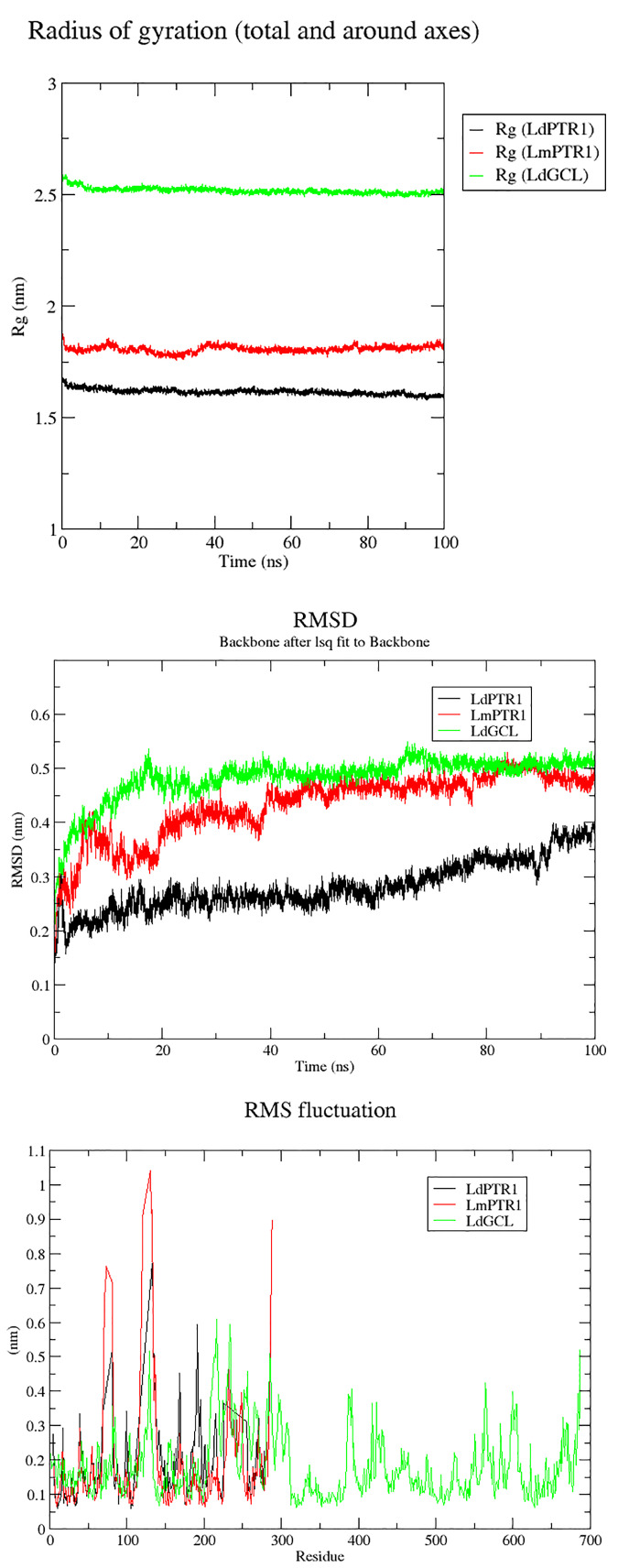
Molecular dynamics simulations over the course of 100 ns were carried out on LdPTR1, LmPTR1 and LdGCL complexed with hardwickiic acid. The stability of the protein-ligand complexes was determined over the 100 ns simulations. The radius of gyration (Rg) which determines its compactness was generated for all the three complexes. A protein structure which is stably folded maintains a fairly steady Rg, while the Rg of an unfolded protein changes significantly over a period. From the Rg plots (Figure 7A), all the three protein-ligand complexes were very stable during the entire simulation. All systems were compact, with LdPTR1 having the least Rg of 1.65 nm. LmPTR1 had an average Rg of 1.8 nm, and LdGCL had the lowest average Rg of 2.5 nm. LdGCL demonstrated the most stability with little fluctuations observed around 0 to 8 ns. LmPTR1 showed the least stability among the three complexes, fluctuated from 0 to about 40 ns, then stabilized thereafter.
Figure 7.
