Abstract
As an intracellular degradation system, autophagy is an essential and defensive cellular program required for cell survival and cellular metabolic homeostasis in response to various stresses, such as nutrient deprivation and the accumulation of damaged organelles. In general, autophagy flux consists of four steps: (1) initiation (formation of phagophore), (2) maturation and completion of autophagosome, (3) fusion of autophagosomes with lysosomes (formation of autolysosome), and (4) degradation of intravesicular components within autolysosomes. The number of genes and reagents that modulate autophagy is increasing. Investigation of their effect on autophagy flux is critical to understanding the roles of autophagy in many physiological and pathological processes. In this review, we summarize and discuss ways to analyze autophagy flux quantitatively and qualitatively with the use of imaging tools. The suggested imaging method can help estimate whether each modulator is an inhibitor or a promoter of autophagy and elucidate the mode of action of specific genes and reagents on autophagy processes.
Keywords: Autolysosome, Autophagosome, Autophagy, Fluorescence Imaging, Quantitative analysis
INTRODUCTION
Autophagy is an intracellular orderly recycling system to degrade unnecessary cytosolic proteins and damaged organelles. It is a dynamic cellular self-eating process that exists in all eukaryotic cells. Autophagy is dependent on lysosomal degradation but is distinct from endocytosis-mediated degradation of plasma-membrane proteins and extracellular components, including bacterial or viral pathogens and apoptotic cells during phagocytosis. Three major types of autophagy are recognized: macroautophagy, microautophagy, and chaperone-mediated autophagy (1, 2). Macroautophagy is responsible for bulky degradation of proteins and organelles of the cytoplasm by sequestering them into a de novo-formed double-layered autophagosome. Microautophagy takes place at the lysosomal surface and engulfs cytoplasmic materials by invagination and septation of the lysosomal membrane (3, 4). Chaperone-mediated autophagy selectively degrades cytosolic proteins that are transported into the lumen of the lysosome (2). In this review, the term “autophagy” indicates macroautophagy.
Autophagy is important for a cell to defend against intracellular and extracellular stresses and maintain metabolic homeostasis. Autophagy has been implicated in many physiological events, including starvation survival, organelle turnover and cellular renewal, cell growth, immunity, animal development, and ageing (5-7). Recent evidence indicates that alterations in autophagy occur frequently in many human diseases, such as neurodegenerative disorders, cancer, and cardiomyopathies (8-10). Although the roles of key proteins in mammalian autophagosome formation have been studied intensively (5, 11) since the discovery of ATG (autophagy-related) genes in yeast (12), their disease-related functions have yet to be elucidated. Currently, more than 230 proteins are recognized by the human autophagy database (from Luxembourg Institute of Health, http://autophagy.lu/index.html) as autophagy-involved human proteins directly or indirectly. A list of 56 compounds that act as an inducer or an inhibitor of autophagy is available, but direct targets of most compounds in the autophagy machinery are still being investigated (13). Searching for new pharmacological agents targeting the autophagy pathway, in addition to known activators and inhibitors of autophagy and evaluation of how each reagent affects autophagy, is critical for appropriate therapeutic application of each compound in different human diseases.
Methods for monitoring mammalian autophagy have been summarized and provided in several review articles (11, 14, 15). Immunoblot analysis of microtubule-associated protein light chain 3 (LC3), one of the mammalian homologs of Atg8 in yeast, shows that the number of autophagosomes increases in proportion to the band intensity of LC3-II, a phosphatidylethanolamine (PE)-conjugated LC3, migrating faster than LC3-I in SDS-PAGE (16). Biochemical analysis of the ratio of LC3-I to LC3-II is sometimes interpreted inappropriately because of degradation of LC3-II itself by autophagy. Detection of p62 protein, also called sequestosome 1, by immunoblot analysis is another useful biochemical method to monitor autophagy activation caused by selective degradation of p62 in the lysosome by autophagy (17, 18). The amount of p62 protein is inversely proportional to the magnitude of autophagy. Biochemical analysis of the ratio of LC3-I to LC3-II or the amount of p62 is limited for quantification of autophagy flux, because measurement of protein-band intensity can be inaccurate. Detection of both LC3 and p62 protein cannot provide information about the step at which a compound inhibits or activates during autophagy. Electron microscopy (EM) is useful for detecting different stages of the autophagy pathway. Early autophagosomes, autolysosomes, and autophagic degradation products are observed and identified as different structures (11, 14), indicating that EM provides qualitative information about autophagy. It is challenging to obtain quantitative information for the number of autophagosomes and autolysosomes. Application of EM to the massive analysis of autophagy has limitations because of the difficulty of sample preparation and observation. Optical imaging using wide-field microscopy, confocal microscopy, and super-resolution microscopy with proper imaging reporters or sensors has provided autophagy researchers with quantitative analytical tools to monitor autophagosomes and autolysosomes.
