Abstract
COVID-19 has become disastrous for world and spread all over. Researchers all around the globe are working to discover a drug to cure from COVID-19. RNA dependent RNA polymerase plays a key role in SARS-CoV-2 replication and thus it could be a potential target for SARS-CoV-2. This study revealed that Protopine, Allocryptopine and (±) 6- Acetonyldihydrochelerythrine could be potential RdRp inhibitors of SARS-CoV-2.
Introduction
Traditional medical science is used as a popular option for the treatment of diseases in developing countries including India, due to the absence or less side effects. Confidence in traditional medicinal system has increased and well established all over the world. Ayurveda of India, the traditional medical science is centuries old. Ancient traditional medicinal practitioners have developed deep understanding responses of plants/plant extracts on the human body and metabolism. Expanding the information of metabolic procedure and human physiology enlarges the scope of utilization of therapeutic plants [1], [2].
Argemone mexicana L., known as Ghamoya and considered as weed commonly found everywhere by fields and wasteland in India. In traditional medicinal system Argemone mexicana L. as a whole plant used to treat oral problem, malignancy, guinea worm disease and diuretic, jaundice, coetaneous infections, itching and dropsy while seeds are used in snake biting, laxative, expectorant and inflammation. Roots are used for antihelmentic, leprosy, and inflammation [3], [4], [5], [6], [7], [8], [9].
In recent years, many scientific investigations have been made using different parts of Argemone mexicana to cure different diseases. The studies revealed anti-HIV, Anxiolytic, antimalerial, immune-modulatory, antispasmodic and Neuropharmacological properties of the plant [10], [11], [12], [13], [14], [15].
December 2019, a new class of coronavirus named SARS-CoV-2 had emerged in Wuhan city of China and quickly spared over the whole world. Without a precise antiviral therapeutics or vaccine, nearly 4 lakh (till 5 June 2020) people have died all over the world so far. The main strategy to treat SARS-CoV-2 is care, supplemented by the combination of antimalerial and anti HIV drugs [16]. Wang M et al. 2020 reported that remdesivir evidently inhibit the contamination of SARS-CoV-2 in Vero E6 cells [17]. At the current situation WHO also concluded that “to date, there is no specific medicine recommended to prevent or treat SARS-CoV-2” [18].
RNA dependent RNA polymerase (RdRp) is one of the most multipurpose enzyme of retro-viruses, it is key enzyme for replicating the genome and for translation. The core structural feature of RdRps are conserved but the disparity in their arrangements are also exist. The structure of RdRp looks like that of a measured right hand and comprises of fingers, palm and thumb subdomains. SARS-CoV-2 is a ss-positive sense RNA. The genome of SARS-CoV-2 encrypts 27 proteins including a RNA-dependent RNA polymerase (RdRP) and four structural proteins. The main cofactor of this complex is the catalytic subunit (nsp12) of a RNA-dependent RNA polymerase (RdRp). Without any other factors, nsp12 has little activity and its abilities require adornment cofactors including nsp7 and nsp8, which increases the RdRp to binding and processivity. RdRp is moreover proposed to be the objective of a class of antiviral medications that are nucleotide analogs, including Remdesivir, which is a prodrug that is changed over to the dynamic medication in the triphosphate structure (RTP) inside cells. All things considered, RdRp has been a subject of serious auxiliary science endeavors [19]
The hypothesis
Information reported by since that antimalerial and anti-HIV drugs are decreasing the load SARS-CoV-2 we have hypothesized that Argemone mexicana can be useful to cure the SARS-CoV-2 by inhibiting the RNA dependent RNA polymerase [19].
Thus, the present research was aimed to investigate RNA dependent RNA polymerase inhibition by some bioactive alkaloids found Argemone mexicana L. in silico.
In silico studies
Software
Python 2.7- language was downloaded from www.python.com, Molecular graphics laboratory (MGL) tools and AutoDock 4.2 was downloaded from www.scripps.edu, Discovery Studio visualizer 4.1 was downloaded from www.accelerys.com.
