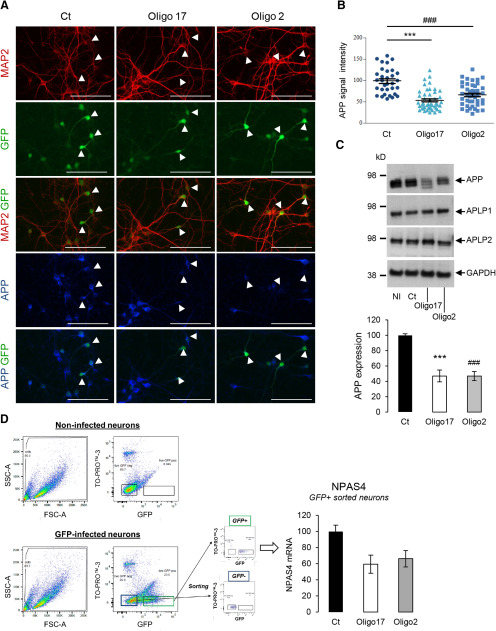
Figure 3.
Decreased NPAS4 expression in APP-silenced primary neurons. APP was knock-down by CRISPR-Cas9 approach in primary neurons cultures. The infectivity and toxicity of lentiviral CRISPR-Cas9 vectors are detailed in Extended Data Figure 3-1. A, APP expression characterization in neurons by immunostaining. Cortical neurons were infected at DIV1 with lentiviruses expressing sgRNAs (Oligo2, Oligo17) or not (Ct). All lentiviruses harbor a GFP expression cassette. Cultures were immunostained for MAP2 (red), APP (blue) and DAPI (light blue) at DIV7. Arrowheads indicate the position of GFP-positive (infected) neurons in each condition. Scale bar: 100 µm. B, Quantification of APP signal in GFP-positive neurons by ImageJ. At least 33 neurons were quantified in two independent experiments for each condition (n = 33 N = 2). Results (mean ± SEM) are given as percentage of control (Ct); ###p < 0.001 (Ct vs Oligo 2) and ***p < 0.001 (Ct vs Oligo17); Kruskal–Wallis test and Dunn’s multiple comparison test. C, upper panel, Representative Western blots showing APP, APLP1, APLP2, and GAPDH protein levels in cortical neurons at DIV7 infected in the same conditions as in Figure 1A. NI = non-infected. Lower panel, Quantification of APP expression in total cell lysates measured by Western blotting. Results (mean ± SEM) are given as percentage of control (Ct); ***p < 0.001 (Ct vs Oligo17), ###p < 0.001 (Ct vs Oligo 2), ANOVA and Bonferroni’s multiple comparison test (n = 6, N = 3). D, Sorting of GFP-expressing neurons (FACS). Scatter plots (FSC vs SSC, left panels) of non-infected and GFP-expressing cells are shown. Dot plots (TOPRO-3, far red vs GFP, right panels) were used to gate (green rectangle) GFP-positive/TOPRO-3 negative cells. RNA was extracted from these cells and NPAS4 mRNA level was quantified by qPCR. Results were obtained from pooled samples (four wells of 4 cm2 each) for each condition (Ct, Oligo2 and Oligo17). Quantification was conducted on two independent experiments (N = 2). Results (mean ± SEM) are expressed as percentage of Ct.