Abstract
Background
The diagnosis performance of B-mode ultrasound (US) for focal liver lesions (FLLs) is relatively limited. We aimed to develop a deep convolutional neural network of US (DCNN-US) for aiding radiologists in classification of malignant from benign FLLs.
Materials and methods
This study was conducted in 13 hospitals and finally 2143 patients with 24,343 US images were enrolled. Patients who had non-cystic FLLs with pathological results were enrolled. The FLLs from 11 hospitals were randomly divided into training and internal validations (IV) cohorts with a 4:1 ratio for developing and evaluating DCNN-US. Diagnostic performance of the model was verified using external validation (EV) cohort from another two hospitals. The diagnosis value of DCNN-US was compared with that of contrast enhanced computed tomography (CT)/magnetic resonance image (MRI) and 236 radiologists, respectively.
Findings
The AUC of ModelLBC for FLLs was 0.924 (95% CI: 0.889–0.959) in the EV cohort. The diagnostic sensitivity and specificity of ModelLBC were superior to 15-year skilled radiologists (86.5% vs 76.1%, p = 0.0084 and 85.5% vs 76.9%, p = 0.0051, respectively). Accuracy of ModelLBC was comparable to that of contrast enhanced CT (both 84.7%) but inferior to contrast enhanced MRI (87.9%) for lesions detected by US.
Interpretation
DCNN-US with high sensitivity and specificity in diagnosing FLLs shows its potential to assist less-experienced radiologists in improving their performance and lowering their dependence on sectional imaging in liver cancer diagnosis.
Keywords: Ultrasound, Convolutional neural network, Focal liver lesions, Diagnosis
Research in context.
Evidence before this study
Literature searches were conducted separately using Medline and ISI Web of Science databases on 12 March 2020 with the terms (“deep learning” OR “radiomic” OR “convolutional neural network” OR “Artificial intelligence” OR “traditional machine learning”) AND (“liver neoplasms”) AND (“ultrasonography”), without date or language restrictions. A total of two studies were published that evaluate the use of radiomics or deep learning to classify the focal liver lesions from B-mode ultrasound. Moreover, none of two studies has an external validation cohort and they are based on single-centre and small sample size. Potential pseudo and specious result caused by overfitting from these studies make it tough for clinical generalization and actual use.
Added value of this study
Database we built in this study is the largest, multicentric and prospective, and standardized ultrasound image data for focal liver lesions, which ensures the quality of ultrasound images, reduces the difference between radiologists and provide the large-scale data basis for deep learning analysis. Based on this database, the deep convolutional neural network model was developed to improve diagnosis power of FLLs and showed satisfied robustness. The diagnosis capability of our model was comparable to contrast enhanced CT and superior to skilled radiologists with 15-year experience in FLLs diagnosis performance. In addition, we showed the diagnosis of model as the attention maps.
Implications of all the available evidence
The high performance of the model for liver lesions will contribute to an increase in ultrasound diagnostic quality, reduce doctor's dependence on CT/MRI and biopsy, facilitate the development of remote medicine, and decrease the costs in the national health care through the early diagnosis of diseases. Furthermore, it has a potential to induce a paradigm shift in the field of diagnosis of liver lesions via image. And the model could be particularly valuable for junior radiologists whose expertise in ultrasound imaging interpretation is insufficient, which will lead to a reduction in misdiagnosis of focal liver lesions by them and lesser dependence on CT/MRI.
Alt-text: Unlabelled box
1. Introduction
Liver cancer is the sixth most prevalent cancer and the fourth most frequent cause of cancer-related death worldwide, with about 841,000 new cases and 782,000 deaths per year, representing a great challenge to health-care systems because of their aggressive presentation [1]. Therefore, early detecting and accurately separating liver malignancy from benign lesions is crucial for prognosis, surveillance and management of patients with focal liver lesions (FLLs) [2]. In clinic practice, B-mode ultrasound (US) is usually the first-line imaging test for FLLs because it is inexpensive and facilitates real time diagnosis without radiation exposure or nephrotoxicity [4], [5], [6]. However, B-mode US is less accurate at diagnosing FLLs compared with tomographic imaging modalities because of high dependence on the quality of the equipment and doctors’ experience, and lack of perfusion information [5,6]. Especially, FLLs of different histological types often display similar appearances on US images [7], whereas FLLs of the same histological type may present completely different US image characteristics because of variances in their disease differentiation and stage [8]. Taking hepatocellular carcinoma (HCC, a growing global public health problem) as an example, the sensitivity of B-mode US for diagnosis is only 46%–63% [6,[9], [10], [11]]. Therefore, US was only recommended as surveillance tool for liver lesion by The American Association for the Study of Liver Diseases (AASLD) and European Association for the Study of the Liver (EASL) guidelines [9,12].
Therefore, more and more physicians and radiologists have been relying on contrast enhanced US (CEUS), computed tomography (CT), and magnetic resonance image (MRI), and even biopsy to obtain accurate diagnosis of FLLs. This is a costly, time-consuming, often subjective, and invasive process that requires substantial experience and expertise among radiologists and pathologists. Further, the unbalanced distribution of medical resources between developing and developed areas has inhibited the application of CEUS/CT/MRI and biopsy. Therefore, timely and effective clinical decisions have been difficult to make, and they have also become more complex, demanding the synthesis of decisions from assessment of large volumes of data that represent clinical information. If the FLL can be well analysed and characterized by US as the routine and primary scanning technology, which may fuel the time-consuming and relatively expensive contrast enhanced imaging to concentrate on complex cases in order to filtrate highly benign or non-urgent cases for clinicians. .