Protein preparation
The three-dimensional crystalline structures of targeted proteins reverase transcriptase (PDB ID: 6 M71) was retrieved from the Protein Data Bank (http://www.rcsb.org/). The coordinates of the structures were complexed with water molecules and other atoms which are responsible for increased resolution and therefore the water molecules and het-atoms were removed using discovery studios and saved in. pdb format. The structures of test compounds Argemexicaine A, Argemexicaine B, Protopine, Allocryptopine were drawn from mole view while (±) 6- Acetonyldihydrochelerythrine obtained from pub chem and stored in .pdb format.
Docking analysis
The docking analysis of reverse transcriptase was carried out using the Autodock tools (ADT) v1.5.4 and autodock v 4.2 programs. Argemexicaine A, Argemexicaine B, Protopine, Allocryptopine and (±) 6- Acetonyldihydrochelerythrine were docked to all the target protein complexes with the molecule considered as a rigid body. The search was carried out with the Lamarckian Genetic Algorithm; populations of 100 individuals with a mutation rate of 0.02 have been evolved for ten generations. The remaining parameters were set as default. The Docked structure was then visualized using Discovery Studio 2016 for obtaining the binding interactions.
Results
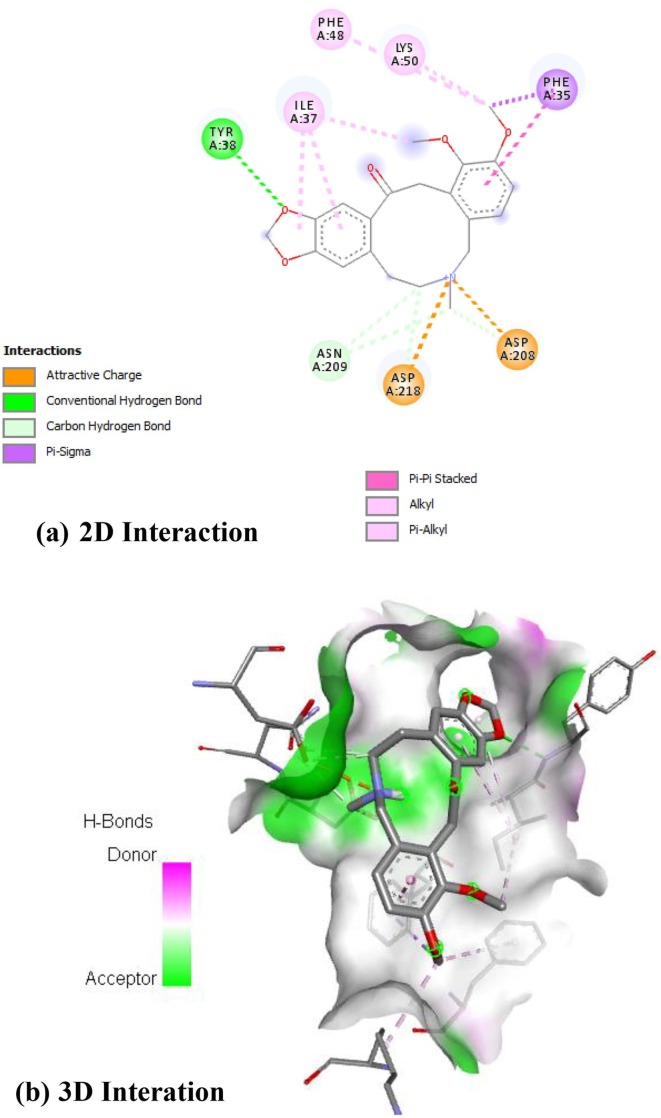
Argemexicaine A showed in Fig. 1 . interacted with A: ASN-209, A: ASP-208 and A: ASP-218 with its oxycycle nucleus with by forming attractive and carbon hydrogen bonds, the bezodioxole and benzene units interacted with A: TYR-38 and A: ILE-37 and A: ILE-37, A: PHE-35 by hydrogen bond, alkyl and alkyl, pi-pi stacked bonds respectively. The bezodioxole group provides quite strong interaction by forming hydrogen bond with A: TYR-38. The alkyl keto group interacts with A: ILE-37, A: PHE-48 and A: LYS-50 by forming pi-pi stacked bonds which provides much stability of complex.
Fig. 1.
Interaction of RdRp on Argemexicaine A [(a) 2D interactions (b) 3D interactions].
