Abstract
Gene expression in HIV-1 is regulated by the promoters in 5’ long-terminal repeat (LTR) element, which contain multiple DNA regulatory elements that serve as binding sites for cellular transcription factors. YY1 could repress HIV-1 gene expression and latent infection. Here, however, we observed that virus production can be increased by YY1 over-expression and decreased under YY1 depleted condition by siRNA treatment. To identify functional domain(s) of YY1 activation, we constructed a number of YY1 truncated mutants. Our data show that full-length YY1 enhances the viral transcription both through U3 and U3RU5 promoters. Moreover, the C-terminal region (296-414 residues) of YY1 is responsible for the transcriptional upregulation, which could be enhanced further in the presence of the viral Tat protein. The central domain of YY1 (155-295 residues) does not affect LTR activity but has a negative effect on HIV-1 gene expression. Taken together, our study shows that YY1 could act as a transcriptional activator in HIV-1 replication, at least in the early stages of infection.
Keywords: HIV-1, Tat, transcription regulation, YY1
INTRODUCTION
Yin Yang 1 (YY1), a ubiquitous and multifunctional member of polycomb group (PcG) of proteins, is known to function in chromatin remodeling (1). This 414 residue-long protein has an N-terminal acidic transcriptional activation domain (1-154 residues), followed by a glycine-lysine rich DNA transcriptional inhibition domain (170-200 residues), a small Recruit Polycomb (REPO) domain (201-225 residues), and a C-terminal DNA binding domain with four GLI-Krüppel-like zinc finger motifs (296-414 residues) that are responsible for DNA binding (2-5).
YY1 was first identified as a transcription factor and cDNA clone was isolated simultaneously by two independent groups (5-7). Multiple diverse functions of YY1 have been identified, such as, regulation of cell proliferation, embryogenesis, differentiation, tumor suppression, oncogenesis, DNA damage and apoptosis (8, 9) and reviewed in (10, 11). Further, YY1 could function as a transcription factor regulating several different eukaryotic and viral genes including those of HIV-1, Adenovirus, adeno-associated virus (AAV), cytomegalovirus, parvovirus and Human papillomavirus (HPV) (reviewed in (10, 12)). The consensus core DNA sequence of YY1 recognition site is known as 5’-CAT-3’s (13). Depending on the promoter and the ability of YY1 to interact with other transcription factors or cofactors, such as histone acetyltransferase (HAT), p300, CREB binding protein (CREBBP), histone deacetylases (HDACs), enhancer of zeste 2 (EZH2) and DNA methyltransferases (DNMTs), YY1 could function as either an activator or a repressor (3,14-18).
HIV-1 has a 5’ long-terminal repeat (LTR) region, regulated by a promoter with multiple DNA regulatory elements that could bind several transcription factors. HIV-1 LTR contains two binding sites for YY1. The first region is at −17 to +27 position of LTR, through which YY1 was shown to repress the LTR-driven reporter gene transcription (19). The repression, however, requires a late SV40 factor (LSF), for which LSF and ZF motifs in the C-terminal domain (308-368 residues) of YY1 are involved. Co-purification of HDAC1 with LTR-LSF-YY1 complex demonstrated the involvement of the central domain (154-199 residues) of YY1 in recruiting HDAC1 for the repression of HIV-1 transcription and thereby promoting establishment and maintenance of its latent infection (16, 20, 21). The second region is at −140 to −120 of LTR. Unlike −17 to +27 region, YY1 binds this region directly in unstimulated conditions and leads to transcriptional repression of LTR. This binding is decreased after T-cell stimulation, thereby releasing the repression (22). Interestingly, YY1 mutant lacking HDAC1-interacting domain (155-198 residues) could bind both −140 to −120 and −17 to +27 regions on LTR but not repress LTR expression. These studies suggested that YY1 functions as a transcriptional repressor of HIV-1 LTR in conjunction with HDAC1 and a regulator leading to the latent infection of HIV-1.
Here, we investigated the effect and role of YY1 on the production of HIV-1 virus. Unprecedentedly, we observed that YY1 could enhance HIV-1 U3 and U3RU5 mediated promoter activity, leading to increased viral transcription and production. Our data strongly indicate that YY1 could function as an activator, at least in the acute infection stage.
