Abstract
Increasing evidence suggests the role of miR-449a in the regulation of tumorigenesis and autophagy. Autophagy plays an important role in the malignancy of T-cell lymphoma. However, it is still unknown whether miR-449a is associated with autophagy to regulate the malignancy of T-cell lymp homa. In this study, we for the first time demonstrated that miR-449a enhanced apoptosis of T-cell lymphoma cells by decreasing the degree of autophagy. Further, miR-449a downregulated autophagy-associated 4B (ATG4B) expression, which subsequently reduced the autophagy of T-cell lymphoma cells. Mechanistically, miR-449a decreased ATG4B protein level by binding to its mRNA 3’UTR, thus reducing the mRNA stability. In addition, studies with nude mice showed that miR-449a significantly inhibited lymphoma characteristics in vivo. In conclusion, our results demonstrated that the “miR-449a/ATG4B/autophagy” pathway played a vital role in the malignancy of T-cell lymphoma, suggesting a novel therapeutic target.
Keywords: ATG4B, Autophagy, MiR-449a, Post-transcriptional regulation, T-cell lymphoma
INTRODUCTION
T-cell lymphoma is one of the most prevalent malignancies worldwide, with a high degree of heterogeneity (1). It represents a type of non-Hodgkin’s lymphoma originating from T cells, and is associated with poor prognosis (1). Several new drugs have been developed, such as histone deacetylase inhibitors, immunoconjugates, CD52 monoclonal antibody, and folic acid antagonists (2, 3). However, the therapeutic outcome and prognosis of patients diagnosed with T-cell lymphoma is still not favorable (4). In the absence of effective therapeutic measures, novel treatment strategies for T-cell lymphoma are imperative.
Autophagy is an evolutionarily conserved mechanism in eukaryotes regulating the turnover of intracellular substances (5). In recent years, the role of autophagy in tumorigenesis has received increasing attention (6). Emerging studies have reported that autophagy plays an important role in the malignancy of lymphoma (7, 8). In T-cell lymphoma, it has been reported that hypoxia-induced autophagy decreases the sensitivity of HuT78 cells to doxorubicin (9). Therefore, we propose that targeting autophagy in T-cell lymphoma may attenuate the malignant progression and enhance the therapeutic efficiency.
MicroRNAs are small RNAs with multiple biological functions discovered in recent years, with a significant role in tumor development (10, 11). Specifically, miR-449a exhibits anti-cancer properties in a variety of tumors (12-14). For example, miR-449a acts as a tumor suppressor by reducing cell proliferation, migration and invasion as well as inducing apoptosis in human glioblastoma cell lines (12). In hepatocellular carcinoma (HCC), miR-449a directly targeted SOX4 and decreased its expression in epithelial-mesenchymal transition (EMT) and HCC metastasis, thus inhibiting TGF-beta-mediated cell migration (13). Recent evidence suggests that miR-449a regulates autophagy level (15, 16). For instance, it has reported that miR-449a induced the knockdown of CISD2, resulting in inhibition of the proliferation of glioma cells by activating beclin1-mediated autophagy (15). These studies revealed that miR-449a plays an important role in tumorigenesis, and is closely related to autophagy. However, whether miR-449a is associated with autophagy in regulating the malignancy of T-cell lymphoma, is still unknown.
