ABSTRACT
Introduction
Bloodstream Infections has become in one of the priorities for the antimicrobial stewardship teams due to their high mortality and morbidity rates. Usually, the first antibiotic treatment for this pathology must be empirical, without microbiology data about the microorganism involved. For this reason, the population studies about the etiology of bacteremia are a key factor to improve the selection of the empirical treatment, because they describe the main microorganisms associated to this pathology in each area, and this data could facilitate the selection of correct antibiotic therapy.
Material and methods
This study describes the etiology of bloodstream infections in the Southeast of Spain. The etiology of bacteremia was analysed by a retrospective review of all age-ranged patients from every public hospital in the Autonomous Community of Valencia (approximately 5,000,000 inhabitants) for five years.
Results
A total of 92,097 isolates were obtained, 44.5% of them were coagulase-negative staphylococci. Enterobacteriales was the most prevalent group and an increase in frequency was observed along the time. Streptococcus spp. were the second microorganisms more frequently isolated. Next, the most prevalent were Staphylococcus aureus and Enterococcus spp., both with a stable incidence along the study. Finally, Pseudomonas aeruginosa was the fifth microorganism more frequently isolated.
Conclusions
These data constitute a useful tool that can help in the choice of empirical treatment for bloodstream infections, since the knowledge of local epidemiology is key to prescribe a fast and appropriate antibiotic therapy, aspect capital to improve survival.
Key-words: Bacteremia, blood cultures, bloodstream infections, etiology
RESUMEN
Introducción
Las bacteriemias se han convertido en una de las prioridades de los Programas de Optimización de uso de Antimicrobianos (PROA) debido a sus altas tasas de morbimortalidad. Normalmente, el tratamiento antibiótico tiene que ser pautado de forma empírica, sin datos del microorganismo implicado. Por esto, los estudios poblacionales sobre la etiología de las bacteriemias son un factor clave para mejorar la elección del tratamiento empírico, ya que describen los principales microorganismos asociados a esta patología en cada área, lo que facilita en gran medida la selección del antibiótico correcto.
Material y métodos
Este estudio describe la etiología de las bacteriemias en el sureste de España durante los años 2013-2017. La etología fue analizada de forma retrospectiva estudiando los microorganismos implicados en todas las bacteriemias diagnosticadas en la Comunidad Valenciana (5.000.000 de habitantes).
Resultados
Se obtuvieron un total de 92.097 aislados clínicos, de los cuales un 44,5% fueron Staphylococcus coagulasa negativos. Las enterobacterias fueron el grupo más prevalente, su frecuencia se incrementó durante el estudio. Los cocos grampositivos, tipo Streptococcus, fueron los siguientes microorganismos que se aislaron de forma más frecuente, su frecuencia disminuyó a lo largo del periodo estudiado. A continuación, Staphylococcus aureus y Enterococcus spp. les siguieron en prevalencia, manteniéndose sus tasas estables a lo largo del estudio. Por último, el quinto microorganismo más prevalente fue Pseudomonas aeruginosa.
Conclusiones
Los datos obtenidos en este estudio constituyen una herramienta que puede facilitar la elección correcta del tratamiento empírico inicial que debe aplicarse en estos procesos.
Palabras clave: Bacteremia, hemocultivos, infecciones del torrente sanguíneo, etiología
INTRODUCTION
Bacteremia diagnosis is one of the priorities of Clinical Microbiology Laboratories due to its high morbility and mortality despite the improvements of antibiotic therapy [1, 2] Owing to this reason, the stewardship groups are choosing this pathology as a prior axis of their performance [3]. Most treatments must be started empirically, with the choice based on the clinical and epidemiological characteristics of the patients, and the local epidemiology. A key point for the manage of this pathology is the availability of empirical therapy guides for each environment, and to achieve this goal it is essential to obtain the etiological data of this pathology in every geographical area [4].
In Autonomous Community of Valencia (AVC, Spain), there is an informatic system to store the etiological data of these processes from all the public hospitals in the region, and so it is possible to perform population studies. The aim of this work was to analyze the etiology of bloodstream infections in Eastern Spain in the period 2013-2017.
MATERIAL AND METHODS
Design. All positive blood cultures from all the public hospitals of the ACV (population: 4,397,476.9) were analyzed from January of 2013 to December of 2017.