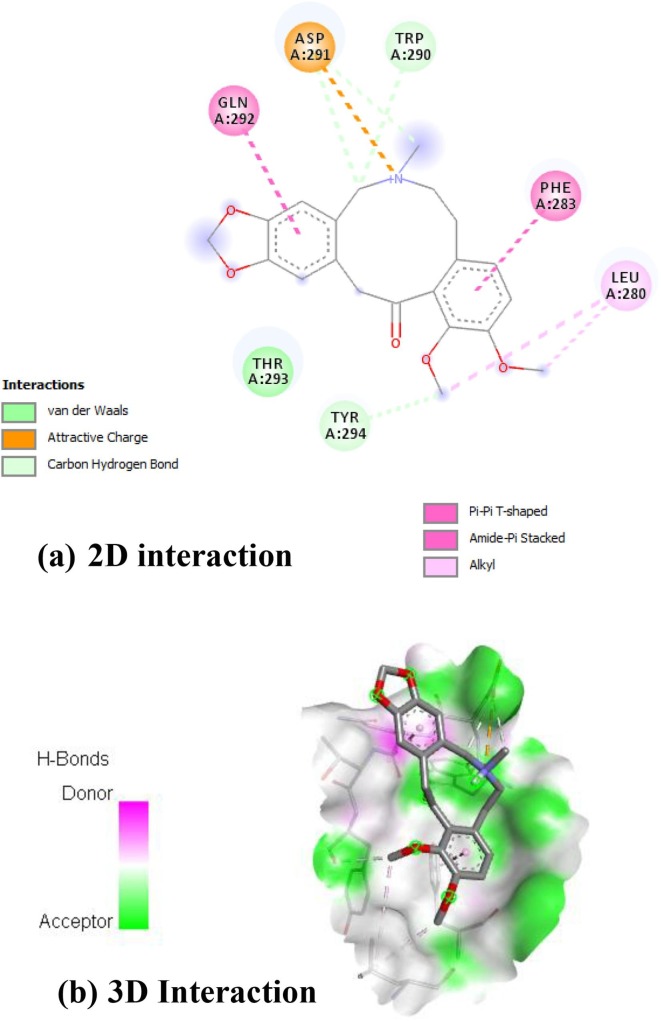
Argemexicaine B showed in Fig. 2 interacted with A: TRP-290, A: ASP-291 by forming van der waals and attractive charge attraction with oxycycle nucleus. The bezodioxole nucleus are failed bond any amino acids whereas benzene units interacted with A: GLN-292, A: PHE-283 by forming amide –pi stacked interation and pi-pi T-shaped interation. The alkyl keto groups interacted with A: LEU-280 and A: TYR-294 to form alkyl and carbon hydrogen bond which provides extra stability of complex.
Fig. 2.
Interaction of RdRp on Argemexicaine B [(a) 2D interactions (b) 3D interactions].
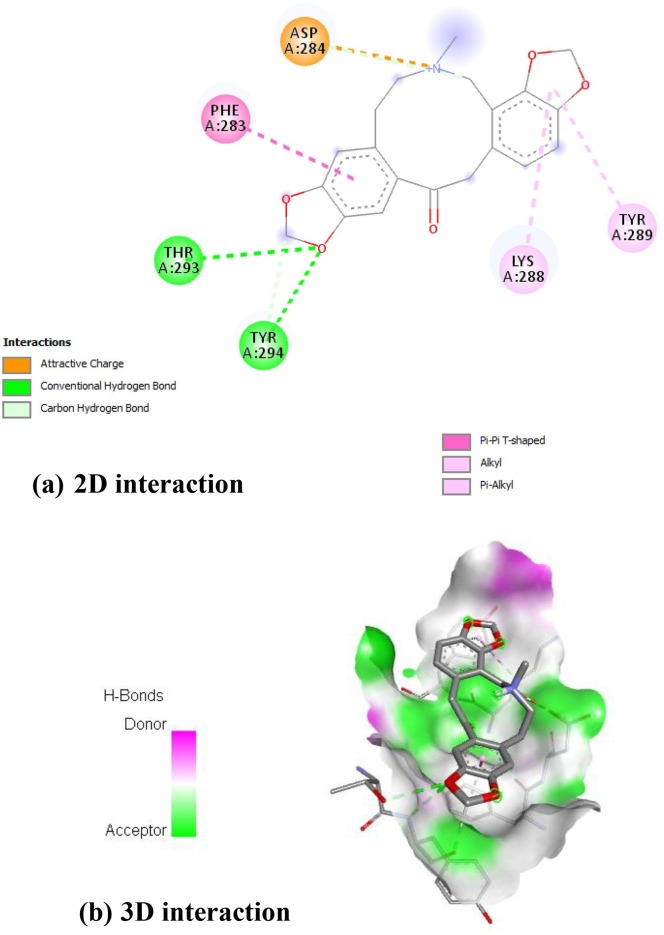
Protopine showed in Fig. 3 interaction with A: ASP-284 and form a bond of attraction with its oxycycle nucleus, the protopine contains two bezodioxole group which interacted with A: THR-293 and A: TYR-294 by forming hydrogen and carbon hydrogen bond while the next bezodioxole group interacted with A: LYS-288 and A: Tyr-289by forming the alkyl bonds. The benzene units interacted with A: PHE-283to form pi-pi T shaped interaction to stabilized the complex with Protopine.
