Abstract
Background and Aims
The genus Allium L., one of the largest monocotyledonous genera and one that includes many economically important crops with nutritional and medicinal value, has been the focus of classification or phylogeny studies for centuries. Recent studies suggested that the genus can be divided into 15 subgenera and 72 sections, which were further classified into three evolutionary lineages. However, the phylogenetic relationships reconstructed by one or two loci showed weaker support, especially for the third evolutionary lineage, which might not show the species relationships very clearly and could hinder further adaptive and evolutionary study.
Methods
In this study, a total of 39 complete chloroplast genomes of Allium (covering 12 Allium subgenera) were collected, and combining these with 125 species of plastomes from 19 other families of monocots, we reconstructed the phylogeny of the genus Allium, estimated the origin and divergence time of the three evolutionary lineages and investigated the adaptive evolution in this genus and related families.
Results
Our phylogenetic analysis confirmed the monophyly and three evolutionary lineages of Allium, while new species relationships were detected within the third evolutionary lineage. The divergence time of the three evolutionary lineages was estimated to be in the early Eocene to the middle Miocene, and numerous positive selected genes (PSGs) and PSGs with high average Ka/Ks values were found in Allium species.
Conclusions
Our results detected a well-supported phylogenetic relationship of Allium. The PSGs and PSGs with high Ka/Ks values, as well as diversified morphologies, complicated chromosome characteristics and unique reproductive modes may play important roles in the adaptation and evolution of Allium species. This is the first study that conducted phylogenetic and evolutionary analyses on the genus Allium combined with the plastome and morphological and cytological data. We hope that this study can contribute to further analysis of Allium for other researchers.
Keywords: Allium, divergence time estimation, gene selection, phylogenetic analysis, monocot evolution
INTRODUCTION
The genus Allium L. (Amaryllidaceae, Allioideae) currently comprises >900 species (Herden et al., 2016), making it one of the largest monocotyledon genera. They are well known (but not always appreciated) for their specific and commonly intense smell and taste, and are characterized by having bulbs enclosed in membranous (sometimes fibrous) tunics, free or almost free tepals, and often a sub-gynobasic style (Friesen et al., 2006). Many economically important species are included, such as garlic, leek, onion and shallot. Two centres for Allium species diversity were suggested in previous studies (Fritsch and Friesen, 2002; Nguyen et al., 2008), one is from the Mediterranean Basin to central Asia and Pakistan, and the other is in North America. It is widely accepted that the Allium species can be classified into 15 subgenera and 72 sections (Friesen et al., 2006). In addition, based on 195 representative species of Allium, Fritsch and Friesen (2002) suggested that the genus Allium was monophyletic and can be differentiated through a number of evolutionary steps into three evolutionary lineages. The taxonomy of Allium was further revised by Li et al. (2010a) based on morphological characters, internal transcribed spacer (ITS) and rps16 sequence data, and the Chinese Allium species were further classified into 13 subgenera and 34 sections. Nonetheless, the phylogenetic relationships reconstructed by ITS sequences showed weaker support, especially for the third evolutionary lineage, and limited rps16 sequences were included in the study by Li et al. (2010a), which might not show the species relationships of Allium very clearly.
In view of the origin and divergence of Allium, unfortunately only one possible Amaryllidaceae fossil that dated to the latest early Eocene was discovered, in Washington (Pigg et al., 2018), and a few fossils of the Asparagales have been reported from the late Eocene (Couper 1960; Herendeen and Crane 1995; Muller 1981); however, they are all too young to calibrate the crown clade of the order (Wikström et al., 2001; Janssen and Bremer 2004). Li et al. (2016b) estimated that the genus Allium originated during the late Eocene [approx. 34.26 million years ago (Mya), highest posterior density (HPD) 95 % 24.29–45.76 Mya] and suggested that this genus originated from eastern Asia and underwent different biogeographical pathways (Li et al., 2010a, 2016b). This is helpful to understand and assess the evolutionary processes and migration history of the Allium. However, the divergence time of some Allium lineages, especially for the important node branches (such as the nodes of the three evolutionary lineages), were not estimated, which might hinder further studies conducted on subgenera or sections of Allium.
Despite the phylogeny, origin and divergence of Allium, species of this genus are naturally distributed only in the Northern Hemisphere and widely spread from the dry sub-tropics to the boreal zone. The habitat of Allium species varies from dry and well-drained soil to moist and organic soil, which can be even in swamps or in water (Block, 2010). Evolution of this genus species was also accompanied by habitat and ecological diversification, and their adaptation to the various habitat environments has resulted in a remarkable morphological polymorphism (e.g. flowers, leaves and bulbs) (Li et al., 2016a). Additionally, the chromosome numbers of Allium are diversified, including basic chromosomes numbers x = 7, 8, 9, 10 or 11, and thus they produce many polyploids (Friesen, 1992; Xu et al., 1998; Zhou et al., 2007; Zhang et al., 2009; Jones, 2012; Li et al., 2017; Peruzzi et al., 2017), which are often considered to have many advantages compared with their diploid progenitors in morphological, physiological, life history characteristics and rates of adaptation (Mayrose et al., 2010). It has been generalized that chromosomal characteristics (e.g. chromosome number, ploidy level, genome size, karyotype asymmetry, etc.) prove to be informative and significant for understanding the relationships, evolution and adaptation of taxa (Sharma and Sharma, 2014). Therefore, Allium is a successful taxon from the point of view of its wide distributions, diversified morphologies and complex chromosome characteristics. Recent transcriptome or genome-wide studies on multiple species that succeed in the evolutionary processes have identified various evolutionary and adaptive processes that may be responsible for adaptation, including for humans (Beall et al., 2010; Simonson et al., 2010; Yi et al., 2010; Peng et al., 2011), animals (Savolainen et al., 2007; Qiu et al., 2012; Ge et al., 2013; Gou et al., 2014) and plants (Leimu and Fischer, 2008; Hancock et al., 2011; Jia et al., 2013; Yates et al., 2014; Zhang et al., 2016b; Zhang et al., 2019). In addition, adaptation to the environment can allow population persistence (Aitken et al., 2008; Franks and Hoffmann, 2012). Local adaptation has become an important component for species responses to changing environments (Davis and Shaw, 2001), and the phenotypic plasticity in a changing environment has received much attention to date (Loarie et al., 2009; Nicotra et al., 2010; Chevin and Hoffmann, 2017; Van Buskirk, 2017; Oostra et al., 2018; Villemereuil et al., 2018). However, several challenges exist in analysing and interpreting the genetic basis of evolution and adaptation of Allium species. For instance, poor genomic information is available in Allium because of the enormous size of the genome (approx. 16.3 Gb) (Duangjit et al., 2013), which is 32 times larger than the genome of rice (Arumuganathan and Earle, 1991). The diploid (2n = 16) genome sizes of garlic (A. sativum) and onion (A. cepa) were estimated to be >30 Gb (Egea et al., 2017; Peska et al., 2019). Although sequencing technologies have undergone rapid development in the past 10 years, analysis of such a complex and huge genome of the Allium species, a non-model plant, remains a Herculean task (Schatz et al., 2012; Nagarajan and Pop, 2013).
Complete chloroplast (cp) genomes are known to be highly conserved in both gene order and gene content (Raubeson and Jansen, 2005), and exhibit a much lower substitution rate than nuclear DNA (Wolfe et al., 1987). Due to their conserved structure, small effective population size, lack of recombination and usually uniparental inheritance (Henry et al., 2016), cp genomes have been extensively used in phylogenetic reconstruction (especially in taxonomically complex groups) (Jansen et al., 2007; Moore et al., 2010; Barrett et al., 2014; Malé et al., 2014; Shaw et al., 2014; Dong et al., 2015; Yu et al., 2017; Ye et al., 2018) and selection pressure analysis (Allen et al., 2011; Carbonell-Caballero et al., 2015; Hu et al., 2015; Xie et al., 2018b). Many cp genomes have been reported in Allium species (Kim and Yoon, 2010; Lee et al., 2017; Filyushin et al., 2018; Jin et al., 2018; Xie et al., 2019a, b; Yang et al., 2019), and it is necessary to performe a comprehensive cp genome analysis on the genus Allium. Here, a total of 39 complete cp genomes of Allium were collected, covering 12 of 15 Allium subgenera. Combining these with 125 species plastomes from another 19 families of monocots, we wish to accomplish the following: (1) to reconstruct the phylogeny of the genus Allium and analyse lineage relationships at the cp genome level; (2) to estimate the origin and divergence time of Allium by using more cp genome regions and more fossils; and (3) to investigate the adaptative evolution of Allium and allied families by using selective pressure analysis and morphological characteristics. Overall, this study will contribute to a comprehensive understanding of plastome evolution in Allium plants.
MATERIALS AND METHODS
Taxon sampling and DNA extraction
A total of 164 species representing 19 families of monocots were included in this study. Thirty-nine species from three evolutionary lineages of Allium (Friesen et al., 2006; Fritsch and Keusgen, 2006) were selected, ranging from the basal to the top lineages of the whole Allium phylogenetic tree, including all types of inflorescences of the genus Allium, and showing the complicated ploidy of chromosome and other morphological characteristics. Thus, we think that these species to some degree can act as representatives of the Allium genus. Among the 164 sampled species, the whole cp genomes of 22 Allium species were reported from our laboratory (Supplementary data Table S1), and the data of the remaining species were downloaded from NCBI (Supplementary data Table S2). Total genomic DNA was extracted from either fresh or silica gel-dried material using the DNeasy Plant Mini Kit following the manufacturer’s protocol (Biomed, Beijing, China). A NanoDrop spectrophotometer (ND-1000; Thermo Fisher Scientific, USA) and agarose gel electrophoresis were used to determine DNA quality and purity.
Sequence assembly and annotation
All examined DNAs of 22 Allium species were sent to Novogene (Beijing, China) for library construction and sequencing. Paired-end libraries were generated on an Illumina Hiseq 2500 platform. The raw reads obtained from Novogene were filtered using Trimmomatic 0.3.2 with default parameters (Bolger et al., 2014). The program MITObim v1.7 (Christoph et al., 2013) was used for plastome assembly, and the whole cp genomes of several Allium species were downloaded from GenBank as references. Reads were assembled de novo using Velvet (Zerbino and Birney, 2008) with k-mer sizes ranging from 27 to 145, and the coverage cut-offs were auto-adjusted. In order to obtain accurate plastomes, each of the species was assembled four times with the reference genomes A. cepa (KM088014), A. sativum (KY085913), A. victorialis (NC_037240) and A. obliquum (LT699701). Gaps that appeared in the assembled cp genomes were further confirmed and corrected by Sanger sequencing and the primers were designed using Lasergene 7.1 (DNASTAR, Madison, WI, USA). The primers and amplifications are shown in Supplementary data Table S3. The modified plastomes were annotated using the program DOGMA (Wyman et al., 2004), and subsequently corrected within GENEIOUS R11 (Biomatters, Ltd, Auckland, New Zealand). Finally, we used OGDRAW (Lohse et al., 2013) to draw circular plastome maps.