The development of an artificial intelligence (AI) framework provides a new opportunity to improve the diagnostic accuracy of FLLs by US imaging. Compared with radiologists reading anatomical images, AI techniques can not only better reflect holistic tumour morphology but also capture granular and task-specific radiological patterns that are difficult to recognize by human vision [13]. Previous studies explored the validity of traditional pattern recognition classifiers and deep convolutional neural networks (DCNNs) in FLL diagnosis via US imaging [[14], [15], [16]]. Nevertheless, these retrospective studies had small sample sizes, did not employ standardized image data, and lacked external validation (EV) to ensure the reliability of their results. Currently, DCNN methods are most widely used in larger-sample based AI studies. Unlike classical radiomics analysis based on hand-designed features, DCNN apply an end-to-end learning strategy, taking image pixels and corresponding class labels from medical image data as inputs to impart enhanced feature learning power. In our previous study, we successfully used a DCNN for liver fibrosis assessment using shear wave elastography with superior accuracy [17].
In this study, we conducted a multicentre study to develop a DCNN-US for classifying of malignant from benign FLLs. Furthermore, we compared the model's results with contrast enhanced CT/MRI and radiologists with different skill levels based on pathological reference.
2. Methods
2.1. Overall design
We developed a ResNet-based [18] convolutional neural network for diagnosis of FLLs using US imaging. Radiomics signatures derived from FLLs and liver, along with ultrasonic features and clinical factors, were incorporated to construct the DCNN-US model. A training cohort was used to determine the radiomic signature. Internal validation (IV) and EV cohorts were used to validate the performance of the generated model. In addition, the EV cohort was also used to compare the performance of contrast enhanced CT/MRI with those of radiologists who had different levels of liver US experience. This multicentre study was approved by the ethics committee of each centre and is registered at ClinicalTrials.gov (NCT03871140). Written informed consent was obtained from all patients in this study. All authors had access to the study data and reviewed and approved the final manuscript.
All diagnoses were confirmed by liver biopsy and/or resection pathology within one month after US scan. The results of histological staining were read by board certified liver pathologists.
2.2. Patients enrollments
All the cohorts used the same inclusion and exclusion criteria. The inclusion criteria were as follows: (1) focal non-cystic liver lesions without previous any local therapy; (2) lesion size >1.0 cm; (3) standard US scan performed less than 1 month before biopsy or surgery; (4) type of FLL confirmed by histologic examination. The exclusion criteria were as follows: (1) liver containing only cystic lesions; (2) any previous local therapy for the index lesion before US scan, including radiotherapy, ablation, and transarterial chemoembolization; (3) unclear US image of liver or lesions or absence of the type of US image that the study required; (4) absence of definite pathological diagnosis; (5) absence of clinical disease history.
2.3. US images acquisition
All US images from the 13 hospitals were in DICOM data format. The US examinations were performed by using 17 devices (Appendix 1.1, Table S2). The criteria for US image acquisition and radiologic feature evaluation of FLLs were established by the study panel, which included 13 radiologists from each centre with >10 years of experience in US-based liver diagnosis. We collected a total of 11 standard US images, for each FLL patient according to the established protocol (Appendix 1.2, Fig. S1). A total of 7 primary ultrasonic features were analysed and summarized in Appendix 1.3, Table S3 and Fig. S2.
2.4. Clinical information acquisition
The demographic and clinical data of all patients including age, gender, history of hepatitis and extra-hepatic tumours, and alpha fetoprotein (AFP) were recorded. AFP was measured within 1 week after US scan. The threshold value for a negative AFP level was ≤200 ng/mL. All US and clinical information were deidentified before they were transferred to investigators.
2.5. Diagnosis model construction by deep learning
The DCNN models were trained on manual planar segmented regions of interest (ROI) from lesion or liver background (LB) images (Appendix 1.4). Three DCNN models named ModelLesion (ModelL), ModelLesion+Background (ModelLB), and ModelLesion+Background+clinic (ModelLBC) with colligated image and clinical information were correspondingly built to analyse their diagnostic capabilities. modelling analysis was performed on python 3.6.5 (https://www.python.org/). The machine learning frameworks used were PyTorch 1.0.0 (https://pytorch.org/) and scikit-learn 0.20.2 (https://scikit-learn.org/stable/).
For ModelL, we applied an 18-layer ResNet [18] pretrained on the ImageNet dataset [19]. The final fully connected layer was removed with replacement by a dropout layer [20,21], a batch normalization [22] layer, and a 512 × 1 fully connected layer to obtain the final predictive score. In addition to binary cross entropy loss, which was commonly used for classification, we also leveraged batch-hard triplet loss [23] to let the network focus on difficult-to-diagnose samples. There were a total of four US images of each lesion, so the lesion-level output was the average of the four predicted image-level outcomes.
For ModelLB, we added an LB branch that shared the same architecture as ModelL but with independent network weights. To merge the features extracted by the two branches, we applied a 1024 × 1 fully connected layer to concatenate the final global features. Ground-truth LB types were used to supervise the LB branch by minimizing the binary cross entropy loss, while the lesion branch and final output were supervised through the benign and malignant classes.
To generate a visual explanation of the model diagnosis process, attention maps were plotted using the GradCAM algorithm [24], which displays the pixels in the ROIs that provide the greatest contribution to the classification output.
To further integrate additional diagnostic factors, we built ModelLBC, which integrates the outcomes of the lesion-level ModelLB and clinical-ultrasonic factors by logistic regression. The clinical-ultrasonic factors used were selected by multivariate analysis. The training details are summarized in appendix 1.5–1.7.
2.6. Nomogram development and validation
To provide a graphical presentation of the DCNN models for convenient clinical use, a nomogram was developed by integrating model outcomes and relative clinical-ultrasonic factors. The calibration curve that measures the concordance of DCNN-predicted outcome and actual histopathological diagnosis of FLLs was plotted. Decision curve analysis was performed to determine the nomograms’ clinical utility.
