Abstract
Objective
Long noncoding RNAs (lncRNAs) are currently considered to have a vital and wide range of biological functions, but the molecular mechanism underlying triglycerides metabolism remains poorly understood. This study aims to identify novel lncRNAs differentially expressed in rat livers with hypertriglyceridemia and elucidated the function role in TG metabolism.
Methods
Differentially expressions of lncRNAs in rat livers with hypertriglyceridemia were identified by transcriptome sequencing and validated by real-time PCR. The role of lnc19959.2 in triglyceride metabolism was assessed both in vitro and in vivo. RNA pulldown and RIP assays were conducted to evaluate the interactions between lnc19959.2 and its target proteins. ChIP and Dual report assays were performed to detect the interactions between transcription factors and promoters of its target genes.
Results
We identified a novel lncRNA, and lnc19959.2 was upregulated in rat livers with hypertriglyceridemia. The knockdown of lnc19959.2 has profound TG lowering effects in vitro and in vivo. Subsequently, the genome-wide analysis identified that the knockdown of lnc19959.2 caused the deregulation of many genes during TG homeostasis. Further mechanism studies revealed that lnc19959.2 upregulated ApoA4 expression via ubiquitinated transcription inhibitor factor Purb, while it specifically interacted with hnRNPA2B1 to downregulate the expression of Cpt1a, Tm7sf2, and Gpam, respectively. In the upstream pathway, palmitate acid upregulated CCAAT/Enhancer-Binding Protein Beta (Cebpb) and facilitated its binding to the promoter of lnc19959.2, which resulted in significant promotion of lnc19959.2 transcriptional activity.
Conclusions
Our findings provide novel insights into a new layer regulatory complexity of an lncRNA modulating triglyceride homeostasis by a novel lncRNA lnc19959.2.
Keywords: LncRNAs, Triglyceride metabolism, Transcriptional regulation, Epigenetics
Graphical abstract
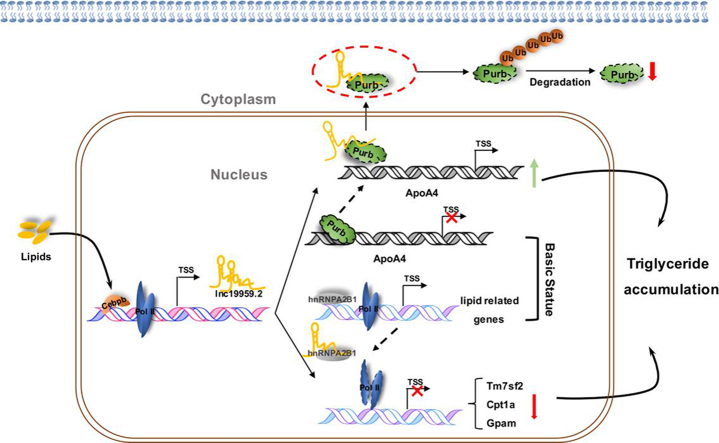
Intracellular high triglyceride status facilitated Cebpb binding to promoter of lnc19959.2, resulted in increasing transcriptional activity of lnc19959.2. On the one hand, Lnc19959.2 up-regulate ApoA4 expression via ubiquitinated transcription inhibitor factor Purb; on the other hand, lnc19959.2-hnRNPA2B1 complex acts on Cpt1a, Tm7sf2 and Gpam in some ways to reduce the efficiency of RNA polymerase II occupy on the target gene promoter. Finally, these processes exacerbate the accumulation of intracellular triglycerides leading to hypertriglyceridemia.
Highlights
-
•
lnc19959.2 was identified as a novel LncRNA in hypertriglyceridemic rat liver.
-
•
lnc19959.2 was involved in triglyceride metabolism in vivo and in vitro.
-
•
lnc19959.2 upregulated ApoA4 expression via ubiquitinated transcription inhibitor factor Purb.
-
•
lnc19959.2 interacted with hnRNPA2B1 and cooperated with RNP II that controls expression of Cpt1a, Tm7sf2 and Gpam.
1. Introduction
Hypertriglyceridemia (HTG) is commonly encountered in the clinic [1]. Of all adults, 27% have HTG (plasma triglycerides level>2 mmol/L) [2]. Increasing production of triglyceride-rich lipoproteins can lead to HTG, resulting in increasing the presence of the remnant particles in the circulation [3]. Recently, a large-scale Mendelian randomization experiment provided compelling consistent evidences that triglycerides are in causal pathways for the development of coronary heart disease [4]. Moreover, elevated triglyceride levels are considered as strong and independent predictors for atherosclerotic cardiovascular disease (ASCVD) and mortality [[5], [6], [7]]. Despite improved ASCVD outcome with statin therapy, residual risk remains [[8], [9], [10]], indicating that triglyceride-rich lipoproteins may be an important additional target for therapy. As expected, patients with elevated triglyceride levels, despite the use of statins, additionally received Icosapent Ethyl, one class of triglyceride-lowering drugs, which can significantly reduce the risk of ischemic events, including cardiovascular death [11,12]. Thus, triglyceride-lowering therapy has gained renewed interest. Given important clinical relevance and therapeutic implications of HTG, clarification of molecular mechanism underlying HTG development and progression is critical to find potential drug targets of HTG.
Long noncoding RNAs (lncRNAs), no-protein-coding transcripts longer than 200 nucleotides (nt), are located within both the cytoplasm and the nucleus [[13], [14], [15]]. Currently, with the development of high-throughput sequencing technology, a large number of lncRNAs have been found and thought to have a vital and wide range of biological functions [16], including chromatin modification [17], genomic imprinting [18], and modulation of post-translational RNA processing [19]. In recent years, despite the fact that many novel lncRNAs have been implicated in the pathogenesis of hepatocyte cholesterol metabolism [20,21], systemic lipid homeostasis [22], and high-fat diet-related obesity [23,24], little is known about the biological function and significance of the potential molecular mechanism about lncRNAs in high-fat diet-induced HTG.
In the current study, in order to investigate the potential role of novel lncRNA in regulating the development of high-fat diet-induced HTG, we used transcriptome profiling to identify a novel lncRNA related to HTG in rats. Our study may provide new clues on the molecular events for potential therapeutic targets of HTG patients.
2. Methods
2.1. Animal models
All experimental procedures involving the use of animals were conducted in accordance with the National Institutes of Health (NIH) and ARRIVE Guidelines for the welfare of animals, and the operation on the laboratory animals was approved by the Animal Care and Use Committee of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China). A total of 24 healthy male Sprague–Dawley rats weighing in around 200 g were obtained from the Animal Development Center (Chinese Academy of Sciences, Shanghai). All rats were acclimated for 1 week before the initiation of the experiment and maintained on a 12/12 h light/dark cycle with free access to food and water. Rats were randomly divided into two groups (n = 12 each): control group (normal diet (ND) group) that received regular rodent chow diet (59.6% carbohydrate, 8.5% fat, and 20.3% protein in total calories) and high-fat diet (HFD) group that were fed with a high-fat diet (43% carbohydrate, 41% fat, and 17% protein in total calories) for 24 weeks. To control the cost of the experiment, three rats were randomly selected from the control group and five rats with higher lipid profiles from the HFD group for subsequent transcriptome sequencing.
2.2. BRL-3A cell line and transient transfections
The BRL-3A rat liver cell line and HEK293T human renal epithelial cell lines (American Type Culture Collection) were cultured in glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Life Technologies, Carlsbad, CA, USA) at 37 °C with humidified air and 5% CO2. All of siRNAs and plasmids were transfected into cells applying Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol. siRNAs were purchased from Riobio Co., Ltd. (Guangzhou, China).
2.3. RNA extraction and transcriptome sequencing
The total RNA was extracted from each individual sample using RNAiso Plus Total RNA extraction reagent (Takara Biotechnology, Dalian, China) following the manufacturer's protocols. The purity and quantity of total RNA were determined by using the NanoDrop ND-2000 (Thermo Fisher, Wilmington, DE, USA). The integrity of RNA was assessed by Agilent 2100 Bioanalyzer system with an RNA 6000 Nano Assay Kit (Agilent Technologies, Santa Clara, CA, USA) as instructed. RNA was further purified by using the RNeasy Micro Kit and RNase-Free DNase set according to the manufacturer's recommendations (QIAGEN, GmBH, Germany).