Sequence divergence analysis
The cp genomes of all 39 Allium species were aligned, and the alignments were visualized using mVISTA (Frazer et al., 2004) with A. altaicum as reference to detect sequence divergence. Furthermore, in order to evaluate the nucleotide diversity (Pi) of each gene, DnaSP version 5.1 (Librado and Rozas, 2009) was used to calculate the nucleotide diversity of genes in LSC (large single copy) regions, SSC (small single copy) regions and inverted repeat (IR) regions.
Phylogenetic analyses
In order to investigate the phylogeny of Allium species and allied families, the 22 assembled Allium species and another 142 allied plastomes were analysed together. First, all single-copy genes (SCGs) of the 164 taxa were extracted and then aligned using MUSCLE v3.6 (Edgar, 2004), manually examined and adjusted. These alignments were then concatenated as a super locus of single-copy genes, which were further used for phylogenetic analysis. Maximum parsimony (MP) was performed using PAUP* version 4.10 (Swofford, 2003). All characters were equally weighted, gaps were treated as missing and character states were treated as unordered. A heuristic search was performed with TBR branch swapping and the Multrees option, and random stepwise addition with 1000 replications. All analyses used the best-fitting models of nucleotide substitutions selected in jModelTest v2.1.4 (Darriba et al., 2012) under the Akaike information criterion (AIC). Maximum likelihood (ML) analyses were conducted using RAxML v8.0 (Stamatakis, 2014) based on the best-fit GTR + G model and 1000 bootstrap replicates. Bayesian analyses were performed with MrBayes v3.2 (Ronquist and Huelsenbeck, 2003). Three independent Markov chain Monte Carlo (MCMC) runs of different lengths, but under the same estimation conditions, were conducted. Each chain ran 1 × 108 generations with the sample frequency of 50, and the initial 20 % of the samples were discarded as burn-in to confirm the stationarity. Tracer v1.5 (Rambaut and Drummond, 2009) was used to assess the quality of the MCMC simulations and stability of runs. Effective sample size (ESS) values were >200 for all parameters, suggesting that sufficient sampling occurred. In addition, phylogenetic analyses were also performed for the coding sequences (CDSs) that were shared in all 164 species.
Molecular dating and fossil calibration
The combined single-copy gene data set was used to estimate the origin times of Allium and other allied families. Bayesian searches for tree topologies and node ages were conducted in BEAST (Drummond and Rambaut, 2007) using a GTR + G substitution model selected by jModelTest 2.1.4 (Darriba et al., 2012) and an uncorrelated log-normal relaxed clock (Drummond et al., 2002). A Yule process was specified as tree prior, and the MCMC algorithm was run for 5 × 107 generations with sampling every 2000 generations, following a burn-in of 10 % of the initial cycles. MCMC samples were inspected in Tracer to confirm sampling adequacy and convergence of the chains to a stationary distribution. Three fossils used to calibrate time in BEAST were as follows. (1) According to the studies of Friis et al. (1994, 1997, 1999), Magallón et al. (2015), Eklund et al. (2004) and Li et al. (2019), 121 Ma was implemented as a minimum age in the penalized likelihood analysis and as the zero offset of a log-normal distribution with log mean of (120.7 + 10 %), and s.d. of 1 in the uncorrelated log-normal analysis. This time is equal to the crown group of the Chloranthaceae. (2) From the references of pollen fossils (Doyle and Hickey, 1976; Doyle and Robbins, 1977; Hickey and Doyle, 1977), we set the minimum age in the penalized likelihood analysis as 112 Ma (calibrate the monocot crown node), and as the zero offset of a log-normally distributed prior with log mean of (112 + 10 %) and s.d. of 1. (3) Based on studies of Bell et al. (2010), Friis (1988) and Magallón et al. (2015), the node of Zingiberales was set as follows: 77.8 Ma was implemented as a minimum age in the penalized likelihood analysis, which equalled the minimum age of the Zingiberales fruits and seeds fossils, and as the zero offset of a log-normal distribution with log mean of (77 + 10 %) and s.d. of 1 in the Bayesian analysis.
Selective pressure analysis
In order to detect the sites that are under positive selection in the protein-coding genes in the plastid genomes of Allium species, an optimized branch-site model (Yang and Dos, 2011) and Bayesian empirical Bayes (BEB) methods (Yang et al., 2005) were conducted. The CDSs of all 164 taxa were extracted and aligned using the software MUSCLE (Edgar, 2004) and the ‘gaps’ in the alignments were further checked. The alignment sequences were further trimmed by Trimal v1.2 (Capellagutiérrez et al., 2009) with parameters Trimal -in $i -out $i.fasta -fasta -noallgaps, and the bona fide alignments were used to perform the positive selection analyses. The Allium lineage was selected as a specifically designated branch to assess potential positive selection in the CODEML program implemented in the PAML package (Yang, 2007). The non-synonymous (Ka) and synonymous (Ks) nucleotide substitution rates and their ratio (ω = Ks/Ks) were used to measure the selective pressure. The ratios ω > 1, ω = 1 and ω < 1 suggest positive selection, neutral selection and negative selection, respectively (Yang and Nielsen, 2002). The log-likelihood values were calculated and tested with a neutral model and an alternative model according to Yang (2007). The right-tailed χ 2 was used to calculate P-values according to the difference in log-likelihood values between the neutral model and alternative model with one degree of freedom to assess the model fit. Afterwards, the BEB method was applied to compute the posterior probabilities of amino acid sites to identify whether these specific sites were under positive selection (codon sites with a high posterior probability) (Yang, 2007; Lan et al., 2017). A gene with a test P-value <0.05 and with positively selected sites was considered as a positively selected gene (PSG). Moreover, in order to compare the differences in selection pressure that were experienced in allied families of Allium, all family lineages included in this study were separately subjectd to positive selection analysis.
Statistics of morphological and chromosomal characteristics
Morphological traits and chromosomal characteristics are important sources for analysing relationships and evolution of taxa (Schneeweiss et al., 2004). Our laboratory members have researched the genus Allium for >20 years, and accumulated a lot of morphological and karyotypic data. Therefore, in order to understand and analyse the species relationships and the evolutionary processes better, we collected and exhibited the morphological and karyotypic traits of the 39 Allium species (the karyotypes are summarized according to previously published references), including the inflorescences, ploidy level, existence or absence of aneuploids chromosome, and so on.
RESULTS
Characteristic of the Allium chloroplast genomes
Chloroplast genome structures are conserved and similar in gene order across 39 Allium species. The genome size ranges from 145 819 to 159 125 bp, and the GC content varied from 36.7 to 37.8 %. The length of the coding regions changes from 64 581 bp to 81 609 bp, and the minimum and maximum lengths were 72 410 bp and 84 711 bp in non-coding regions. Information on LSC, SSC and IR regions, and gene number is given in Table 1 and Supplementary data Table S4.
Table 1.
Summary of major characteristics of Allium plastomes, including genome size, GC content and gene number.