In this review, we describe confocal imaging methods with various fluorescent reporters to monitor autophagy flux and discuss their strengths and weaknesses. A brief quantification of imaging data for each method is described. We also provide background and general experimental procedures in a mammalian cell selected herein. Depending on the cell type of interest, stress conditions, and treated compounds, experimental procedures and interpretation of data will differ.
OVERVIEW OF AUTOPHAGY
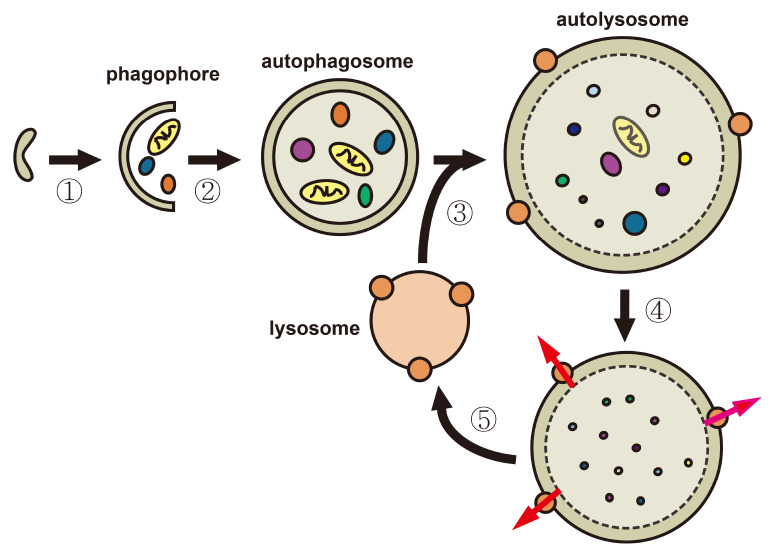
Autophagy consists of several dynamic steps but sequential measurement of each step is difficult (11, 14, 15). In mammalian cells, autophagy begins with a phagophore (an isolation membrane) (Fig. 1, step 1), which expands to engulf and sequester the cargo in a double-membraned autophagosome, which is loaded with protein aggregates and various organelles (Fig. 1, step 2). The autophagosome fuses with an endosome to form an amphisome (an acidic late autophagosome) (19). In the next step, the autophagosome fuses with a lysosome (Fig. 1, step 3) to form an autolysosome and promote degradation of the inner membrane of the autophagosome and the loaded materials by lysosomal hydrolases in the autolysosome (Fig. 1, step 4). The autolysosome is ready for the next cycle once degraded monomeric units are exported to the cytosol (Fig. 1, step 5) (20, 21). Some lysosomes are formed through the initiation of proto-lysosomal tubules and vesicles that originated from the autolysosomes and subsequent maturation with the supply of hydrolases (22).
Fig. 1.
Schematic overview of autophagy in mammalian cells. (1) Initiation of macroautophagy (herein, autophagy): An isolation membrane develops at the ER or other endomembrane and encloses cytosolic materials, including damaged organelles. (2) Maturation and completion of an autophagosome: The cytosolic compartments are sequestered in the double-membraned autophagosome. (3) Fusion of the autophagosome with the lysosomes: The outer membrane of the autophagosome fuses with the lysosomal membrane to form an autolysosome. (4) Degradation of intravesicular components within autolysosomes: Internal materials are degraded by various acidic hydrolases, and the degraded monomeric units, such as amino acids and fatty acids, are exported to the cytosol through channels (brown circles) for reuse. (5) Formation of a recycled lysosome: An autophagic lysosome is reformed from an autolysosome.