Initially, a total amount of 3 μg RNA per sample was used as a genetic material for the RNA sample preparations. Ribosomal RNA was removed by using the Ribo-Zero rRNA Removal Kit (Epicenter, USA), and ribosomal RNA-free residue was cleaned up by using ethanol precipitation. Sequencing libraries were generated using TruSeq RNA Sample Preparation Kit v2 (Illumina, San Diego, CA, USA) following the manufacturer's instructions. Subsequently, the products of libraries were purified using AMPure XP system (Beckman Coulter, Beverly, CA, USA). Finally, the quantity and quality of each library were assessed by Qubit® 2.0 Fluorimeter (Invitrogen Life Technologies, Carlsbad, USA) and Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA), respectively. All the sequencing libraries' construction and transcriptome sequencing were performed at the Shanghai Biotechnology Corporation (Shanghai, China) on an Illumina Hissed 2500 platform (Illumina, San Diego, CA, USA) according to the Selex transcriptome sequencing protocols. All of the RNA transcriptome sequencing data have been uploaded to the GEO database.
2.4. Quantitative real-time PCR validation
The total RNA was extracted from rats' liver tissues using TRIZOL reagent (Life Technologies, Carlsbad, CA) and then the 1ug of total RNA was converted to cDNA in 20 μL of reverse transcriptase reaction with PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara Biotechnology, Dalian, China) according to the manufacturer's instructions. Real-time PCR was performed with the SYBR Green (Takara Biotechnology, Dalian, China) detected by a 7900HT FAST Real-Time PCR System (Life Technologies, Carlsbad, CA, USA). For quantitative analysis, the relative expression level of lncRNA or mRNA was normalized to an internal invariant control, U6 small nucleolar RNA, or GAPDH. Each reaction was performed in triplicate, and the expression of each lncRNA or mRNA was represented as fold change by using the 2-▵▵CT method. Primers used in the present study were listed in Supplementary Table 2.
2.5. Northern Blot
Total RNAs were extracted from BRL-3A cells, and agarose gel electrophoresis was applied to censor the quality of total RNAs at the same time. Then, we identified the expression of lnc19959.2 through Northern Blot with DIG Northern Starter Kit (Roche, USA). To prepare the probes by PCR for lnc19959.2, formaldehyde was used to denature gel electrophoresis overnight and transfer it to the nylon membrane (NC). Prehybridization of cross-linked membranes was performed with DIG Easy Hyb, and lnc19959.2 expression was detected with probes (F: 5′ TACCTCAGATACTCAGCCACA 3′, R: 5′ GCAGGGTGCAGACTTCC 3’). Finally the Negative Control (NC) membrane was washed and the signal was detected and exposed in the darkroom with X-ray film using a Digoxin Hybridization Test Kit II (Roche, USA).
2.6. RACE
Rapid amplification of cDNA ends (RACE) assays were rapid amplification of cDNA fragments from low-abundance transcripts to obtain a full-length cDNA with 5′ and 3′ ends. PCR experiments were performed with SMARTer RACE cDNA Amplification Kit (Clontech, USA) according to the manufacturer's protocol. The PCR products were detected by agarose gel electrophoresis and sequenced by Sanger Sequencing. The 5′ and 3′ RACE Gene Special Primers (GSPs) were listed in Supplementary Table 3.
2.7. RNA pulldown assays
RNA pulldown assays were performed with Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, USA) according to the manufacturer's protocol. Sense and antisense transcript of NONRATT019959.5 was labeled by Biotin RNA Labeling Mix (Roche Diagnostics, USA). RNA was incubated with DNase I at 37 °C, for 15 min, mixed with cell lysates at 4 °C for 1 h in rotation, and then 30 μL magnetic beads were added (Thermo Fisher Scientific, USA) combined with proteins. Complexes were detected by mass spectrometry and confirmed by western blot. The primers sequence was listed in Supplementary Table 5 and the proteins pulled down by lnc19959.2 were listed in Supplementary Table 6.
2.8. RIP
RNA immunoprecipitation (RIP) assays were conducted using the Magan RIPTMRNA–Binding Protein Immunoprecipitation Kit (Millipore, USA). Anti-Purb, Anti-hnRNPA2B1 (Abcam, Hong Kong, China), and Anti-IgG (Abnova Corporation, Taiwan, China) were immunoprecipitated with lysed cell extracts according to the manufacturer's protocol. Immunoprecipitated RNA was quantified by RT-qPCR. Each reaction was performed in triplicate.
2.9. ChIP
Chromatin immunoprecipitation (ChIP) assays were conducted with ChIP Assay Kit (Beyotime Institute of Biotechnology, Nanjing, China) according to the manufacturer's protocol. We fixed cells using paraformaldehyde to crosslink chromatin DNA and sonicated it to 200–800 bp fragments. Anti-Purb, anti-Cebpb, anti-RNA polymerase II (RNP II), and anti-hnRNPA2B1 (Abcam Hong Kong, China) and anti-IgG (Abnova Corporation, Taiwan, China) were used to immunoprecipitate DNA. The primer sequence was listed in Supplementary Table 7.
2.10. Quantification and statistical analysis
All data were presented as mean ± standard error of the mean (SEM). Significance was assessed by one-way ANOVA followed by Tukey's test and LSD for multiple comparisons. A Chi-square test was used to assess the significance of variables in the contingency table. The correct P values for these multiple tests were calculated by false discovery rate (FDR), unless otherwise indicated. All the statistics were performed in SPSS 22.0 (SPSS Inc., Chicago, Illinois, USA) for Windows (Microsoft Corp, Redmond, Washington, USA). A P value < 0.05 was considered statistically significant. All probability values were 2-sided.
2.11. Data and software availability
2.11.1. Accession number
The accession number for the microarray published here is GEO: GSE137712, GSE99557, and GSE137131. All informatics analysis was performed by using the R language platform, a well-known open-source software environment for statistical computing and graphics.
3. Results
3.1. lncRNA lnc19959.2 was upregulated during hepatocyte triglyceride metabolism in vivo and in vitro
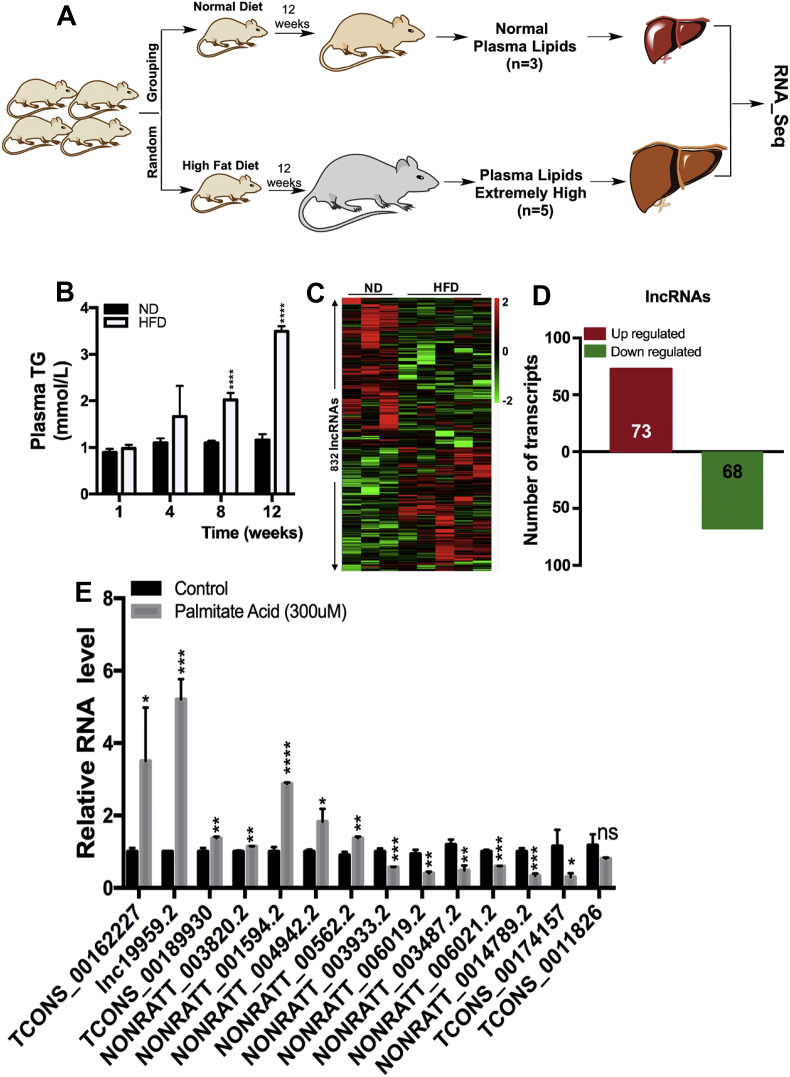
In an attempt to discover the functional lncRNAs associated with hepatocyte triglyceride metabolism in vivo, we performed RNA-seq analysis of ribosomal RNA-depleted total RNA in liver tissues from Sprague–Dawley (SD) rats fed with or without HFD (Figure 1A,B and Supplementary Figs. 1A–1C; ND (n = 3) versus HFD (n = 5), ∼1 mmol/L versus ∼3.5 mmol/L), a widely used animal model for hypertriglyceridemia. Global lncRNA profile analysis revealed that 832 out of 31244 lncRNAs were significantly differentially expressed (Figure 1C). Among these differentially expressed lncRNAs, 68 lncRNAs were downregulated and 73 lncRNAs were upregulated (P < 0.05; fold change>2.0, Figure 1D). An upregulated and uncharacterized lncRNA, termed lnc19959.2 (gene symbol NONRATT019959.2), was successfully validated by RT-qPCR and further confirmed the upregulation in BRL-3A rat liver cell lines by stimulating with palmitic acid (PA) (Figure 1E) which was especially upregulated TG level (Supplementary Figs. 1D and 1E). Given that relevancy of lnc19959.2 in hepatocyte triglycerides metabolism in vitro and in vivo, it was chosen for subsequent functional study.