| Taxon | Total genome | LSC length (bp) | SSC length (bp) | IR length (bp) | Gene number | Protein coding | tRNAs | rRNAs | Coding region | Non-coding region | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | GC (%) | Length (bp) | GC (%) | length (bp) | GC (%) | ||||||||
| Allium altaicum | 153 129 | 36.8 | 82 197 | 17 912 | 26 510 | 131 | 85 | 38 | 8 | 78 183 | 37.3 | 74 946 | 36.3 |
| Allium ampeloprasum | 152 732 | 36.7 | 81 775 | 17 905 | 26 526 | 127 | 81 | 38 | 8 | 77 289 | 37.2 | 75 443 | 36.2 |
| Allium caeruleum | 153 267 | 36.8 | 82 389 | 18 058 | 26 410 | 131 | 85 | 38 | 8 | 79 195 | 37.2 | 74 072 | 36.4 |
| Allium cepa | 153 440 | 36.8 | 82 543 | 17 929 | 26 485 | 132 | 86 | 38 | 8 | 79 263 | 37.3 | 74 177 | 36.3 |
| Allium chinense | 152 525 | 36.8 | 81 324 | 18 205 | 26 498 | 126 | 87 | 31 | 8 | 80 115 | 37.3 | 72 410 | 36.2 |
| Allium chrysanthum | 153 621 | 36.8 | 82 744 | 17 985 | 26 446 | 132 | 86 | 38 | 8 | 79 269 | 37.2 | 74 352 | 38.4 |
| Allium chrysocephalum | 153 710 | 36.8 | 82 688 | 17 998 | 26 512 | 132 | 86 | 38 | 8 | 79 280 | 37.2 | 74 430 | 38.4 |
| Allium cyathophorum | 154 174 | 36.8 | 83 359 | 17 881 | 26 467 | 132 | 86 | 86 | 8 | 79 382 | 37.3 | 74 792 | 36.3 |
| Allium fetisowii | 154 018 | 36.9 | 83 657 | 17 941 | 26 210 | 132 | 86 | 86 | 8 | 79 300 | 37.3 | 74 718 | 36.5 |
| Allium fistulosum | 152 859 | 36.9 | 81 930 | 17 921 | 26 504 | 132 | 86 | 86 | 8 | 79 307 | 37.3 | 73 552 | 36.5 |
| Allium forrestii | 153 186 | 36.8 | 82 339 | 17 959 | 26 444 | 132 | 86 | 86 | 8 | 79 194 | 37.2 | 73 992 | 36.4 |
| Allium herderianum | 153 605 | 36.8 | 82 658 | 17 983 | 26 482 | 132 | 86 | 38 | 8 | 79 276 | 37.2 | 74 329 | 38.4 |
| Allium macleanii | 152 633 | 36.9 | 82 890 | 17 213 | 26 265 | 131 | 85 | 38 | 8 | 78 988 | 37.3 | 73 645 | 36.5 |
| Allium macranthum | 152 876 | 37.1 | 83 600 | 18 959 | 25 095 | 132 | 86 | 38 | 8 | 78 723 | 37.6 | 74 153 | 36.6 |
| Allium mairei | 152 913 | 36.9 | 82 493 | 18 762 | 25 829 | 132 | 86 | 38 | 8 | 78 914 | 37.3 | 73 999 | 36.5 |
| Allium maowenense | 153 608 | 36.8 | 82 668 | 18 000 | 26 470 | 132 | 86 | 38 | 8 | 79 256 | 37.2 | 74 352 | 38.4 |
| Allium monanthum | 154 804 | 37.0 | 83 835 | 18 007 | 24 551 | 132 | 86 | 38 | 8 | 79 305 | 37.5 | 75 499 | 36.5 |
| Allium nanodes | 154 077 | 37.0 | 84 274 | 20 075 | 24 864 | 131 | 85 | 38 | 8 | 70 029 | 37.3 | 84 048 | 36.8 |
| Allium neriniflorum | 154 280 | 37.0 | 83 131 | 18 191 | 26 479 | 132 | 86 | 38 | 8 | 79 532 | 37.5 | 74 748 | 36.5 |
| Allium nutans | 153 456 | 36.9 | 82 533 | 17 951 | 26 486 | 132 | 86 | 38 | 8 | 79 299 | 37.2 | 74 157 | 36.6 |
| Allium obliquum | 152 597 | 36.8 | 81 798 | 18 059 | 26 370 | 132 | 86 | 38 | 8 | 79 325 | 37.2 | 73 272 | 36.4 |
| Allium oschanini | 153 580 | 36.8 | 82 522 | 18 030 | 26 514 | 132 | 86 | 38 | 8 | 79 311 | 37.3 | 74 269 | 36.3 |
| Allium paradoxum | 145 819 | 37.1 | 80 919 | 13 504 | 25 698 | 133 | 86 | 39 | 8 | 64 581 | 37.3 | 81 238 | 36.9 |
| Allium platyspathum | 152 529 | 36.8 | 81 544 | 17 955 | 26 515 | 131 | 85 | 38 | 8 | 78 514 | 37 | 74 015 | 36.6 |
| Allium polyrhizum | 153 086 | 36.9 | 82 438 | 19 188 | 25 730 | 132 | 86 | 38 | 8 | 79 127 | 37.3 | 73 959 | 36.5 |
| Allium praemixtum | 153 226 | 36.8 | 82 163 | 18 041 | 26 511 | 132 | 86 | 38 | 8 | 79 302 | 37.3 | 73 924 | 36.3 |
| Allium prattii | 154 482 | 37.0 | 83 428 | 18 056 | 26 499 | 131 | 85 | 38 | 8 | 69 771 | 37.3 | 84 711 | 36.8 |
| Allium przewalskianum | 153 245 | 36.9 | 82 428 | 18 703 | 26 057 | 132 | 86 | 38 | 8 | 79 239 | 37.3 | 74 006 | 36.5 |
| Allium_pskemense | 153 788 | 36.7 | 82 721 | 18 033 | 26 517 | 132 | 86 | 38 | 8 | 79 197 | 37.2 | 74 591 | 36.2 |
| Allium rude | 153 697 | 36.7 | 82 815 | 17 978 | 26 452 | 132 | 86 | 38 | 8 | 79 284 | 37.2 | 74 413 | 38.2 |
| Allium sativum | 153 118 | 36.7 | 82 048 | 17 986 | 26 542 | 128 | 83 | 37 | 8 | 77 808 | 37.2 | 75 310 | 36.2 |
| Allium schoenoprasides | 153 583 | 36.7 | 82 551 | 18 082 | 26 475 | 132 | 86 | 38 | 8 | 79 540 | 37.2 | 74 043 | 36.2 |
| Allium schoenoprasum | 153 014 | 36.8 | 82 059 | 17 937 | 26 509 | 132 | 86 | 38 | 8 | 79 296 | 37.2 | 73 718 | 36.4 |
| Allium spicatum | 153 225 | 36.9 | 82 382 | 17 930 | 26 456 | 132 | 86 | 38 | 8 | 79 184 | 37.3 | 74 041 | 36.5 |
| Allium strictum | 152 962 | 36.8 | 82 880 | 20 562 | 24 760 | 132 | 86 | 38 | 8 | 79 169 | 37.2 | 73 793 | 36.4 |
| Allium tuberosum | 157 735 | 36.9 | 86 472 | 19 079 | 26 092 | 132 | 86 | 38 | 8 | 81 609 | 37.4 | 76 126 | 36.4 |
| Allium ursinum | 159 125 | 37.3 | 88 056 | 18 105 | 26 482 | 131 | 84 | 39 | 8 | 77 859 | 37.5 | 81 266 | 37.1 |
| Allium victorialis | 154 074 | 37.0 | 83 173 | 17 853 | 26 526 | 131 | 85 | 38 | 8 | 78 996 | 37.5 | 75 078 | 36.5 |
| Allium xichuanense | 153 673 | 36.7 | 82 797 | 17 950 | 26 463 | 132 | 86 | 38 | 8 | 79 269 | 37.2 | 74 404 | 38.4 |
Sequence divergence
The sequence divergence analysis showed high sequence similarity across the Allium plastid genomes (Supplementary data Fig. S1). In addition, the IR regions and coding regions were more conserved than the LSC, SSC and non-coding regions (Supplementary data Fig. S2). The nucleotide diversity values of the LSC regions ranged from 0 to 0.02525 with a mean value of 0.00963 (the values varied from 0.00354 to 0.02584 with an average value of 0.01493 in SSC regions), while the values were from 0.0000 to 0.00765 with a mean value of 0.00229 in the IR regions (Supplementary data Fig. S3). Ten genes with high nucleotide diversity (>0.02) were detected, namely ndhK, ndhE, ndhA, rps16, matK, psaI, rpl22, ndhF, rpl32 and trnK-UUU.
Morphological and chromosomal characteristics
In order to exhibited the morphological and chromosomal traits reasonably and aesthetically, we combined them with the phylogenetic results and they are presented in Fig. 1. Most of the species possess an umbel inflorescence with various flower densities, while individual species have a spike inflorescence (A. spicatum) and an inconspicuous umbel inflorescence (A. mairei). In view of the ploidy levels of the species, the basic chromosome number of 37 Allium species is x = 8, and only in A. macranthum and A. ursinums is it x = 7. In addition to diploidy, many Allium species have multiple ploidies, such as A. monanthum (2–4x, x = 8), A. tuberosum (2–4x, 8x, x = 8), A. ampeloprasum (2–6x, x = 8) and A. nutans (2–10x, 13x, x = 8). Moreover, some Allium species are aneuploid, such as A. victorialis, A. nutans, A. strictum and A. schoenoprasum (Fig. 1).
Fig. 1.
Phylogenetic relationships, flower morphologies and chromosome characteristics of Allium species collected in this study. The tree was constructed by maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) based on 48 shared SCGs. Support values marked above the branches follow the order MP (bootstrap values)/BS (bootstrap support)/PP (posterior probability); *Maximum support in all three analyses. Three evolutionary lineages (L1–L3) are marked with different colours: L1, pink; L2, purple; L3, red. The chromosome ploidies are marked using different coloured circles.
Characteristics of the SCG and CDS data sets and phylogenetic analysis
In order to test whether additional characters or taxa were responsible for any changes observed in resolution and support of the integrated phylogenies, the 48 SCGs and the 43 CDSs were used to perform phylogenetic analysis, respectively. Alignments of the SCG data set showed a length of 32 055 bp with 14 336 variable sites (44.72 %), and 12 025 parsimony-informative characters (PICs; 37.51 %). The CDS data set possesses 30 093 characters with 13 276 variable sites (44.12 %) and 11 152 PICs (37.06 %).
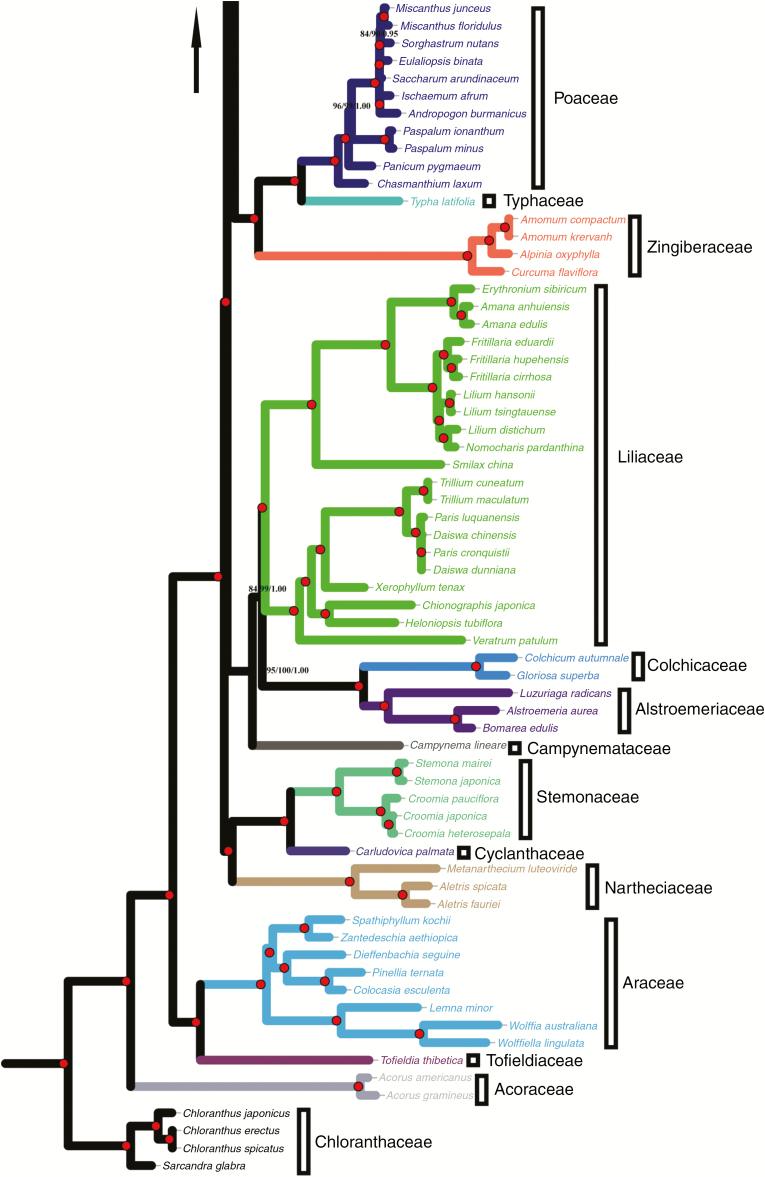
Bayesian inference, MP and ML analyses of the SCG and CDS data sets shared between the 164 plastomes (four species from family Chloranthaceae were set as outgroups: Sarcandra glabra, Chloranthus japonicus, C. erectus and C. spicatus) generated almost identical topologies with generally high bootstrap support and posterior probability (Fig. 2; Supplementary data Fig. S4). Monophyly of each family was strongly confirmed, and Narcissus poeticus and Agapanthus coddii showed a close relationship with the genus Allium (Figs 1 and 2). The family Asparagaceae was identified as being closest to the Amaryllidaceae, and the Acoraceae was located at the stem of the phylogeny followed by the Tofieldiaceae and the Araceae. Following the studies of Friesen et al. (2006) and Li et al. (2010a), the Allium species were divided into three lineages (L1–L3; Figs 1 and 2; Supplementary data Fig. S4): L1 was composed of A. monanthum, A. macranthum, A. paradoxum and A. ursinum; L2 consisted of A. nanodes, A. prattii, A. victorialis, A. macleanii, A. fetisowii and A. neriniflorum; and L3 was formed by the remaining 29 Allium species. The flower morphologies of 39 Allium species and the chromosome characteristics of all collected Allium species are shown in Fig. 1 and Supplementary data Table S5.