Initiation of autophagy
Formation of phagophores requires class III phosphoinositide 3-kinase Vps34 activation by means of a macromolecular complex, including Vps34, Beclin-1 (mammalian Atg6), Atg14, and Vps15 (23-25). Vps34 generates phosphatidylinositol(3)phosphate (PtdIns(3)P; PI(3)P), which is critical for recruitment of effectors such as double FYVE-containing protein 1 (DFCP1) and WD-repeat domain phosphoinositide-interacting (WIPI) family proteins (5, 26), thereby promoting development of the isolation membrane or autophagosome (27). Imaging of fluorescent protein with PI3P-binding domain (two FYVEs of DFCP1, WIPI, or others) is useful for monitoring phagophore formation (26, 28, 29).
Maturation and completion of autophagosome
Autophagosome formation results from the sequestration of cytoplasmic compartments by the phagophore. At the late stage of phagophore formation, conjugation of the Atg8 family proteins―microtubule-associated protein 1 LC3, GABARAP, and GATE16―to the PE are produced by Atg7 and Atg3, and their conjugated Atg proteins are integrated into the membrane of autophagosomes (12, 30). LC3 without PE and with PE are described as LC3-I and LC3-II, respectively. LC3-II is known to function as a receptor for a selective substrate, p62/sequestosome 1, on the autophagosomal inner membrane (31). Imaging of fluorescent LC3-II is used to monitor autophagosomes or autolysosomes.
Fusion of autophagosomes with lysosomes
Autophagosomes containing cytosolic materials fuse with lysosomes that are responsible for massive cellular degradation to form autolysosomes (21). Autolysosome formation requires active microtubule organization during the fusion process (32). This process can be monitored by the analysis of colocalization of fluorescent LC3-II with lysosome markers, such as lysosomal-associated membrane protein 1 (LAMP1) and LAMP2 proteins. Inhibition of the fusion decreases the colocalization level of LC3 and LAMP1/2 proteins.
Degradation of intravesicular components within autolysosomes
Lysosomal hydrolases, which are active at an acidic pH (about 4.7) (33), degrade the cytoplasm-derived compartments, including the inner membrane of autophagosomes. A defect in lysosomal enzymes or inhibition of acidification of the luminal space of the lysosome by a V-ATPase inhibitor induces the accumulation of autolysosomes with undegraded cytosolic materials derived from autophagosomes (34, 35). Imaging analysis of autolysosome size is useful for monitoring the accumulation of autolysosomes, which lose degradation capacity.
FLUORESCENCE IMAGING ANALYSIS WITH MCHERRY-GREEN FLUORESCENT PROTEIN (GFP)-LC3 REPORTER
Background
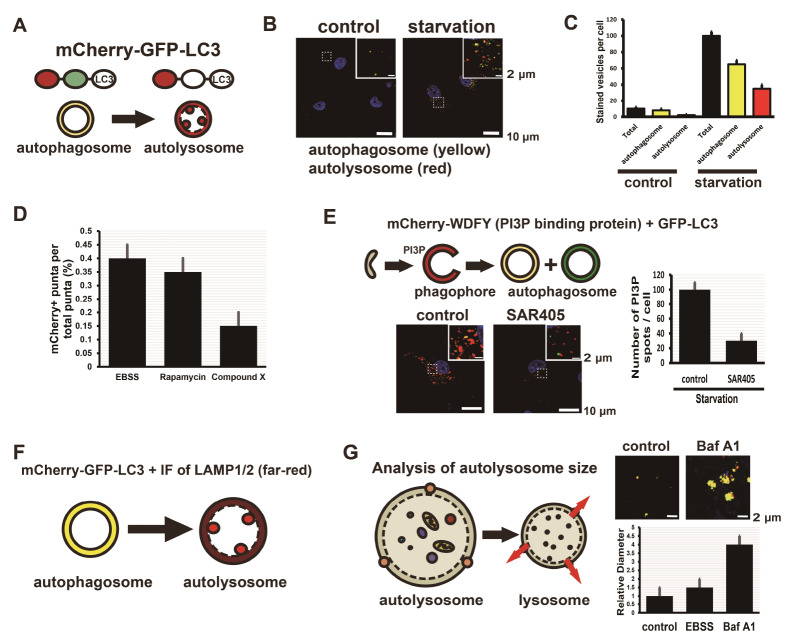