RESULTS
MiR-449a enhances the apoptosis of cells in T-cell lymphoma
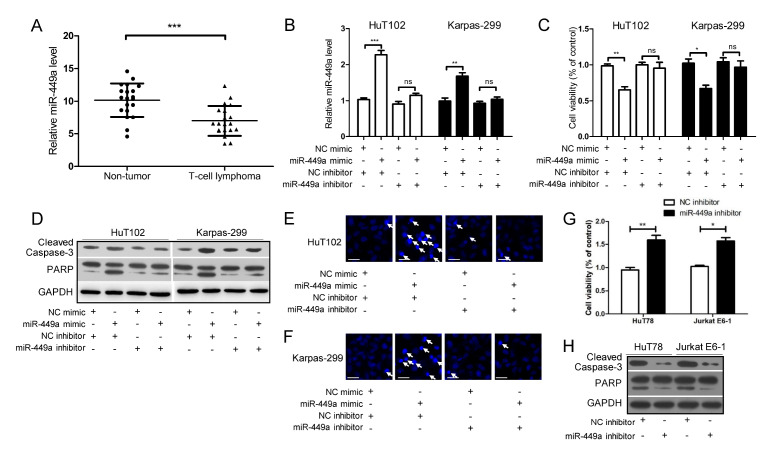
First, the miR-449a level in T-cell lymphoma tissues was lower than in non-cancerous lymph node tissues (Fig. 1A). Next, as shown in Supplementary Fig. 1, the miR-449a level was relatively high in H19, HuT78 and Jurkat E6-1 cell lines, and relatively low in HuT102 and Karpas-299 cell lines. Therefore, the cell lines HuT102 and Karpas-299 with a relatively low expression of miR-449a were selected to perform the following overexpression experiments. Subsequently, the miR-449a mimic and inhibitor were used in this study. Fig. 1B showed that treatment with miR-449a mimic elevated the miR-449a level, while simultaneous transfection miR-449a inhibitor abrogated the effect of miR-449a mimic. Further, as shown in Fig. 1C-F the miR-449a mimic decreased the cell viability (Fig. 1C), increased the levels of cleaved Caspase-3 and PARP (Fig. 1D) and promoted the release of apoptotic bodies (Fig. 1E and F), which were abolished by simultaneous treatment with miR-449a inhibitor (Fig. 1C-F). Meanwhile, the cell lines HuT78 and Jurkat E6-1 expressing high levels of miR-449a were used for the following silencing experiments. As shown in Fig. 1G and 1H, exposure to miR-449a inhibitor increased the cell viability (Fig. 1G), and decreased the levels of cleaved Caspase-3 and PARP (Fig. 1H) in HuT78 and Jurkat E6-1 cells. These data indicate that overexpression of miR-449a strengthens the apoptosis of cells in T-cell lymphoma, and knockdown of miR-449a attenuated the cellular apoptosis.
Fig. 1.
MiR-449a enhances cellular apoptosis in T-cell lymphoma. (A) Analysis of miR-449a level in 20 T-cell lymphoma tissues and the corresponding adjacent non-cancerous lymph node tissues. (B-F) HuT102 and Karpas-299 cells were transfected with miR-449a mimic (or NC mimic) and treated with miR-449a inhibitor (or NC inhibitor) for 24 h. The levels of miR-449a (B), the degree of cell viability (C), the levels of cleaved Caspase-3/PARP (D), and the number of apoptotic cells (marked with white arrows) were detected (scale bar, 20 μm) (E, F). (G, H) HuT78 and Jurkat E6-1 cells were treated with miR-449a inhibitor (or NC inhibitor) for 24 h. Cell viability (G) and the level of cleaved Caspase-3/PARP (H) were evaluated. *P < 0.05, **P < 0.01, ***P < 0.001, ns: no significance.