RESULTS
YY1 is important for HIV-1 replication
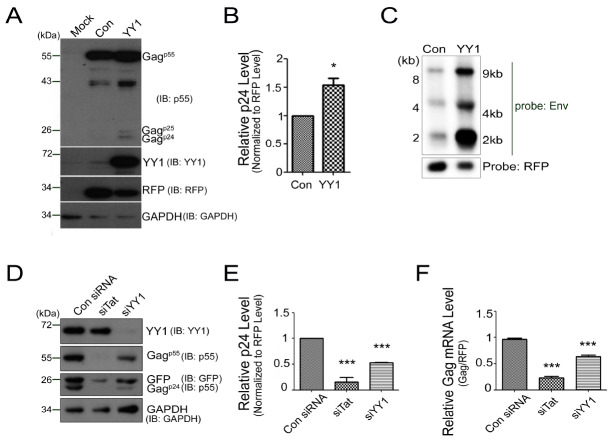
To investigate the effect of YY1 on HIV-1 replication and production, 293T cells were co-transfected with HIV-1 proviral DNA, pNL4-3GFP, and a YY1 expression plasmid. HIV-1 production was determined at 24 h post-transfection by measuring the amount of Gag protein production in cell lysate and viral production in cell supernatant using Western blotting and p24Gag ELISA analysis, respectively. Overexpression of YY1 enhanced the Gag protein production in cells and virus in cell supernatant (Fig. 1A and 1B). An RFP expression vector was included as a normalization control in all our transfection assays. We constructed an mCherry-tagged YY1 expression plasmid and used in co-transfection with the pNL4-3GFP to rule out transfection efficiency based experimental bias. Intensity of GFP was measured in mCherry+ GFP+ cells by FACS analysis. YY1-mCherry expressing cells showed higher GFP intensity than mCherry control (Supplementary Fig. 1). These data indicated clearly that YY1 could increase HIV-1 gene expression, resulting in an enhancement of HIV-1 production. Since YY1 is a well-known transcription factor, we verified the level of HIV-1 mRNA by northern blotting analysis. Production of all HIV-1 mRNA derivatives, such as un-spliced (9 kb), single spliced (4 kb) and multiple spliced (2 kb) RNAs, were increased in YY1 overexpressed cells (Fig. 1C).
Fig. 1.
YY1 enhances HIV-1 replication in 293T cell. (A-C) 293T cells were transfected pCMV/Con or pCMV/HA-YY1 with pNL4-3GFP and pDs-Red. Cell lysate were subjected to Western blotting (A) and for quantification of p24 levels, the viral supernatants were analyzed by p24 ELISA, *P < 0.05, Student’s t test, obtained from three independent experiments (B). HIV-1 mRNA was determined by Northern blotting (C). (D-F) 239T cells were transfected with either control siRNA, siTat or siYY1 with pNL4-3GFP and pDs-Red. Western blotting of cell lysate (D), p24 ELISA of viral supernatant (E) and qRT-PCR of Gag RNA (F). The asterisks indicate a significant difference from control, one-way ANOVA with Dunnett’s multiple comparisons test, ***P < 0.0001.
Further, using RNA interference, we investigated the effect of YY1 on HIV-1 production. After 24 h post-transfection with either siYY1 or siTat (as a positive control for knock-down experiment), resulting cells and viral supernatants were harvested, and the virus level was measured. Western blotting showed that transfection with siYY1, which repressed YY1 by over 80%, resulted in a concomitant decrease in levels of Gag and GFP proteins compared to that of control siRNA treated cells (Fig. 1D). Further, p24Gag ELISA showed that knock-down of YY1 caused about 50% reduction of the virus production compared to the control (Fig. 1E). Treatment with siTat, which targets HIV-1 specific transcriptional activator, further supported the effect of siYY1 on virus production. Quantitation by qRT-PCR showed about 40% decrease in Gag mRNA (9 kb) in the presence of siYY1 (Fig. 1F). Altogether, these results suggest that YY1 has a positive role in HIV-1 transcription and virus production.