Fig. 3.
Interaction of RdRp on Protopine [(a) 2D interactions (b) 3D interactions].
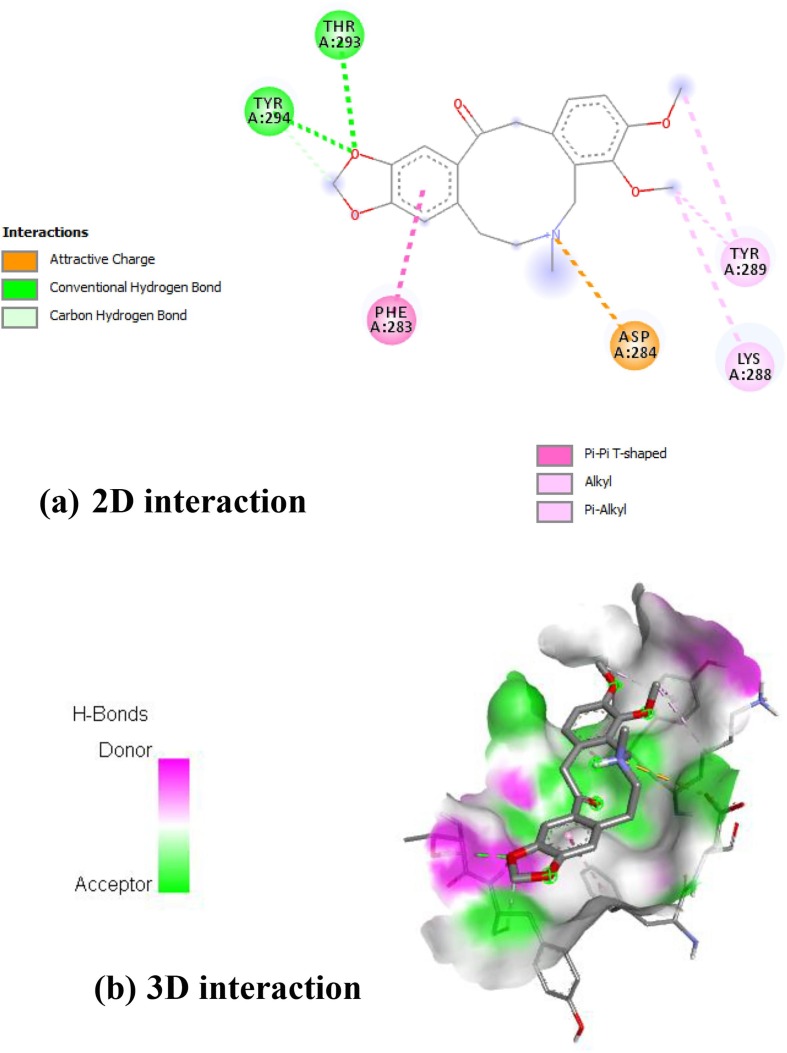
Allocryptopine showed in Fig. 4 interaction with A: ASP-284 to form an attractive bond with oxycycle nucleus. The bezodioxole unit forms the hydrogen bond with A: TYR-294 and A: THR-293. The benzene units interacted with A: PHE-283 to form pi-pi T shaped interaction. The alkyl keto groups interacted with A: LYS-288 and A: TYR-289 by forming the alkyl and pi-alkyl interaction.