Figure 1.
Lnc19959.2 is a novel lncRNA highly expressed in rat fatty livers. (A) Schematic diagram shows rat livers (ND, n = 3; HFD, n = 5) selected for RNA-seq. (B) Serum total triglyceride (TG) levels in normal diet (ND) and high-fat diet (HFD) rats. Serum TG was significantly increased after 8 weeks in HFD rats. (C) Heatmap of 832 different expression lncRNAs (ND, n = 3; HFD, n = 5). (D) The number of significantly changed lncRNAs (red indicates the upregulated lncRNAs, with fold change > 2.0; green indicates the downregulated lncRNAs, with fold change<0.5). (E) RT-qPCR analysis the lncRNA expressions after treating BRL-3A cells with palmitate acid (300 μM) for 24 h. The CT value of GAPDH was 13.483 ± 1.192 (n = 3) and lnc19959.2 was 21.791 ± 0.171 (n = 3, mean ± SD). Unpaired t-test was used to measure the statistical significance; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001. Results are mean ± SEM of three independent experiments with duplicate wells.
3.2. Identification and characterization of lnc19959.2 as a long-chain noncoding RNA
Initially, northern blot with overlap probe detected two transcripts (Supplementary Fig. 2A): one transcript for Slc35c2 (1859 nt less than 2027 nt) and the other transcript for lnc19959.2 (more than 2332 nt), respectively. To further identify the exact sequence and full length of lnc19959.2, we applied the SMART RACE technology to get its 3′ and 5’ end sequences (Supplementary Figs. 2B and 2C) and found that the total length of lnc19959.2 transcript was 2501 nt (Supplementary Fig. 2D). Subsequently, using a BLAST search of the lnc19959.2 sequence retrieved from the NONCODE database, we identified that this lncRNA was located on a modestly conserved locus chromosome 12.3q (Supplementary Figs. 2E and 2F) and partially overlapped with Slc35c2 gene exons 8, 9, and 10. Like other lncRNAs, lnc19959.2 was transcribed with a polyA + tail (Supplementary Fig. 2G) but was without coding capacity (Supplementary Fig. 2K–2L). Lnc19959.2 was mainly located in the nucleus but not the cytoplasmic region, which is different from the Slc35c2 gene (Supplementary Fig. 2H). The analysis of the expression of lnc19959.2 in different rat major tissues revealed that it was widely distributed and showed no more tissue-restricted expression patterns (Supplementary Figs. 2I and 2J).
3.3. Knockdown of lnc19959.2 has profound TG lowering effects in vitro and in vivo
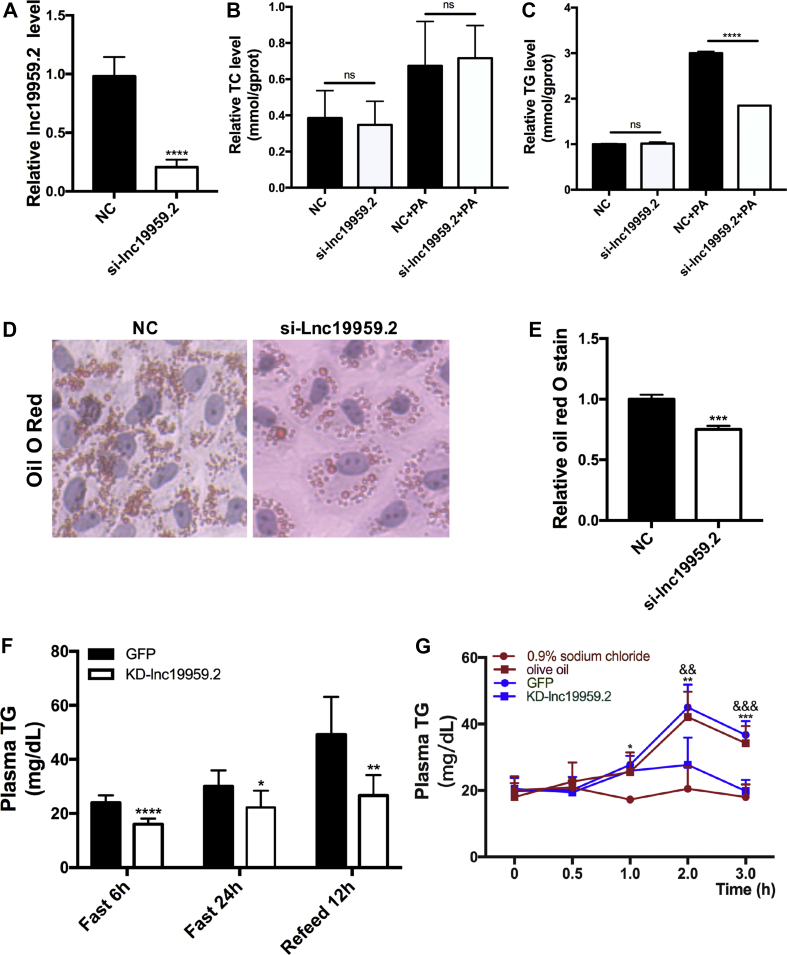
To explore the functional role of lnc19959.2 in the regulation of hepatocyte lipid metabolism in vitro (Supplementary Fig. 3A), we tested and identified a smart silence RNA that could specifically suppress the expression of lnc19959.2 but not Slc35c2 gene for more than 80% (Figure 2A). Remarkably, the knockdown of lnc19959.2 decreased triglycerides but not cholesterol levels along with PA physiological simulation in BRL-3A cells (Figure 2B,C). Consistent with this interpretation, we observed that fewer lipid droplets accumulate in the lnc19959.2 knockdown status (Figure 2D,E). By contrast, similar results cannot be found in the Slc35c2 knockdown status (Supplementary Figs. 3D and 3E).
Figure 2.
Knockdown lnc19959.2 reduces plasma TG levels in rats. (A) RT-qPCR analysis lnc19959.2 expression level following transient transfection of BRL-3A cells with smart siRNA of lnc19959.2. (B–C) The TC and TG levels detected in BRL-3A cells after transient transfection siRNA of lnc19959.2 with or without PA. (D–E) Oil Red O-hematoxylin staining of BRL-3A cells after transient transfection siRNA of lnc19959.2 with PA. (F) Plasma TG levels in high-fat diet (HFD) rats of control (GFP, n = 6) and lnc19959.2 KD (KD-lnc19959.2, n = 9) after a 6-hour fast (Fast 6 h), a 24-hour fast (Fast 24 h), or 12-hour refeeding after a 24-hour fast (Refeed 12 h). (G) Blood triglyceride levels detected at different time points (0, 0.5, 1, 2, and 3 h) after receiving olive oil by oral gavage in ND rats which are injected with adenovirus and, for another group, after receiving olive oil and 0.9% sodium chloride, separately (∗P < 0.05 means 0.9% sodium chloride versus olive oil, and P < 0.05 means GFP versus KD-lnc19959.2). Unpaired t-test was used to measure the statistical significance; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
We further assess the impact of the knockdown of lnc19959.2 in the regulation of TG metabolism in vivo by using different rat models. As expected, efficiently knockdown of lnc19959.2 with adenovirus dramatically reduced serum TG levels in all three feeding conditions (Figure 2F and Supplementary Figs. 3F–3H). In addition, we observed that the knockdown of lnc19959.2 showed a marginal increase in their circulating TG that quickly returned to baseline in 3 h of lipid load assay with orally delivering a dose of olive oil (Figure 2G) in the ND rat model. To determine whether lnc19959.2 is a novel candidate therapeutic target lncRNA for improving the conditions of hypertriglyceridemia, we administrated lnc19959.2 knockdown adenoviruses into a high-fat-fed rat with hypertriglyceridemia. Plasma TG level was significantly reduced by 30%∼50% but not in the TC level on the lnc19959.2 knockdown rats.