GFP-LC3 is used commonly as an imaging marker for autophagosomes (31) but not for autolysosomes, because GFP possesses a pKa of 6.0 and is acid-labile (36). The mCherry protein is very acid-stable because it has a pKa of <4.5 (37). Imaging of the mCherry-GFP-LC3 protein shows the fluorescence difference between autophagosomes and autolysosomes in a cell (18). The mCherry-GFP-LC3 proteins seen in neutral vesicles display both green and red fluorescence, but in acidic vesicles display red fluorescence only (Fig. 2A). In HepG2 cells expressing mCherry-GFP-LC3, nutrient starvation elicits an increase in the number of autophagosomes and autolysosomes (Fig. 2B). Quantification of the number of spots (yellow or red) shows the total number of autophagosomes and autolysosomes and their ratio (Fig. 2C). Analysis of the spot number ratio of autolysosomes (mCherry+GFP-) to autophagosomes (mCherry+GFP+) is useful for estimation of the transition from autophagosome to autolysosome. Treatment with compound X is suggested to inhibit the fusion of autophagosomes with lysosomes or to increase pH in the lumen of autolysosomes (Fig. 2D). Treatment with rapamycin, which induces autophagy by inhibiting the mechanistic target of rapamycin complex 1 (mTorc1) (38, 39), does not inhibit the fusion of autophagosomes with lysosomes.
Fig. 2.
Confocal imaging of mCherry-GFP-LC3 vesicles, mCherry-WDFY vesicles, and immunofluorescence staining for LAMP1 or LAMP2 vesicles shows a quantification of autophagosomes, autolysosome, and phagophores. (A) mCherry-GFP-LC3 protein is useful for distinguishing between autophagosomes and autolysosomes because of the difference of pH in the lumens. mCherry and GFP proteins are stable in the neutral pH of the lumen of autophagosomes, whereas GFP protein is acid-labile and mCherry protein is acid-stable in the autolysosome. (B) Nutrient-deprived HepG2 cells show more yellow-stained autophagosomes and red-stained autolysosomes in confocal microscopy. (C) The total number of autophagic vesicles increases in cells without nutrients (starvation). (D) The ratio of the number of autolysosome spots (mCherry+GFP-) to total spots (autophagosomes, mCherry+GFP+ and autolysosomes, mCherry+GFP-) is proportional to the fusion of autophagosomes with lysosomes. Treatment with Rapamycin, an inducer of autophagy, shows values similar to starvation (EBSS, Earle’s balanced salt solution) and treatment with compound X inhibits the fusion of autophagosomes with lysosomes. Quantitative data are presented as means ± SD from three independent experiments. (E) HepG2 cells expressing mCherry-WDFY (PI3P-binding protein) and GFP-LC3 indicate phagophores (red, mCherry+GFP-), early autophagosomes (yellow, mCherry+GFP+), and late autophagosomes (green, mCherry-GFP+). Treatment with SAR405, an inhibitor of Vps34, decreases the number of phagophores. Quantitative data are presented as means ± SD from three independent experiments. (F) Immunofluorescence staining of HepG2 cells expressing mCherry-GFP-LC3 with antibodies to LAMP1 or LAMP2 lysosomal membrane protein indicates autophagosomes (yellow, mCherry+GFP+) and autolysosomes (red spots in a far-red vesicle, mCherry+GFP- spots in a vesicle containing LAMP1/2). (G) HepG2 cells expressing mCherry-GFP-LC3 were observed using confocal microscopy and the size of autolysosome vesicles was calculated by measuring the diameter. Treatment with Baf A1 inhibits degradation of cytosolic compartments derived from autophagosomes in lysosomes and increases in the number of enlarged autolysosomes. Quantitative data are presented as means ± SD from three independent experiments.
