Summary
We explored the relationships between lymphocyte subsets, cytokines, pulmonary inflammation index (PII) and disease evolution in patients with (corona virus disease 2019) COVID‐19. A total of 123 patients with COVID‐19 were divided into mild and severe groups. Lymphocyte subsets and cytokines were detected on the first day of hospital admission and lung computed tomography results were quantified by PII. Difference analysis and correlation analysis were performed on the two groups. A total of 102 mild and 21 severe patients were included in the analysis. There were significant differences in cluster of differentiation 4 (CD4+ T), cluster of differentiation 8 (CD8+ T), interleukin 6 (IL‐6), interleukin 10 (IL‐10) and PII between the two groups. There were significant positive correlations between CD4+ T and CD8+ T, IL‐6 and IL‐10 in the mild group (r 2 = 0·694, r 2 = 0·633, respectively; P < 0·01). After ‘five‐in‐one’ treatment, all patients were discharged with the exception of the four who died. Higher survival rates occurred in the mild group and in those with IL‐6 within normal values. CD4+ T, CD8+ T, IL‐6, IL‐10 and PII can be used as indicators of disease evolution, and the PII can be used as an independent indicator for disease progression of COVID‐19.
Keywords: coronavirus infected disease, lymphocyte subsets, cytokines, pulmonary inflammatory index, disease classification
In December 2019, a pneumonia epidemic caused by a new coronavirus occurred in Wuhan, China, and the World Health Organization (WHO) temporarily named the virus 2019‐nCoV. On 11 February 2020, the Director General of the WHO (Tedros Adhanom Ghebreyesus) announced that the official name of the disease caused by the new coronavirus was ‘COVID‐19’. The main sources of infection are patients with pneumonia infected by the new coronavirus. Respiratory droplets are the main route of transmission, and it can also be transmitted by direct contact. At present, the occurrence, development, mechanism of prognosis, and immune status of patients with COVID‐19 are unclear [1, 2, 3, 4]. In the present study, the lymphocyte subsets and cytokines in the peripheral blood of 123 patients with COVID‐19 infection were detected by flow cytometry. The results of lung computed tomography (CT) scans were quantified using the pulmonary inflammation index (PII). The purpose of the present study was to explore the relationship between lymphocyte subsets, cytokines, PII, and COVID‐19 disease progression.
Patients and methods
Patients
This study was approved by the Ethics Committee of Chongqing Three Gorges Central Hospital (No. 2020‐3), and informed consent was obtained from each patient. From 26 January to 4 February 2020, 123 inpatients diagnosed with COVID‐19 were studied. The epidemiological history of each patient was investigated and physical examinations were performed. All patients were confirmed to be positive for the new coronavirus nucleic acid by real‐time fluorescent reverse transcriptase‐polymerase chain reaction (RT‐PCR). Patients were diagnosed according to the WHO interim guidance for COVID‐19 [5, 6, 7]. Patients were divided into mild (including normal and mild) and severe (including severe and critical) groups as follows:
Mild: mild clinical symptoms and imaging showing no pneumonia.
Normal: symptoms of fever, respiratory tract inflammation, imaging showing pneumonia.
Severe: meeting any of the following: respiratory distress, respiratory rate (RR) ≥30 breaths/min in the resting state, mean oxygen saturation ≤93%, arterial blood oxygen partial pressure (PaO2)/fraction of inspired oxygen concentration (FiO2) ≤300 mmHg.
Critical: respiratory failure requiring mechanical ventilation, shock occurs; combined failure of other organs requiring Intensive Care Unit (ICU) monitoring and treatment.
Blood sampling
Blood samples were collected by a nurse according to doctor instructions. None of the patients were treated before blood sampling, and they had not received the standardised treatment according to the diagnosis and treatment scheme of COVID‐19 [5, 7]. Lymphocyte subsets and cytokines of all patients were detected by flow cytometry on the first day after admission.
Lymphocyte subsets detection
Two test tube blood samples, labelled A and B, were taken for each patient. A 5 μl sample of CD3/CD8/CD45/CD4 antibody (Beijing Tongshengshidai Biotechnology Co., Ltd, Beijing, China) was added to tube A, and 5 μl of CD16+ 56/CD45/CDl9 antibody was added to tube B. After adding 50 μl of ethylenediamine tetra‐acetic acid (EDTA) anti‐coagulated whole blood to each tube, the tubes were vortexed and kept at room temperature for 15 min in darkness. Then, the samples were analysed by four‐colour fluorescence labelled flow cytometry (Mindray BriCyte E6; Mindray, Shenzhen, China). The normal ranges of CD4+ T, CD8+ T, B cell, natural killer (NK) cell, CD4+ T/CD8+ T are 410–1590, 190–1140, 90–660 (× 106/l), 90–590 (× 106/l), 0·7–2·87, respectively.