Source of information. Data were obtained from “RedMIVA”, a microbiological network that connects and unifies the information provided by Microbiological laboratories of Public Hospitals [5]. One isolated was studied per patient.
Statistical analyses. We expressed categorical variables as counts (percentage) and continuous variables as mean and standard deviation (SD) or median and InterQuartile Range (IQR), where appropriate. We assessed statistical differences between groups using the chi-square or Fisher’s exact test, for categorical variables. We used the TStudent’s test for the continuous variables. To estimate the rate per 100,000 inhabitants/year, the overall population was obtained from official statistic census. The alpha error was set at 0.05, and all p values were two-tailed. We conducted all statistical analyses using IBM SPSS Statistics version 22.0.
RESULTS
During the period studied (2013-2017) 92,097 clinical isolates were obtained. Global data were analyzed excluding coagulase-negative staphylococci (CNS), because in this study it was not possible to differentiate between culture contamination and true bacteremia caused by this microorganism. The data obtained for CNS were analysed separately.
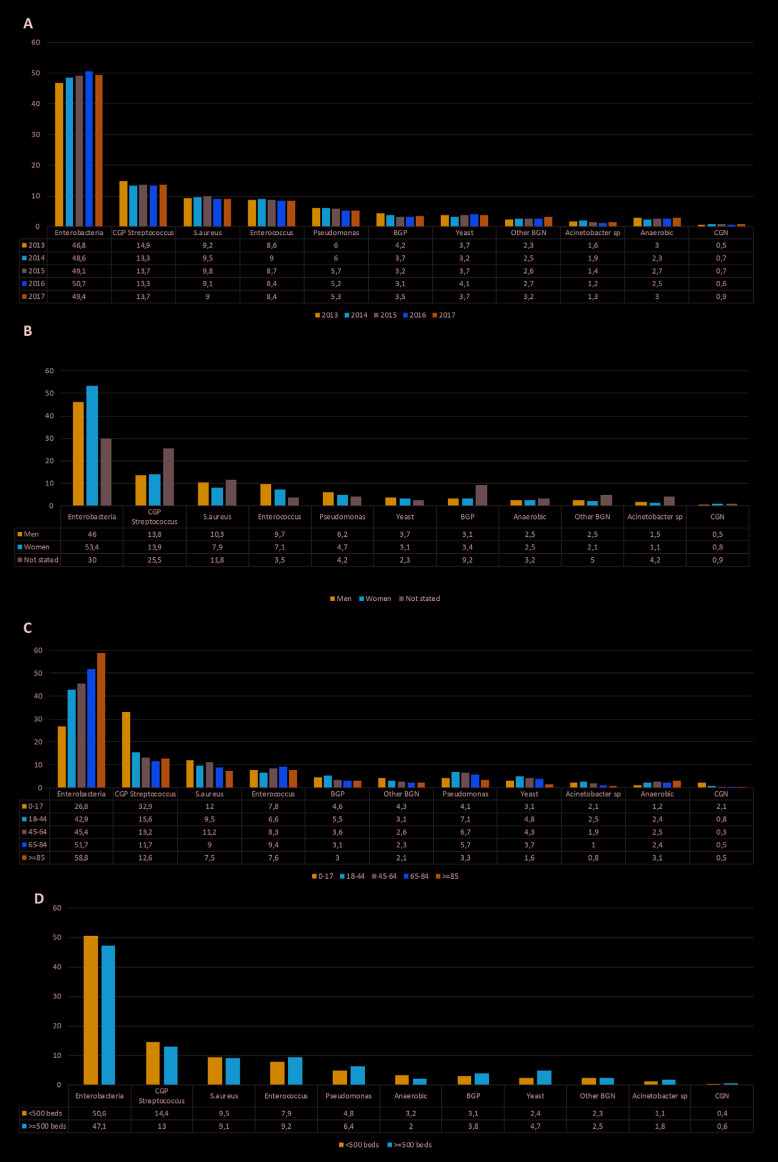
Global rate and prevalence of the main microorganisms and their clusters. The distribution of the main microorganisms classified by year, age, sex and type of hospital is shown in figure 1. The overall incidence rates of bloodstream infections per 100,000 inhabitants caused by bacteria and yeast are shown in table 1, the rates of bacteremia classified by microorganism are shown in table 2.
Figure 1.
Distribution of the main microorganisms by year (A), sex (B), age (C) and type of hospital (D).
Table 1.