YY1 has a positive effect on HIV-1 replication in CD4+ T cells
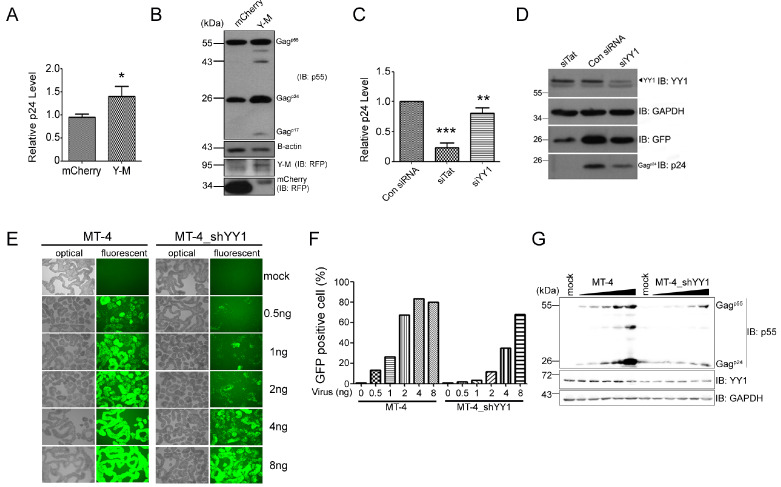
Further, to confirm the positive effect of YY1 on virus production, as observed in proviral DNA transfection system, we investigated the effect of YY1 on HIV-1 infection cycle, using a CD4+ T cell line MT-4 cells. At 24 h post-transfection of MT-4 cells with either YY1-mCherry expression vector or siYY1, the cells were infected with HIV-1 NL4-3GFP viruses. Three days post-infection, we observed that MT-4 cells expressing the YY1-mCherry yielded more viruses than the cells expressing mCherry alone (Fig. 2A and 2B). Conversely, when YY1 was depleted by siYY1 RNA, virus production became lower than in the case of control siRNA (Fig. 2C and 2D). Notably, however, that effect of overexpression or knock-down of YY1 in MT-4 cells is lesser than that observed in 293T cells, due to lower transfection efficiency of MT-4 cells.
Fig. 2.
YY1 enhances HIV-1 replication in MT-4 cell. (A, B) MT-4 cells were transfected pCDNA3/mCherry or pCDNA3/YY1-mCherry (Y-M). 24 h later, HIV-1NL4-3GFP infected and incubated for three days. (A) p24 ELISA result of viral supernatant. *P < 0.05, Student’s t test, obtained from three independent experiments. (B) Western blotting of cell lysate. (C, D) MT-4 cells were transfected either consiRNA, siTat or siYY1 then HIV-1NL4-3GFP infected and incubated as described above. (C) p24 ELISA result of viral supernatant. (D) Western blotting of cell lysate. The asterisks indicate a significant difference from control, one-way ANOVA with Dunnett’s multiple comparisons test, **P < 0.01, ***P < 0.001. (E-G) MT-4 or MT-4_shYY1 cells were infected HIV-1NL4-3GFP with indicated concentration then incubated for three days. (E) HIV-1 infected cell captured by fluorescent microscope, (F) GFP positive cell were analyzed by FACS, (G) Western blotting. All experiments are performed more three independent experiments.
To verify the requirement and positive effect of YY1 on HIV-1 virus production in MT-4 cells, we constructed and established a YY1 stably knocked-down MT-4 cell line, named MT-4_shYY1. Equal numbers of MT-4 and MT-4_shYY1 cells were infected by different MOI of HIV-1 NL4-3GFP viruses, as indicated in Fig. 2E. Three days post-infection, virus infected cells, expressing GFP, were captured by fluorescent microscope and analyzed by FACS analysis (Fig. 2E and 2F) and Western blotting (Fig. 2G). Similar to siRNA treatment, MT-4_shYY1 cells showed a lower level of virus production than control MT-4 cells. Quantification of virus level 3 h after infection (Supplementary Fig. 2) confirmed that same level of virus entered the cells, indicating that the lower level of virus production in MT-4_shYY1 cells, compared to MT-4 cells, is not due to different numbers of virus entered. These data altogether demonstrated that YY1 also functions positively in the virus infection cycle in CD4 + T cell.