Cytokine detection
The cytokine detection reagent was provided by Qingdao Raisecare Biotechnology Co., Ltd (Shandong, China; lot number: 20190801). Interleukin 4 (IL‐4), IL‐6, IL‐10, IL‐17, tumour necrosis factor (TNF), and interferon (IFN) were detected by multiple microsphere flow immunofluorescence. After the blood sample and the corresponding flow tube were numbered 101, 102, 103, 104, and 105, EDTA‐K2 anti‐coagulated whole blood was centrifuged at 1000 g for 30 min. Then, 25 μl of experimental buffer, 25 μl of centrifuged plasma, 25 μl of capture microsphere antibody, and 25 μl of detection antibody were added to the corresponding flow tubes. After incubation at room temperature for 2 h in darkness with gentle shaking, 25 μl of streptavidin‐phycoerythrin (SA‐PE) was added to the flow tubes and incubation was continued for an additional 30 min. Subsequently, diluted wash buffer (1:10) was added. After a few seconds of vortex shaking, the flow tube was centrifuged at 432 g for 5 min. The liquid was slowly poured out, and the flow tube was inverted on absorbent paper. Then, 100 μl of diluted washing buffer (1:10) was added to the flow tube, which was shaken for 10 s followed by detection. The normal ranges of IL‐4, IL‐6, IL‐10, IL‐17, TNF, and IFN are 0–8·56, 0–5·4, 0–12·9, 0–21·4, 0–16·5, 0–23·1 pg/ml.
CT acquisition
Chest CT scans were performed on all patients with suspected COVID‐19 infection using two 16‐row multi‐slice CT scanners (Somatom Sensation 16, Siemens, Erlangen, Germany; uCT 510, United Imaging, China). Patients were placed in a supine position and advanced head, continuous spiral scanning was performed from the lung top to the lung bottom. For CT acquisition, the tube voltage was 120 kVp with automatic tube current modulation and pitch 0·99–1·45 mm. From the raw data, CT images were reconstructed on a matrix of 512 × 512 and a field of view 350 × 350 mm as axial images (section thickness of 10 mm) The thickness of the axial reconstruction layer was 1·0 mm, the window width was 1000–2000 HU, and the window level was 700–500 HU. Quantitative results were individually assessed by two senior radiologists, and discussions were used to resolve differences (not common) in data interpretation.
Quantification of lung CT lesions by PII
A modified semi‐quantitative scoring system based on commercial Lung Quantitative Software (Siemens) was used to quantitatively estimate the pulmonary involvement of all patients according to the distribution and size of the lesions. The lesion distribution score was based on the segment of the lung where the lesion was located. There were 20 segments in total for the left and right lobes, 1 point for each segment of lung, and 20 points for the left and right lungs. The lesion size score was based on whether the lesion occupied >50% of the lung segment. The score was 1 for ≥50% involvement and zero for <50%, with a maximum score of 20. The PII scores were distributed between zero and 100%. A larger value indicated a more severe inflammatory load. PII = lesion distribution score + lesion size score/total score (40 points) × 100%.
Statistical analysis
All statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS®), version 22.0 (SPSS Inc., Chicago, IL, USA). Descriptive analyses were performed for categorical variables such as gender. Continuous variables such as inspection results were expressed as the mean ± SD, tested for normality, and compared using the independent samples t‐test. Otherwise, the Mann–Whitney test was used. The proportions of categorical variables were compared using a chi‐squared test. Pearson product‐moment analysis was used to evaluate associations between variables and the statistic was r 2 and larger r 2 values indicated a better linear correlation. A P < 0·05 was considered statistically significant.
Results
Baseline data
A total of 102 patients with COVID‐19 (55 males and 47 females) with a mean (SD; range) age of 43·05 (13·12; 15–82) years in the mild group and 21 patients (11 males and 10 females) with a mean (SD; range) age of 61·29 (15·55; 34–79) years in the severe group were enrolled in the study (Table 1). The main symptoms at the onset of illness were fever, cough, sore throat, and poor appetite. The duration of symptoms in the severe group were longer than in the mild group. Morbidities were more common in the severe group than in the mild group. The white blood cell, neutral cell, lymphocyte, platelet, and haemoglobin counts of the severe group were lower than those in the mild group.
Table 1.
The basic information of the 123 patients with COVID‐19.
| Characteristic | Mild group (n = 102) | Severe group (n = 21) | P |
|---|---|---|---|
| Age, years, n (mean [SD]) | 102 (43·05 [13·12]) | 21 (61·29 [15·55]) | <0·0001 |
| Fever, n (mean [SD]) | 74 (6·12 [3·78]) | 21 (7·26 [4·60]) | 0·216 |
| Cough, n (mean [SD]) | 63 (7·37 [6·10]) | 16 (12·08 [15·31]) | 0·358 |
| Sore throat, n (mean [SD]) | 13 (7·70 [4·52]) | 0 | 0·183 |
| Poor appetite and fatigue, n (mean [SD]) | 6 (6·03 [4·14]) | 4 (6·43 [0·612]) | 0·061 |
| Other, n (mean [SD]) | 12 (7·73 [3·38]) | 1 (12·80 [0]) | 0·037 |
| Any comorbidity, n (%) | 13 (13) | 15 (71) | – |
| Diabetes, n (%) | 3 (3) | 5 (24) | – |
| Cardiovascular disease, n (%) | 2 (2) | 6 (28) | – |
| Hypertension, n (%) | 6 (6) | 3 (14) | – |
| Chronic obstructive, n (%) | 2 (2) | 1 (5) | – |
| Laboratory findings | |||
| White cell count, ×109/l, median (IQR) | 5·4 (4·0–7·8) | 5·1 (5·0–6·9) | 0·715 |
| <3·5, n (%) | 27 (26) | 3 (14) | – |
| 3·5–9·5, n (%) | 70 (68) | 16 (76) | 0·0036 |
| >9·5, n (%) | 5 (6) | 2 (9·5) | – |
| Neutrophil count, 109/l, median (IQR) | 3·4 (2·8–3·7) | 4·5 (3·1–5·7) | 0·0023 |
| Lymphocyte count, ×109/l, median (IQR) | 1·3 (0·7–1·5) | 0·9 (0·6–1·1) | <0·0001 |
| <1·1, n (%) | 41 (40) | 18 (86) | 0·3104 |
| ≥1·1, n (%) | 61 (60) | 3 (14) | – |
| Haemoglobin, g/l, median (IQR) | 102 (120–148) | 21 (120–143) | 0·1012 |
| Platelet count, ×109/l, median (IQR) | 102 (129–236) | 21 (113–209) | 0·0312 |
| <125, n (%) | 13 (12·7) | 7 (33) | 0·7891 |
| ≥125, n (%) | 89 (88·3) | 14 (67) | 0·462 |
| Viral load type, n (%) | |||
| N | 87 (85) | 19 (90) | 0·7346 |
| ORFlab | 5 (5) | 1 (5) | – |
| nCoV | 10 (10) | 1 (5) | – |
Continuous variables were described using the mean, median, and interquartile range (IQR) values or mean ± SD.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Lymphocyte subsets in peripheral blood of patients with COVID‐19
The results of each index, CD4+ T, CD8+ T, B cell, NK cell, CD4+ T/CD8+ T were divided into below normal value, within normal value, and above normal value groups. The corresponding quantities and proportions were calculated (Table 2).