Overall rate of bloodstream infections per 100,000 inhabitantsyear by year (excluding coagulase-negative staphylococci).
| Year | |||||
|---|---|---|---|---|---|
| 2013 | 2014 | 2015 | 2016 | 2017 | |
| Number of isolates | 9,450 | 9,737 | 10,203 | 10,573 | 11,177 |
| ACV total population | 5,117,190 | 5,129,266 | 5,011,797 | 5,004,171 | 4,974,769 |
| Overall rate of bacteremia | 184.67 | 189.83 | 203.58 | 211.28 | 224.67 |
| Overall rate of fungemia | 6.40 | 6.16 | 6.06 | 8.27 | 7.67 |
AVC: Autonomous Community of Valencia.
Table 2.
Overall rate of bloodstream infection per 100,000 inhabitants-year due to main Gram-positive and Gram-negative microorganisms by year.
| Year | |||||||
|---|---|---|---|---|---|---|---|
| 2013 | 2014 | 2015 | 2016 | 2017 | Average | ||
| Gram-positive | |||||||
| Streptococcus spp. | 27.45 | 25.30 | 27.79 | 28.17 | 30.85 | 27.91 | |
| S. aureus | 17.04 | 17.95 | 19.85 | 19.24 | 20.26 | 18.87 | |
| Enterococcus spp. | 15.9 | 17.13 | 17.61 | 17.72 | 18.79 | 17.43 | |
| E. faecalis | 9.92 | 11.60 | 11.59 | 10.91 | 11.81 | 11.17 | |
| S. pneumonia | 4.80 | 6.10 | 6.48 | 6.71 | 7.83 | 6.38 | |
| E. faecium | 4.80 | 4.38 | 4.66 | 5.55 | 5.66 | 5.01 | |
| Gram-positive bacillli | 7.62 | 6.99 | 6.50 | 6.37 | 7.61 | 7.02 | |
| Gram-negative | |||||||
| Enterobacterales | 86.43 | 92.29 | 99.94 | 107.07 | 111.04 | 99.35 | |
| E. coli | 54.05 | 55.03 | 59.16 | 62.74 | 67.07 | 59.61 | |
| K. pneumoniae | 12.15 | 15.28 | 18.03 | 21.02 | 20.28 | 17.35 | |
| Pseudomonas spp. | 11.04 | 11.36 | 11.59 | 10.89 | 11.87 | 11.35 | |
| P. aeruginosa | 6.40 | 6.16 | 6.96 | 8.27 | 7.67 | 7.09 | |
| Anaerobes | 5.10 | 4.40 | 4.92 | 5.17 | 6.53 | 5.22 | |
| Gram-negative bacilli | 4.06 | 4.36 | 4.90 | 5.37 | 6.47 | 5.03 | |
| Acinetobacter spp. | 3.06 | 3.33 | 2.71 | 2.19 | 2.47 | 2.75 | |
| Gram-negative cocci | 0.50 | 0.50 | 0.75 | 0.77 | 1.06 | 0.72 | |
Excluding coagulase-negative staphylococci, Enterobacterales was the most frequently isolated family (49% of the cases) for the period studied and this group showed a trend to increase along the years (p<0.0001). Streptococcus spp. were the second group isolated most often. However, the percentage of this group decreased (p<0.0001). The next most frequently isolated microorganism was Staphylococcus aureus followed by Enterococcus spp. both maintained their prevalence along the period studied. Pseudomonas aeruginosa decreased slightly in the last years (p<0.0001) (figure 1A).
When the global data were classified by sex, Enterobacterales were frequently isolated in women (p<0.0001), while S. aureus (p<0.0001), Enterococcus spp. (p<0.0001), and P. aeruginosa (p<0.0001) were more frequently isolated in men (figure 1B).
By age, the isolates of Enterobacterales reached 26.8% in people between 0 and 17 years old, versus 58.8% in people older than 85 years and Streptococcus spp. represented the 12.5% in people older than 85 years versus 32.9% in people between 0 and 17 years (p<0.0001) (figure 1C). In hospitals with less than 500 beds, Enterobacterales was the most prevalent family (p <0.0001) (figure 1D). Yeast were more prevalent in hospitals with more than 500 beds, 4.7% against to 2.4% in the smaller hospitals (p<0.0001).