Rg, RMSDs and RMSFs plots for the protein–hardwickiic acid complex MDs undertaken in 100 ns: (A) Rg plots of the three proteins in complex with hardwickiic acid. (B) The plot of RMSDs over 100 ns for the three complexes. (C) RMS Fluctuations (RMSFs) of amino acid residues pertaining to the three proteins in complex with hardwickiic acid. LdPTR1, LmPTR1, and LdGCL complexes are illustrated as black, red, and green colors, respectively.
The RMSDs were computed to determine the stability of the complexes which is shown by fluctuations. During the equilibration phase, RMSDs of the complexes increased and then converged 20 ns later. From Figure 7B, the three complexes (backbone) showed stability after a gradual rise. The LdPTR1-hardwickiic acid complex increased in RMSD from 0 ns until about 20 ns, after which it appeared fairly stable. After about 40 ns, the RMSD of LmPTR1-HA complex averaged around 0.45 nm. The RMSD of LdGCL-HA complex was the most stable form. It was observed to rise during the equilibration phase from 0.2 nm to about 0.5 nm during the first 20 ns.
To determine protein regions which exhibited higher structural flexibility, the root mean square fluctuations (RMSF) per residue was determined (Figure 7C). LdPTR1 and LmPTR1 showed similar trends by exhibiting greater fluctuations with the highest RMSFs. This corroborated the high structural identity of the two protein structures. For both LdPTR1 and LmPTR1, the high fluctuation regions were within residue index 60 to 80 and 100 to 140 with 100 to 140 being the highest. LdPTR1 also exhibited high fluctuation from residues 180 to 200. The active sites of both PTR1 proteins were around residue index 100 to 140 and 180 to 200. It was highly possible that key residues Asn109 and Ser111 caused the fluctuations from 100 to 140, while Asp181, Tyr191, Tyr194, and Lys198 were responsible for the fluctuations from 180 to 200. LdGCL demonstrated the lowest RMSF values as compared to LdPTR1 and LmPTR1. The major fluctuations for LdGCL were observed in the regions with residue index 105 to 140 and 200 to 230. It could be possible that Thr101, Pro102, Asp103, and Pro105 which had contacts with hardwickiic acid were responsible for the high spike exhibited around residues 105 to 140.
In Silico Pharmacokinetic Properties Prediction
All the four compounds were predicted to have moderately soluble with low ESOL logS (Table 4). ESOL logS quantifies the aqueous solubility of a molecule (Daina et al., 2017). HA and curcumin were predicted to possess good gastrointestinal (GI) absorption implying a high possibility of enhanced absorption of orally administered drugs into the intestinal tract and the bloodstream (Kwofie et al., 2019). The Abbot Bioavailability scores of the compounds were also determined. HA was predicted to possess the highest value (0.56), whereas amphotericin B was predicted to possess the lowest (0.17). Both miltefosine and curcumin had a bioavailability score of 0.55. The Abbot Bioavailability score denotes oral bioavailability of at least 10% in rat or measurable permeability in Caco-2. It relies on total charge, and topological polar surface area (TPSA) together with violations of Lipinski's rules to classify compounds into four categories with probabilities of 11%, 17%, 56%, or 85% (Martin, 2005).
Table 4.
Prediction of pharmacokinetic properties for HA, miltefosine, amphotericin B and curcumin using SwissADME.
| Compound | Molecular Weight | ESOL LogS | ESOL Class | GI Absorption | BBB Permeant | Pgp substrate | Bioavailability score |
|---|---|---|---|---|---|---|---|
| HA | 316.4 | -5.25 | Moderately soluble | High | Yes | No | 0.56 |
| Miltefosine | 407.6 | -5.32 | Moderately soluble | Low | No | Yes | 0.55 |
| Amphotericin B | 924.1 | -5.37 | Moderately soluble | Low | No | Yes | 0.17 |
| Curcumin | 368.4 | -3.94 | Moderately soluble | High | No | No | 0.55 |
Properties determined include P-glycoprotein substrate (Pgp), Gastrointestinal absorption (GI), Blood Brain Barrier permeation (BBB), Estimated Solubility (ESOL), and Bioavailability score.
Since HA and curcumin were also predicted as non-substrate of P-gp, they can be distributed effectively within the circulatory system (Table 4). The P-glycoprotein (P-gp) acts as physiological filter by eliminating unwanted materials including toxins (Lin and Yamazaki, 2003). The Blood Brain Barrier (BBB) permeation denotes the effectiveness of molecules to permeate the barrier to initiate signaling after binding to key receptors. Also, a drug must permeate this barrier in order to have pharmacological effect on the brain parenchyma (Suenderhauf et al., 2012). Only Hardwickiic acid was predicted to have the ability to permeate the barrier, (Table 4).