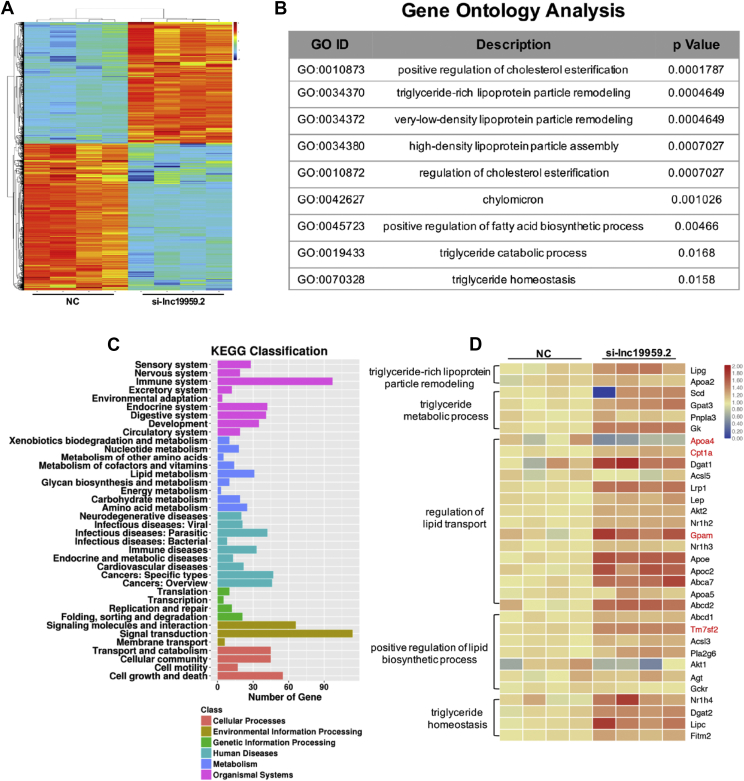
3.4. Lnc19959.2 is required to maintain the expression of TG-Associated genes
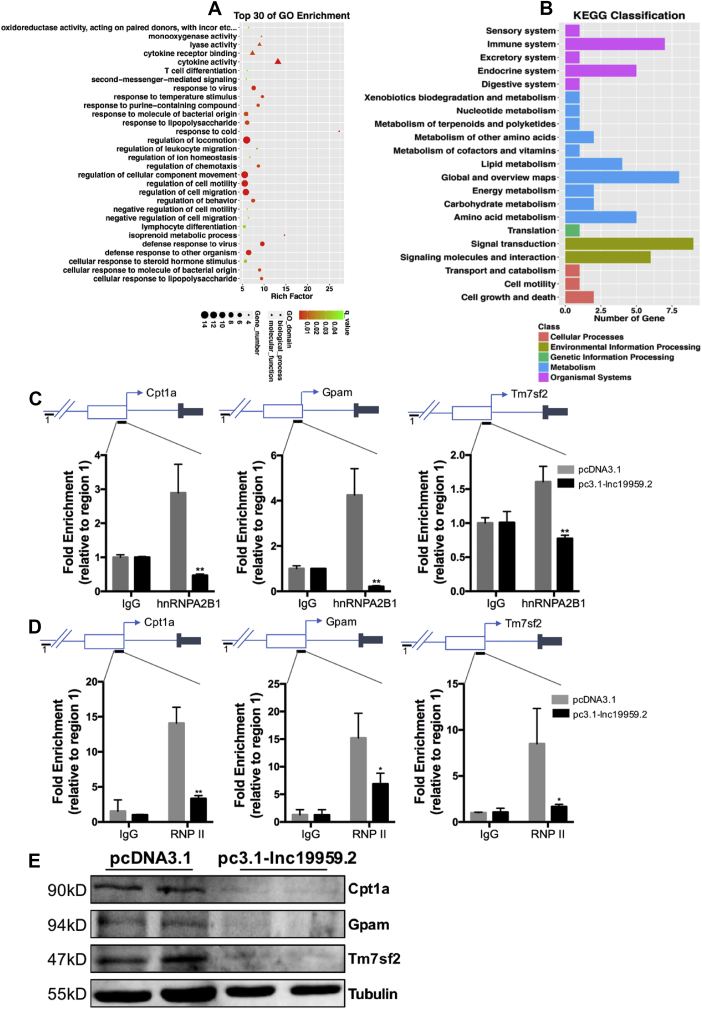
To better decipher the contributions of lnc19959.2 to hepatocyte triglyceride metabolism, a global gene expression profiling analysis was performed in lnc19959.2-silenced BRL-3A rat liver cell lines (Figure 3A and Supplementary Fig. 4A). Initially, gene ontology analysis differentially expressed genes showed enrichment in lipid, especially in triglyceride metabolic processes (Figure 3B,C and Supplementary Fig. 4C). Subsequently, we used RT-qPCR analysis to confirm that these genes from the gene ontology (GO) pathways were regulated partially. It is noteworthy that ApoA4 was statistically downregulated, while Cpt1a, Tm7sf2, and Gpam were upregulated in the lnc19959.2-silenced BRL-3A cell line (Figure 3D).
Figure 3.
lnc19959.2 participates in many metabolism pathways and affects a series of TG-associated genes. (A) Heatmap of mRNA expression profile sequence, after knockdown of lnc19959.2 expressions in BRL-3A cells. (B) Gene ontology analysis of microarray in BRL-3A cell treatment of siRNA of lnc19959.2 and Negative Control (NC). (C) The top 30 KEGG pathways correlated with the cellular process, environmental information processing, genetic information processing, human diseases, metabolism, and organismal systems. (D) RT-qPCR detected a subset of the lipid-related pathway gene expressions.
3.5. Identification of Purb and hnRNPA2B1 as binding partners of lnc19959.2
We next sought to determine the underlying molecular mechanism by which lnc19959.2 regulates triglycerides-associated genes. Owing to lnc19959.2 mainly present in the nucleus without the evidence of translation ability, we proposed that lnc19959.2 acts as a nuclear RNA. Because the knockdown lnc19959.2 had no effect on the expression of adjacent protein-coding genes (Supplementary Figs. 5A and 5B), we excluded the possibility that lnc19959.2 acts by influencing its adjacent genes in cis-regulation.
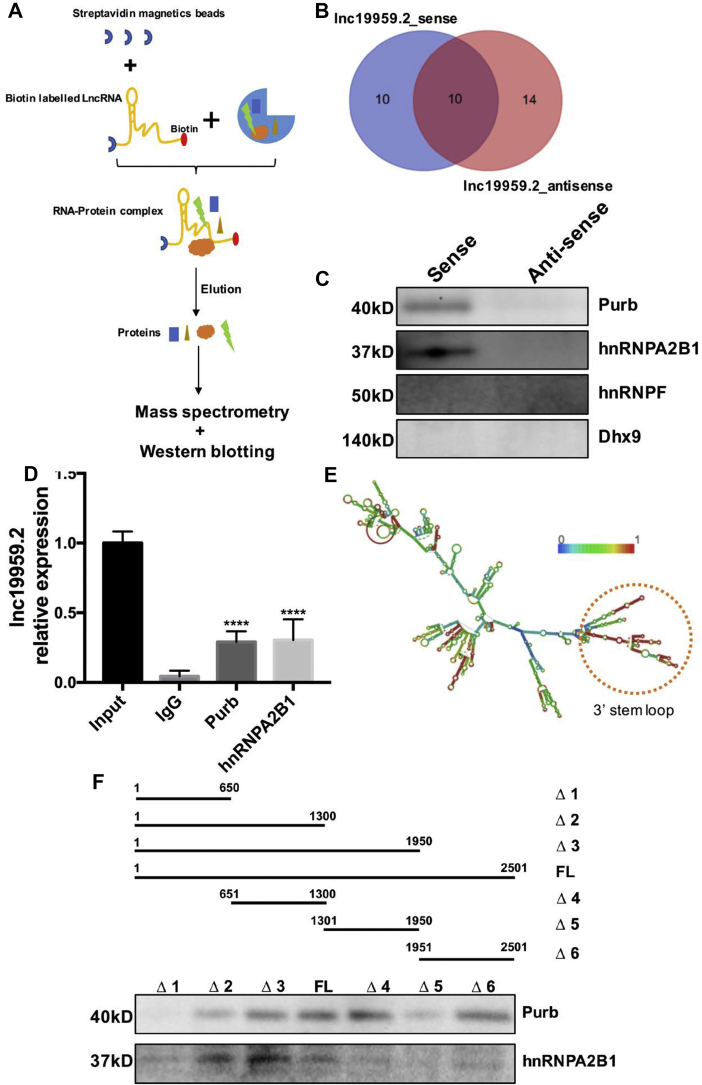
As some nuclear lncRNAs have been shown to affect transcription by physical interaction with intracellular protein [[25], [26], [27]], we next conducted RNA pulldown assays to capture potential interacting proteins. As shown in the flow diagram (Figure 4A), sense and antisense of lnc19959.2 were labeled with biotin, and then the captured and eluted proteins were determined by mass spectrometry (MS) (Figure 4B). Interestingly, Purb and hnRNPA2B1, two nuclear-localized proteins, were identified as binding partners of lnc19959.2 (Figure 4C). Subsequently, RIP assays further confirmed the specificity of these interactions (Figure 4D). RNA folding analysis identified a stable stem-loop structure within the 3′ region of lnc19959.2, indicating the necessary spatial conformation for the interaction (Figure 4E). In addition, we also predicted the protein-RNA interaction signals between 3′ and 5’ regions of lnc19959.2 with Purb and hnRNPA2B1, respectively (Supplementary Fig. 5C). To further explore that the definite region genuinely interacts with lnc19959.2, we analyzed a series of truncated sequences and found that most of the 3′ fragment and a small portion of the 5′ fragment of lnc19959.2 are involved in the interaction with Purb and hnRNPA2B1 (Figure 4F).