Fig. 2.
Phylogenetic relationships inferred from 164 species based on 48 shared SCGs. Tree constructed by maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) based on 48 shared SCGs. Support values marked above the branches follow the order MP (bootstrap values)/BS (bootstrap support)/PP (posterior probability); red circles in nodes represent maximum support in all three analyses. Accessions from different families are written using different colours and labelled at the top of the tree. *Species sequenced by our laboratory research group.
Estimation of divergence time
Divergence time analyses based on fossils indicated that the crown groups of monocots arose 148.989 Mya (95 % HPD: 108.006–166.103 Mya; Fig. 3; Table 2; Supplementary data Fig. S5). In addition, the divergence time of the Amaryllidaceae was estimated as approx. 49.399 Mya (95 % HPD: 48.165–50.588 Mya). The genus Allium originated approx. 41.932 Mya (95 % HPD: 34.549–47.605 Mya) and then diverged into three lineages (L1–L3; Fig. 3, Table 2). The L1 lineage separated around 22.184 Mya (95 % HPD: 15.232–34.812 Mya) and the divergence time of L2 and L3 was approx. 19.655 Mya (95 % HPD: 13.491–31.145 Mya). From the results, we found that most of the Allium species differentiated in the late Tertiary to middle Miocene (approx. 10–15 Mya).
Fig. 3.
Divergence time estimation based on 48 shared SCGs. (A) The maximum credibility tree from the divergence times estimated with BEAST. The 95 % highest posterior density (HPD) estimates for each well-supported clade are represented by bars, and white triangles with a black outline represent compressed clades. Letters (A–C) in a black circle represent fossil calibration points (see the Materials and Methods). The node ages are given for each node. (B) Phylogeny of three evolutionary lineages of Allium; L1–L3: the first to the third evolutionary lineages, the origin time of Allium and three evolutionary lineages are shown with arrows, and red background represents the main periods of time during which Allium species differentiated.
Table 2.
Age estimates for Allium and allied families based on the combined single-copy gene data sets
| Number | Median (Ma) | 95 % HPD | Node labels | |
|---|---|---|---|---|
| Lower (Mya) | Upper (Mya) | |||
| 1 | 49.399 | 48.165 | 50.588 | Asparagaceae |
| 2 | 49.399 | 48.165 | 50.588 | Amaryllidaceae |
| 3 | 41.932 | 34.549 | 47.605 | Allium |
| 4 | 22.184 | 15.232 | 34.812 | Allium (Line 1) |
| 5 | 19.655 | 13.491 | 31.145 | Allium (Line 2–Line 3) |
| 6 | 54.225 | 50.408 | 60.443 | Asphodelaceae |
| 7 | 62.724 | 58.043 | 67.976 | Iridaceae |
| 8 | 74.633 | 70.766 | 78.327 | Orchidaceae |
| 9 | 64.151 | 34.63 | 69.312 | Poaceae |
| 10 | 64.151 | 34.63 | 69.312 | Typhaceae |
| 11 | 78.509 | 76.553 | 81.051 | Zingiberaceae |
| 12 | 70.428 | 44.204 | 89.435 | Liliaceae |
| 13 | 46.541 | 34.959 | 65.071 | Colchicaceae |
| 14 | 46.541 | 34.959 | 65.071 | Alstroemeriaceae |
| 15 | 81.281 | 67.609 | 98.668 | Campynemataceae |
| 16 | 53.764 | 39.71 | 71.134 | Stemonaceae |
| 17 | 53.764 | 39.71 | 71.134 | Cyclanthaceae |
| 18 | 101.205 | 67.051 | 124.253 | Nartheciaceae |
| 19 | 110.645 | 82.94 | 131.2 | Araceae |
| 20 | 110.645 | 82.94 | 131.2 | Tofieldiaceae |
| 21 | 148.989 | 108.006 | 166.103 | Acoraceae |
| 22 | 162.446 | 120.369 | 174.875 | Chloranthaceae |
Selective pressure analysis
A total of 43 CDSs that were shared by 164 taxa were eventually used for positive selection analysis, and the branch site model in CODEML was independently performed for Allium species and allied families. For the Allium lineage, ten genes with at least one positively selected site were detected from the BEB test, and seven of them showed a significant P-value (P < 0.05). For family Amaryllidaceae, 27 genes were found with positively selected sites and eight of them have significant P-values. Among the 19 families, the Orchidaceae possessed the maximum number of genes (i.e. 36 genes) that had sites under positive selection, and nine of them had significant P-values. The Typhaceae possessed the minimum number of genes (seven genes) that have positively selected sites, but none of them had a significant P-value. The detailed results of positive selective analysis are shown in Fig. 4A and Supplementary data Table S6.
Fig. 4.
Frequency of genes that possessed positively selected sites and the curve of the average Ka, Ks and Ka/Ks values of each gene. (A) The frequency of genes with positively selected sites in each family. Each grey square indicates the positively selected gene which occurs in the corresponding family, and black squares indicate genes whose positively selected sites are significant (P < 0.05. (B) The curve of the average Ka, Ks and Ka/Ks values of each gene; different colours of the bars are consistent with the curve colour of Ka, Ks and Ka/Ks.
In terms of the gene (owning positively selected sites) frequency that was detected in genus Allium and the 19 families, we found that atpA and psbC possessed the highest frequency and appeared in 17 families, following by rpl11 (15 families), rpoC2 (14 families), petA (14 families), psaB (12 families) and atpI (12 families). The detailed gene frequencies are listed in Fig. 4A and Supplementary data Table S6.
The average Ka, Ks and Ka/Ks values of the 43 CDSs were calculated. The average Ka value of psaA (0.38991) was highest, following by rpoC2 (0.38348), rpoC1 (0.3581), atpA (0.3254), atpB (0.172), rpoA (0.15243), rps11 (0.13878) and matK (0.13379). psaA (0.54487) possessed the highest average value of Ks, followed by petN (0.53934), psbI (0.5232), rpoC1 (0.46461) and rpoC2 (0.4294). The average Ka/Ks ratio was highest in psbC (6.31579), followed by rps11 (2.03818), psaI (1.27620), rpoC2 (0.89306) atpA (0.78090) and rpoC1 (0.77075) (Fig. 4B; Supplementary data Table S7).
DISCUSSION
The plastome variation of the genus Allium
Recently, cp genomes have been used to evaluate the genetic variation in genera or families (e.g. Myrtaceae, Corylus, Podophylloideae and Apodanthaceae) (Bayly et al., 2013; Bellot and Renner, 2016; Ye et al., 2018; Yang et al., 2018). The cp genome size, gene order and structure of the 22 Allium species were similar to those reported in previous Allium plastid genomes (Table 1), with sizes ranging from 145 to 160 kb (Kim and Yoon, 2010; Lee et al., 2017; Jin et al., 2018; Yang et al., 2019). Additionally, the GC contents of the Allium species varied from 36.7 to 37.8 %, and the GC contents in non-coding intergenic regions were much lower than those in coding regions, which is similar to the case in most land plants (Bock, 2007). The overall cp genome assemblies obtained from 39 Allium species indicated that there is a high sequence similarity across the Allium cp genomes (Supplementary data Fig. S2), which also suggested that the Allium cp genomes are relatively well conserved. In addition, the sequence variations were more conserved in the IR regions than in the LSC and SSC regions, and similar results have been found in most angiosperms (Khakhlova and Bock, 2006; Zhang et al., 2016a; Wu et al., 2018). Ten genes (rpl32, trnK-UUU, ndhF, rpl22, psaI, matK, rps16, ndhA, ndhE and ndhK) with nucleotide diversity of >0.0200 were detected (Supplementary data Fig. S3). Among them, ndhF, rpl22, matK, rps16 and ndhE have been reported as highly variable regions in many plants (Fu et al., 2017; Fan et al., 2018; Wu et al., 2018; Ye et al., 2018). These genes with high nucleotide diversity may be good sources for interspecies phylogenetic analysis in the future.