Specific distribution of the most clinically relevant microorganisms. The prevalence of Escherichia coli decreased during the studied. This microorganism was more frequently isolated in women (p<0.0001), it was also the most frequently isolated in older patients (p<0.0001). On the other hand, E. coli was also more frequently isolated in hospitals with less than 500 beds than in larger hospitals (p<0.0001). It was more frequently isolated in summer, and spring, and less in winter(p<0.0001).
Klebsiella pneumoniae increased from 2013 to 2016, in contrast, in 2017 its prevalence decreased until 17.8%. It was more prevalent in men when compared to women (p<0.0001); in patients between 45 and 64 years old, and in hospitals with more than 500 beds (p<0.0001). It was more frequently isolated in autumn and summer, and less in winter (p<0.0001).
P. aeruginosa was more frequently isolated in men than women (p<0.0001); in patients between 18 and 44 years old when compared to patients older than 80 years old (p<0.0001); and in hospitals with more than 500 beds (p<0.0001). It was more frequent in summer than in winter (p<0.0001).
When the main Gram-positive microorganisms were analyzed, a decrease of Streptococcus pneumoniae annual prevalence was observed. This microorganism was more frequently isolated in men, in young patients (p<0.0001), and in hospitals with less than 500 beds (p<0.0001). It was more frequently isolated in winter than in summer (p<0.0001).
Enterococcus faecalis showed a small decrease along the study (p<0.0001), while Enterococcus faecium isolation rate remained constant. Both microorganisms were more prevalent in men (p<0.0001, both). E. faecalis was more frequently isolated in young patients than in older patients (p<0.0001). On the other side E. faecium was more prevalent in patients between 45 and 64 years. Both were more frequently isolated in hospitals with more than 500 beds (p<0.0001).
S. aureus showed minimal changes along the study. Its prevalence was higher in men than women (p<0.0001), in patients younger than 17 years old, and in patients between 45 and 64 years old (p<0.0001). It was more frequent in winter and autumn. These data are shown in table 3.
Table 3.
Distribution of the most clinical significance microorganisms by year, sex, age, hospital size and season. Data are expressed in percentage.
| Escherichia coli | Klebsiella pneumoniae | Enterococcus faecalis | Streptococcus pneumoniae | Enterococcus faecium | Salmonella | ||
|---|---|---|---|---|---|---|---|
| Year | 2013 | 61.1 | 13.7 | 11.2 | 7.3 | 5.5 | 1.4 |
| 2014 | 58.9 | 16.4 | 12.4 | 6.5 | 4.6 | 1.1 | |
| 2015 | 58.6 | 18.0 | 11.5 | 6.4 | 4.6 | 0.9 | |
| 2016 | 58.3 | 19.5 | 10.1 | 6.2 | 5.3 | 0.7 | |
| 2017 | 58.9 | 17.8 | 10.4 | 6.9 | 4.9 | 1.1 | |
| Sex | Men | 53.1 | 19.5 | 13.3 | 7.0 | 5.9 | 1.5 |
| Women | 66.6 | 14.2 | 8.4 | 6.3 | 3.8 | 0.7 | |
| No stated | 50.3 | 26.7 | 6.0 | 10.3 | 6.0 | 1.5 | |
| Age | 0-17 | 37.3 | 14.9 | 20.7 | 15.5 | 3.7 | 2.4 |
| 18-44 | 40.3 | 14.6 | 8.8 | 7.7 | 5.6 | 1.8 | |
| 45-64 | 40.6 | 20.6 | 9.5 | 6.1 | 6.2 | 0.9 | |
| 65-84 | 45.3 | 12.6 | 9.9 | 5.6 | 4.9 | 0.9 | |
| ≥85 | 70.1 | 9.5 | 8.6 | 5.4 | 3.3 | 0.5 | |
| Hospital size | <500 beds | 62.2 | 15.4 | 10.4 | 7.1 | 3.8 | 1.3 |
| ≥500 beds | 55.4 | 19.3 | 11.9 | 6.2 | 6.4 | 1.1 | |
| Seasonally | Spring | 17.0 | 4.1 | 3.3 | 1.9 | 1.4 | 0.3 |
| Summer | 17.3 | 5.3 | 3.1 | 0.6 | 1.4 | 0.4 | |
| Autumn | 16.6 | 5.5 | 3.0 | 1.8 | 1.3 | 0.3 | |
| Winter | 14.3 | 4.0 | 2.8 | 3.1 | 1.4 | 0.1 |
Analysis of coagulase-negative staphylococci. During the study period (2013-2017) 40,957 isolates (44.5%) of coagulase-negative staphylococci were obtained. In every year, the percentage of the isolates of these microorganisms remained above 40% but decreasing (p<0.0001). The presence of these microorganisms was age-related, it represented 58.1% between 0-17 years, and 39.4% in older than 85 years (p<0.0001). Besides, it is important to remark that these microorganisms were more prevalent in hospitals with more than 500 beds (48.3% vs 40.5%; p<0.0001) (table 4).