Procedure
HepG2 cells were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco) and penicillin-streptomycin (Hyclone) at 37°C in a humidified atmosphere of 5% CO2. Cells were cultured on a 12-well plate containing cover slips (diameter, 18 mm,) coated with poly-L-lysine for both live-cell imaging and fixed-cell staining. Cells were infected with mCherry-GFP-LC3 adenovirus overnight (16-20 hours; infection with the virus for more than 20 h might be toxic to cells). Cells expressing mCherry-GFP-LC3 were incubated under selected conditions for 8 h (control, standard media; starvation, Earle's balanced salt solution [EBSS]). Cells were treated with 0.25 μM rapamycin for 8 h for autophagy induction. Cells were fixed for 10 min with 4% formaldehyde in phosphate-buffered saline (PBS), exposed for 30 min at room temperature to PBS containing 5% horse serum (Gibco-BRL) and 0.1% Triton X-100. The cells were also stained with 4’6’-diamidino-2-phenylindole (0.2 μg/ml) to detect DNA. We did confocal microscopy as described previously (40) with minor modifications and acquired the images using a 60× Plan Apochromat VC objective, NA 1.40, by illuminating with a 488-nm multi-Ar laser (for excitation of GFP fluorochrome) or with a 561-nm diode-pumped solid-state laser (for excitation of mCherry fluorochrome) with a microscope (A1R, Nikon or LSM880 Airy, Zeiss) equipped with a galvano detector. Images were processed with NIS-Elements AR 3.0 software (from Nikon) or Zen software (from Zeiss). For live-cell imaging, cells were maintained in an incubation chamber (Chamlide TC; Live Cell Instrument) at 37°C under an atmosphere of 5% CO2 in air.
Quantification of autophagy flux
For spot count, we analyzed each z-stack image to avoid miscounting overlapping autophagosome and autolysosome spots. For measurements with NIS or Zen software, we deleted background signals for the same value in all images to measure only the exact spots. Spots remaining on the deleted background can be measured. As a result, spots that have values greater than a certain fluorescence intensity were quantified. Spots with both mCherry (red) and GFP (green) were counted as autophagosomes and spots with only mCherry (red) were counted as autolysosomes.
Note
The mCherry-positive and GFP-positive spots indicate not only autophagosomes but also autolysosomes that have a neutral pH because of a malfunction of vacuolar-type H+-ATPase (V-ATPase). To distinguish between the two, further characterization experiments, such as a colocalization assay of mCherry-GFP-LC3 proteins with lysosome marker proteins, are required.
FLUORESCENCE IMAGING ANALYSIS WITH PTDINS(3)P REPORTER
Background
PtdIns(3)P is associated with endosomal functions and is mainly localized in the early endosomes (41). Accumulation of PtdIns(3)P by Vps34, a Class III PI(3)K, is critical for the initiation of autophagy (23, 24). PtdIns(3)P-binding proteins, including Atg18 (WD-repeat domain phosphoinositide-interacting 2, WIPI2) and DFCP1, are recruited to autophagic membranes and regulate LC3 lipidation, thereby promoting phagophore formation (27, 29). WD repeat and FYVE domain-containing 3 (WDFY3), known as autophagy-linked FYVE, is associated with autophagic membranes as a PtdIns(3)P-binding protein (42). Imaging of mCherry-WDFY (a PtdIns(3)P sensor) and GFP-LC3 is useful for monitoring early stages of autophagy (Fig. 2E). Phagophore-containing PtdIns(3)P is shown in red, early autophagosomes with both PtdIns(3)P and GFP-LC3 are shown in yellow, and late autophagosomes with only GFP-LC3 are shown in green. Selective Vps34 inhibitors such as SAR405 (43) inhibit early autophagy by restraining phagophore formation (Fig. 2E).