Table 2.
Distribution of lymphocyte subsets in peripheral blood of the 123 patients with novel coronavirus (2019‐nCoV)–infected pneumonia (COVID‐19).
| Groups | Groups | CD4+ T | CD8+ T | B cell | NK cell | CD4+ T/CD8+ T |
|---|---|---|---|---|---|---|
| Mild group | Below normal values, n (%) | 54 (52·90) | 29 (28·40) | 26 (25·49) | 35 (34·31) | 3 (2·94) |
| Within normal values, n (%) | 48 (47·10) | 73 (71·60) | 75 (73·50) | 66 (64·71) | 95 (93·14) | |
| Above normal values, n (%) | – | – | 1 (0·01) | 1 (0·01) | 4 (3·92) | |
| Severe group | Below normal values, n (%) | 20 (95·24) | 13 (61·90) | 6 (28·57) | 10 (47·62) | 2 (9·53) |
| Within normal values, n (%) | 1 (4·76) | 8 (38·10) | 15 (71·43) | 11 (52·38) | 18 (85·71) | |
| Above normal values, n (%) | – | – | – | – | 1 (4·76) |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Cytokines in peripheral blood of patients with COVID‐19
Peripheral blood IL‐4, IL‐6, IL‐10, IL‐17, TNF, and IFN were divided into values of zero, within the normal value (normal values other than zero), and above the normal value according to the results of various indicators. The corresponding quantities and proportions are listed in Table 3.
Table 3.
Cytokine status in peripheral blood of the 123 patients with novel coronavirus (2019‐nCoV)‐infected pneumonia (COVID‐19).
| Groups | Groups | IL‐4 | IL‐6 | IL‐10 | IL‐17 | TNF | IFN |
|---|---|---|---|---|---|---|---|
| Mild group | 0, n (%) | – | 57 (55·88) | – | – | – | 5 (4·90) |
| Within normal values, n (%) | 102 (100·00) | 14 (13·73) | 102 (100·00) | 102 (100·00) | 100 (98·04) | 92 (90·20) | |
| Above normal values, n (%) | – | 31 (30·39) | – | – | 2 (1·96) | 5 (4·90) | |
| Severe group | 0, n (%) | – | 3 (14·29) | – | – | – | – |
| Within normal values, n (%) | 21 (100·00) | 2 (9·52) | 21 (100·00) | 21 (100·00) | 21 (100·00) | 20 (95·24) | |
| Above normal values, n (%) | – | 16 (76·19) | – | – | – | 1 (4·76) |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
All of the 102 (100%) patients in the mild group had IL‐4, IL‐10, and IL‐17 results within normal values; 57 (55·88%) patients had IL‐6 values of zero and 14 (13·73%) within normal values, 31 (30·39%) were higher than normal; 100 (98·04%) of the patients had TNF‐α values within the normal values, two (1·96%) had higher than normal values; 92 (90·20%) patients had normal IFN values, five (4·90%) had IFN values of zero, and five (4·90%) were higher than normal.
In the severe group, IL‐4, IL‐10, IL‐17, and TNF were all within the normal values in all of the patients; IL‐6 was zero in three patients (14·29%), two (9·52%) were within the normal values, 16 (76·19%) were higher than the normal values; IFN was within the normal values in 20 patients (95·24%), and one (4·76%) had a value higher than normal.
Pulmonary CT changes and PII in patients with COVID‐19
Pulmonary CT images were obtained. When the patient had a mild condition, patchy ground glass opacities were seen in the apical and posterior segments of the right upper lobe, the posterior basal segment of the right lower lobe, the medial segment of the right middle lobe, the lingual segment of the left upper lobe, the anterior inner basal segment of the left lower lobe, the posterior basal segment and the subpleural area of the outer basal segment of the left lower lobe (Fig. 1). When the patient was in a severe condition, a strip‐like consolidation shadow and patchy ground glass‐like density shadow were seen in the apical and posterior segments of the upper lobe of both lungs, tongue segment of the left upper lobe, and lower lobe of both lungs with a fuzzy boundary. These shadows were mainly distributed under the pleura of both lungs (Fig. 2).