Fig. 4.
Interaction of RdRp on Allocryptopine [(a) 2D interactions (b) 3D interactions].
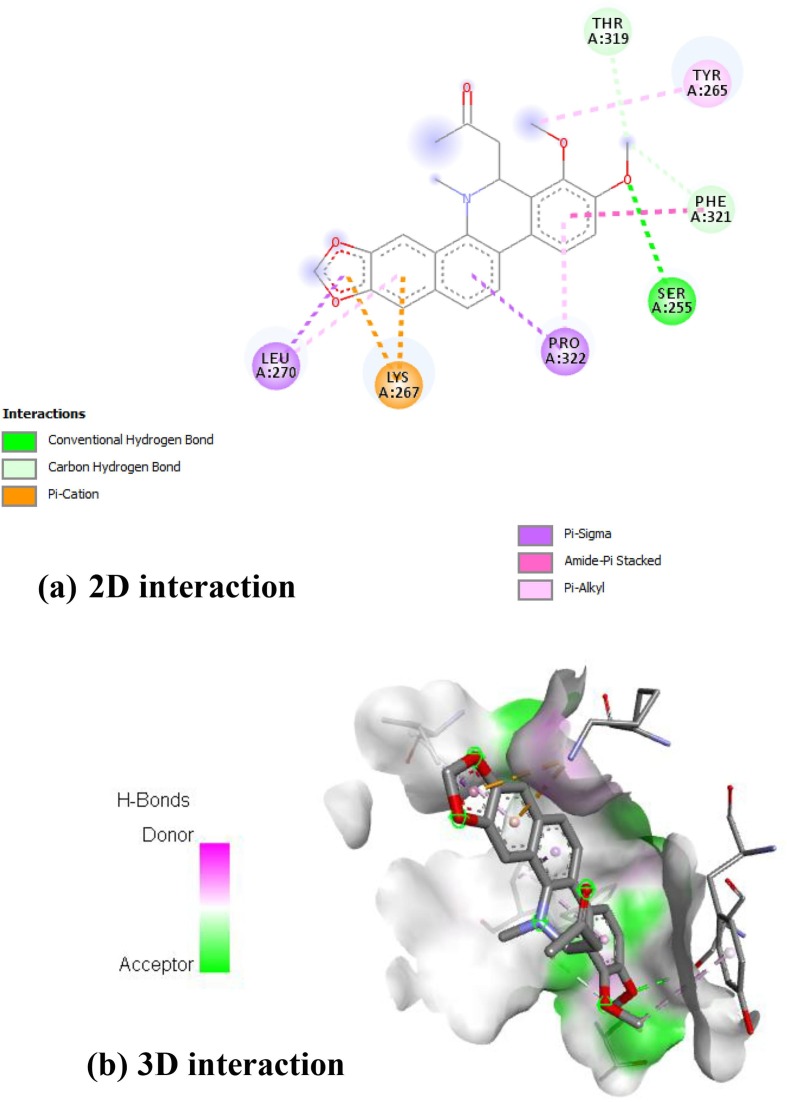
(±) 6- Acetonyldihydrochelerythrine showed in Fig. 5 . Interacted with A: LEU-270 by forming pi-sigma bond of benzodioxole unit. The benzene units interacted with A: LEU-270, A: LYS-267, A: PRO-322 and A: PHE-321 by forming pi-sigma, pi-cation, pi-sigma, pi-alkyl and amide pi stacked bonds respectively. The alkyl keto group interacted with A: SER-255, A: TYR-265, A: THR-319, A: PHE-321 by forming hydrogen bond, pi-alkyl and last two amino acids form carbon hydrogen bond respectively and stabilized the complex.
Fig. 5.
Interaction of RdRp on (±) 6- Acetonyldihydrochelerythrine [(a) 2D interactions (b) 3D interactions].
Table 1 provides the idea of binding energy, inhibition constant and hydrogen bond of ligands. The lowest binding energy of ligand has the ability to bind strongly with the receptor. The ligand Protopine have the lowest binding energy while Argemexicaine A have the highest binding energy with the RNA dependent RNA polymerase.
Table 1.