Procedure
To image PtdIns(3)P during phagophore formation, we used the expression plasmid containing cDNA that encodes a PtdIns(3)P-binding protein (or domain), such as DFCP1 (26), UVRAG (44), or WDFY (42). HepG2 cells were cultured on a 12-well plate containing cover slips (diameter, 18 mm) coated with poly-L-lysine and transfected with GFP-LC3 and mCherry-WDFY (PI3P reporter). Cells were treated with 500 nM SAR405 (VPS34 inhibitor) under starvation (EBSS) for 8 h. Co-expression of mCherry-WDFY and GFP-LC3 can distinguish between early autophagosomes (WDFY+LC3+) and mature autophagosomes (WDFY-LC3+).
Quantification of autophagy flux
We did quantification as described for imaging analysis with mCherry-GFP-LC3 reporter. We calculated the number of PtdIns(3)P spots per cell from WDFY positive (red) spots per cell. From analysis in cells expressing both mCherry-WDFY and GFP-LC3, early autophagosomes were quantified from WDFY+LC3+ spots (yellow) and late autophagosomes from WDFY-LC3+ spots (green).
Note
Expression of fluorescent PtdIns(3)P-binding proteins, such as DFCP1, UVRAG, and WDFY, shows somewhat basal structures, like ER, that are not involved in autophagy depending on their initial locations. During induced autophagy, newly formed PtdIns(3)P spots should be counted as phagophores or early autophagosomes. In normal conditions (for example, nutrient-rich media), PtdIns(3) spots seem to be localized on the endosomes, the ER, and the Golgi.
FLUORESCENCE IMAGING ANALYSIS WITH MCHERRY-GFP-LC3 AND LYSOSOMAL MARKER PROTEIN
Background
It is difficult to monitor the process of fusion of autophagosomes with lysosomes by using fluorescence imaging. Mature autophagosomes require a specific SNARE Syntaxin 17 on their outer membranes to fuse with lysosomes (45). To distinguish autophagosomes before and after their fusion with lysosomes, we quantify the fusion process by measuring what portion of LC3 positive spots are colocalized with lysosomal markers. Immunofluorescence of LAMP1 or LAMP2 proteins in cells expressing mCherry-GFP-LC3 shows that autophagosomes appear as a yellow circular vesicle (mCherry+GFP+LC3) and autolysosomes appear as red spots (mCherry+GFP–LC3) inside a circular far-red vesicle (LAMP1/2+) (Fig. 2F).
Procedure
HepG2 cells were cultured on a 12-well plate containing cover slips (diameter, 18 mm,) coated with poly-L-lysine for both live-cell imaging and fixed-cell staining. Cells were infected with mCherry-GFP-LC3 adenovirus overnight. For immunofluorescence staining of cells for LAMP1 and LAMP2, cells were fixed with 100% cold methanol and incubated with 5% horse serum (Gibco-BRL), 0.1% Triton X-100 in PBS for 30 min and then incubated for 30 min at room temperature with primary antibodies to LAMP1 (#9091, Cell Signaling) or LAMP2 in the same solution. After washing with PBS, cells were incubated for 30 min at room temperature with Alexa Fluor 633– conjugated goat secondary antibodies (Invitrogen) at a 1:1,000 dilution in PBS containing 5% horse serum and 0.1% Triton X-100. Cells were also stained with 0.2 μg/ml 4’6’-diamidino-2-phenylindole to detect DNA. We acquired confocal images by illuminating with a 488-nm, a 561-nm, or a 640-nm laser with a microscope (A1R, Nikon) equipped with a galvano detector, and processed images with NIS-Elements AR 3.0 software.
Quantification of autophagy flux
We did quantification as described for imaging analysis with mCherry-GFP-LC3 reporter, with some modifications. We used a colocalization module to measure the number of autolysosomes by quantifying overlapping spots from both mCherry+ GFP-LC3 and LAMP1/2+. The number of autophagosomes was measured by counting mCherry+GFP-LC3 spots.
Note
Treatment with an inhibitor of lysosome function, such as Bafilomycin A (a V-ATPase inhibitor), induces accumulation of enlarged autolysosomes. Red spots (mCherry+GFP-LC3) inside a circular far-red vesicle (LAMP1/2+) are counted as autolysosomes.