Table 4.
Distribution of coagulasenegative staphylococci by year, age and hospital size.
| Year | ||||
|---|---|---|---|---|
| 2013 | 2014 | 2015 | 2016 | 2017 |
| 46.1% | 46.3% | 44.3% | 43.5% | 42.4% |
| Age | ||||
| 0-17 | 18-44 | 45-64 | 65-84 | ≥80 |
| 58.1 | 50.0 | 45.2 | 42.2 | 39.4 |
| Hospital Size | ||||
| <500 | ≥500 | |||
| 40.5 | 48.3 | |||
DISCUSSION
Bloodstream infections are associated to high mortality and morbidity rates, generating a high cost [6]. However, despite their relevance, there is a low number of population studies that determine the etiology of these processes in wide geographic areas. Studies performed in the 70’s revealed rates of these pathologies between 80 and 189 cases per 100,000 inhabitants/year, and were higher in industrialized countries, in line with population aging and increased healthcare needs [7]. These prevalence rates are lower than the rates found in this study, that shows a rate of bloodstream infections above 200 cases per 100,000 inhabitants/year.
A great variability has been observed between geographic areas and calendar year due to population demographic changes, antibiotic prophylaxis, climatic reasons, socioeconomic status or type of study [8, 9]. Classically, it has been considered that the three most common etiologies are E. coli, S. aureus, and S. pneumoniae, which occur at approximate rates of 35, 25 and 10 per 100,000 inhabitants/year, respectively. In our environment, the prevalence rates of these pathogens were 59.61, 18.87 y 6.38 cases per 100,000 inhabitants/ year. S. pneumoniae seems to be displaced by Enterococcus spp. (16.18 cases per 100,000 inhabitants/year) probably due to great number of nosocomial bloodstream infections and the use of anti-pneumococcal vaccine [10, 11].
In our area, Enterobacterales (99.35 cases per 100,000 inhabitants/year) are the microorganisms most frequently involved in this pathology, for that reason it is mandatory to consider the inclusion of drugs active against these pathogens in empiric treatments. It was the most frequently isolated pathogen in women and older patients, probably due to bacteremia with urinary origin, and in small hospitals. K. pneumoniae was more prevalent in hospitals with more than 500 beds, probably because the important number of nosocomial infections caused by this pathogen, and its association to ICU admission [12]. This phenomenon is also characteristic of P. aeruginosa, which has a high mortality rate (29.1%) related to previous comorbidity [13]. These data suggest that empirical treatment for bacteremia in large hospitals should include drugs against these pathogens.
On the main Gram-positive microorganism, it was observed a decrease in the prevalence of S. pneumoniae. Despite of this decrease, S. pneumoniae is a pathogen to consider in the empirical treatment because of the invasive pneumococcal disease has a high prevalence in specific collectives as immunosuppressed patients, particularly in patients with HIV infection (331 cases per 100,000 inhabitants/year); in patients with an autologous or allogeneic stem cell transplantation (812 cases per 100,000 inhabitants/year), with a solid organ transplantation (465 per 100,000), and in patients with chronic inflammatory diseases (65 per 100,000), and is associated with high morbidity and mortality [14].
S. pneumoniae were more prevalent in winter, as expected due to the fact that these microorganisms are associated to respiratory pathologies, it has been published that the estimated proportion of pneumococcal pneumonia that is bacteremic is 24.8% [15]. S. pneumoniae was more frequently isolated in young patients due to its high incidence in children, in fact this pathogen is the most prevalent in this age-range (30.5%) [16]. These results show that the empirical treatment for young people in winter with bacteremia derived from pneumonia must be treated with antibiotics against this pathogen.