YY1 upregulates the HIV-1 promoter activity and trans-acting activity of HIV-1 Tat
We observed that overexpression of YY1 increased mRNA level of HIV-1 (Fig. 1C) and knockdown of YY1 resulted in the opposite (Fig. 1F). Thus, to investigate the direct influence of YY1 on HIV-1 promoter further, we constructed a firefly luciferase (Fluc) expression plasmid driven by U3, U3R, and U3RU5 (also referred to as LTR) region of HIV-1. U3 contains the modulatory region (nt −454 to −104), a core enhancer (nt −105 to −79), and basal promoter (nt −78 to −1) (23, 24). R (+1 to +82) contains transactivation response RNA structure (TAR) (+19 to +44), which is a Tat protein binding region, and Tat-TAR interaction leads to enhance the efficiency of viral transcription and elongation (25, 26). Moreover, two binding sites for YY1 were reported in HIV-1 LTR, one resides in U3 region (−120 to −140) and the other in R region (−17 to +27).
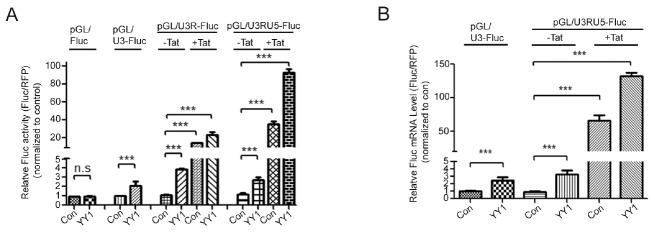
YY1 increased U3, U3R and U3RU5 driven Fluc reporter gene expression over three-fold (Fig. 3A). Since U3R and U3RU5 contain the TAR, Fluc expression is considerably increased in the presence of Tat as shown earlier (Fig. 3A). Unexpectedly, YY1 elevates the Fluc expression under the condition already activated by Tat protein. To confirm if increased Fluc level is due to increased RNA level, qRT-PCR analyses were performed. Similar to Fluc protein expression, increase in YY1 increases Fluc RNA level (Fig. 3B). These data indicate role of YY1 as a transcription activator in both conditions, in the absence and presence of HIV-1 Tat protein.
Fig. 3.
YY1 up-regulates HIV-1 promoter activity and trans-acting activity of Tat protein. 293T cells were transfected pCMV/Con or pCMV/HA-YY1 with pGL/Fluc or pGL/U3-Fluc or pGL/U3R-FLuc or pGL/U3RU5-Fluc, pDs-Red and pCMV-Tat (in the case mentioned). 24h later, cells were harvested and performed luciferase assay (A) and qRT-PCR (B). The asterisks indicate a significant difference from control, one-way ANOVA with Dunnett’s multiple comparisons test, *P < 0.05, **P < 0.01, ***P < 0.0001 and n.s indicate non- significant, obtained from more three independent experiments.
YY1 has two regions with different activities
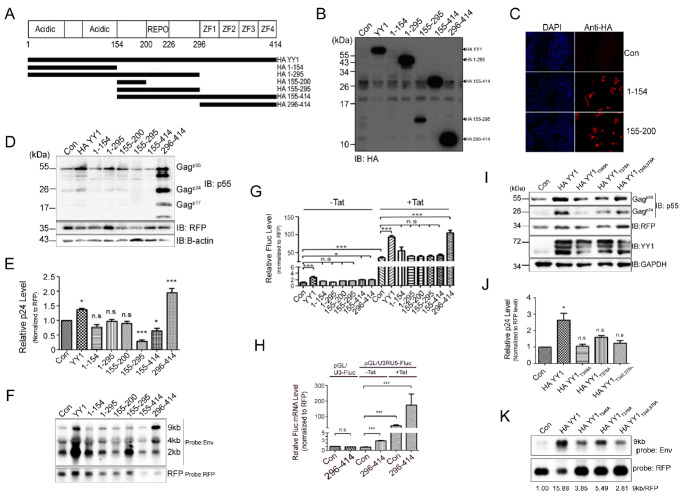
In order to determine the functional domains of YY1 responsible for the positive effect on HIV-1 virus production, we constructed various truncated mutants of YY1 (Fig. 4A). The expression levels of all the mutants were confirmed by Western blotting (Fig. 4B). Expression of most of the mutants was detected by Western blotting except of 1-154 and 155-200 domains. Therefore, expression of 1-154 and 155-200 domains was confirmed by immunostaining (Fig. 4C).