Phylogenetics of Allium
Appropriate and multiple gene combination is one of the most important determinants of accurate phylogenetic estimation. The nuclear ribosomal DNA genes [e.g. ITS and external transcribed spacer (ETS)] and many cpDNA fragments (e.g. matK, rps16 and trnL–trnF) have been used to infer the phylogeny of Allium (Friesen et al., 2006; Li et al., 2010a, 2016b; Huang et al., 2014; Herden et al., 2016; Li et al., 2016a; Hauenschild et al., 2017). Li et al. (2010a) reconstructed the phylogeny of Allium based on ITS and rps16; however, the phylogenetic relationships reconstructed by ITS showed weaker support, especially for the third evolutionary lineage, and limited rps16 sequences were included in that study. Here, our plastome phylogenomic analysis of the monocots, based on the shared SCGs and CDSs, provided strong support for the monophyly of Allium and Amaryllidaceae (Figs 1 and 2; Supplementary data Fig. S4), in agreement with previous molecular evidence (Friesen et al., 2006; Li et al., 2010a). This phylogeny also confirmed the three evolutionary lineags of Allium (L1, L2 and L3) that were provided by Friesen et al. (2006), but detected new species relationships within the third evolutionary lineage with high support values. The phylogenetic placements of some species (such as A. caeruleum, A. schoenoprasoides and A. platyspathum), however, differed from the results that were found in the study of Li et al. (2010a), and a similar phenomenon was also revealed in the species A. forrestii, A. strictum and A. obliquum. Notably, in previous phylogenetic analyses, some species of Allium formed weakly supported clades (bootstrap support/posterior probability <50 %), particularly in the third evolutionary lineage (L3) (Friesen et al., 2006; Li et al., 2010a), which was also consistent with the study of Hauenschild et al. (2017), who suggested that some of the subgenera in the third evolutionary clade are not monophyletic. Allium species possess greatly varied morphologies, and, of them, flower changes are most conspicuous (Fig. 1). Previous studies suggested that the diversity of flower form can be attributed to a range of evolutionary novelties that change the appearance of the flower in ways that influence their perceptions by animal pollinators (Darwin, 1859; Moyroud and Glover, 2017). However, changes in flower morphologies are not always driven by pollinators (e.g. Polemonium viscosum) (Galen and Butchart, 2003), which may hint that interspecies relationships are not always consistent with morphologies, such as A. cyathophorum and A. spicatum; the inflorescence of the former is an umbel, and that of the latter is a spike, but they show close a relationship in phylogeny analysis (Figs 1 and 2) (Friesen et al., 2000). All these results may indicate that species relationships of Allium are complex. Although we detected some new species relationships and obtained high support for each branch in this study, relationships among species of Allium are still not well resolved (especially for species in the third evolutionary lineage), and more extensive geographic and genomic sampling for further resolution is required in the future. Here, we conducted the first cp genome analysis on the whole of the Allium genus; we hope this study can contribute to the further analysis on Allium for other researchers.
Divergence time analysis
We estimated that the Amaryllidaceae origin occurred 49.399 Mya (95 % HPD 48.165–50.588 Mya) and the genus Allium originated during the late Eocene [41.932 Mya (95 % HPD: 34.549–47.605 Mya)] (Fig. 3; Table 2; Supplementary data Fig. S5), which was roughly the same time scale as in the study of Li et al. (2016b), who estimated that Allium originated during the late Eocene [34.26 Mya (95 % HPD: 24.29–45.76 Ma)]. This time was also consistent with the possible fossil time of Allium or Amaryllidaceae (49.42 ± 0.54 Mya) that was found in Washington (Pigg et al., 2018). Although the divergence time of many subgenera in Allium has been estimated (Herden et al., 2016; Li et al., 2016b; Xie et al., 2018a), hitherto no study has been conducted on the origin times of the three evolutionary lineages. In this study, we first detected the divergence times of three evolutionary lineages, which are in the early Eocene to the middle Miocene, and most of the Allium species differentiated in the late Tertiary to middle Miocene (approx. 10–15 Ma). Many species divergence or speciation events occurred during this time (Li et al., 2010b; Xu et al., 2010; Zhang and Fritsch, 2010; Gao et al., 2013; Qin et al., 2013; Yu et al., 2014; Zhang et al., 2015) due to factors such as orogeny and climatic oscillations; this may suggest that these factors also play important roles in species divergence of Allium during that time. Moreover, our study suggests that crown group monocots arose in the lower Cretaceous [148.989 Mya (95 % HPD: 108.006–166.103 Mya)]; this estimate is generally congruent with several estimates from other relaxed molecular clock analyses, for example 156 Mya (139–167 Mya) (Smith et al., 2010), 161 Mya (141–176 Mya) (Foster et al., 2017) and 154 Mya (131–184 Mya) (Li et al., 2019), although younger estimates such as 134 Mya (125–145 Mya) (Magallón et al., 2013) and 138 Mya (127–149 Mya) (Zeng et al., 2014) have also been suggested. Despite possible limitations, this analysis provided new insights into the divergence and origin of the genus Allium and other allied families. The detected divergence time of Allium and three evolutionary lineages (L1–L3) may contribute to future studies on subgenera or sections of the genus Allium.
Evolution and positive selection of Allium
Adaptative evolution is a process enabling an organism to fit its habitat better by means of natural selection (Lan et al., 2017). By positive selection analysis, we detected seven significant PSGs (P < 0.05) in the Allium lineage, which is just lower than in the Orchidaceae (nine PSGs) (Fig. 4A; Supplementary data Table S6). As we know, the Orchidaceae have successfully colonized almost every habitat on Earth; the number of its species accounts for approx 10 % of flowering plant species and it has been regarded as a taxon which is highly evolutionary and with great adaptability (Roberts and Dixon, 2008; Givnish et al., 2015). In consideration of the numerous PSGs found in the Allium lineage, its wide species distributions (De Wilde-Duyfjes, 1976; Choi and Oh, 2011; Govaerts et al., 2016), diversified habitat and morphologies (Fig. 1) (Fritsch and Friesen, 2002; Friesen et al., 2006; Block, 2010) and complex chromosome characteristics (Friesen, 1992; Xu et al., 1998; Zhou et al., 2007; Zhang et al., 2009; Jones, 2012; Li et al., 2017; Peruzzi et al., 2017), it may be suggested that Allium is also a successful taxon (the same as the Orchidaceae) in evolution and adaptation, and underwent strong positive natural selection.
By analysing the functions of these PSGs, we found that three significant PSGs (rpoA, rpoB and rpoC2) are associated with RNA polymerase, one gene rpl20 is associated with the large subunit of ribosomal proteins (LSU), and another three genes, petD, rbcL and matK, are related to the subunits of the cytochrome b/f complex, the large subunit of Rubisco and maturase, respectively (Fig. 4A; Supplementary data Table S4). A previous study revealed that RNA polymerase can control the process of gene transcription and affect the pattern of gene expression, thereby allowing species to adapt to a changing environment, and maintain basic metabolic processes necessary for survival (Ishihama, 2000). Furthermore, petD and rbcL are essential in the electron transport chain for generation of ATP and play important roles in plant photosynthesis (Weiss et al., 1991; Allahverdiyeva et al., 2005; Cramer et al., 2011; Xiao et al., 2012). rpl20 is involved in translation, which is an important part of protein synthesis (Krause, 1995), and the matK gene encodes a maturase that is involved in splicing type II introns from RNA transcripts and has been recommended frequently in phylogenetics and barcoding (Hilu, 2000; Hilu et al., 2003; Dunning and Savolainen, 2010). Most of the genes mentioned above have been reported to be under positive selection in previous studies (Dong et al., 2013; Carbonell-Caballero et al., 2015; Ivanova et al., 2017; Xie et al., 2018b; Ye et al., 2018). We also found that most of the genes in Allium with non-significant (P > 0.05) positively selected sites are associated with photosynthesis (e.g. psbC, psbE, petG, petL and atpB) and self-replication (e.g. rps11, rps14 and rps18), which are extremely important processes for plant growth and development (Bryant and Frigaard, 2006; Ewaschuk and Turney, 2006). Therefore, all these genes with positively selected sites may have played key roles in the adaptation of Allium species during the evolutionary process.
Additionally, we found that most of the genes with higher frequencies in these 19 families tend to possess higher average Ka/Ks values (Fig. 4; Supplementary data Table S7). For instance, psbC is present in 17 families and has the highest value (Ka/Ks = 6.31579), followed by rps11, which appeared in 15 families and had the second highest Ka/Ks value (2.03818). Such genes were also detected in the Allium lineage (e.g. psbC, rps11 and rpoC2). A previous study suggested that adaptive evolution may preferentially occur at the molecular level, expressed by an increased value of Ka/Ks (Bakewell et al., 2007). Many studies have confirmed that the higher the Ka/Ks ratio, the stronger the positive selection that species underwent (Hurst, 2002; Fay and Wu, 2003; Ai et al., 2015; Yang et al., 2015). Thus, those genes with high a Ka/Ks rate may play important roles in the adaptation and evolution of Allium.
Moreover, what needs to be pointed out are the chromosome characteristics of Allium species. It has been generalized that chromosome characteristics (chromosome number, ploidy level, karyotype asymmetry, etc.) are crucial to investigate the species relationships and evolution (Sharma and Sharma, 2014). According to previous studies (see the statistical results of Fig. 1 and Supplementary data Table S5), polyploidization is a common karyological feature for Allium species (Ohri et al., 1998; Jones, 2012; Peruzzi et al., 2017). Wu et al. (2010) suggested that the tetraploids of A. przewalskianum arose independently from diploids at least eight times, and that those in A. mairei arose at least three times (Yang, 2010). It has been considered that polyploids possess many advantages compared with their diploid progenitors in morphological, physiological, and life history characteristics, and rates of adaptation (Ramsey and Schemske, 2002; Mayrose et al., 2010). In addition, it has been reported that about 20 out of 97 Allium species with B chromosomes are polyploids (Vujošević et al., 2013), and the B chromosomes were considered to play important roles for species to adapt to harsh environments (e.g. cold, drought or high elevation) (Plowman and Bougourd, 1994, Chen et al., 2005, Wu et al., 2010). Hong (1990) considered that the growth habit and breeding system are the main factors that influence the polyploid frequency of a species. However, a reduction in fertility or even infertility may have occurred when changes in chromosome number generated aneuploids or odd chromosomes (e.g. triploid and pentaploid) (Hong, 1990). Species in Allium are perennial and bulbiferous herbs, and sometimes possess well-developed rhizomes, which can combine asexual and sexual propagation very well (Xu and Kamelin, 2000), and thereby overcome the disadvantages from their chromosomal abnormalities (Zhang et al., 2009). Therefore, the polyploidization, B chromosome and unique breeding system are very important for Allium species to adapt to various environments in the evolutionary processes.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following.
Figure S1: gene maps of the Allium species’ chloroplast genomes.
Figure S2: alignment of 39 Allium chloroplast genome sequences.
Figure S3: the nucleotide diversity of genes in the whole chloroplast genomes of the Allium species.
Figure S4: phylogenetic tree reconstruction based on the 43 shared CDSs of the 164 species.
Figure S5: detailed results of divergence time based on 48 shared SCGs.
Table S1: information for sample collection.