S. aureus were isolated more frequently in hospitals with less than 500 beds, probably because of the changes in nosocomial infection rates related to this pathogen. Our area rate (18.87 cases per 100,000 inhabitants/year) is in line with the range previously published in occidental countries (16-41 cases per 100,000 inhabitants/year). Other geographical areas have communicated an increase of this microorganism, as has occurred in our study [17].
Globally, the data obtained in this study are in line with a population study performed in Denmark. E. coli is also the most prevalent microorganism, followed by S. aureus, K. pneumoniae, E. faecium, E. faecalis, P. aeruginosa and Candida albicans. Besides, results are in agreement with the seasonality of S. pneumoniae, and the association between men patients and bacteremia due to S. aureus, Enterococcus spp. and K. pneumoniae [18].
Our data revealed many isolates of coagulase-negative staphylococci, but a lot of them are associated with patients’ skin microbiome contamination during sample collection. Previous studies, which use the Laboratory-Confirmed Bloodstream Infection (LCBI) criteria, showed that 85% of isolates are contaminants [19]. Despite this limitation we think that there is a high incidence of contaminated blood-cultures in our environment, and for that reason it is mandatory to start formative programs focused on the control of this phenomenon, as our percentage of isolations is higher than the reported in other geographical areas (37.1%) [20, 21].
In the age of antibiotic resistance, this study reveals unknown data to date about the situation of bloodstream infections in our geographical area. These results are useful for designing antibiotic policies in stewardship programs and remark the importance of knowing this type of information in every geographic area. Countries as Denmark have already started strategies in this line by the development of The Danish Collaborative Bacteremia Network (DACOBAN) research database, which is able to report data from 1.7 million residents (32% of the Danish population) [22].
FUNDING
This article has been funded by a grant (IISP 57739) from “MERCK & CO., INC” to Juan Carlos Rodriguez.
CONFLICTS OF INTEREST
The authors declare that they have no competing interest.
References
- 1.Kirn TJ, Weinstein MP. Update on blood cultures: how to obtain, process, report, and interpret. Clin Microbiol Infect 2013;19:513-20. doi: 10.1111/1469-0691.12180. [DOI] [PubMed] [Google Scholar]
- 2.Vila J, Gómez MD, Salavert M, Bosch J. Métodos de diagnóstico rápido en microbiología clínica: necesidades clínicas. Enferm Infecc Microbiol Clin 2017;35:41-6. doi/ 10.1016/j.eimc.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Merino E, Caro E, Ramos JR, Boix V, Gimeno A, Rodríguez JC, et al. . Impact of a stewardship program on bacteraemia in adult inpa-tients. Rev Esp Quimioter 2017;30:257-63. PMid: [PubMed] [Google Scholar]
- 4.Blot S. Setting the baseline to fight Gram-negative bacteraemia: the necessity of epidemiological insights. Infect Dis (Auckl) 2018:1-3. doi: 10.1080/23744235.2018.1492150. [DOI] [PubMed] [Google Scholar]
- 5.Muñoz I, Vanaclocha H, González F. [The importance of microbio-logical surveillance networks in monitoring resistant bacteria. Red-MIVA]. Rev Esp Quimioter 2007;20:193-202. PMid: . [PubMed] [Google Scholar]
- 6.Riu M, Chiarello P, Terradas R, Sala M, Castells X, Knobel H, et al. . Impacto económico de las bacteriemias nosocomiales. Com-paración de tres metodologías de cálculo. Enferm Infecc Microbiol Clin 2016;34:620-5. doi: 10.1016/j.eimc.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Poolman JT, Anderson AS. Escherichia coli and Staphylococcus aureus : leading bacterial pathogens of healthcare associated in-fections and bacteremia in older-age populations. Expert Rev Vac-cines 2018;17:607-18. doi: 10.1080/14760584.2018.1488590. [DOI] [PubMed] [Google Scholar]
- 8.Storm L, Schnegelsberg A, Mackenhauer J, Andersen LW, Jessen MK, Kirkegaard H. Socioeconomic status and risk of intensive care unit admission with sepsis. Acta Anaesthesiol Scand 2018;62:983-92. doi: 10.1111/aas.13114. [DOI] [PubMed] [Google Scholar]