MiR-449a strengthens the apoptosis by attenuating the autophagy of T-cell lymphoma cells
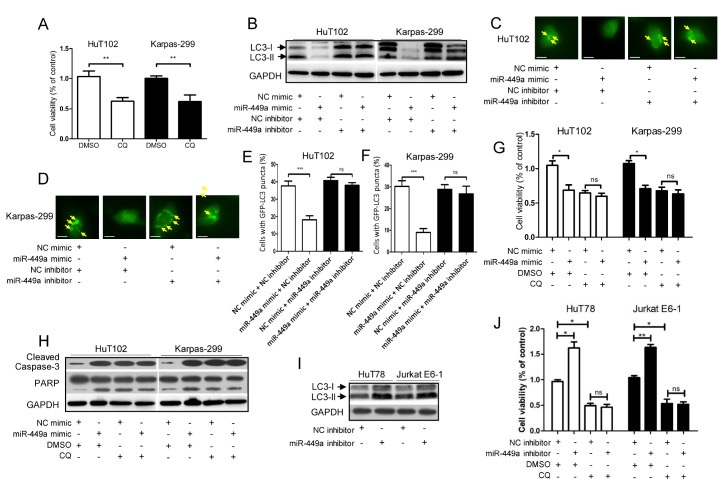
As shown in Fig. 2A, treatment with autophagy inhibitor chloroquine (CQ) remarkably weakened the viability of HuT102 and Karpas-299 cells, indicating that autophagy contributed to the malignancy of T-cell lymphoma cells. In addition, as shown in Fig. 2B-F, miR-449a mimic decreased the LC3-II protein level (Fig. 2B) and the formation of GFP-LC3 puncta (Fig. 2C-F) in HuT102 and Karpas-299 cells, which was abolished by simultaneous treatment with an miR-449a inhibitor (Fig. 2B-F). Furthermore, as shown in Fig. 2G and H treatment with miR-449a mimic or chloroquine alone in HuT102 and Karpas-299 cells dramatically reduced the cell viability (Fig. 2G) and enhanced the levels of cleaved Caspase-3 and PARP (Fig. 2H). However, the simultaneous application of miR-449a mimic and chloroquine did not further decrease the cell viability (Fig. 2G) or increase the levels of cleaved Caspase-3 and PARP (Fig. 2H). We also performed the silencing experiments involving HuT78 and Jurkat E6-1 cells. As shown in Fig. 2I, miR-449a inhibitor increased the protein expression of LC3-II. Furthermore, application of CQ did not enhance the effect of miR-449a inhibitor in increasing the cell viability of HuT78 and Jurkat E6-1 cells (Fig. 2J). These results indicated that the expression of mir-449a was negatively correlated with autophagy level, and the effect of miR-449a mimic on apoptosis increase was mediated via attenuation of autophagy in T-cell lymphoma cells.
Fig. 2.
MiR-449a strengthens apoptosis by attenuating the autophagy of T-cell lymphoma cells. (A) HuT102 and Karpas-299 cells were treated with autophagy inhibitor CQ (20 μM) or DMSO (0.1%) for 24 h. The cell viability was measured. (B-F) HuT102 and Karpas-299 cells were transfected with miR-449a mimic (or NC mimic) and miR-449a inhibitor (or NC inhibitor) in the absence (B) or presence (C-F) of GFP-LC3 expression vector for 24 h. The LC3 level (B), and the GFP-LC3 puncta (marked with yellow arrows) were determined (Scale bar, 5 μm) (C, D). Data (C, D) were quantified and expressed as the percentage of cells containing five or more GFP-LC3 puncta (E, F). (G, H) After transfection with miR-449a mimic or NC mimic for 24 h, the HuT102 and Karpas-299 cells were treated with CQ (20 μM) or DMSO (0.1%). The cell viability (G), and the levels of cleaved Caspase-3/PARP (H) were evaluated. (I) HuT78 and Jurkat E6-1 cells were treated with miR-449a inhibitor (or NC inhibitor) for 24 h, and the LC3 level was determined. (J) After exposure to miR-449a inhibitor or NC inhibitor for 24 h, the HuT78 and Jurkat E6-1 cells were treated with CQ (20 μM) or DMSO (0.1%). The cell viability was determined. CQ: chloroquine. *P < 0.05, **P < 0.01, ***P < 0.001, ns: no significance.