Table S2: the GenBank accessions of all 164 taxa cp genome sequences used this study.
Table S3: primers used for gap closure in this study.
Table S4: gene content in Allium species’ cp genomes.
Table S5: chromosome data statistics of Allium species collected in this study.
Table S6: the potential positive selection test on Allium and allied families.
Table S7: the average values of Ka, Ks and Ka/Ks for each gene.
ACKNOWLEDGEMENTS
The authors thank Yi-Qi Deng, Jun Wen, Xian-Lin Guo and Juan Li for their help in material collection and data analysis.The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
FUNDING
This work was supported by the National Natural Science Foundation of China (grant nos 31872647, 31570198 and 31700176), and the Chinese Ministry of Science and Technology through the ‘National Science and Technology Infrastructure Platform’ project (grant no. 2005DKA21403-JK) and the fourth national survey of traditional Chinese medicine resources (grant no. 2019PC002).
LITERATURE CITED
- Ai B, Gao Y, Zhang XL, Tao JJ, Kang M, Huang HW. 2015. Comparative transcriptome resources of eleven Primulina species, a group of ‘stone plants’ from a biodiversity hotspot. Molecular Ecology Resources 15: 619–632. [DOI] [PubMed] [Google Scholar]
- Aitken SN, Yeaman S, Holliday JA, Wang TL, Curtis-McLane S. 2008. Adaptation, migration or extirpation: climate change outcomes for tree populations. Evolutionary Applications 1: 95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Mamedov F, Mäenpää P, Vass I, Aro EM. 2005. Modulation of photosynthetic electron transport in the absence of terminal electron acceptors: characterization of the rbcL deletion mutant of tobacco. Biochimica et Biophysica Acta 1709: 69–83. [DOI] [PubMed] [Google Scholar]
- Allen JF, de Paula WB, Puthiyaveetil S, Nield J. 2011. A structural phylogenetic map for chloroplast photosynthesis. Trends in Plant Science 16: 645–655. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. 1991. Nuclear DNA content of some important plant species. Plant Molecular Biology Reporter 9: 208–218. [Google Scholar]
- Bakewell MA, Shi P, Zhang J. 2007. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proceedings of the National Academy of Sciences, USA 104: 7489–7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, Specht CD, Leebens-Mack J, Stevenson DW, Zomlefer WB, Davis JI. 2014. Resolving ancient radiations: can complete plastid gene sets elucidate deep relationships among the tropical gingers (Zingiberales)? Annals of Botany 113: 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly MJ, Rigault P, Spokevicius A, Ladiges PY, Tibbits J. 2013. Chloroplast genome analysis of Australian eucalypts – Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae). Molecular Phylogenetics and Evolution 69: 704–716. [DOI] [PubMed] [Google Scholar]
- Beall CM, Cavalleri GL, Deng LB, et al. 2010. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proceedings of the National Academy of Sciences, USA 107: 11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CD, Soltis DE, Soltis PS. 2010. The age and diversification of the angiosperms re-revisited. American Journal of Botany 97: 1296–1303. [DOI] [PubMed] [Google Scholar]
- Bellot S, Renner SS. 2016. The plastomes of two species in the Endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biology and Evolution 8: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block E. 2010. Garlic and other Alliums. The lore and the science. Cambridge: Royal Society of Chemistry. [Google Scholar]
- Bock R. 2007. Structure, function, and inheritance of plastid genomes. In: Bock R, ed. Cell and molecular biology of plastids. Berlin: Springer, 29–63. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DA, Frigaard NU. 2006. Prokaryotic photosynthesis and phototrophy illuminated. Trends in Microbiology 14: 488–496. [DOI] [PubMed] [Google Scholar]
- Capellagutiérrez S, Sillamartínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Caballero J, Alonso R, Iba VE, Terol J, Talon M, Dopazo J. 2015. A phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus Citrus. Molecular Biology and Evolution 32: 2015–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XL, Shi DY, Deng GB, Chen SN. 2005. A preliminary study of B-chromosome and physiological resistance characteristics of Allium mairei. Acta Botanica Boreali-Occidentalia Sinica 25: 1238–1241. [Google Scholar]
- Chevin LM, Hoffmann AA. 2017. Evolution of phenotypic plasticity in extreme environments. Philosophical Transactions of the Royal Society B: Biological Sciences 372: 20160138. doi: 10.1098/rstb.2016.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Oh BU. 2011. A partial revision of Allium (Amaryllidaceae) in Korea and north-eastern China. Botanical Journal of the Linnean Society 167: 153e211. doi: 10.1111/j.1095-8339.2011.01166.x [Google Scholar]
- Christoph H, Lutz B, Bastien C. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Research 41: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couper RA. 1960. New Zealand Mesozoic and Cainozoic plant microfossils. New Zealand Geological Survey. Paleontological Bulletin 32. New Zealand Department of Scientific and Industrial Research. [Google Scholar]
- Cramer WA, Yamashita E, Baniulis D, Hasan SS. 2011. The cytochrome b6f complex of oxygenic photosynthesis. Chichester, UK: John Wiley Sons, Ltd. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1859. On the origins of species by means of natural selection. London: Murray. [Google Scholar]
- Davis MB, Shaw RG. 2001. Range shifts and adaptive responses to Quaternary climate changes. Science 292: 673–679. [DOI] [PubMed] [Google Scholar]
- De Wilde-Duyfjes BEE. 1976. A revision of the genus Allium L. (Liliaceae) in Africa. Belmontia 7: 75–78. [Google Scholar]
- Dong W, Xu C, Li C. et al. 2015. Ycf1, the most promising plastid DNA barcode of land plants. Scientific Reports 5: 8348. doi: 10.1038/srep08348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong WP, Chao X, Tao C, Zhou SL. 2013. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLoS One 8: e77965. doi: 10.1371/journal.pone.0077965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JA, Hickey LJ. 1976. Pollen and leaves from the mid-Cretaceous Potomac Group and their bearing on early angiosperm evolution. In: Beck C, ed. Origin and early evolution of angiosperms. New York: Columbia University Press, 139–206. [Google Scholar]
- Doyle J, Robbins EI. 1977. Angiosperm pollen zonation of the continental Cretaceous of the Atlantic Coastal Plain and its application to deep wells in the Salisbury Embayment. Palynology 1: 41–78. [Google Scholar]
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161: 1307–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duangjit J, Bohanec B, Chan A, Town C, Havey M. 2013. Transcriptome sequencing to produce SNP-based genetic maps of onion. Theoretical and Applied Genetics 126: 2093–2101. [DOI] [PubMed] [Google Scholar]
- Dunning LT, Savolainen V. 2010. Broad-scale amplification of matK for DNA barcoding plants, a technical note. Botanical Journal of the Linnean Society Society 164: 1–9. [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea LA, Merida-Garcia R, Kilian A, Hernandez P, Dorado G. 2017. Assessment of genetic diversity and structure of large garlic (Allium sativum) germplasm bank, by diversity arrays technology ‘genotyping-by-sequencing’ platform (DArTseq). Frontiers in Genetics 8: 98. doi: 10.3389/fgene.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund H, Doyle JA, Herendeen PS. 2004. Morphological phylogenetic analysis of living and fossil Chloranthaceae. International Journal of Plant Sciences 165: 107–151. [Google Scholar]
- Ewaschuk R, Turney PD. 2006. Self-replication and self-assembly for manufacturing. Artificial Life 12: 411–433. [DOI] [PubMed] [Google Scholar]
- Fan WB, Wu Y, Yang J, Shahzad K, Li ZH. 2018. Comparative chloroplast genomics of dipsacales species: insights into sequence variation, adaptive evolution, and phylogenetic relationships. Frontiers in Plant Science 9: 689. doi: 10.3389/fpls.2018.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu C. 2003. Sequence divergence, functional constraint, and selection in protein evolution. Annual Review of Genomics and Human Genetics 4: 213–235. [DOI] [PubMed] [Google Scholar]
- Filyushin AM, Beletsky AV, Mazur AM, Kochieva EZ. 2018. Characterization of the complete plastid genome of lop-sided onion Allium obliquum L. (Amaryllidaceae). Mitochondrial DNA Part B: Resources 3: 393–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CSP, Sauquet H, Marlien VDM, Mcpherson H, Rossetto M, Ho SYW. 2017. Evaluating the impact of genomic data and priors on Bayesian estimates of the angiosperm evolutionary timescale. Systematic Biology 66: 338–351. [DOI] [PubMed] [Google Scholar]
- Franks SJ, Hoffmann AA. 2012. Genetics of climate change adaptation. Annual Review of Genetics 46: 185–208. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Research 32: W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen N. 1992. Systematics of the Siberian polyploid complex in subgenus Rhizirideum (Allium). In: Hanelt P, Hammer K, Knüpffer oder Knuepffer H eds. The genus Allium: taxonomic problems and genetic resources. Proceedings of an international symposium held at Gatersleben, Germany, 11–13 June 1991. Gatersleben, Germany: Institut fur Pflanzengenetikund Kulturpflanzenforschung, 55–66. [Google Scholar]
- Friesen N, Fritsch RM, Pollner S, Blattner FR. 2000. Molecular and morphological evidence for an origin of the aberrant genus Milula within Himalayan species of Allium (Alliacae). Molecular Phylogenetics and Evolution 17: 209–218. [DOI] [PubMed] [Google Scholar]
- Friesen N, Fritsch RM, Blattner FR. 2006. Phylogeny and intrageneric classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences. Aliso 22: 372–395. [Google Scholar]
- Friis EM. 1988. Spirematospermum chandlerae sp. Nov, an extinct species of Zingiberaceae from the North American Cretaceous. Tertiary Research 9: 7–12. [Google Scholar]
- Friis EM, Pedersen KR, Crane PR. 1994. Angiosperm floral structures from the Early Cretaceous of Portugal. Plant Systematics and Evolution 8: 31–49. [Google Scholar]
- Friis EM, Crane PR, Pedersen KR. 1997. Anacostia, a new basal angiosperm from the Early Cretaceous of North America and Portugal with trichotomocolpate/monocolpate pollen. Grana 36: 225–244. [Google Scholar]
- Friis EM, Pedersen KR, Crane PR. 1999. Early angiosperm diversification: the diversity of pollen associated with angiosperm reproductive structures in Early Cretaceous floras from Portugal. Annals of the Missouri Botanical Garden 86: 259–296. [Google Scholar]
- Fritsch RM, Friesen N. 2002. Evolution, domestication and taxonomy. In: Rabinowitch HD, Currah L, eds. Allium crop science: recent advances. Wallingford, UK: CABI Publishing, 5–30. [Google Scholar]
- Fritsch RM, Keusgen M. 2006. Occurrence and taxonomic significance of cysteine sulphoxides in the genus Allium L. (Alliaceae). Phytochemistry 67: 1127–1135. [DOI] [PubMed] [Google Scholar]