Biological Activity Prediction for Hardwickiic Acid
The activity of HA was predicted using features of molecules with known activities by implementing a Bayesian algorithm (Parasuraman, 2011). Biological activity is based on the predicted probable inactivity (Pi) and probable activity (Pa) of a molecule (Jamkhande et al., 2016). PASS predicted HA to be Caspase-3 stimulant with Pa 0.757 and Pi 0.008; and Caspase-8 stimulant with Pa 0.688 and Pi 0.004. HA was also predicted to possess antiprotozoal activity (specifically, antileishmanial) with Pa of 0.434 and Pi 0.037. From recent studies, Leishmania infections have been reported to delay host cell apoptosis by inhibiting caspase-3 activity (Ruhland et al., 2007; Abhishek et al., 2018). Caspase-8 inhibition following L. major infection has also been shown to decrease the expression of interferon gamma by CD4 and CD8 T cells (Pereira et al., 2008). Therefore, HA maybe implicated in the apoptotic mechanisms. For a given compound activity, whenever the Pa > Pi, the predicted biological activity can be pursued experimentally (Jamkhande and Barde, 2014; Kwofie et al., 2018). Therefore, HA is an attractive antileishmanial candidate, which reinforces the results of the in vitro studies.
Conclusion
The study evaluated the leishmanicidal potential of HA characterised from the stembark of Croton sylvaticus against Leishmania donovani and L. major promastigotes. HA exhibited strong antileishmanial activity on L. donovani promastigotes with an IC50 of 31.57 ± 0.06 µM when compared to amphotericin B with an IC50 of 3.35 ± 0.14 µM. Also, the cytotoxic activity of CC50 = 247.83 ± 6.32 µM was obtained against 29.99 ± 2.82 µM for curcumin with selectivity index of SI = 7.85. Molecular modelling and docking studies revealed that HA had binding affinity greater than -7.0 kcal/mol with all 6 Leishmania receptors used in this study. HA exhibited the highest binding affinity (-8.0 kcal/mol) with LmGCL. HA was also observed to interact with critical residues Lys16, Ser111 and Arg17 required for LdPTR1 binding. Moreover, HA was predicted as Caspase-3 and Caspase-8 stimulants and possesses antileishmanial activity. Therefore, HA may play a critical role in caspase mediated apoptotic mechanisms. The potential apoptotic role was further corroborated experimentally since HA was shown to induce parasite death by loss of membrane integrity. HA is an attractive antileishmanial candidate worthy of further in vivo experimentation.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
DO-S, LY, and SK conceptualised the project. DO-S, JC, and BA were responsible for isolation and characterisation of HA. LY and JT conducted the biological assays while SK and EB undertook the cheminformatics. All authors co-wrote the first draft of the manuscript and proofread the submitted manuscript.
Funding
Bill and Melinda Gates Foundation supported the postdoctoral fellowship (OPP52155) of LRTY at NMIMR, Ghana. This work was supported by funds from a World Bank African Centres of Excellence grant (ACE02-WACCBIP: Awandare) and a DELTAS Africa grant (107755/Z/15/Z: Awandare). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [107755/Z/15/Z: Awandare] and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Emeritus Professor Ivan Addae-Mensah, Department of Chemistry, University of Ghana, for providing the plant extract for the analysis. We would like to thank the University of Ghana's West African Centre for Cell Biology of Infectious Pathogens for providing us free high performance computing time on Zuputo.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.00753/full#supplementary-material
References
- Abhishek K., Das S., Kumar A., Kumar A., Kumar V., Saini S., et al. (2018) Leishmania donovani induced Unfolded Protein Response delays host cell apoptosis in PERK dependent manner. PloS Neglected Trop. Dis. 12 (7), e0006646. 10.1371/journal.pntd.0006646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., et al. (2015). Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Alvar J., Vélez I. D., Bern C., Herrero M., Desjeux P., Cano J., et al. (2012). Leishmaniasis worldwide and global estimates of its incidence. PloS One 7 (5), e35671. 10.1371/journal.pone.0035671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara B. M. R., Wimalasiri W. R., Bandara K. A. N. (1987). Isolation and insecticial activity of (-)-hardiwickii acid from Croton aromaticus. Planta Med. 6, 575. 10.1055/s-2006-962818 [DOI] [PubMed] [Google Scholar]