2.7. Stratified analysis to assess the diagnostic accuracy
Lesion size and LB echo are two important factors that affect FLL diagnosis by radiologists. Stratification analysis was performed to verify the diagnostic power of the proposed DCNN-US model. Patients were stratified into several subgroups according to lesion size (1.1–2.0 cm, 2.1–5.0 cm, and >5.0 cm) and the LB (normal, hepatic steatosis, and hepatic fibrosis).
2.8. Comparative evaluation of diagnostic performance of DCNN model
We compared the performance of DCNN-US with those of radiologists with different levels of liver US experience (1–5 years, 5–10 years, 10–15 years, and >15 years), and then the model and contrast enhanced CT/MRI were compared in terms of diagnostic power for the index FLLs analysed by DCNN. In total, 236 radiologists independently viewed anonymized videos (10–20 s) of the US data of 338 FLLs from the EV cohort that contained the target tumour and whole LB, with the radiologists blinded to the histological results (Appendix 1.8). These skilled radiologists each had >15 years’ experience of performing and interpreting US and had been certified by a board. All contrast enhanced CT/MRI images were interpreted by radiologists with >5 years’ experience in liver diagnosis, who were provided with the same clinical information as used in the model analysis (Appendix 1.9–1.10).
2.9. Statistical analysis
Statistical analysis was performed using the R (Version 3.4.1; www.R-project.org) and PASW Statistics (version 18.0; SPSS Inc., Chicago, IL, USA) software packages. The AUC, accuracy, sensitivity, specificity, PPV and NPV of DCNN-US were calculated on per-patient basis. The Youden index was used to set the cut-off for the predicted score during training. Two-sided Delong tests were used to estimate whether statistical differences exist between AUC values. Mann-Whitney U test was used to explore the differences in diagnostic accuracy, sensitivity, and specificity between the radiologists and DCNN-US model. Univariate and multivariate analyses were performed by the logistic regression model, and p<0.05 was considered statistically significant.
3. Results
3.1. Clinical characteristics
Up to 2263 potentially eligible patients were enrolled in this cohort between May 2018 and June 2019. According to the enrollment criteria, 122 patients were excluded because of unqualified US imaging (unclear, unstandardized data), small lesion size (maximum diameter ≤1.0 cm), or indeterminate histological diagnosis. Finally, 24,343 US images of 2213 nodules from 13 Chinese hospitals were enrolled for analysis (Fig. 1). Among them, a total of 1815 patients from 11 hospitals were randomly divided into training (16,500 images of 1500 lesions) and IV (4125 images of 375 lesions) cohorts with a 4:1 ratio of the respective numbers of FLLs. The additional 3718 images of 338 FLLs from another two hospitals were enrolled as an independent EV cohort after the establishment of the DCNN models. In total, 16 types of FLLs were enroled in this study (Table S4). The three cohorts’ patient characteristics are shown in Table 1.
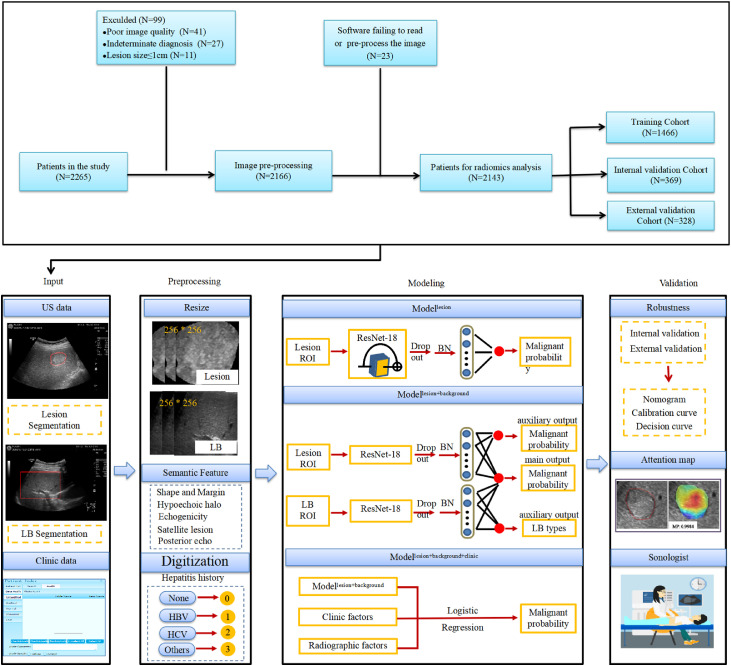
Fig. 1.
Flowchart: development and validation of DCNN-US for diagnosis of focal liver lesions. US=Ultrasound, LB=Liver background, HBV=Heaptitis B virus, HCV=Heaptitis C virus, ROI=Region of interest, BN=Batch normalization, CT=Computed tomography, MRI=Magnetic resonance image, DCNN-US=Deep convolutional neural network of ultrasound.
Table 1.