Fig 1.
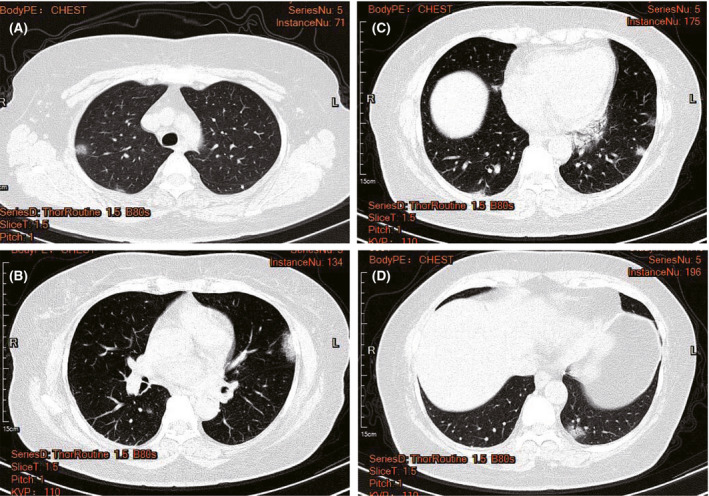
Typical CT images of patients with mild 2019 novel coronavirus pneumonia (COVID‐19). Patchy ground glass opacities can be seen in the apical and posterior segments of the right upper lobe, the posterior basal segment of the right lower lobe, the medial segment of the right middle lobe, the lingual segment of the left upper lobe, the anterior inner basal segment of the left lower lobe, the posterior basal segment, and the subpleural of outer basal segment of the left lower lobe. [Colour figure can be viewed at http://www.wileyonlinelibrary.com/]
Fig 2.
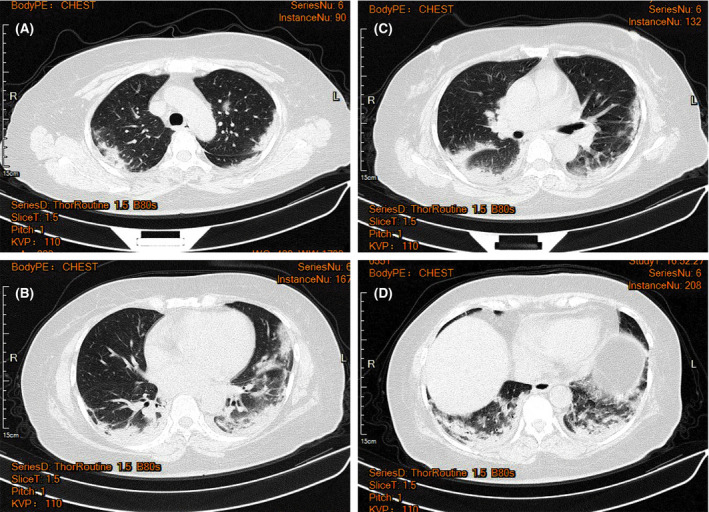
Typical CT images of patients with severe 2019 novel coronavirus pneumonia (COVID‐19). Strip‐like consolidation shadows and patchy ground glass‐like density shadows can be seen in the apical and posterior segment of the upper lobe of both lungs, tongue segment of the left upper lobe, and the lower lobe of both lungs with a fuzzy boundary, mainly distributed under the pleura of both lungs when the patient was in severe condition. [Colour figure can be viewed at http://www.wileyonlinelibrary.com/]
The PII was used to quantify the lung lesions of patients. There was a significantly higher PII in the severe group compared with the mild group (P < 0·05, Table 4).
Table 4.
Difference analysis of lymphocyte subsets, cytokines and pulmonary inflammatory index between the two groups.
| Items | Mild group, n (mean [SD]) | Severe group, n (mean [SD]) | P |
|---|---|---|---|
| CD4+ T | 102 (451·3 [23·0]) | 21 (263·2 [28·83]) | 0·0005 |
| CD8+ T | 102 (288·6 [14·23]) | 21 (179 [23·87]) | 0·0013 |
| B cell | 102 (166 [11·98]) | 21 (125·3 [13·49]) | 0·1375 |
| NK cell | 102 (147 [10·36]) | 21 (119·6 [16·500]) | 0·258 |
| CD4+ T/CD8+ T | 102 (1·671 [0·059]) | 21 (1·509 [0·170]) | 0·2857 |
| IL‐4 | 102 (1·69 [0·070]) | 21 (1·83 [0·185]) | 0·4317 |
| IL‐6 | 45 (13·41 [1·84]) | 21 (37·77 [7·801]) | <0·0001 |
| IL‐10 | 102 (2·464 [0·085]) | 21 (4·59 [0·378]) | <0·0001 |
| IL‐17 | 102 (1·095 [0·0226]) | 21 (1·16 [0·0571]) | 0·2463 |
| TNF | 102 (4·077 [1·588]) | 21 (2·948 [0·443]) | 0·7486 |
| IFN | 97 (5·132 [0·841]) | 21 (6·904 [1·247]) | 0·3533 |
| PII | 102 (0·129 [0·039]) | 21 (0·508 [0·179]) | <0·0001 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Analysis of lymphocyte subsets, cytokines, and PII differences between the two groups
Data (57 cases of IL‐6 and three cases of IFN in the mild group) with an index value of zero in the two groups were excluded from the analysis. Two independent samples t‐tests were performed, with α = 0·05 as the inspection level. Significant differences were observed in CD4+ T, CD8+ T, IL‐6, IL‐10, and PII between the two groups (P < 0·05), while no significant difference was detected in B cell, NK cell, CD4+ T/CD8+ T, IL‐4, IL‐17, TNF, and IFN (P > 0·05, Table 4.