Figure 4.
lnc19959.2 interacts with Purb and hnRNPA2B1. (A) Experimental design for RNA pulldown assay to identify the cellular proteins associating with lnc19959.2. lnc19959.2 was transcription-labeled with biotin in vitro, refolded, and incubated with BRL-3A total cell lysates. (B) Venn diagram showing the number of different proteins binding with lnc19959.2_sense and lnc19959.2_antisense, separately, according to the MS analysis. And 10 proteins were overlapped. (C) Four of the candidate proteins filtered from mass spectrometry. Purb and hnRNPA2B1 were confirmed that they were specifically bound to lnc19959.2. (D) RIP assay analysis of Purb and hnRNPA2B1 associated with lnc19959.2. BRL-3A cell lysates were immunoprecipitated with anti-Purb and anti-hnRNPA2B1 or control mouse IgG antibody. RT-qPCR was used to validate the presence of lnc19959.2 in the complexes. (E) The total length of lnc19959.2 minimum free energy (MFE) secondary structure. This structure is colored by base-pairing probabilities. And red shows high confidence and blue is just the opposite. (F) The different length (full-length (FL) and truncated lnc19959.2 (Δ1: 1–650, Δ2: 1–1300, Δ3: 1–1950, FL: 1–2501, Δ4: 651–1300, Δ5: 1301–1950, Δ6: 1951–2501)) of lnc19959.2 were biotinylated in RNA pulldown assays and western blot was used to detect Purb and hnRNPA2B1. Unpaired t-test was used to measure the statistical significance; ∗∗∗∗P < 0.0001.
3.6. Lnc19959.2 upregulates ApoA4 expression via ubiquitinated transcription inhibitor factor Purb
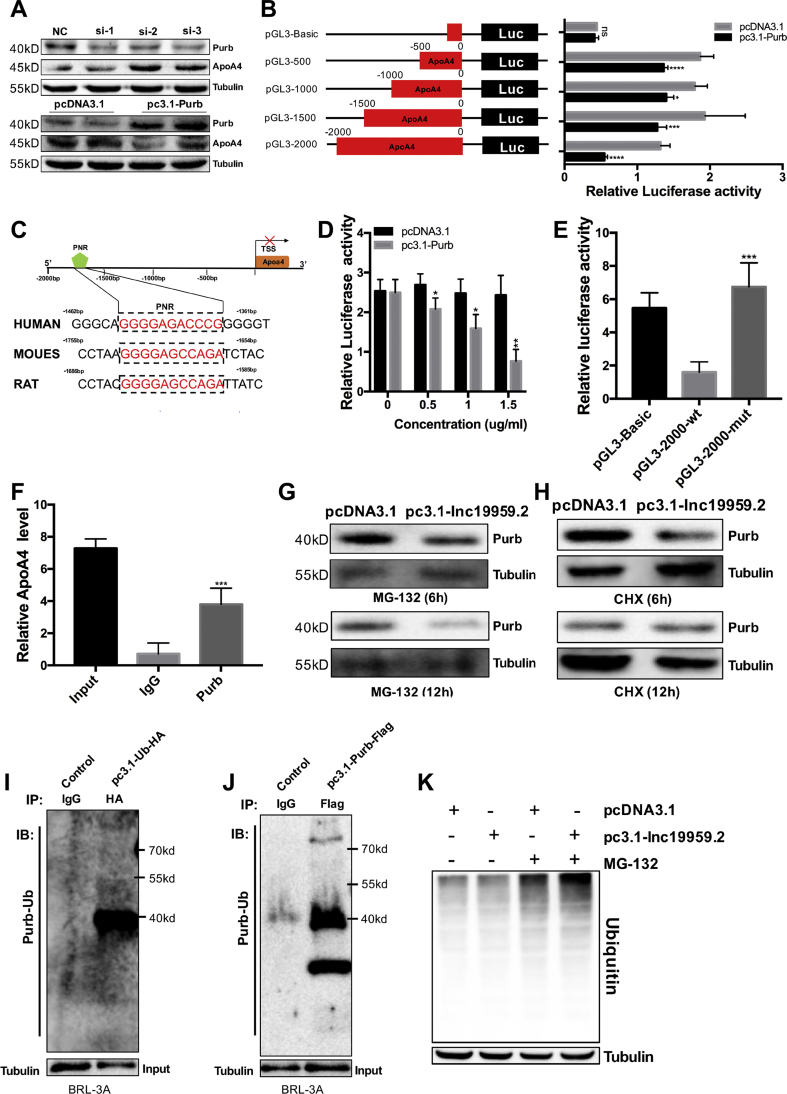
Owing to the fact that Purb is a transcription inhibitor factor, we hypothesized that lnc19959.2 modulates the expression of TG-associated genes via Purb protein. Notably, knocked down Purb increased while the overexpression of Purb decreased ApoA4 expression (Figure 5A and Supplementary Figs. 6B and 6C). To further determinate the mechanism by which Purb regulated ApoA4 expression, we performed sequence analysis and identified that five PNRs (Purb binding sites) were located at −2.0 kb of ApoA4 promoter, respectively (Supplementary Fig. 6A). Of them, PNR at −2000∼-1500 bp is most conserved in rats, mice, and humans (Figure 5C). Notably, much higher basal and Purb inhibited luciferase activities were observed on Luc-ApoA4-2000 (Figure 5B), indicating that the suppressed promoter and Purb responsive elements are within this region. Moreover, the inhibitory efficiency showed a dose-dependent manner with the increment of Purb (Figure 5D). In contrast, when we deleted 11 bp nucleotides including Purb site at −2000∼-1500 bp from Luc-ApoA4-2000, the promoter lost both basal, and Purb inhibited activities (Figure 5E). To determine whether Purb proteins bound to the ApoA4 promoter in intact cells, we applied chromatin immunoprecipitation (ChIP) assay and detected Purb binding to the −2000∼-1500 bp regions of the ApoA4 promoter in BRL-3A cells (Figure 5F).
Figure 5.