Binding energy and inhibition concentration of ligands.
| S.N | Name of Ligand(s) | Binding Energy Kcal/mole | Inhibitory Constant (μM) | Hydrogen Bond |
|---|---|---|---|---|
| 1 | Argemexicaine A | −5.57 | 82.05 | A:TYR-38 |
| 2 | Argemexicaine B | −5.62 | 75.89 | – |
| 3 | Protopine | −6.07 | 36.03 | A:THR-293, A: TYR-294 |
| 4 | Allocryptopine | −5.75 | 61.33 | A:THR-293, A: TYR-294 |
| 5 | (±) 6- Acetonyldihydrochelerythrine | −5.66 | 71.55 | A: SER-255 |
Discussion
At present, COVID-19 is a worldwide challenge for scientific communities as its pandemic mentality is hazardously influencing a huge number of individuals and taking a large number of lives regular. Be that as it may, until this point in time, no acceptable advancement has been made in the treatment of Covid-19 [20], [21], [22]. A few endeavors have been made to treat this illness yet these medication up-and-comers stay faulty attributable to low adequacy [23].
Computational methodologies for drug repurposing techniques could be a powerful way to deal with this COVID-19 pandamic. In this investigation, a few polymerase inhibitors focusing on RdRp of SARS-CoV-19 were contemplated, as RdRp had just demonstrated to be a powerful target of viral drug focus for different viral pathogens, for example, Hepatitis C Virus, HIV, Zika virus and so on [24], [25].
Here, the natural drugs repurposing along with molecular docking was utilized for the screening and examination of the drug candidates against RdRp of SARS-CoV-2. Natural compounds from Argemona mexicana were screened out which could be developed as a new drug to treat COVID-19.
RdRp assumes irreplaceable roles in the existence pattern of RNA viruses. RNA viruses start RNA synthesis by virus polymerase using primer independent and primer dependent mechanism. In addition, RdRp based RNA synthesis doesn't happen in the mammalian cells offering a chance to configuration drugs explicitly acting against RNA viruses. Moreover, the protein structure of RdRp in RNA viruses is seen as astoundingly conserved. Different antiviral drugs have been fashioned focusing on this enzyme for the treatment of diseases brought about by RNA viruses and they are working viably [26], [27]. Therefore, this current investigation intended to recognize potential medications focusing on this compound for the treatment of COVID-19.
Molecular docking analysis revealed that Protopine, Allocryptopine and (±) 6- Acetonyldihydrochelerythrine could be potential RdRp inhibitors of SARS-CoV-2.
Conclusion
COVID-19 has made disastrous worldwide and affecting individuals consistently, having just asserted absences of lives, and seriously hampered the worldwide economy. The current examination planned to battle this worldwide emergency by recommending potential medication contender for the treatment of COVID-19 and found that protopine is the best inhibitor among all alkaloids.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mehy.2020.109905.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mehy.2020.109905.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Appendix C. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Pandeya K.B., Tripathi I.P., Mishra M.K., Dwivedi N., Pardhi Y., Kamal A. A critical review on traditional herbal drugs: an emerging alternative drug for diabetes. Int J Org Chem. 2013;3(1):1–22. [Google Scholar]
- 2.Haidan Y., Qianqian M., Li Y., Guangchun P. The traditional medicine and modern medicine from natural products. Molecules. 2016;21(5):559. doi: 10.3390/molecules21050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhalke R.D., Gosavi S.A. Anti-stress and antiallergic effect of Argemone mexicana stems in asthma. Arch Pharm Sci Res. 2009;1(2):127–129. [Google Scholar]
- 4.Ibrahim H.A., Ibrahim H. Phytochemical screening and toxicity evaluation on the leaves of Argemone mexicana Linn. (Papaveraceae) Int J App Sci. 2009;3:39–43. [Google Scholar]
- 5.Brahmachari G., Gorai D., Roy R. Argemone mexicana: Chemical and pharmacological aspects. Revista Brasileira de Farmacognosia. 2013;23(3):559–575. [Google Scholar]