ANALYSIS OF AUTOPHAGY FLUX WITH AUTOLYSOSOME SIZE
Background
At the final stage of autophagy, cytosolic materials in the autolysosome are degraded by lysosomal hydrolases. A defect in lysosome function increases the number of autolysosomes that are larger than normal (34, 46). The difference between inhibition of autophagosome–lysosome fusion and a defect in the autolysosomal degradation can be perceived, because the fusion of autophagosome with lysosome is independent of V-ATPase-mediated acidification in the lysosome (35, 46). Careful analysis of enlarged autolysosomes shows malfunction of the degradation process in the autolysosome (Fig. 2G). In cells expressing mCherry-GFP-LC3, the size of mCherry+GFP+ spots (autolysosomes) increases during treatment with Baf A1 (V-ATPase inhibitor).
Procedure
HepG2 cells were infected with mCherry-GFP-LC3 adenovirus overnight. Infected cells were incubated with 100 nM Baf A1 for 8 h, fixed, and observed using confocal microscopy as described for imaging analysis with mCherry-GFP-LC3 reporter.
Quantification of autophagy flux
In measuring autolysosome size, we measured mCherry+GFP- spots in control cells (without treatment). Because Baf A1 blocks degradation of mCherry-GFP-LC3 in autolysosomes by inhibiting lysosomal acidification, Baf A1-treated cells have mCherry+GFP+ spots, indicating both autophagosomes and autolysosomes. We analyzed the size of the spots by measuring the diameter or circumference of each spot with NIS-Elements AR 3.0 software.
Note
Quantification of the size of autophagosomes or autolysosomes has not been the focus of autophagy researchers looking at autophagy flux analysis. This size analysis is useful to estimate degradation and lysosome reuse processes from autolysosomes.
ASSAY OF AUTOPHAGY REGULATORS WITH FLUORESCENCE IMAGING
Confocal imaging using fluorescent reporters together with immunostaining has allowed the identification of autophagy regulators. Here, we summarize current fluorescence imaging methods to analyze autophagy flux. Careful quantification of imaging data is required to reveal whether a candidate compound is an inhibitor or a promoter of autophagy flux (Table 1). The number of PtdIns(3)P spots indicates the initial stages of autophagy. The total number of autophagosomes and autolysosomes and their ratio imply several processes during the transition of early autophagosomes to autolysosomes. Colocalization of autophagosomes with lysosomes shows fusion of autophagosomes and autolysosomes. Analysis of the size of autolysosomes indicates degradation processes in autolysosomes. We review modified steps of autophagy with the use of five quantitative imaging analyses (Table 1). When treatment with an unknown compound or malfunction of a gene shows a phenotype that is categorized into a specific family, its effect on autophagy flux will be proposed.
Table 1.
Expectation of inhibition or promotion of autophagy flux with imaging analysis
| Modified step of autophagy processes | Number of PI3P spots | Total number of autophagosomes & autolysosomes | Ratio of autolysosomes to autophagosomes | Size of autolysosomes | Colocalization of autophagosomes with lysosomes |
|---|---|---|---|---|---|
| Inhibition of formation of phagophores | + | + | ND | ND | ND |
| Inhibition of maturation of autophagosomes | ++ | + | ND | ND | ND |
| Inhibition of formation of autolysosomes | ++ | +++ | + | ++ | + |
| Inhibition of degradation process in lysosomes | ++ | +++ | +++ | +++ | +++ |
| Promotion of autophagy | +++ | +++ | ++ | ++ | ++ |
Five different imaging methods are used to evaluate effects of specific compounds on autophagy flux. +: low, ++: average, +++: high, ND: non-determined.
CONCLUSION
Methods to analyze autophagy in mammalian cells using imaging have improved substantially with regard to development of imaging reporters and instrumentation. A variety of tools have been introduced and summarized in many review articles (11, 14, 15, 47). In this review, we propose that comprehensive understanding of confocal imaging data of PtdIns(3)P, mCherry-GFP-LC3, lysosome markers, and autolysosome size sheds light on new regulators of autophagy and modes of action of a candidate compound. Especially, accurate quantification of autophagic vesicles using fluorescence imaging requires the reliable, uniform expression of reporters among cells. Development of stable cells expressing reporters and production of viruses to carry reporters are suggested solutions. We anticipate improvements to imaging instrumentation (structured illumination microscopy, SIM; stochastic optical reconstruction microscopy, STORM; stimulated emission depletion microscopy, STED microscopy; confocal microscopy with Airyscan) with super-resolution below the optical diffraction limit of ∼250 nm and imaging analysis programs to process and quantify large datasets. We hope these improvements motivate autophagy researchers to screen and characterize pharmacologic agents targeting autophagy to treat human diseases such as cancer and neurodegenerative disorders.