- Fu CN, Li HT, Milne R, et al. 2017. Comparative analyses of plastid genomes from fourteen Cornales species: inferences for phylogenetic relationships and genome evolution. BMC Genomics 18: 956. doi: 10.1186/s12864-017-4319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galen C, Butchart B. 2003. Ants in your plants: effects of nectar thieves on pollen fertility and seed-siring capacity in the alpine wildflower, Polemonium viscosum. Oikos 101: 521–528. [Google Scholar]
- Gao YD, Harris A, Zhou SD, He XJ. 2013. Evolutionary events in Lilium (including Nomocharis, Liliaceae) are temporally correlated with orogenies of the Q–T Plateau and the Hengduan mountains. Molecular Phylogenetics and Evolution 68: 443–460. [DOI] [PubMed] [Google Scholar]
- Ge RL, Cai QL, Shen YY, et al. 2013. Draft genome sequence of the Tibetan antelope. Nature Communication 4: 1858. doi: 10.1038/ncomms2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ, Spalink D, Ames M, et al. 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversifcation. Proceedings of the Royal Society B: Biological Sciences 282: 1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, Wang Z, Li N, et al. 2014. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Research 24: 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts R, Friesen N, Fritsch R, et al. 2016. World checklist of Allium. Kew: Royal Botanic Gardens; http://apps.kew.org/wcsp/. [Google Scholar]
- Hancock AM, Brachi B, Faure N, et al. 2011. Adaptation to climate across the Arabidopsis thaliana genome. Science 334: 83–86. [DOI] [PubMed] [Google Scholar]
- Hauenschild F, Favre A, Schnitzler J, Michalak I, Freiberg M, Muellner-Riehl AN. 2017. Spatio-temporal evolution of, Allium, L. in the Qinghai–Tibet-Plateau region: immigration and, in situ, radiation. Plant Diversity 39: 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry D, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biology 17: 134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herden T, Hanelt P, Friesen N. 2016. Phylogeny of Allium L. subgenus Anguinum (G. Don. ex W.D.J. Koch) N. Friesen (Amaryllidaceae). Molecular Phylogenetics and Evolution 95: 79–93. [DOI] [PubMed] [Google Scholar]
- Herendeen PS, Crane PR. 1995. The fossil history of the monocotyledons. In: Rudall PJ, Cribb PJ, Cutler DF, Humphries CJ, eds. Monocotyledons: systematics and evolution. Kew: Royal Botanic Gardens, 1–21. [Google Scholar]
- Hickey LK, Doyle JA. 1977. Early Cretaceous fossil evidence for angiosperm evolution. The Botanical Review 43: 3–104. [Google Scholar]
- Hilu KW. 2000. Phylogeny of Poaceae inferred from matK sequences. Annals of the Missouri Botanical Garden 86: 835–851. [Google Scholar]
- Hilu KW, Borsch T, Muller K, et al. 2003. Angiosperm phylogeny based on matK sequence information. American Journal of Botany 90: 1758–1776. [DOI] [PubMed] [Google Scholar]
- Hong DY. 1990. Plant cytotaxonomy. Beijing: Science Press. [Google Scholar]
- Hu SL, Sablok G, Wang B, et al. 2015. Plastome organization and evolution of chloroplast genes in Cardamine species adapted to contrasting habitats. BMC Genomics 16: 306. doi: 10.1186/s12864-015-1498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. 2002. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends in Genetics 18: 486–487. [DOI] [PubMed] [Google Scholar]
- Huang DQ, Li QQ, Zhou CJ, Zhou SD, He XJ. 2014. Intraspecific differentiation of Allium wallichii (Amaryllidaceae) inferred from chloroplast DNA and internal transcribed spacer fragments. Journal of Systematics and Evolution 52: 341–354. [Google Scholar]
- Ishihama A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annual Review of Microbiology 54: 499–518. [DOI] [PubMed] [Google Scholar]
- Ivanova Z, Sablok G, Daskalova E, et al. 2017. Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Frontiers in Plant Science 8: 204. doi: 10.3389/fpls.2017.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proceedings of the National Academy of Sciences, USA 104: 19369–19374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen T, Bremer K. 2004. The age of major monocot groups inferred from 800+ rbcL sequences. Botanical Journal of the Linnean Society Society 146: 385–398. [Google Scholar]
- Jia JZ, Zhao SC, Kong XY, et al. 2013. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95. [DOI] [PubMed] [Google Scholar]
- Jin FY, Xie DF, Zhou SD, He XJ. 2018. Characterization of the complete chloroplast genome of Allium prattii. Mitochondrial DNA Part B: Resources 3: 153–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N. 2012. B chromosomes in plants. Plant Biosystems 146: 727–737. [Google Scholar]
- Khakhlova O, Bock R. 2006. Elimination of deleterious mutations in plastid genomes by gene conversion. The Plant Journal 46: 85–94. [DOI] [PubMed] [Google Scholar]
- Kim S, Yoon MK. 2010. Comparison of mitochondrial and chloroplast genome segments from three onion (Allium cepa L.) cytoplasm types and identification of a trans-splicing intron of cox2. Current Genetics 56: 177–188. [DOI] [PubMed] [Google Scholar]
- Krause M. 1995. Transcription and translation. Methods in Cell Biology 48: 483. [PubMed] [Google Scholar]
- Lan Y, Sun J, Tian R, et al. 2017. Molecular adaptation in the world’s deepest-living animal: insights from transcriptome sequencing of the hadal amphipod Hirondellea gigas. Molecular Ecology 26: 3732–3743. [DOI] [PubMed] [Google Scholar]
- Lee J, Chon JK, Lim JS, Kim EK, Nah G. 2017. Characterization of complete chloroplast genome of Allium victorialis and its application for barcode markers. Plant Breed Biotechnology 5: 221–227. [Google Scholar]
- Leimu R, Fischer M. 2008. A meta-analysis of local adaptation in plants. PLoS One 3: e4010. doi: 10.1371/journal.pone.0004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HT, Yi TS, Gao LM, et al. 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants 5: 461–470. [DOI] [PubMed] [Google Scholar]
- Li MJ, Tan JB, Xie DF, Huang DQ, Gao YD, He XJ. 2016a. Revisiting the evolutionary events in Allium subgenus Cyathophora (Amaryllidaceae): insights into the effect of the Hengduan Mountains Region (HMR) uplift and quaternary climatic fluctuations to the environmental changes in the Qinghai–Tibet Plateau. Molecular Phylogenetics and Evolution 94: 802–813. [DOI] [PubMed] [Google Scholar]
- Li MJ, Guo XL, Li J, Zhou SD, Liu Q, He XJ, 2017. Cytotaxonomy of Allium (Amaryllidaceae) subgenera Cyathophora and Amerallium sect. Bromatorrhiza. Phytotaxa 331: 185–198. [Google Scholar]
- Li QQ, Zhou SD, He XJ, Yu Y, Zhang YC, Wei XQ. 2010a. Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Annals of Botany 106: 709–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QQ, Zhou SD, Huang DQ, He XJ, Wei XQ. 2016b. Molecular phylogeny, divergence time estimates and historical biogeography within one of the world’s largest monocot genera. AoB Plants 8: plw041. doi: 10.1093/aobpla/plw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Stocks M, Hemmilä S, et al. 2010b. Demographic histories of four spruce (Picea) species of the Qinghai–Tibetan Plateau and neighboring areas inferred from multiple nuclear loci. Molecular Biology and Evolution 27: 1001–1014. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Loarie SR, Duffy PB, Hamilton H, Asner GP, Field CB, Ackerly DD. 2009. The velocity of climate change. Nature 462: 1052–1055. [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research 41: W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S, Hilu KW, Quandt D. 2013. Land plant evolutionary timeline: gene effects are secondary to fossil constraints in relaxed clock estimation of age and substitution rates. American Journal of Botany 100: 556–573. [DOI] [PubMed] [Google Scholar]
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández TA. 2015. Metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Malé PG, Bardon L, Besnard G, et al. 2014. Genome skimming by shotgun sequencing helps resolve the phylogeny of a pantropical tree family. Molecular Ecology Resources 14: 966–975. [DOI] [PubMed] [Google Scholar]
- Mayrose I, Barker MS, Otto SP. 2010. Probabilistic models of chromosome number evolution and the inference of polyploidy. Systematic Biology 59: 132–144. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proceedings of the National Academy of Sciences, USA 107: 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyroud E, Glover BJ. 2017. The evolution of diverse floral morphologies. Current Biology 27: R941–R951. [DOI] [PubMed] [Google Scholar]
- Muller J. 1981. Fossil pollen of extant angiosperms. The Botanical Review 47: 1–142. [Google Scholar]
- Nagarajan N, Pop M. 2013. Sequence assembly demystified. Nature Reviews. Genetics 14: 157–167. [DOI] [PubMed] [Google Scholar]
- Nguyen NH, Driscoll HE, Specht CD. 2008. A molecular phylogeny of the wild onions (Allium; Alliaceae) with a focus on the western north American center of diversity. Molecular Phylogenetics and Evolution 47: 1157–1172. [DOI] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, et al. 2010. Plant phenotypic plasticity in a changing climate. Trends in Plant Science 15: 684–692. [DOI] [PubMed] [Google Scholar]
- Ohri D, Fritsch RM, Hanelt P. 1998. Evolution of genome size in Allium (Alliaceae). Plant Systematics and Evolution 210: 57–86. [Google Scholar]
- Oostra V, Saastamoinen M, Zwaan BJ, Wheat CW. 2018. Strong phenotypic plasticity limits potential for evolutionary responses to climate change. Nature Communication 9: 1005. doi: 10.1038/s41467-018-03384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Yang Z, Zhang H, et al. 2011. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Molecular Biology and Evolution 28: 1075–1081. [DOI] [PubMed] [Google Scholar]
- Peruzzi L, Carta A, Altinordu F. 2017. Chromosome diversity and evolution in Allium (Allioideae, Amaryllidaceae). Plant Biosystems 151: 212–220. [Google Scholar]
- Peska V, Mandakova T, Ihradska V, Fajkus J. 2019. Comparative dissection of three giant genomes: Allium cepa, Allium sativum, and Allium ursinum. International Journal of Molecular Sciences 20: E733. doi: 10.3390/ijms20030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigg KB, Bryan FA, DeVore ML. 2018. Paleoallium billgenseli Gen et sp. Nov: fossil monocot remains from the latest early Eccene republic flora, Northeastern Washington state, USA. International Journal of Plant Sciences 179: 477–486. [Google Scholar]
- Plowman AB, Bougourd SM. 1994. Selectively advantageous effects of B chromosomes on germination behavior in Allium schoenoprassum L. Heredity 72: 587–593. [Google Scholar]
- Qin AL, Wang MM, Cun YZ, et al. 2013. Phylogeographic evidence for a link of species divergence of Ephedra in the Qinghai–Tibetan Plateau and adjacent regions to the Miocene Asian aridification. PLoS One 8: e56243. doi: 10.1371/journal.pone.0056243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q, Zhang GJ, Ma T, et al. 2012. The yak genome and adaptation to life at high altitude. Nature Genetics 44: 946–949. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. 2009. Tracer v1.5 http://beast.bio.ed.ac.uk/Tracer.