- Barrack K. L., Tulloch L. B., Burke L. A., Fyfe P. K., Hunter W. N. (2011). Structure of recombinant Leishmania donovani pteridine reductase reveals a disordered active site. Acta Crystallogr. Sect. F.: Struct. Biol. Cryst. Commun. 67, 33–37. 10.1107/S174430911004724X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates P. A. (2007). Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int. J. Parasitol. 37 (10), 1097–1106. 10.1016/j.ijpara.2007.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., et al. (2000). The protein data bank. Nucleic Acids Res. 28 (1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal F. A., Coy-Barrera E. (2014). In-silico analyses of sesquiterpene-related compounds on selected Leishmania enzyme-based targets. Molecules 19 (5), 5550–5569. 10.3390/molecules19055550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert S., Waterhouse A., De Beer T. A. P., Tauriello G., Studer G., Bordoli L., et al. (2017). The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 45 (D1), D313–D319. 10.1093/nar/gkw1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkowski T. A., Naghibzadeh S., Liang J. (2003). CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res. 31 (13), 3352–3355. 10.1093/nar/gkg512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biterova E. I., Barycki J. J. (2009). Mechanistic details of glutathione biosynthesis revealed by crystal structures of Saccharomyces cerevisiae glutamate cysteine ligase. J. Biol. Chem. 284 (47), 32700–32708. 10.1074/jbc.M109.025114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson E. E. (2010). Natural products as chemical probes. ACS Chem. Biol. 5 (7), 639–653. 10.1021/cb100105c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzuti A., Paglietti G., Hunter W. N., Gamarro F., Piras S., Loriga M., et al. (2008). Discovery of potent pteridine reductase inhibitors to guide antiparasite drug development. PNAS 105 (5), 1448–1453. 10.1073/pnas.0704384105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaichantipyuth C., Muangsin N., Chaichit S., Roengsumran S., Petsom A., Watanabe T., et al. (2004). Crystal structure of (-)-hardwickiic acid, C19H27OCOOH. NCS 219 (2), 111–113. 10.1524/ncrs.2004.219.2.111 [DOI] [Google Scholar]
- Chang M. W., Lindstrom W., Olson A. J., Belew R. K. (2007). Analysis of HIV wild-type and mutant structures via in silico docking against diverse ligand libraries. J. Chem. Inf. Model. 47 (3), 1258–1262. 10.1021/ci700044s [DOI] [PubMed] [Google Scholar]
- Chawla B., Madhubala R. (2010). Drug targets in Leishmania. J. Parasitic Dis. 34 (1), 1–13. 10.1007/s12639-010-0006-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colotti G., Baiocco P., Fiorillo A., Boffi A., Poser E., Chiaro F., et al. (2013). Structural insights into the enzymes of the trypanothione pathway: Targets for antileishmaniasis drugs. Future Med. Chem. 5 (15), 1861–1875. 10.4155/fmc.13.146 [DOI] [PubMed] [Google Scholar]
- Corrales R. M., Sereno D., Mathieu-Daudé F. (2010). Immunology & medical microbiology. FEMS Immunol. Med. Microbiol. 58 (1), 27–38. 10.1111/j.1574-695X.2009.00608.x [DOI] [PubMed] [Google Scholar]
- Crowther G. J., Shanmugam D., Carmona S. J., Doyle M. A., Hertz-Fowler C., Berriman M., et al. (2010) Identification of attractive drug targets in neglected- disease pathogens using an in Silico approach. PloS Neglected Trop. Dis. 4 (8), e804. 10.1371/journal.pntd.0000804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V., Brooks C. L., Huang R. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717. 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., Olson A. J. (2015). Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 1263, 243–250. 10.1007/978-1-4939-2269-7_19 [DOI] [PubMed] [Google Scholar]
- de Vries H. J. C., Reedijk S. H., Schallig H. D. F. H. (2015). Cutaneous Leishmaniasis: Recent Developments in Diagnosis and Management. Am. J. Clin. Dermatol. 16 (2), 99–109. 10.1007/s40257-015-0114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newslett. Protein Crystallogr. 40, 82–92. [Google Scholar]