Fig. 4.
YY1 have two motifs with antagonistic roles on HIV-1 replication. (A) Scheme of YY1 and its truncated mutants used in this study. (B, C) A confirmation of protein expression by Western blotting (B) and Immunofluorescent analysis (C). (D-F) 293T cells were transfected pCMV/Con or indicated YY1 derivatives with pNL4-3GFP and pDs-Red. Western blotting of cell lysate (D), p24 ELISA of viral supernatant (E). Northern blotting of HIV-1 mRNA (F). (G) 293T cells were transfected pCMV/Con or indicated YY1 derivatives with pGL/U3RU5-Fluc, pDs-Red and pCMV-Tat. 24 h later, luciferase assay were performed. (H) 293T cells were transfected pCMV/Con or HA-296-414 with pGL/U3-Fluc or pGL/U3RU5-Fluc, pDs-Red and pCMV-Tat. 24 h later, qRT-PCR.were performed. (I-K) 293T cells were transfected pCMV/Con or indicated YY1 derivatives. Western blotting of cell lysate (I), p24 ELISA of viral supernatant (J) Northern blotting of HIV-1 mRNA. The relative ratio (9kb/RFP) was calculated by measuring the band intensity using Image J software (NIH) (K). The asterisks indicate a significant difference from control, one-way ANOVA with Dunnett’s multiple comparisons test, *P < 0.05, **P < 0.01, ***P < 0.0001 and n.s indicate non-significant, obtained from more three independent experiments.
Further, each of the truncated mutants was then co-transfected individually with pNL4-3GFP and pDs-red, and the virus production levels were measured. Among the mutants, while the 296-414 domain alone showed a significant enhancement of virus production, the 155-295 domain resulted in a decrease (Fig. 4D and 4E). Northern blotting indicated that the increase of virus production with the 296-414 domain is due to up-regulation of viral transcription as observed with wild type YY1 expression (Fig. 4F). However, an apparent decrease in the viral production with the 155-295 domain of YY1 did not correlate with the viral mRNA level. Measurement of Fluc activity of U3RU5-Fluc with YY1 truncated mutants showed that in the absence of Tat protein, only full-length YY1 significantly enhanced the Fluc activity, but in the presence of Tat protein, 296-414 domain activated the Fluc activity as much as full-length YY1 (Fig. 4G). Transcription activation via 296-414 domain was confirmed by qRT-PCR (Fig. 4H). The 296-414 domain did not affect U3 and slightly increased the U3RU5. Similar to protein levels, Fluc mRNA levels were increased in the presence of Tat (Fig. 4G), indicating that the increased virus production via 296-414 domain occurred in a Tat-dependent manner.
A previous study showed that phosphorylation of threonine residues at 348 and 378 abolish DNA binding activity of YY1 (27). To analyze the importance of DNA binding activity on HIV-1 production, we constructed the alanine substitution mutants on these sites, YY1T348A, YY1T378A and a double mutation mutant YY1T348,378A. We then determined the effect of these mutants on virus production via co-transfection with pNL4-3GFP into the 293T cells. Western blotting and p24 ELISA show that all three mutants have abolished the activation ability of virus production (Fig. 4I and 4J) and transcription activation (Fig. 4K), suggesting that the Threonine residues at 348 and 378 in 296-414 domain of YY1 are critical for a positive transcriptional activation of YY1 on HIV-1 production.
These data suggest that YY1 might have bifunctional properties in HIV-1 production. While 296-414 domain of YY1 has a transcriptional activator activity especially in the presence of Tat, 155-295 domain may have some other role(s), like post-transcriptional or translational repressor activity which needs to be elucidated further. Taken together, the data presented here suggest strongly that full-length YY1 could function as HIV-1 transcriptional activator.