MiR-449a binds to ATG4B mRNA 3’UTR, thus decreasing ATG4B expression and attenuating the autophagy of T-cell lymphoma cells
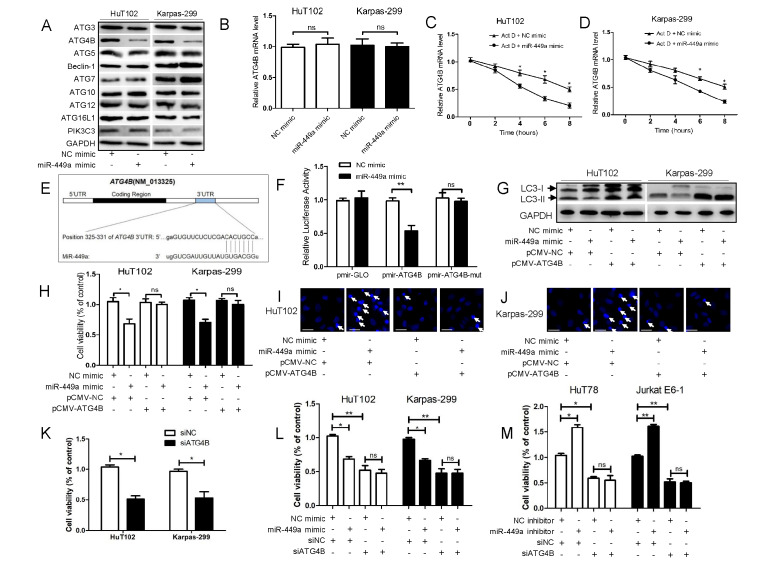
The protein expression of several autophagy related genes (ATGs) was detected. As shown in Fig. 3A, the miR-449a mimic decreased the expression of ATG4B and ATG5 in HuT102 and Karpas-299 cells. The ATG4B expression was most prominently altered and was further investigated. As shown in Supplementary Fig. 2, treatment with miR-449a inhibitor increased the protein level of ATG4B in HuT78 and Jurkat E6-1 cells. As shown in Supplementary Fig. 3, ATG4B expression was relatively lower in H19, HuT78 and Jurkat E6-1 cells, and relatively higher in HuT102 and Karpas-299 cells, suggesting that the expression of miR-449a and ATG4B was negatively correlated. Next, we found that the miR-449a mimic had no effect on the ATG4B mRNA level (Fig. 3B). The actinomycin D (Act D, a transcription inhibitor) assays were performed. As shown in Fig. 3C and D, miR-449a enhanced the degradation of ATG4B mRNA in the presence of Act D, indicating that miR-449a attenuated the ATG4B mRNA stability. Furthermore, bioinformatic analysis predicted that the binding site of miR-449a in the 3’UTR of ATG4B mRNA is located at residues 325-331 (CACUGCC) (Fig. 3E). The results of luciferase reporter assays showed that miR-449a significantly reduced the activity of pmir-ATG4B rather than pmir-ATG4B-mut in HuT102 cells (Fig. 3F).
Fig. 3.
MiR-449a binds to ATG4B mRNA 3’UTR, thereby decreasing ATG4B expression and attenuating the autophagy level of T-cell lymphoma cells. (A, B) HuT102 and Karpas-299 cells were transfected with miR-449a mimic or NC mimic for 24 h. The protein levels of nine ATGs (A) and the ATG4B mRNA level (B) were assayed. (C, D) HuT102 (C) and Karpas-299 (D) cells were transfected with miR-449a mimic or NC mimic for 24 h, followed by treatment with Act D (5 μg/mL). The mRNA level of ATG4B was detected. (E) Schematic representation of a predicted binding site of miR-449a in the 3’UTR of human ATG4B mRNA. (F) HuT102 cells were co-transfected with the reporter plasmids (pmir-ATG4B, pmir-ATG4B-mut or empty pmir-GLO) and RNA oligonucleotides (miR-449a mimic or NC mimic) for 24 h. The luciferase activity was determined. (G-J) HuT102 and Karpas-299 cells were transfected with miR-449a mimic (or NC mimic) and pCMV-ATG4B (or pCMV-NC) for 24 h. Subsequently, the levels of LC3 (G), the degree of cell viability (H), and the number of apoptotic cells (marked with white arrows) (I, J) were determined (scale bar, 20 μm). (K) HuT102 and Karpas-299 cells were transfected with siATG4B (or siNC) for 24 h, and the cell viability was determined. (L and M) HuT102 and Karpas-299 cells were co-transfected with miR-449a mimic (or NC mimic) and siATG4B (or siNC) (L), while HuT78 and Jurkat E6-1 cells were co-transfected with miR-449a inhibitor (or NC inhibitor) and siATG4B (or siNC) (M). After 24 h, the cell viability was determined. ATGs: autophagy related genes, Act D: actinomycin D, pmir-ATG4B: luciferase reporter plasmids containing the wild-type 3’UTR of ATG4B, pmir-ATG4B-mut: luciferase reporter plasmids containing the mutant 3’UTR of ATG4B. pCMV-ATG4B: the ATG4B expression plasmid without 3’UTR. siATG4B: siRNA for ATG4B, siNC: negative control siRNA. *P < 0.05, **P < 0.01, ns: no significance.