- Dostálová A., Volf P. (2012). Leishmania development in sand flies: Parasite-vector interactions overview. Parasites Vectors. 5, 276. 10.1186/1756-3305-5-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas C., Ouellette M., Tovar J., Cunningham M. L., Fairlamb A. H., Tamar S., et al. (1997). Disruption of the trypanothione reductase gene of Leishmania decreases its ability to survive oxidative stress in macrophages. EMBO J. 16 (10), 2590–2598. 10.1093/emboj/16.10.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006). CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–W118. 10.1093/nar/gkl282. (WEB. SERV. ISS.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M., et al. (2006). Comparative Protein Structure Modeling Using Modeller. Current Protocols in Bioinformatics Supplement 15, 5.6.1–5.6.30. 10.1002/0471250953.bi0506s15 [DOI] [PMC free article] [PubMed]
- Feasey N., Wansbrough-Jones M., Mabey D. C. W., Solomon A. W. (2010). Neglected tropical diseases. Br. Med. Bull. 93 (1), 179–200. 10.1093/bmb/ldp046 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Morandi F., Motiejunas D., Nerini E., Henrich S., Luciani R., et al. (2011). Virtual screening identification of nonfolate compounds, including a CNS drug, as antiparasitic agents inhibiting pteridine reductase. J. Med. Chem. 54 (1), 211–221. 10.1021/jm1010572 [DOI] [PubMed] [Google Scholar]
- Fiser A., Šali A. (2003). MODELLER: Generation and Refinement of Homology-Based Protein Structure Models. Methods Enzymol. 374, 461–491. 10.1016/S0076-6879(03)74020-8 [DOI] [PubMed] [Google Scholar]
- Georgiadou S. P., Makaritsis K. P., Dalekos G. N. (2016). Leishmaniasis revisited: Current aspects on epidemiology, diagnosis and treatment. J. Trans. Internal Med. 3 (2), 43–50. 10.1515/jtim-2015-0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbani M., Farhoudi R. (2018). Leishmaniasis in humans: Drug or vaccine therapy? Drug Design Dev. Ther. 10.2147/DDDT.S146521 [DOI] [PMC free article] [PubMed]
- Grondin K., Haimeur A., Mukhopadhyay R., Rosen B. P., Ouellette M. (1997). Co-amplification of the γ-glutamylcysteine synthetase gene gsh1 and of the ABC transporter gene pgpA in arsenite-resistant Leishmania tarentolae. EMBO J. 10.1093/emboj/16.11.3057 [DOI] [PMC free article] [PubMed]
- Huang X., Miller W. (1991). A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 10.1016/0196-8858(91)90017-D [DOI]
- Jamkhande P. G., Barde S. R. (2014). Evaluation of anthelmintic activity and in silico PASS assisted prediction of Cordia dichotoma (Forst.) root extract. Ancient Sci. Life 34 (1), 39–43. 10.4103/0257-7941.150779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamkhande P. G., Pathan S. K., Wadher S. J. (2016). In silico PASS analysis and determination of antimycobacterial, antifungal, and antioxidant efficacies of maslinic acid in an extract rich in pentacyclic triterpenoids. Int. J. Mycobacteriol. 5 (4), 417–425. 10.1016/j.ijmyco.2016.06.020 [DOI] [PubMed] [Google Scholar]
- Jassbi A. R. (2006). Chemistry and biological activity of secondary metabolites in Euphorbia from Iran. Phytochemistry. 10.1016/j.phytochem.2006.06.030 [DOI] [PubMed]
- Kamhawi S. (2006). Phlebotomine sand flies and Leishmania parasites: friends or foes? Trends Parasitol. 10.1016/j.pt.2006.06.012 [DOI] [PubMed]
- Kaur J., Sundar S., Singh N. (2010). Molecular docking, structure-activity relationship and biological evaluation of the anticancer drug monastrol as a pteridine reductase inhibitor in a clinical isolate of Leishmania donovani. J. Antimicrob. Chemother. 10.1093/jac/dkq189 [DOI] [PubMed]
- Khademvatan S., Gharavi M. J., Rahim F., Saki J. (2011). Miltefosine-Induced Apoptotic Cell Death on Leishmania major, and L. tropica Strains. Korean J. Parasitol. 10.3347/kjp.2011.49.1.17 [DOI] [PMC free article] [PubMed]
- Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009). The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37 (SUPPL. 1), D387–D392. 10.1093/nar/gkn750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Kumar A., Verma S. S., Dwivedi N., Singh N., Siddiqi M. I., et al. (2008). Leishmania donovani pteridine reductase 1: Biochemical properties and structure-modeling studies. Exp. Parasitol. 10.1016/j.exppara.2008.05.005 [DOI] [PubMed]