- 6.Chopra R.N., Nayar S.L., Chopra I.C. Council of Industrial Research; New Delhi: 1986. Glossary of Indian Medicinal Plants. [Google Scholar]
- 7.Jyothi A.C., Santosh J.P., Ganes P.B. Screening of aqueous plant extracts against Beauveria Bassiana infection to 5th instar larvae of Bombyx mori L. J Med Plants Res. 2011;5(16):3936–3939. [Google Scholar]
- 8.Osho A., Adentunji T. Antimicrobial activity of the essential oil of Argemone mexicana Linn. J Med Plant Res. 2010;4(1):19–22. [Google Scholar]
- 9.Ganesan G. Traditional oral care medicinal plants survey of Tamil Nadu. Nat Product Radiance. 2008;7:166–172. [Google Scholar]
- 10.Chang Y.C., Hsieh P.W., Chang F.R., Wu R.R., Liaw C.C., Lee K.H. Two new protopines argemexicaines A and B and the anti-HIV alkaloid 6-acetonyldihydrochelerythrine from formosan Argemone mexicana. Planta Med. 2003;69(2):148–152. doi: 10.1055/s-2003-37710. [DOI] [PubMed] [Google Scholar]
- 11.Arcos-Martínez A.I., Muñoz-Muñiz O.D., Domínguez-Ortiz M.A., Saavedra-Vélez M.V., Vázquez-Hernández M., Alcántara-López M.G. Anxiolytic-like effect of ethanolic extract of Argemone mexicana and its alkaloids in Wistar rats. Avicenna J Phytomed. 2016;6(4):476–488. [PMC free article] [PubMed] [Google Scholar]
- 12.Willcox M.L., Graz B., Falquet J., Sidibe O., Forster O., Diallo D. Argemone mexicana decoction for the treatment of uncomplicated falciparum malaria. Trans R Soc Trop Med Hyg. 2007;101(12):1190–1198. doi: 10.1016/j.trstmh.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Goel A., Kumar D., Bhatia A.K. Modulation of immune responses by aqueous extract of Argemone Mexicana leaves. J Immunol Immunopathol. 2008;10(1):65–69. [Google Scholar]
- 14.Anarthe S., Chaudhari S. Neuropharmacological study of Argemone mexicana Linn. J Appl Pharm Sci. 2011;01(04):121–126. [Google Scholar]
- 15.Singla D., Sharma A., Kaur J., Panwar B., Raghava G.P.S. BIAdb: a curated database of benzylisoquinoline alkaloids. BMC Pharmacol Toxicol. 2010;10(1):1–8. doi: 10.1186/1471-2210-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance.
- 17.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo H., Tang Q.L., Shang Y.X., Liang S.B., Yang M., Robinson N. Can Chinese Medicine Be Used for Prevention of Corona Virus Disease 2019 (COVID-19)? A review of historical classics, research evidence and current prevention programs. Chin J Integr Med. 2020;26:243–250. doi: 10.1007/s11655-020-3192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin W, Mao C, Luan X, Shen DD, Shen Qingya, Su H, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020; 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed]
- 20.Lake M.A. What we know so far: COVID-19 current clinical knowledge and research. Clin Med. 2020;20:124–127. doi: 10.7861/clinmed.2019-coron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuen K.S., Ye Z.W., Fung S.Y., Chan C.P., Jin D.Y. SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci. 2020;10:40. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19), Drug Discov Ther 2020:14;58–60. https://doi.org/10.5582/ddt.2020.01012. [DOI] [PubMed]
- 23.Zhou D., Dai S.-M., Tong Q. COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J Antimicrob Chemother. 2020;dkaa114 doi: 10.1093/jac/dkaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Clercq E. The design of drugs for HIV and HCV. Nat Rev Drug Discov. 2007;6:1001–1018. doi: 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- 25.Xu H.-T., Hassounah S.A., Colby-Germinario S.P., Oliveira M., Fogarty C., Quan Y. Purification of Zika virus RNA-dependent RNA polymerase and its use to identify small-molecule Zika inhibitors. J Antimicrob Chemother. 2017;72:727–734. doi: 10.1093/jac/dkw514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elfiky A.A., Ribavirin Remdesivir, Sofosbuvir Galidesivir. and Tenofovir against SARSCoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;117592 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganeshpurkar A, Gutti G, Singh SK, RNA-dependent RNA polymerases and their emerging roles in antiviral therapy. In: Viral Polym, Elsevier, (2019) 1–42. https://doi.org/10.1016/b978-0-12-815422-9.00001-2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.