Next, the ATG4B expression plasmid (without 3’UTR) was constructed. The overexpression efficiency of ATG4B is shown in Supplementary Fig. 4. Fig. 3G-J showed that miR-449a mimic decreased the LC3-II protein level (Fig. 3G), suppressed the cell viability (Fig. 3H) and elevated the number of apoptotic bodies (Fig. 3I and J), which was abolished by simultaneous overexpression of ATG4B (Fig. 3G-J). Moreover, we also silenced ATG4B in HuT78 and Jurkat E6-1 cells. As shown in Fig. 3K, silencing of ATG4B decreased the cell viability of HuT78 and Jurkat E6-1 cells. Silencing of ATG4B did not further decrease the cell viability of HuT102 and Karpas-299 cells by miR-449a mimic (Fig. 3L), and treatment with miR-449a inhibitor no longer contributed to further cell viability of HuT78 and Jurkat E6-1 cells (Fig. 3M). These data suggested that the effect of miR-449a on decreased cell viability was mediated by regulating ATG4B expression. Collectively, these results showed that miR-449a binds to the 325-331 region of ATG4B mRNA 3’UTR, to downregulate the ATG4B protein level, and ultimately reduced the autophagy level of T-cell lymphoma cells.
Overexpression of miR-449a inhibits the growth of T-cell lymphoma xenograft tumors in vivo
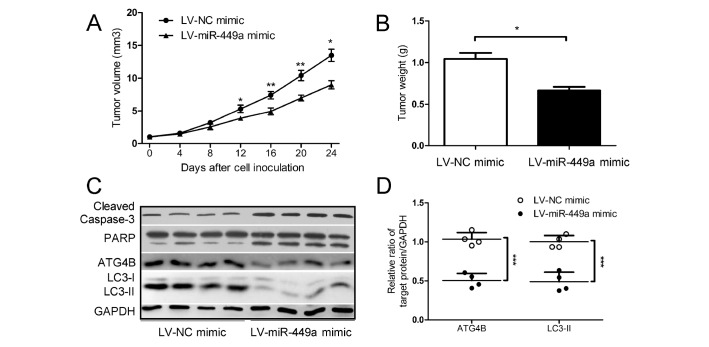
The above studies showed that miR-449a enhanced the apoptosis of T-cell lymphoma cells in vitro. Subsequent studies investigated the effect of miR-449a on xenograft tumor growth in vivo. The HuT102 cells with or without miR-449a showed a stable overexpression, and animal experiments were conducted. As shown in Fig. 4A and B, the miR-449a mimic dramatically repressed the growth of T-cell lymphoma xenograft tumors in nude mice, evidenced by the tumor volume (Fig. 4A) and weight (Fig. 4B). Additionally, the miR-449a mimic markedly reduced the protein levels of ATG4B and LC3-II (Fig. 4C and D), and increased the protein levels of cleaved Caspase-3 and PARP (Fig. 4C). These results indicated that overexpression of miR-449a attenuated the autophagy level and promoted the apoptosis of T-cell lymphoma cells, thus ultimately diminishing the growth of T-cell lymphoma xenograft tumors in vivo.
Fig. 4.