- Kwofie S., Dankwa B., Odame E., Agamah F., Doe L., Teye J., et al. (2018). In Silico Screening of Isocitrate Lyase for Novel Anti-Buruli Ulcer Natural Products Originating from Africa. Molecules 23 (7), 1550. 10.3390/molecules23071550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwofie S. K. S. K., Broni E., Teye J., Quansah E., Issah I., Wilson M. D. M. D., et al. (2019). Pharmacoinformatics-based identification of potential bioactive compounds against Ebola virus protein VP24. Comput. Biol. Med. 113, 103414. 10.1016/j.compbiomed.2019.103414 [DOI] [PubMed] [Google Scholar]
- Kyriazis I. D., Koutsoni O. S., Aligiannis N., Karampetsou K., Skaltsounis A. L., Dotsika E. (2016). The leishmanicidal activity of oleuropein is selectively regulated through inflammationand oxidative stress-related genes. Parasites Vectors. 10.1186/s13071-016-1701-4 [DOI] [PMC free article] [PubMed]
- Lagunin A., Stepanchikova A., Filimonov D., Poroikov V. (2000). PASS: prediction of activity spectra for biologically active substances. Bioinf. (Oxford England) 16 (8), 747–748. 10.1093/bioinformatics/16.8.747 [DOI] [PubMed] [Google Scholar]
- Lahlou M. (2013). The Success of Natural Products in Drug Discovery. Pharmacol. Pharm. 10.4236/pp.2013.43a003 [DOI]
- Lamotte S., Späth G. F., Rachidi N., Prina E. (2017). The enemy within: Targeting host–parasite interaction for antileishmanial drug discovery. PloS Neglected Trop. Dis. 10.1371/journal.pntd.0005480 [DOI] [PMC free article] [PubMed]
- Laskowski R. A., Swindells M. B. (2011). LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51 (10), 2778–2786. 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- Lima G. S., Castro-Pinto D. B., MacHado G. C., Maciel M. A. M., Echevarria A. (2015). Antileishmanial activity and trypanothione reductase effects of terpenes from the Amazonian species Croton cajucara Benth (Euphorbiaceae). Phytomedicine. 10.1016/j.phymed.2015.08.012 [DOI] [PubMed]
- Lin J. H., Yamazaki M. (2003). Role of P-glycoprotein in pharmacokinetics: Clinical implications. Clin. Pharmacokinet. 42 (1), 59–98. 10.2165/00003088-200342010-00003 [DOI] [PubMed] [Google Scholar]
- Magrane M., Consortium U. P. (2011). UniProt Knowledgebase: A hub of integrated protein data. Database. 10.1093/database/bar009 [DOI] [PMC free article] [PubMed]
- Mandal G., Mandal S., Sharma M., Charret K. S., Papadopoulou B., Bhattacharjee H., et al. (2015). Species-Specific Antimonial Sensitivity in Leishmania Is Driven by Post-Transcriptional Regulation of AQP1. PloS Neglected Trop. Dis. 10.1371/journal.pntd.0003500 [DOI] [PMC free article] [PubMed]
- Martin Y. C. (2005). A bioavailability score. J. Med. Chem. 10.1021/jm0492002 [DOI] [PubMed]
- McChesney J., Clark A. M., Silveira E. R. (1991). Antimicrobial diterpenes of Croton sonderianus, 1. Hardwickiic an 3,4-secotrachylobanoic acids. J. Natural Prod. 54, 1625–1633. 10.1021/np50078a021 [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Roy G., Guimond C., Ouellette M. (2009). The γ-glutamylcysteine synthetase gene of Leishmania is essential and involved in response to oxidants. Mol. Microbiol. 10.1111/j.1365-2958.2009.06907.x [DOI] [PubMed]
- Mwangi J. W., Thoithi G. N., Addae-Mensah I., Achenbach H., Lwande W., Hassanali A. (1998). Aromatic plants of Kenya III: Volatile and some non-volatile constituents of Croton sylvaticus Hochst. East Cent. Afri. J. Pharm. Sci. 1, 41–43. [Google Scholar]
- Nare B., Hardy L. W., Beverley S. M. (1997. a). The roles of pteridine reductase 1 and dihydrofolate reductase- thymidylate synthase in pteridine metabolism in the protozoan parasite Leishmania major. J. Biol. Chem. 10.1074/jbc.272.21.13883 [DOI] [PubMed]
- Nare B., Luba J., Hardy L. W., Beverley S. (1997. b). New approaches to Leishmania chemotherapy: pteridine reductase 1 (PTR1) as a target and modulator of antifolate sensitivity. Parasitology. 10.1017/s0031182097001133 [DOI] [PubMed]
- Ndunda B. (2014). Phytochemistry and bioactivity investigations of three Kenyan croton species. [PhD Thesis]. Nairobi: University of Nairobi.