Purb acts as a transcriptional repressor to inhibit ApoA4 expression. (A) Western blot detected the ApoA4 and Purb expression levels after knockdown or overexpression of Purb in BRL-3A cells. (B) Dual-luciferase reporter assay detected relative luciferase activities after cotransfection of four truncated ApoA4 promoters with pcDNA3.1 vector or pc3.1-Purb in HEK293T cells. (C) The schematic diagram shows that Purb binding site (PNR) contained at 2000 bp of ApoA4 promoter was well conserved. (D) Dual-luciferase reporter assay detected relative luciferase activities after cotransfection of ApoA4 promoters of 2000 bp with pcDNA3.1 vector or pc3.1-Purb in different doses (range: 0–1.5 μg/mL) in HEK293T cells. (E) Dual-luciferase reporter assay detected relative luciferase activities after cotransfected with wt/mutApoA4 promoters of 2000 bp with pc3.1-Purb in HEK293T cells. (F) ChIP analysis of Purb interaction with the ApoA4 promoter. BRL-3A cells lysates were immunoprecipitated with anti-Purb or control mouse IgG antibody. (G–H) Overexpression of lnc19959.2 and control in BRL-3A cells, respectively, were incubated with the proteasome inhibitor MG-132 (10 μM) or the protein synthesis inhibitor cycloheximide (CHX, 10 μg/mL) for 6 or 12 h. The protein level of Purb extracted from the whole cell was detected by western blot. (I–J) BRL-3A cells lysates were immunoprecipitated with an HA or Flag-specific antibody in BRL-3A cell lysates which were stably expressing ubiquitin with C-terminal HA tag or Purb with C-terminal Flag tag, respectively. And then they were analyzed by western blot with anti-Purb or anti-Ubiquitin. Bottom, the input of the cell lysates. (K) Western blot detects the ubiquitination levels in BRL-3A cells. Unpaired t-test was used to measure the statistical significance; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
Next, we want to explore the molecular consequences of the interaction between lnc19959.2 and Purb. Overexpression of lnc19959.2 dramatically diminished the level of Purb protein. Reciprocally, the knockdown of lnc19959.2 increased the level of Purb protein (Supplementary Fig. 6F). However, we did not observe a reduction in Purb transcription level (Supplementary Fig. 6D). To further determine the mechanism by which lnc19959.2 reduced Purb protein stability, we treated lnc19959.2 overexpressing and control BRL-3A cells with the protein synthesis inhibitor cycloheximide (CHX) or the proteasome inhibitor MG-132. As shown in Figure 5G,H, only MG-132 abolished the downregulation of Purb protein level in BRL-3A cells when overexpressing lnc19959.2. Indicating lnc19959.2 may downregulate Purb protein abundance via the ubiquitin-proteasome pathway (Supplementary Fig. 6G). To further confirm our study, we conducted the immunoprecipitation assays to detect whether Purb and ubiquitin interacted with each other (Figure 5I,J and Supplementary Figs. 6H and 6I). Indeed, the overexpression of lnc19959.2 dramatically ubiquitinated Purb protein in BRL-3A cells (Figure 5K).
3.7. Lnc19959.2 interacts with hnRNPA2B1 to downregulate Cpt1a, Tm7sf2, and Gpam expressions
To further explore the function of the interaction between hnRNPA2B1 and lnc19959.2, we examined the regulatory effects of hnRNPA2B1 on the expression of lnc19959.2 target genes in BRL-3A cells. After the siRNA-mediated knockdown of hnRNPA2B1, we observed significant changes in many metabolism pathways (Figure 6A,B). In addition to preferentially binding RNA, hnRNPA2B1 also specifically binds with multiple promoter DNA sequences and acts as a regulatory factor, thereby participating in the regulation of transcription [28]. Thus, we hypothesize that hnRNPA2B1 may perform this function at lnc19959.2 target gene locus. In support of this idea, the overexpression of lnc19959.2 in BRL-3A cells remarkably reduced the hnRNPA2B1 engagement in promoters of its target genes, including Cpt1a, Tm7sf2, and Gpam (Figure 6C). Given that lnc19959.2 was a nuclear lncRNA, we also tested its ability to affect RNA polymerase II-dependent transcription. Similar results could be found in the RNA polymerase II (RNPII) engagement in the setting of lnc19959.2 expressions (Figure 6D). In addition, Cpt1a, Tm7sf2, and Gpam protein expression levels were reduced particularly in the lnc19959.2 overexpression panel (Figure 6E). Based on these findings, we proposed a model that lnc19959.2 reduced the occupancy of hnRNPA2B1 along with RNA polymerase II in the promoter of its target genes, which result in inactivation of these gene expressions.
Figure 6.
Cpt1a, Tm7sf2, and Gpam are coregulatory by lnc19959.2-hnRNPA2B1. (A–B) The top 30 gene ontology (GO) related to biological process (BP), cellular component (CC), and molecular function (MF) and the top 30 KEGG pathways correlated with the cellular process, environmental information processing, genetic information processing, human diseases, metabolism, and organismal systems in hnRNPA2B1 knockdown BRL-3A cells. (C) ChIP-qPCR analysis recruitment of hnRNPA2B1 to promoter regions determined in BRL-3A cells transduced with control (pcDNA3.1) or lnc19959.2 overexpression (pc3.1-lnc19959.2). The results retrieved and normalized by an upstream site (region 1) are shown as a percent of input (n = 3 per group). (D) ChIP-qPCR analysis recruitment of RNA polymerase II (RNPII) to promoter regions determined in BRL-3A cells transduced with control (pcDNA3.1) or lnc19959.2 overexpressing plasmid (pc3.1-lnc19959.2). Results retrieved and normalized by an upstream site (region1) are shown as a percent of input (n = 3 per group). (E) Western blot detected Cpt1a, Tm7sf2, and Gpam protein expression levels. Unpaired t-test was used to measure the statistical significance; ∗∗P < 0.01.
3.8. Lnc19959.2 is activated by the transcription factor of CCAAT/Enhancer-Binding Protein Beta (Cebpb) under palmitate acid stimulation
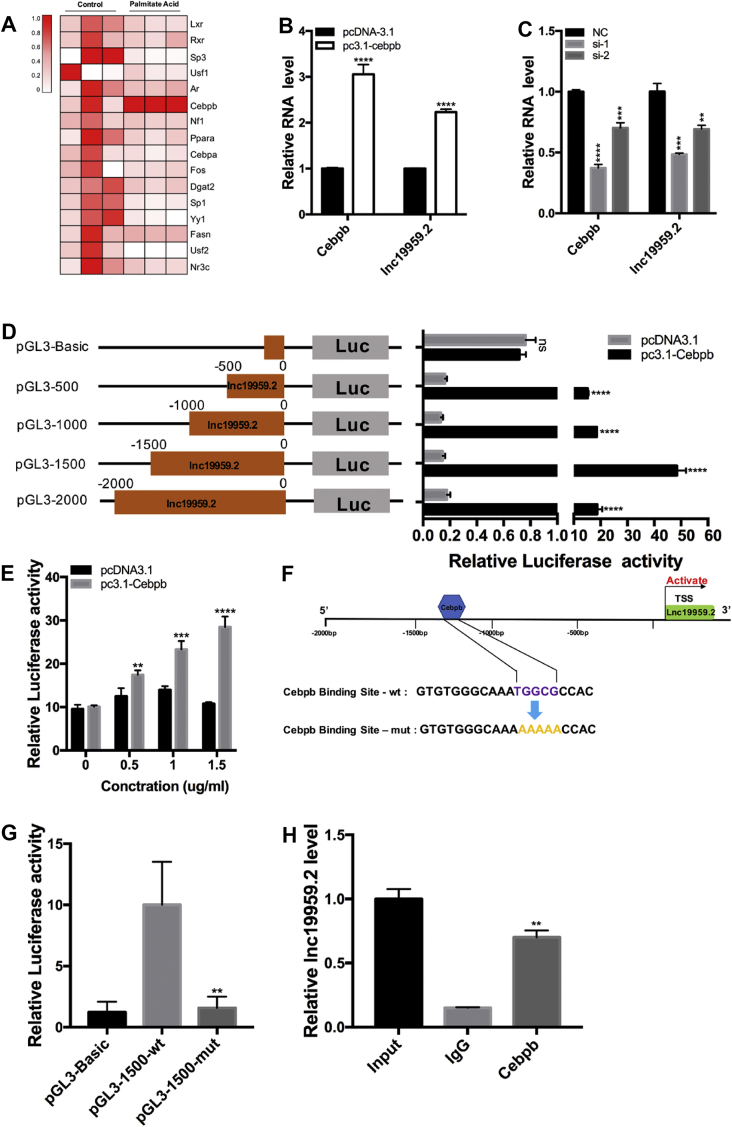
To elucidate the mechanism by which lnc19959.2 was upregulated in HFD-rat livers, we investigated the transcription factors (TFs) that had a response for the expression of lnc19959.2 in the BRL-3A cell line with the treatment of PA. Sequence analysis revealed a number of TFs within the lnc19959.2 promoter region (Supplementary Fig. 7A). A series of RT-qPCR was used to identify the dramatic changes of these TF expressions with the treatment of PA (Figure 7A). Remarkably, Cebpb, which acts as a classical transcription factor involved in several cellular processes and in various diseases, appeared more frequently in website predictions (Supplementary Fig. 7B). Thus, we assumed that Cebpb was most likely a transcriptional activator of lnc19959.2.
Figure 7.