- 9.Montassier E, Batard E, Gastinne T, Potel G, Cochetière MF. Recent changes in bacteremia in patients with cancer: a systematic review of epidemiology and antibiotic resistance. Eur J Clin Microbiol In-fect Dis 2013;32:841-50. doi: 10.1007/s10096-013-1819-7. [DOI] [PubMed] [Google Scholar]
- 10.Laupland KB. Incidence of bloodstream infection: a review of population-based studies. Clin Microbiol Infect 2013;19:492-500. doi: 10.1111/1469-0691.12144. [DOI] [PubMed] [Google Scholar]
- 11.Pai S, Enoch DA, Aliyu SH. Bacteremia in children: epidemiology, clinical diagnosis and antibiotic treatment. Expert Rev Anti Infect Ther 2015;13:1073-88. doi: 10.1586/14787210.2015.1063418. [DOI] [PubMed] [Google Scholar]
- 12.Chetcuti Zammit S, Azzopardi N, Sant J. Mortality risk score for Klebsiella pneumoniae bacteraemia. Eur J Intern Med 2014;25:571-6. doi: 10.1016/j.ejim.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Callejas-Díaz A, Fernández-Pérez C, Ramos-Martínez A, Múñez-Ru-bio E, Sánchez-Romero I, Vargas Núñez JA. Impacto de la bacter-iemia por Pseudomonas aeruginosa en un hospital de tercer nivel: mortalidad y factores pronósticos. Med Clin (Barc) 2019;152:83-9. doi: 10.1016/j.medcli.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 14.van Aalst M, Lötsch F, Spijker R, van der Meer JTM, Langendam MW, Goorhuis A, et al. . Incidence of invasive pneumococcal disease in immunocompromised patients: A systematic review and me-ta-analysis. Travel Med Infect Dis 2018;24:89-100. doi: 10.1016/j.tmaid.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Said MA, Johnson HL, Nonyane BAS, Deloria-Knoll M, O'Brien KL, AGEDD Adult Pneumococcal Burden Study Team, et al. . Estimating the Burden of Pneumococcal Pneumonia among Adults: A System-atic Review and Meta-Analysis of Diagnostic Techniques. PLoS One 2013;8:e60273. doi: 10.1371/journal.pone.0060273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gangoiti I, Valle JR, Sota M, Martinez-Indart L, Benito J, Mintegi S. Characteristics of children with microbiologically confirmed inva-sive bacterial infections in the emergency department. Eur J Emerg Med 2018;25:274-80. doi: 10.1097/MEJ.0000000000000453. [DOI] [PubMed] [Google Scholar]
- 17.Asgeirsson H, Thalme A, Weiland O. Staphylococcus aureus bacte-raemia and endocarditis–epidemiology and outcome: a review. Infect Dis (Auckl) 2018;50:175-92. doi: 10.1080/23744235.2017.1392039. [DOI] [PubMed] [Google Scholar]
- 18.Gubbels S, Nielsen J, Voldstedlund M, Kristensen B, Schønheyder HC, Vandenbroucke-Grauls CMJE, et al. . Utilization of blood cul-tures in Danish hospitals: a population-based descriptive analy-sis. Clin Microbiol Infect 2015;21:344.e13-344.e21. doi: 10.1016/j.cmi.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 19.Karakullukçu A, Kuşkucu MA, Ergin S, Aygün G, Midilli K, Küçükbas-maci Ö. Determination of clinical significance of coagulase-neg-ative staphylococci in blood cultures. Diagn Microbiol Infect Dis 2017;87:291-4. doi: 10.1016/j.diagmicrobio.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Garcia RA, Spitzer ED, Beaudry J, Beck C, Diblasi R, Gilleeny-Bla-bac M, et al. . Multidisciplinary team review of best practices for collection and handling of blood cultures to determine effective interventions for increasing the yield of true-positive bacteremi-as, reducing contamination, and eliminating false-positive cen-tral line–associated bloodstream infections. Am J Infect Control 2015;43:1222-37. doi: 10.1016/j.ajic.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 21.Laupland KB, Niven DJ, Pasquill K, Parfitt EC, Steele L. Culturing rate and the surveillance of bloodstream infections: a popula-tion-based assessment. Clin Microbiol Infect 2018;24:910.e1-910. e4. doi: 10.1016/j.cmi.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 22.Gradel K, Arpi M, Knudsen J, Schonheyder H, Ostergaard C, Sogaard M. The Danish Collaborative Bacteraemia Network (DACOBAN) da-tabase. Clin Epidemiol 2014;6:301. doi: 10.2147/CLEP.S66998. [DOI] [PMC free article] [PubMed] [Google Scholar]