- Parasuraman S. (2011). Prediction of activity spectra for substances. J. Pharmacol. Pharmacotherapeutics 2 (1), 52–53. 10.4103/0976-500X.77119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira W. F., Guillermo L. V. C., Ribeiro-Gomes F. L., Lopes M. F. (2008). Inhibition of caspase-8 activity reduces IFN-γ expression by T cells from Leishmania major infection. Anais Da Academia Bras. Cienc. 10.1590/S0001-37652008000100008 [DOI] [PubMed]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., et al. (2004). UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 25 (13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Ready P. D. (2013). Biology of Phlebotomine Sand Flies as Vectors of Disease Agents. Annu. Rev. Entomol. 10.1146/annurev-ento-120811-153557 [DOI] [PubMed]
- Rojas R., Segovia C., Trombert A. N., Santander J., Manque P. (2014). The effect of tunicamycin on the glucose uptake, growth, and cellular adhesion in the protozoan parasite crithidia fasciculata. Curr. Microbiol. 10.1007/s00284-014-0620-x [DOI] [PubMed]
- Rose P. W., Prlić A., Altunkaya A., Bi C., Bradley A. R., Christie C. H., et al. (2017). The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 45 (D1), D271–D281. 10.1093/nar/gkw1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhland A., Leal N., Kima P. E. (2007). Leishmania promastigotes activate PI3K/Akt signalling to confer host cell resistance to apoptosis. Cell. Microbiol. 10.1111/j.1462-5822.2006.00769.x [DOI] [PubMed]
- Süzgeaç-Selaçuk S., Meriaçli A. H., Güven K. C., Kaiser M., Casey R., Hingley-Wilson S., et al. (2011). Evaluation of Turkish seaweeds for antiprotozoal, antimycobacterial and cytotoxic activities. Phytother. Res. 10.1002/ptr.3330 [DOI] [PubMed]
- Saccoliti F., Angiulli G., Pupo G., Pescatori L., Madia V. N., Messore A., et al. (2017). Inhibition of Leishmania infantum trypanothione reductase by diaryl sulfide derivatives. J. Enzyme Inhib. Med. Chem. 10.1080/14756366.2016.1250755 [DOI] [PMC free article] [PubMed]
- Salatino A., Salatino M. L. F., Negri G. (2007). Traditional uses, chemistry and pharmacology of Croton species (Euphorbiaceae). J. Braz. Chem. Soc. 10.1590/S0103-50532007000100002 [DOI]
- Sasidharan S., Chen Y., Saravanan D., Sundram K. M., Yoga Latha L. (2011). Extraction, isolation and characterization of bioactive compounds from plants' extracts. Afr. J. Tradit. Complementary Altern. Medicines. 10.4314/ajtcam.v8i1.60483 [DOI] [PMC free article] [PubMed]
- Schüttelkopf A. W., Van Aalten D. M. F. (2004). PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. Sect. D.: Biol. Crystallogr. 10.1107/S0907444904011679 [DOI] [PubMed]
- Schüttelkopf A. W., Hardy L. W., Beverley S. M., Hunter W. N. (2005). Structures of Leishmania major pteridine reductase complexes reveal the active site features important for ligand binding and to guide inhibitor design. J. Mol. Biol. 10.1016/j.jmb.2005.06.076 [DOI] [PubMed]
- Schmidt T. J., Nour A. M. M., Khalid S. A., Kaiser M., Brun R. (2009). Quantitative structure - Antiprotozoal activity relationships of sesquiterpene lactones. Molecules. 10.3390/molecules14062062 [DOI] [PMC free article] [PubMed]
- Schwede T., Kopp J., Guex N., Peitsch M. C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 10.1093/nar/gkg520 [DOI] [PMC free article] [PubMed]
- Shen M., Sali A. (2006). Statistical potential for assessment and prediction of protein structures. Protein Sci. 10.1110/ps.062416606 [DOI] [PMC free article] [PubMed]
- Shi Q. W., Su X. H., Kiyota H. (2008). Chemical and pharmacological research of the plants in genus Euphorbia. Chem. Rev. 10.1021/cr078350s [DOI] [PubMed]