Characteristics of patients in three cohorts.
| Parameters | All patients (N = 2143) | Training cohort (N = 1446) | Internal validation cohort (N = 369) | External validation cohort (N = 328) |
|---|---|---|---|---|
| Mean age±(SD) (years) | 55.7 ± 12.4 | 55.6 ± 12.3 | 56.0 ± 12.5 | 55.9 ± 12.9 |
| Tumour size (cm)* | 3.7 (1.1–19.6) | 3.6 (1.1–19.6) | 3.7 (1.1–17.3) | 4.0 (1.1–16.7) |
| Gender (%) | ||||
| Male | 1460 (68.1) | 995 (68.8) | 240 (65.0) | 225 (68.6) |
| Female | 683 (31.9) | 451 (31.2) | 129 (35.0) | 103 (31.4) |
| History of hepatitis (%) | ||||
| No | 1172 (54.7) | 766 (53.0) | 214 (58.0) | 192 (58.5) |
| Yes | 971 (45.3) | 680 (47.0) | 155 (42.0) | 136 (41.5) |
| AFP (ng/mL) (%) | ||||
| Missing | 137 (6.4) | 75 (5.2) | 16 (4.3) | 46 (14.0) |
| ≤200 | 1732 (80.8) | 1180 (81.6) | 310 (84.0) | 242 (73.8) |
| >200 | 274 (12.8) | 191 (13.2) | 43 (11.7) | 40 (12.2) |
| History of extra-hepatic tumours (%) | ||||
| No | 1634 (76.2) | 1122 (77.6) | 275 (74.5) | 237 (72.3) |
| Yes | 509 (23.8) | 324 (22.4) | 94 (25.5) | 91 (27.7) |
| Features of the liver background (%) | ||||
| Normal | 1010 (47.1) | 655 (45.3) | 175 (47.4) | 180 (54.9) |
| Hepatic steatosis | 184 (8.6) | 127 (8.8) | 41 (11.1) | 16 (4.9) |
| Hepatic fibrosis | 949 (44.3) | 664 (45.9) | 153 (41.5) | 132 (40.3) |
| Lymph-node metastasis (%) | ||||
| No | 1965 (91.7) | 1348 (93.2) | 344 (93.2) | 274 (83.5) |
| Yes | 177 (8.3) | 98 (6.8) | 25 (6.8) | 54 (16.5) |
| Vascular invasion (%) | ||||
| No | 1982 (92.5) | 1340 (92.7) | 345 (93.5) | 297 (90.5) |
| Yes | 161 (7.5) | 106 (7.3) | 24 (6.5) | 31 (9.5) |
| Ascites (%) | ||||
| No | 2016 (94.1) | 1369 (94.7) | 347 (94.0) | 300 (91.5) |
| Yes | 127 (5.9) | 77 (5.3) | 22 (6.0) | 28 (8.5) |
| Tumour number (%)* | ||||
| Malignant | 1786 (80.7) | 1221 (81.4) | 303 (80.8) | 262 (77.5) |
| Benign | 427 (19.3) | 279 (18.6) | 72 (19.2) | 76 (22.5) |
| Tumour shape (%)* | ||||
| Circular | 106 (4.8) | 82 (5.5) | 13 (3.5) | 11 (3.3) |
| Ellipse | 857 (38.7) | 564 (37.6) | 127 (33.9) | 166 (49.1) |
| Irregular | 1250 (56.5) | 853 (56.9) | 235 (62.6) | 161 (47.6) |
| Tumour margin (%)* | ||||
| Smooth | 1313 (59.3) | 882 (58.8) | 208 (55.5) | 223 (66.0) |
| Non-smooth | 900 (40.7) | 618 (41.2) | 167 (44.5) | 115 (34.0) |
| Tumour echogenicity (%)* | ||||
| Hyper- | 452 (20.4) | 294 (19.6) | 75 (20.0) | 83 (24.6) |
| Iso- | 80 (3.6) | 57 (3.8) | 14 (3.7) | 9 (2.7) |
| Hypo- | 920 (41.6) | 597 (39.8) | 141 (37.5) | 182 (53.8) |
| Heterogeneous | 761 (34.4) | 552 (36.8) | 145 (38.7) | 64 (18.9) |
| Intratumoral vascularity (%)* | ||||
| No | 1021 (46.1) | 666 (44.4) | 176 (46.9) | 179 (53.0) |
| Yes | 1192 (53.9) | 834 (55.6) | 199 (53.1) | 159 (47.0) |
| Posterior acoustic enhancement (%)* | ||||
| Absent | 1412 (63.8) | 972 (64.8) | 232 (61.9) | 208 (61.5) |
| Present | 801 (36.2) | 528 (35.2) | 143 (38.1) | 130 (38.5) |
| Hypoechoic halo (%)* | ||||
| No | 1439 (65.0) | 937 (62.5) | 232 (61.9) | 270 (79.9) |
| Yes | 774 (35.0) | 563 (37.5) | 143 (38.1) | 68 (20.1) |
| Peritumoral satellite lesion (%)* | ||||
| Absent | 2182 (98.6) | 1476 (98.4) | 370 (98.7) | 336 (99.4) |
| Present | 31 (1.4) | 24 (1.6) | 5 (1.3) | 2 (0.6) |
| Diagnostic method (%)* | ||||
| Biopsy | 1394 (63.0) | 936 (62.4) | 227 (60.5) | 231 (68.3) |
| Resection | 819 (37.0) | 564 (37.6) | 148 (39.5) | 107 (31.7) |
Note. Qualitative variables are expressed as n (%), and quantitative variables are expressed as Mean±SD or median, as appropriate.
date calculated based on tumour number.
3.2. Univariate and multivariate analysis
In this study, a total of 14 factors (7 US features and 7 clinical factors) were analysed with univariate analysis, and those that achieved statistical significance by univariate analysis were included in the multivariable analysis, including 2 US features and 4 clinical factors (Table S5). The results of multivariable logistic regression analyses showed that hypoechoic halo (OR, 18.389; 95% CI, 9.921–34.084; p<0.001), history of extrahepatic tumours (OR, 16.166; 95% CI, 9.311–28.065; p<0.001), history of hepatitis (OR, 11.736; 95% CI, 7.857–17.529; p<0.001), older age of patients (OR, 3.323; 95% CI, 2.096–5.269; p<0.001), male sex (OR, 2.303; 95% CI, 1.629–3.256; p<0.001), and intratumoural vascularity (OR, 1.911; 95% CI, 1.344–2.717; p<0.001) were independent predictors associated with malignant FLLs.