DISCUSSION
Here, using 293T cells in transfection assays and using MT-4 cell-line infection assays, we report that overexpressing YY1 increased HIV-1 production and found that such an increase came from transcriptional activation of HIV-1 (Fig. 1 and 2). Effect of YY1 on HIV-1 LTR promoter was verified using constructs with U3, U3R and U3RU5 driven Fluc reporter gene assay. We observed that YY1 enhances the U3 promoter activity, but higher enhancement was observed with U3R and U3RU5 forms. The latter activity was also increased in the presence of Tat transcriptional activator (Fig. 3). However, direct or indirect binding of YY1 to LTR has been shown previously to repress LTR activity through the recruitment of HDAC1. The contradicting observations might be due to the following: A) Unlike the previous studies, we employed RFP expression plasmid as an internal control in our assays, which enabled relatively accurate measurements in the transfection assays (28). B) We observed a decrease in the virus production when YY1 is depleted as knock-down of YY1 decreased the virus production (Fig. 1 and 2). In the only knock-down experiment on latently HIV infected cell lines, 2D10 and J89, the authors hypothesized that YY1 depletion leads to active transcription of HIV in both the cell lines. However, because that was occurred only in 2D10 cells, the study concluded that maintenance of HIV-1 latency is complex, involving multiple restrictive mechanisms (29). C) We determined transient virus production within 24-48 h post-transfection of proviral plasmid or 72 h post-virus infection. However, previous studies have determined the virus production 5 days (or even longer) after either transfection or infection (16, 19, 21).
Having established this phenomenon of YY1, we constructed various truncated or point mutants of YY1 and analyzed their effect on HIV-1 replication. YY1 was shown to possess two domains acting functionally different (Fig. 4). That is, regions spanning 155-295 and 296-414 residues of YY1 have negative and positive effect on HIV-1 replication, respectively. The 155-295 segment contains a REPO domain (201-225) which is known to recruit the PcG proteins onto DNA for transcriptional repression (4). However, we did not observe the transcription repression upon over-expression of YY1 155-295 segment, other than a little increase of 2 kb mRNA, a multiple spliced RNA of HIV-1 (Fig. 4F). However, although there is no detectable decrease of 9 kb mRNA expressing Gag protein in 155-295 segment over-expressing cells, a decrease in the Gag protein level in both cell and media was observed (Fig. 4D and 4E). Therefore, 155-295 segment might affect the post-transcription step(s) of HIV-1 production. However, 296-414 segment increased 9 kb mRNA level and thereby the virus production. U3RU5 reporter gene analysis depicts that such an elevation is resulted from the overexpression of 296-414 segment, as shown by enhanced Tat-dependent Fluc activity (Fig. 4G and 4H). Therefore, apparently, YY1 is unable to increase the virus production as high as 296-414, due to the inhibitory effect of 155-295 domain, which appears to be acting after the post-transcriptional step.
A previous study showed that 170-200 reside region of YY1 is acetylated by p300 and PCAF and 261-414 region of YY1 is acetylated by PCAF. They showed that acetylation sites are for binding of HDAC proteins (HDAC1, HDAC2 and HDAC3). When YY1 is acetylated, interaction with HDACs proteins is increased and HDACs deacetylate 170-200 region but not 261-414 region (15). To find out whether acetylation of 155-295 region would decrease the HIV-1 production, we predicted the acetylation site(s) using Acetylation prediction software (GPS-PAIL 2.0, http://pail.biocuckoo.org/online.php), which predicted highest probability of acetylation on Lys178 followed by Lys179, and four acetylation sites in 160-181 region. Therefore, we constructed 155-295K178A,K179A and 155-295D160-181. Further, 155-295K283A,K286A,K288A was constructed since 155-200 region does not have any effect on HIV production (Fig. 4). Co-transfection with the said constructs and pNL4-3GFP was performed and the virus production was measured. We observed that these mutants resulted in further decrease in the virus production than wild type (data not shown). These results suggested that acetylation does not interfere with repression activity of 155-295 region, although some other sites might be acetylated.
Tat protein is acetylated at Lys50 residue by p300 Histone acetyltransferase (30). A previous study showed that acetylated Tat interacts with PCAF and this interaction synergistically elevates the HIV promoter activity (31). Although we did not confirm the physical interaction between and YY1-PCAF, it is possible that the region 296-414 reinforces the Tat-PCAF interaction.
In summary, our data show clearly that a cellular factor, YY1, could have a positive effect on HIV-1 replication, at least in HIV-1 actively replication cell or early infection. Therefore, further studies are necessary to find out the precise requirement for switching the role of YY1 to a repressor of HIV-1 transcription for the viral latency, as reported previously.