- Sippl M. J. (1993). Recognition of errors in three-dimensional structures of proteins. Proteins: Struct. Funct. Genet. 17 (4), 355–362. 10.1002/prot.340170404 [DOI] [PubMed] [Google Scholar]
- Sousa M. C., Varandas R., Santos R. C., Santos-Rosa M., Alves V., Salvador J. A. R. (2014). Antileishmanial activity of semisynthetic lupane triterpenoids betulin and betulinic acid derivatives: Synergistic effects with miltefosine. PloS One. 10.1371/journal.pone.0089939 [DOI] [PMC free article] [PubMed]
- Suenderhauf C., Hammann F., Huwyler J. (2012). Computational prediction of blood-brain barrier permeability using decision tree induction. Molecules. 10.3390/molecules170910429 [DOI] [PMC free article] [PubMed]
- Tiwari K., Dubey V. K. (2018). Leishmania donovani asparaginase variants exhibit cytosolic localization. Int. J. Biol. Macromol. 10.1016/j.ijbiomac.2018.03.071 [DOI] [PubMed]
- Tiwari N., Gedda M. R., Tiwari V. K., Singh S. P., Singh R. K. (2017). Limitations of Current Therapeutic Options, Possible Drug Targets and Scope of Natural Products in Control of Leishmaniasis. Mini-Rev. Med. Chem. 10.2174/1389557517666170425105129 [DOI] [PubMed]
- Tovar J., Wilkinson S., Mottram J. C., Fairlamb A. H. (1998). Evidence that trypanothione reductase is an essential enzyme in Leishmania by targeted replacement of the tryA gene locus. Mol. Microbiol. 10.1046/j.1365-2958.1998.00968.x [DOI] [PubMed]
- Trott O., Olson A. J. (2010). AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, EfficientOptimization, and Multithreading. J. Comput. Chem. 31 (2), 455–461. 10.1002/jcc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcano L., Torrente E., Missineo A., Andreini M., Gramiccia M., Di Muccio T., et al. (2018). Identification and binding mode of a novel Leishmania Trypanothione reductase inhibitor from high throughput screening. PloS Neglected Trop. Dis. 10.1371/journal.pntd.0006969 [DOI] [PMC free article] [PubMed]
- Turner P. (2005). XMGRACE, Version 5.1. 19 (Beaverton: Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology; ). 10.1163/_q3_SIM_00374 [DOI] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. C. (2005). GROMACS: Fast, flexible, and free. J. Comput. Chem. 10.1002/jcc.20291 [DOI] [PubMed]
- Wang Y., Xiao J., Suzek T. O., Zhang J., Wang J., Bryant S. H. (2009). PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 10.1093/nar/gkp456 [DOI] [PMC free article] [PubMed]
- WHO (2018). Neglected Tropical Diseases Through Changes on the Skin: A Training Guide for Front-Line Health Workers. pp 2-5. WHO; Retrieved September 18, 2019, from https://www.who.int/neglected_diseases/resources/9789241513531/en/. [Google Scholar]
- WHO (2019). Leishmaniasis, Retrieved September 18, 2019, from https://www.who.int/en/news-room/fact-sheets/detail/leishmaniasis.
- Wiederstein M., Sippl M. J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–W410. 10.1093/nar/gkm290. (Web Server). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong I. L. K., Chan K. F., Chen Y. F., Lun Z. R., Chan T. H., Chow L. M. C. (2014). In vitro and in vivo efficacy of novel flavon`oid dimers against cutaneous leishmaniasis. Antimicrob. Agents Chemother. 10.1128/AAC.02425-13 [DOI] [PMC free article] [PubMed]
- Xu W. H., Liu W. Y., Liang Q. (2018). Chemical constituents from croton species and their biological activities. Molecules. 10.3390/molecules23092333 [DOI] [PMC free article] [PubMed]
- Yahiaoui B., Taibi A., Ouaissi A. (1996). A Leishmania major protein with extensive homology to silent information regulator 2 of Saccharomyces cerevisiae. Gene. 10.1016/0378-1119(95)00785-7 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.