Correlation analysis of CD4+ T, CD8+ T, IL‐6, IL‐10, and PII in the two groups
A pairwise correlation analysis was performed on the indicators (CD4+ T, CD8+ T, IL‐6, IL‐10, and PII) according to the results of the difference analysis. The indicators with correlations were analysed by multiple linear regression analysis. There were significant positive correlations between CD4+ T and CD8+ T, IL‐6 and IL‐10 in the mild group (r 2 = 0·694, r 2 = 0·633, respectively; P < 0·01), and moderate correlations were found between CD4+ T and CD8+ T, CD8+ T and IL‐6, IL‐6 and IL‐10 in the severe group (r 2 = 0·407, r 2 = 0·311, r 2 = 0·633, respectively) (Table 5).
Table 5.
Correlation analysis of CD4+ T, CD8+ T, IL‐6, IL‐10 and pulmonary inflammatory index (PII) in the two groups.
| Items | CD4+ T | CD8+ T | IL‐6 | IL‐10 | ||||
|---|---|---|---|---|---|---|---|---|
| Mild group (n = 45) | Severe group (n = 18) | Mild group (n = 102) | Severe group (n = 21) | Mild group (n = 102) | Severe group (n = 21) | Mild group (n = 102) | Severe group (n = 21) | |
| CD4+ T | – | – | 0.694** | 0·407 | 0·021 | −0·1 | −0·13 | −0·211 |
| CD8+ T | 0.694** | 0·407 | – | – | −0·018 | 0·311 | −0·125 | 0·24 |
| IL‐6 | 0·021 | −0·1 | −0·018 | 0·311 | – | – | 0·116 | 0.633** |
| IL‐10 | −0·13 | −0·211 | −0·125 | 0·24 | 0·116 | 0.633** | – | – |
| PII | −0·144 | −0·197 | −0·1 | 0·106 | 0·143 | 0·105 | 0·029 | 0·218 |
Data in the table are the correlation coefficient values r 2;
Significant correlation at the 0·01 level.
P < 0·01.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Disease evolution and prognosis of the patients with COVID‐19
A total of 123 patients were followed until discharge, and the longest follow‐up time was 20 days. The diagnosis time, time of treatment in hospital, nucleic acid conversion to negative time, total hospitalisation time, and disease evolution (including mild to severe, severe to mild, mortality, and discharge) of all patients are presented in Table 6. In Table 7, we supplemented the data of CD4+ T, CD8+ T, IL‐6, IL‐10 at the time of admission and before discharge (the test time point was 1–3 days before discharge). The results showed that there was no significant difference between CD4+ T, IL‐6, and IL‐10 at the time of admission and before discharge, while the CD8+ T before discharge was higher than that at admission in the mild group, and the difference was statistically significant. The CD4+ T, CD8+ T, and IL‐6 in patients in the severe group improved before discharge, suggesting that the cellular immune function of the patients had been restored. The IL‐10 values before discharge were not statistically different from those at admission, and may be related to the testing time point. The CD4+ T, IL‐6, and IL‐10 levels in the four dead patients were significantly different from those at admission, suggesting that a strong inflammatory response occurred before the patients died.
Table 6.
Time of diagnosis, time of treatment in hospital, time of nucleic acid conversion to negative and total time of hospitalisation of the 123 COVID‐19 patients.
| Characteristic | Mild group | Severe group | P |
|---|---|---|---|
| Time of diagnosis, days, n (mean [SD]) | 102 (5·64 [4·12]) | 21 (7·80 [3·66]) | 0·161 |
| Time to treatment in hospital, days, n (mean [SD]) | 102 (7·89 [4·16]) | 21 (9·80 [3·64]) | 0·425 |
| Time of nucleic acid conversion to negative, days, n (mean [SD]) | 102 (7·78 [2·56]) | 17 (12·60 [5·87]) | 0·126 |
| Total time of hospitalisation, days, n (mean [SD]) | 102 (10·46 [2·91]) | 21 (13·60 [3·77]) | 0·041 |
| Disease evolution, n (%) | |||
| Mild to severe | 14 (11·38) | – | – |
| Severe to mild | – | 14 (66·67) | – |
| Mortality rate | 0 (0) | 4 (19·05) | – |
| Cure rate | 102 (100) | 17 (80·95) | 0·0007 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Table 7.