3.3. Diagnostic performance of the DCNN-US models
The diagnostic performance of the three DCNN models is shown in Fig. 2, S4 and table S6. ModelLBC achieved optimal diagnostic power, with AUC values of 0.925 (95% CI: 0.886–0.963), and 0.924 (95% CI: 0.889–0.959) in the IV and EV cohorts, respectively, followed by ModelLB and ModelL, which had relatively decreased AUCs. The AUC values of ModelLBC in the EV cohort on lesions 1.1–2.0 cm, 2.1–5.0 cm, and >5.0 cm were 0.926 (95% CI: 0.815–1.000), 0.899 (95% CI: 0.838–0.960) (p = 0.6783, 1.1–2.0 cm vs 2.1–5.0 cm), and 0.962 (95% CI: 0.933–0.991) (p = 0.5389, 1.1–2.0 cm vs >5.0 cm), respectively (Table 2 and Fig. S5). Stratification analysis according to lesion size showed no statistical differences between the IV and EV cohorts (all p>0.05). The diagnostic results of ModelLBC for FLLs with normal, hepatic steatosis, and hepatic fibrosis backgrounds are shown in table S7 and Fig. S5.
Fig. 2.
Model robustness analysis. (a-c) represents 1st fold in training, internal validation and external validation cohort, respective. The green, blue, red line represents the ROC curve of ModelL, ModelLB, ModelLBC, respectively. ROC=Receiver operating characteristic, ModelL=Modellesion, ModelLB=Modellesion+background, ModelLBC=Modellesion+background+clinic.
Table 2.
Stratification analysis of ModelLBC according to tumour size.
| Tumour size |
Training cohort |
Internal validation cohort |
External validation cohort |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | ACC | SE | SP | AUC | ACC | SE | SP | AUC | ACC | SE | SP | |
| (95%CI) | (95%CI) | (95%CI) | (95%CI) | (95%CI) | (95%CI) | (95%CI) | (95%CI) | (95%CI) | (95%CI) | (95%CI) | (95%CI) | |
| 1.1–2.0 cm | 0.925 | 0.908 | 0.962 | 0.594 | 0.938 | 0.860 | 0.875 | 0.500 | 0.926 | 0.941 | 1.000 | 0.500 |
| (N = 403) | (0.879–0.971) | (0.880–0.935) | (0.941–0.984) | (0.438–0.719) | (0.806–1.000) | (0.780–0.940) | (0.792–0.938) | (0.000–1.000) | (0.815–1.000) | (0.902–0.980) | (1.000–1.000) | (0.167–0.833) |
| 2.1–5.0 cm | 0.947 | 0.936 | 0.956 | 0.813 | 0.872 | 0.875 | 0.906 | 0.684 | 0.899 | 0.861 | 0.891 | 0.757 |
| N = 1078) | (0.919–0.975) | (0.920–0.953) | (0.939–0.970) | (0.733–0.880) | (0.795–0.950) | (0.831–0.919) | (0.863–0.949) | (0.526–0.842) | (0.838–0.960) | (0.819–0.904) | (0.845–0.938) | (0.649–0.865) |
| >5.0 cm | 0.992*,@ | 0.944 | 0.925 | 0.991 | 0.956 | 0.863 | 0.855 | 0.879 | 0.962 | 0.835 | 0.795 | 0.939 |
| (N = 732) | (0.985–0.999) | (0.922–0.964) | (0.898–0.953) | (0.972–1.000) | (0.917–0.994) | (0.811–0.916) | (0.774–0.919) | (0.788–0.970) | (0.933–0.991) | (0.777–0.884) | (0.727–0.864) | (0.848–1.000) |
Note. ModelLBC=ModelLesion+Background+Clinic, AUC=Area under the Curve, ACC=Accuracy, SE=Sensitivity, SP=Specificity, 95%CI=Confidence interval of 95%.
Comparisons the AUCs of ModelLesion+Background+Clinic amongst three subgroups were performed by Delong test.
p< 0.05 for comparison of ‘1.1–2.0 cm’ subgroup with the ‘>5.0 cm’ subgroup in the same cohort.
p< 0.05 for comparison of ‘2.1–5.0 cm’ subgroup with the ‘>5.0 cm’ subgroup in the same cohort.
3.4. Diagnostic robustness of the proposed models
Five-fold cross validation was performed in the training and IV cohort, which produced 5 independent models validated in IV cohorts. The Delong tests showed there were no statistical difference among the 5-fold models (all the p>0.05). In addition, the AUC of ModelLBC in the EV cohort was 0.924 (95% CI: 0.889–0.959), which showed no statistical difference with that of IV cohort (0.925, 95% CI: 0.886–0.963) (p = 0.7761) (Fig. S4). The results manifested satisfied robustness of the DCNN network.
3.5. Development and validation of nomogram
A nomogram was developed based on ModelLBC to provide the predicted probability of malignant FLLs for each individual (Fig. 3). The nomogram calibration curves showed good consistency across the training (p = 0.6573), IV (p = 0.3680), and EV (p = 0.4276, Fig. S5) cohorts. The decision curve analysis in Fig. 3 shows that with a threshold of 2%, the net benefit of ModelLBC for diagnosis of FLLs was 0.809, providing a better decision strategy than ModelLB, the “all malignant FLLs strategy”, or the “all benign FLLs strategy”.
Fig. 3.