Cebpb activates the lnc19959.2 expression. (A) Heatmaps present gene expressions selected from sequence prediction software. (B–C) RT-qPCR analysis of lnc19959.2 expression levels in BRL-3A cells in the condition of knockdown or overexpression of Cebpb. (D) Dual-luciferase reporter assay detected relative luciferase activities after cotransfection of four truncated lnc19959.2 promoters with pcDNA3.1 vector or pc3.1-Cebpb in HEK293T cells. (E) Dual-luciferase reporter assay detected relative luciferase activities after cotransfection of lnc19959.2 promoters of 1500 bp with pcDNA3.1vector or pc3.1-Cebpb in different doses (range: 0–1.5 μg/mL) in HEK293T cells. (F) Schematic diagram shows that the part of lnc19959.2 promoter sequence of wt and mut of Cebpb binding site is located in −1500~-1000 bp. (G) Dual-luciferase reporter assay detected relative luciferase activities after being cotransfected with wt/mutlnc19959.2 promoters of 1500 bp with pc3.1-Cebpb in HEK293T cells. (H) ChIP analysis of Cebpb interacted with the lnc19959.2 promoter. BRL-3A cells lysates were immunoprecipitated with anti-Cebpb or control mouse IgG antibody. Unpaired t-test was used to measure the statistical significance; ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
As expected, the expression of lnc19959.2 was downregulated or upregulated by interfering with or overexpressing Cebpb in BRL-3A cell lines, respectively (Figure 7B,C). Furthermore, dual luciferase assay showed that the transcriptional activity of lnc19959.2 was activated after cotransfecting with Cebpb expressing plasmids (Figure 7D). Activation of lnc19959.2 promoters was most evident in the −1500 bp region, and the relative fluorescence activity showed a dose-dependent increase when the Cebpb plasmid concentration increased (Figure 7E). In contrast, the luciferase activity was significantly attenuated when we mutated the Cebpb binding site of lnc19959.2 upstream sequence (Figure 7F,G). Finally, ChIP assays confirmed that Cebpb actually bound to the promoter region of lnc19959.2 (Figure 7H). In addition, the LXRα agonist T0901317 first activated Cebpb and subsequently upregulated the lnc19959.2 expression (Supplementary Figs. 7C and 7D).
4. Discussion
Here, we proposed the functional actions of lnc19959.2 in modulating triglycerides metabolism, as shown in the graphical abstract. In hepatic cells, the transcriptional factor Cebpb was initially activated when exposed to high lipid condition; after that, it directly bound to the promoter of lnc19959.2 and then promoted its expression. lnc19959.2 was mainly located in the nucleus; it specifically bound to Purb and hnRNPA2B1. Intriguingly, it acted in a distinctively different manner on modulating TG metabolism-associated gene expressions when they interacted with these two nuclear proteins. On the one hand, lnc19959.2-mediated ubiquitination increased the degradation of Purb protein, a transcriptional repressor, and in turn attenuated its binding to the promoter of ApoA4 gene, resulting in an increased expression of ApoA4; on the other hand, lnc19959.2 interacted with hnRNPA2B1, a transcriptional coactivator, and cooperated with RNA polymerase II that controls the basal and inducible expression of Cpt1a, Tm7sf2, and Gpam. All in all, we outlined lnc19959.2, induced by high lipid condition, participated in modulating the TG association gene expression, and causally linked to hyperlipidemia. Our work demonstrated a new layer regulatory complexity of an lncRNA that modulates triglyceride metabolism and also broadens the known mechanisms of lncRNA actions.
Recently, a genome-wide association study and subsequent resequencing study have successfully identified genetic variants within the APOA5/APOA4/APOC3/APOA1 cluster associated with TG levels [29,30]. Despite microRNAs [31,32], a number of lncRNAs, such as APOA1-AS and APOA4-AS within these well-known gene clusters, participated in TG regulation [33,34]. Of important note, APOA4-AS is coexpressed with APOA4 and concordantly and specifically regulates the expression of APOA4, which is well known to have important roles in enhancing hepatic TG secretion and increasing plasma TG levels [35]. In contrast to APOA4-AS that acts as in cis-regulation, lnc19959.2 acts as decoy mode in trans to upregulate the ApoA4 expression and finally participates in TG metabolism. Conversely, several key genes known to regulate TG metabolism were also downregulated by lnc19959.2, including Cpt1a, Gpam, and Tm7sf2. Cpt1a is reported to be involved in long-chain fatty acid and triglyceride metabolic process [36], while Gpam and Tm7sf2 can regulate SREBP and are involved in lipid and lipoprotein metabolism [[37], [38], [39]]. Given that these target mRNAs mentioned above are majorly involved in TG metabolism, it is not surprising that dysregulation of lnc19959.2 level could be related to TG metabolism.
HnRNPA2B1 belongs to the A/B subfamily of ubiquitously expressed heterogeneous nuclear ribonucleic proteins (hnRNPs) [28]. It acts as an RNA binding protein and plays very important roles in multiple aspects of RNA metabolism [40], including transcription [28], alternative splicing [41], mRNA stability [41], and mRNA degradation [42]. In the present study, we performed RNA-seq in which hnRNPA2B1 knockdown samples showed significant enrichment in lipid metabolic processes. In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG) classification related to genes includes lipid and energy metabolism pathways. In accordance with the established roles in lipid metabolism, the knockdown of hnRNPA2B1 regulated the expression of Cyp7a1 and Abca1 in hepatocyte cholesterol metabolism as mentioned in previous reports [20]. Intriguingly, hnRNPA2B1 showed the ability to modulate the target gene expression in distinctive manners, such as inactive transcription in TG metabolism (our study), and to promote RNA degradation in cholesterol metabolism as previously described [20]. Moreover, another hnRNP, named RALY, is mediated by LeXis and acts as a transcriptional cofactor for cholesterol biosynthetic genes in the mouse liver [21]. These results support the notion that an lncRNA-hnRNP-target gene axis is operational in rats/mice and may have relevance in dyslipidemia. Therefore, it will be interesting to identify additional novel lncRNAs that interacted with hnRNP to regulate its target genes and ultimately coordinate other lipids metabolism in the future.
The current study has several important limitations which should be acknowledged. Firstly, it was worthy noticing that there was no clear evidence that lnc19959.2 has conserved RNA transcript in human beings. Thus, further studies in humans are needed to explore this in future. Secondly, lnc19959.2 was located in both the nucleus and cytoplasm, indicating that it controls triglyceride metabolism on multiple levels. Although our present study demonstrated that lnc19959.2 works as a functional nuclear transcript, we cannot exclude a possibility that it is also regulated through cytosolic events, which warrant further study. Thirdly, one potential pitfall of our study is that it is very difficult to model the genuine liver biology role of lnc19959.2 in the regulation of hepatocyte TG metabolism by using hepatic cell lines, and in the future, this may be further explored in a transgenic rat model. However, the successful replication of effect on TG regulation in two rat live cell lines (BRL-3A and CBRH-7919) and subsequent hub gene expression signals (ApoA4, Cpt1a, Tm7sf2, and Gpam) and the interactions among Purb and hnRNPA2B1 with lnc19959.2 were additionally tested and confirmed in the high-fat fed rat liver model by using lnc19959.2 knockdown adenoviruses in vivo that improves the plausibility of our study (Supplementary Fig. 8).
In summary, our work revealed the noncoding RNA lnc19959.2 as an additional mediator of the complex effects on hepatic TG metabolism. These data demonstrated that lnc19959.2 contributes to the ability similar to PA to promote TG metabolism. The findings of the present study also suggest that the inhibition of lnc19959.2 elicits a lipid lowering effect in the high lipid environment that may contribute to a potential therapeutic benefit in the future.
Authors’ contributions
D. W. W. and H. D. conceived and supervised the project. J. W., D. W. W., and H. D. wrote the manuscript. D. T. S. assisted in the interpretation of data and writing of the manuscript. J. W., D. X., S. M., Y. C. J., D. T. S., D. H., S. Y. L., and H. H. L. carried out the experiments. J. W., S. M., C. C., and H. D. performed all bioinformatics analyses. D. T. S. aided in the analyses of transcriptome sequencing data. J. W., D. W. W., Y. W., and H. D. discussed the results. All authors read and approved the final manuscript.
Acknowledgments
This study was funded by the research grants from the National Natural Science Foundation of China (Nos. 81670215, 81974047, 81470379, 91839302, and 81400184) and National Key R&D Program of China (No. 2017YFC0909400).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.100996.
Contributor Information
Dao Wen Wang, Email: dwwang@tjh.tjmu.edu.cn.