Overexpression of miR-449a inhibits the growth of T-cell lymphoma xenograft tumors in vivo. (A-D) HuT102 cells with or without miR-449a stable overexpression (LV-miR-449a mimic or LV-NC mimic) were injected into the axillary regions of nude mice (n = 4 per group). The tumor size was monitored every four days (volume = width2 × length × 1/2) (A). On day 24 after cell inoculation, the tumors were harvested and weighted (B). The levels of ATG4B, LC3 and cleaved Caspase-3/PARP in the xenograft tumors were determined (C). The ratios of ATG4B/GAPDH (or LC3-II /GAPDH) based on the results shown in (C) were analyzed and normalized against that of the corresponding control (D). *P < 0.05, **P < 0.01, ***P < 0.001.
DISCUSSION
T-cell lymphoma is a very heterogeneous group of tumors (17, 18), lacking in effective treatment strategies (19). Recent reports demonstrate that the histone deacetylase inhibitor-chidamide improves the therapeutic effect of T-cell lymphomas (2, 3). CD30 monoclonal antibody-brentuximab vedotin has also been reported to be effective in the initial treatment of T-cell lymphoma (20). However, the overall therapeutic efficiency of T-cell lymphoma remains unsatisfactory clinically. In this study, we found that autophagy inhibition enhanced the apoptosis of T cells, which provides a new perspective for the treatment of T-cell lymphoma.
ATG4B, an autophagy-related protein, plays a vital role in the formation of autophagosomes (21, 22). Studies have shown that ATG4B significantly enhances the autophagy level in cancer cells, and promotes the malignant progression of tumors (21, 23, 24). For instance, osteosarcoma Saos-2 cells lacking ATG4B are defective in autophagy and fail to form tumors in mouse models (24). Another report showed that inhibition of ATG4B activities using genetic approaches or an ATG4B inhibitor decreased the autophagy level and suppressed the tumorigenicity of glioblastoma cells (25). In our previous studies, we also found that inhibition of ATG4B attenuated autophagy, enhanced cell death and apoptosis in epirubicin-treated HCC cells (26). In this study, we first demonstrated the role of ATG4B in T-cell lymphoma cells. The overexpression of ATG4B enhanced the autophagy level of T-cell lymphoma cells. Meanwhile, the overexpression of ATG4B abrogated the miR-449a-induced reduction of autophagy in T-cell lymphoma cells. However, a further study is needed to explore the mechanism by which overexpression of ATG4B resulted in elevation of autophagy in T-cell lymphoma.
Current studies have reported that multiple microRNAs are involved in autophagy (27-29). Among them, miR-449a, a microRNA transcribed from the long arm of chromosome 5, has been associated with autophagy (15, 16). For example, miR-449a enhanced autophagy in glioma cells (15) and in silica-induced pulmonary fibrosis (16). Interestingly, in our study involving T-cell lymphoma cell lines, miR-449a downregulated the autophagy level. MiR-449a has different effects on autophagy, probably because its target molecules exhibit different effects on autophagy. For example, miR-449a activates autophagy in glioma cells by targeting CISD2 and decreasing its expression (15), which acts as an autophagy suppressor. In our study, miR-449a attenuated the autophagy of T-cell lymphoma cell lines by decreasing ATG4B protein expression associated with autophagy. In the present study, we evaluated the protein levels of several ATGs upon overexpression of miR-449a. In addition to ATG4B, ATG5 also showed a slight downward trend by miR-449a mimic. Our team will further explore the role of ATG5 in the regulation of autophagy in lymphoma. Aside from ATGs, it remains unknown whether miR-449a regulates the autophagy of T-cell lymphoma cells via non-ATG levels. Further investigations are needed.
In addition to serving as a target for cancer therapy, microRNAs can also be used as molecular markers for early screening and prognostic evaluation of tumors (30, 31). A recent study reported that cyclic serum miRNA profiles in sarcoma patients provide an early and accurate method of sarcoma detection, which may lead to cure and extended survival (32). In the present study, we demonstrated that miR-449a may be targeted to attenuate autophagy and enhance apoptosis of T-cell lymphoma cells. It is still unclear whether miR-449a can be used as a prognostic indicator, or whether additional microRNAs are needed to construct a prediction system. Further in-depth studies and investigations are needed.