Individualized predictive graphical presentation for clinical use. (a) Nomogram for the DCNN-US model. By integrating the predicted score, clinical factors including gender, age, tumour history, hepatitis history, hypoechoic halo, and vascularity of lesion and the final diagnostic outcome are presented on the bottom line of the nomogram along with malignancy probability. Gender, 0: female, 1: male; Tumour history, 0: no, 1: yes; Hepatitis history, 0: none, 1: HBV, 2: HCV, 3: HBV+HCV; Hypoechoic halo, 0: absent, 1 present; Vascularity, 0: absent, 1: present. (b) Calibration curve of the training cohort's nomogram. (c) Calibration curve of the internal validation cohort's nomogram. (d) Calibration curve of the external validation cohort's nomogram. Calibration curves indicate the consistency between histological diagnosis and predicted malignancy scores. The blue dotted line represents a perfect prediction by an ideal model. The pink solid line represents the nomogram's performance. A closer distance of the pink line to the blue line represents a better prediction. The p value of the Hosmer–Lemeshow test was greater than 0.05 for both the training and internal validation cohorts, showing good calibration between predictive outcome and histological diagnosis. (e) Decision curve analysis. The y-axis represents net benefit. The yellow and green lines measure the benefit obtained from ModelLB and ModelLBC, respectively. The blue and black lines measure the benefit of using the “all malignant FLLs” and “all benign FLLs” strategies, respectively. DCNN-US=Deep convolutional neural network of ultrasound, FLLs=Focal liver lesions, ModelLB=Modellesion+background, ModelLBC=Modellesion+background+clinic.
3.6. DCNN-US model versus the radiologists and contrast enhanced CT/MRI
Comparison of ModelLB in EV cohort with the judgement of 236 US radiologists (accessible to clinical information), each having 15 years’ experience showed that the diagnostic accuracy and sensitivity of ModelLB were similar (76.0% vs 76.0%, p = 0.8220 and 76.1% vs 77.4%, p = 0.940) (Fig. 4 and Table S8–9). However, the ModelLBC showed better diagnostic accuracy, sensitivity, and specificity than those of 15-year skilled radiologists (84.7% vs 76.0%, 86.5% vs 76.1%, and 85.5% vs 76.9%, respectively) (all p<0.01, Fig. 4 and Table S8–9).
Fig. 4.
Classification performance of the DCNN-US model and radiologists on focal liver lesions. (a-c) represents accuracy, sensitivity and specificity comparison between 15-year skilled radiologists and DCNN-US model, respectively. All p values were performed by non-parametric test. (d) Bar graph shows the diagnostic performance of the DCNN-US model, contrast enhanced CT and MRI. CT=Computed tomography, MRI=Magnetic resonance imaging, DCNN-US=Deep convolutional neural network of ultrasound.
In addition, compared with the contrasted enchanced CT, ModelLBC showed comparable accuracy (both 84.7%) but slightly inferior to contrast enhanced MRI for lesions detected by US (87.9%) (Fig. 4 and Table S8).
3.7. Visual interpretation of the model
Fig. 5 presents attention maps of eight cases that had lesions that were difficult to distinguish between malignant and benign by naked vision. For FLLs with the same histology but different B-mode US features or different histological features but the same B-mode US features, DCNN-US provided accurate diagnostic outcomes, with the attention maps illustrating distinguishable colour patterns. Attention maps were drawn to interpret the diagnostic mechanism of the neural network; these quantified each pixel's contribution to accurate diagnosis by analysing the lesion ROI. The red parts of the map indicated areas that provided more malignancy-related information during the network's diagnostic process (Fig. 5).
Fig. 5.
Attention maps of model on the benign and malignant lesions. Colours from warm to cold represent the degree of pixels’ contribution to FLL diagnosis. Red indicates the areas that contributed most, and blue areas contributed least. The number in the picture indicates the malignancy probability predicted by the model. (a) Hepatocellular carcinoma with different ultrasound appearance (Left MP: 0.9988, Right MP: 0.9989). (b) Metastatic cancer with different ultrasound appearance (Left MP: 0.9497, Right MP: 0.9997). (c) Hepatocellular carcinoma (MP: 0.9981) with similar ultrasound appearance to hepatic adenoma (MP: 0.2099). (d) Cholangiocellular carcinoma (MP: 0.8686) with similar ultrasound appearance to focal nodular hyperplasia (MP: 0.1393). FLL=Focal liver lesion, MP=malignancy probability predicted by the model.
4. Discussion
In this multicentre study, our DCNN-US model was tested in two validation cohorts and achieved good performance in FLL diagnosis. Our model's AUC for classifying malignant from benign lesions after EV reached 0.924 (95% CI: 0.889–0.959). Its accuracy, sensitivity, and specificity were higher than those of 15-year skilled radiologists. Further, its accuracy and sensitivity were comparable to the diagnostic performance of contrast enhanced CT for lesions detected by US.
In clinical practice, US is an indispensable and the most commonly used imaging modality in the work-up, management, and follow up of patients with FLLs [3,4,9]. High dependence on doctors’ experience lead to difficulties with accurate recognition of malignant characteristics among complex lesion types by radiologists, especially when the lesions occurred in the liver with cirrhosis or steatosis, as shown by varying diagnostic rates between the radiologists in our study. AI approaches provide an inspiring opportunity to recognize the diverse image characteristics of FLLs that are difficult to identify by naked vision. The DCNN technique we used automatically extracts FLLs mapping features through large cohort-based data mining. Subtle textural patterns that contributed to the diagnosis of FLLs could be explored and found via the neural network, thus improving its diagnostic power.