Changes in CD4+ T, CD8+ T, IL‐6, and IL‐10 on admission and before discharge in the two groups.
| Items | Detect time | N (mean [SD]) | P |
|---|---|---|---|
| Mild group | |||
| CD4+ T | On admission | 102 (451·3 [23·04]) | 0·4666 |
| Before discharge | 102 (476·1 [24·88]) | ||
| CD8+ T | On admission | 102 (288·6 [14·23]) | 0·0004 |
| Before discharge | 102 (388·1 [23·6]) | ||
| IL‐6 | On admission | 102 (2·664 [0·09]) | 0·637 |
| Before discharge | 102 (2·724 [0·09]) | ||
| IL‐10 | On admission | 102 (6·164 [1·08]) | 0·9003 |
| Before discharge | 102 (6·314 [0·51]) | ||
| Severe group who survived | |||
| CD4+ T | On admission | 17 (284·2 [30·2]) | 0·006 |
| Before discharge | 17 (437·2 [42·35]) | ||
| CD8+ T | On admission | 17 (170·1 [21·03]) | 0·0108 |
| Before discharge | 17 (297·1 [56·85]) | ||
| IL‐6 | On admission | 17 (28·78 [7·33]) | 0·036 |
| Before discharge | 17 (10·2 [3·28]) | ||
| IL‐10 | On admission | 17 (4·452 [0·38]) | 0·0867 |
| Before discharge | 17 (3·579 [0·27]) | ||
| Severe group who dead | |||
| CD4+ T | On admission | 4 (247·5 [22·91]) | 0·0286 |
| Before discharge | 4 (179·8 [6·86]) | ||
| CD8+ T | On admission | 4 (125·3 [16·39]) | >0·9999 |
| Before discharge | 4 (128·5 [29·23]) | ||
| IL‐6 | On admission | 4 (25·95 [6·69]) | 0·0286 |
| Before discharge | 4 (827 [577·1]) | ||
| IL‐10 | On admission | 4 (4·828 [0·58]) | 0·0286 |
| Before discharge | 4 (20·38 [7·51]) | ||
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
By the end of 4 February 2020, all patients in the mild group had been discharged and four patients in the severe group had died. These deaths occurred at 2, 9, 12, and 13 days after hospitalisation. There was a significant difference in survival rate between the mild and severe groups. Additionally, we selected a statistically significant index of IL‐6 and divided the patients into a within normal values IL‐6 group (n = 76) and an above normal values group (n = 47) and analysed the survival rate. There were no deaths in the within normal group, and four deaths occurred in the above normal group. There was a significant difference in the survival rate of the two groups (Fig. 3). The overall survival (OS) of the severe group and above normal IL‐6 group were the same, the median value and 95% CI was 10·5 (7·563–14·44) day. The progression‐free survival (PFS) of the mild group and within normal IL‐6 group were 5 (3·692–6·736) and 5 (3·565–7·149) day, respectively.
Fig 3.
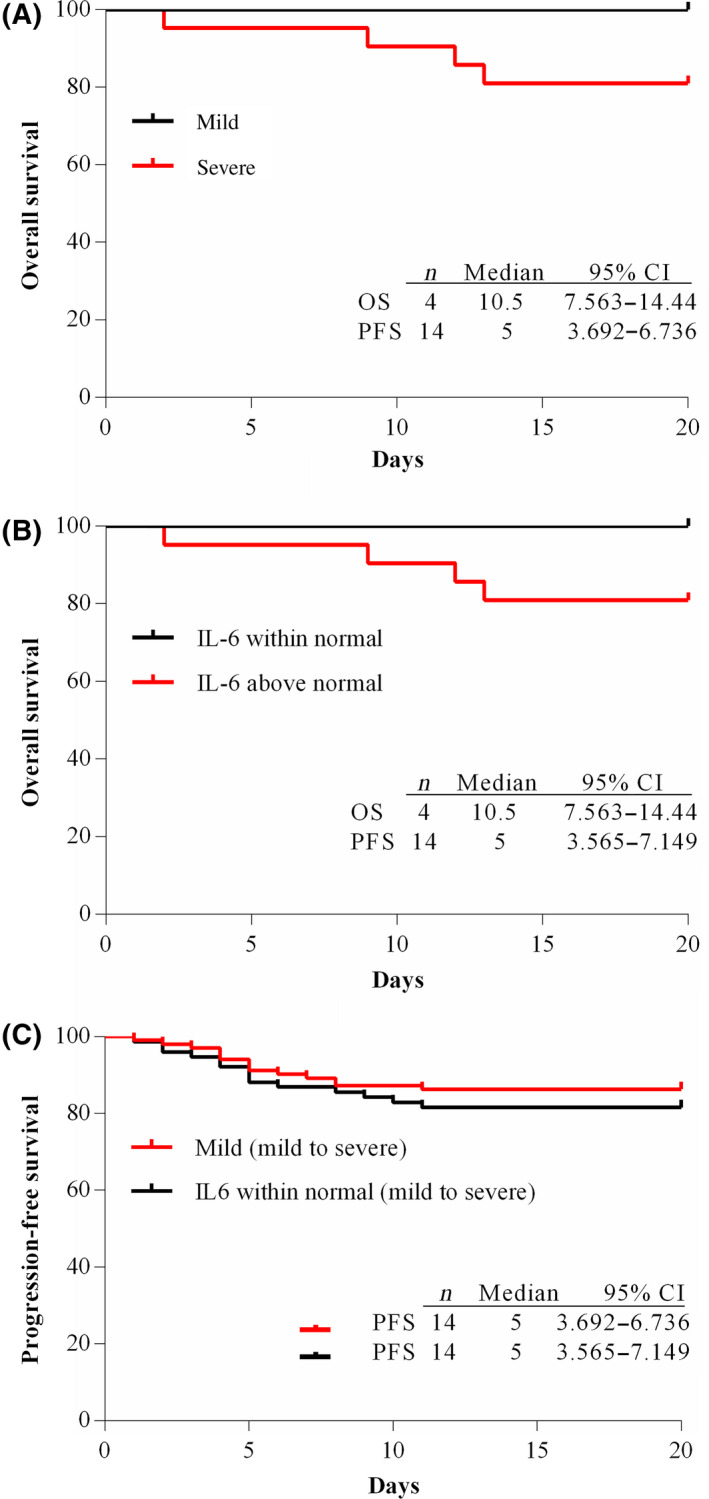
Kaplan–Meier curves of mild and severe patients, within normal and above normal values IL‐6 patients. (A) Kaplan–Meier curve of mild and severe group; (B) Kaplan–Meier curve of within normal and above normal IL‐6 group; (C) Progression‐free curve of the mild group and within normal IL‐6 group. [Colour figure can be viewed at http://www.wileyonlinelibrary.com/]
Discussion
Lymphocyte subsets play an important role in cellular immune regulation with each cell restricting and regulating one another. We found that CD4+ T and CD8+ T in the severe group had greater reductions than those in the mild group. This suggested that T lymphocytes provide an important defence against COVID‐19, which is consistent with the conclusions of Chen [8] involving the severe acute respiratory syndrome (SARS) coronavirus. The human body responds in a similar way when coping with homologous coronavirus infection. There was no significant difference between the mild and severe groups in B cells and NK cells, suggesting that they had a less important role in the disease evolution [9]