- Ramsey J, Schemske DW. 2002. Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics 33: 589–639. [Google Scholar]
- Raubeson LA, Jansen RK. 2005. Chloroplast genomes of plants. In: Henry RJ, ed. Diversity and evolution of plants – genotypic and phenotypic variation in higher plants. Wallingford, UK: CABI Publishing, 45–46. [Google Scholar]
- Roberts DL, Dixon KW. 2008. Orchids. Current Biology 18: R325–R329. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Savolainen O, Pyhajarvi T, Knurr T. 2007. Gene flow and local adaptation in trees. Annual Review of Ecology Evolution and Systematics 38: 595–619. [Google Scholar]
- Schatz MC, Witkowski J, McCombie WR. 2012. Current challenges in de novo plant genome sequencing and assembly. Genome Biology 13: 243. doi: 10.1186/gb-2012-13-4-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeweiss GM, Palomeque T, Colwell AE, Weiss-Schneeweiss H. 2004. Chromosome numbers and karyotype evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. American Journal of Botany 91: 439–448. [DOI] [PubMed] [Google Scholar]
- Sharma G, Sharma N. 2014. Cytology as an important tool for solving evolutionary problem in angiosperms. Proceedings of the National Academy of Sciences, India-Section B: Biological Sciences 84: 1–7. [Google Scholar]
- Shaw J, Shafer HL, Leonard OR, Kovach MJ, Schorr M, Morris AB. 2014. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. American Journal of Botany 101: 1987–2004. [DOI] [PubMed] [Google Scholar]
- Simonson TS, Yang YZ, Huff CD, et al. 2010. Genetic evidence for high-altitude adaptation in Tibet. Science 329: 72–75. [DOI] [PubMed] [Google Scholar]
- Smith SA, Beaulieu JM, Donoghue MJ. 2010. An uncorrelated relaxed clock analysis suggests an earlier origin for flowering plants. Proceedings of the National Academy of Sciences, USA 107: 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony, version 4b10. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Van Buskirk J. 2017. Spatially heterogeneous selection in nature favors phenotypic plasticity in Anuran larvae. Evolution 71: 1670–1685. [DOI] [PubMed] [Google Scholar]
- Villemereuil PD, Mouterde M, Gaggiotti OE, Till-Bottraud I. 2018. Patterns of phenotypic plasticity and local adaptation in the wide elevation range of the alpine plant Arabis alpina. Journal of Ecology 106: 1952–1971. [Google Scholar]
- Vujoševiæ M, Jovanoviæ V, Blagojeviæ J. 2013. Polyploidy and B chromosomes in Allium flavum from Serbia. Archives of Biological Sciences 65: 23–32. [Google Scholar]
- Weiss H, Friedrich T, Hofhaus G, Preis D. 1991. The respiratory-chain NADH dehydrogenase (complex I) of mitochondria. Berlin, Heidelberg: Springer. [DOI] [PubMed] [Google Scholar]
- Wikström N, Savolainen V, Chase MW. 2001. Evolution of the angiosperms: calibrating the family tree. Proceedings of the Royal Society B: Biological Sciences 268: 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences, USA 84: 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LL, Cui XK, Milne RI, Sun YS, Liu JQ. 2010. Multiple autopolyploidizations and range expansion of Allium przewalskianum Regel. (Alliaceae) in the Qinghai–Tibetan plateau. Molecular Ecology 19: 1691–1704. [DOI] [PubMed] [Google Scholar]
- Wu Y, Liu F, Yang DG, et al. 2018. Comparative chloroplast genomics of Gossypium species: insights into repeat sequence variations and phylogeny. Frontiers in Plant Science 9: 376. doi: 10.3389/fpls.2018.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li J, Ouyang M, et al. 2012. DAC is involved in the accumulation of the cytochrome b6/f complex in Arabidopsis. Plant Physiology 160: 1911–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Xie DF, Zhong Y, et al. 2018a. The effect of Hengduan Mountains Region (HMR) uplift to environmental changes in the HMR and its eastern adjacent area: tracing the evolutionary history of Allium section Sikkimensia. Molecular Phylogenetics and Evolution 130: 380–396. [DOI] [PubMed] [Google Scholar]
- Xie DF, Yu Y, Deng YQ, et al. 2018b. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. International Journal of Molecular Sciences 19: E1847. doi: 10.3390/ijms19071847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DF, Jin FY, Yang X, Li H, He XJ. 2019a The complete chloroplast genome of a wild onion species Allium monanthum (Alliaceae). Mitochondrial DNA B: Resources 4: 854–855. [Google Scholar]
- Xie DF, Yu HX, Price M, et al. 2019. b Phylogeny of Chinese Allium species in Section Daghestanica and adaptive evolution of Allium (Amaryllidaceae, Allioideae) species revealed by complete chloroplast genome. Frontiers in Plant Science 10: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JM, Kamelin RV. 2000. Allium L. In: Wu ZY, Raven PH, eds. Flora of China, Vol. 24 Beijing: Science Press, Missouri Botanical Garden Press, 165–202. [Google Scholar]
- Xu JM, Yang L, He XJ. 1998. A study on karyotype differentiation of Allium fasciculatum (Liliaceae). Acta Phytotaxonomica Sinica 36: 346–352. [Google Scholar]
- Xu TT, Abbott RJ, Milne RI, et al. 2010. Phylogeography and allopatric divergence of cypress species (Cupressus L.) in the Qinghai–Tibetan Plateau and adjacent regions. BMC Evolutionary Biology 10: 194. doi: 10.1186/1471-2148-10-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wang Y, Zhang Z, He S. 2015. Comprehensive transcriptome analysis reveals accelerated genic evolution in a tibet fish, Gymnodiptychus pachycheilus. Genome Biology and Evolution 7: 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Xie DF, Zhou SD, He XJ. 2019. Characterization of the complete chloroplast genome of Allium kingdonii. Mitochondrial DNA B: Resources 4: 868–869. [Google Scholar]
- Yang Y. 2010. Cytotype distribution and polyploid origins of Allium mairei Lévl. (Alliaceae). China: Kunming Institute of Botany, Chinese Academy of Sciences. [Google Scholar]
- Yang Z, Zhao TT, Ma QH, Liang LS, Wang GX. 2018. Comparative genomics and phylogenetic analysis revealed the chloroplast genome variation and interspecific relationships of Corylus (Betulaceae) species. Frontiers in Plant Science 9: 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Dos RM. 2011. Statistical properties of the branch-site test of positive selection. Molecular Biology and Evolution 28: 1217–1228. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Molecular Biology and Evolution 19: 908–917. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Wong WSW, Nielsen R. 2005. Bayes Empirical Bayes Inference of amino acid sites under positive selection. Molecular Biology and Evolution 22: 1107–1118. [DOI] [PubMed] [Google Scholar]
- Yates SA, Swain MT, Hegarty MJ, et al. 2014. De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genomics 15: 453. doi: 10.1186/1471-2164-15-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye WQ, Yap ZY, Li P, Comes HP, Qiu YX. 2018. Plastome organization, genome-based phylogeny and evolution of plastid genes in Podophylloideae (Berberidaceae). Molecular Phylogenetics and Evolution 127: 978–987. [DOI] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329: 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XQ, Maki M, Drew BT, et al. 2014. Phylogeny and historical biogeography of Isodon (Lamiaceae): rapid radiation in south-west China and Miocene overland dispersal into Africa. Molecular Phylogenetics and Evolution 77: 183–194. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Gao LM, Soltis DE, et al. 2017. Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytologist 215: 1235–1248. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Sun R, Kong H, Zhang N, Ma H. 2014. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nature Communications 5: 4956. doi: 10.1038/ncomms5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ML, Fritsch PW. 2010. Evolutionary response of Caragana (Fabaceae) to Qinghai–Tibetan Plateau uplift and Asian interior aridification. Plant Systematics and Evolution 288: 191–199. [Google Scholar]
- Zhang ML, Wen ZB, Hao XL, Byalt VV, Sukhorukov AP, Sanderson SC. 2015. Taxonomy, phylogenetics and biogeography of Chesneya (Fabaceae), evidenced from data of three sequences, ITS, trnS–trnG, and rbcl. Biochemical Systematics and Ecology 63: 80–89. [Google Scholar]
- Zhang TC, Qiao Q, Novikova PY, et al. 2019. Genome of Crucihimalaya himalaica, a close relative of Arabidopsis, shows ecological adaptation to high altitude. Proceedings of the National Academy of Sciences, USA 116: 7137–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Zhou SD, Ren HY, et al. 2009. Karyotype in 20 populations belonging to 10 species of Allium from Southwest China. Journal of Wuhan Botanical Research 27: 351–360. [Google Scholar]
- Zhang YJ, Du LW, Liu A, et al. 2016a. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Frontiers in Plant Science 7: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZF, Li YY, Xiao BZ. 2016b. Comparative transcriptome analysis highlights the crucial roles of photosynthetic system in drought stress adaptation in upland rice. Scientific Reports 6: 19349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SD, He XJ, Yu Y, Xu JM. 2007. Karyotype studies on twenty-one populations of eight species in Allium section Rhizirideum. Acta Phytotaxonomica Sinica 45: 207–216. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.