Clinical information is a vital referent for radiologists to make right diagnoses after analysis of image characteristics. Different from previous published three studies [14-16], we developed three DCNN-US models that incorporate lesions, the LB image signature, and seven easily acquired clinical factors in this study. We achieved improved diagnostic power for diagnosis of FLLs with strict validations and large sample analysis, which could increase the efficiency of full excavation of the neural network. In this study, patients were enrolled from 13 different centres and US images were acquired from 17 different devices, so the DCNN was forced to learn centre- and device- invariant features and rules for diagnosis of FLLs, which improved generalization of data-driven algorithms. The effective performance of the EV cohort also verified the feasibility of our idea. In addition, DCNN modelling can convert obscure and unexplainable derived imaging features into comprehensible attention maps, which renders radiomics no longer a black box but a visual tool that can provide highly suspected malignant regions to which radiologists can refer, especially for heterogeneous nodules. Particularly, in our research, we leveraged batch-hard triplet loss to mine difficult samples and forced the network to pay more attention to the differentiation of those samples, thus further improving the performance of the DCNN model to a level that surpassed previous studies [21,24,25].
When our model was constructed by mining the diverse image characteristics of lesion and liver, the diagnostic power of ModelLB could reached the level of 15-year skilled radiologist but is inferior to contrast enhanced CT/MRI. Once the clinic information were fed into ModelLB, its diagnostic ability exceeded that of 15-year skilled radiologists and was comparable to that of contrast enhanced CT for lesions detected by US. Even for lesions ≤2.0 cm, ModelLBC also obtained satisfactory diagnostic performance, which overcome the limitation of US-based diagnosis of small lesions [3,6]. All these satisfied results were attribute to standardized image acquisition criteria in the study, the automated quantification of large numbers of image features from multicentre database and model design advatages. It is worth mentioning that our FLLs US database is unique and valuable in that a total of 2213 lesions confirmed by pathology with 24,343 standardized images from multicentres were used to develop the DCNN model. It is a challenge for AI study in US of liver although it is not as enough as AI studies on MRI/CT [26,27].
The DCNN-US model has a high potential to maximize healthcare resources and narrow the gap in FLLs diagnosis between radiologists with different experience levels, decrease the dependence on sectional imaging and rich-experienced radiologists. Some patients with benign conditions may even be able to bypass the contrast enhanced CT/MRI evaluation and be referred for routine surveillance or short-term follow-up, while for patients with malignant FLLs may be earlier detected and timely given a privilege to further evaluation, which could be considered as cost-effective in view of the high expenditure for treating liver cancer. Once the DCNN-US model with convincing results is translated into the clinic in the future, operators would only need to perform a selection of DCNN-US ROIs in the daily workflow of B-mode US to conduct this analysis and acquire a second opinion proposed by the model, which is potentially easy for clinical application.
Our study has some limitations. First, only 19.3% of this study's enroled lesions were benign lesions. The unbalanced nature of the data may compromise the efficacy of DCNN-US to some extent. The main reason is that a tight US follow-up is often recommended clinically for patients with imaging possible benign FLLs, which decreased the proportion of enrollment of benign FLLs with pathological diagnosis. Second, we only achieved favourable ability in diagnosing benign and malignant FLLs for not enough lesion numbers for AI analysis. Classification of different type of FLLs will be our future effort. Third, for the lesions detected by US, our model was comparable to contrast enhanced CT in identifying benign and malignant FLLs, therefore our research suffered from some degree of bias due to the missed detection of US and detection ability of AI on US is also our future research effort. Finally, the DCNN-US developed here classifies FLLs solely on the basis of B-mode US imaging data. In the future, we anticipate that an AI system based on CEUS data can be constructed for more precise FLLs classification. And the observe if radiologists' diagnostic performance and detection ability for FLLs will be improved with use of proposed model as an effective tool to aid radiologists in current clinical settings need to be tested.
In conclusion, this study suggests that DCNN-US shows improved sensivity and specificity in identifying benign and malignant FLLs, which is superior to skilled radiologists and comparable to that of contrast enhanced CT. The advanced technical performance obtained by the DCNN-US model suggests the potential of this non-invasive, cheap, and convenient method to proceed and be tested in clinical trials.
Contributors
Conception and design: P. Liang, J.Tian, J.Yu, and R.Zheng.
Collection and assembly of data: Q.Yang, J,Wei, X.Hao, D.Kong, X.Yu, T.Jiang, J.Xi, W.Cai, Y.Luo, X.Jing, Y.Yang, Z.Cheng, J.Wu, H.Zhang, J.Liao, P.Zhou, Y.Song, Y.Zhang, Z.Han, W.Cheng, L.Tang, F.Liu, J.Dou, P.Liang, J.Yu, R.Zheng
Data analysis and interpretation: Q.Yang, J.Yu, J.Wei and X.Hao
Administrative support: P.Liang, J.Yu, X.Yu and J.Tian.
Manuscript writing: Q.Yang, J.Yu, J.Wei, X.Hao and D.Kong
Final approval of manuscript: All authors
Declaration of Competing Interest
The authors report no conflicts of interest.
Acknowledgments
Funding
A full list of funding agencies that contributed to this study can be found in the acknowledgements section.
Acknowledgements
This work was supported by Grants 81627803, 81971625, 91859201, 81227901 and 81527805 from the National Scientific Foundation Committee of China, Grant JQ18021 from the National Scientific Foundation Committee of Beijing, Grant NCRCG-PLAGH-2019011 from Fostering Funds for National Distinguished Young Scholar Science Fund and the National Clinical Research Centre for Geriatric Diseases of Chinese PLA General Hospital, Grant 2018ZX10723-204 from the National Key R&D Program of Ministry of Science and Technology of China, Grants GJJSTD20170004 and QYZDJ-SSW-JSC005 from Chinese Academy of Sciences, Grant Z171100000117023 from Beijing Municipal Science & Technology Commission and Grant XDBS01000000 from the Strategic Priority Research Program of Chinese Academy of Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102777.