Huang et al. [10] reported that the levels of IL‐2, IL‐7, IL‐10, TNF‐α, granulocyte‐colony stimulating factor (G‐CSF), inducible protein 10 (IP‐10), monocyte chemoattractant protein‐1 (MCP‐1), and macrophage inflammatory protein 1‐alpha (MIP‐1α) were significantly higher in 2019‐nCoV‐infected patients in the ICU than non‐ICU patients. The incidence of acute respiratory distress syndrome, secondary infection, shock, acute heart and kidney injury was significantly higher in ICU patients than in non‐ICU patients. The results of the present study demonstrated that there was no significant difference in IL‐4, IL‐17, and TNF between the severe and the mild groups. The IL‐6 and IL‐10 levels in the severe group were significantly higher than those in the mild group. In the mild group, 55·88% of the patients had an IL‐6 detection value of zero. The reason for this is unclear, but it may be related to the inhibition of T helper cell type 2 (Th2) cells involved in humoral immunity in the early stage of infection. This suggests that IL‐6 and IL‐10 can be used to predict the transition from mild to severe infection in patients. If so, this would be consistent with the concept of ‘cytokine storm’ presented by Liu et al. [11]
After entering the lungs by respiration, 2019‐nCoV activates immune cells, cytokines, and other pathogen‐resistance systems. In the struggle between the virus and the human host, the immunity of the patient decreases and the virus virulence increases. This leads to congestion and oedema of the lung tissue, thickening of the interstitial lung, and increased exudation in the alveolar space to form a transparent membrane‐like structure. CT is very important because it is the main tool for screening, primary diagnosis, and evaluation of disease severity of COVID‐19. We used the PII to quantify the lung lesions in patients with COVID‐19 and found that the PII was significantly higher in the severe group compared with the mild group. The PII was positively correlated with IL‐6 and IL‐10 in the mild group, but not in the severe group, suggesting the value of the PII as an independent factor to predict the transition from mild to severe infection. Although the PII can independently predict the transition from mild to severe infection in patients, the high costs of CT and radiation indicate the more economical protocol of checking patients every 3–5 days. Compared with CT, cytokines and lymphocyte subsets are easier to obtain, and the present study found that they can also predict the transition from mild to severe infection. Therefore, in clinical practice, we suggest that the PII be combined with examination of cytokines and lymphocyte subsets to accurately determine changes in the patient’s condition. This will allow prevention and advance treatment to improve the cure rate and reduce the mortality rate.
Our hospital has formulated a ‘five‐in‐one’ diagnosis and treatment plan, and established a multi‐disciplinary team (MDT) team to optimise the treatment scheme and improve the curative effect based on the results of IL‐6, IL‐10, and PII. Specific measures include: (i) Western medicine: lopinavir–ritonavir/abidol, IFN, chloroquine are mainly used [12, 13, 14]; (ii) Traditional Chinese Medicine (TCM): lianhuaqingwen, Chinese medicine granules and decoctions as well as COVID‐19 mixtures were prescribed according to patient symptoms. The participation rate of TCM was 90·82%, with a total of 450 interventions [15, 16]; (iii) Psychology: patient psychology was evaluated and psychological counselling was conducted to help patients cope with their emotions and to reduce stress; (iv) Nutrition: nutritional assessment and treatments were provided for patients with COVID‐19 who could not eat. There have been >100 nutrition intervention patients and >90% of these patients had improved nutritional indicators [17]; (v) Rehabilitation: mild patients were recommended to have reduced bedtime and to receive early respiratory rehabilitation treatment (mainly Trinity breathing exercises) [18]. For patients with poor physical fitness and inability to stand, a change of position and appropriate activities are suggested to prevent further body damage caused by remaining in bed and inactivity.