In summary, this study demonstrated that miR-449a downregulates ATG4B in T-cell lymphoma cells by binding to its mRNA 3’UTR, which attenuated the autophagy level and subsequently promoted apoptosis of T-cell lymphoma cells. Moreover, targeting the “miR-449a/ATG4B/ autophagy” pathway enhanced the apoptosis of T cells in lymphoma, suggesting that this pathway may serve as a novel therapeutic target in T-cell lymphoma.
MATERIALS AND METHODS
Detailed information is provided in the Supplementary Information.
Supplemental Materials
ACKNOWLEDGEMENTS
The assistance provided by all participants and contributors to this study is gratefully acknowledged.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Sermer D, Pasqualucci L, Wendel HG, Melnick A, Younes A. Emerging epigenetic-modulating therapies in lymphoma. Nat Rev Clin Oncol. 2019;16:494–507. doi: 10.1038/s41571-019-0190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y, Dong M, Hong X, et al. Results from a multicenter, open-label, pivotal phase II study of chidamide in relapsed or refractory peripheral T-cell lymphoma. Ann Oncol. 2015;26:1766–1771. doi: 10.1093/annonc/mdv237. [DOI] [PubMed] [Google Scholar]
- 3.Ji MM, Huang YH, Huang JY, et al. Histone modifier gene mutations in peripheral T-cell lymphoma not otherwise specified. Haematologica. 2018;103:679–687. doi: 10.3324/haematol.2017.182444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Arnam JS, Lim MS, Elenitoba-Johnson KSJ. Novel insights into the pathogenesis of T-cell lymphomas. Blood. 2018;131:2320–2330. doi: 10.1182/blood-2017-11-764357. [DOI] [PubMed] [Google Scholar]
- 5.Pfister AS. Emerging Role of the Nucleolar Stress Response in Autophagy. Front Cell Neurosci. 2019;13:156. doi: 10.3389/fncel.2019.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poillet-Perez L, White E. Role of tumor and host autophagy in cancer metabolism. Genes Dev. 2019;33:610–619. doi: 10.1101/gad.325514.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, McCarty N. Tampering with cancer chemoresistance by targeting the TGM2-IL6-autophagy regulatory network. Autophagy. 2017;13:627–628. doi: 10.1080/15548627.2016.1271516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Zhou X, Zhang Y, et al. CUL4B regulates autophagy via JNK signaling in diffuse large B-cell lymphoma. Cell Cycle. 2019;18:379–394. doi: 10.1080/15384101.2018.1560718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan J, Zhang J, Zhang X, et al. AEG-1 is involved in hypoxia-induced autophagy and decreases chemosensitivity in T-cell lymphoma. Mol Med. 2018;24:35. doi: 10.1186/s10020-018-0033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 11.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Y, Ma J, Xue Y, et al. MiR-449a exerts tumor-suppressive functions in human glioblastoma by targeting Myc-associated zinc-finger protein. Mol Oncol. 2015;9:640–656. doi: 10.1016/j.molonc.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandbothe M, Buurman R, Reich N, et al. The microRNA-449 family inhibits TGF-beta-mediated liver cancer cell migration by targeting SOX4. J Hepatol. 2017;66:1012–1021. doi: 10.1016/j.jhep.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Li F, Liang J, Bai L. MicroRNA-449a functions as a tumor suppressor in pancreatic cancer by the epigenetic regulation of ATDC expression. Biomed Pharmacother. 2018;103:782–789. doi: 10.1016/j.biopha.2018.04.101. [DOI] [PubMed] [Google Scholar]