Hu Ding, Email: dingo8369@163.com.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Brahm A., Hegele R.A. Hypertriglyceridemia. Nutrients. 2013;5(3):981–1001. doi: 10.3390/nu5030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordestgaard B.G. Triglyceride-rich lipoproteins and atherosclerotic cardiovascular disease: new insights from epidemiology, genetics, and biology. Circulation Research. 2016;118(4):547–563. doi: 10.1161/CIRCRESAHA.115.306249. [DOI] [PubMed] [Google Scholar]
- 3.Tenenbaum A., Klempfner R., Fisman E.Z. Hypertriglyceridemia: a too long unfairly neglected major cardiovascular risk factor. Cardiovascular Diabetology. 2014;13:159. doi: 10.1186/s12933-014-0159-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarwar N., Sandhu M.S., Ricketts S.L., Butterworth A.S., Di Angelantonio E., Boekholdt S.M. Triglyceride-mediated pathways and coronary disease: collaborative analysis of 101 studies. Lancet. 2010;375(9726):1634–1639. doi: 10.1016/S0140-6736(10)60545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordestgaard B.G., Varbo A. Triglycerides and cardiovascular disease. Lancet. 2014;384(9943):626–635. doi: 10.1016/S0140-6736(14)61177-6. [DOI] [PubMed] [Google Scholar]
- 6.Nordestgaard B.G., Benn M., Schnohr P., Tybjaerg-Hansen A. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. Journal of the American Medical Association. 2007;298(3):299–308. doi: 10.1001/jama.298.3.299. [DOI] [PubMed] [Google Scholar]
- 7.Bansal S., Buring J.E., Rifai N., Mora S., Sacks F.M., Ridker P.M. Fasting compared with nonfasting triglycerides and risk of cardiovascular events in women. Journal of the American Medical Association. 2007;298(3):309–316. doi: 10.1001/jama.298.3.309. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz G.G., Abt M., Bao W., DeMicco D., Kallend D., Miller M. Fasting triglycerides predict recurrent ischemic events in patients with acute coronary syndrome treated with statins. Journal of the American College of Cardiology. 2015;65(21):2267–2275. doi: 10.1016/j.jacc.2015.03.544. [DOI] [PubMed] [Google Scholar]
- 9.Holmes M.V., Millwood I.Y., Kartsonaki C., Hill M.R., Bennett D.A., Boxall R. Lipids, lipoproteins, and Metabolites and Risk of myocardial Infarction and stroke. Journal of the American College of Cardiology. 2018;71(6):620–632. doi: 10.1016/j.jacc.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toth P.P., Granowitz C., Hull M., Liassou D., Anderson A., Philip S. High triglycerides are associated with increased cardiovascular events, medical costs, and resource use: a real-world administrative claims analysis of statin-treated patients with high residual cardiovascular risk. Journal of the American Heart Association. 2018;7(15) doi: 10.1161/JAHA.118.008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhatt D.L., Steg P.G., Miller M., Brinton E.A., Jacobson T.A., Ketchum S.B. Cardiovascular risk reduction with icosapent Ethyl for hypertriglyceridemia. New England Journal of Medicine. 2019;380(1):11–22. doi: 10.1056/NEJMoa1812792. [DOI] [PubMed] [Google Scholar]
- 12.Bhatt D.L., Steg P.G., Miller M., Brinton E.A., Jacobson T.A., Ketchum S.B. Effects of icosapent Ethyl on total ischemic events: from REDUCE-IT. Journal of the American College of Cardiology. 2019;73(22):2791–2802. doi: 10.1016/j.jacc.2019.02.032. [DOI] [PubMed] [Google Scholar]
- 13.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nature Reviews Genetics. 2016;17(10):601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 14.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 15.Mercer T.R., Dinger M.E., Sunkin S.M., Mehler M.F., Mattick J.S. Specific expression of long noncoding RNAs in the mouse brain. Proceedings of the National Academy of Sciences of the U S A. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nature Reviews Genetics. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 17.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J.T., Bartolomei M.S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 19.Kretz M., Webster D.E., Flockhart R.J., Lee C.S., Zehnder A., Lopez-Pajares V. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes & Development. 2012;26(4):338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan X., Yan J., Ren J., Zhong B., Li J., Li Y. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology. 2016;64(1):58–72. doi: 10.1002/hep.28391. [DOI] [PubMed] [Google Scholar]
- 21.Sallam T., Jones M.C., Gilliland T., Zhang L., Wu X., Eskin A. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016;534(7605):124–128. doi: 10.1038/nature17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P., Ruan X., Yang L., Kiesewetter K., Zhao Y., Luo H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metabolism. 2015;21(3):455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt E., Dhaouadi I., Gaziano I., Oliverio M., Klemm P., Awazawa M. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nature Communications. 2018;9(1):3622. doi: 10.1038/s41467-018-05933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X.Y., Xiong X., Liu T., Mi L., Peng X., Rui C. Long noncoding RNA licensing of obesity-linked hepatic lipogenesis and NAFLD pathogenesis. Nature Communications. 2018;9(1):2986. doi: 10.1038/s41467-018-05383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Q., Hao Q., Prasanth K.V. Nuclear long noncoding RNAs: key regulators of gene expression. Trends in Genetics. 2018;34(2):142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munschauer M., Vogel J. Nuclear lncRNA stabilization in the host response to bacterial infection. The EMBO Journal. 2018;37(13) doi: 10.15252/embj.201899875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vance K.W., Ponting C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends in Genetics. 2014;30(8):348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y., Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cellular and Molecular Life Sciences. 2009;66(7):1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N.P., Rieder M.J. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nature Genetics. 2008;40(2):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansen C.T., Wang J., Lanktree M.B., Cao H., McIntyre A.D., Ban M.R. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nature Genetics. 2010;42(8):684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui G., Li Z., Li R., Huang J., Wang H., Zhang L. A functional variant in APOA5/A4/C3/A1 gene cluster contributes to elevated triglycerides and severity of CAD by interfering with microRNA 3201 binding efficiency. Journal of the American College of Cardiology. 2014;64(3):267–277. doi: 10.1016/j.jacc.2014.03.050. [DOI] [PubMed] [Google Scholar]
- 32.Hu S.L., Cui G.L., Huang J., Jiang J.G., Wang D.W. An APOC3 3'UTR variant associated with plasma triglycerides levels and coronary heart disease by creating a functional miR-4271 binding site. Scientific Reports. 2016;6:32700. doi: 10.1038/srep32700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halley P., Kadakkuzha B.M., Faghihi M.A., Magistri M., Zeier Z., Khorkova O. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Reports. 2014;6(1):222–230. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin W., Li X., Xie L., Li S., Liu J., Jia L. A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Research. 2016;44(13):6423–6433. doi: 10.1093/nar/gkw341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.VerHague M.A., Cheng D., Weinberg R.B., Shelness G.S. Apolipoprotein A-IV expression in mouse liver enhances triglyceride secretion and reduces hepatic lipid content by promoting very low density lipoprotein particle expansion. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33(11):2501–2508. doi: 10.1161/ATVBAHA.113.301948. [DOI] [PubMed] [Google Scholar]
- 36.Dekkers K.F., van Iterson M., Slieker R.C., Moed M.H., Bonder M.J., van Galen M. Blood lipids influence DNA methylation in circulating cells. Genome Biology. 2016;17(1):138. doi: 10.1186/s13059-016-1000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vickers K.C., Shoucri B.M., Levin M.G., Wu H., Pearson D.S., OseiHwedieh D. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57(2):533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiavoni G., Bennati A.M., Castelli M., Della Fazia M.A., Beccari T., Servillo G. Activation of TM7SF2 promoter by SREBP-2 depends on a new sterol regulatory element, a GC-box, and an inverted CCAAT-box. Biochimica et Biophysica Acta. 2010;1801(5):587–592. doi: 10.1016/j.bbalip.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Dihingia A., Bordoloi J., Dutta P., Kalita J., Manna P. Hexane-Isopropanolic Extract of Tungrymbai, a North-East Indian fermented soybean food prevents hepatic steatosis via regulating AMPK-mediated SREBP/FAS/ACC/HMGCR and PPARalpha/CPT1A/UCP2 pathways. Scientific Reports. 2018;8(1):10021. doi: 10.1038/s41598-018-27607-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Contreras R., Cloutier P., Shkreta L., Fisette J.F., Revil T., Chabot B. hnRNP proteins and splicing control. Advances in Experimental Medicine & Biology. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 41.Goodarzi H., Najafabadi H.S., Oikonomou P., Greco T.M., Fish L., Salavati R., Cristea I.M. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature. 2012;485(7397):264–268. doi: 10.1038/nature11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geissler R., Simkin A., Floss D., Patel R., Fogarty E.A., Scheller J. A widespread sequence-specific mRNA decay pathway mediated by hnRNPs A1 and A2/B1. Genes & Development. 2016;30(9):1070–1085. doi: 10.1101/gad.277392.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
2.11.1. Accession number
The accession number for the microarray published here is GEO: GSE137712, GSE99557, and GSE137131. All informatics analysis was performed by using the R language platform, a well-known open-source software environment for statistical computing and graphics.