Autophagy flux is a dynamic process consisting of several intermediate membraned structures, such as phagophores, autophagosomes, and autolysosomes. Accumulation of specific intermediates results from not only inhibition but also activation of autophagy (11, 48). Careful investigations to distinguish specific intermediate vesicles among them, as briefly summarized in this review, are required for identifying modulators of autophagy flux and elucidating modes of action of specific modulators.
ACKNOWLEDGEMENTS
This study was supported by grants from the National Research Foundation of Korea (2019R1A6C1010020 and 2018R1D1A1 A02049371) and from the Ministry of Food and Drug Safety in 2016 (16173MFDS009) in Korea.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 3.Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melendez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 10.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 13.Vakifahmetoglu-Norberg H, Xia HG, Yuan J. Pharmacologic agents targeting autophagy. J Clin Invest. 2015;125:5–13. doi: 10.1172/JCI73937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshii SR, Mizushima N. Monitoring and Measuring Autophagy. Int J Mol Sci. 2017;18:1–13. doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 17.Bjorkoy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 19.Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun. 1988;151:40–47. doi: 10.1016/0006-291X(88)90556-6. [DOI] [PubMed] [Google Scholar]
- 20.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 22.Yu L, McPhee CK, Zheng L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 25.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 26.Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polson HE, de Lartigue J, Rigden DJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 28.Noda T, Matsunaga K, Taguchi-Atarashi N, Yoshimori T. Regulation of membrane biogenesis in autophagy via PI3P dynamics. Semin Cell Dev Biol. 2010;21:671–676. doi: 10.1016/j.semcdb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb JL, Ravikumar B, Rubinsztein DC. Microtubule disruption inhibits autophagosome-lysosome fusion: implications for studying the roles of aggresomes in polyglutamine diseases. Int J Biochem Cell Biol. 2004;36:2541–2550. doi: 10.1016/j.biocel.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Kielian MC, Cohn ZA. Intralysosomal accumulation of polyanions. II. Polyanion internalization and its influence on lysosomal pH and membrane fluidity. J Cell Biol. 1982;93:875–882. doi: 10.1083/jcb.93.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chevrier M, Brakch N, Celine L, et al. Autophagosome maturation is impaired in Fabry disease. Autophagy. 2010;6:589–599. doi: 10.4161/auto.6.5.11943. [DOI] [PubMed] [Google Scholar]
- 35.Mauvezin C, Neufeld TP. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. 2015;11:1437–1438. doi: 10.1080/15548627.2015.1066957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 37.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 38.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 39.Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 40.Lim JM, Lee KS, Woo HA, Kang D, Rhee SG. Control of the pericentrosomal H2O2 level by peroxiredoxin I is critical for mitotic progression. J Cell Biol. 2015;210:23–33. doi: 10.1083/jcb.201412068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicot AS, Laporte J. Endosomal phosphoinositides and human diseases. Traffic. 2008;9:1240–1249. doi: 10.1111/j.1600-0854.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simonsen A, Birkeland HC, Gillooly DJ, et al. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 43.Ronan B, Flamand O, Vescovi L, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 44.He S, Ni D, Ma B, et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol. 2013;15:1206–1219. doi: 10.1038/ncb2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Ueno K, Saito M, Nagashima M, et al. V-ATPase-dependent luminal acidification is required for endocytic recycling of a yeast cell wall stress sensor, Wsc1p. Biochem Biophys Res Commun. 2014;443:549–555. doi: 10.1016/j.bbrc.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Li Y, Wei F, Duan Y. Optical Imaging Paves the Way for Autophagy Research. Trends Biotechnol. 2017;35:1181–1193. doi: 10.1016/j.tibtech.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]