- 15.Sun AG, Meng FG, Wang MG. CISD2 promotes the proliferation of glioma cells via suppressing beclin1mediated autophagy and is targeted by microRNA449a. Mol Med Rep. 2017;16:7939–7948. doi: 10.3892/mmr.2017.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Y, Jiang C, Zhu J, et al. miR-449a inhibits cell proliferation, migration, and inflammation by regulating high-mobility group box protein 1 and forms a mutual inhibition loop with Yin Yang 1 in rheumatoid arthritis fibroblast-like synoviocytes. Arthritis Res Ther. 2019;21:134. doi: 10.1186/s13075-019-1920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorigue M, Sancho JM. Recent landmark studies in follicular lymphoma. Blood Rev. 2019;35:68–80. doi: 10.1016/j.blre.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Argnani L, Broccoli A, Zinzani PL. Cutaneous T-cell lymphomas: Focusing on novel agents in relapsed and refractory disease. Cancer Treat Rev. 2017;61:61–69. doi: 10.1016/j.ctrv.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Horwitz S, O'Connor OA, Pro B, et al. Brentuximab vedotin with chemotherapy for CD30-positive peripheral T-cell lymphoma (ECHELON-2): a global, double-blind, randomised, phase 3 trial. Lancet. 2019;393:229–240. doi: 10.1016/S0140-6736(18)32984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Li J, Ouyang L, Liu B, Cheng Y. Unraveling the roles of Atg4 proteases from autophagy modulation to targeted cancer therapy. Cancer Lett. 2016;373:19–26. doi: 10.1016/j.canlet.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 22.Fu Y, Huang Z, Hong L, et al. Targeting ATG4 in Cancer Therapy. Cancers (Basel) 2019;11:649. doi: 10.3390/cancers11050649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurdi A, Cleenewerck M, Vangestel C, et al. ATG4B inhibitors with a benzotropolone core structure block autophagy and augment efficiency of chemotherapy in mice. Biochem Pharmacol. 2017;138:150–162. doi: 10.1016/j.bcp.2017.06.119. [DOI] [PubMed] [Google Scholar]
- 24.Akin D, Wang SK, Habibzadegah-Tari P, et al. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10:2021–2035. doi: 10.4161/auto.32229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang T, Kim CK, Alvarez AA, et al. MST4 Phosphorylation of ATG4B Regulates Autophagic Activity, Tumorigenicity, and Radioresistance in Glioblastoma. Cancer Cell. 2017;32:840–855.e848. doi: 10.1016/j.ccell.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang N, Wu Y, Lyu X, et al. HSF1 upregulates ATG4B expression and enhances epirubicin-induced protective autophagy in hepatocellular carcinoma cells. Cancer Lett. 2017;409:81–90. doi: 10.1016/j.canlet.2017.08.039. [DOI] [PubMed] [Google Scholar]
- 27.Yao L, Zhu Z, Wu J, et al. MicroRNA-124 regulates the expression of p62/p38 and promotes autophagy in the inflammatory pathogenesis of Parkinson's disease. FASEB J. 2019;33:8648–8665. doi: 10.1096/fj.201900363R. [DOI] [PubMed] [Google Scholar]
- 28.Sun W, Yi Y, Xia G, et al. Nrf2-miR-129-3p-mTOR Axis Controls an miRNA Regulatory Network Involved in HDACi-Induced Autophagy. Mol Ther. 2019;27:1039–1050. doi: 10.1016/j.ymthe.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Su Y, Xu A, et al. miR-221/222-Mediated Inhibition of Autophagy Promotes Dexamethasone Resistance in Multiple Myeloma. Mol Ther. 2019;27:559–570. doi: 10.1016/j.ymthe.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu A, Sun M, Xu L, et al. Quantitative zeptomolar imaging of miRNA cancer markers with nanoparticle assemblies. Proc Natl Acad Sci U S A. 2019;116:3391–3400. doi: 10.1073/pnas.1810764116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bahmanpour Z, Sheervalilou R, Choupani J, Shekari Khaniani M, Montazeri V, Mansoori Derakhshan S. A new insight on serum microRNA expression as novel biomarkers in breast cancer patients. J Cell Physiol. 2019;234:19199–19211. doi: 10.1002/jcp.28656. [DOI] [PubMed] [Google Scholar]
- 32.Asano N, Matsuzaki J, Ichikawa M, et al. A serum microRNA classifier for the diagnosis of sarcomas of various histological subtypes. Nat Commun. 2019;10:1299. doi: 10.1038/s41467-019-09143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.