The present study has several limitations. First, even though the hospital is the largest diagnosis and treatment centre for patients with COVID‐19 in the Chongqing area, the hospital only had 123 patients on 4 February 2020. This sample size was small compared with Wuhan, where the disease was first identified. This may have affected the results. Second, the humoral immunity level of the patients was not monitored, so there was a deficiency in evaluation of the immune system. Third, due to the large‐scale outbreak of the epidemic restricting the flow of people, data on healthy, negative control patients are lacking.
Future studies will use data collected from healthy patients as negative controls. Multicentre co‐operative research will be carried out with other designated treatment units. These studies will include more patients with confirmed COVID‐19 infections, have negative controls, and use a randomised study design.
Funding information
Project No. 2020CDJGRH‐YJ03 supported by the Fundamental Research Funds for the Central Universities.
Author contributions
Jinglong Lv conceived of, and designed, the study. He had full access to all study data and assumes responsibility for data integrity and the accuracy of the data analysis. Suxin Wan, Shibing Fan, Xianxiang Zhang, Qing Xiao, Kaihu Xiao, Jianglin Xiang, Bangshuo Zhang, and Yongping Chen contributed to writing the report. Lian Guo and Chunhui Lang were involved in critical revision of the report. Qingjie Yi, Zhengjun Yi, Mao Qiang, and Cailiang Gao contributed to the statistical analysis. All authors contributed to data acquisition, data analysis, or data interpretation, and reviewed and approved the final version.
Conflict of interest
All authors declare no conflicts of interest.
Acknowledgements
We thank Lian Guo, director of the Teaching Department of the Chongqing Three Gorges Central Hospital, and Chunhui Lang, director of the Foreign Affairs Department of Scientific Research, for their support. We thank LetPub (http://www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
References
- 1. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim JY, Choe PG, Oh Y, Oh KJ, Kim J, Park SJ, et al. The first case of 2019 novel coronavirus pneumonia imported into Korea from Wuhan, China: implication for infection prevention and control measures. J Korean Med Sci. 2020;35:e61. DOI: 10.3346/jkms.2020.35.e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020. [Epub ahead of print]. DOI: 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu P, Zhu J, Zhang Z, Han Y, Huang L. A familial cluster of infection associated with the 2019 novel coronavirus indicating potential person‐to‐person transmission during the incubation period. J Infect Dis. 2020. [Epub ahead of print]. DOI: 10.1093/infdis/jiaa077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. General Office of the National Health and Family Planning Commission . Diagnosis and treatment of pneumonia with a new coronavirus infection (trial version 5). Chin J Integr Tradit West Med. 2020:1–3. [Google Scholar]
- 6. Phan LT, Nguyen TV, Luong QC, Nguyen TV, Nguyen HT, Le HQ, et al. Importation and human‐to‐human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382:872–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. World Health Organization . Clinical management of severe acute respiratory infection when novel coronavirus (2019‐nCoV) infection is suspected: interim guidance, 28 January 2020. World Health Organization, 2020. Available at: https://apps.who.int/iris/handle/10665/330893. Accessed March 2020. [Google Scholar]
- 8. Chen H. Study on immune response of SARS coronavirus‐specific cytotoxic T lymphocytes. Shanghai, China: Second Military Medical University, 2005. [Google Scholar]
- 9. Xue K. Effect of atypical influenza virus infection on NK cell activity. Liaoning, China: China Medical University, 2014. [Google Scholar]
- 10. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu J, Li S, Liu J, Liang B, Wang X, Li W, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS‐CoV‐2 infected patients. 2020. DOI: 10.1101/2020.02.16.20023671 [DOI] [PMC free article] [PubMed]
- 12. Luo Y, Wang CZ, Hesse‐Fong J, Lin JG, Yuan CS. Application of Chinese medicine in acute and critical medical conditions. Am J Chin Med. 2019;47:1223–5. [DOI] [PubMed] [Google Scholar]
- 13. Shang L, Zhao J, Hu Y, Du R, Cao B. On the use of corticosteroids for 2019‐nCoV pneumonia. Lancet. 2020;395:683–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Z, Chen X, Lu Y, Chen F, Zhang W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci Trends. 2020;14:64–8. [DOI] [PubMed] [Google Scholar]
- 15. Jin R, Kong L, Yan D. Consensus on the safe and rational use of early intervention Chinese patent medicines in the novel corona‐virus pneumonia (COVID‐19) (Beijing). Chin J Hosp Pharm. 2020;1–11. [Google Scholar]
- 16. Ni L, Tao H, Yang X, Zhang J, Ni X. Analysis and strategy on prevention plan of corona virus disease 2019 by traditional Chinese Medicine. Chin Arch Tradit Chin Med. 2020;1–13. [Google Scholar]
- 17. CSPEN, C.S.f.P.a.E.N . Expert advice of medical nutritional treatment for novel coronavirus‐caused pneumonia patients. Chin Arch Gen Surg. 2020;14:1. [Google Scholar]
- 18. Liu X, Liu L, Lu Y, Feng L, Zhao F, Wu X, et al. Guidance and suggestions on rehabilitation training of integrated traditional Chinese and western medicine for functional recovery of patients with novel coronavirus pneumonia. Shanghai J Tradit Chin Med. 2020b;3:9–13. [Google Scholar]