MATERIALS AND METHODS
See supplementary information for Material and Methods.
Supplemental Materials
ACKNOWLEDGEMENTS
This work was supported by a research foundation of Korea (NRF) Grant funded by the Korean Govermment (2016R1D1A1A02937509 and 2017R1A5A1015366).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushmeyer S, Park K, Atchison ML. Characterization of functional domains within the multifunctional transcription factor, YY1. J Biol Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- 3.Austen M, Luscher B, Luscher-Firzlaff JM. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J Biol Chem. 1997;272:1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci U S A. 2006;103:19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 6.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 7.Park K, Atchison ML. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3' enhancer and the immunoglobulin heavy-chain mu E1 site. Proc Natl Acad Sci U S A. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/MCB.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S, Shi Y, Mulligan P, et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat Struct Mol Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 11.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/S0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–66. doi: 10.1016/S0304-419X(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 13.Hyde-DeRuyscher RP, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JS, Galvin KM, See RH, et al. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 15.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coull JJ, Romerio F, Sun JM, et al. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol. 2000;74:6790–6799. doi: 10.1128/JVI.74.15.6790-6799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satijn DP, Hamer KM, den Blaauwen J, Otte AP. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko CY, Hsu HC, Shen MR, Chang WC, Wang JM. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J Biol Chem. 2008;283:30919–30932. doi: 10.1074/jbc.M804029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margolis DM, Somasundaran M, Green MR. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol. 1994;68:905–910. doi: 10.1128/JVI.68.2.905-910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He G, Margolis DM. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol Cell Biol. 2002;22:2965–2973. doi: 10.1128/MCB.22.9.2965-2973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romerio F, Gabriel MN, Margolis DM. Repression of human immunodeficiency virus type 1 through the novel cooperation of human factors YY1 and LSF. J Virol. 1997;71:9375–9382. doi: 10.1128/JVI.71.12.9375-9382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernhard W, Barreto K, Raithatha S, Sadowski I. An upstream YY1 binding site on the HIV-1 LTR contributes to latent infection. PLoS One. 2013;8:e77052. doi: 10.1371/journal.pone.0077052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaynor R. Cellular transcription factors involved in the regulation of HIV-1 gene expression. AIDS. 1992;6:347–363. doi: 10.1097/00002030-199204000-00001. [DOI] [PubMed] [Google Scholar]
- 24.Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakobovits A, Smith DH, Jakobovits EB, Capon DJ. A discrete element 3' of human immunodeficiency virus 1 (HIV-1) and HIV-2 mRNA initiation sites mediates transcriptional activation by an HIV trans activator. Mol Cell Biol. 1988;8:2555–2561. doi: 10.1128/MCB.8.6.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boris-Lawrie KA, Brady JN, Kumar A. Sequences within the R region of the long terminal repeat activate basal transcription from the HIV-1 promoter. Gene Expr. 1992;2:215–230. [PMC free article] [PubMed] [Google Scholar]
- 27.Rizkallah R, Hurt MM. Regulation of the transcription factor YY1 in mitosis through phosphorylation of its DNA-binding domain. Mol Biol Cell. 2009;20:4766–4776. doi: 10.1091/mbc.e09-04-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ylisastigui L, Kaur R, Johnson H, et al. Mitogen-activated protein kinases regulate LSF occupancy at the human immunodeficiency virus type 1 promoter. J Virol. 2005;79:5952–5962. doi: 10.1128/JVI.79.10.5952-5962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barton K, Margolis D. Selective targeting of the repressive transcription factors YY1 and cMyc to disrupt quiescent human immunodeficiency viruses. AIDS Res Hum Retroviruses. 2013;29:289–298. doi: 10.1089/aid.2012.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ott M, Schnolzer M, Garnica J, et al. Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr Biol. 1999;9:1489–1492. doi: 10.1016/S0960-9822(00)80120-7. [DOI] [PubMed] [Google Scholar]
- 31.Dorr A, Kiermer V, Pedal A, et al. Transcriptional synergy between Tat and PCAF is dependent on the binding of acetylated Tat to the PCAF bromodomain. EMBO J. 2002;21:2715–2723. doi: 10.1093/emboj/21.11.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.