Contributor Information
Rongqin Zheng, Email: zhengrq@mail.sysu.edu.cn.
Jie Yu, Email: jiemi301@163.com.
Jie Tian, Email: jie.tian@ia.ac.cn.
Ping Liang, Email: liangping301@hotmail.com.
Appendix. Supplementary materials
References
- 1.Bray Freddie, Ferlay Jacques, Soerjomataram Isabelle. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2018;0:1–31. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Amarapurkar Deepak, Han Kwang-Hyub, Chan Henry Lik-Yuen. Application of surveillance programs for hepatocellular carcinoma in the Asia–Pacific Region. J Gastroenterol Hepatol. 2009;24:955–961. doi: 10.1111/j.1440-1746.2009.05805.x. [DOI] [PubMed] [Google Scholar]
- 3.Marrero J.A., Ahn J., Rajender R.K. ACG clinical guideline: the diagnosis and management of focal liver lesions. Am J Gastroenterol. 2014;109(9):1328–1347. doi: 10.1038/ajg.2014.213. [DOI] [PubMed] [Google Scholar]
- 4.Heimbach Julie K., Kulik Laura M., Finn Richard S. AASLD Guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–380. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 5.Tchelepi H., Ralls P.W. Ultrasound of focal liver masses. Ultrasound Q. 2004;20(4):155–169. doi: 10.1097/00013644-200412000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Yu N.C., Chaudhari V., Raman S.S. CT and MRI improve detection of hepatocellular carcinoma, compared with ultrasound alone, in patients with cirrhosis. Clin Gastroenterol Hepatol. 2011;9(2):161–167. doi: 10.1016/j.cgh.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Xu M., Pan F.S., Wang W. The value of clinical and ultrasound features for the diagnosis of infantile hepatic hemangioma: comparison with contrast-enhanced CT/MRI. Clin Imaging. 2018;51:311–317. doi: 10.1016/j.clinimag.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Grazioli L., Ambrosini R., Frittoli B., Grazioli M., Morone M. Primary benign liver lesions. Eur J Radiol. 2017;95:378–398. doi: 10.1016/j.ejrad.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 9.EASL Clinical Practice Guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 10.Kinkel K., Lu Y., Both M. Detection of hepatic metastases from cancers of the gastrointestinal tract by using noninvasive imaging methods (US, CT, MR imaging, PET): a meta-analysis. Radiology. 2002;224(3):748–756. doi: 10.1148/radiol.2243011362. [DOI] [PubMed] [Google Scholar]
- 11.Vecchiato F., D'Onofrio M., Malagò R. Detection of focal liver lesions: from the subjectivity of conventional ultrasound to the objectivity of volume ultrasound. Radiol Med. 2009;114(5):792–801. doi: 10.1007/s11547-009-0421-7. [DOI] [PubMed] [Google Scholar]
- 12.Tan C.H., Low S.C., Thng C.H. APASL and AASLD consensus guidelines on imaging diagnosis of hepatocellular carcinoma: a review. Int J Hepatol. 2011;2011 doi: 10.4061/2011/519783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar V., Gu Y., Basu S. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30(9):1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ta Casey N., Kono Yuko, Eghtedari Mohammad. Focal liver lesions: computer-aided diagnosis by using contrast-enhanced US cine recordings. Radiology. 2018;0:1–10. doi: 10.1148/radiol.2017170365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao Z., Dong Y., Wu G. Preoperative diagnosis and prediction of hepatocellular carcinoma: radiomics analysis based on multi-modal ultrasound images. BMC Cancer. 2018;18(1):1089. doi: 10.1186/s12885-018-5003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmauch B., Herent P., Jehanno P. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging. 2019;0:1–7. doi: 10.1016/j.diii.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Wang K., Lu X., Zhou H. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2018;0:1–13. doi: 10.1136/gutjnl-2018-316204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He K, Zhang X, Ren S, et al. 2015. Deep residual learning for image recognition. arXiv preprint arXiv:151203385.
- 19.Deng J, Dong W, Socher R, et al. ImageNet: A large-scale hierarchical image database. Proceedings of the IEEE conference on computer vision and pattern recognition. 2009. pp. 248-255.
- 20.Hinton GE, Srivastava N, Krizhevsky A, et al. 2012. Improving neural networks by preventing co-adaptation of feature detectors. arXiv preprint arXiv:12070580.
- 21.Srivastava N., Hinton G.E., Krizhevsky A. Dropout A simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014:1929–1958. [Google Scholar]
- 22.Ioffe S78, Szegedy C. 2015. Batch normalization Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:150203167.
- 23.Hermans A, Beyer L, Leibe B. 2017. In defense of the triplet loss for person Re-Identification. arXiv preprint arXiv:170307737.
- 24.Ramprasaath R. Selvaraju, Michael Cogswell, Abhishek Das, et al. 2016. Grad-CAM visual explanations from deep networks via gradient-based localization. arXiv preprint arXiv:161002391.
- 25.Li X., Zhang S., Zhang Q. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: a retrospective, multicohort, diagnostic study. Lancet Oncol. 2019;20(2):193–201. doi: 10.1016/S1470-2045(18)30762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mokrane F.Z., Lu L., Vavasseur A., Otal P., Peron J.M., Luk L. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30(1):558–570. doi: 10.1007/s00330-019-06347-w. [DOI] [PubMed] [Google Scholar]
- 27.Park H.J., Lee S.S., Park B., Yun J., Sung Y.S., Shim W.H. Radiomics analysis of Gadoxetic acid-enhanced MRI for staging liver fibrosis. Radiology. 2019;290(2):380–387. doi: 10.1148/radiol.2018181197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.